Abstract
Immunotherapy with CD123-specific T-cell engager proteins or with T cells expressing CD123-specific chimeric antigen receptors is actively being pursued for acute myeloid leukemia. T cells secreting bispecific engager molecules (ENG-T cells) may present a promising alternative to these approaches. To evaluate therapeutic potential, we generated T cells to secrete CD123/CD3-bispecific engager molecules. CD123-ENG T cells recognized primary acute myeloid leukemia (AML) cells and cell lines in an antigen-dependent manner as judged by cytokine production and/or tumor killing, and redirected bystander T cells to AML cells. Infusion of CD123-ENG T cells resulted in regression of AML in xenograft models conferring a significant survival advantage of treated mice in comparison to mice that received control T cells. At high effector to target ratios, CD123-ENG T cells recognized normal hematopoietic stem and progenitor cells (HSPCs) with preferential recognition of HSPCs from cord blood compared to bone marrow. We therefore introduced the CD20 suicide gene that can be targeted in vivo with rituximab into CD123-ENG T cells. The expression of CD20 did not diminish the anti-AML activity of CD123-ENG T cells, but allowed for rituximab-mediated ENG-T cell elimination. Thus, ENG-T cells coexpressing CD20 suicide and CD123 engager molecules may present a promising immunotherapeutic approach for AML.
Introduction
The outcome for pediatric and adult patients with acute myeloid leukemia (AML) remains poor, particularly in those with high risk or relapsed disease.1,2,3 Additionally, current treatment protocols heavily rely on chemotherapeutic agents whose use commonly leads to serious acute and long-term toxicities. Given this, there is a need to develop novel targeted therapies that improve outcomes and reduce treatment-related complications of current therapies.
The ex vivo preparation of antigen-specific T cells followed by their adoptive transfer is one attractive strategy to improve outcomes for hematological malignancies, since T-cell killing does not rely on the broadly cytotoxic mechanisms of conventional therapies.4,5,6,7 Indeed the adoptive transfer of T cells that are genetically modified with CD19-specific chimeric antigen receptors (CARs) has resulted in impressive clinical responses; especially in patients with acute lymphoblastic leukemia.8,9,10,11,12,13,14,15 However, for AML, there has been limited success. Lewis Y (LeY)-specific CAR T cells have been tested so far in one clinical study without robust response.16 In addition, CD33-specific CAR T cells were evaluated in a single patient with limited success.17 Several groups have explored interleukin-3 receptor alpha (IL3Rα, CD123)-specific CAR T cells for AML in preclinical models, and while these cells had potent antitumor activity, one group demonstrated that normal hematopoietic stem and progenitor cells (HSPCs) are also eliminated.18,19,20,21,22
We and others have developed an alternative strategy to generate tumor-specific T cells by genetic modification with diabodies,23 or secretable, bispecific T-cell engager molecules, which consist of two single chain variable fragments (scFVs) specific for a tumor-associated antigen and CD3ɛ (ENG-T cells).24 These T cells not only recognize and kill tumor cells in a tumor-associated antigen-dependent manner, but also have the unique ability to redirect bystander T cells to tumor cells.24 Consistent synthesis of engagers by adoptively transferred T cells should be superior to the direct infusion of the recombinant bispecific antibody, because these typically have short half-lives and do not accumulate at tumor sites.
Here, we report the development of CD123-ENG T cells and demonstrate that these ENG-T cells recognize and kill CD123-positive target cells in vitro, redirect bystander T cells, and have potent antitumor activity in vivo. Since CD123-ENG T cells recognized normal HSPCs at high effector to target ratios, we further genetically modified CD123-ENG T cells to also include the CD20 suicide gene (CD20.CD123-ENG T cells).25,26,27 CD20.CD123-ENG T cells had equivalent effector function in comparison to CD123-ENG T cells and were readily killed in the presence of rituximab and complement making them a promising T-cell therapy “candidate” for future clinical development.
Results
Generation of CD123-ENG T cells
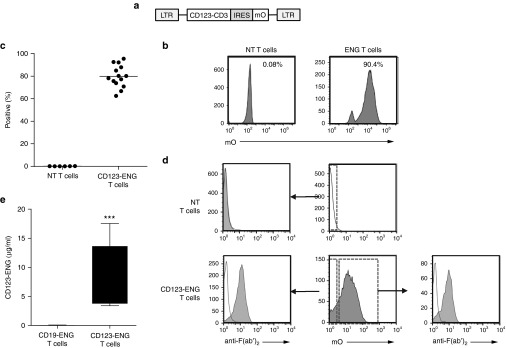
CD123-ENG T cells were generated by transduction with an RD114-pseudotyped retroviral vector encoding a bispecific CD123-CD3 engager molecule, an internal ribosomal entry site (IRES), and mOrange (Figure 1a). Transduction of T cells was confirmed by fluorescence-activated cell sorting (FACS) analysis for mOrange expression. Mean transduction efficiency was 79.2% (range: 62.5–95.5%; n = 14; Figure 1b,c). Phenotypic analysis of transduced T cells revealed a mixture of CD4- and CD8-positive T cells, with reproducible percentages of naive, central memory, and effector memory cell populations (Supplementary Figure S1, n = 5). Transduction of cells and expression of CD123-ENG did not alter the T-cell phenotype in comparison to nontransduced (NT) T cells activated and expanded in parallel. CD123-ENG secretion and binding to both transduced and NT T cells was confirmed by FACS analysis using an anti-mouse F(ab')2 (Figure 1d). To quantify CD123-ENG protein in cell culture media, we developed an enzyme-linked immunosorbent assay (ELISA) using recombinant CD123 T-cell ENG protein as a standard (Supplementary Figure S2). CD123 T-cell ENG protein was readily detected in medium conditioned by CD123-ENG T cells (mean: 7.5 µg/ml, 95% CI: 4.0–11.1 µg/ml) in contrast to medium conditioned by T cells expressing CD19 T-cell ENG protein (CD19-ENG T cells; mean: 9.8 ng/ml, 95% CI: 0–26.06 ng/ml) confirming specificity of the developed assay (Figure 1e).
Figure 1.
Generation of CD123-ENG T cells. (a) Schematic of retroviral vector encoding CD123-ENG and mOrange. (b,c) Representative FACS diagram and summary data (CD123-ENG T cells (n = 14), NT T cells (n = 6) of mOrange expression post-transduction. (d) A mouse F(ab')2 antibody was used to detect cell surface-bound CD123 T-cell ENG protein. mOrange-positive and -negative T cells stained positive (filled curve) for CD123 T-cell ENG in contrast to samples that were stained with isotype alone (open curve). NT T cells cultured without CD123-ENG T cells did not stain positive with the mouse F(ab')2 antibody, confirming specificity. (e) Detection of CD123 T-cell ENG protein in media of CD123-ENG and CD19-ENG T cells after 24 hours of culture (n = 4, performed in triplicates, box graph, whiskers: min, max, CD123-ENG versus CD19-ENG T cells P < 0.001).
CD123-ENG T cells recognize and kill CD123-positive AML cells in vitro
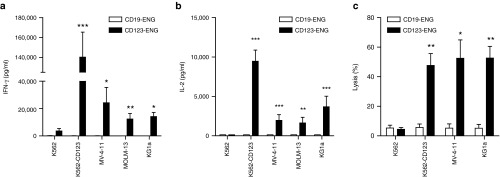
To demonstrate the specificity of CD123-ENG T cells against tumor cells expressing the antigen target, we used a panel of CD123-positive (K562-CD123, KG1a, MV-4–11, MOLM-13) and CD123-negative (K562) cell lines (Supplementary Figure S3). We performed coculture assays with these tumor cells and ENG T cells targeting CD123 or an irrelevant antigen not expressed on cell targets (CD19). CD123-ENG T cells secreted significant amounts of IFNγ and IL-2 when exposed to cells expressing CD123 (Figure 2a,b). In contrast, coculture with CD123-negative cells did not induce cytokine production. Similarly, CD19-ENG T cells did not produce cytokine when cocultured with target cells (Figure 2a,b). To further confirm specificity, we assessed the killing potential of CD123-ENG T cells against CD123-positive target cells in a 4-hour chromium release cytotoxicity assay. CD123-ENG T cells effectively lysed AML cells expressing CD123, but did not kill the CD123-negative K562 cell line (Figure 2c). CD19-ENG T cells did not have any cytolytic activity against the tested cell lines, confirming specificity.
