Abstract
Streptococcus gordonii is a commensal inhabitant of human oral biofilms. Previously, we identified an enzyme called SdbA that played an important role in biofilm formation by S. gordonii. SdbA is thiol-disulfide oxidoreductase that catalyzes disulfide bonds in secreted proteins. Surprisingly, inactivation of SdbA results in enhanced biofilm formation. In this study we investigated the basis for biofilm formation by the ΔsdbA mutant. The results revealed that biofilm formation was mediated by the interaction between the CiaRH and ComDE two-component signalling systems. Although it did not affect biofilm formation by the S. gordonii parent strain, CiaRH was upregulated in the ΔsdbA mutant and it was essential for the enhanced biofilm phenotype. The biofilm phenotype was reversed by inactivation of CiaRH or by the addition of competence stimulating peptide, the production of which is blocked by CiaRH activity. Competition assays showed that the enhanced biofilm phenotype also corresponded to increased oral colonization in mice. Thus, the interaction between SdbA, CiaRH and ComDE affects biofilm formation both in vitro and in vivo.
Introduction
Streptococcus gordonii is a commensal inhabitant of human oral cavity. It is non-cariogenic, and its presence is associated with oral health [1,2]. Colonization with S. gordonii is beneficial because it can neutralize the surrounding pH to mitigate damage from cariogenic species, in addition to directly inhibiting the growth of some pathogens by secreting substances such as bacteriocins and hydrogen peroxide [3–7]. More passively, it also occupies space in the oral biofilm that would otherwise be available to cariogenic species, such Streptococcus mutans.
As a pioneer colonizer, S. gordonii colonizes early in life and is able to bind directly to salivary proteins on the tooth surface, forming the base of oral biofilms [8,9]. The ability to adhere and form biofilms in the host is crucial for persistence in the oral cavity; otherwise S. gordonii would be washed away. In addition, biofilms increase fitness by facilitating natural genetic transformation and by providing a protective niche in the continually fluctuating environment of the oral cavity [8,10,11]. Biofilm formation by S. gordonii is a complex process involving adhesins, signalling systems, ABC-transporters, and glycosyltransfrases among other factors [12–16]. These factors cooperate to maintain biofilms in the competitive and stressful environment of the oral cavity.
Recently we found that an enzyme required for disulfide bond formation, SdbA, played a role in biofilm formation [17]. SdbA is a thiol-disulfide oxidoreductase that catalyzes disulfide bond formation in extracytoplasmic proteins [17,18]. These bonds are important for the folding and stability of certain proteins, and ΔsdbA mutants are unable to form disulfide bonds. This generates a stress signal that triggers activation of the two-component signalling system CiaRH, presumably in response to an accumulation of misfolded proteins [19]. CiaH is a histidine kinase located at the membrane that activates the response regulator CiaR, which then drives the expression of multiple proteins including DegP (HtrA), a quality control protease that degrades aberrant proteins at the cell surface [19,20].
Inactivation of sdbA generates a pleiotropic phenotype [20]. The mutants are deficient in genetic competence, bacteriocin production, and extracellular DNA (eDNA) production, and autolysis, yet, somewhat surprisingly inactivation of sdbA enhances biofilm formation. Some of these phenotypes are a direct result of inactivation of sdbA. For example, the major autolysin AtlS is a natural substrate of SdbA, and is therefore inactive in the ΔsdbA mutant. Other phenotypes, such as the loss of bacteriocin production, are a stress response mediated by CiaRH. Bacteriocin production in S. gordonii is regulated by the ComDE quorum-sensing system. The histidine kinase ComD is activated when it senses an accumulation of secreted competence-stimulating peptide (CSP), and upon activation, it phosphorylates the response regulator ComE. This ultimately leads to expression of the bacteriocin genes, as well as genetic competence. However, activation of CiaRH in the ΔsdbA mutant eliminates CSP production, effectively shutting down the ComDE pathway and bacteriocin production [19]. Thus the pleiotropic phenotype of the ΔsdbA mutant involves multiple mechanisms, some of which are not directly related to disulfide bond formation.
The basis for enhanced biofilm formation by the ΔsdbA mutant is unknown. In this study, we sought to investigate how inactivation of sdbA leads to the hyperbiofilm phenotype, and to determine the effect of SdbA on oral colonization in mice. Our results reveal that biofilm formation by the ΔsdbA mutant is mediated by the CiaRH two-component signalling system, and the ability of CiaRH to repress production of competence stimulating peptide (CSP).
Results
CiaRH expression in biofilms
Previously, we found that expression of the two-component signalling system CiaRH is upregulated in the ΔsdbA mutant [19]. Although the role of CiaRH in biofilm formation by S. gordonii has not been investigated, CiaRH is required for biofilm formation and colonization in other streptococci, including S. pneumoniae [21–25], S. mutans [26], and group B Streptococcus [27]. This suggested that upregulation of CiaRH in the ΔsdbA mutant might contribute to its enhanced biofilm phenotype.
Our previous investigation of ciaRH expression in the ΔsdbA mutant examined cultures grown in BHI to the early exponential growth phase, which coincides with bacteriocin production and natural genetic competence. To determine if ciaRH expression is also upregulated when the ΔsdbA mutant is grown in biofilms, we tested expression using cells prepared from either the biofilm inoculum or 24 h biofilms.
Biofilms were grown following the protocol described by Loo et al., (42), which was designed to optimize S. gordonii biofilm formation in vitro. This approach uses cells grown in a rich medium as the inoculum, and a defined biofilm medium (BM) for subsequent biofilm growth. To investigate ciaRH expression and biofilm formation by the ΔsdbA mutant, biofilm inoculum was prepared from cells grown to the early stationary phase in HTVG medium. Analysis of the inoculum revealed that ciaR expression was upregulated by an average of >80-fold in the ΔsdbA mutant compared to the parent (Fig 1a). Expression of degP, a serine protease regulated by CiaRH, was also upregulated by an average of 30-fold, supporting the notion that the CiaRH system is activated in the ΔsdbA mutant (Fig 1b). After the cells had been grown in biofilms for 24 h, ciaR and degP expression in the ΔsdbA mutant biofilms had decreased to a level that was not significantly different from the parent, although expression remained 2 to 3-fold higher in the ΔsdbA mutant compared to the parent (Fig 1c and 1d). This suggested that CiaRH signalling might influence biofilm formation by the ΔsdbA mutant, particularly during the initial phases of biofilm development.
Fig 1. CiaRH expression in the S. gordonii biofilms.
Expression of the cia-induced genes ciaR and degP in the biofilm inoculum and in 24 h biofilms. (a) Expression of ciaR in the parent, ΔsdbA mutant, and sdbA-complemented mutant (+SdbA) in the biofilm inoculum. RNA was isolated from cells grown to early stationary phase in HTVG medium. (b) Expression of degP in the biofilm inoculum. Biofilms were grown in a defined biofilm medium (BM), and RNA was isolated from biofilm cells grown in BM medium for 24 h. (c) Expression of ciaR and (d) degP in 24 h biofilms. Results are means ± SD of three experiments. Data were analyzed by one-way ANOVA and asterisks indicate a significant difference from the parent (**P < 0.01).
