Abstract
Insight into mechanisms that link the actions of 1,25-dihydroxyvitamin D3 (1,25(OH)2D3) to the regulation of gene expression has evolved extensively since the initial discovery of a nuclear protein known as the vitamin D receptor (VDR). Perhaps most important was the molecular cloning of this receptor which enabled its inclusion within the nuclear receptor gene family and further studies of both its structure and regulatory function. Current studies are now refocused on the vitamin D hormone's action at the genome, where VDR together with other transcription factors coordinates the recruitment of chromatin active coregulatory complexes that participate directly in the modification of gene output. These studies highlight the role of chromatin in the expression of genes and the dynamic impact of the epigenetic landscape that contextualizes individual gene loci thus influencing the VDR's transcriptional actions. In this chapter, we summarize advances made over the past few years in understanding vitamin D action on a genome-wide scale, focusing on overarching principles that have emerged at this level. Of particular significance is the finding that dynamic changes that occur to the genome during cellular differentiation at both genetic and epigenetic levels profoundly alter the ability of 1,25(OH)2D3 and its receptor to regulate gene expression. We address the broad impact of differentiation on specific epigenetic histone modifications that occur across the genome and the ability of the VDR to influence this activity at selected gene loci as well. These studies advance our understanding of not only vitamin D action but also of the complex and dynamic role played by the genome itself as a major determinant of VDR activity.
1. INTRODUCTION
A binding protein eventually designated the vitamin D receptor (VDR) was discovered first in the intestine (Brumbaugh & Haussler, 1974a, 1974b) and then in other tissues including the parathyroid glands, kidney, and bone (Pike, 1991). This protein’s biochemical features, including its retention on chromatin (Haussler & Norman, 1969) and subsequently its ability to bind to DNA (Pike & Haussler, 1979), suggested that it was similar to that of other steroid hormone receptors and that it might play a role in transcriptional regulation. The DNA-binding capacity of the VDR enabled investigators to purify the protein, to generate both polyclonal and monoclonal antibodies useful in the protein’s characterization (Dame, Pierce, Prahl, Hayes, & DeLuca, 1986; Pike, Donaldson, Marion, & Haussler, 1982), and beginning in 1987 to clone the chicken (McDonnell, Mangelsdorf, Pike, Haussler, & O’Malley, 1987) and subsequently the human (Baker et al., 1988) and rat (Burmester, Maeda, & DeLuca, 1988) genes as well. These latter achievements and the domain structure that was revealed from subsequent studies (McDonnell, Pike, & O’Malley, 1988; McDonnell, Scott, Kerner, O’Malley, & Pike, 1989) confirmed that 1,25(OH)2D3 was a true steroid hormone and that the receptor was a bona fide member of the steroid receptor gene family (Evans, 1988). They also enabled subsequent studies in patients with hypocalcemic rickets and other clinical features that identified a series of mutations within the VDR gene itself that was causative for hereditary 1,25(OH)2D3-resistant rickets (HVDRR) (Feldman & Malloy, 1990; Forghani et al., 2010; Hughes et al., 1988). This syndrome was first identified by Bell and colleagues in 1978 (Brooks et al., 1978) and proved to be due to defects in the VDR protein (Eil, Liberman, Rosen, & Marx, 1981; Marx et al., 1978; Pike et al., 1984). This discovery, the first for any member of the nuclear receptor family, confirmed the integral and essential role for the VDR as the mediator of the activities of the vitamin D hormone. Importantly, the human phenotype of this disease has been recapitulated in mice through genetic deletion of key elements of the VDR gene from the mouse genome (Bouillon et al., 2008; Li et al., 1997; Yoshizawa et al., 1997). More recently, advanced studies of the VDR gene from both mouse and humans have defined the genetic loci spanning these two genes and determined the locations of key regulatory elements that function to modulate VDR gene output in response to hormones such as the glucocorticoids, retinoic acid, and 1,25(OH)2D3 itself (Zella, Kim, Shevde, & Pike, 2006; Zella et al., 2010). The ability of a transgene that contained either the mouse or the human version of the VDR gene to recapitulate the tissue-specific expression of the VDRin the mouse and to rescue the phenotype of the VDR-null mouse has provided final confirmation of the role of the VDR in 1,25(OH)2D3 action (Lee, Bishop, Goellner, O’Brien, & Pike, 2014). It has also enabled the creation of a humanized mouse model that replicates a particular syndromic subset of HVDRR patients, wherein a VDR molecule incapable of binding 1,25(OH)2D3 is able to prevent the development of alopecia that is seen in mice that do not express the VDR; this activity appears to be 1,25(OH)2D3 independent (Lee, Goellner, O’Brien, & Pike, 2014). These up-to-date studies conclusively demonstrate the importance of the VDR as the mediator of all of the known actions of 1,25(OH)2D3 in disease.
2. GENOME-WIDE ANALYSIS REVEALS NEW CONCEPTS IN VITAMIN D ACTION
Traditional studies over several decades using reporter plasmid analyses facilitated the conclusion that 1,25(OH)2D3 is capable of regulating many vitamin D target genes including osteocalcin (Kerner, Scott, & Pike, 1989; Ozono, Liao, Kerner, Scott, & Pike, 1990), osteopontin (Nilsson et al., 2005), and Cyp24a1 (Ohyama et al., 1994; Zierold, Darwish, & DeLuca, 1995). These studies coupled with the extensive use of electrophoretic mobility shift and other analyses identified key features of the VDR’s DNA-binding sites, termed vitamin D response elements or VDREs, and the participation of RXR as a heterodimer partner essential for adequate DNA-binding affinity (Pike & Meyer, 2014; Pike et al., 2010). However, despite the fact that many of these interactions at target genes have been confirmed via the application of chromatin immunoprecipitation Features of Gene Regulation by Vitamin D analysis (ChIP) (Kim, Shevde, & Pike, 2005), this latter technique was unable to provide sought after confirmation for many gene promoters and more importantly failed to identify mechanisms that mediated regulation for genes such as Tnfsf11 (RANKL), Vdr, and numerous others as well. Accordingly, it was the development of genome-wide methods such as ChIP-chip (tiled microarrays) and then ChIP-seq (DNA sequencing) analyses that extended the technical reach of ChIP analysis to resolve many of the unknown mechanisms, eventually enabling the quantification of transcription factor-binding sites across entire cellular genomes. Importantly, these techniques were also used to acquire genome-wide data sets for coregulatory factors, chromatin modifiers, and for the presence of epigenetic modifications on both DNA and histones as well. Indeed, any feature for which an antibody could be developed was a potential target. This largely unbiased approach to transcriptional regulation has fundamentally revolutionized our approach to the study of genetic and epigenetic components that are essential for gene regulation, simultaneously revealing an abundance of new insights. Indeed, we have used ChIP-chip and subsequently ChIP-seq analyses to gain a genome-wide perspective through which 1,25(OH)2D3 and its receptor mediate the regulation of cellular transcriptomes in numerous cell types (Lee et al., 2015; Meyer, Benkusky, Lee, & Pike, 2014; Meyer, Benkusky, & Pike, 2014; Meyer, Goetsch, & Pike, 2010b, 2012; Meyer & Pike, 2013; St John, Bishop, et al., 2014). A list of the overarching principles that have emerged is presented in Table 1 (Pike, Lee, & Meyer, 2014). These studies indicated that between 2000 and 8000 VDR-binding sites are detected following activation by 1,25(OH)2D3 in a cell-type-dependent quantitation and that these sites are highly enriched for a DNA sequence found previously in representative genes such as osteocalcin and osteopontin. Furthermore, the majority of these sites are co-occupied by RXR, thereby confirming this principle on a genome-wide scale. Interestingly, we also discovered that while the DNA-binding activity of the VDR at these cellular genomes was largely dependent upon the presence of 1,25(OH)2D3, a significant number of sites were fully occupied by the receptor even in the absence of ligand. The basis for this type of DNA binding is unknown, but does not appear to be due to the absence of RXR, which frequently occupied most of the VDR-binding sites regardless of the presence of 1,25(OH)2D3. This latter finding was accompanied by the discovery that most regulatory regions were located within introns and intergenic regions highly distal to the genes they regulate. These observations will be discussed in the next sections, but provide both novel insight into vitamin D action and explain the difficulties that emerged early on in identifying gene regulatory mechanisms when the focus was limited technically to regions located near gene promoters. The summary provided in Table 1 is supported in part by additional studies that have been conducted by other investigators in the vitamin D field (Ding et al., 2013; Heikkinen et al., 2011; Ramagopalan et al., 2010; Sherman et al., 2014).
Table 1.