Figure 2.
CD123-ENG T cells recognize and kill CD123-positive acute myeloid leukemia cells. (a, b) CD123-ENG or CD19-ENG T cells were cocultured with CD123-positive (K562-CD123, MV-4-11, MOLM-13, KG1a) or -negative (K562) cell lines. After 24 hours, (a) IFNγ or (b) IL-2 was determined by ELISA (n = 3–4, assay performed in duplicates; CD123-ENG versus CD19-ENG: *P < 0.05, **P < 0.01, ***P < 0.001). (C) Cytotoxicity assays were performed using CD123-ENG or CD19-ENG T cells as effectors and CD123-positive (K562-CD123, MV-4-11, KG1a) or -negative (K562) cell lines as targets at a E:T ratio of 10:1 (mean + SD; n = 4; assay was performed in triplicates, *P < 0.02, **P < 0.002).
CD123-ENG T cells redirect bystander T cells to CD123-positive AML cells in vitro
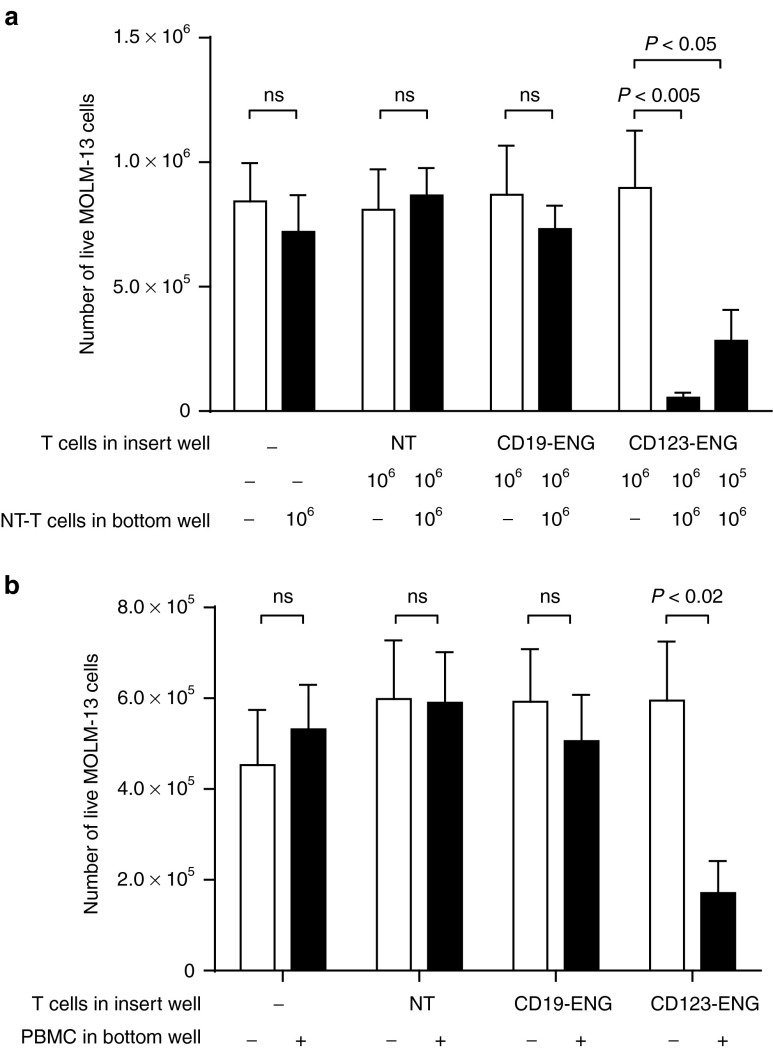
To examine the potential of ENG T cells to activate bystander cells in vitro, we performed transwell assays with insert wells that allow the diffusion of molecules but not the migration of cells. ENG T cells were plated in the insert well and KG1a target cells with NT T cells in the bottom well. Only CD123-ENG T cells, in contrast to NT T cells and CD19-ENG T cells, were able to induce significant target killing by NT T cells (Figure 3a). No tumor cell killing was observed in wells that contained only target cells without T cells, indicating that engager protein alone is inherently nontoxic. We confirmed the ability of CD123-ENG T cells to redirect T cells that had not been expanded ex vivo by repeating the experiment with peripheral blood mononuclear cells (PBMCs) as a source of bystander T cells in the bottom well (Figure 3b).
Figure 3.
CD123-ENG T cells activate bystander T cells and freshly isolated peripheral blood mononuclear cells (PBMCs) against CD123-positive target cells. (a) 1 × 106 MOLM-13.GFP.ffLuc cells were plated in the bottom well with or without 1 × 106 NT T cells. Control (NT, CD19-ENG) or CD123-ENG T cells were plated in the insert well at the indicated T-cell dose. Following 24-hour incubation, viable MOLM-13.GFP.ffLuc cells were quantified by luciferase assay (n = 3; assay was performed in duplicates; for CD123-ENG: 106 ENG-T cells in insert, 106 NT T cells in bottom well versus 106 ENG-T cells in insert, no NT T cells in bottom well: P < 0.005, 105 ENG-T cells in insert, 106 NT T cells in bottom well versus 106 ENG-T cells in insert, no NT T cells in bottom well: P < 0.05). (b) 1 × 106 MOLM-13.GFP.ffLuc cells were plated in the bottom well with or without 5 × 106 PBMCs. 1 × 106 autologous NT, CD19-ENG, or CD123-ENG T cells were plated in the insert well. Following 24-hour incubation, viable MOLM-13.GFP.ffLuc cells were quantified by luciferase assay (n=3; assay was performed in triplicates; for CD123-ENG: PBMCs versus no PBMCs: P < 0.02).
CD123-ENG T cells kill primary AML blasts in vitro
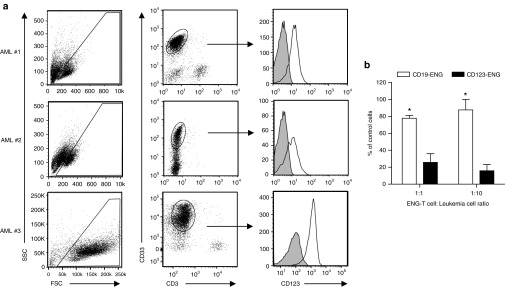
To further assess the potential translational impact of CD123-ENG T cells as a treatment for AML patients, we tested their cytolytic activity against primary AML blasts. Pediatric AML samples were collected, and first evaluated for CD123 expression (Figure 4a). The blast percentage recorded by our clinical hematopathology laboratory and further described in the methods section was AML#1: 33%, AML#2: 90%, and AML#3: 65%. Primary samples were then treated with CD123- or CD19-ENG T cells at an E:T ratio of 1:1 or 1:10 prior to plating in semisolid media with growth factors present. Untreated samples served as controls. After 10–14 days of culture, cells were counted. At both E:T ratios tested CD123-ENG T cells induced significant AML blast killing in comparison to CD19-ENG T cells (Figure 4b, P < 0.05).
Figure 4.
Primary AML cells are killed by CD123-ENG T cells in vitro. (a) FACS analysis of pediatric primary acute myeloid leukemia (AML) samples for CD123 expression. AML blasts,(CD33+, CD3-, CD19- (data not shown for CD19)) were analyzed for CD123 expression (filled curve: isotype control; open curve: CD123 MAb). (b) Primary AML samples were treated with CD19-ENG or CD123-ENG T cells at E:T ratios of 1:1 and 1:10 for 6 hours. Following coculture, cells were plated in MethoCult media and incubated for 10–14 days. Final cell counts are displayed as percentage of control; n = 3; CD19-ENG versus CD123-ENG T cells: *P < 0.05.