Biofilm formation by the ΔsdbA mutant is mediated by CiaRH
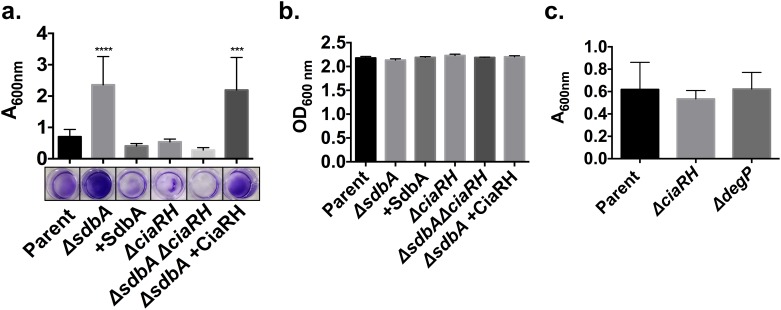
To determine if upregulation of ciaRH in the ΔsdbA mutant contributed to biofilm formation, we tested biofilm formation in CiaRH-deficient mutants. Analysis of 24 h biofilms revealed that CiaRH was critical for biofilm formation by the ΔsdbA mutant. Mutation of ciaRH abolished the enhanced biofilm phenotype in the ΔsdbA mutant, while complementation with a functional ciaRH system reversed the phenotype (Fig 2a) (P <0.0001). Although the ΔsdbAΔciaRH mutant formed very little biofilm, it did not have an obvious growth defect, and the total growth (planktonic plus biofilm cells) was similar across all of the mutants (Fig 2b).
Fig 2. CiaRH is required for biofilm formation by the ΔsdbA mutant.
(a) Crystal violet staining of 24 h biofilms grown in 24-well plates. Biofilms were grown with the parent, ΔsdbA, sdbA-complemented mutant (+SdbA), ΔciaRH, ΔsdbAΔciaRH double mutant, and ΔsdbA ciaRH-complemented mutant (ΔsdbA + CiaRH). Results are means ± SD of at least three experiments. The lower panel shows representative wells after staining. (b) In parallel to the biofilm formation assay, three additional wells for each strain were tested for total growth. The optical density was measured for the combined biofilm and planktonic cells for each mutant. (c) Biofilm formation of single deletion mutants for ciaRH and degP in the parent strain. Biofilms were grown for 24 h in 24-well plates prior to staining. Data were analyzed by one-way ANOVA and asterisks indicate a significant difference from the parent (***P ≤ 0.001, ****P ≤ 0.0001).
Surprisingly, however, inactivation of ciaRH and degP in the parent did not affect biofilm formation (Fig 2a and 2c). This suggested that the basal level of CiaRH activity in the parent is not a major contributor to biofilm formation, whereas upregulation of ciaRH is crucial for biofilm formation by the ΔsdbA mutant.
Biofilm formation by S. gordonii ΔsdbA and ΔsspAB mutants involve different mechanisms
Reports of mutations that enhance biofilm formation in S. gordonii are rare. One exception are mutants that lack the antigen I/II proteins SspA and SspB. Similar to the ΔsdbA mutant, the ΔsspAΔsspB mutant displays increased biofilm formation and initial attachment [14]. The enhanced biofilm formation by the ΔsspAΔsspB mutant has been attributed in part to upregulation of the surface lipoprotein ScaA, a substrate binding protein, which also enhances biofilm formation by S. gordonii in response to nicotine [14,28]. Initially we hypothesized that a similar scenario could be playing out in the ΔsdbA mutant, however, we found that scaA expression was unchanged in the ΔsdbA mutant (S1a Fig). In addition, we found that inactivation of the ciaRH system did not affect biofilm formation by the ΔsspAΔsspB mutant (S1b Fig). Thus, despite their similar phenotypes, biofilm formation by the ΔsdbA mutant involves a different mechanism.
Multiple factors contribute to biofilm formation, including extracellular polysaccharides, eDNA, and proteins [12]. To get clues to the nature of the ΔsdbA biofilms, we tested total carbohydrate production, surface charge, and sensitivity to DNase I and trypsin. Surface charge and carbohydrate production of cells from the biofilm inoculum and 24 h biofilms was similar in all strains (Fig 3). DNase I treatment produced a modest decrease in the amount of biofilm, suggesting a minor role for eDNA in biofilm stability (Fig 3d). In contrast, trypsin significantly reduced biofilm formation in both the parent and the ΔsdbA mutant. Thus, proteins appear to be the most important factor contributing to biofilm formation and stability in S. gordonii (Fig 3d). The nature of the biofilm matrix appears to be similar between the ΔsdbA mutant and the parent, except that the total amount of surface attachment and biofilm formation is greater in the mutant. Although speculative, it is possible that certain proteins contributing to biofilm formation might be upregulated in response to the sdbA mutation.
Fig 3. Characterization of ΔsdbA biofilms.
(a) Alcian blue binding assay for surface charge in cells grown to early stationary phase in HTVG medium (Inoculum), and (b) in cells grown in 24 h biofilms (Biofilm). Bars represent the percentage of unbound dye. (c) Total carbohydrate production. (d) Sensitivity to DNase I and trypsin. Biofilms were grown for 24 h prior to the addition of either DNase I or trypsin, and incubated for an additional 1 h before staining with crystal violet. Data were analyzed by one-way ANOVA and asterisks indicate a significant difference from the control biofilm for each strain (***P ≤ 0.001**P < 0.01, *P < 0.05).
CiaRH mediated repression of the Com system contributes to biofilm formation by the ΔsdbA mutant
Previously we have shown that CiaRH represses the ComDE quorum-sensing system in the ΔsdbA mutant. ComD is a sensor histidine kinase that, along with its response regulator ComE, regulates expression over 150 genes in S. gordonii, including the genes for genetic competence and bacteriocin production [29]. ComDE is activated by secreted CSP, which is encoded by comC. Considering the relationship between SdbA, CiaRH, and ComDE, we hypothesized that these systems might contribute to biofilm production in the ΔsdbA mutant.
CiaRH represses the ComDE quorum sensing system in the ΔsdbA mutant by inhibiting production of the CSP autoinducer [19]. Accordingly, de-repression of ComDE can be achieved in two ways: (1) inactivate the CiaRH system or (2) expose cells to exogenous CSP [19]. Given that inactivation of CiaRH reduced biofilm formation in the ΔsdbA mutant, we asked whether exogenous CSP would produce a similar effect. In a previous study, we demonstrated that exogenous CSP induces comCDE expression in the ΔsdbA mutant to a level comparable to the parent [19].
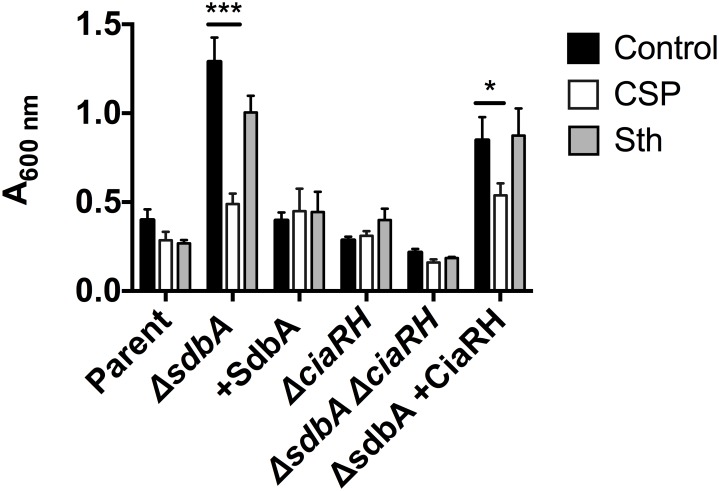
To test the effect of the CSP on biofilm formation, biofilms were grown for 24 h in the presence of 0.5 μg/ml CSP and quantified by crystal violet staining. Exogenous CSP did not affect biofilm formation by the parent, which is consistent with previous findings [30]. However, when the ΔsdbA mutant was grown with exogenous CSP biofilm formation was reduced to a level that was not significantly different from the parent (Fig 4) (P <0.001). This effect was specific to the ΔsdbA mutant, and required a functional and activated CiaRH system. Consequently, CSP did not affect biofilm formation by the sdbA-complemented mutant or the ΔsdbAΔciaRH mutant. Similarly, exogenous CSP did not affect biofilm formation by the ΔciaRH single mutant. Complementation of ciaRH into the ΔsdbAΔciaRH background restored sensitivity to CSP.