Overarching Principles of 1,25(OH)2D3-Mediated Gene Regulation in Target Cells
|
VDR-binding sites (the cistrome): 2000–8000 1,25(OH)2D3-sensitive binding sites/genome whose number and location are determined by cell type |
| Active transcription unit: The VDR/RXR heterodimer |
|
Distal-binding site location: dispersed in cis-regulatory modules (CRMs or enhancers) across the genome; located in a cell-type-specific manner near promoters, but predominantly within introns and distal intergenic regions; frequently located in clusters of elements |
|
VDR/RXR-binding site sequence (VDRE): induction mediated by classic hexameric half-sites (AGGTCA) separated by three base pairs; repression mediated by divergent sites |
| Mode of DNA binding: predominantly, but not exclusively, 1,25(OH)2D3 dependent |
|
Modular features: CRMs contain binding sites for multiple transcription factors that facilitate both independent or synergistic interaction |
|
Epigenetic CRM signatures: defined by the dynamically regulated posttranslational histone H3 and H4 modifications and selectively regulated by 1,25(OH)2D3 |
|
VDR cistromes are highly dynamic: cistromes change during cell differentiation, maturation, and disease activation and thus have consequential effects on gene expression |
Principles in bold represent those previously defined and now confirmed by genome-wide analysis.
Principles in italics represent newly defined genome-wide features of vitamin D action.
3. NOVEL PRINCIPLES OF VITAMIN D ACTION
The observation that DNA binding of the VDR occurs in both a 1,25(OH)2D3-dependent and -independent fashion and that the sites to which the receptor binds occur most frequently many kilobases distal to genetic start sites has profound implications for vitamin D action. Perhaps the most interesting principle has been the discovery that the VDR cistrome is highly dynamic during the course of cellular differentiation resulting in a striking change in the gene expression profile that characterizes the more mature cell. We consider these three issues in the context of vitamin D action in the next sections.
3.1 Modes of DNA Binding and Implications for the Regulatory Activity of the VDR
As indicated above, ChIP-seq analysis revealed that while VDR-binding site occupancy on cellular genomes is highly dependent upon 1,25(OH)2D3, a number of sites were found to contain prebound VDR and RXR prior to ligand activation (Meyer, Benkusky, Lee, et al., 2014; Meyer et al., 2012). This novel observation highlights at least two modes of VDR DNA binding and raises questions as to both the nature of the underlying mechanism through which the VDR binds to DNA in the absence of activation by the hormone and whether this interaction is capable of regulating transcriptional activity independent of 1,25(OH)2D3. Although the mechanism remains obscure, emerging evidence suggests that the VDR may regulate the expression of specific genes and their associated biology through at least two different mechanisms that likely involve VDR DNA binding yet are independent of 1,25(OH)2D3. It is worth noting that despite several unique biochemical properties inherent to the VDR, this latter type of activity is not unexpected given the complex nature of the transcriptional actions of other nuclear receptors.
3.1.1 Ligand-Independent Function of the VDR in the Hair Cycle
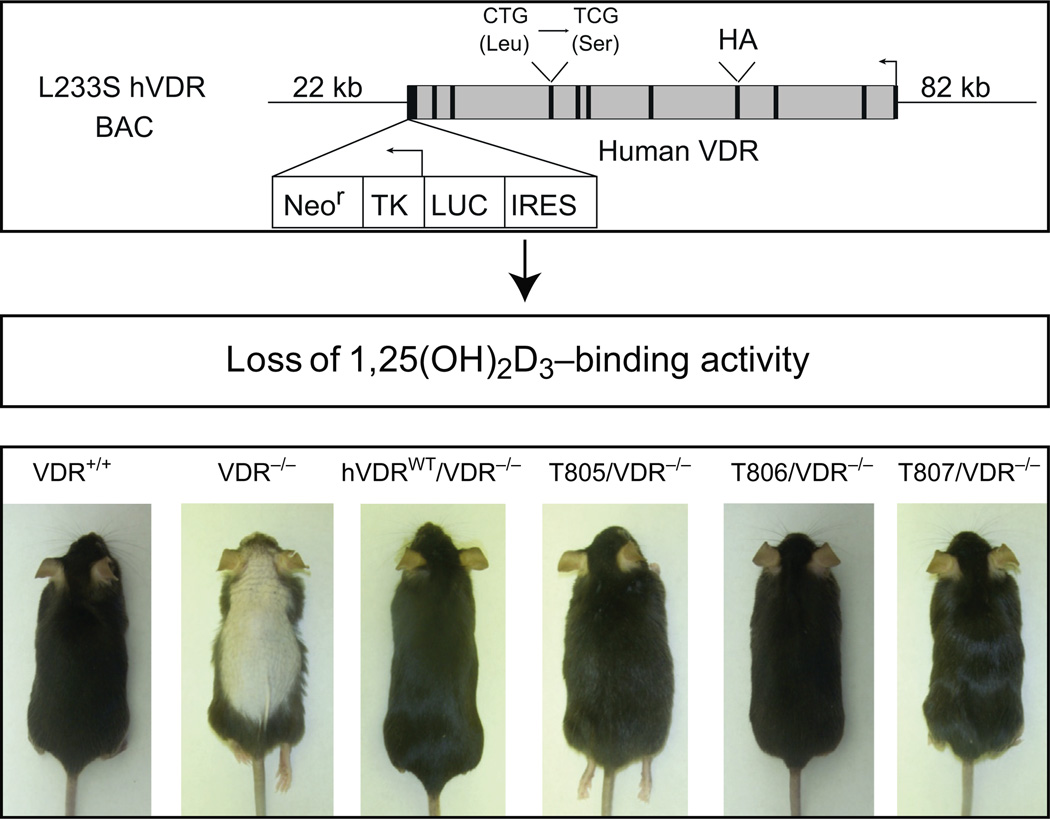
As discussed earlier, the syndrome of HVDRR in humans is due to a wide variety of mutations within the VDR gene that results in the production of a receptor that is unable to regulate gene expression (Malloy, Pike, & Feldman, 1999; Malloy et al., 2014). These molecular defects lead to a broad disease phenotype that is particularly evident at the skeleton. Only a subset of these patients display alopecia, however, and this feature was ultimately linked to VDR gene mutations that compromise the overall expression of the VDR rather than to mutations that alter its functional capability to bind to and interact with either 1,25(OH)2D3 or DNA, or to recruit coregulatory proteins essential for gene regulation. Further studies using mouse models supported these observations in humans; VDR-null mice become alopecic whereas Cyp27b1-deleted mice unable to produce 1,25(OH)2D3 are unaffected, prompting the emerging hypothesis that control of the hair cycle in the skin could be the result of a 1,25(OH)2D3-independent function of the VDR (Cianferotti, Cox, Skorija, & Demay, 2007; Demay et al., 2007; Li et al., 1997; Panda et al., 2001). While it is known that this biological activity of the VDR involves interaction with components of the Wnt-β catenin and Hedgehog signaling pathways, the mechanism of this regulation remains unclear (Demay et al., 2007). To address this issue directly, we recently developed genetic models in which mice express either the human wild-type VDR or a mutant form that is incapable of binding 1,25(OH)2D3 from transgenes that are comprised of the natural human VDR gene locus (Lee, Bishop, et al., 2014; Lee, Goellner, et al., 2014). Following introduction into the VDR-null mouse background, both transgenes recapitulate expression of the endogenous mouse VDR gene in tissues in which the VDR is known to be expressed. However, while the wild-type human VDR was able to rescue both the deranged skeletal phenotype of the VDR-null mouse and to prevent alopecia, the 1,25(OH)2D3-binding-deficient form of the human VDR could only rescue the alopecia (Lee, Goellner, et al., 2014; Fig. 1). This study confirmed a previous observation involving the expression of a mutant VDR in keratinocytes and established the fundamental paradigm that the VDR is capable of 1,25(OH)2D3-independent actions (Chen, Sakai, & Demay, 2001; Skorija et al., 2005). Interestingly, the VDR displays numerous 1,25(OH)2D3-dependent activities in the skin as well suggesting that both processes can occur simultaneously (Bikle, 2004, 2012). It is perhaps of relevance that loss of the VDR in mice potentiates tumor development in the skin whereas loss 1,25(OH)2D3 production does not.
Figure 1.
A mutant human VDR deficient in 1,25(OH)2D3-binding activity restores hair follicle cycling in VDR-null mice and prevents alopecia. Minigenes containing the human VDR gene locus were introduced into a VDR-null mouse background to create humanized mice expressing either wild-type (hVDR) or mutant (hVDR-L233S) VDR proteins. Both hVDR-WT and hVDR-L233S rescue the alopecia observed in VDR-null mice. VDR+/+, unmodified normal mice; hVDR-L233S 805, 806, and 807 represent three rescued mouse strains that express increasing concentrations of VDR protein in all appropriate tissues. See Lee, Goellner, et al. (2014).