CD123-ENG T cells have anti-AML activity in vivo
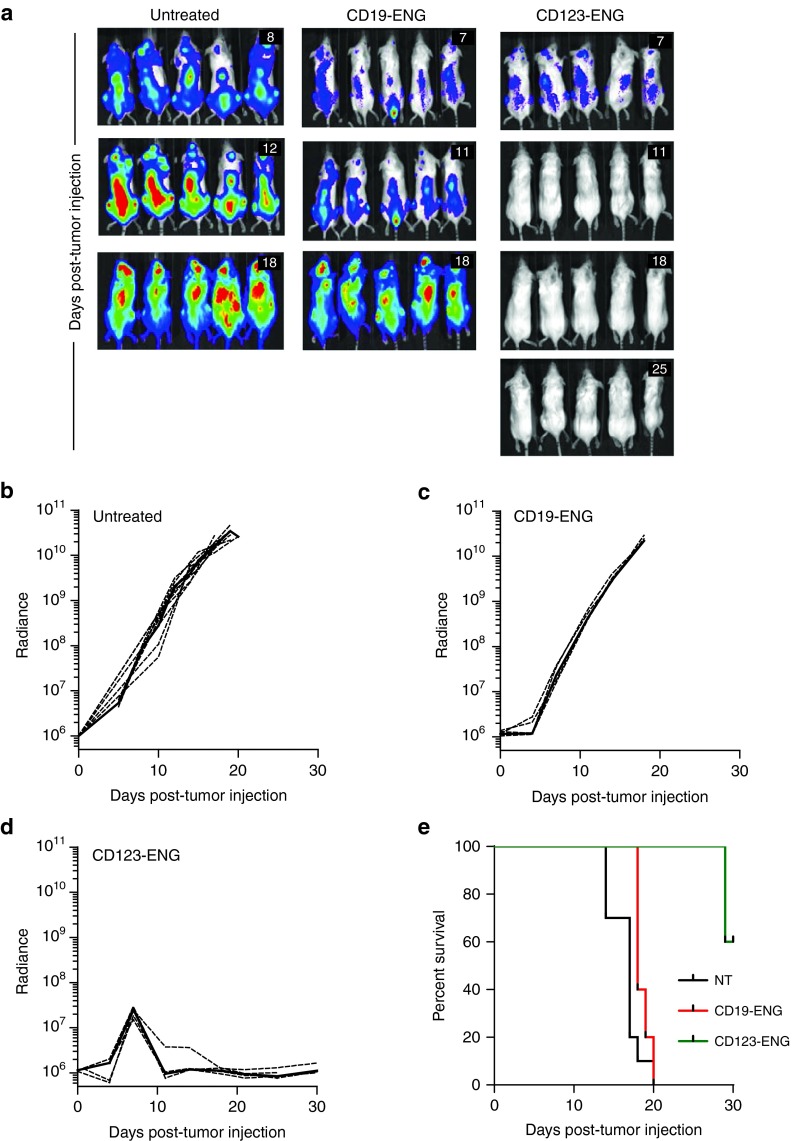
We evaluated the anti-AML activity of CD123-ENG T cells in our MOLM-13 AML NSG xenograft model. Post sublethal irradiation, mice were injected with MOLM-13.GFP.ffLuc cells and on day 7 and 14 received an intravenous injection of CD19-ENG or CD123-ENG T cells. Untreated mice served as controls. Within 4 days post the first T-cell injection mice treated with CD123-ENG T cells had a significant decline in their bioluminescence signal (P < 0.001) with subsequent leukemia eradication in all treated mice (Figure 5a,d). In contrast, untreated mice and mice treated with CD19-ENG T cells had a significant increase (P < 0.002) in their bioluminescence signal (Figure 5b,c) and died of progressive disease by day 20 (Figure 5e). While none of the CD123-ENG T-cell treated mice developed recurrent leukemia, 4/5 mice died 29 (×2), 46, and 67 days post-tumor cell injection. The cause of death was xenogeneic graft versus host disease (GVHD) as commonly reported with adoptive transfer of human T cells in the NSG murine model.28,29 We confirmed this for 1/4 mice by demonstrating human T-cell infiltration throughout the liver (Supplementary Figure S4A).
Figure 5.
CD123-ENG T cells have potent anti-acute myeloid leukemia activity in vivo. Antitumor activity of CD123-ENG T cells in MOLM-13 leukemia NSG xenograft model. MOLM-13.GFP.ffLuc-bearing mice received an i.v. dose of 1 × 107 CD123-ENG (n = 5) or CD19-ENG T cells (n = 5) on day 7 and 14 post-tumor cell injection. Untreated animals served as controls (n = 10). Tumor growth was followed by bioluminescence imaging. (a) Representative images of animals (day post-tumor cell injection is shown in the upper right corner of images). (b–d) Quantitative bioluminescence imaging results (dotted lines: individual mice; solid lines: median; radiance=photons/sec/cm2/sr). Starting day 3 post-first T-cell injection for CD123-ENG versus CD19-ENG T cells, and CD123-ENG T cells versus untreated: P < 0.001). (e) Kaplan-Meier survival curve (control versus CD123-ENG T cells: P = 0.0004; control versus CD19-ENG T cells: NS; CD123-ENG versus CD19-ENG T cells: P = 0.0015).
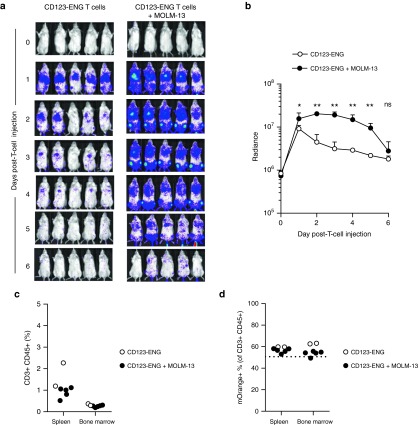
To assess the contribution of target antigen-specific CD123-ENG T-cell stimulation in inducing GVHD, MOLM-13 bearing and control mice were injected on day 7 with 1 × 107 CD123-ENG T cells that were also genetically modified to express GFP.ffLuc. While in control mice there was a rapid, significant decline (P < 0.05) of infused T cells, no significant decline of CD123-ENG T cells was observed for 5 days postinfusion in MOLM-13 bearing mice (Figure 6a,b). Mice were euthanized on day 6 to determine the presence of human T cells (human CD3 and CD45 positive) and CD123-ENG expressing human T cells (mOrange positive) in spleen and bone marrow. The average frequency of human T cells was 1.1% (range: 0.52–2.27%) in spleens and 0.3% (range: 0.27–0.37%) in bone marrows with no significant difference between control and MOLM-13 bearing mice (Figure 6c). In the infused CD123-ENG T-cell product 50.7% of cells were mOrange positive. On average 56.3% (range: 53.0–59.8%) of human T cells from spleens and 57.3% (range: 54.3–63.2%) of human T cells from bone marrows were positive for mOrange (CD123-ENG T cells) with no significant difference between control and MOLM-13 bearing mice, and no significant difference to the infused T-cell product (Figure 6d). Residual MOLM-13 cells were not detectable in spleens and bone marrows of mice that had been injected with MOLM-13 as judged by FACS analysis of CD123 (data not shown).
Figure 6.
Antigen-specific persistence of CD123-ENG T cells in vivo. MOLM-13 bearing or control mice were injected I.V. with 1 × 107 CD123-ENG.mOrange T cells that were also genetically modified to express GFP.ffLuc (n = 5 per group). (a) Images of individual mice. (b) Quantitative bioluminescence imaging results (radiance = photons/sec/cm2/sr, mean and SD is plotted, *P < 0.001, **P < 0.0001). (c, d) On day 6 post-T-cell injection, mice were euthanized and spleens and bone marrows (both femurs) of all (5) acute myeloid leukemia (AML)-bearing mice and of two control mice that received CD123-ENG T cells were processed for FACS analysis. (c) Percentage of human T cells (CD45-positive, CD3-positive) gated on live cells (AML-bearing mice versus control mice: P = NS). (d) mOrange-positive cells (%) gated on human T cells (CD45-positive, CD3-positive); dotted line: mOrange-positive cells (%) in infused CD123-ENG T cells (AML- bearing mice versus control mice: P = NS; % mOrange positive cells in infused CD123-ENG T cells versus % mOrange-positive T cells on day 6 postinfusion: P = NS).
Toxicity of CD123-ENG T cells against hematopoietic stem and progenitor cells
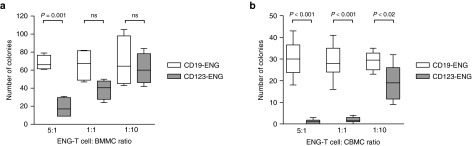
Given the reported low level of expression of CD123 on HSPCs, we sought to determine if CD123-ENG T cells recognize this cell population.30,31 We used cord blood or bone marrow as a source of HSPCs. Samples were treated with CD123- or CD19-ENG T cells at E:T ratios of 5:1, 1:1, or 1:10 prior to plating in semisolid media with growth factors present. Untreated samples served as controls. After 10–14 days, colony forming unit (CFUs) were counted. At an E:T ratio of 5:1, significant toxicity was observed (Figure 7a,b). At an E:T ratio of 1:1 or 1:10, toxicity was dependent on the stem cell source with umbilical cord- derived HSPCs being more sensitive to CD123-ENG T cells than bone marrow-derived HSPCs.
