Abstract
A system based on two designed peptides, namely the cationic peptide K, (KIAALKE)3, and its complementary anionic counterpart called peptide E, (EIAALEK)3, has been used as a minimal model for membrane fusion, inspired by SNARE proteins. Although the fact that docking of separate vesicle populations via the formation of a dimeric E/K coiled-coil complex can be rationalized, the reasons for the peptides promoting fusion of vesicles cannot be fully explained. Therefore it is of significant interest to determine how the peptides aid in overcoming energetic barriers during lipid rearrangements leading to fusion. In this study, investigations of the peptides’ interactions with neutral PC/PE/cholesterol membranes by fluorescence spectroscopy show that tryptophan-labeled K∗ binds to the membrane (KK∗ ∼6.2 103 M−1), whereas E∗ remains fully water-solvated. 15N-NMR spectroscopy, depth-dependent fluorescence quenching, CD-spectroscopy experiments, and MD simulations indicate a helix orientation of K∗ parallel to the membrane surface. Solid-state 31P-NMR of oriented lipid membranes was used to study the impact of peptide incorporation on lipid headgroup alignment. The membrane-immersed K∗ is found to locally alter the bilayer curvature, accompanied by a change of headgroup orientation relative to the membrane normal and of the lipid composition in the vicinity of the bound peptide. The NMR results were supported by molecular dynamics simulations, which showed that K reorganizes the membrane composition in its vicinity, induces positive membrane curvature, and enhances the lipid tail protrusion probability. These effects are known to be fusion relevant. The combined results support the hypothesis for a twofold role of K in the mechanism of membrane fusion: 1) to bring opposing membranes into close proximity via coiled-coil formation and 2) to destabilize both membranes thereby promoting fusion.
Introduction
The fusion of biological membranes is a central step in manifold biological processes such as neurotransmitter release or viral infection. The membrane merging is catalyzed by diverse fusion machineries comprising several proteins such as SNARE (soluble N-ethylmaleimide-sensitive factor attachment receptor) or viral fusion proteins. The fusion processes share common features on the membrane level: two opposing membranes specifically approach closely and merge via a stalk intermediate and a fusion pore is opened (1). The natural fusion machineries are thought to stabilize the intermediate states of lipid rearrangement and/or aid in overcoming the energetic barriers between them, employing concerted protein-protein and protein-membrane interactions (1, 2, 3). From this perspective it appears intriguing that simple synthetic molecules have been successfully designed to yield specific membrane fusion as well (4, 5).
A simple model system that has been used in numerous membrane fusion studies was inspired by the SNARE proteins (6, 7, 8, 9, 10, 11, 12, 13). It is based on two complementary coiled-coil forming peptides with the sequences (EIAALEK)3 and (KIAALKE)3 named E and K, respectively (Fig. 1). These water soluble peptides can be covalently linked to DOPE lipid moieties via a polyethylene glycol (PEG12) chain on the N-termini of the peptides, yielding two complementary lipopeptides called LPE and LPK. These molecules can be readily incorporated into lipid bilayers giving fusogenic vesicles. Formation of the heterodimeric coiled-coil complex E/K is thought to be the initial step in the fusion mechanism bringing the vesicles into close proximity. Little is known about the details of the subsequent processes; however, fusion proceeds effectively, specifically, and leakage free, which are important hallmarks of biological systems. Also, it has been shown that the E/K system and its derivatives can target living cells in vitro and in vivo (14, 15, 16). Thus, we aim to understand the biophysical details of this process.
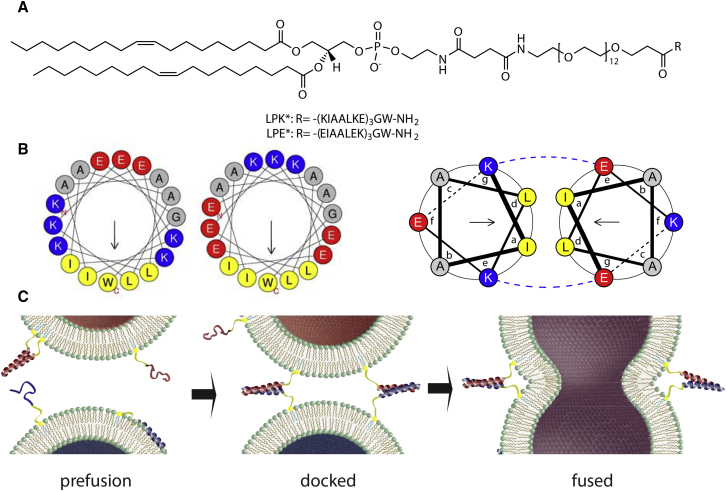
Figure 1.
(A) Chemical structures of the lipopeptides LPK∗ and LPE∗ used in this study. (B) Helical wheel projections (17, 18) of the 18 C-terminal amino acid residues of K∗ and E∗ are shown: (left) hydrophobic moments (arrows) of monomeric helical K∗ and E∗ indicate that binding to lipid monolayers may occur with the hydrophobic leucine and isoleucine residues (yellow) inserting into the hydrophobic part of the monolayer; (right) in the E∗/K∗ complex, coiled-coil binding is achieved by hydrophobic leucine and isoleucine residues, which are covered from the solvent in the core of the complex; blue dashed lines indicate supporting electrostatic interactions. (C) Model of fusion between LPE∗ (red) and LPK∗ (blue) bearing vesicles, showing putative peptide states (19, 20).
Recent infrared spectroscopic studies of LPE and LPK containing lipid mono- or bilayers revealed differing behaviors of the two peptides before the fusion commences (19, 20). The negatively charged E shows relatively weak interactions with the lipid interface, staying in the water phase in the form of homomeric coiled-coil dimers E/E. In contrast, the positively charged K interacts more strongly with the lipid interfaces and incorporates into vesicle bilayers. The membrane incorporation is accompanied by an increase in the helicity of the peptide. Recent molecular dynamics (MD) simulation studies support the different binding affinities (21, 22, 23).
The current model of lipopeptide-mediated fusion starts with the docking of two opposing vesicles, which is triggered by the formation of the E/K coiled-coil complex (Fig. 1 C). It has been proposed that in this docked state monomeric K molecules interact with the membrane they are bound to as well as with the opposing bilayer. However, it has not yet been proven that heterodimeric coiled-coil formation and K membrane interaction coincide on the membrane interface, because the peptide state could only be determined unambiguously in systems where only E or only K were present. The increase in helicity that accompanies both coiled-coil formation and membrane incorporation hampered an unequivocal interpretation of infrared and circular dichroism spectra in vesicle systems with both peptides present (20). Thus, it remains unclear if one peptide state—membrane-bound or coiled-coil—is predominant on the membrane during the course of fusion and afterwards.
The further progress of the fusion mechanism after the docking state remains a matter of speculation. It was hypothesized that the direct K-membrane interactions lead to distortion of the bilayers or increased local curvature and thereby promote the merging of lipid bilayers (19, 20). In this model the two peptides E and K have different functions apart from coiled-coil formation, leaving multiple tasks to K. However, very little is known about the structural consequences of the insertion of the amphipathic helix of K into lipid membranes. In lipid monolayers the incorporation is accompanied by an increase in the surface pressure, probably because of a steric compression of adjacent lipids. Coarse-grained (CG) MD studies implied local enrichment of DOPE (21) or cholesterol (22) around adsorbed K molecules. Apart from that no experimental data on the consequences of peptide insertion for the bilayer structure, the peptide penetration depth, or the creation of local defects and curvature is currently available. Accordingly, the molecular events leading to membrane merging after vesicle docking by LPE and LPK remain unclear.
In our study, we address these unresolved points: the peptide states during and after fusion, the penetration depth, and the bilayer structure upon peptide incorporation. Analogs of E, K, LPE, and LPK are prepared by extension of the peptide C termini with Gly-Trp, yielding the fluorescent E∗, K∗, LPE∗, and LPK∗, respectively (Fig. 1). Fluorescence together with circular dichroism (CD) spectroscopy, allows an unambiguous determination of the peptide state in systems with one or both complementary peptides present. Beyond that, the penetration depth of the free and tethered peptides into the bilayer is estimated by means of depth-dependent fluorescence quenching using brominated lipids as quenchers.
Solid-state NMR spectroscopic techniques are employed to study the impact of peptides on the distribution and structural changes of membrane lipids and their supramolecular arrangement. Particularly, the 100% natural abundance of the 31P isotope makes it possible to measure with high sensitivity the peptide mediated effects on the lipid head group dynamics and orientational distribution as well as membrane macroscopic phase properties (24).
The molecular picture is completed by CG and atomistic MD simulations, which allow protein-membrane interactions to be studied at a high spatiotemporal resolution (25). Earlier MD studies of the LPE/LPK system used relatively limited bilayer patches concentrating on local peptide membrane interactions (21, 22, 23). Here, we aim to study long-range structural consequences of the peptide membrane interactions such as curvature formation that is only observable in larger bilayers.
Materials and Methods
Materials
Fmoc-protected amino acids and Sieber amide resin for peptide synthesis were purchased from Merck-Millipore (Darmstadt, Germany). The lipids 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), the 1-palmitoyl-2-stearoyl(m,n)dibromo-sn-glycero-3-phosphocholines (di-BrPC, m = 6, 9, 11, n = 7, 10, 12) and cholesterol were purchased from Avanti Polar Lipids (Alabaster, AL). Solvents, buffer salts, and acrylamide were purchased from Sigma-Aldrich (Zwijndrecht, the Netherlands). All water was ultrapure with resistance ≥ 18 MΘ cm−1 and TOC ≤ 2 ppm produced from a MilliQ Reference A+ purification system. All experiments were carried out in phosphate buffered saline (PBS) of the following composition: 150 mM NaCl, 20 mM PO43− in H2O, pH 7.4.