Fig 4. CSP diminishes biofilm formation by the ΔsdbA mutant.
Crystal violet staining of 24 h biofilms grown in the presence of either CSP, Sth1 bacteriocin (Sth), or without added peptide (Control). Biofilms were grown with the parent, ΔsdbA, sdbA-complemented mutant (+SdbA), ΔciaRH, ΔsdbAΔciaRH double mutant, and ΔsdbA ciaRH-complemented mutant (ΔsdbA +CiaRH). Results are means ± SD of three experiments. Asterisks indicate a significant difference from the control biofilm for each strain, as determined by one-way ANOVA (****P < 0.0001, *P < 0.05).
CSP is a small charged peptide with 19 amino acids and a pI of 10.28. To determine if the biofilm reducing effect was specific to CSP, we tested the effect of another peptide, the S. gordonii bacteriocin Sth1 (17 amino acids, pI 12.01). Growth with Sth1 did not produce a significant decrease in biofilm formation by any of the strains (Fig 4). This indicates that the effect is specific to CSP, and suggests that CiaRH mediated repression of CSP production is required for biofilm formation by the ΔsdbA mutant.
Inactivation of SdbA enhances oral colonization in mice
Although in vitro biofilm formation can vary depending on growth conditions and medium, it has been demonstrated to be correlated with in vivo colonization in mice in S. pneumoniae, including the role of CiaRH [21]. As an inhabitant of oral biofilms, biofilm formation is integral to S. gordonii colonization. The enhanced biofilm phenotype of the ΔsdbA mutant suggested that it might also affect colonization.
To determine how SdbA affects colonization, we ran a competitive assay between the parent and ΔsdbA mutant in a mouse model of oral colonization. We did not pursue colonization assays using the ΔsdbAΔciaRH mutant given its inability to form biofilm in vitro. Mice were inoculated with equal amounts of the parent and the ΔsdbA mutant. Following colonization, bacteria were recovered from the oral cavity and enumerated by growing with antibiotics that allowed differentiation of the two strains. Samples were plated on medium with tetracycline and spectinomycin to prevent growth of other oral bacteria, and with or without erythromycin to select for the sdbA mutant. The number of CFU for the parent was determined by subtracting the erythromycin resistant colonies from the total. Consistent with the ability of the ΔsdbA mutant to form more biofilm in vitro, the ΔsdbA mutant exhibited enhanced colonization after 24 h, with an average of 9.3 x 103 CFU/ml compared to 2 x 103 CFU/ml for the parent (P = 0.006) (Fig 5). By day 7, the difference was less pronounced and S. gordonii numbers were lower overall, however, the ΔsdbA mutant continued to outcompete the parent with an average of 2.8 x 103 CFU/ml compared to 6.8 x 102 CFU/ml (P = 0.039). Thus, the ΔsdbA mutant exhibits enhanced biofilm formation both in vitro and in vivo.
Fig 5. The ΔsdbA mutant outcompetes the parent an oral colonization model.
Mice were inoculated with a 1:1 mixture consisting of ~109 CFU of the parent and ΔsdbA mutant. Bacteria were recovered by swabbing oral surfaces after 1 and 7 days and enumerated using selective agar. Points represent the CFU/ml for the parent and the ΔsdbA mutant recovered from each mouse. Asterisks indicate a significant difference from zero using a one-sample T-test (**P < 0.01).
Discussion
In this study we investigated the basis for enhanced biofilm formation by the S. gordonii ΔsdbA mutant. SdbA catalyzes disulfide bond formation in secreted proteins, but it was not clear how this impacted biofilm formation. The role of SdbA in biofilm formation appears to be primarily indirect, and mediated by the interaction between the CiaRH two-component signalling system and the Com quorum-sensing system.
Since disulfide bonds are important for the folding and stability of extracytoplasmic proteins, inactivation of a disulfide catalyst can affect multiple proteins on the cell surface. This has the potential to alter surface adhesion and biofilm initiation, in addition to creating a stress response. For example, inactivation of the disulfide bond forming enzymes DsbA or DsbB eliminates biofilm formation in pathogenic E. coli and Pseudomonas aeruginosa [31–33]. In some instances, however, biofilm formation can be enhanced, such as in Pseudomonas putida where DsbA mutants form more biofilm due to increased EPS production [34], or in Salmonella enterica DsbA mutants, which have enhanced biofilm formation as a result of increased production of the fimbriae protein CsgA [35]. Although the mechanisms are not fully understood, the increased protein production and biofilm formation appear to be a stress response involving stress-induced signalling systems [35].
We found that a similar situation occurs in the S. gordonii ΔsdbA mutant. Inactivation of SdbA generated a signal that triggered the stress-related two-component regulatory system, CiaRH, which in turn stimulated biofilm formation. The CiaRH system is found throughout the genus Streptococcus and is best characterized in S. pneumoniae where it contributes to cell wall homeostasis, antibiotic resistance, and repression of genetic competence [20,36–38]. CiaRH is also important for biofilm formation in S. pneumoniae and S. mutans, and similar to the findings presented here, upregulation of CiaRH in S. pneumoniae variants results in enhanced biofilm formation [21,24,26]. The system is not as well characterized in S. gordonii, although it generally appears to have similar properties, and has been shown to contribute to acid stress resistance and competence repression [19,39]. In addition, ciaR (SGO_1072) has been found to be upregulated when S. gordonii is grown in mixed biofilms with Fusobacterium nucleatum [40]. Our results suggest that CiaRH was crucial for biofilm formation by the ΔsdbA mutant, but was dispensable in the parent. A possible explanation for this observation is that CiaRH has a greater influence on biofilm formation under adverse conditions, such as in the fluctuating environment of the oral cavity, and additional investigation is required to better understand the role of CiaRH in the parent.
In a previous study, we found that ciaRH expression was upregulated approximately 4-fold in the ΔsdbA mutant when grown under conditions that induce bacteriocin production, which occurs during a brief window in the early exponential growth phase [19]. Here, we found that the ΔsdbA mutant upregulates ciaRH expression by approximately 80-fold compared to the parent when grown to early stationary phase in HTVG medium. This increase appeared to be critical for biofilm formation, although ciaRH expression levelled off once the cells were in biofilms, suggesting that expression is influenced by the growth phase and the medium used in the experiment. ciaRH expression in S. pneumoniae is influenced by growth conditions, and has been demonstrated to peak during the early stationary growth phase [36]. The results presented here indicate that ciaRH expression in S. gordonii might follow a similar pattern.
Our analysis of the ΔsdbA mutant biofilms suggested that the surface charge and EPS production is similar to the parent. Although CiaRH regulates dlt expression in S. pneumoniae, which affects the surface charge through D-alanylation of lipoteichoic acid, ciaRH does not affect dlt expression in S. gordonii [40]. Our results support this conclusion, since even a dramatic increase in ciaRH expression did not have a significant impact on surface charge. Similarly, eDNA was not a major contributor to the ΔsdbA mutant biofilm, which is consistent with our earlier observation that the ΔsdbA mutant produces less eDNA than the parent [17]. Rather, the enhanced biofilm phenotype of the ΔsdbA mutant was primarily dependent on extracytoplasmic protein (Fig 3c).