Figure 7.
CD123-ENG T cells recognize HSPCs at high effector to target ratios. Bone marrow mononuclear cells (BMMCs) or cord blood mononuclear cells (CBMCs) were cultured of E:T ratios of 5:1, 1:1, and 1:10 with CD19-ENG or CD123-ENG T cells for 6 hours. Cells were plated in MethoCult media and CFUs were counted after 10–14 days. Box graph, whiskers: min, max. (a) BMMCs: n = 4; CD123-ENG versus CD19-ENG T cells: 5:1 P = 0.001, 1:1 P = ns, and 1:10 P = ns. (b) CBMCs: n = 4; CD123-ENG versus CD19- ENG T cells: 5:1 P < 0.001, 1:1 P < 0.001, and 1:10 P < 0.02.
CD123-ENG T cells expressing the CD20 suicide gene retain their anti-AML activity and are eliminated in the presence of rituximab and complement
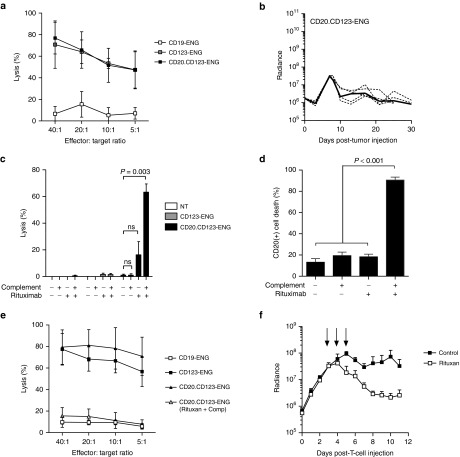
While CD123-ENG T cells had potent anti-AML activity, they induced killing of normal HSPCs at high E:T ratios, indicating that CD123-ENG T cells should also express a suicide gene to allow their selective depletion in the event of unwanted side effects. We focused on transgenic expression of CD20, which allows the depletion of T cells by the FDA-approved CD20 monoclonal antibody rituximab via complement-mediated or antibody-dependent cell-mediated cytotoxicity. To coexpress CD20 and CD123 T-cell ENG molecules in T cells, we generated a retroviral vector encoding CD20, a 2A sequence, and our CD123-ENG (CD20.CD123-ENG; Supplementary Figure S5A). CD20.CD123 ENG T cells were generated by retroviral transduction and a mean of 60.3% (range: 42.5–73.5%, n = 8, Supplementary Figure S5B) of T cells were genetically modified as judged by FACS analysis for CD20. There was no change in the percentage of CD20-positive T cells early (day 3–5) versus late (day 13–15) post-transduction, indicating that transgene expression is stable over time (Supplementary Figure S5C). We observed no difference in the cytolytic activity between CD123-ENG T cells and CD20.CD123-ENG T cells (Figure 8a, P = 0.450). In vivo, in the MOLM-13 AML NSG xenograft model, a single dose of 1 × 107 CD20.CD123-ENG T cells had similar antitumor activity (Figure 8b) as CD123-ENG T cells in the previously performed experiment (Figure 5d).
Figure 8.
CD20 gene-modified CD123-ENG T cells retain their effector function against CD123-positive acute myeloid leukemia and are effectively eliminated by rituximab. (a) Cytotoxicity assays were performed using CD123-ENG, CD20.CD123-ENG, or CD19-ENG T cells as effectors and KG1a as CD123-positive target. (n = 3; assay was performed in triplicates, P = 0.450, CD123-ENG versus CD20.CD123-ENG T cells). (b) MOLM-13.GFP.ffLuc-bearing mice received an i.v. dose of 1 × 107 CD20.CD123-ENG T cells on day 7 post-tumor cell injection. Quantitative bioluminescence imaging results (dotted lines: individual mice; solid line: median; radiance = photons/sec/cm2/sr). (c) CD20.CD123-ENG, CD123-ENG, or NT T cells were labeled with 51Chromium and treated with rituximab and/or complement in a standard cytotoxicity assay. (n = 4; assay was performed in triplicate; for CD20.CD123-ENG T cells: untreated versus rituximab and complement: P = 0.003). (d, e) CD20.CD123-ENG T cells were treated with rituximab, complement, or rituximab and complement, and cultured for 7 days. (D) FACS analysis for CD20-positive cells. Data is presented as percent cell death of CD20-positive cells 7 days post-treatment (n = 5; untreated, rituximab treated, complement treated vs rituximab and complement treated cells: P < 0.001). (e) Cytotoxicity assays using untreated CD123-ENG, CD19-ENG, CD20.CD123-ENG, or rituximab and complement treated CD20.CD123-ENG T-cell lines as effectors and KG1a cells as targets. n = 3; assay performed in triplicates; untreated versus treated CD20.CD123-ENG T cells: P < 0.001; untreated CD20.CD123-ENG T cells versus CD19-ENG T cells: P < 0.001; treated CD20.CD123-ENG T cells versus CD19-ENG T cells: P = ns for all E:T ratios tested. (f) Mice engrafted with MOLM-13 were treated with 3x106 CD20.CD123-ENG/GFP.ffLuc T cells on Day 7 post-leukemia injection. On Days 3–5 post-T-cell injection mice (n = 4) received 250 µg rituximab IP. Untreated mice (n = 3) served as controls. Quantitative bioluminescence imaging results (radiance = photons/sec/cm2/sr, mean and SD is plotted; P < 0.05 starting 2 days post-first dose of rituximab).
To evaluate the functionality of the CD20 suicide gene, we performed a cytotoxicity assay with 51Cr-labeled NT, CD123-ENG, or CD20.CD123-ENG T cells in the presence of rituximab and/or complement. While complement or rituximab alone induced <20% killing of CD20.CD123-ENG T cells, ~60% killing was observed in the presence of rituximab and complement in a 4-hour chromium release assay (Figure 8c). Since the mean transduction efficiency of CD20.CD123-ENG T-cell lines was 60.3% (range: 42.5–73.5%) in the performed assays, these results demonstrate >95% killing of transduced T cells in the presence of rituximab and complement. These findings were confirmed in a 4-hour chromium release assay using CD20-selected CD20.CD123-ENG T cells (Supplementary Figure S6A,B). In contrast, rituximab and complement had no cytolytic activity against NT or CD123-ENG T cells. To determine the durability of rituximab/complement-mediated killing of CD20.CD123-ENG T cells in a cell culture assay, CD20.CD123-ENG T cells were treated with rituximab/complement for two hours, washed, and the surviving T cells cultured for 7 days. Greater than 90% of CD20.CD123-ENG T cells were killed as judged by FACS analysis (Figure 8d, n = 5, P < 0.001). In contrast, no significant killing of CD20-positive cells was observed in the presence of rituximab or complement alone, when compared to untreated cells. Concurrent with FACS analysis, a standard chromium release cytotoxicity assay was performed using KG1a as targets and untreated or rituximab/complement-treated CD20.CD123-ENG T-cell lines as effectors, one week after treatment. CD19-ENG T-cell lines served as negative, and CD123-ENG T-cell lines as positive controls. While untreated CD20.CD123-ENG T-cell lines readily killed KG1a cells, rituximab/complement-treated CD20.CD123-ENG T-cell lines did not, indicating successful depletion of CD20.CD123-ENG T cells (Figure 8e). Having established the functionality of the CD20 suicide gene in vitro, we assessed the ability of rituximab to deplete CD20.CD123-ENG T cells in vivo. Post sublethal irradiation, mice were injected with MOLM-13 cells, and on day 7 received 3 × 106 CD20.CD123-ENG T cells that were also genetically modified with GFP.ffLuc, and selected for CD20 expression (Supplementary Figure S6A). Starting day 3 post-T-cell infusion, mice received intraperitoneal injection (IP) of 250 µg rituximab for 3 consecutive days. Rituximab administration resulted in a greater than 90% decrease in the bioluminescence signal, demonstrating the effectiveness of rituximab to destroy CD20.CD123-ENG T cells in vivo. (Figure 8f, rituximab versus no treatment: P < 0.05 starting day 2 post-first dose of rituximab).