Peptide synthesis
The peptides E: Ac-(EIAALEK)3-NH2, E∗: Ac-(EIAALEK)3GW-NH2, K: Ac-(KIAALKE)3-NH2, K∗: Ac-(KIAALKE)3GW-NH2, and [15N-Ala10]-K∗ were synthesized using standard Fmoc-chemistry on a Biotage Syro I and purified by RP-high-performance liquid chromatography to yield a purity > 95% based on high-performance liquid chromatography. Identity of the peptides was determined by liquid chromatography-mass spectrometry. The lipopeptides were synthesized and purified as described elsewhere (7, 9). Peptide stock solutions in PBS were prepared ∼2 mg/ml and the concentration was determined by ultraviolet absorbance at 280 nm, for peptides without tryptophan the concentration was based on the mass. Lipopeptide stock solutions were prepared in CHCl3:MeOH 3:1 solution and added to the lipids before solvent evaporation.
Vesicle preparation
Lipid stock solutions of the compositions DOPC:DOPE:cholesterol (2:1:1) and DOPC:di-BrPC:DOPE:cholesterol (1:1:1:1) were prepared in CHCl3:MeOH 3:1. For experiments with lipopeptides stock solutions were mixed with LPX∗ (X = E or K, respectively) stock solutions to yield mixtures with 1 mol% LPX∗. Lipid films were created by slow evaporation of the solvents under N2 stream and kept under vacuum overnight. The films were rehydrated with PBS yielding final lipid concentrations of typically 1 or 10 mM for vesicle titration experiments. Large unilamellar vesicles (LUVs) were formed by sonication at 55°C for ∼15 min. The size of the vesicles was tested by dynamic light scattering using a Malvern Zetasizer Nano-S (Worcestershire, UK) and was typically found to be ∼100 nm.
Fluorescence spectroscopy
Tryptophan fluorescence emission was measured using a Tecan Infinite M1000 pro plate reader (Männedorf, Switzerland). Excitation was 280 nm and emission was recorded in the range 300–450 nm using steps of 1 nm and bandwidths of 5 and 10 nm for excitation and emission. For each measurement three scans at 25°C were averaged and corrected for scattering. Black Greiner 96 well plates were used.
For experiments with increasing lipid-to-peptide (L:P) ratio mixtures of peptides at constant concentrations of tryptophan containing peptides [X∗] = 2.5 μM and increasing concentrations of lipid up to 5 mM were prepared with constant volume of 200 μl. The complexes E∗/K and K∗/E were used at molar ratios 1:1 with [X∗] = 2.5 μM. For determination of the highest partition coefficient of [K∗], additional data points were collected with lipid concentrations up to 1.25 mM. The maxima of the emission λmax were determined from adjacent average smoothed spectra. The data was interpreted as partition equilibrium (26, 27, 28, 29). The fluorescence intensities, F, in presence of vesicles were used to calculate the concentration of membrane-bound peptide, Pb, by the following:
| (1) |
with the concentration of the tryptophan containing peptide, CX∗, and F0 the fluorescence intensity without vesicles. The fluorescence intensity when all peptide is membrane-bound, F∞, was obtained from extrapolation of a double reciprocal plot of F, versus the lipid concentration to 1/[lipid] = 0. To obtain the partition coefficient, Kp, the data was linearly fitted to the following:
| (2) |
where Xb∗ is the molar ratio of bound peptide per accessible (outer leaflet ∼60%) lipid and Cf is the concentration of free (unbound) peptide.
For acrylamide quenching, peptides in absence or presence of vesicles [lipid]:[X∗] = 1000:1, were mixed with increasing concentrations of quencher yielding constant concentrations [X∗] = 2.5 μM and V = 200 μl and increasing [acrylamide] = 0…150 μM in each well. In quenching experiments with LPE∗ and LPK∗ concentrations were [LPX∗] = 2.5 μM and [lipid]:[LPX∗] = 100:1. The fluorescence emission in presence, F, and in absence, F0, of quencher was fitted to yield the Stern-Vollmer constants KSV by the following:
| (3) |
For quenching with brominated lipids vesicles were composed of DOPC:diBr-PC:DOPE:cholesterol (1:1:1:1). F0 was measured in DOPC:DOPE:cholesterol (2:1:1). Experiments were done at [LPX∗] = 10 μM, [lipid]:[LPX∗] = 100:1 and [X∗] = 2.5 μM [lipid]:[X∗]=1000:1. The quenching efficiency, QE, was calculated by the following:
| (4) |
Circular dichroism spectroscopy
CD spectra were measured using a Jasco J815 CD spectrometer equipped with a Jasco PTC 123 Peltier temperature controller (Easton, MD). Quartz cuvettes with path lengths, l, of 1, 2, and 5 mm were used. Peptide solutions or peptide vesicle mixtures were prepared with [lipid]:[peptide] = 100:1 and 5 μM total peptide concentration. Spectra of the lipopeptides tethered to vesicle were measured at [lipid]:[LPX∗] = 100:1 in 250 μM total lipid concentration and complementary peptide E or K was added [LPX∗]:[Y∗] = 1:1. The relative α-helicity, rh, was calculated from the ellipticity at 222 nm, [θ]222, by the following (30, 31):
| (5) |
Oriented solid-state NMR sample preparations
The lipids and cholesterol were codissolved in chloroform. The chloroform was removed under an N2 stream and the sample dried in vacuum overnight. Buffer (10 mM TES pH 7 100 mM NaCl) was added and five freeze-thaw cycles were performed. The peptide was added in powdered form and additional two freeze-thaw cycles were performed. The sample was centrifuged at 20,000 g overnight. The precipitant was deposited onto ultrathin glass plates (00, 8×22 mm; Paul Marienfeld, Lauda Königshofen, Germany), dried overnight under air and rehydrated at 93% relative helicity (air in contact with a saturated KNO3 solution). Finally, samples were stacked and sealed with Escal neo plastic sheets (Mitsubishi Gas Chemical, Tokyo, Japan).
Samples oriented from organic solvents
Peptides and lipids were codissolved in chloroform. The chloroform was removed under an N2 stream until ∼500 μl was left over. The solution was deposited onto ultrathin glass plates as described above.
NMR measurements
31P, 2H, and 15N solid-state NMR spectra were acquired on a 300 MHz and 750 MHz (15N) Advance spectrometer (Bruker Biospin, Rheinstetten, Germany) using a static triple resonance probe. For recording phosphorus spectra a Hahn echo pulse sequence was applied using a 31P B1 field of 50 kHz and an echo time of 10 μs with continuous wave proton decoupling. Two-dimensional (2D) exchange 31P-NMR experiments were performed using a NOESY type pulse sequence (32). The 31P B1 field of 50 kHz and a first delay of 10 μs were applied. All 31P solid-state NMR experiments were performed with a recycle delay of 3 s. An exponential apodisation function corresponding to a line broadening of 50 Hz was applied before Fourier transformation. Spectra were referenced relative to external concentrated (85 weight%) phosphoric acid (0 ppm).
2H solid-state NMR measurements were performed using a quadrupolar echo sequence (solid echo) (33). The 2H B1 field was 62.5kHz and the echo delay was 10 μs with a recycle delay of 0.5 s. An exponential apodisation function corresponding to a line broadening of 100 Hz was applied before Fourier transformation. D2O was used as an external reference (0 ppm).
15N solid-state NMR spectra were recorded using a cross-polarization sequence followed by a Hahn echo. The echo delay was 40 μs the cp time 1000 μs and the power level during cross-polarization was 25 kHz. Recycle delay was 3 s. An exponential apodisation function corresponding to a line broadening of 150 Hz was applied before Fourier transformation. The spectra were referenced relative to external 15N-NH4Cl (39.3 ppm) (34).
Coarse-grained MD simulations
All CG systems were generated by multiplication of a smaller system from our earlier work (DOPC:DOPE:cholesterol 2:1:1 (22),) which was either peptide-free or contained one preadsorbed fully helical peptide K, thus yielding membrane patches with peptide to lipid ratios ranging from 0:1150 to 1:35. To create a finite patch, a simulation box larger than the membrane patch was created and the patch was solvated by water with 150 mM NaCl. The polarizable Martini force field (35, 36) was used as this force field was shown to properly reflect the binding propensities of peptide E and K to lipid bilayers (22). The peptides were modeled as helices, the secondary structure was kept fixed throughout the CG simulations. All simulations were performed using GROMACS 4.6 (37). For further details on simulation parameters see the Supporting Material and our previous work (22).
All-atom MD simulations
The atomistic systems were prepared as follows. First, one system in CG representation from our previous work (22) was converted to the atomistic resolution (CHARMM36 force field (38, 39)) using backward (40). In the next step, the amount of water and ions was reduced to correspond to the experimental water layer thickness between lipid layers of ∼1 nm (lipid:water ratio 1:17, 100 mM NaCl). This simulation system is denoted as a single-peptide simulation. After equilibration simulation of 300 ns, the system was doubled by rotating one copy by 180° around the y axis and translating along the x axis by the size of the box. By merging these single-peptide K-containing systems, a simulation system with two peptides bound to individual bilayer leaflets arose. This system, termed a double-peptide simulation, was simulated for 450 ns. The peptide-free system was prepared by peptide deletion in the single-peptide simulation and equilibrated for 300 ns. For details on simulation parameters used see (22). The analysis of the curvature succeeded by the program g_lomepro (41). Lipid is considered as protruding if at least one carbon of one of the lipid tails is found more outside the bilayer than the phosphorus atom of the given lipid.