Biofilm formation by S. gordonii involves multiple factors, including the glucosyltransferase GtfG [41], two-component signalling systems [16,42], ABC-transporters [16], and multiple surface adhesins that facilitate binding to host surfaces and salivary proteins [14]. In some instances, deletion of certain surface proteins can lead to enhanced biofilm and colonization, such as has been observed in mutants lacking SspAB [14] and AbpA [41,43]. Although the surface lipoprotein ScaA has been linked to enhanced biofilm formation, scaA expression was not upregulated in the ΔsdbA mutant. Similarly, we did not detect differences between the protein profiles of the ΔsdbA mutant and the biofilm defective ΔsdbAΔciaRH mutant that would suggest changes in the abundance of a large surface adhesin (data not shown).
Instead, the mechanism of CiaRH mediated biofilm formation by the ΔsdbA mutant involves the Com quorum-sensing system. CiaRH negatively regulates the Com system in the ΔsdbA mutant by inhibiting CSP production, however the system remains functional and can respond to exogenous CSP [19]. The results presented here show that exogenous CSP inhibits biofilm formation by the ΔsdbA mutant. Previously, we demonstrated that exogenous CSP does not decrease CiaRH expression in the ΔsdbA mutant, and therefore biofilm inhibition by CSP is not related to altered CiaRH levels [19]. Although we can only speculate as to how CSP affects biofilm formation, there are at least two plausible mechanisms. First, it could be a direct effect of the CSP protein. Charged peptides can bind to the cell surface or to matrix components resulting in altered adhesion [44], and pneumococcal CSP was recently shown to be retained at the cell surface by ComD [45]. A second possible mechanism is that CSP activates Com signalling, which in turn affects expression of other factors that mediate biofilm formation. Considering that CSP influences expression of >100 genes, we think that this is the most probable scenario [29].
In addition to regulating genetic competence, ComDE is a global regulator that plays an important role in stress responses. An analysis of the transcriptional response to CSP in S. gordonii revealed 162 upregulated and 89 downregulated genes, many of which function in processes outside of the competence pathway [29]. The ComDE quorum-sensing system has been linked to biofilm formation and colonization in multiple species of streptococci, although its precise role remains unclear. For example, mutation of comD inhibits biofilm formation and colonization by S. gordonii, S. mutans, and S. pneumoniae [22,42,46], whereas mutation of comE alone actually enhanced colonization by S. pneumoniae [22]. It was hypothesized that the unphosphorylated form of comE inhibited attachment, and therefore attachment was increased in the ΔcomE mutant [22]. There is also evidence to suggest that CSP affects biofilm formation. For example, CSP enhances biofilm formation by Streptococcus intermedius [47]. In both S. gordonii and S. mutans, mutation of the ΔcomCDE operon, or ΔcomC (which encodes CSP), altered the biofilm density and architecture [30,46]. Although the S. gordonii ΔcomC mutation did not effect the total biomass of S. gordonii monoculture biofilms, the mutant formed more biofilm when grown in mixed cultures with the fungus Candida albicans [30]. Thus ComDE can clearly influence biofilm formation, but the relationship is complex, and not yet fully understood.
The enhanced biofilm phenotype of the ΔsdbA mutant appears to require a coordinated increase in CiaRH activity and repression of comDE. Although the mechanisms remain unclear, the interconnectedness of the CiaRH and ComDE systems, particularly in relation to stress responses, has been observed previously in multiple streptococci including S. gordonii [22,38,39,48,49]. This is exemplified by S. pneumoniae, where activation of genetic competence in CiaRH mutants causes the cells to lyse [38]. Although the degree of activation varies with growth conditions, we found that the CiaRH system is consistently upregulated in S. gordonii ΔsdbA mutants. CiaRH is associated with cell wall integrity, and provides protection from conditions that perturb the cell wall such as acid stress, β-lactam antibiotics, mutations in penicillin-binding proteins [24,26,39,49]. In the absence of SdbA, substrates requiring disulfide bonds will remain in a reduced state and are vulnerable to oxidation and misfolding. Since an accumulation of misfolded protein at the cell surface is a potentially lethal situation, upregulation of ciaRH, and the CiaR-regulated protease DegP, likely serves to mitigate stress from misfolded proteins in the ΔsdbA mutant.
In addition to a general stress response to misfolded proteins, CiaRH might also respond more directly to inactivation of the major autolysin AtlS, which is a natural SdbA substrate [17]. The ΔsdbA mutant produces AtlS protein, but it lacks autolytic activity and shows altered processing compared to the parent [17]. At this point, we can only speculate as to how mutation of SdbA triggers the activation of the CiaRH system, however, the loss of AtlS activity would be a plausible mechanism given its role in cell wall turnover. Notably, AtlS contributes to cell surface biogenesis and biofilm formation, and its expression is induced by exogenous CSP [50]. Thus, AtlS is a possible link between cell wall integrity, biofilm formation, and CSP. However, additional investigation will be required to understand how the inactive form of AtlS produced by the ΔsdbA mutant affects the cell, and whether it contributes to CiaRH mediated biofilm formation
Consistent with the enhanced biofilm formation observed in vitro, the ΔsdbA mutant also outcompeted the parent in a mouse oral colonization assay. We do not know if cross-contamination of CSP produced by the parent affected colonization by the ΔsdbA mutant in the competition assay, but it is plausible that most of the CSP would be washed away by saliva. Regardless, the mutant outcompeted the parent after 1 day, and continued to outnumber the parent after 7 days.
In summary, we investigated how mutation of a disulfide bond forming catalyst, SdbA, leads to enhanced biofilm formation by S. gordonii. The mechanism appears to be a general stress response mediated by the CiaRH two-component signalling system. Upregulation of CiaRH represses expression of the ComDE quorum-sensing system, which in turn leads to enhanced biofilm formation. Conversely, exposure to CSP reverses the phenotype. The interplay between the CiaRH and Com systems enhances both in vitro biofilm formation and in vivo colonization by the ΔsdbA mutant. S. gordonii is associated with oral health, and it has potential biotechnological value as a live vaccine vector, or possibly even as a probiotic [51]. The enhanced biofilm formation and colonization of the ΔsdbA mutant suggests that it could be particularly useful for such applications.
The interaction between multiple regulatory systems highlights the complexity of biofilm formation by S. gordonii. Understanding these pathways can provide a basis for investigating more complex biofilm communities and understanding how this commensal bacterium contributes to oral health.