Discussion
Herein, we describe the generation of CD123-ENG T cells and demonstrate that these cells recognize and kill CD123-positive target cells in an antigen-dependent manner, redirect “bystander” T cells to CD123-positive target cells, and have potent antitumor activity in vivo. We also observed killing of HSPCs at high effector to target ratios, and therefore combined the expression of CD123-ENG molecules with a CD20 suicide gene in T cells. CD20.CD123-ENG T cells retained their antileukemia activity and could be readily depleted in the presence of rituximab and complement.
T-cell immunotherapy as a less toxic, precision therapy for the treatment of leukemia is a therapeutic approach that has enjoyed recent popularity due to the success of CD19-CAR T cells administered for refractory acute lymphoblastic leukemia.8,9,10,11,12,13,14,15 In contrast to CAR T-cell therapy for acute lymphoblastic leukemia, CAR T-cells targeting AML have not been extensively tested in the clinic. However, several antigens are currently being actively explored in preclinical studies including CD33, CD44v6, CD123, and LeY.16,17,18,19,20,21,22,32–36 While all of these antigens have been targeted with CAR T cells, none has been targeted with ENG T cells, which are a new class of T cells with the unique ability to redirect bystander T cells to tumor cells in an antigen dependent fashion.24
CD123-ENG T cells not only killed AML cell lines but also primary AML blasts in vitro, confirming studies by others that CD123-positive AML cells can be targeted with genetically modified T cells.18,19,20,21,22 While all previous studies used T cells expressing CD123-CARs, which cannot activate bystander T cells against AML cells, we show here that ENG T cells readily redirect unmodified bystander T cells to CD123-positive target cells. Since the efficacy of CAR T-cell therapy relies on significant in vivo expansion of T cells, the observed “bystander effect” might allow for disease control with limited in vivo expansion of adoptively transferred T cells. We have shown in vivo that the short-term persistence of CD123-ENG T cells is significantly impacted by the presence of the antigen target. In our model system, the ratio of CD123-ENG expressing to unmodified human T cells is maintained, suggesting equal stimulation of CD123-ENG and unmodified cells. This indicates that ENG T cells have the capability of activating 100% of infused T cells, potentially overcoming lower transduction efficiencies of patients' T cells. We observed xenogenic GVHD in treated mice. In our in vivo CD123-ENG T-cell persistence studies we only observed a significant difference in the frequency of CD123-ENG T cells between MOLM-13 bearing mice and control mice for the first 5 days after injection. Thus GVHD in our model is most likely due to nonspecific xenogenic T-cell stimulation at later time points post-infusion as observed by others.28,29
CD123-ENG molecules induced significant IL-2 production in an antigen-dependent manner without having a costimulatory domain. While AML blasts do not express standard costimulatory molecules such as CD80,37 they express for example NKG2D ligands,38 which can provide costimulation39 explaining the observed IL-2 production. One potential advantage of ENG T cells versus the infusion of bispecific antibodies is that T cells can traffic to tumor sites and produce ENG molecules locally. We could not demonstrate this advantage in the described systemic AML model, but have shown ENG T-cell homing to tumor sites in a local B-cell malignancy model.40 Bispecific antibodies such as blinatumomab that consist of 2 scFvs have a half-life of 2 hours in vivo.41 Since half-life depends on molecular weight, we expect that scFv-based ENG molecules secreted from T cells will have a similar half-life.
CD123 is expressed at low levels on plasmacytoid dendritic cells, basophils, monocytes, endothelial cells, and HSPCs.30,42,43 Controversy exists in regards to the recognition of normal HSPCs by CD123-CAR T cells. While one group demonstrated that the infusion of CD123-CAR T cells inhibited the engraftment of fetal liver- or bone marrow-derived HSPCs in NSG mice,21,22 two groups reported limited toxicity of CD123-CAR T cells.18,19,20 Limited toxicity has also been observed for a CD123/CD3-specific dual affinity retargeting recombinant protein. In a non-human primate model, CD123/CD3-specific dual affinity retargeting induced a cytokine release syndrome (CRS) with the first infusion and a transient decrease in hematocrit after multiple infusions.44 Here we show that the toxicity of CD123-ENG T cells depends on the used E:T ratio as well as the source of the HSPCs in CFU assays. We did not differentiate between lineage-specific CFUs since we wanted to assess the ‘global toxicity' on normal HSPCs of CD123-ENG T cells. We are planning to determine the effects on lineage-specific CFUs in the future. We observed that HSPCs derived from cord blood were more sensitive to CD123-ENG T-cell mediated killing than HSPCs derived from bone marrow. Others have reported that the percentage of HSPCs expressing CD123 as well as the expressed level of CD123 is higher in cord blood than bone marrow derived HSPCs, which supports our findings.45 While high ratios caused significant killing of HSPCs, limited killing was observed at low ratios at which AML blasts were still killed. These results suggest that there may be a “therapeutic window”, which would allow the killing of AML blasts, while sparing normal HSPCs.
Since it is impossible to predict the effective E:T ratio in vivo, we reasoned that introducing a ‘suicide gene' into CD123-ENG T cells would be advisable for future clinical development. We first evaluated the inducible caspase 9 suicide gene in CD123-ENG T cells, but observed baseline toxicity (Supplementary Figure S7). We next focused on expressing CD20 in T cells in combination with rituximab administration as a “suicide gene” approach,25,26,27 since antibodies can travel freely through the sinusoidal clefts that are present in bone marrow,46 the site of potential HSPC toxicity. CD20.CD123-ENG T cells had stable CD20 expression, comparable antitumor activity in vitro and in vivo, and were readily eliminated by rituximab in the presence of complement in vitro. We also showed that rituximab administration in vivo resulted in a greater than 90% depletion of CD20.CD123-ENG/GFP.ffLuc T cells as judged by bioluminescence imaging in MOLM-13-bearing NSG mice. Of note, NSG mice have deficient complement pathways.47 Thus the observed in vivo T-cell killing in these mice relies mainly on FC-receptor mediated killing. In humans, which have an intact complement pathway as well as FC-receptor mediated killing, we expect rituximab to even be more effective in depleting CD20-positive T cells. Thus, our in vitro and in vivo results indicate that CD20 expression in combination with rituximab administration is a suitable suicide gene method to eliminate CD123-ENG T cells. Nevertheless, to move our approach to clinical application, we propose to perform first-in-human studies in the pretransplant setting. We aim to use CD20.CD123-ENG T cells to induce complete remission in refractory AML patients who have an identified stem cell donor, before proceeding to an allogeneic stem cell transplant. Our approach is comparable to the strategy taken by other investigators who have open clinical studies with CD123-CAR T cells (NCT02159495, NCT02623582), and is expected to mitigate the risk of long-term aplasia and graft failure from all CD123-targeted T-cell therapies, including CD123-ENG T cells.
In summary, our study demonstrates that CD20.CD123-ENG T cells have potent antitumor activity, redirect bystander T cells to CD123-positive AML, and are readily eliminated by rituximab/complement. Thus, CD20.CD123-ENG T cells may be a promising alternative to current CD123-targeted immunotherapy approaches that either rely on the continuous infusion of recombinant protein or the adoptive transfer of CAR T cells.
Materials and Methods
Cells and culture conditions. 293T, K562, KG1a, and MV-4-11 cell lines were purchased from the American Type Culture Collection (ATCC, Manassas, VA) and cultured in Dulbecco's Modified Eagle Medium (DMEM, ThermoScientific, Waltham, MA) supplemented with 2 mmol/l Glutamax (Invitrogen, Carlsbad, CA) and 10% Fetal Bovine Serum (FBS, ThermoScientific (293T)) or Roswell Park Memorial Institute (RPMI, ThermoScientific), supplemented with 2 mmol/l Glutamax and 10% (K562) or 20% (MV-4–11) FBS or scove's Modified Dulbecco's Medium (IMDM, ThermoScientific) supplemented with 2 mmol/l Glutamax and 20% Fetal Bovine Serum (FBS) FBS (KG1a). MOLM-13 cell line was purchased from the Leibniz Institute (DSMZ, German Collection of Microoganisms and Cell Cultures, Braunschweig, Germany) and cultured in RPMI supplemented with 2 mmol/l Glutamax and 10% FBS. All cells were maintained in a humidified atmosphere containing 5% CO2 at 37 °C. K562 cells expressing CD123 (K562.CD123) were generated by transducing K562 cells with a lentiviral vector encoding CD123 and a green fluorescent protein/puromycin resistance gene (GFPpuro; pCDH.CMV.CD123.EF1.GFPpuro). MOLM-13 cells expressing an enhanced GFP firefly luciferase fusion gene (MOLM-13.GFP.ffLuc) were generated by transducing MOLM-13 cells with a retroviral vector encoding GFP.ffLuc.48 GFP-positive cells were sorted and maintained in RPMI supplemented with 2 mmol/l Glutamax and 10% FBS. Luciferase expression was confirmed using D-luciferin.