3.1.2 Ligand-Independent Suppression of Gene Expression by the VDR
Is there evidence that the VDR functions in a ligand-independent manner in additional biologic processes? Interestingly, the expression of a 1,25(OH)2D3-binding-defective VDR in mice as described above has provided additional circumstantial evidence. For example, while the mutant receptor is unable to rescue systemic features of altered mineral homeostasis, several of these parameters such as the level of PTH appear to be exaggerated relative of their VDR-null mouse counterparts and certain features of the skeletal phenotype appear to be exacerbated as well (Lee, Goellner, et al., 2014). Interestingly, these skeletal abnormalities were documented early on in the Cyp27b1-null mouse. Whether these aberrations are due to reverse VDR activity (suppression) in the absence of ligand remains to be determined. Interestingly, recent studies suggest that in the absence of 1,25(OH)2D3, the VDR may suppress the expression of genes that are normally induced by the receptor in the presence of ligand (Lee, Goellner, et al., 2014). This may indicate that the unliganded VDR can exert transcriptional effects on target genes that are opposite to those identified following 1,25(OH)2D3 activation. Much additional research will be required to prove this hypothesis, however, as at present virtually all of the fundamental mechanistic support for this potential set of VDR activities remains to be delineated. Nevertheless, the role of the VDR in the hair follicle to regulate the hair cycle provides strong conceptual framework that the VDR may regulate gene expression in the absence of 1,25(OH)2D3.
3.2 1,25(OH)2D3 Regulates Transcription via Multiple Enhancers Located at Sites Distal to Gene Promoters
A striking result of extensive unbiased genome-wide ChIP-seq analysis of transcription factor localization across multiple genomes has been the discovery that many if not most genes are regulated by multiple enhancers that are not positioned near promoter regions, but rather within intronic and/or intergenic regions 10s if not 100s of kilobases distal to their transcriptional start sites (TSSs) (Meyer, Benkusky, Lee, et al., 2014; Meyer et al., 2010b, 2012). Indeed, it has been estimated that most genes are regulated by an average of 10 enhancers and that the average distance of an enhancer from its promoter target is greater than 250 kb (Dunham et al., 2012; Gerstein et al., 2012). These findings suggest that the identification of promoter proximal elements near genes based upon the transfected plasmid approach is highly biased, at best incomplete, and often incorrect. A major consequence of results emerging from extensive ChIP-seq analyses is that it is no longer reasonable to explore regulatory mechanisms based upon these earlier molecular biologic approaches with any expectation that an understanding of the regulatory features of a gene will be forthcoming. Significant additional problems with the traditional approaches have also emerged; in the absence of the entire gene locus as well as an appropriate chromatin context, the contribution and interaction of multiple enhancers and the myriad of chromatin regulatory proteins that impact the architecture of the gene locus through both DNA and histone modifications are largely negated. As a consequence, while the results of unbiased ChIP-seq analyses have provided a better understanding of the mechanisms that regulate the output of individual genes, they have at the same time made it much more difficult to define the genetic and epigenetic contributors to such regulation.
3.2.1 Defining Gene Regulation from Distal Sites
Cistromic analysis in many cell types has now revealed the presence of multiple VDR-binding sites within genetic loci that are frequently dispersed to distal intronic and intergenic sites. Each enhancer site may contain one or more VDREs that can be located within a few nucleotides of each other or more than 200 bp (> a nucleosome) from each other. While hundreds of vitamin D target genes are configured in this manner, specific examples include Cyp24a1, Vdr, Cbs, Tnfsf11, c-FOS, Spp1, Runx2, Cdon, Mmp13, Col2a, Trpv6, S100g, and others as well. In the case of Cyp24a1, while the promoter proximal element defined in earlier studies (Zierold et al., 1995) was confirmed, a downstream cluster of regulatory elements has also been identified more recently (Meyer, Goetsch, & Pike, 2010a). In genes such as the Vdr (Zella et al., 2006, 2010), Mmp13 (Meyer, Benkusky, Lee, et al., 2014; Meyer, Benkusky, & Pike, 2015), and many others, the presence of proximal elements in earlier reports was not confirmed, although more distal sites of VDR actions have been delineated. In other cases, VDR-binding sites at genes for which the regulatory activity of 1,25(OH)2D3 had previously been unknown were clarified, frequently located many kilobases distal to the gene’s TSS. These studies of the VDR support the concept that has emerged for most transcription factors that the enhancers to which they bind are often found not only distal to genetic start sites but may contain unregulated genes that are dispersed between the enhancer and the gene it actually regulates.
3.2.2 Confirming Enhancer Function at Target Genes
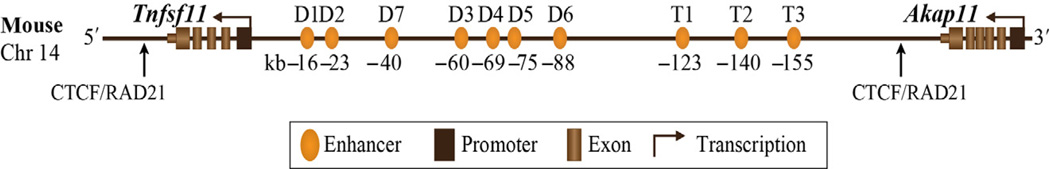
A major consequence of the above arrangement of regulatory enhancers is that while enhancers can be readily located, it is no longer possible to identify the genes to which they are linked without direct experimentation conducted in an endogenous gene-like chromatin context. Accordingly, we have examined enhancer/target gene relationships by stably integrating recombinantly modified bacterial artificial chromosomes (BAC clones) containing extended gene loci for the Vdr, Tnfsf11, and Cyp24a1 genes, for example, into the genomes of cells in vitro and comparing 1,25(OH)2D3 regulatory activities of both wild-type BAC and mutant clones that contain specific enhancer deletions (Lee, Goellner, et al., 2014; Meyer et al., 2010a; Onal et al., 2014; Zella et al., 2006, 2010). Most importantly, these minigenes have also been inserted into the mouse genome as well, verifying that the clones contain sufficient genetic information to direct not only appropriate tissue-specific expression of the gene but a level of expression capable of rescuing the phenotype of mice that are null for the gene product complimented through the transgene (Lee, Bishop, et al., 2014; Onal et al., 2014). A distinct advantage of this in vivo approach is that both the basal and the regulatory features of the transgene of interest can be examined in a wide variety of tissues known to express the gene rather than from a single cell type in culture. A successful rescue permits the conclusion that the transgene contains the fully complement of features that control expression of the gene in its original endogenous setting. The consequence of enhancer deletion on the regulated expression of the transgene as well as the expression of mutant forms of the transcriptional product can then be evaluated as well, the latter highlighted for the human VDR gene in the above section which essentially humanizes the mouse (Lee, Bishop, et al., 2014). We have also deleted putative gene enhancers within the mouse genome and evaluated the consequence of this action on subsequent target gene activity in selected tissues as well (Galli et al., 2008). This has been particularly informative for the mouse Tnfsf11 gene, which contains at least 10 distinct regulatory regions that differentially control the expression of RANKL in osteoblast-lineage cells and in hematopoietic immune cells (Bishop, Coy, Nerenz, Meyer, & Pike, 2011; Bishop, Meyer, & Pike, 2009; Bishop et al., 2014; Kim, Yamazaki, Shevde, & Pike, 2007; Kim, Yamazaki, Zella, Shevde, & Pike, 2006; Fig. 2). Finally, we have begun to explore the role of enhancers in diseased tissue as well, such as in the atherosclerotic plaques that emerge in ApoE-null mice fed a high-fat diet. In this case, mice containing enhancer deletions that compromise the expression of specific genes are crossed into the ApoE-null background and the consequence of enhancer-mediated gene misexpression examined in the high-fat diet-induced atherosclerotic plaques. In another vein, assessing enhancer/target gene relationship for genes that are suppressed by 1,25(OH)2D3 is particularly complex as many features that are routinely apparent and responsible for gene activation frequently do not apply. For example, while VDR-binding sites can be found near a subset of genes that are repressed by 1,25(OH)2D3, many repressed genes do not contain these adjacent VDR-binding sites (Meyer, Benkusky, Lee, et al., 2014; St John, Bishop, et al., 2014). Thus, the mechanisms associated with repression continue to remain obscure. The distal nature of regulatory elements also suggests a requirement for DNA looping as assessed by chromosome conformation capture (3C) technology to bring multiple distal sites into proximity with the active transcriptional centers associated with individual genes.