Results and Discussion
Membrane binding of K before and after vesicle docking
The membrane affinity of the fluorescent peptides E∗ and K∗, alone and mixed with nonfluorescent E and K was measured to study the peptide membrane interactions in the prefusion and docked states. First, it was ensured that the structural and thermodynamic properties of the tryptophan labeled E∗ and K∗ were not altered compared with the unlabeled E and K and that coiled-coil formation does not influence the fluorescence spectra (see Supporting Material). From earlier studies it is known that labeling does not influence the fusogenic activity (7). Vesicles of ∼100 nm diameter were titrated into the peptide solutions and the tryptophan fluorescence emission was measured. The emission depends on the microenvironment of tryptophan (42, 43) and allows for quantitative studies of the membrane binding of peptides.
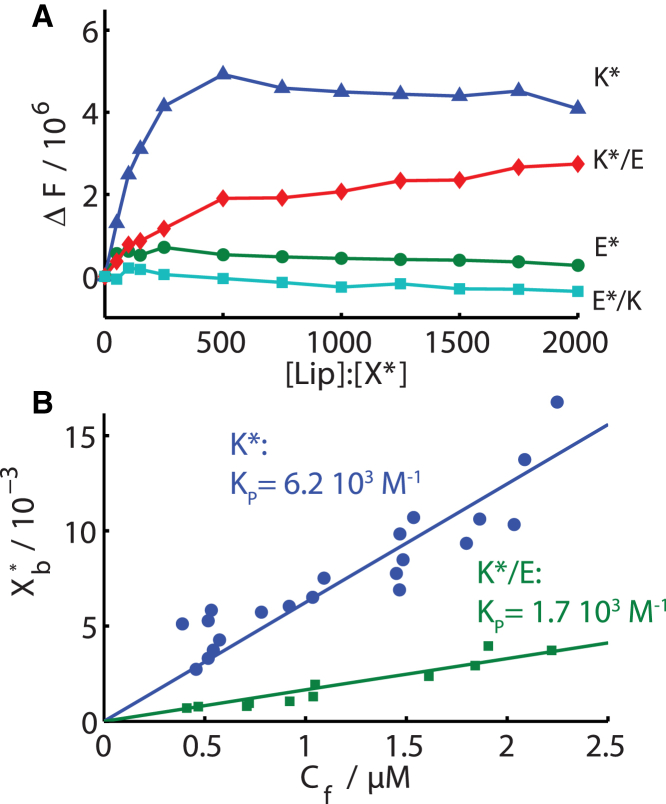
Fig. 2 A displays the change of fluorescence intensity upon varying the lipid to peptide ratio by addition of vesicles of the composition DOPC:DOPE:cholesterol (molar ratio: 2:1:1), as used in recent vesicle fusion studies (5, 6, 7, 8, 9, 10, 16, 44, 45). For comparative purposes, the total concentration of the tryptophan bearing peptide (X∗, X = K or E, respectively) was kept constant. The intensity of the emission of K∗ alone or when added as the complex K∗/E increased with increasing lipid concentration. The spectra, at maximum lipid concentration, reveal an additional blue shift of the fluorescence emission maxima of K∗ and K∗/E by ∼13 nm (Fig. S2). In contrast the emission spectra of E∗ and the complex E∗/K were not significantly influenced by the presence of vesicles showing the lack of membrane-association of E∗. The intensity increase for K∗ and K∗/E with increasing L:P ratio together with the spectral blue shift indicates a change of the environment of the tryptophan residue to a more apolar environment (42, 43). Hence the C-terminal tryptophan residue of K∗ inserts into the hydrophobic part of the lipid bilayer.
Figure 2.
Increase of (A) fluorescence intensity, ΔF, and maximum of (B) fluorescence emission, λmax, versus L:P ratio for peptides E∗, K∗, and the complexes E∗/K, K∗/E mixed with vesicles. Apparent binding isotherms of K∗ and K∗/E binding to DOPC:DOPE:cholesterol (2:1:1). Xb∗ is the molar ratio of bound peptide to accessible total lipid and Cf the concentration of free peptide. Straight lines are linear fits of the data yielding as slope the displayed apparent partition coefficients, Kp. To see this figure in color, go online.
Strikingly, the change to a less polar environment was observed for K∗/E but not for E∗/K, meaning the tryptophan labels on E∗ and K∗ experience significantly different changes in their environment, although both residues are located very closely on the C termini of the parallel coiled-coil complex (46). Thus, the coiled-coil does not partition into the membrane as a complex, because hydrophobic amino acids are shielded in the core of the coiled-coil being unavailable for hydrophobic membrane interactions (Fig. 1 B). Instead, coiled-coil formation in solution competes with the membrane-K∗ interaction. Furthermore, the intensity increase is smaller for K∗/E then for K∗, showing that the active concentration of K∗ available for membrane binding is reduced, due to the K∗/E complex formation. This competition between membrane-bound and coiled-coil-bound K∗ is also illustrated in another titration experiment, showing that in the presence of vesicles the emission maximum shifts back to higher wavelengths upon increasing molar ratio of [E]:[K∗] (Fig. S2 C). This red shift, i.e., the change in the environment of the tryptophans to a more polar environment, is due to the increase of membrane unbound K∗ molecules upon increased coiled-coil binding.
The increase in fluorescence intensity was used to calculate the partition coefficient, KP, between the bilayer and the aqueous environment according to Eq. 2 in the experimental section (Fig. 2 B). The partition coefficient of K∗ was found to be ∼6.2 × 103 M−1, which is in the same order of magnitude as coefficients found for the partitioning of other positively charged amphipathic α-helices into neutral membranes such as model class A peptides (29) or magainin 2 (47). The apparent partition coefficient of K∗/E (1.7 × 103 M−1) is significantly lower than that found for K∗ alone. The binding of K∗ in the coiled-coil complex therefore reduces the amount of free peptide available for membrane binding, leading to a lower apparent partition coefficient.
The binding experiments described above with varying L:P of free E∗ and K∗ cannot be realized easily for the vesicle tethered LPE∗ and LPK∗. Hence, with the lipopeptides, acrylamide quenching experiments had to be performed at constant L:P to indirectly determine the peptide state. Acrylamide quenches the fluorescence of tryptophan residues only if they are not hydrophobically shielded. The binding of K∗ to the vesicles was saturated at L:P of 1000:1 (Fig. 2 A). Therefore for further quenching measurements at constant lipid concentrations, this ratio was chosen. For measurements with LPE∗ and LPK∗, a ratio of 100:1 was chosen because this reflects the conditions of typical vesicle fusion experiments (5, 6, 7, 8, 9, 10, 16, 44, 45).
Linear Stern-Vollmer plots showed that acrylamide readily quenches the fluorescence of free K∗ and E∗ in solution (Supporting Material; Fig. S3). The Stern-Vollmer constants (KSV) were calculated to summarize these results (Table 1). Simultaneously the relative helicity of peptide vesicle mixtures were also measured using CD spectroscopy (Table 1). Additionally, CD spectra of the studied system were recorded. Both, coiled-coil complex formation and peptide membrane interaction was accompanied by CD spectra that show two minima at ∼208 and ∼222 nm, which is typical for α-helical peptides (Figs. S1 and S4). The ellipticity at 222 nm was used to calculate the relative helicity of the peptides (Table 1), which complements the fluorescence data with information about changes in the secondary structure of the peptides.
Table 1.
Stern-Vollmer Constants KSV from Acrylamide Quenching Experiments and Helicities from CD Spectroscopy of Peptides and Lipopeptides in PBS and Mixed with Vesicles
|
KSV (M−1) |
Helicity (%) |
|||
|---|---|---|---|---|
| PBSa | Vesiclesa,b | PBSc | Vesiclesc,d | |
| K∗ | 33.4 | 8.2 | 23 | 46 |
| K∗/E | 30.1 | 12.5 | 63 | 64 |
| E∗ | 31.7 | 26.0 | 23 | 23 |
| E∗/K | 28.0 | 27.7 | 61 | 59 |
| KSV (M−1)e | Helicity (%)e | |||
| LPK∗ | 7.5 | 44 | ||
| LPK∗/E | 8.0 | 42 | ||
| LPE∗ | 26.5 | 41 | ||
| LPE∗/K | 23.2 | 52 | ||
X stands for E or K, respectively.
[X∗] = 2.5 μM.
[lipid]:[X∗] = 1000:1.
[total peptide] = 5 μM.
[lipid]:[peptide] = 100:1.
[LPX∗] = 2,5 μM, [lipid]:[LPX∗] = 100:1.
KSV values at ∼30 M−1 confirm good accessibilities of the aromatic side chains in E∗ and K∗, solvated in PBS buffer (Table 1). Upon vesicle addition LPK∗ and K∗ were quenched significantly less effectively (i.e., KSV decreased) due to hydrophobic shielding of the tryptophan residues by the bilayer. In contrast the tryptophan moieties in E∗ and LPE∗ are only slightly less accessible for the quencher in the presence of vesicles. In general, the E∗ and LPE∗ are therefore not embedded in the membrane. Whereas E∗ showed no increased helicity, the enhanced helicity of LPE∗ in these systems can be attributed to the known tendency of E∗ to form homodimers at increased local concentration (20).
