To the Editor:
Genome editing technologies, such as clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) systems, are being widely applied in biomedical research with the potential of developing novel therapeutics.1,2 To date, a variety of viral systems have been used to deliver CRISPR reagents, including lentiviruses (LVs), adenoviruses (AdVs), and adeno-associated viruses (AAVs). Although viral vectors offer high efficiency of delivery of CRISPR systems in cultured cells or local tissues in vivo, long-term constitutive expression of CRISPRs in cells and tissues for months or even longer raises the concern that any potential off-target effects could be exacerbated. Use of nonintegrating viral vectors could address this issue, but the duration of CRISPR expression via nonintegrating LVs, AdVs, or AAVs could still last for weeks to months. Delivery of CRISPRs in the format of Cas9 messenger RNA (mRNA) with guide RNA as mRNA/single guide RNA (sgRNA) complexes or Cas9 recombinant protein with guide RNA as ribonucleoproteins (RNPs) provides potential solutions to reduce off-target effects.3 The use of engineered nanoparticles for in vivo delivery of Cas9 mRNA/sgRNA complexes or RNPs represents an important delivery strategy for potential therapeutics.4 However, the efficacy of these strategies is largely constrained by poor in vivo delivery efficiency, especially for large molecules such as Cas9 mRNA, the lack of cell/tissue specificity, and the relatively compromised stability of mRNA/sgRNA or RNP complexes after delivery. To promote the delivery efficiency and to reduce the duration of CRISPR expression via viral vector systems, we report here the development of a self-restricted CRISPR system that can be used to shorten Cas9 expression duration within a lentiviral vector format to a couple of days. Our approach contributes to a significant reduction in off-target effects compared to the standard lentiCRISPR system.
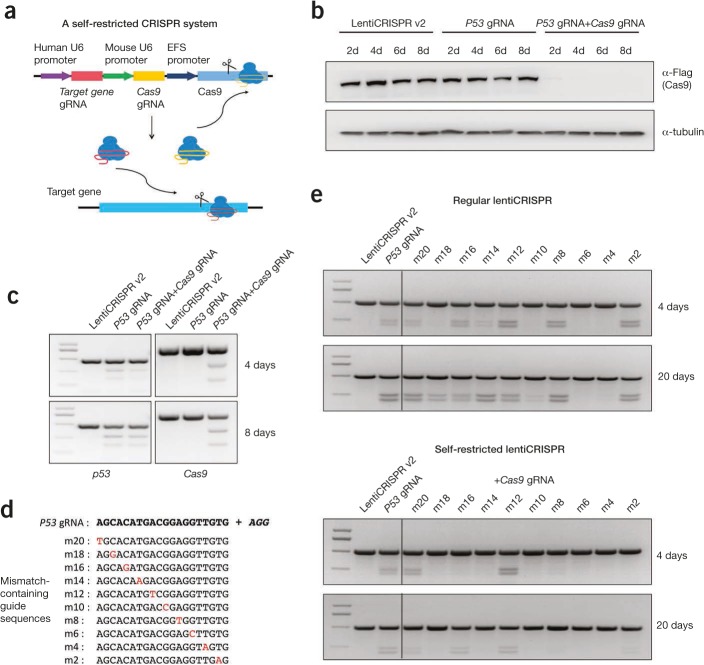
In this study, we re-engineered the lentiCRISPR v2 plasmid5 by inserting a second guide RNA expression cassette using a mouse U6 promoter. From this cassette, a guide RNA targeting Cas9 itself was co-expressed with the guide RNA that recognized the target gene. Therefore, we hypothesized that the Cas9 expression cassette within the viral vector would be simultaneously targeted and destroyed along with the target gene, resulting in a much reduced expression duration of Cas9. Thus, we named this modified CRISPR system a “self-restricted CRISPR system” (Figure 1a). We first screened for an efficient guide RNA targeting Cas9 (targeting sequences: GGAAGGACTCTTCCAGTCTG + TGG), and in a proof-of-principle experiment, generated lentiviruses carrying the CRISPR targeting the P53 gene (targeting sequences: AGCACATGACGGAGGTTGTG + AGG) using both the regular lentiCRISPR and self-restricted CRISPR systems. A human hepatoma cell line Huh7 was respectively transduced with lentiCRISPR v2 viruses (without guide RNA), lentiCRISPR v2 viruses carrying guide RNA targeting P53 gene (regular lentiCRISPR), and lentiCRISPR v2 viruses carrying guide RNAs targeting both P53 and Cas9 genes (self-restricted CRISPR). Cells were harvested at the indicated days post-infection for subsequent analysis. Western blot analysis showed that Cas9 protein expression was significantly reduced in cells modified with the self-restricted CRISPR system at as early as day 2 post-infection compared to the control groups (Figure 1b).