Figure 2.
Schematic linear structure of the mouse Tnfsf11 gene locus with associated regulatory enhancers located on chromosome 14. The gene spans over 220 kb and is defined by two CTCF/RAD21 sites that serve as boundary elements for the gene. The Tnfsf11 exons are defined as brown rectangles, and enhancers are defined by orange ovals. Arrows define the direction of transcription. Both the Tnfsf11 and neighboring Akap11 genes are located on the reverse strand. D1–D7 represent enhancers that regulate Tnfsf11 expression in osteoblast-lineage cells, whereas T1–T3 represent enhancers that regulate Tnfsf11 expression in hematopoietic B and T cells. See Onal et al. (2014).
3.2.3 Application of the CRISPR/Cas9 Method to the Study of Gene Regulation
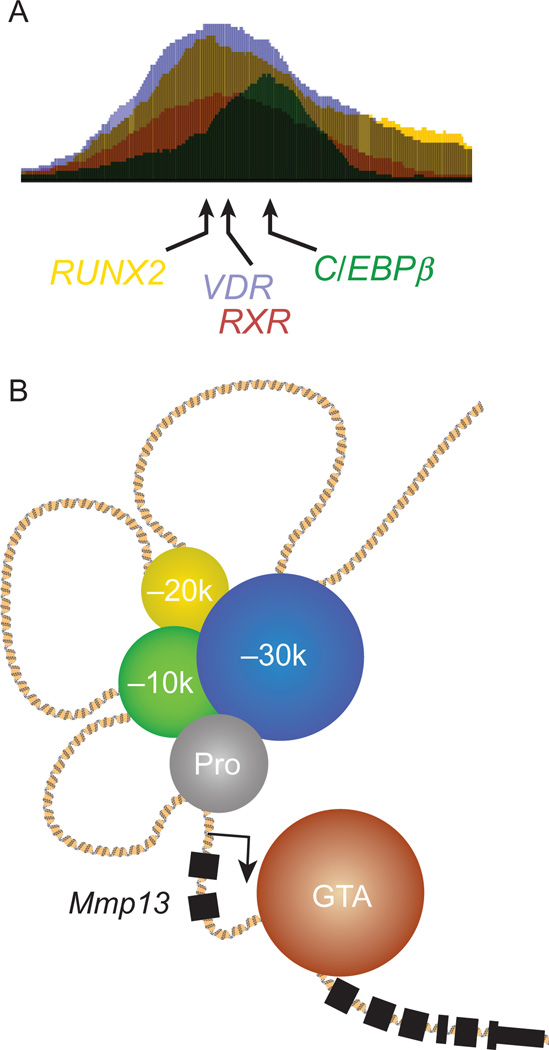
Current studies are now focused on using genome-editing methods employing CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9 to establish the relevance of specific regulatory elements both in cells in culture and in mice in vivo (Cong et al., 2013). Indeed, a detailed examination of the regulation of Mmp13 expression by 1,25(OH)2D3 in osteoblast-lineage cells has revealed the presence of three enhancers located −10, −20, and −30 kb upstream of the Mmp13 TSS (Meyer et al., 2015; Fig. 3). While the promoter region of the Mmp13 manifested no vitamin D-sensitive activity in regulating Mmp13 mRNA transcript production, the enhancer located at −10 kb not only bound the VDR but mediated all of the actions of 1,25(OH)2D3 on Mmp13 mRNA expression. The more distal enhancers mediated the regulation of this gene by other transcription factors including RUNX2 and C/EBPβ. Further dissection of the activities of these enhancers using deletion directed by the CRISPR/Cas9 system revealed unique activities inherent to each enhancer in the osteoblast and the fact that despite their linear distances from each other, the loss of key regulatory capability derived from the most distal enhancer (−30 kb) conferred a profound impact on the binding activities of the other transcription factors at the remaining sites and at the promoter. These activities via genetic dissection by the CRISPR/Cas9 system are likely to provide the preferred methodological approach to dissecting transcriptional regulation and enhancer function in the near future in exquisite detail. Indeed, we have already deleted a series of enhancers from vitamin D target genes in mice in vivo and shown that these mutations have a striking impact of the expression of the genes to which they are linked.
Figure 3.
Organization of osteoblast enhancer complexes (OEC) that bind RUNX2, C/EBPβ, and the VDR. (A) Consolidated OEC-binding arrangement for RUNX2, C/EBPβ, and VDR at the Spp1 (osteopontin) gene as defined by ChIP-seq analysis. (B) Dispersed OEC-binding arrangement for RUNX2, C/EBPβ, and VDR at the Mmp13 gene locus as defined by ChIP-seq and other functional analyses. Schematic depiction of the individual RUNX2 (blue), C/EBPβ (yellow), and VDR (green) enhancers shown clustered and interacting upstream of the Mmp13 gene locus with the promoter proximal region (gray) and the general transcription assembly (brown) located near the transcriptional start site. See Meyer, Benkusky, and Pike (2015).
4. THE INFLUENCE OF CELLULAR DIFFERENTIATION ON VITAMIN D ACTIVITY
Perhaps the most important observation made regarding the actions of 1,25(OH)2D3 on a genome-wide scale has been the discovery that cellular differentiation exerts a dramatic quantitative and qualitative impact on the hormone’s ability to regulate gene expression (Meyer, Benkusky, Lee, et al., 2014; Meyer, Benkusky, & Pike, 2014; St John, Bishop, et al., 2014; St John, Meyer, et al., 2014). It has been known since early times that the effects of 1,25(OH)2D3 on osteoblast-lineage cells differ significantly depending upon the cellular state of differentiation, although these conclusions were drawn largely from studies of single genes such as osteocalcin (Lian et al., 1989). In recent studies that have highlighted this concept more broadly, we observed that the treatment of early osteoblasts with 1,25(OH)2D3 resulted in a transcriptome that was more expansive and strikingly different than that observed in mineralizing osteoblasts following differentiation (Meyer, Benkusky, Lee, et al., 2014). While an extensive overlap of genes was noted in both cell types, the transcriptome itself was quantitatively reduced in more mature cells, in part because expression of the VDR itself was downregulated. Indeed, some genes were no longer responsive to 1,25(OH)2D3. The most striking observation, however, was the finding that despite a significant reduction in VDR expression, the basal levels of many vitamin D target genes were altered due to differentiation and the response to 1,25(OH)2D3 was qualitatively altered as well. In some cases, the gene exhibited an increase in response to 1,25(OH)2D3 and in others a decrease; in some cases, the directionality of regulation was reversed. These findings highlight the highly dynamic nature of the biologic activities of 1,25(OH)2D3 in cells that are closely related and raise the important underlying question of how the differentiation process is able to impact response to 1,25(OH)2D3 mechanistically.
4.1 Differentiation Is Accompanied by Direct Alterations in the VDR Cistrome
A comparison of the VDR/RXR cistrome in early precursors and late mineralizing osteoblasts using ChIP-seq analysis revealed a striking reduction in the number of binding sites for the VDR in more mature cells and a significant reduction in their location as well (Meyer, Benkusky, Lee, et al., 2014). As expected, VDR binding was frequently lost at sites near genes that were no longer responsive to 1,25(OH)2D3. In contrast, however, although frequently reduced following differentiation, VDR binding was retained at many sites near genes that remained responsive or showed increased response to 1,25(OH)2D3, although in many cases the level of receptor binding activity was significantly reduced as assessed by ChIP-seq analysis. This observation suggests the possibility that transcription factors other than the VDR that are either recruited to or enriched at sites near these genes following differentiation may contribute in enhancing response to 1,25(OH)2D3. It is also possible that these factors may influence the epigenetic landscape that defines functional features of the target gene, facilitating further the ease with which 1,25(OH)2D3 and its receptor may be able to modify the gene’s expression. This hypothesis is supported by the observation that the basal levels of expression of many of these genes are often changed following cellular differentiation.