Methods and Materials
Bacterial strains and growth conditions
Experiments were carried out using S. gordonii SecCR1 as the parent strain. S. gordonii SecCR1 is a derivative of S. gordonii Challis DL-1 that secretes a single-chain variable fragment antibody against complement receptor 1 (CR1), which is a protein that requires disulfide bonds for stability [52]. This strain has been used in our previous studies of disulfide bond formation in S. gordonii, and the S. gordonii SecCR1 ΔsdbA mutant has the same phenotype as the S. gordonii Challis DL-1 ΔsdbA mutant [19]. Additional strains and mutants are described in Table 1. S. gordonii was grown in either HTVG medium (0.5% (wt/vol) glucose, 3.5% (wt/vol) tryptone, 100 mM HEPES, 0.29 μM p-aminobenzoic acid, 0.59 μM thiamine-HCl, 8.2 μM nicotinamide, and 0.53 μM riboflavin, pH 7.6) [53], or in a semi-defined Biofilm Medium (BM; 58 mM K2HPO4, 15 mM KH2PO4, 10 mM (NH4)2SO4, 35 mM NaCl, 0.8% (wt/vol) glucose, 0.2% (wt/vol) Casamino Acids, 25 μM MgCl27H2O, 2 mM MgSO2 7H2O, 0.04 mM nicotinic acid, 0.1 mM pyridoxine HCl, 0.01 mM pantothenic acid, 1 μM riboflavin, 0.3 μM thiamin HCl, 0.05 μM D-biotin, 4 mM L-glutamic acid, 1 mM L-arginine HCl, 1.3 mM L-cysteine HCl, 0.1 mM L-tryptophan, filter sterilized) [42]. For experiments that required growth on solid medium, cultures were plated on Brain Heart Infusion agar (BHI, Difco) containing the appropriate antibiotics. Cultures were grown at 37°C, 5% CO2, without shaking. Antibiotics were used at the following concentrations: erythromycin 10 μg/ml, tetracycline 10 μg/ml, spectinomycin 250 μg/ml, chloramphenicol 5 μg/ml, and kanamycin 250 μg/ml.
Table 1. Bacterial strains and primers used in this study.
| Strains | Relevant characteristics | Source |
|---|---|---|
| S. gordonii SecCR1 | hppG::tet, secretes anti-CR1 scFv, TetR, SpecR | 49 |
| ΔsdbA | SecCR1, sdbA::ermAM, TetR, SpecR, ErmR | 20 |
| +SdbA | ΔsdbA, sdbA-complemented on chromosome, TetR, SpecR, KanR | 20 |
| ΔdegP | degP::ermAM, TetR, SpecR, ErmR | 19 |
| ΔciaRH | SecCR1, ciaRH::aphA3, TetR, SpecR, KanR | 19 |
| ΔsdbAΔciaRH | ΔsdbA, ciaRH::aphA3, TetR, SpecR, KanR, ErmR | 19 |
| ΔsdbA + CiaRH | ΔsdbAΔciaRH, ciaRH-complemented on chromosome, TetR, SpecR, ErmR, CmR | 19 |
| OB219 | sspAsspB::ermAM, ErmR | 53 |
| OB219 ΔciaRH | ΔsspAΔsspB, ciaRH::aphA3, KanR, ErmR | This study |
Biofilm formation
Biofilms were grown as described by Loo et al. (42), with the following modifications. S. gordonii was grown in HTVG for 12 h to an optical density (OD) of approximately 1.2 at 600 nm in 15 ml conical tubes (Sarstedt, model #62.554.205). Cells were harvested by centrifugation (3000 x g for 10 min) and the spent medium was discarded. The cells were then gently suspended in BM to an OD600 of 0.250 in the same tube. All biofilms were grown using BM. Flat bottom 24-well plates (Falcon, model #353047) were inoculated with 1 ml per well of the cell suspension, and the plates were incubated for either 3 or 24 h at 37°C, 5% CO2. Following incubation, the medium was removed and the wells were washed twice with 1 ml phosphate buffered saline (PBS) to remove loosely attached cells. The plates were air dried for 15 min and fixed with 10% (v/v) formaldehyde and 5% (v/v) acetic acid in PBS for 15 min. The fixing solution was removed and the wells were washed 3 times with 1 ml PBS. The biofilms were then stained with 0.5 ml of 0.1% crystal violet. After 15 min, the wells were rinsed three times and the bound stain was solubilized in 1 ml of acetone/ethanol solution (1:1). The liquid was transferred to a new microtiter plate and the optical density was measured at 600 nm in a microplate reader. The biofilm assays were carried out in triplicate with three or more separate experiments.
To test the effect of peptides on biofilm formation, biofilms were prepared as described above except that 0.5 μg of either CSP (DVRSNKIRLWWENIFFNKK; Biomatik, Cambridge, ON, Canada) or a fragment of Sth1 (AGFTGGIAVGLNRVNRK; Biomatik) was added to each well during the initial biofilm setup. The biofilms were then incubated for 24 h at 37°C, 5% CO2, and stained with crystal violet as described above.
Sensitivity to trypsin and DNase I (Sigma-Aldrich, Oakville, ON, Canada) was tested by adding the enzymes to pre-formed biofilms. Biofilms were grown for 24 h prior to the addition of 100 μg of enzyme per well. The plates were incubated at 37°C for an additional 1 h and subsequently processed and stained as described above.
To test the total growth of biofilm and planktonic cells, biofilms were removed by scraping and vigorous pipetting to remove attached cells from bottom of the well. The optical density of the combined biofilm and planktonic cells was measured at 600 nm for triplicate wells for each mutant. Crystal violet staining confirmed that the biofilms were successfully detached.
Quantitative real-time PCR (qPCR)
Expression of ciaR and degP was measured in the biofilm inoculum and in biofilm cells. For the inoculum, RNA was isolated from cultures of the parent, ΔsdbA, and sdbA-complemented mutant grown in HTVG for 12 h to an OD600 of ~1.0–1.2. Total RNA was isolated using the hot acid phenol method, as described previously [54]. The RNA (1 μg) was treated with 1 unit of amplification grade DNase I (Life Technologies Inc., Waltham, MA) for 15 min at room temperature, and removal of DNA was confirmed by PCR using the 16S rRNA primers (SL525/697). cDNA synthesis was carried out using random primers and SuperScript II reverse transcriptase (Life Technologies Inc.) according to the manufacturer’s directions.
Biofilms for RNA isolation were grown in BM as described above. Cells from were suspended in BM to an OD600 of 0.250, and 15 ml suspensions were used to inoculate tissue culture treated petri dishes (100 mm x 20 mm, Corning, model #353003). The plates were incubated at 37°C, 5% CO2 for 24 h. Planktonic cells and spent medium were then removed by pipetting and the biofilms were washed once with 10 ml phosphate buffered saline (PBS). The biofilm cells were scraped from the petri dish and suspended in 1.5 ml PBS using a sterile swab. RNA isolation and cDNA synthesis was carried out as described above.
qPCR to amplify ciaR, degP, and scaA was carried out using the primers listed in Table 2 with iTaq Universal SYBR Green Supermix (Bio-Rad Laboratories, Mississauga, ON, Canada) according to the manufacturer’s directions. The reactions were performed using a 7900 HT Fast Real-Time PCR system (Applied Biosystems) at 95°C for 30 s, followed by 40 cycles of 95°C for 15 s and 60°C for 60 s. The cycle threshold (CT) was calculated using SDS 2.2.2 software (Applied Biosystems). The relative expression was calculated using the comparative CT method [55] using 16S rRNA as an internal control gene. Each reaction was performed in duplicate using cDNA from at least three biological replicates.