Flow cytometry. Fluorochrome conjugated isotype controls, anti-CD123, anti-CD3, anti-CD4, anti-CD8, anti-CD33, anti-CD34, anti-CD45, anti-CD45RA, and anti-CCR7 were purchased from BD Biosciences (San Jose, CA). CD123-specific scFvs were detected with an AlexaFluor 647-conjugated goat anti-mouse F(ab')2 (Jackson Immunoresearch, West Grove, PA). Expression of mOrange was detected by FACS. Analysis was performed on at least 20,000 cells per sample using a FACSCalibur instrument and Cell Quest software (BD Biosciences).
Construction of retroviral vectors. The construction of the retroviral vector encoding the CD19-specific engager molecule has been previously reported.24 The CD123-specific engager molecule containing the immunoglobulin heavy-chain leader peptide, the CD123-specific scFv (26292),48 a short serine-glycine linker, and a CD3-specific scFV derived from OKT3 was synthesized by ThermoFisher Scientific (Grand Island, NY), and subcloned into pSFG-IRES-mOrange. The retroviral vector encoding full length CD20, a 2A sequence, and the CD123 engager molecule was generated by sublconing a minigene encoding CD20.2A.CD123-ENG (ThermoFisher Scientific) into a pSFG retroviral vector. RD114-pseudotyped retroviral particles were generated as previously described.50 pCDH.CMV.hCD123.EF1.GFPpuro was generated by cloning a codon optimized cDNA (ThermoFisher Scientific) of human CD123 into the multiple cloning site (MCS) of pCDH.CMV.MCS.EF1.GFPpuro. VSVG-pseudotyped lentiviral particles were generated according to the manufacturer's instructions (System Biosciences, Inc., Mountain View, CA).
Generation of Engager-T cells. PBMCs were obtained from healthy donors under a Baylor College of Medicine Institutional Review Board (IRB)-approved protocol, after informed consent was obtained in accordance to the Declaration of Helsinki. Cells were activated by stimulation on OKT3 (CRL-8001, ATCC) and anti-CD28 (Becton Dickinson, Mountain View, CA) coated non-tissue culture-treated 24-well plates. Recombinant human interleukin (IL)-7 (10 ng/µl, R&D Systems, Minneapolis, MN) and IL-15 (5 ng/µl, R&D Systems) were added to cultures on day 2, and the following day cells were transduced with retroviral particles immobilized on RetroNectin (Clontech Laborotories, Mountain View, CA). T cells were maintained and expanded in the presence of IL-7 and IL-15. Cells were analyzed for expression of mOrange by FACS 5 to 7 days post-transduction.
CD123-ENG ELISA assay. Human recombinant CD123 (R&D Systems) was plated at 10 ng/well on a 96-well non- tissue culture treated plate. Media from ENG-T cells (CD123 and control: CD19) was plated and allowed to incubate for 1 hour at room temperature (RT). Goat anti-mouse F(ab') (Jackson Immunoresearch) was added, and incubated at RT for 1 hour. The plate was washed, and secondary anti-goat Horseradish Peroxidase (HRP) antibody (Jackson Immunoresearch) was added. After 1 hour incubation at RT, the plate was washed and developing agent was added (tetramethylbenzidine (TMB) substrate, Sigma-Aldrich). Absorbance was read at 450 nm. A standard curve was generated using recombinant CD123 T-cell ENG protein (custom synthesis, ThermoFisher Scientific; Supplementary Figure S2).
Cytokine secretion assay. ENG-T cells were plated with target cells at a 2:1 ratio. Following 24 hours of culture, supernatant was harvested and analyzed for the presence of interferon (IFN) γ or IL-2 using ELISA kits (R&D systems) according to the manufacturer's instruction.
Chromium release assay. Target cells were labeled with 0.1 mCi 51Chromium(Cr) and incubated with nontransduced (NT) or ENG-T cells at 40:1, 20:1, 10:1, and 5:1 effector to target (E:T) ratios for 4 hours. Targets in media alone or 1% Triton X-100 were used for spontaneous and maximum 51Cr release, respectively. Supernatants were collected and radioactivity measured on a gamma counter. Mean percentage of specific lysis of triplicate samples was calculated as 100* (experimental release – spontaneous release)/(maximum release – spontaneous release).
Transwell assay. MOLM-13.GFP.ffLuc cells were plated in the bottom well of a 24-well tissue culture plate, and NT T cells, PBMCs, or media were added. Media, NT T cells, or ENG-T cells were added to the insert well (0.4 µm pore, Corning, Corning, NY). After 24 hours, viable tumor cells were quantified by luciferase assay.
Primary leukemia sample cytotoxicity assay. Pediatric AML samples were obtained under a Baylor College of Medicine IRB approved protocol, after informed consent was obtained in accordance to the Declaration of Helsinki. The clinical flow laboratory uses a multi-parametric flow cytometric assay with a set of 33 antibodies. The blast population of each sample was essentially identified using CD45 and side scatter, with almost all myeloid clones expressing dim to moderate CD45 compared to normal cell populations (lymphocytes and monocytes). The blast percentage in each specimen was calculated by gating on CD45/SSC-A along-with one or more distinct immunophenotypes expressed on the blasts. The blast percentage in the specimens used in this study ranged from 33–90%. Cells were incubated with media alone or ENG-T cells at indicated E:T ratios for 6 hours, then plated in MethoCult (Classic) media (Stemcell Technologies, Vancouver, BC, Canada) per manufacturer's instruction. After 10–14 days, media was resuspended with dilution into RPMI and cells were manually counted with a hemocytometer and trypan blue exclusion.
CFU assays. Bone marrow and cord blood units were obtained under a Baylor College of Medicine IRB approved protocol in accordance to the Declaration of Helsinki. Bone marrow or cord blood mononuclear cells (BMMCs, CBMCs) were isolated by standard density gradient centrifugation and cyropreserved. For CFU assays, CBMCs or BMMCs were incubated with media alone or ENG-T cells at indicated E:T ratios for 6 hours, then plated in MethoCult (Classic) media (Stemcell Technologies, Vancouver, BC, Canada) per manufacturer's instruction. After 10–14 days, colonies were counted.
Complement-dependent cytotoxicity assay. T cells transduced with CD20.CD123-ENG were labelled with 0.1 mCi 51Cr and then treated with 10 µg/ml rituximab (Roche, San Francisco, CA), 10% baby rabbit complement (Cedarlane Labs, Burlington, NC), or 10 µg/ml rituximab and 10% baby rabbit complement. Cells were incubated for 4 hours. Targets in media alone or 1% Triton X-100 were used for spontaneous and maximum 51Cr release, respectively. Supernatants were collected and radioactivity measured on a gamma counter. Mean percentage of specific lysis of triplicate samples was calculated as 100* (experimental release – spontaneous release)/(maximum release – spontaneous release).
FACS-based cytotoxicity assay. CD20.CD123-ENG T cells were treated with 10 µg/mL rituximab, 10% baby rabbit complement, or 10 µg/ml rituximab and 10% baby rabbit complement and incubated for 2 hours. Following incubation, the remaining T cells were expanded for one week in IL-7 and IL-15 before determining the presence of CD20-positive cells by FACS analysis.
Xenograft NSG model. All animal experiments were performed on a protocol approved by the Baylor College of Medicine Institutional Animal Care and Use Committee in accordance to the American Association for Laboratory Animal Science. NSG mice (NOD.Cg- Prkdcscid/Il2rgtm1Wjl/SzJ, Bar Harbor, ME) were sub-lethally irradiated with 200 cGy 24 hours prior to tail vein injection with 5x104 MOLM-13.GFP.ffLuc cells. Mice were treated with ENG-T cells intravenously at day 7 or at days 7 and 14 days after MOLM-13.GFP.ffLuc injection. Untreated mice served as controls. In experiments designed to track T-cell expansion and persistence in vivo, CD123-ENG T cells were modified with GFP.ffLuc. Unmodified MOLM-13 AML cells were used as described. To test the ability of rituximab to deplete T cells in vivo, mice were sub-lethally irradiated with 200 cGy 24 hours prior to tail vein injection with 5 × 104 MOLM-13 cells. Mice received on day 7 post-MOLM-13 injection 3 × 106 CD20.CD123-ENG-T cells. CD20.CD123-ENG T cells also expressed GFP.ffLuc. On day 3 post-T-cell injection, mice received 250 µg of rituximab IP for 3 consecutive days. Untreated mice served as controls. Bioluminescent imaging was performed on an IVIS system (IVIS, Xenogen, Alameda, CA) as previously described.51 Euthanasia was performed at prior determined time points or when animals met euthanasia criteria in accordance with Baylor College of Medicine's Center for Comparative Medicine.