4.2 The Impact of Osteoblast Differentiation on Master Regulatory Factor Distribution, Histone Modifying Activity, and Response to 1,25(OH)2D3
To explore the idea elaborated above, we first used ChIP-seq analysis to determine the distribution of binding sites for two key osteoblast-lineage determining factors RUNX2 and C/EBPβ across the osteoblast genome before and after differentiation (Meyer, Benkusky, & Pike, 2014). Importantly, while the level of expression of these individual factors changed only modestly following differentiation, both the level of their occupancy at existing sites as well as their accumulation (or loss thereof) at new sites of action was significantly altered. An examination of the effects of 1,25(OH)2D3 on these cistromes revealed that in addition to the hormone’s known direct inhibitory role on RUNX2 and C/EBPβ expression, the hormone also caused a modest redistribution of RUNX2 and C/EBPβ DNA binding in early osteoblasts suggesting that the hormone was likely capable of affecting the presence of RUNX2 and C/EBPβ concentrations at selected sites as well (Meyer, Benkusky, Lee, et al., 2014). Given the role of these factors in the recruitment of chromatin regulatory modifiers, these results suggest further that significant quantitative and/or qualitative changes in the levels of epigenetic modification might also be detectable at genes whose expression levels were altered as a function of osteoblast differentiation both in the absence and presence of 1,25(OH)2D3. Interestingly, we found that while the epigenetic landscape was generally unchanged when examined using all genes that were expressed in both precursor cell and their differentiated counterpart, significant changes were observed when the analysis was restricted to genes whose expression patterns were altered as a result of the differentiation process (Meyer, Benkusky, & Pike, 2014; St John, Bishop, et al., 2014). Of particular importance were the changes observed at H3K4me1, H3K4me2, H3K9ac, H3K27ac, and H4K5ac, modifications that denote the locations of regulatory enhancers or that highlight variations in chromatin decondensation and accessibility. Numerous changes were also noted atH3K4me3, a mark that specifies the location of gene promoters, and at H3K36me3, H4K20me1, and H4K5ac, marks that identify genomic regions spanning the transcription units (exons and introns) of genes. Interestingly, bioinformatic examination also revealed that most of the changes in the levels of signature marks at enhancers were quantitative (St John, Bishop, et al., 2014). Thus, only a few changes in histone modification that denoted the novel appearance of a newly minted enhancer (or its loss) could uniquely regulate the expression of the gene to which it was linked were observed. These results suggest broadly that programmed creation of the vast majority of regulatory enhancers in cells of the osteoblast lineage likely occurs early in the mesenchymal progression, and that alterations in transcription factor expression and activity (i.e., RUNX2 and C/EBPβ) may be dominant at influencing the epigenetic landscape surrounding regulated genes, could influence gene output, and likely modulate response to 1,25(OH)2D3 and other systemic regulatory hormones.
4.3 Identification and Structure of the Osteoblast Enhancer Complex
The potential influence of both RUNX2 and C/EBPβ activity on cellular response to 1,25(OH)2D3 together with a preliminary bioinformatic analysis that suggested the enriched presence of DNA-binding motifs for these latter two factors at sites of VDR binding on a genome-wide scale prompted us to examine the potential relationship between the VDR cistrome and those for RUNX2 and C/EBPβ directly. This analysis confirmed that either RUNX2 or C/EBPβ or both were frequent occupants at active VDR DNA-binding sites (Meyer, Benkusky, Lee, et al., 2014). Further inspection revealed that 70% of the 4174 VDR/RXR-binding sites that were identified in early osteoblasts also contained RUNX2, while 42% contain both RUNX2 and C/EBPβ. A more detailed examination identified an even closer physical relationship between RUNX2, C/EBPβ, and VDR/RXR, prompting its description as a consolidated “osteoblast enhancer complex” (Fig. 3A). Indeed, we found that RUNX2 and C/EBPβ bind bidirectionally 8 and 9 bp on average from the VDR/RXR peak center, respectively. As RUNX2 and C/EBPβ are independently active in the regulation of gene expression in osteoblast-lineage cells, these findings support the idea that enhancers of this type are likely capable of mediating both the independent actions of these specific transcription factors and of the actions of the VDR, and perhaps in some cases of integrating the actions of all three as well. Interestingly, other transcription factor arrangements for VDR/RXR, RUNX2, and C/EBPβ are also apparent. Thus, many genes including Mmp13 are regulated by set of dispersed enhancers that bind RUNX2, C/EBPβ, or the VDR individually (Meyer, Benkusky, Lee, et al., 2014; Meyer et al., 2015). Given the linear distances between each enhancer in these examples, we speculate that the activities of each of these regulatory modules are likely integrated collectively at the promoters for target genes via complex DNA looping in a manner that is reminiscent of that seen for the consolidated osteoblast enhancer complex. Our recent studies of the Mmp13 gene support this concept through the demonstration that targeted CRISPR/Cas9-mediated deletion of several of the genes enhancers impacts not only the overall expression of Mmp13 but influences binding activity of transcription factors such as RUNX2, C/EBPβ, and the VDR at the enhancers that remain in the Mmp13 locus (Meyer et al., 2015; Fig. 3B). The prebound nature of both RUNX2 and C/EBPβ on DNA and their broad master regulatory properties in osteoblasts suggest that they may play an instrumental role in establishing and maintaining enhancers for genes that are not only relevant to the osteoblast lineage but that their actions at these enhancers may facilitate the availability of sites to which the VDR and other secondary regulators can be recruited. If this hypothesis is correct, while the VDR is a primary determinant of vitamin D action, both RUNX2 and C/EBPβ and likely others that operate in a lineage-dependent fashion are also determinants of the quantitative and qualitative nature of the response, in part by contributing to processes such as histone modification that control the output of gene expression.
5. VDR MODULATES HISTONE ACETYLATION AT TARGET GENES
Initial studies of vitamin D action revealed that VDR binding at the proximal elements associated with Spp1 and Cyp24a1 regulation resulted in a differential increase in the level of histone H3 and H4 acetylation at these genes, suggesting the existence of a chromatin response to the actions of 1,25 (OH)2D3 that might be gene-selective (Kim et al., 2005). Subsequent studies of the genes for Vdr, Tnfsf11, and others support this view (Kim et al., 2006; Zella et al., 2006). Consistent with these observations, we subsequently discovered that the effects of VDR binding on a genome-wide scale in osteoblasts and osteocytes also reflect this premise (Meyer, Benkusky, Lee, et al., 2014; St John, Bishop, et al., 2014). Accordingly, while acetylation levels of H3K9, H4K5, and H3K27 were increased at sites of VDR action near many genes, sites in other genes were unaffected. It has long been known that one of the functions of the VDR in gene activation is to initiate the recruitment of coregulatory factors that include CBP, p300, and the SRC family of histone acetyltransferases and several histone deacetyltransferases as well (Sutton & MacDonald, 2003). It is clear that the actions of these enzymes at the histones associated with many genes likely account for the changes in acetylation that are observed, although the mechanism that underlies this site-selectivity is not understood. It seems likely that the requirement for gene activation differs among individual genes, perhaps based upon the nature of the residual expression level of the gene in question and the presence of additional transcription factors that contribute to this level of expression.
Acetylation levels represent a hallmark of chromatin decondensation and transcription factor accessibility to binding sites on DNA, particularly if access to those sites is restricted due to nucleosome positioning (Shahbazian & Grunstein, 2007). Alternative explanations as to the role of increased acetylation as well as methylation include the possibility that site-specific increases lead to the recruitment of additional chromatin regulators that are necessary for nucleosomal redistribution, eviction, or exchange, thereby enabling enhancer/promoter engagement through DNA reorganization (Berger, 2007). Separate studies using 3C analysis have shown, for example, that the presence of estrogen and the estrogen receptor at distal enhancers facilitates this type of DNA reorganization (Pan et al., 2008). Interestingly, while our studies of this event at the Tnfsf11 (RANKL) and Cyp24a1 genes have shown linkage between the promoters for these genes and their associated distal enhancers, they do not appear to be influenced by 1,25(OH)2D3 (Bishop et al., 2011; Meyer et al., 2010a). Increased methylation at specific sites on histones likewise precipitates changes in gene output, likely due in this case to the selective recruitment of chromatin regulators known as “readers” whose downstream actions are currently being characterized (Calo & Wysocka, 2013; Ruthenburg, Allis, & Wysocka, 2007). Future studies will be required to delineate the consequence of increased acetylation and methylation by VDR at the molecular level and identify the specific players that are involved. Nevertheless, the observation that activated VDR initiates enhanced expression of specific chromatin regulators as well as their recruitment to genes provides an initial starting point.