Table 2. Primers used in this study.
| Primer | Gene | Direction | Description | Sequence |
|---|---|---|---|---|
| SL1178 | sgo.1071 | For | CiaRH mutant | AAAACGCTGCAAAATAATCA |
| SL1221 | sgo.1074 | Rev | CiaRH mutant | TTCAACCAATTCGCTAAATC |
| SL697 | 16S | For | qPCR | ATTTATTGGGCGTAAAGCGAGCGC |
| SL525 | 16S | Rev | qPCR | GAATTAAACCACATGCTCCACCGC |
| SL1214 | degP | For | qPCR | TGGGAATAAGGTTCCTGGTG |
| SL1215 | degP | Rev | qPCR | CGGCAGGAATTCTGACTACAG |
| SL1216 | ciaR | For | qPCR | CATGCAGGTTTTTGATGGTG |
| SL1217 | ciaR | Rev | qPCR | TCAGGAAGCATCAGATCCAG |
Genetic manipulations
A ΔciaRH mutant was constructed in the ΔsspAΔsspB mutant, OB219 [56], by creating a clean deletion of ciaRH and replacing the genes with a kanamycin resistance cassette (aphA3) [57]. Polymerase chain reaction (PCR) was carried out using Phusion high-fidelity DNA polymerase (New England Biolabs, Whitby, ON, Canada) to amplify a construct from the ΔciaRH mutant in the S. gordonii SecCR1 background. The construct consisted of the aphA3 gene flanked by a 425 bp fragment of the gene upstream from ciaR, sgo.1071, and a 525 bp fragment of the downstream gene sgo.1074, and was amplified using the primer pair SL1178 and SL1221 (Table 2). The resulting PCR product was used to transform S. gordonii OB219 as described previously [52]. Transformants were selected on BHI containing kanamycin, and insertion of aphA3 was confirmed by PCR.
Carbohydrate quantification and surface charge
Total carbohydrates were measured using the phenol-sulfuric acid method [58]. Carbohydrate production was measured using cells grown for 12 h in HTVG (biofilm inoculum) and in 24 h biofilm cells. Biofilm cells were prepared from biofilms grown in petri dishes, as described above. Cells were suspended in distilled water and standardized to an OD600 of 2.0. The cell suspensions (200 μl) were mixed with an equal volume of 5% (vol/vol) phenol (Sigma-Aldrich), followed by the addition of 1 ml concentrated H2SO4. The reactions were incubated for 10 min at ambient temperature, cooled in a water bath for 5 min, and the optical density was measured at 490 nm. Total carbohydrates were calculated from a standard curve prepared with glucose. Surface charge was analyzed using the cationic dye Alcian Blue 8GX (Sigma-Aldrich). An Alcian Blue binding assay was carried out as described previously [59], using for cells from the biofilm inoculum and from 24 h biofilms. Briefly, the cells were washed with PBS and used to prepare aliquots standardized to an OD600 of 0.5. The cells were mixed with 65 μg/ml Alcian Blue and incubated at room temperature for 10 min with rotation to allow the dye to bind. The cells were removed by centrifugation at 10 000 x g, 5 min and 150 μl of supernatant was transferred to a 96-well plate. The optical density was read at 650 nm and the percentage of unbound dye was calculated as ((A650 Control—A650 Sample) / A650 Control) x 100%.
Competition assay
This study was carried out in strict accordance with the Guidelines of the Canadian Council on Animal Care. The protocol was approved by the local ethics committee (University Committee on Laboratory Animals of Dalhousie University). Mice were housed in groups of 5 in conventional type II cages containing nesting materials along with water and food supply. Animal euthanasia was performed via isoflurane inhalation followed by CO2.
Oral colonization was tested as described previously, with modifications [60]. BALB/c mice (female, 6 weeks old, Charles River Laboratory, St. Constant, Qc, Canada) were fed aqueous kanamycin (500 μg/ml) for 2 days prior to colonization to reduce background levels of natural microflora. The S. gordonii parent and ΔsdbA mutant were grown for 18 h and then dechained by vigorously passing the cells through a 26 G needle. Microscopy was used to confirm dechaining. Cultures were standardized by their optical density, and the parent and the mutant were then mixed in a 1:1 ratio (50 μl total volume). The inoculum was also plated for enumeration, and contained 5.2 x 109 CFU/ml of the parent and 1.8 x 109 CFU/ml of the ΔsdbA mutant.
The mice were sedated with ketamine-xylazine and inoculated with 10 μl intranasally and 40 μl orally. After 24 h (n = 4) and 7 days (n = 6), the mice were euthanized. Sterile swabs were used to recover bacteria from the oral cavity by swabbing the tooth surfaces, tongue, buccal mucosa, and throat to the trachea. The swabs were then vortexed with 1 ml PBS and serially diluted. Aliquots were plated on selective agar that allowed for differentiation of the parent and mutant. All media included spectinomycin and tetracycline to select for the S. gordonii parent and ΔsdbA mutant, while preventing growth of other bacteria. To differentiate the parent and the ΔsdbA mutant, bacteria were plated on media with and without erythromycin. The ΔsdbA mutant was quantified as the erythromycin resistant colonies, while the parent was quantified by subtracting the erythromycin resistant colonies from the total CFU. The recovered CFU were standardized to the initial inoculum density, and data were analyzed by Student’s t-test.
Statistical analysis
Results were analyzed by one-way ANOVA or student’s t test using GraphPad Prism version 6 (GraphPad Software Inc., La Jolla, California).
Supporting Information
(a) Quantitative PCR analysis of scaA expression in 24 h biofilm cells. RNA was prepared from the parent, the ΔsdbA mutant, and the sdbA-complemented mutant (+SdbA). (b) Biofilm formation by the parent, the ΔsdbA mutant, and the ΔsspAB mutant (OB219) with and without a functional CiaRH two-component signaling system. Biofilms were grown for 24 h and stained with crystal violet.
(TIFF)
Data Availability
All relevant data are within the paper and its Supporting Information file.