To determine the peptide folding in the docked state during fusion, additional experiments with K∗/E and E∗/K were performed (Table 1; Fig. S3). Upon coiled-coil formation from K∗ to K∗/E and E∗ to E∗/K, the helicity increases significantly, with and without vesicles. In presence of vesicles, the KSV, i.e., the tryptophan accessibility is slightly increased from K∗ to K∗/E, because an increased amount of K∗ resides in the aqueous environment due to the concurrent coiled-coil complex formation in the solution. In contrast, the accessibility for tryptophan in E∗-vesicle mixtures does not change upon E∗/K complex formation, because it is already situated in the solution, beforehand. For LPK∗ neither the tryptophan accessibility, nor the helicity changed due to LPK∗/E complex formation. Thus the majority of the peptides remain bound to the vesicle rather than forming a coiled-coil complex in this experiment. Apparently, the tethered peptide in LPK∗ is bound stronger to the vesicle than the untethered K∗. Similar effects were also observed earlier in a study of the interactions of K, E, LPK, and LPE with lipid monolayers (19).
Taken together the fluorescence data shown here consolidate earlier findings showing the membrane incorporation of K and LPK during prefusion states whereas LPE and E lack membrane interactions. The addition of the tryptophan did not alter these qualitative results. Moreover, it is revealed that the membrane interaction of monomeric helical K is in competition with the E/K complex formation in mixed systems. Thus this equilibrium also affects the vesicle membranes in the docked state during the fusion process. Because of the close proximity of the two prefusion membranes in the docked state, monomeric K might interact with both its own and the opposing bilayer. Thus the influence of this insertion on the membrane structure is of direct relevance to the progress of the fusion mechanism, which we investigate below.
Penetration depth of peptide K
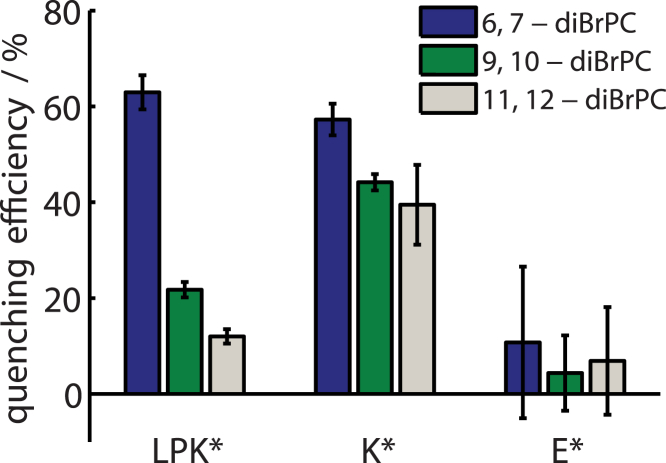
Next, we studied the penetration depth of K∗ and LPK∗ into the bilayer, which is important to assess structural membrane alterations. Depth-dependent fluorescence quenching experiments were performed using 1-palmitoyl-2-(dibromo)stearoyl-PC lipids modified with bromine atoms at different positions on the acetyl chain (6, 7-; 9, 10-;11, 12-diBrPC). Both LPK∗ and K∗ are quenched most efficiently by bilayers containing the lipids with the bromine incorporated at a relatively shallow position, that is, 6,7-diBrPC (Fig. 3), which indicates that the tryptophan moieties are incorporated closest to this position. As expected tryptophan quenching in E∗ by the diBr-PCs was minor, because this peptide does not interact with membranes.
Figure 3.
Tryptophan quenching efficiency of LPK∗, K∗, and E∗ mixed with vesicles containing di-BrPC. Error bars represent 95% confidence intervals. Quenching vesicles were of the composition DOPC:di-BrPC:DOPE: cholesterol 1:1:1:1. [lipid]:[K∗] = 1000:1; [lipid]:[LPK∗] = 100:1. To see this figure in color, go online.
The depth-dependent quenching data was analyzed by two methods: the parallax and the distribution analysis methods (48, 49, 50). Here, only results from the latter are reported as this method is considered to better reflect the physics of the quenching, because of the disordered nature of the lipid bilayer. A detailed discussion of the data using both methods is given in the Supporting Material (Fig. S5). The distribution analysis reveals that the most probable insertion depth, i.e., the center of the depth distributions of indole rings of K∗ and LPK∗, is not deeper than 11 Å from the bilayer center. Furthermore, K∗ shows a broader distribution of possible penetration depths that may indicate more conformational freedom of the indole rings of K∗ compared with LPK∗. The penetration depth of unlabeled K or LPK might slightly deviate from this result because of the influence of the tryptophan label. However, both LPK and LPK∗ trigger membrane fusion and both show comparable membrane affinity that is opposed by LPE and LPE∗. Thus we assume the possible differences in penetration depth to be of little relevance for the membrane fusion and the peptide membrane affinity.
This shallow membrane incorporation of the tryptophan centered at a depth of ∼11 Å from the bilayer center is in line with a peptide insertion model with the helical peptide parallel to the membrane surface and with the hydrophobic face penetrating into the hydrophobic core of the bilayer. Assuming an α-helical structure with a distance of the indole ring to the helical axis of ∼8 Å, the helix would be centered at a distance of 19 Å from the bilayer center (49), which agrees extremely well with the 19–21 Å distance obtained from a recent CG simulation of K in bilayers of this composition (22), as well as with this study’s CG simulations of lipid vesicles with adsorbed peptides K (17–19 Å) and atomistic simulations (20,2 Å). Based on the estimated phosphate-to-phosphate distance for the bilayer under study of 40 Å, this means the helix center is in close proximity to the glycerol and phosphate groups of the lipids.
Membrane disturbing effects of peptide K
The insertion of K∗ close to the lipid head group is expected to affect the bilayer structure, which was studied with 31P-solid-state NMR. This method yields valuable insights into the macroscopic properties of lipid bilayers (24, 51) such as changes in curvature of the membrane (52) and conformational changes of the phospholipid head groups (53). In particular, in supported lipid bilayers the orientation dependence of the anisotropic 31P-chemical shift of the phosphate group is a sensitive indicator of local and global changes in the membrane geometry (52, 54) and head group conformation (53).
Initially, samples for oriented 31P-NMR experiments were prepared composed of DOPC:DOPE:cholesterol (2:1:1) however, no stable and well-aligned lipid bilayers could be obtained. Therefore, membranes were prepared from the closely related POPC:POPE:cholesterol system because the 1-palmitoyl-2-oleoyl-phospholipids are commonly used for NMR studies of both vesicular and oriented states of lipids.
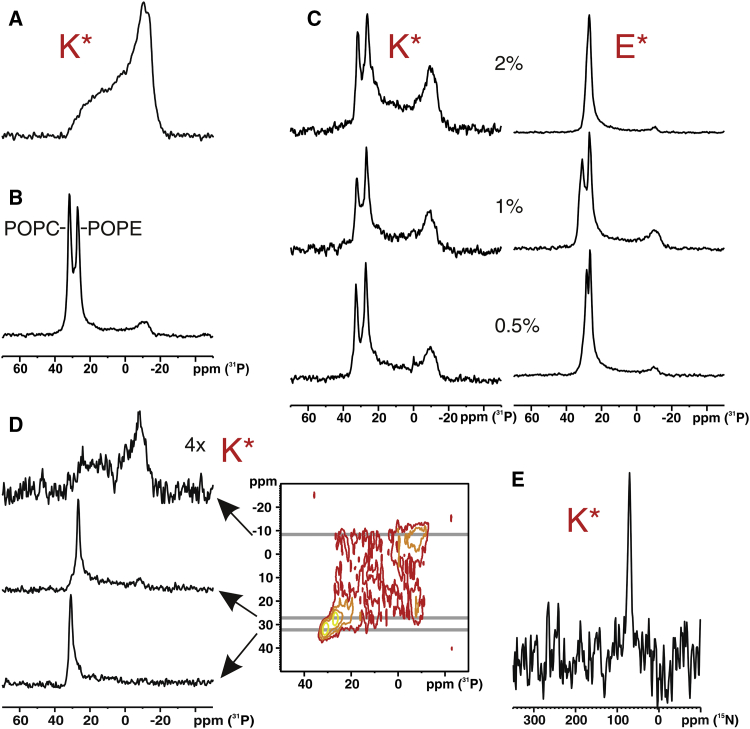
The static 31P-NMR measurements from large multilamellar vesicles made from this lipid mixture (at 300 K) indicate closely related Lα-phase spectra in the absence or presence of K∗ (Figs. 4 A and S6). However, when uniaxial-oriented membranes were investigated, considerable disruptive effects on the alignment of the lipids and/or the head group conformation became apparent. In the absence of peptide, two predominant and well-resolved resonances are shown in Fig. 4 B at 30 and 25 ppm, which were attributed to POPC and POPE lipids, respectively (Fig. S7). The chemical shift anisotropy of isolated PE (Δ = 38.8 ppm)5 and PC (Δ = 45 ppm) suggests liquid crystalline bilayers. These chemical shift positions are indicative of phospholipids oriented with their long axes parallel to the magnetic field direction that in this experiment coincides with the sample normal. Furthermore, they show that motional averaging around the membrane normal occurs, as typically observed for liquid crystalline bilayers.
Figure 4.
Solid-state NMR spectra (at 300 K) of E∗ and K∗ comprising POPC:POPE:cholesterol (2:1:1) membranes. (A) Proton-decoupled 31P-solid-state NMR spectrum of vesicular membranes in the presence of 2 mol% K∗. 31P-solid-state spectra of oriented samples in the (B) absence and (C) presence of increasing amounts of K∗ and E∗. (D) 2D 31P-solid-state NMR exchange spectrum in the presence of 2 mol% K∗ (mixing time 400 ms). (E) Proton-decoupled 15N solid-state NMR spectrum of [15N-Ala10]-K∗, reconstituted into uniaxially oriented membranes. To see this figure in color, go online.