6. SUMMARY
Recent studies have revealed a set of general overarching principles through which 1,25(OH)2D3 and its receptor regulate the expression of genes in cellular targets. These findings confirm and extend many of the general features that have been identified for vitamin D action over the past several decades, but have also revealed important new concepts as well. These include the finding that while VDR binds to DNA largely in response to activation by 1,25(OH)2D3, a subset of genetic targets contain prebound VDR even in the absence of the ligand. This finding highlights the possibility that the VDR may have a more ubiquitous 1,25(OH)2D3-independent function in gene regulation, an activity that appears to be illustrated by VDR actions in the hair cycle. An additional discovery is the finding that regulatory enhancers for the VDR as well as for most other transcription factors are often multiple at single gene loci and more importantly located at sites that are frequently remote relative to the genes they regulate. This finding highlights the likely role of DNA looping that enables these distal enhancers to contact the transcriptional machinery near the promoter and to impact transcriptional output. Finally, our results suggest that the process of differentiation is capable of altering the transcription factor milieu at important target genes and directly affecting the epigenetic landscape surrounding these genes as well. This combination of effects is capable of altering the DNA-binding activity of secondary regulatory factors such as the VDR and influencing in both a positive and negative manner the protein’s transcriptional activity as well. These results illustrate the highly dynamic nature of VDR action within tissues thereby adding a new dimension to our understanding of the role of the genome in defining cellular context that ultimately impacts hormonal response. This molecular and regulatory complexity and the certain consequence of disease on these events at the genomic levels are likely to be significant in vivo.
Acknowledgments
We acknowledge the helpful contributions of members of the Pike laboratory to this review and financial support from National Institutes of Health grants from NIDDK (DK-072281, DK-074993) and NIAMS (AR-045173 and AR-062442) to J.W.P.
Footnotes
Disclosure: The authors declare no conflict of interest.
REFERENCES
- Baker AR, McDonnell DP, Hughes M, Crisp TM, Mangelsdorf DJ, Haussler MR, et al. Cloning and expression of full-length cDNA encoding human vitamin D receptor. Proceedings of the National Academy of Sciences of the United States of America. 1988;85(10):3294–3298. doi: 10.1073/pnas.85.10.3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447(7143):407–412. doi: 10.1038/nature05915. http://dx.doi.org/10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- Bikle DD. Vitamin D regulated keratinocyte differentiation. Journal of Cellular Biochemistry. 2004;92(3):436–444. doi: 10.1002/jcb.20095. [DOI] [PubMed] [Google Scholar]
- Bikle DD. Vitamin D and the skin: Physiology and pathophysiology. Reviews in Endocrine & Metabolic Disorders. 2012;13(1):3–19. doi: 10.1007/s11154-011-9194-0. http://dx.doi.org/10.1007/s11154-011-9194-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop KA, Coy HM, Nerenz RD, Meyer MB, Pike JW. Mouse Rankl expression is regulated in T cells by c-Fos through a cluster of distal regulatory enhancers designated the T cell control region. The Journal of Biological Chemistry. 2011;286(23):20880–20891. doi: 10.1074/jbc.M111.231548. http://dx.doi.org/10.1074/jbc.M111.231548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop KA, Meyer MB, Pike JW. A novel distal enhancer mediates cytokine induction of mouse RANKl gene expression. Molecular Endocrinology. 2009;23(12):2095–2110. doi: 10.1210/me.2009-0209. doi:me.2009-0209 [pii]10.1210/me.2009-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop KA, Wang X, Coy HM, Meyer MB, Gumperz JE, Pike JW. Transcriptional regulation of the human TNFSF11 gene in t cells via a cell type-selective set of distal enhancers. Journal of Cellular Biochemistry. 2014;116(2):320–330. doi: 10.1002/jcb.24974. http://dx.doi.org/10.1002/jcb.24974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouillon R, Carmeliet G, Verlinden L, van Etten E, Verstuyf A, Luderer HF, et al. Vitamin D and human health: Lessons from vitamin D receptor null mice. Endocrine Reviews. 2008;29(6):726–776. doi: 10.1210/er.2008-0004. doi:er.2008-0004 [pii] 10.1210/er.2008-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks MH, Bell NH, Love L, Stern PH, Orfei E, Queener SF, et al. Vitamin-D-dependent rickets type II. Resistance of target organs to 1,25-dihydroxyvitamin D. The New England Journal of Medicine. 1978;298(18):996–999. doi: 10.1056/NEJM197805042981804. http://dx.doi.org/10.1056/NEJM197805042981804. [DOI] [PubMed] [Google Scholar]
- Brumbaugh P, Haussler M. 1 Alpha,25-dihydroxycholecalciferol receptors in intestine. I. Association of 1 alpha,25-dihydroxycholecalciferol with intestinal mucosa chromatin. The Journal of Biological Chemistry. 1974a;249(4):1251–1257. [PubMed] [Google Scholar]
- Brumbaugh PF, Haussler MR. 1a,25-dihydroxycholecalciferol receptors in intestine. II. Temperature-dependent transfer of the hormone to chromatin via a specific cytosol receptor. The Journal of Biological Chemistry. 1974b;249(4):1258–1262. [PubMed] [Google Scholar]
- Burmester JK, Maeda N, DeLuca HF. Isolation and expression of rat 1,25-dihydroxyvitamin D3 receptor cDNA. Proceedings of the National Academy of Sciences of the United States of America. 1988;85(4):1005–1009. doi: 10.1073/pnas.85.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo E, Wysocka J. Modification of enhancer chromatin: What, how, and why? Molecular Cell. 2013;49(5):825–837. doi: 10.1016/j.molcel.2013.01.038. http://dx.doi.org/10.1016/j.molcel.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CH, Sakai Y, Demay MB. Targeting expression of the human vitamin D receptor to the keratinocytes of vitamin D receptor null mice prevents alopecia. Endocrinology. 2001;142(12):5386–5389. doi: 10.1210/endo.142.12.8650. [DOI] [PubMed] [Google Scholar]
- Cianferotti L, Cox M, Skorija K, Demay MB. Vitamin D receptor is essential for normal keratinocyte stem cell function. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(22):9428–9433. doi: 10.1073/pnas.0702884104. http://dx.doi.org/10.1073/pnas.0702884104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. http://dx.doi.org/10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dame M, Pierce E, Prahl J, Hayes C, DeLuca H. Monoclonal antibodies to the porcine intestinal receptor for 1,25-dihydroxyvitamin D3: Interaction with distinct receptor domains. Biochemistry. 1986;25(16):4523–4534. doi: 10.1021/bi00364a011. [DOI] [PubMed] [Google Scholar]
- Demay MB, MacDonald PN, Skorija K, Dowd DR, Cianferotti L, Cox M. Role of the vitamin D receptor in hair follicle biology. The Journal of Steroid Biochemistry and Molecular Biology. 2007;103(3–5):344–346. doi: 10.1016/j.jsbmb.2006.12.036. http://dx.doi.org/10.1016/j.jsbmb.2006.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding N, Yu RT, Subramaniam N, Sherman MH, Wilson C, Rao R, et al. A vitamin D receptor/SMAD genomic circuit gates hepatic fibrotic response. Cell. 2013;153(3):601–613. doi: 10.1016/j.cell.2013.03.028. http://dx.doi.org/10.1016/j.cell.2013.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. http://dx.doi.org/10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eil C, Liberman UA, Rosen JF, Marx SJ. A cellular defect in hereditary vitamin-D-dependent rickets type. II: Defective nuclear uptake of 1,25-dihydroxyvitamin D in cultured skin fibroblasts. The New England Journal of Medicine. 1981;304(26):1588–1591. doi: 10.1056/NEJM198106253042608. http://dx.doi.org/10.1056/NEJM198106253042608. [DOI] [PubMed] [Google Scholar]
- Evans R. The steroid and thyroid hormone receptor superfamily. Science. 1988;240(4854):889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman D, Malloy P. Hereditary 1,25-dihydroxyvitamin D resistant rickets: Molecular basis and implications for the role of 1,25(OH) 2D3 in normal physiology. Molecular and Cellular Endocrinology. 1990;72(3):C57–C62. doi: 10.1016/0303-7207(90)90137-w. [DOI] [PubMed] [Google Scholar]