Figure 1.
A self-restricted CRISPR system to reduce off-target effects. (a) A schematic diagram of the self-restricted CRISPR system. LentiCRISPR v2 construct was modified by inserting a second guide RNA cassette targeting the Cas9 gene driven by a mouse U6 promoter. (b) Exogenous Cas9 protein levels in Huh7 cells as determined by western blot using Flag antibody. Endogenous tubulin expression was used as an internal control of protein loading. Huh7 cells were transduced with different lentiviruses and harvested at different days post-infection for analysis. (c) T7E1 assays performed at both the P53 and Cas9 targeting loci with genomic DNA extracted from Huh7 cells. Cells were transduced with different lentiviruses and harvested at the indicated days post-transfection. (d) Guide RNAs with single-point mutations were generated to evaluate the effect of spacer–protospacer mismatch at the P53 gene locus in regular and self-restricted lentiCRISPR systems. (e) T7E1 assays performed at the P53 targeting locus with genomic DNA from Huh7 cells transduced with respective lentiviruses carrying various guide RNAs and harvested at the indicated days post-infection. Primers used in T7E1 assays at the Cas9 targeting locus are: 5'-CGTGATCACCGACGAGTACA -3' and 5'-AGGTTTCCGAACAGGCCATT-3'; Primers used in T7E1 assays at the P53 targeting locus are 5'-AGAGACCCCAGTTGCAAACC-3'and 5'-TTCTTTGCTGCCGTCTTCCA-3'. EFS, elongation factor-1α, short form; gRNA, guide RNA; m2, m4, etc., mismatch-containing guide sequences.
To determine the targeting efficiency at the P53 gene locus, T7E1 analysis in Huh7 cells harvested at days 4 and 8 displayed a significant and comparable genome editing activity at the P53 gene locus with specific cleavage activity at the Cas9 gene locus in the self-restricted CRISPR system (Figure 1c). In fact, we noticed a slight increase in cleavage activity at day 8 compared to day 4 at both P53 and Cas9 gene loci. We suspected that this was a result of residual Cas9 expression after 4 days in the self-restricted CRISPR system, even though the signal was not detected by western blot, possibly as a result of limited sensitivity.
We next confirmed whether the reduced off-target effect was a result of a significantly shortened duration of the Cas9 protein expression in the self-restricted CRISPR system. Huh7 cells were therefore transduced with lentiviruses carrying the respective guide RNAs and were then collected for analysis at days 4 and 20 with prolonged expression of Cas9. We first assessed potential off-target mutagenesis at 10 sites that are mostly matched to the P53 on-target site by genome-wide prediction using the CRISPR Design Server (http://crispr.mit.edu/) (Supplementary Table S1). We found no evidence of significant off-target mutagenesis for either the regular CRISPR or self-restricted CRISPR systems with this specific guide RNA within the limit of detection by the T7E1 assay (Supplementary Figure S1).
As off-target effects for each guide RNA vary with the target sequence, we attempted to investigate whether potential off-target effects are caused by close similarity of sequences between on-target and off-target sites. We therefore designed 10 guide RNA sequences containing a single-nucleotide mismatch that was introduced between the spacer and protospacer target at the P53 gene locus with varying distances from 5́ to the protospacer adjacent motif (Figure 1d). Potential off-target effects using the two CRISPR systems were then examined at days 4 and 20 post viral infection. When using the regular lentiCRISPR system, we found clear editing activity mediated by several guide RNAs harboring a single-nucleotide mismatch at day 4, including one with a mismatch that was proximal to the protospacer adjacent motif (m2). Moreover, almost all guide RNAs with single-nucleotide mismatches displayed significant editing activity at day 20, indicating that single-base mismatches still contribute to efficient genomic cleavage activity of Cas9, at least with these specific guide RNA sequences. Therefore, our results showed that prolonged exposure to Cas9 could contribute to significant off-target effects (Figure 1e). On the other hand, with the self-restricted CRISPR system, the cleavage activity of several guide RNAs was clearly reduced compared to the regular lentiCRISPR system, especially at day 20, suggesting that reduced expression duration of Cas9 when using the self-restricted CRISPR significantly improved the specificity of genome editing.
In summary, we designed a self-restricted CRISPR system by co-expressing an additional guide RNA targeting the Cas9 gene itself. We found that this system limited the expression duration of the Cas9 protein to days even in a genome integrating lentiviral vector, resulting in a significant reduction in off-target effects. Viral vectors are commonly used for delivery of CRISPR materials. For instance, CRISPRs expressed from lentiviral vectors have frequently been used to generate mutant cell lines and for genome-wide screening. CRISPRs expressed from AdVs or AAVs have been used for in vivo genome editing to develop potential therapeutics.