Fig. 4 C shows that addition of 0.5% K∗ (L:P ∼1:160) induced the appearance of spectral intensities reaching up to a second maximum at ∼–10 ppm with a line shape closely resembling that of a 31P powder spectrum. Because we did not observe much static and dynamic disorder of the acyl chains (Supporting Material S4), the broad line reflects an orientational distribution that is mainly restricted to the phospholipid head groups. There are few other 31P-NMR studies reporting on fusogenic peptides in oriented membranes (55, 56, 57). All 31P-NMR spectra show broad lines in this region, whose amplitude has been related to the level of fusion activity of the peptides (56). However, a conclusion based on 31P-NMR alone should be drawn with caution, because membrane-perforating antibiotic peptides also showed related 31P-NMR line shapes. However, in contrast with our observations (Supporting Material S4) these antibiotics caused significant static and dynamic disorder of the acyl chains evidenced by the 2H-NMR spectra (58, 59).
Integration of the signals yields the relative amount of oriented and disordered fractions. Some degree of misalignment is often observed for supported lipid bilayer stacks prepared from vesicle suspensions as shown by the control spectrum in the absence of peptide (Fig. 4 B). This contribution has been taken into consideration in our analysis. After background correction the total amount of perturbation amounts to ∼20%. The fraction rises with peptide concentration, but in a nonlinear manner. This nonlinearity might be the consequence of K∗ forming highly polar homo coiled-coil aggregates (folding constant 3400 M−1) (60).
In the case of E∗, the peak of the aligned POPC molecules shifts downfield with increasing peptide concentration, finally leading to one slightly broadened signal that coincides with the 31P-NMR signal of POPE (Fig. 4 C). Notably, a similar intense broad component as observed for K∗ is absent. A small broad peak at –10 ppm remains constant upon increasing the peptide concentration and is caused by the same misalignment of lipids observed in the control spectrum. The absence of a peptide dependent broad line can be likely explained by the low membrane affinity of this peptide as was found in our membrane-binding experiments (Fig. 2) and can be understood to be caused by a poor conformational stability (22.6% helicity, Table 1). Evidently, the E∗ molecules remain in the thin water layers separating the multilamellar bilayers. This asymmetric behavior of E and K peptides in planar membranes agrees well with the observations described above for the interaction with vesicular membranes. Thus, the role of E in fusion is likely restricted to E/K coiled-coil formation to bring the membranes into close proximity.
In a previous investigation a similarly broad 31P line shape was attributed to macroscopically sized cylindrically curved bilayers (58). Such a binding mode implies a large distribution of peptide orientations that can be manifested by a broad range of 15N-NMR chemicals shifts when the corresponding membrane-bound peptide (labeled with 15N at one of its peptide bonds) is investigated. The 15N chemical shift provides a direct indicator of the alignment of the peptide relative to the membrane surface (61, 62). Therefore, a membrane sample of uniaxial orientation was prepared with 2% K∗ carrying a 15N label at the alanine 10 position. The proton-decoupled 15N solid-state NMR spectrum of this sample is depicted in Fig. 4 E. Because the sample was oriented with its normal parallel to the magnetic field, the chemical shift of 70 ppm measured for [15N-Ala10]-K∗ indicates an alignment of the peptide parallel to the membrane normal (transmembrane helices would exhibit 15N chemical shifts at ∼200 ppm). This observation contradicts cylindrical (58) or vesicular membrane binding modes (63) but agrees perfectly with the proposed shallow incorporation of helical K∗ in the vesicular membrane, as was found in the fluorescence experiments. The insertion of the peptide into the membrane should be accompanied with a redistribution of the phosphate groups. Thus we investigated further the ability of peptide K to perturb the membrane in addition to its function to form coiled-coils.
First, a homo-nuclear 2D-31P-NMR exchange experiment was conducted. If lateral diffusion within the mixing time of the 2D-NMR experiment covers a significant distance on a curved surface, the realignment of the molecule will be accompanied with a change in the chemical shift relative to the magnetic field direction. This will manifest as off-diagonal intensities in the 2D NMR spectra. Therefore, 2D exchange spectra were recorded for bilayers in the presence of 2% K∗ at different mixing times (1, 5, 10, 50, and 400 ms). The off-diagonal intensity measured for a tm of 400 ms, depicted in Fig. 4 D, indicates that a molecular reorientation of the lipids takes place within the applied mixing time. Thus, the lipid layers are curved because of the presence of peptide K. According to the 2D-NMR experiment, only the PE chemical anisotropy is involved in exchange, whereas the PC chemical shift anisotropy is not affected (cf. expanded cross sections in Fig. 4 D). This observation is likely attributed to a selective PE enrichment near the peptide-induced local curvature.
Because we did not observe any phase transition, the peptide-induced membrane curvature is best explained by considering the fact that the peptides are bound at both sides of the multilamellar bilayers. From the molecular point of view, it is evident that membrane insertion of 2–3 nm sized, cylindrical-shaped helices would affect the distribution of the local headgroup conformations and the composition of the lipid mixture as well. From the macroscopic point of view, the membrane immersion of peptides is accompanied by local membrane deformations, which are manifested by an altered bilayer curvature. The locally altered membrane perturbation would further propagate from one lipid bilayer to another in these tightly stacked bilayer systems (64, 65).
Thus, the initially created distortions of the membrane structure would finally lead to undulations of a wave-like and macroscopic membrane distribution pattern. To reveal more details about such a complex peptide-induced membrane deformation, MD simulations were performed.
Peptide K promotes positive curvature in coarse-grained MD simulations
Membrane curvature is in silico most effectively studied in CG MD simulations of a metastable finite bilayer patch that minimizes the suppression of curved structures in the typically employed periodic setups and also allows for vesicle formation (66). A peptide-free bilayer patch (DOPC:DOPE:cholesterol 2:1:1, in total 1150 molecules, Fig. S9) was found to form a vesicle within 140–170 ns in a random direction due to spontaneous fluctuations (Table S2). A single adsorbed K induced spontaneous positive curvature and accelerated vesicle formation (85–126 ns). Accordingly, peptide K was finally bound on the outer layer of the vesicle. Adsorption of multiple peptides K (8 or 16) to one side of the patch further accelerated the vesicle formation (∼65 ns), whereas the presence of peptides K on both sides of the patch prevented it on a timescale of 400 ns. In summary, adsorbed peptides K induce a positive curvature of lipid bilayers in MD simulations. The speed and the strength of this spontaneous curvature depends on the L:P ratio.
Peptide K alters the bilayer structure in all-atom MD simulations
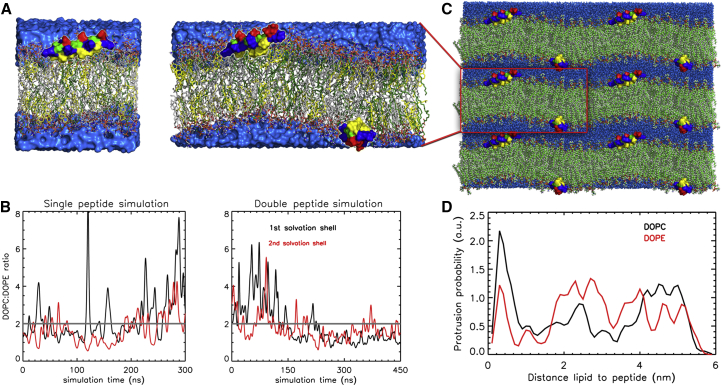
We used atomistic MD simulations to obtain atomistic insight into the K-membrane interactions, complementary to our NMR investigations. In total, three set-ups of DOPC:DOPE:cholesterol 2:1:1 bilayers were simulated. Namely, a peptide free lipid bilayer, a lipid bilayer with one, and a lipid bilayer with two peptides K adsorbed. The latter double-peptide simulation box had an extended length in one direction and the two peptides were adsorbed on different leaflets. In the smaller peptide-free and single-peptide simulation systems, the periodic boundary conditions hamper the evolution of membrane curvature. Hence, the extended double-peptide system is more suitable to estimate the ability of peptide K to induce a spontaneous curvature since the effects of both peptides on the curvature will add together. A snapshot of this system illustrating the helicity and orientation of the peptide is given in the Supporting Material (Fig. S11). During the whole simulation of 0.4 μs, the double-peptide system waves with curvature of ∼1/20 nm−1 were induced by adsorbed Ks. The induced waves were stable in terms of both the curvature radius and the wave position (see Fig. 5, A and C). Both K molecules were localized on the top of the positively curved lipid bilayer and were found to stabilize it in agreement with the predictions made by CG simulations on free bilayer patches (see above). The radius of ∼20 nm is smaller than the experimentally observed one, possibly because of natural irregularities and smearing out in the experimental system.
Figure 5.
(A) Snapshots at 300 ns of atomistic simulations showing the positions and orientation of the adsorbed peptides K (glutamic acids in red, lysines in blue, alanine in green, and hydrophobic leucines and isoleucines in yellow) on the bilayer in the single peptide (left) and double peptide (right) simulations. Water is shown as blue surface and lipids (DOPC in gray, DOPE in dark green, and cholesterol in yellow) in stick representation. (B) DOPE accumulation in the vicinity of K shown as a ratio between DOPC and DOPE lipids found in the individual solvation shell. The total ratio of DOPC and DOPE in the simulation system is shown as a gray line. In the double peptide simulation the number of DOPE gets comparable with the number of DOPC (mainly in the first solvation shell). (C) Double-peptide simulation system with its periodic neighbors. (D) Lipid protrusion probability in dependence of the distance to K.
A comparison of this atomistic simulation with a simulation of a system containing a pure bilayer enabled us to deduce additional effects of K on the membrane structure. First, the PE and PC head groups surrounding the adsorbed peptide were tilted more parallel to the membrane normal by 8.2° and 10.2°, respectively. Such a “narrowing” of lipid head groups has been described before for membranes surrounded by positively charged ions (53).