- Forghani N, Lum C, Krishnan S, Wang J, Wilson D, Blackett P, et al. Two new unrelated cases of hereditary 1,25-dihydroxyvitamin D-resistant rickets with alopecia resulting from the same novel nonsense mutation in the vitamin D receptor gene. Journal of Pediatric Endocrinology & Metabolism. 2010;23(8):843–850. doi: 10.1515/jpem.2010.136. [DOI] [PubMed] [Google Scholar]
- Galli C, Zella L, Fretz J, Fu Q, Pike J, Weinstein R, et al. Targeted deletion of a distant transcriptional enhancer of the receptor activator of nuclear factor-kappaB ligand gene reduces bone remodeling and increases bone mass. Endocrinology. 2008;149(1):146–153. doi: 10.1210/en.2007-0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, Cheng C, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489(7414):91–100. doi: 10.1038/nature11245. http://dx.doi.org/10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haussler MR, Norman AW. Chromosomal receptor for a vitamin D metabolite. Proceedings of the National Academy of Sciences of the United States of America. 1969;62(1):155–162. doi: 10.1073/pnas.62.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heikkinen S, Väisänen S, Pehkonen P, Seuter S, Benes V, Carlberg C. Nuclear hormone 1α,25-dihydroxyvitamin D3 elicits a genome-wide shift in the locations of VDR chromatin occupancy. Nucleic Acids Research. 2011;39(21):9181–9193. doi: 10.1093/nar/gkr654. http://dx.doi.org/10.1093/nar/gkr654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes MR, Malloy PJ, Kieback DG, Kesterson RA, Pike JW, Feldman D, et al. Point mutations in the human vitamin D receptor gene associated with hypocalcemic rickets. Science. 1988;242(4886):1702–1705. doi: 10.1126/science.2849209. [DOI] [PubMed] [Google Scholar]
- Kerner SA, Scott RA, Pike JW. Sequence elements in the human osteocalcin gene confer basal activation and inducible response to hormonal vitamin D3. Proceedings of the National Academy of Sciences of the United States of America. 1989;86(12):4455–4459. doi: 10.1073/pnas.86.12.4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Shevde N, Pike J. 1,25-Dihydroxyvitamin D3 stimulates cyclic vitamin D receptor/retinoid X receptor DNA-binding, co-activator recruitment, and histone acetylation in intact osteoblasts. Journal of Bone and Mineral Research. 2005;20(2):305–317. doi: 10.1359/JBMR.041112. [DOI] [PubMed] [Google Scholar]
- Kim S, Yamazaki M, Shevde NK, Pike JW. Transcriptional control of receptor activator of nuclear factor-kappaB ligand by the protein kinase A activator forskolin and the transmembrane glycoprotein 130-activating cytokine, oncostatin M, is exerted through multiple distal enhancers. Molecular Endocrinology. 2007;21(1):197–214. doi: 10.1210/me.2006-0315. [DOI] [PubMed] [Google Scholar]
- Kim S, Yamazaki M, Zella LA, Shevde NK, Pike JW. Activation of receptor activator of NF-kappaB ligand gene expression by 1,25-dihydroxyvitamin D3 is mediated through multiple long-range enhancers. Molecular and Cellular Biology. 2006;26(17):6469–6486. doi: 10.1128/MCB.00353-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SM, Bishop KA, Goellner JJ, O’Brien CA, Pike JW. Mouse and human BAC transgenes recapitulate tissue-specific expression of the vitamin D receptor in mice and rescue the VDR-null phenotype. Endocrinology. 2014;155(6):2064–2076. doi: 10.1210/en.2014-1107. http://dx.doi.org/10.1210/en.2014-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SM, Goellner JJ, O’Brien CA, Pike JW. A humanized mouse model of hereditary 1,25-dihydroxyvitamin D-resistant rickets without alopecia. Endocrinology. 2014;155(11):4137–4148. doi: 10.1210/en.2014-1417. http://dx.doi.org/10.1210/en.2014-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SM, Riley EM, Meyer MB, Benkusky NA, Plum LA, DeLuca HF, et al. 1,25-dihydroxyvitamin D3 controls a cohort of vitamin D receptor target genes in the proximal intestine that is enriched for calcium-regulating components. The Journal of Biological Chemistry. 2015;290(29):18199–18215. doi: 10.1074/jbc.M115.665794. http://dx.doi.org/10.1074/jbc.M115.665794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YC, Pirro AE, Amling M, Delling G, Baron R, Bronson R, et al. Targeted ablation of the vitamin D receptor: An animal model of vitamin D-dependent rickets type II with alopecia. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(18):9831–9835. doi: 10.1073/pnas.94.18.9831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian J, Stewart C, Puchacz E, Mackowiak S, Shalhoub V, Collart D, et al. Structure of the rat osteocalcin gene and regulation of vitamin D-dependent expression. Proceedings of the National Academy of Sciences of the United States of America. 1989;86(4):1143–1147. doi: 10.1073/pnas.86.4.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malloy PJ, Pike JW, Feldman D. The vitamin D receptor and the syndrome of hereditary 1,25-dihydroxyvitamin D-resistant rickets. Endocrine Reviews. 1999;20(2):156–188. doi: 10.1210/edrv.20.2.0359. [DOI] [PubMed] [Google Scholar]
- Malloy PJ, Tasic V, Taha D, Tütüncüler F, Ying GS, Yin LK, et al. Vitamin D receptor mutations in patients with hereditary 1,25-dihydroxyvitamin D-resistant rickets. Molecular Genetics and Metabolism. 2014;111(1):33–40. doi: 10.1016/j.ymgme.2013.10.014. http://dx.doi.org/10.1016/j.ymgme.2013.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx SJ, Spiegel AM, Brown EM, Gardner DG, Downs RW, Attie M, et al. A familial syndrome of decrease in sensitivity to 1,25-dihydroxyvitamin D. The Journal of Clinical Endocrinology and Metabolism. 1978;47(6):1303–1310. doi: 10.1210/jcem-47-6-1303. [DOI] [PubMed] [Google Scholar]
- McDonnell DP, Mangelsdorf DJ, Pike JW, Haussler MR, O’Malley BW. Molecular cloning of complementary DNA encoding the avian receptor for vitamin D. Science. 1987;235(4793):1214–1217. doi: 10.1126/science.3029866. [DOI] [PubMed] [Google Scholar]
- McDonnell D, Pike J, O’Malley B. The vitamin Dreceptor: A primitive steroid receptor related to thyroid hormone receptor. Journal of Steroid Biochemistry. 1988;30(1–6):41–46. doi: 10.1016/0022-4731(88)90074-x. [DOI] [PubMed] [Google Scholar]
- McDonnell D, Scott R, Kerner S, O’Malley B, Pike J. Functional domains of the human vitamin D3 receptor regulate osteocalcin gene expression. Molecular Endocrinology. 1989;3(4):635–644. doi: 10.1210/mend-3-4-635. [DOI] [PubMed] [Google Scholar]
- Meyer MB, Benkusky NA, Lee CH, Pike JW. Genomic determinants of gene regulation by 1,25-dihydroxyvitamin D3 during osteoblast-lineage cell differentiation. The Journal of Biological Chemistry. 2014;289(28):19539–19554. doi: 10.1074/jbc.M114.578104. http://dx.doi.org/10.1074/jbc.M114.578104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer MB, Benkusky NA, Pike JW. The RUNX2 cistrome in osteoblasts: Characterization, down-regulation following differentiation, and relationship to gene expression. The Journal of Biological Chemistry. 2014;289(23):16016–16031. doi: 10.1074/jbc.M114.552216. http://dx.doi.org/10.1074/jbc.M114.552216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer MB, Benkusky NA, Pike JW. Selective distal enhancer control of the Mmp13 gene identified through clustered regularly interspaced short palindromic repeat (CRISPR) genomic deletions. The Journal of Biological Chemistry. 2015;290(17):11093–11107. doi: 10.1074/jbc.M115.648394. http://dx.doi.org/10.1074/jbc.M115.648394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer MB, Goetsch PD, Pike JW. A downstream intergenic cluster of regulatory enhancers contributes to the induction of CYP24A1 expression by 1alpha,25-dihydroxyvitamin D3. The Journal of Biological Chemistry. 2010a;285(20):15599–15610. doi: 10.1074/jbc.M110.119958. http://dx.doi.org/10.1074/jbc.M110.119958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer MB, Goetsch PD, Pike JW. Genome-wide analysis of the VDR/RXR cistrome in osteoblast cells provides new mechanistic insight into the actions of the vitamin D hormone. The Journal of Steroid Biochemistry and Molecular Biology. 2010b;121(1–2):136–141. doi: 10.1016/j.jsbmb.2010.02.011. doi:S0960-0760(10)00065-8 [pii]10.1016/j.jsbmb.2010.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer MB, Goetsch PD, Pike JW. VDR/RXR and TCF4/β-catenin cistromes in colonic cells of colorectal tumor origin: Impact on c-FOS and c-MYC gene expression. Molecular Endocrinology. 2012;26(1):37–51. doi: 10.1210/me.2011-1109. doi:me.2011-1109 [pii]10.1210/ me.2011-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer MB, Pike JW. Corepressors (NCoR and SMRT) as well as coactivators are recruited to positively regulated 1α,25-dihydroxyvitamin D3-responsive genes. The Journal of Steroid Biochemistry and Molecular Biology. 2013;136:120–124. doi: 10.1016/j.jsbmb.2012.08.006. http://dx.doi.org/10.1016/j.jsbmb.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson SK, Johnston HM, Whitty GA, Williams B, Webb RJ, Denhardt DT, et al. Osteopontin, a key component of the hematopoietic stem cell niche and regulator of primitive hematopoietic progenitor cells. Blood. 2005;106(4):1232–1239. doi: 10.1182/blood-2004-11-4422. doi:2004-11-4422 [pii]10.1182/blood-2004-11-4422. [DOI] [PubMed] [Google Scholar]