Statistical analysis. GraphPad Prism 5 software (GraphPad software) was used for statistical analysis. Measurement data were presented as mean ± standard error. For comparison between two groups, two-tailed t-test was used. For comparisons of three or more groups, the values were analyzed by one-way analysis of variance with Bonferroni's post-test. For the mouse experiments, survival, determined from the time of tumor cell injection, was analyzed by the Kaplan–Meier method and by the log-rank test.
SUPPLEMENTARY MATERIAL Figure S1. CD123-ENG T cells exhibit similar T-cell phenotype to non-transduced, ex-vivo expanded T cells. Figure S2. Standard curve of developed ELISA to detect CD123 T-cell ENG protein. Figure S3. Myeloid leukemia cell lines express CD123. Figure S4. Human T-cell infiltration into liver of NSG mice treated with CD123-ENG T cells. Figure S5. Generation of CD20.CD123-ENG T cells. Figure S6. Selection and in vitro elimination of CD20.CD123-ENG T cells. Figure S7. Expression of inducible caspase 9 (iC9) in CD123 ENG T cells is toxic.
Author Contribution
C.L.B. and S.G. designed the study. C.L.B., A.S., D.T., N.J., M.P.V., K.I., A.G., P.N., and C.A. performed experiments. All authors contributed to data analysis and manuscript preparation.
Acknowledgments
We thank Cliona Rooney, Malcolm Brenner, and Helen Heslop for helpful discussions and advice. This work was supported by The Leukemia & Lymphoma Society, Ladies Leukemia League, Alex's Lemonade Stand Foundation for Childhood Cancer, When Everyone Survives Foundation, and a philanthropic gift to fund AML research at the Center for Cell and Gene Therapy. C.L.B. was supported by NIH grant 5T32HL092332-12. M.P.V. is a St. Baldrick's Foundation fellow. C.A. is an ASH Research Scholar. The Center for Cell and Gene Therapy had a research collaboration with Celgene and Bluebird Bio. K.I., M.P.V., C.A., and S.G. have patent applications in the field of T cell and gene-modified T-cell therapy for cancer.
Supplementary Material
References
- Löwenberg, B, Downing, JR and Burnett, A (1999). Acute myeloid leukemia. N Engl J Med 341: 1051–1062. [DOI] [PubMed] [Google Scholar]
- Woods, WG (2006). Curing childhood acute myeloid leukemia (AML) at the half-way point: promises to keep and miles to go before we sleep. Pediatr Blood Cancer 46: 565–569. [DOI] [PubMed] [Google Scholar]
- Gorman, MF, Ji, L, Ko, RH, Barnette, P, Bostrom, B, Hutchinson, R et al. (2010). Outcome for children treated for relapsed or refractory acute myelogenous leukemia (rAML): a Therapeutic Advances in Childhood Leukemia (TACL) Consortium study. Pediatr Blood Cancer 55: 421–429. [DOI] [PubMed] [Google Scholar]
- Manzo, T, Heslop, HE and Rooney, CM (2015). Antigen-specific T cell therapies for cancer. Hum Mol Genet 24(R1): R67–R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichs, CS and Rosenberg, SA (2014). Exploiting the curative potential of adoptive T-cell therapy for cancer. Immunol Rev 257: 56–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Themeli, M, Rivière, I and Sadelain, M (2015). New cell sources for T cell engineering and adoptive immunotherapy. Cell Stem Cell 16: 357–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- June, CH, Maus, MV, Plesa, G, Johnson, LA, Zhao, Y, Levine, BL et al. (2014). Engineered T cells for cancer therapy. Cancer Immunol Immunother 63: 969–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalos, M, Levine, BL, Porter, DL, Katz, S, Grupp, SA, Bagg, A et al. (2011). T cells with chimeric antigen receptors have potent antitumor effects and can establish memory in patients with advanced leukemia. Sci Transl Med 3: 95ra73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochenderfer, JN, Dudley, ME, Feldman, SA, Wilson, WH, Spaner, DE, Maric, I et al. (2012). B-cell depletion and remissions of malignancy along with cytokine-associated toxicity in a clinical trial of anti-CD19 chimeric-antigen-receptor-transduced T cells. Blood 119: 2709–2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochenderfer, JN, Dudley, ME, Carpenter, RO, Kassim, SH, Rose, JJ, Telford, WG et al. (2013). Donor-derived CD19-targeted T cells cause regression of malignancy persisting after allogeneic hematopoietic stem cell transplantation. Blood 122: 4129–4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brentjens, RJ, Davila, ML, Riviere, I, Park, J, Wang, X, Cowell, LG et al. (2013). CD19-targeted T cells rapidly induce molecular remissions in adults with chemotherapy-refractory acute lymphoblastic leukemia. Sci Transl Med 5: 177ra38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz, CR, Micklethwaite, KP, Savoldo, B, Ramos, CA, Lam, S, Ku, S et al. (2013). Infusion of donor-derived CD19-redirected virus-specific T cells for B-cell malignancies relapsed after allogeneic stem cell transplant: a phase 1 study. Blood 122: 2965–2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maude, SL, Frey, N, Shaw, PA, Aplenc, R, Barrett, DM, Bunin, NJ et al. (2014). Chimeric antigen receptor T cells for sustained remissions in leukemia. N Engl J Med 371: 1507–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochenderfer, JN, Dudley, ME, Kassim, SH, Somerville, RP, Carpenter, RO, Stetler-Stevenson, M et al. (2015). Chemotherapy-refractory diffuse large B-cell lymphoma and indolent B-cell malignancies can be effectively treated with autologous T cells expressing an anti-CD19 chimeric antigen receptor. J Clin Oncol 33: 540–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, DW, Kochenderfer, JN, Stetler-Stevenson, M, Cui, YK, Delbrook, C, Feldman, SA et al. (2015). T cells expressing CD19 chimeric antigen receptors for acute lymphoblastic leukaemia in children and young adults: a phase 1 dose-escalation trial. Lancet 385: 517–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie, DS, Neeson, PJ, Khot, A, Peinert, S, Tai, T, Tainton, K et al. (2013). Persistence and efficacy of second generation CAR T cell against the LeY antigen in acute myeloid leukemia. Mol Ther 21: 2122–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, QS, Wang, Y, Lv, HY, Han, QW, Fan, H, Guo, B et al. (2015). Treatment of CD33-directed chimeric antigen receptor-modified T cells in one patient with relapsed and refractory acute myeloid leukemia. Mol Ther 23: 184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardiros, A, Dos Santos, C, McDonald, T, Brown, CE, Wang, X, Budde, LE et al. (2013). T cells expressing CD123-specific chimeric antigen receptors exhibit specific cytolytic effector functions and antitumor effects against human acute myeloid leukemia. Blood 122: 3138–3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettamanti, S, Marin, V, Pizzitola, I, Magnani, CF, Giordano Attianese, GM, Cribioli, E et al. (2013). Targeting of acute myeloid leukaemia by cytokine-induced killer cells redirected with a novel CD123-specific chimeric antigen receptor. Br J Haematol 161: 389–401. [DOI] [PubMed] [Google Scholar]
- Pizzitola, I, Anjos-Afonso, F, Rouault-Pierre, K, Lassailly, F, Tettamanti, S, Spinelli, O et al. (2014). Chimeric antigen receptors against CD33/CD123 antigens efficiently target primary acute myeloid leukemia cells in vivo. Leukemia 28: 1596–1605. [DOI] [PubMed] [Google Scholar]