Additionally, DOPE molecules accumulated in the vicinity of K in curved bilayers in both CG (Supporting Material S5; Fig. S10) and atomistic simulations. In the latter, in the interval of 200–450 ns the number of DOPE increased by 31% in the first and 25% in the second solvation shell compared with the overall ratio of PC:PE (2:1) in the system (Fig. 5 B). However, DOPE accumulated in the vicinity of peptide K only in the curved system, which is unexpected because with its small head group and bulky tails, DOPE typically partitions into areas of negative curvature. This surprising result can be understood considering that the conical shape of DOPE is well-suited to the space requirements of the adsorbed peptide: it fills the space underneath the peptide while offering enough room in between the lipid head groups for the peptide (67).
Interestingly, we observed that the cholesterol concentration in the peptide vicinity also depends on the membrane curvature. In our previous CG investigations of K adsorption on flat bilayers, an increased cholesterol content near the peptide was measured (22) whereas for the curved bilayers studied here this was decreased by 60% with respect to the average membrane composition. This dramatic decrease probably has two reasons: 1) K seems to prefer DOPE over other membrane components, and 2) cholesterol is known to accumulate in areas of negative curvature (68), which was suggested to be fusion relevant (69).
Other fusion-relevant effects of fusion peptides on the membrane structure are membrane dehydration and increased lipid tail protrusion. These effects were shown to be connected (70) and their importance in membrane fusion has been elucidated before (71, 72). Moreover, splaying lipids were shown to reduce the energy barrier between the lamellar and stalk phases during membrane fusion (73, 74). Lipid protrusions were shown to be increased in the vicinity of both SNAREs and influenza fusion peptides (75, 76).
K was found to only slightly (by 2%–4%) dehydrate the membrane in the simulations. On the other hand, the effect on lipid protrusions is much more pronounced as K enhanced the lipid protrusion in terms of both the average protrusion time, which increased by 30%–60%, and the number of protrusions that increased by up to 44% (see Table S3). Interestingly, different reasons for increased protrusion probability in flat and curved bilayers were observed. In the case of a small flat bilayer with one K adsorbed to one of the membrane leaflets, mostly the protrusion time was enhanced, whereas the number of protrusion events for DOPC even slightly decreased. The influence on DOPE was larger than on DOPC. In the case of the curved system with peptides adsorbed to both leaflets, the number of protrusion events significantly increased for DOPC lipids only, resulting in a similar protrusion probability for DOPC and DOPE lipids. It has to be noted that in case of a pure bilayer, DOPE protrudes with 48% higher probability than DOPC, probably because of its smaller head group. In the curved system, the protrusion times increased for both DOPC and DOPE lipids by 35% and 37%, respectively, relative to the pure bilayer. Moreover, for DOPE the protrusion probability depends less on the distance to the peptides than for DOPC. The latter protrudes preferentially if in direct contact with the peptide (Fig. 5 D).
Combining the NMR and MD data reported in this study, a new qualitative interpretation, to our knowledge, of the oriented 31P NMR spectrum of a fusion peptide-carrying membrane is provided. The broad 31P NMR line at ∼–15 ppm represents a more or less random distribution of the head groups near the membrane-bound peptide. The combined fluorescence, 15N-NMR, and MD data agree with a K bound parallel to the membrane surface. However, in contrast to our fluorescence-binding experiments using vesicular membranes (Fig. 2 B), the membrane perturbation monitored by 31P-NMR experiments of oriented membranes does not show a linear dependence on peptide concentration. The decrease of membrane binding affinity is accompanied with an increasing concentration of cationic homo-K dimers. These might be stabilized by the anionic Glu side chains pointing to the water phase from other membrane-bound K monomers (Fig. 5 A).
The one-dimensional and 2D 31P NMR data indicate that the membrane-immersed K is capable of altering the bilayer curvature. The ability of K to induce curvature is confirmed by both CG and atomistic MD simulations. The latter systems also indicate a wave-like alteration of the bilayer curvature, a model that can explain the 31P NMR data. Additionally, in MD simulations the direction of the induced curvature can be seen: K induces positive curvature and is thus located at the peak of the bilayer wave. The random peptide adsorption on the bilayers’ upper and lower leaflet in NMR samples will lead to formation of local positive curvature where K is adsorbed to the upper leaflet and negative curvature, in spots where K is attached to the lower leaflet. Thus a wave-like pattern is created. Moreover, because of the repulsion between the peptide’s negatively charged amino acids pointing to the water phase and the neighboring lipid bilayer being separated by only a small water layer, the wave is suggested to propagate to the next lipid layer. Such a propagation can explain the broadening of the oriented lines in the 31P NMR.
These results show that the study of multilamellar lipid systems by NMR techniques can serve as a tool to study the role of fusion peptides during the very early fusion stages when two approaching membranes are still separated by a very thin water layer.
Implications for the mechanism of coiled-coil mediated fusion
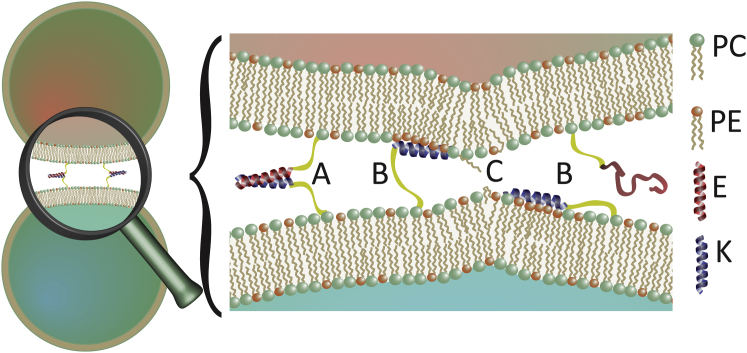
It is generally anticipated that the first step in fusion triggered by LPE and LPK is a vesicle docking due to the formation of one or more LPE-LPK complexes (6, 7, 8, 9, 10). The tryptophan fluorescence data shows that the E/K complex is in equilibrium with the membrane-incorporated monomeric K (Fig. 6, A and B). This means that 1) K molecules bound in the coiled-coil complex can dissociate and bind to the outer sides of both prefusion membranes, and 2) membrane-bound K molecules can remain stable in their state in the interstitial region between the membranes for a distinct time. Thus, K can adopt multiple roles to promote fusion. In its role as partner for E, it adopts a role similar to SNARE proteins with the function to tether the two membranes specifically together to adopt a state comparable with the primed vesicle at the neuronal membrane (77). In the docked state K can bind as monomer to the membranes and adopt its second role. It acts as a membrane active agent supporting the membrane deformation and rupture, comparable with the suggested role of certain fusion peptides during viral entry (78). Unlike in neuronal vesicle fusion where primed, i.e., docked vesicles proceed rapidly to full fusion specifically triggered by Ca2+ influx, we propose that in the E/K system the slow accumulation of membrane-bound K at the docking site by diffusion and/or dissociation of the complex causes the further progression to full fusion nonspecifically. Our results revealed several effects with the potential to facilitate membrane fusion, namely, curvature creation, increased lipid tail protrusion, and PE accumulation.
Figure 6.
Model of multiple roles of peptide K in the docked prefusion state: (A) initial vesicle docking by coiled-coil formation; (B) membrane binding of monomeric K to its own and the opposing membrane and local PE enrichment; and (C) creation of local positive curvature and increase of lipid protrusions.
Local positive curvature created by the insertion of K at the docking site, might lead to a further reduction of the intermembrane distance due to membrane buckling (Fig. 6 C) (79, 80). Dehydration of the polar lipid interface and protrusion of single lipid acyl chains are considered as the main energetic barriers toward the formation of the stalk intermediate during fusion (73, 74, 75). The increase of protrusion probability in the vicinity of incorporated K revealed by our atomistic MD simulations (Fig. 6 C), implies that this effect may catalyze the stalk formation. Both MD and NMR results imply the accumulation of PE lipids at the adsorbed K molecule (Fig. 6 B). Apparently, the smaller size of the PE head group provides the necessary space for insertion of the peptide. Because of their relatively small head groups, PE lipids aid the formation of negatively curved membranes. In the position found in our simulation, on the top of the curved membrane in the prefusion state, PE could enlighten the transition to the stalk state (1, 81, 82, 83), where the peptide and its surroundings are temporarily found in an area of negative curvature.
Conclusions
This study has shown that peptide K can interact with lipid membranes in the course of fusion. Its amphipathic helix is stabilized by the hydrophobic part of the membrane and penetrates shallowly into the membrane. This interaction is in equilibrium with the heterodimeric coiled-coil formation with its partner E and thus also occurs when the vesicles are docked together in close proximity.
From the combined NMR and MD data reported in this study, a new qualitative interpretation, to our knowledge, of the oriented 31P NMR spectrum of membranes interacting with fusion peptides is provided. The results obtained for the interaction of the K-peptide with planar multilamellar membranes comprise the first strong evidence, to our knowledge, for a K-induced PE-enriched positive membrane curvature with the peptide bound at the top of the curved bilayer. Furthermore, atomistic MD simulations imply an increased lipid acyl chain protrusion in the vicinity of inserted K. The dual role of K in the fusion process comprises hallmarks of SNARE mediated fusion, i.e., the vesicle docking by coiled-coils as well as hallmarks of viral fusion, namely the membrane perturbation by peptide incorporation. Thus, contrary to earlier postulations the E/K system might not be seen as a minimal model just for SNARE mediated fusion (7), but moreover as a minimal model for protein mediated fusion in general. Thus, this system poses a facile approach to experimentally study the basic mechanisms of fusion on the membrane level. Furthermore, the insight that membrane disturbing effects enhance artificial fusion just as natural fusion should be considered to improve the design of future artificial membrane fusion systems.