- Ohyama Y, Ozono K, Uchida M, Shinki T, Kato S, Suda T, et al. Identification of a vitamin D-responsive element in the 5′-flanking region of the rat 25-hydroxyvitamin D3 24-hydroxylase gene. The Journal of Biological Chemistry. 1994;269(14):10545–10550. [PubMed] [Google Scholar]
- Onal M, Bishop KA, St John HC, Danielson AL, Riley EM, Piemontese M, et al. A DNA segment spanning the mouse Tnfsf11 transcription unit and its upstream regulatory domain rescues the pleiotropic biologic phenotype of the RANKL null mouse. Journal of Bone and Mineral Research. 2014;30(5):855–868. doi: 10.1002/jbmr.2417. http://dx.doi.org/10.1002/jbmr.2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozono K, Liao J, Kerner SA, Scott RA, Pike JW. The vitamin D-responsive element in the human osteocalcin gene. Association with a nuclear proto-oncogene enhancer. The Journal of Biological Chemistry. 1990;265(35):21881–21888. [PubMed] [Google Scholar]
- Pan YF, Wansa KD, Liu MH, Zhao B, Hong SZ, Tan PY, et al. Regulation of estrogen receptor-mediated long range transcription via evolutionarily conserved distal response elements. The Journal of Biological Chemistry. 2008;283(47):32977–32988. doi: 10.1074/jbc.M802024200. http://dx.doi.org/10.1074/jbc.M802024200. [DOI] [PubMed] [Google Scholar]
- Panda DK, Miao D, Tremblay ML, Sirois J, Farookhi R, Hendy GN, et al. Targeted ablation of the 25-hydroxyvitamin D 1alpha -hydroxylase enzyme: Evidence for skeletal, reproductive, and immune dysfunction. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(13):7498–7503. doi: 10.1073/pnas.131029498. http://dx.doi.org/10.1073/pnas.131029498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike J. Vitamin D3 receptors: Structure and function in transcription. Annual Review of Nutrition. 1991;11:189–216. doi: 10.1146/annurev.nu.11.070191.001201. [DOI] [PubMed] [Google Scholar]
- Pike J, Dokoh S, Haussler M, Liberman U, Marx S, Eil C. Vitamin D3-resistant fibroblasts have immunoassayable 1,25-dihydroxyvitamin D3 receptors. Science. 1984;224(4651):879–881. doi: 10.1126/science.6326262. [DOI] [PubMed] [Google Scholar]
- Pike J, Donaldson C, Marion S, Haussler M. Development of hybridomas secreting monoclonal antibodies to the chicken intestinal 1 alpha,25-dihydroxyvitamin D3 receptor. Proceedings of the National Academy of Sciences of the United States of America. 1982;79(24):7719–7723. doi: 10.1073/pnas.79.24.7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike JW, Haussler MR. Purification of chicken intestinal receptor for 1,25-dihydroxyvitamin D. Proceedings of the National Academy of Sciences of the United States of America. 1979;76(11):5485–5489. doi: 10.1073/pnas.76.11.5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike JW, Lee SM, Meyer MB. Regulation of gene expression by 1,25-dihydroxyvitamin D3 in bone cells: Exploiting new approaches and defining new mechanisms. Bonekey Report. 2014;3:482. doi: 10.1038/bonekey.2013.216. http://dx.doi.org/10.1038/bonekey.2013.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike JW, Meyer MB. Fundamentals of vitamin D hormone-regulated gene expression. The Journal of Steroid Biochemistry and Molecular Biology. 2014;144PA:5–11. doi: 10.1016/j.jsbmb.2013.11.004. http://dx.doi.org/10.1016/j.jsbmb.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike JW, Meyer MB, Martowicz ML, Bishop KA, Lee SM, Nerenz RD, et al. Emerging regulatory paradigms for control of gene expression by 1,25-dihydroxyvitamin D3. The Journal of Steroid Biochemistry and Molecular Biology. 2010;121(1–2):130–135. doi: 10.1016/j.jsbmb.2010.02.036. http://dx.doi.org/10.1016/j.jsbmb.2010.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramagopalan SV, Heger A, Berlanga AJ, Maugeri NJ, Lincoln MR, Burrell A, et al. A ChIP-seq defined genome-wide map of vitamin D receptor binding: Associations with disease and evolution. Genome Research. 2010;20(10):1352–1360. doi: 10.1101/gr.107920.110. http://dx.doi.org/10.1101/gr.107920.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: Intricacy of writing and reading a single epigenetic mark. Molecular Cell. 2007;25(1):15–30. doi: 10.1016/j.molcel.2006.12.014. doi:S1097-2765(06)00872-0 [pii]10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annual Review of Biochemistry. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. http://dx.doi.org/10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- Sherman MH, Yu RT, Engle DD, Ding N, Atkins AR, Tiriac H, et al. Vitamin D receptor-mediated stromal reprogramming suppresses pancreatitis and Features of Gene Regulation by Vitamin D enhances pancreatic cancer therapy. Cell. 2014;159(1):80–93. doi: 10.1016/j.cell.2014.08.007. http://dx.doi.org/10.1016/j.cell.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skorija K, Cox M, Sisk JM, Dowd DR, MacDonald PN, Thompson CC, et al. Ligand-independent actions of the vitamin D receptor maintain hair follicle homeostasis. Molecular Endocrinology. 2005;19(4):855–862. doi: 10.1210/me.2004-0415. http://dx.doi.org/10.1210/me.2004-0415. [DOI] [PubMed] [Google Scholar]
- St John HC, Bishop KA, Meyer MB, Benkusky NA, Leng N, Kendziorski C, et al. The osteoblast to osteocyte transition: Epigenetic changes and response to the vitamin D3 hormone. Molecular Endocrinology. 2014;28(7):1150–1165. doi: 10.1210/me.2014-1091. http://dx.doi.org/10.1210/me.2014-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St John HC, Meyer MB, Benkusky NA, Carlson AH, Prideaux M, Bonewald LF, et al. The parathyroid hormone-regulated transcriptome in osteocytes: Parallel actions with 1,25-dihydroxyvitamin D3 to oppose gene expression changes during differentiation and to promote mature cell function. Bone. 2014;72C:81–91. doi: 10.1016/j.bone.2014.11.010. http://dx.doi.org/10.1016/j.bone.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton AL, MacDonald PN. Vitamin D: More than a “bone-a-fide” hormone. Molecular Endocrinology. 2003;17(5):777–791. doi: 10.1210/me.2002-0363. [DOI] [PubMed] [Google Scholar]
- Yoshizawa T, Handa Y, Uematsu Y, Takeda S, Sekine K, Yoshihara Y, et al. Mice lacking the vitamin D receptor exhibit impaired bone formation, uterine hypoplasia and growth retardation after weaning. Nature Genetics. 1997;16(4):391–396. doi: 10.1038/ng0897-391. http://dx.doi.org/10.1038/ng0897-391. [DOI] [PubMed] [Google Scholar]
- Zella LA, Kim S, Shevde NK, Pike JW. Enhancers located within two introns of the vitamin D receptor gene mediate transcriptional autoregulation by 1,25-dihydroxyvitamin D3. Molecular Endocrinology. 2006;20(6):1231–1247. doi: 10.1210/me.2006-0015. http://dx.doi.org/10.1210/me.2006-0015. [DOI] [PubMed] [Google Scholar]
- Zella LA, Meyer MB, Nerenz RD, Lee SM, Martowicz ML, Pike JW. Multifunctional enhancers regulate mouse and human vitamin D receptor gene transcription. Molecular Endocrinology. 2010;24(1):128–147. doi: 10.1210/me.2009-0140. doi:me.2009-0140 [pii] 10.1210/me.2009-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zierold C, Darwish HM, DeLuca HF. Two vitamin D response elements function in the rat 1,25-dihydroxyvitamin D 24-hydroxylase promoter. The Journal of Biological Chemistry. 1995;270(4):1675–1678. doi: 10.1074/jbc.270.4.1675. [DOI] [PubMed] [Google Scholar]