- Gill, S, Tasian, SK, Ruella, M, Shestova, O, Li, Y, Porter, DL et al. (2014). Preclinical targeting of human acute myeloid leukemia and myeloablation using chimeric antigen receptor-modified T cells. Blood 123: 2343–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenderian, SS, Ruella, M, Shestova, O, Klichinsky, M, Aikawa, V, Morrissette, JJ et al. (2015). CD33-specific chimeric antigen receptor T cells exhibit potent preclinical activity against human acute myeloid leukemia. Leukemia 29: 1637–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compte, M, Blanco, B, Serrano, F, Cuesta, AM, Sanz, L, Bernad, A et al. (2007). Inhibition of tumor growth in vivo by in situ secretion of bispecific anti-CEA x anti-CD3 diabodies from lentivirally transduced human lymphocytes. Cancer Gene Ther 14: 380–388. [DOI] [PubMed] [Google Scholar]
- Iwahori, K, Kakarla, S, Velasquez, MP, Yu, F, Yi, Z, Gerken, C et al. (2015). Engager T cells: a new class of antigen-specific T cells that redirect bystander T cells. Mol Ther 23: 171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Introna, M, Barbui, AM, Bambacioni, F, Casati, C, Gaipa, G, Borleri, G et al. (2000). Genetic modification of human T cells with CD20: a strategy to purify and lyse transduced cells with anti-CD20 antibodies. Hum Gene Ther 11: 611–620. [DOI] [PubMed] [Google Scholar]
- Serafini, M, Manganini, M, Borleri, G, Bonamino, M, Imberti, L, Biondi, A et al. (2004). Characterization of CD20-transduced T lymphocytes as an alternative suicide gene therapy approach for the treatment of graft-versus-host disease. Hum Gene Ther 15: 63–76. [DOI] [PubMed] [Google Scholar]
- Griffioen, M, van Egmond, EH, Kester, MG, Willemze, R, Falkenburg, JH and Heemskerk, MH (2009). Retroviral transfer of human CD20 as a suicide gene for adoptive T-cell therapy. Haematologica 94: 1316–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, MA, Covassin, L, Brehm, MA, Racki, W, Pearson, T, Leif, J et al. (2009). Human peripheral blood leucocyte non-obese diabetic-severe combined immunodeficiency interleukin-2 receptor gamma chain gene mouse model of xenogeneic graft-versus-host-like disease and the role of host major histocompatibility complex. Clin Exp Immunol 157: 104–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali, N, Flutter, B, Sanchez Rodriguez, R, Sharif-Paghaleh, E, Barber, LD, Lombardi, G et al. (2012). Xenogeneic graft-versus-host-disease in NOD-scid IL-2Rγnull mice display a T-effector memory phenotype. PLoS One 7: e44219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz, L, Nomdedéu, JF, López, O, Carnicer, MJ, Bellido, M, Aventín, A et al. (2001). Interleukin-3 receptor alpha chain (CD123) is widely expressed in hematologic malignancies. Haematologica 86: 1261–1269. [PubMed] [Google Scholar]
- Manz, MG, Miyamoto, T, Akashi, K and Weissman, IL (2002). Prospective isolation of human clonogenic common myeloid progenitors. Proc Natl Acad Sci USA 99: 11872–11877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinert, S, Prince, HM, Guru, PM, Kershaw, MH, Smyth, MJ, Trapani, JA et al. (2010). Gene-modified T cells as immunotherapy for multiple myeloma and acute myeloid leukemia expressing the Lewis Y antigen. Gene Ther 17: 678–686. [DOI] [PubMed] [Google Scholar]
- Casucci, M, Nicolis di Robilant, B, Falcone, L, Camisa, B, Norelli, M, Genovese, P et al. (2013). CD44v6-targeted T cells mediate potent antitumor effects against acute myeloid leukemia and multiple myeloma. Blood 122: 3461–3472. [DOI] [PubMed] [Google Scholar]
- Testa, U, Pelosi, E and Frankel, A (2014). CD 123 is a membrane biomarker and a therapeutic target in hematologic malignancies. Biomark Res 2: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hear, C, Heiber, JF, Schubert, I, Fey, G and Geiger, TL (2015). Anti-CD33 chimeric antigen receptor targeting of acute myeloid leukemia. Haematologica 100: 336–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley, SA and Walter, RB (2015). Update on antigen-specific immunotherapy of acute myeloid leukemia. Curr Hematol Malig Rep 10: 65–75. [DOI] [PubMed] [Google Scholar]
- Chan, L, Hardwick, N, Darling, D, Galea-Lauri, J, Gäken, J, Devereux, S et al. (2005). IL-2/B7.1 (CD80) fusagene transduction of AML blasts by a self-inactivating lentiviral vector stimulates T cell responses in vitro: a strategy to generate whole cell vaccines for AML. Mol Ther 11: 120–131. [DOI] [PubMed] [Google Scholar]
- Schlegel, P, Ditthard, K, Lang, P, Mezger, M, Michaelis, S, Handgretinger, R et al. (2015). NKG2D Signaling Leads to NK Cell Mediated Lysis of Childhood AML. J Immunol Res 2015: 473175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markiewicz, MA, Carayannopoulos, LN, Naidenko, OV, Matsui, K, Burack, WR, Wise, EL et al. (2005). Costimulation through NKG2D enhances murine CD8+ CTL function: similarities and differences between NKG2D and CD28 costimulation. J Immunol 175: 2825–2833. [DOI] [PubMed] [Google Scholar]
- Velasquez, MP, Torres, D, Iwahori, K, Kakarla, S, Arber, C, et al. (2016). T cells expressing CD19-specific Engager Molecules for the Immunotherapy of CD19-positive Malignancies. Sci Rep 6: 27130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman, LM and Gore, L (2014). Blinatumomab, a bi-specific anti-CD19/CD3 BiTE(®) antibody for the treatment of acute lymphoblastic leukemia: perspectives and current pediatric applications. Front Oncol 4: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taussig, DC, Pearce, DJ, Simpson, C, Rohatiner, AZ, Lister, TA, Kelly, G et al. (2005). Hematopoietic stem cells express multiple myeloid markers: implications for the origin and targeted therapy of acute myeloid leukemia. Blood 106: 4086–4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpelainen, EI, Gamble, JR, Vadas, MA and Lopez, AF (1996). IL-3 receptor expression, regulation and function in cells of the vasculature. Immunol Cell Biol 74: 1–7. [DOI] [PubMed] [Google Scholar]
- Chichili, GR, Huang, L, Li, H, Burke, S, He, L, Tang, Q et al. (2015). A CD3xCD123 bispecific DART for redirecting host T cells to myelogenous leukemia: preclinical activity and safety in nonhuman primates. Sci Transl Med 7: 289ra82. [DOI] [PubMed] [Google Scholar]
- Taussig, DC, Pearce, DJ, Simpson, C, Rohatiner, AZ, Lister, TA, Kelly, G et al. (2005). Hematopoietic stem cells express multiple myeloid markers: implications for the origin and targeted therapy of acute myeloid leukemia. Blood 106: 4086–4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan, AM, Weinstein, JN, Carrasquillo, JA, Bunn, PA Jr, Reynolds, JC, Foon, KA et al. (1987). Immunolymphoscintigraphy and the dose dependence of 111In-labeled T101 monoclonal antibody in patients with cutaneous T-cell lymphoma. Cancer Res 47: 6093–6099. [PubMed] [Google Scholar]
- Shultz, LD, Schweitzer, PA, Christianson, SW, Gott, B, Schweitzer, IB, Tennent, B et al. (1995). Multiple defects in innate and adaptive immunologic function in NOD/LtSz-scid mice. J Immunol 154: 180–191. [PubMed] [Google Scholar]
- Vera, J, Savoldo, B, Vigouroux, S, Biagi, E, Pule, M, Rossig, C et al. (2006). T lymphocytes redirected against the kappa light chain of human immunoglobulin efficiently kill mature B lymphocyte-derived malignant cells. Blood 108: 3890–3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, X, Ho, M and Pastan, I (2007). New immunotoxins targeting CD123, a stem cell antigen on acute myeloid leukemia cells. J Immunother 30: 607–613. [DOI] [PubMed] [Google Scholar]
- Chow, KK, Naik, S, Kakarla, S, Brawley, VS, Shaffer, DR, Yi, Z et al. (2013). T cells redirected to EphA2 for the immunotherapy of glioblastoma. Mol Ther 21: 629–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinn, KR, Chaudhuri, TR, Szafran, AA, O'Quinn, D, Weaver, C, Dugger, K et al. (2008). Noninvasive bioluminescence imaging in small animals. ILAR J 49: 103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.