Author Contributions
M.R., A.K., and J.R. conceived the overall study. A.L.B. and D.F.B. synthesized the peptides and lipopeptides. M.R. and V.d.W. performed the fluorescence and CD study. C.A. and B.B. performed the NMR study. K.P., S.A.K., and R.A.B. performed the MD study. M.R., J.R., C.A., B.B., and K.P. wrote the manuscript. All authors reviewed the manuscript.
Acknowledgments
M.R., A.L.B., and A.K. acknowledge the financial support of the European Research Council via an ERC starting grant (contract 240391), the Netherlands Organisation for Scientific Research (NWO) via a VICI grant to A.K. (contract 724.014.001), and a VENI grant to A.L.B. (contract 722.015.006). B.B. and C.A. gratefully acknowledge financial contributions of the Agence Nationale de la Recherche (MemPepSyn 14-CE34-0001-01 and LabEx Chemistry of Complex Systems 10-LABX-0026_CSC), the University of Strasbourg, the CNRS, and the RTRA International Center of Frontier Research in Chemistry. K.P, S.A.K, and R.A.B acknowledge support by the German Science Foundation (DFG) within the project BO2963/2-1 and the Research Training Group “Dynamic Interactions at Biological Membranes from Single Molecules to Tissue” (RTG 1962). Computer time was provided by the computer center (RRZE) of the Friedrich-Alexander University of Erlangen-Nürnberg.
Editor: Kalina Hristova
Footnotes
Martin Rabe’s present address is Department of Interface Chemistry and Surface Engineering, Max-Planck-Institut für Eisenforschung GmbH, Düsseldorf, Germany.
Supporting Materials and Methods, Supporting Discussion, eleven figures, and three tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)30937-7.
Contributor Information
Martin Rabe, Email: m.rabe@mpie.de.
Burkhard Bechinger, Email: bechinger@unistra.fr.
Supporting Citations
References (84, 85, 86, 87, 88, 89, 90, 91, 92) appear in the Supporting Material.
Supporting Material
References
- 1.Chernomordik L.V., Kozlov M.M. Protein-lipid interplay in fusion and fission of biological membranes. Annu. Rev. Biochem. 2003;72:175–207. doi: 10.1146/annurev.biochem.72.121801.161504. [DOI] [PubMed] [Google Scholar]
- 2.Jahn R., Lang T., Südhof T.C. Membrane fusion. Cell. 2003;112:519–533. doi: 10.1016/s0092-8674(03)00112-0. [DOI] [PubMed] [Google Scholar]
- 3.Martens S., McMahon H.T. Mechanisms of membrane fusion: disparate players and common principles. Nat. Rev. Mol. Cell Biol. 2008;9:543–556. doi: 10.1038/nrm2417. [DOI] [PubMed] [Google Scholar]
- 4.Ma M., Bong D. Controlled fusion of synthetic lipid membrane vesicles. Acc. Chem. Res. 2013;46:2988–2997. doi: 10.1021/ar400065m. [DOI] [PubMed] [Google Scholar]
- 5.Marsden H.R., Tomatsu I., Kros A. Model systems for membrane fusion. Chem. Soc. Rev. 2011;40:1572–1585. doi: 10.1039/c0cs00115e. [DOI] [PubMed] [Google Scholar]
- 6.Marsden H.R., Korobko A.V., Kros A. Controlled liposome fusion mediated by SNARE protein mimics. Biomater. Sci. 2013;1:1046–1054. doi: 10.1039/c3bm60040h. [DOI] [PubMed] [Google Scholar]
- 7.Marsden H.R., Elbers N.A., Kros A. A reduced SNARE model for membrane fusion. Angew. Chem. Int. Ed. 2009;48:2330–2333. doi: 10.1002/anie.200804493. [DOI] [PubMed] [Google Scholar]
- 8.Tomatsu I., Marsden H.R., Kros A. Influence of pegylation on peptide-mediated liposome fusion. J. Mater. Chem. 2011;21:18927–18933. [Google Scholar]
- 9.Versluis F., Voskuhl J., Kros A. In situ modification of plain liposomes with lipidated coiled coil forming peptides induces membrane fusion. J. Am. Chem. Soc. 2013;135:8057–8062. doi: 10.1021/ja4031227. [DOI] [PubMed] [Google Scholar]
- 10.Zheng T., Voskuhl J., Kros A. Controlling the rate of coiled coil driven membrane fusion. Chem. Commun. (Camb.) 2013;49:3649–3651. doi: 10.1039/c3cc38926j. [DOI] [PubMed] [Google Scholar]
- 11.Kong L., Askes S.H.C., Campbell F. Temporal control of membrane fusion through photolabile PEGylation of liposome membranes. Angew. Chem. Int. Ed. Engl. 2016;55:1396–1400. doi: 10.1002/anie.201509673. [DOI] [PubMed] [Google Scholar]
- 12.Mora N.L., Bahreman A., Kros A. Targeted anion transporter delivery by coiled-coil driven membrane fusion. Chem. Sci. (Camb.) 2016;7:1768–1772. doi: 10.1039/c5sc04282h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwenen L.L.G., Hubrich R., Steinem C. Resolving single membrane fusion events on planar pore-spanning membranes. Sci. Rep. 2015;5:12006. doi: 10.1038/srep12006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang J., Bahreman A., Kros A. ACS Central Science; Washington, DC: 2016. Drug Delivery via Cell Membrane Fusion Using Lipopeptide Modified Liposomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang J., Shimada Y., Kros A. Application of coiled coil peptides in liposomal anticancer drug delivery using a Zebrafish xenograft model. ACS Nano. 2016;10:7428–7435. doi: 10.1021/acsnano.6b01410. [DOI] [PubMed] [Google Scholar]
- 16.Zope H.R., Versluis F., Kros A. In vitro and in vivo supramolecular modification of biomembranes using a lipidated coiled-coil motif. Angew. Chem. Int. Ed. Engl. 2013;52:14247–14251. doi: 10.1002/anie.201306033. [DOI] [PubMed] [Google Scholar]
- 17.Gautier R., Douguet D., Drin G. HELIQUEST: a web server to screen sequences with specific α-helical properties. Bioinformatics. 2008;24:2101–2102. doi: 10.1093/bioinformatics/btn392. [DOI] [PubMed] [Google Scholar]
- 18.Grigoryan G., Keating A.E. Structural specificity in coiled-coil interactions. Curr. Opin. Struct. Biol. 2008;18:477–483. doi: 10.1016/j.sbi.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rabe M., Schwieger C., Kros A. Membrane interactions of fusogenic coiled-coil peptides: implications for lipopeptide mediated vesicle fusion. Langmuir. 2014;30:7724–7735. doi: 10.1021/la500987c. [DOI] [PubMed] [Google Scholar]
- 20.Rabe M., Zope H.R., Kros A. Interplay between lipid interaction and homo-coiling of membrane-tethered coiled-coil peptides. Langmuir. 2015;31:9953–9964. doi: 10.1021/acs.langmuir.5b02094. [DOI] [PubMed] [Google Scholar]
- 21.Bulacu M., Sevink G.J.A. Erratum. Computational insight in the role of fusogenic lipopeptides at the onset of liposome fusion. Biochim. Biophys. Acta. 2015;1848:1716–1725. doi: 10.1016/j.bbamem.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 22.Pluhackova K., Wassenaar T.A., Böckmann R.A. Spontaneous adsorption of coiled-coil model peptides K and E to a mixed lipid bilayer. J. Phys. Chem. B. 2015;119:4396–4408. doi: 10.1021/acs.jpcb.5b00434. [DOI] [PubMed] [Google Scholar]
- 23.Woo S.Y., Lee H. All-atom simulations and free-energy calculations of coiled-coil peptides with lipid bilayers: binding strength, structural transition, and effect on lipid dynamics. Sci. Rep. 2016;6:22299. doi: 10.1038/srep22299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bechinger B., Salnikov E.S. The membrane interactions of antimicrobial peptides revealed by solid-state NMR spectroscopy. Chem. Phys. Lipids. 2012;165:282–301. doi: 10.1016/j.chemphyslip.2012.01.009. [DOI] [PubMed] [Google Scholar]
- 25.Pluhackova K., Böckmann R.A. Biomembranes in atomistic and coarse-grained simulations. J. Phys. Condens. Matter. 2015;27:323103. doi: 10.1088/0953-8984/27/32/323103. [DOI] [PubMed] [Google Scholar]
- 26.Schwarz G., Stankowski S., Rizzo V. Thermodynamic analysis of incorporation and aggregation in a membrane: application to the pore-forming peptide alamethicin. Biochim. Biophys. Acta. 1986;861:141–151. doi: 10.1016/0005-2736(86)90573-0. [DOI] [PubMed] [Google Scholar]
- 27.Beschiaschvili G., Seelig J. Melittin binding to mixed phosphatidylglycerol/phosphatidylcholine membranes. Biochemistry. 1990;29:52–58. doi: 10.1021/bi00453a007. [DOI] [PubMed] [Google Scholar]
- 28.Gazit E., Boman A., Shai Y. Interaction of the mammalian antibacterial peptide cecropin P1 with phospholipid vesicles. Biochemistry. 1995;34:11479–11488. doi: 10.1021/bi00036a021. [DOI] [PubMed] [Google Scholar]
- 29.Mishra V.K., Palgunachari M.N. Interaction of model class A1, class A2, and class Y amphipathic helical peptides with membranes. Biochemistry. 1996;35:11210–11220. doi: 10.1021/bi960760f. [DOI] [PubMed] [Google Scholar]
- 30.Gans P.J., Lyu P.C., Kallenbach N.R. The helix-coil transition in heterogeneous peptides with specific side-chain interactions: theory and comparison with CD spectral data. Biopolymers. 1991;31:1605–1614. doi: 10.1002/bip.360311315. [DOI] [PubMed] [Google Scholar]