Funding Statement
This work was funded by Natural Sciences and Engineering Research Council of Canada, grant #183712, to SL. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Marchant S, Brailsford SR, Twomey AC, Roberts GJ, Beighton D. The predominant microflora of nursing caries lesions. Caries Res. 2001;35: 397–406. 10.1159/000047482 [DOI] [PubMed] [Google Scholar]
- 2.Gross EL, Beall CJ, Kutsch SR, Firestone ND, Leys EJ, Griffen AL. Beyond Streptococcus mutans: Dental caries onset linked to multiple species by 16s rrna community analysis. PLoS One. 2012;7: e47722 10.1371/journal.pone.0047722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang BY, Kuramitsu HK. Interactions between oral bacteria: Inhibition of Streptococcus mutans bacteriocin production by Streptococcus gordonii. Appl Environ Microbiol. 2005;71: 354–362. 10.1128/AEM.71.1.354-362.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang B, Deutch A, Hong J, Kuramitsu H. Proteases of an early colonizer can hinder Streptococcus mutans colonization in vitro. J Dent Res. 2011;90: 501–505. 10.1177/0022034510388808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kreth J, Zhang Y, Herzberg MC. Streptococcal antagonism in oral biofilms: Streptococcus sanguinis and Streptococcus gordonii interference with Streptococcus mutans. J Bacteriol. 2008;190: 4632–4640. 10.1128/JB.00276-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jakubovics NS, Gill SR, Vickerman MM, Kolenbrander PE. Role of hydrogen peroxide in competition and cooperation between Streptococcus gordonii and Actinomyces naeslundii. FEMS Microbiol Ecol. 2008;66: 637–644. 10.1111/j.1574-6941.2008.00585.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jakubovics NS, Robinson JC, Samarian DS, Kolderman E, Yassin SA, Bettampadi D, et al. Critical roles of arginine in growth and biofilm development by Streptococcus gordonii. Mol Microbiol. 2015;97: 281–300. 10.1111/mmi.13023 [DOI] [PubMed] [Google Scholar]
- 8.Kolenbrander PE, Palmer RJ, Periasamy S, Jakubovics NS. Oral multispecies biofilm development and the key role of cell-cell distance. Nat Rev Microbiol. 2010;8: 471–480. 10.1038/nrmicro2381 [DOI] [PubMed] [Google Scholar]
- 9.Dû LD, Kolenbrander PE. Identification of saliva-regulated genes of Streptococcus gordonii DL1 by differential display using random arbitrarily primed PCR. Infect Immun. 2000;68: 4834–4837. 10.1128/IAI.68.8.4834-4837.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petersen FC, Tao L, Scheie AA. DNA binding-uptake system: a link between cell-to-cell communication and biofilm formation. J Bacteriol. 2005;187: 4392–4400. 10.1128/JB.187.13.4392-4400.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Molin S, Tolker-Nielsen T. Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure. Curr Opin Biotechnol. 2003;14: 255–261. 10.1016/S0958-1669(03)00036-3 [DOI] [PubMed] [Google Scholar]
- 12.Nobbs AH, Lamont RJ, Jenkinson HF. Streptococcus adherence and colonization. Microbiol Mol Biol Rev. 2009;73: 407–450. 10.1128/MMBR.00014-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chaudhuri B, Paju S, Haase EM, Vickerman MM, Tanzer JM, Scannapieco FA. Amylase-binding protein B of Streptococcus gordonii is an extracellular dipeptidyl-peptidase. Infect Immun. 2008;76: 4530–4537. 10.1128/IAI.00186-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, Lei Y, Nobbs A, Khammanivong A, Herzberg MC. Inactivation of Streptococcus gordonii SspAB alters expression of multiple adhesin genes. Infect Immun. 2005;73: 3351–3357. 10.1128/IAI.73.6.3351-3357.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jakubovics NS, Brittan JL, Dutton LC, Jenkinson HF. Multiple adhesin proteins on the cell surface of Streptococcus gordonii are involved in adhesion to human fibronectin. Microbiology. 2009;155: 3572–3580. 10.1099/mic.0.032078-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Whiteley M, Kreth J, Lei Y, Khammanivong A, Evavold JN, et al. The two-component system BfrAB regulates expression of ABC transporters in Streptococcus gordonii and Streptococcus sanguinis. Microbiology. 2009;155: 165–173. 10.1099/mic.0.023168-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davey L, Ng CKW, Halperin SA, Lee SF. Functional analysis of paralogous thiol-disulfide oxidoreductases in Streptococcus gordonii. J Biol Chem. 2013;288: 16416–16429. 10.1074/jbc.M113.464578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davey L, Cohen A, LeBlanc J, Halperin SA, Lee SF. The disulfide oxidoreductase SdbA is active in Streptococcus gordonii using a single C-terminal cysteine of the CXXC motif. Mol Microbiol. 2016;99: 236–253. 10.1111/mmi.13227 [DOI] [PubMed] [Google Scholar]
- 19.Davey L, Halperin SA, Lee SF. Mutation of the thiol-disulfide oxidoreductase SdbA activates the CiaRH two-component system, leading to bacteriocin expression shutdown in Streptococcus gordonii. J Bacteriol. 2016;198: 321–331. 10.1128/JB.00800-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Halfmann A, Kovács M, Hakenbeck R, Brückner R. Identification of the genes directly controlled by the response regulator CiaR in Streptococcus pneumoniae: Five out of 15 promoters drive expression of small non-coding RNAs. Mol Microbiol. 2007;66: 110–126. 10.1111/j.1365-2958.2007.05900.x [DOI] [PubMed] [Google Scholar]
- 21.Blanchette-Cain K, Hinojosa C a, Akula Suresh Babu R, Lizcano A, Gonzalez-Juarbe N, Munoz-Almagro C, et al. Streptococcus pneumoniae biofilm formation is strain dependent, multifactorial, and associated with reduced invasiveness and immunoreactivity during colonization. MBio. 2013;4: 1–11. 10.1128/mBio.00745-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kowalko JE, Sebert ME. The Streptococcus pneumoniae competence regulatory system influences respiratory tract colonization. Infect Immun. 2008;76: 3131–3140. 10.1128/IAI.01696-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sebert ME, Palmer LM, Rosenberg M, Weiser JN. Microarray-based identification of htra, a Streptococcus pneumoniae gene that is regulated by the CiaRH two-component system and contributes to nasopharyngeal colonization. Infect Immun. 2002;70: 4059–4067. 10.1128/IAI.70.8.4059-4067.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trappetti C, Ogunniyi AD, Oggioni MR, Paton JC. Extracellular matrix formation enhances the ability of Streptococcus pneumoniae to cause invasive disease. PLoS One. 2011;6: e19844 10.1371/journal.pone.0019844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Throup JP, Koretke KK, Bryant AP, Ingraham K a, Chalker AF, Ge Y, et al. A genomic analysis of two-component signal transduction in Streptococcus pneumoniae. Mol Microbiol. 2002;35: 566–576. 10.1046/j.1365-2958.2000.01725.x [DOI] [PubMed] [Google Scholar]
- 26.Ahn S-J, Wen ZT, Burne RA. Multilevel Control of Competence Development and stress tolerance in Streptococcus mutans UA159. Infect Immun. 2006;74: 1631–1642. 10.1128/IAI.74.3.1631-1642.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quach D, van Sorge NM, Kristian SA, Bryan JD, Shelver DW, Doran KS. The CiaR response regulator in Group B Streptococcus promotes intracellular survival and resistance to innate immune defenses. J Bacteriol. 2009;191: 2023–2032. 10.1128/JB.01216-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang R, Li M, Ye M, Yang K, Xu X, Gregory RL. Effects of nicotine on Streptococcus gordonii growth, biofilm formation, and cell aggregation. Appl Environ Microbiol. 2014;80: 7212–7218. 10.1128/AEM.02395-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vickerman MM, Iobst S, Jesionowski A, Gill SR. Genome-wide transcriptional changes in Streptococcus gordonii in response to competence signaling peptide. J Bacteriol. 2007;189: 7799–7807. 10.1128/JB.01023-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jack AA, Daniels DE, Jepson MA, Margaret Vickerman M, Lamont RJ, Jenkinson HF, et al. Streptococcus gordonii comCDE (competence) operon modulates biofilm formation with Candida albicans. Microbiology. 2015;161: 411–421. 10.1099/mic.0.000010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee Y, Kim Y, Yeom S, Kim S, Park S, Jeon CO, et al. The role of disulfide bond isomerase A (DsbA) of Escherichia coli O157:H7 in biofilm formation and virulence. FEMS Microbiol Lett. 2008;278: 213–22. 10.1111/j.1574-6968.2007.00993.x [DOI] [PubMed] [Google Scholar]