To address the potential issue that long-term constitutive expression of CRISPRs in cells and tissues when using viral vectors promotes more severe off-target effects, researchers have developed different inducible expression systems to control Cas9 protein expression, such as the intein-Cas9 variants that can be triggered by small molecules,6 as well as the doxycycline-inducible Cas9 viral vectors.7 Indeed, considerably decreased off-target effects without influence on on-target efficiency have been observed using these systems, a finding that could support the working strategy that Cas9 expression duration can be manipulated to achieve higher specificity. However, there is potential toxicity associated with small molecular inducers. Moreover, an inducible/repressible expression system requires co-expression of additional genes (e.g., rtTA), and residual transgene expression often exists when turned off (referred to as leakiness). Altogether, these approaches may complicate the system and thus limit potential application for in vivo therapeutics.
In the present study, we demonstrated that the self-restricted CRISPR system enabled us to control off-target effects while maintaining the advantages of using viral vectors that permit highly efficient CRISPR genome editing as well as the ease of generating CRISPR constructs based on conventional strategies. Although we have only utilized the self-restricted CRISPR system in a lentiviral vector targeting a single gene in vitro, we anticipate that this system might also be applied in a genome-wide gene knockout screening platform, for instance, using the GeCKO libraries5 to reduce potential off-target effects that could affect accuracy of the screen. Moreover, the self-restricted CRISPR system can also be applied to other viral vectors including AdVs and AAVs, and can be used in combination with currently available strategies to reduce the off-target effects including the use of Cas9 nickases,8 Cas9-FokI chimeric proteins,9 or proper modifications to residues of the Cas9 protein.10,11 Altogether, our approach could greatly increase specificity for genome editing by controlling Cas9 expression, paving the way for more complexed in vivo genome editing studies for developing novel therapeutics.
SUPPLEMENTARY MATERIAL Table S1. Mutagenesis analysis at the ten top genome-wide off-target sites used in this study. Figure S1. Top ten genome-wide off-target sites used in this study.
Acknowledgments
We thank Dr. Xianfeng Chen for discussion with him that led to the inspiration of this work. This work was supported by the Hundred Talents Program of the Chinese Academy of Sciences (Q.D.), the National Youth 1000 Talents Program (Q.D.), the Shanghai Pujiang Program (15PJ1409200 to Q.D.), the Shanghai Institutes for Biological Sciences Fellowship (Y5Y1X41491) (Y.Z.), the National Natural Science Foundation of China (81500614 to Y.Z.), the Research Grants Council (24110515 to K.O.L.), and the Health and Medical Research Fund of Hong Kong (03140346 to K.O.L.).
Supplementary Material
References
- Hsu, PD, Lander, ES and Zhang, F (2016). Development and applications of CRISPR-Cas9 for genome engineering. Cell 157: 1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder, ML and Gersbach, CA (2016). Genome-editing technologies for gene and cell therapy. Mol Ther 24: 475–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S, Kim, D, Cho, SW, Kim, J and Kim, JS (2014). Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res 24: 1012–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, H, Song, CQ, Dorkin, JR, Zhu, LJ, Li, Y, Wu, Q et al. (2016). Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat Biotechnol 34: 328–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana, NE, Shalem, O and Zhang, F (2014). Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11: 783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, KM, Pattanayak, V, Thompson, DB, Zuris, JA and Liu, DR (2015). Small molecule-triggered Cas9 protein with improved genome-editing specificity. Nat Chem Biol 11: 316–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, J, Wu, L, Zhang, SM, Lu, M, Cheung, WK, Cai, W et al. (2016). An easy and efficient inducible CRISPR/Cas9 platform with improved specificity for multiple gene targeting. Nucleic Acids Res; e-pub ahead of print 25 July 2016. [DOI] [PMC free article] [PubMed]
- Ran, FA, Hsu, PD, Lin, CY, Gootenberg, JS, Konermann, S, Trevino, AE et al. (2013). Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154: 1380–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, SQ, Wyvekens, N, Khayter, C, Foden, JA, Thapar, V, Reyon, D et al. (2014). Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol 32: 569–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaymaker, IM, Gao, L, Zetsche, B, Scott, DA, Yan, WX and Zhang F (2016). Rationally engineered Cas9 nucleases with improved specificity. Science 351: 84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver, BP, Pattanayak, V, Prew, MS, Tsai, SQ, Nguyen, NT, Zheng, Z et al. (2016). High- fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529: 490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.