- 32.Heras B, Shouldice SR, Totsika M, Scanlon MJ, Schembri MA, Martin JL. DSB proteins and bacterial pathogenicity. Nat Rev Microbiol. 2009;7: 215–225. 10.1038/nrmicro2087 [DOI] [PubMed] [Google Scholar]
- 33.Arts IS, Ball G, Leverrier P, Garvis S, Nicolaes V, Vertommen D, et al. Dissecting the machinery that introduces disulfide bonds in Pseudomonas aeruginosa. MBio. 2013;4: e00912–13. 10.1128/mBio.00912-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee Y, Oh S, Park W. Inactivation of the Pseudomonas putida KT2440 dsbA gene promotes extracellular matrix production and biofilm formation. FEMS Microbiol Lett. 2009;297: 38–48. 10.1111/j.1574-6968.2009.01650.x [DOI] [PubMed] [Google Scholar]
- 35.Anwar N, Rouf SF, Römling U, Rhen M. Modulation of biofilm-formation in Salmonella enterica serovar typhimurium by the periplasmic DsbA/DsbB oxidoreductase system requires the GGDEF-EAL domain protein STM3615. PLoS One. 2014;9: 1–12. 10.1371/journal.pone.0106095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Halfmann A, Schnorpfeil A, Müller M, Marx P, Günzler U, Hakenbeck R, et al. Activity of the two-component regulatory system CiaRH in Streptococcus pneumoniae R6. J Mol Microbiol Biotechnol. 2011;20: 96–104. 10.1159/000324893 [DOI] [PubMed] [Google Scholar]
- 37.Schnorpfeil A, Kranz M, Kovács M, Kirsch C, Gartmann J, Brunner I, et al. Target evaluation of the non-coding csRNAs reveals a link of the two-component regulatory system CiaRH to competence control in Streptococcus pneumoniae R6. Mol Microbiol. 2013;89: 334–349. 10.1111/mmi.12277 [DOI] [PubMed] [Google Scholar]
- 38.Dagkessamanskaia A, Moscoso M, Overweg K, Reuter M, Martin B, Wells J, et al. Interconnection of competence, stress and CiaR regulons in Streptococcus pneumoniae: competence triggers stationary phase autolysis of ciaR mutant cells. Mol Microbiol. 2004;51: 1071–1086. [DOI] [PubMed] [Google Scholar]
- 39.Liu Y, Burne RA. Multiple two-component systems modulate alkali generation in Streptococcus gordonii in response to environmental stresses. J Bacteriol. 2009;191: 7353–62. 10.1128/JB.01053-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hendrickson EL, Wang T, Dickinson BC, Whitmore SE, Wright CJ, Lamont RJ, et al. Proteomics of Streptococcus gordonii within a model developing oral microbial community. BMC Microbiol. 2012;12: 211 10.1186/1471-2180-12-211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chaudhuri B, Rojek J, Vickerman M, Tanzer JM, Scannapieco FA. Interaction of salivary alpha-amylase and amylase-binding-protein A (AbpA) of Streptococcus gordonii with glucosyltransferase of S. gordonii and Streptococcus mutans. BMC Microbiol. 2007;7: 60 10.1186/1471-2180-7-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Loo CY, Corliss DA, Ganeshkumar N. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J Bacteriol. 2000;182: 1374–1382. 10.1128/JB.182.5.1374-1382.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanzer JM, Grant L, Thompson A, Li L, Rogers JD, Haase EM, et al. Amylase-binding proteins A (AbpA) and B (AbpB) differentially affect colonization of rats’ teeth by Streptococcus gordonii. Microbiology. 2003;149: 2653–2660. 10.1099/mic.0.26022-0 [DOI] [PubMed] [Google Scholar]
- 44.Batoni G, Maisetta G, Esin S. Antimicrobial peptides and their interaction with biofilms of medically relevant bacteria. Biochim Biophys Acta. 2015;1858: 1044–1060. 10.1016/j.bbamem.2015.10.013 [DOI] [PubMed] [Google Scholar]
- 45.Prudhomme M, Berge M, Martin B, Polard P. Pneumococcal Competence coordination relies on a cell-contact sensing mechanism. PLoS Genet. 2016;12: e1006113 10.1371/journal.pgen.1006113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y, Tang N, Aspiras MB, Lau PCY, Lee JH, Ellen RP, et al. A quorum-sensing signaling system essential for genetic competence in streptococcus mutans is involved in biofilm formation. J Bacteriol. 2002;184: 2699–2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Petersen FC, Pecharki D, Scheie AA. Biofilm mode of growth of Streptococcus intermedius favored by a competence-stimulating signaling peptide. J Bacteriol. 2004;186: 6327–6331. 10.1128/JB.186.18.6327-6331.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Piñas GE, Cortes PR, Albarracín Orio AG, Echenique J. Acidic stress induces autolysis by a CSP-independent ComE pathway in Streptococcus pneumoniae. Microbiology. 2008;154: 1300–1308. 10.1099/mic.0.2007/015925-0 [DOI] [PubMed] [Google Scholar]
- 49.Mascher T, Heintz M, Zähner D, Merai M, Hakenbeck R. The CiaRH system of Streptococcus pneumoniae prevents lysis during stress induced by treatment with cell wall inhibitors and by mutations in pbp2x involved in β-lactam resistance. J Bacteriol. 2006;188: 1959–1968. 10.1128/JB.188.5.1959-1968.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Y, Burne RA. The major autolysin of Streptococcus gordonii is subject to complex regulation and modulates stress tolerance, biofilm formation, and extracellular-DNA release. J Bacteriol. 2011;193: 2826–2837. 10.1128/JB.00056-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee SF. Oral colonization and immune responses to Streptococcus gordonii: Potential use as a vector to induce antibodies against respiratory pathogens. Curr Opin Infect Dis. 2003;16: 231–235. 10.1097/01.qco.0000073772.11390.ae [DOI] [PubMed] [Google Scholar]
- 52.Knight JB, Halperin SA, West KA, Lee SF. Expression of a functional single-chain variable-fragment antibody against complement receptor 1 in Streptococcus gordonii. Clin Vaccine Immunol. 2008;15: 925–931. 10.1128/CVI.00500-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burne RA, Wen ZT, Chen YM, Penders JEC. Regulation of expression of the fructan hydrolase gene of Streptococcus mutans GS-5 by induction and carbon catabolite repression. J Bacteriol. 1999;181: 2863–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tremblay YDN, Lo H, Li YH, Halperin SA, Lee SF. Expression of the Streptococcus mutans essential two-component regulatory system VicRK is pH and growth-phase dependent and controlled by the LiaFSR three-component regulatory system. Microbiology. 2009;155: 2856–2865. 10.1099/mic.0.028456-0 [DOI] [PubMed] [Google Scholar]
- 55.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3: 1101–1108. 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- 56.Love RM, McMillan MD, Jenkinson HF. Invasion of dentinal tubules by oral streptococci is associated with collagen recognition mediated by the antigen I/II family of polypeptides. Infect Immun. 1997;65: 5157–5164. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=175743&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dunny GM, Lee LN, LeBlanc DJ. Improved electroporation and cloning vector system for Gram-positive bacteria. Appl Environ Microbiol. 1991;57: 1194–201. Available: Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=182867&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.DuBois M, Gilles KA, Hamilton JK, Rebers PA, Smith F. Colorimetric method for determination of sugars and related substances. Anal Chem. 1956;28: 350–356. 10.1021/ac60111a017 [DOI] [Google Scholar]
- 59.Davis E, Kennedy D, Halperin SA, Lee SF. Role of the cell wall microenvironment in expression of a heterologous SpaP-S1 fusion protein by Streptococcus gordonii. Appl Environ Microbiol. 2011;77: 1660–1666. 10.1128/AEM.02178-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee SF, Halperin SA, Wang H, MacArthur A. Oral colonization and immune responses to Streptococcus gordonii expressing a pertussis toxin S1 fragment in mice. FEMS Microbiol Lett. 2002;208: 175–178. Available: http://www.ncbi.nlm.nih.gov/pubmed/11959433 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(a) Quantitative PCR analysis of scaA expression in 24 h biofilm cells. RNA was prepared from the parent, the ΔsdbA mutant, and the sdbA-complemented mutant (+SdbA). (b) Biofilm formation by the parent, the ΔsdbA mutant, and the ΔsspAB mutant (OB219) with and without a functional CiaRH two-component signaling system. Biofilms were grown for 24 h and stained with crystal violet.
(TIFF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information file.