- 31.Chen Y.-H., Yang J.T., Chau K.H. Determination of the helix and β form of proteins in aqueous solution by circular dichroism. Biochemistry. 1974;13:3350–3359. doi: 10.1021/bi00713a027. [DOI] [PubMed] [Google Scholar]
- 32.Jeener J., Meier B.H., Ernst R.R. Investigation of exchange processes by two-dimensional NMR spectroscopy. J. Chem. Phys. 1979;71:4546–4553. [Google Scholar]
- 33.Metzger D.S., Gaines J.R. Nuclear magnetic resonance studies of solidified H2–D2 mixtures. II. Pulsed techniques. Phys. Rev. 1966;147:644–657. [Google Scholar]
- 34.Bertani P., Raya J., Bechinger B. 15N chemical shift referencing in solid state NMR. Solid State Nucl. Magn. Reson. 2014;61-62:15–18. doi: 10.1016/j.ssnmr.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 35.Yesylevskyy S.O., Schäfer L.V., Marrink S.J. Polarizable water model for the coarse-grained MARTINI force field. PLoS Comp. Biol. 2010;6:e1000810. doi: 10.1371/journal.pcbi.1000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Jong D.H., Singh G., Marrink S.J. Improved parameters for the Martini coarse-grained protein force field. J. Chem. Theory Comput. 2013;9:687–697. doi: 10.1021/ct300646g. [DOI] [PubMed] [Google Scholar]
- 37.Hess B., Kutzner C., Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 38.Best R.B., Zhu X., Mackerell A.D., Jr. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. J. Chem. Theory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klauda J.B., Venable R.M., Pastor R.W. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wassenaar T.A., Pluhackova K., Tieleman D.P. Going backward: a flexible geometric approach to reverse transformation from coarse grained to atomistic models. J. Chem. Theory Comput. 2014;10:676–690. doi: 10.1021/ct400617g. [DOI] [PubMed] [Google Scholar]
- 41.Gapsys V., de Groot B.L., Briones R. Computational analysis of local membrane properties. J. Comput. Aided Mol. Des. 2013;27:845–858. doi: 10.1007/s10822-013-9684-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lakowicz J.R. Springer; Heidelberg, Germany: 2007. Principles of Fluorescence Spectroscopy. [Google Scholar]
- 43.Eftink M.R. Methods of Biochemical Analysis. Wiley; New York: 2006. Fluorescence techniques for studying protein structure; pp. 127–205. [DOI] [PubMed] [Google Scholar]
- 44.Voskuhl J., Wendeln C., Kros A. Immobilization of liposomes and vesicles on patterned surfaces by a peptide coiled-coil binding motif. Angew. Chem. Int. Ed. Engl. 2012;51:12616–12620. doi: 10.1002/anie.201204836. [DOI] [PubMed] [Google Scholar]
- 45.Versluis F., Dominguez J., Kros A. Coiled-coil driven membrane fusion: zipper-like vs. non-zipper-like peptide orientation. Faraday Discuss. 2013;166:349–359. doi: 10.1039/c3fd00061c. [DOI] [PubMed] [Google Scholar]
- 46.Zheng T., Boyle A., Kros A. Probing coiled-coil assembly by paramagnetic NMR spectroscopy. Org. Biomol. Chem. 2015;13:1159–1168. doi: 10.1039/c4ob02125h. [DOI] [PubMed] [Google Scholar]
- 47.Wieprecht T., Beyermann M., Seelig J. Binding of antibacterial magainin peptides to electrically neutral membranes: thermodynamics and structure. Biochemistry. 1999;38:10377–10387. doi: 10.1021/bi990913+. [DOI] [PubMed] [Google Scholar]
- 48.Chattopadhyay A., London E. Parallax method for direct measurement of membrane penetration depth utilizing fluorescence quenching by spin-labeled phospholipids. Biochemistry. 1987;26:39–45. doi: 10.1021/bi00375a006. [DOI] [PubMed] [Google Scholar]
- 49.London E., Ladokhin A.S. Measuring the depth of amino acid residues in membrane-inserted peptides by fluorescence quenching. In: Sidney T.J.M., Simon A., editors. Current Topics in Membranes. Academic Press; San Diego, CA: 2002. pp. 89–115. [Google Scholar]
- 50.Ladokhin A.S. Measuring membrane penetration with depth-dependent fluorescence quenching: distribution analysis is coming of age. Biochim. Biophys. Acta. 2014;1838:2289–2295. doi: 10.1016/j.bbamem.2014.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cullis P.R., De Kruijff B. Polymorphic phase behaviour of lipid mixtures as detected by 31P NMR. Evidence that cholesterol may destabilize bilayer structure in membrane systems containing phosphatidylethanolamine. Biochim. Biophys. Acta. 1978;507:207–218. doi: 10.1016/0005-2736(78)90417-0. [DOI] [PubMed] [Google Scholar]
- 52.Kim C., Spano J., Wi S. Evidence of pores and thinned lipid bilayers induced in oriented lipid membranes interacting with the antimicrobial peptides, magainin-2 and aurein-3.3. Biochim. Biophys. Acta. 2009;1788:1482–1496. doi: 10.1016/j.bbamem.2009.04.017. [DOI] [PubMed] [Google Scholar]
- 53.Scherer P.G., Seelig J. Electric charge effects on phospholipid headgroups. Phosphatidylcholine in mixtures with cationic and anionic amphiphiles. Biochemistry. 1989;28:7720–7728. doi: 10.1021/bi00445a030. [DOI] [PubMed] [Google Scholar]
- 54.Wi S., Kim C. Pore structure, thinning effect, and lateral diffusive dynamics of oriented lipid membranes interacting with antimicrobial peptide protegrin-1: 31P and 2H solid-state NMR study. J. Phys. Chem. B. 2008;112:11402–11414. doi: 10.1021/jp801825k. [DOI] [PubMed] [Google Scholar]
- 55.Gabrys C.M., Yang R., Weliky D.P. Nuclear magnetic resonance evidence for retention of a lamellar membrane phase with curvature in the presence of large quantities of the HIV fusion peptide. Biochim. Biophys. Acta. 2010;1798:194–201. doi: 10.1016/j.bbamem.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Agrawal P., Kiihne S., de Groot H. Solid state NMR investigation of the interaction between biomimetic lipid bilayers and de novo designed fusogenic peptides. ChemBioChem. 2007;8:493–496. doi: 10.1002/cbic.200600518. [DOI] [PubMed] [Google Scholar]
- 57.Grage S.L., Afonin S., Ulrich A.S. Interaction of the fusogenic peptide B18 in its amyloid-state with lipid membranes studied by solid state NMR. Chem. Phys. Lipids. 2004;132:65–77. doi: 10.1016/j.chemphyslip.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 58.Buffy J.J., McCormick M.J., Hong M. Solid-state NMR investigation of the selective perturbation of lipid bilayers by the cyclic antimicrobial peptide RTD-1. Biochemistry. 2004;43:9800–9812. doi: 10.1021/bi036243w. [DOI] [PubMed] [Google Scholar]
- 59.Salnikov E.S., Mason A.J., Bechinger B. Membrane order perturbation in the presence of antimicrobial peptides by (2)H solid-state NMR spectroscopy. Biochimie. 2009;91:734–743. doi: 10.1016/j.biochi.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 60.Rabe M., Boyle A., Kros A. Determination of oligomeric states of peptide complexes using thermal unfolding curves. Biopolymers. 2015;104:65–72. doi: 10.1002/bip.22598. [DOI] [PubMed] [Google Scholar]
- 61.Bechinger B., Sizun C. Alignment and structural analysis of membrane polypeptides by 15N and 31P solid-state NMR spectroscopy. Conc. Magn. Reson. A. 2003;18A:130–145. [Google Scholar]
- 62.Bechinger B., Resende J.M., Aisenbrey C. The structural and topological analysis of membrane-associated polypeptides by oriented solid-state NMR spectroscopy: established concepts and novel developments. Biophys. Chem. 2011;153:115–125. doi: 10.1016/j.bpc.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 63.Wasniewski C.M., Parkanzky P.D., Weliky D.P. Solid-state nuclear magnetic resonance studies of HIV and influenza fusion peptide orientations in membrane bilayers using stacked glass plate samples. Chem. Phys. Lipids. 2004;132:89–100. doi: 10.1016/j.chemphyslip.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 64.Tayebi L., Ma Y., Parikh A.N. Long-range interlayer alignment of intralayer domains in stacked lipid bilayers. Nat. Mater. 2012;11:1074–1080. doi: 10.1038/nmat3451. [DOI] [PubMed] [Google Scholar]
- 65.Bechinger B. Lipid multilayers: domains stack up. Nat. Mater. 2012;11:1005–1006. doi: 10.1038/nmat3507. [DOI] [PubMed] [Google Scholar]
- 66.Pannuzzo M., Raudino A., Böckmann R.A. Peptide-induced membrane curvature in edge-stabilized open bilayers: a theoretical and molecular dynamics study. J. Chem. Phys. 2014;141:024901. doi: 10.1063/1.4885340. [DOI] [PubMed] [Google Scholar]
- 67.Bechinger B. Rationalizing the membrane interactions of cationic amphipathic antimicrobial peptides by their molecular shape. Curr. Opin. Colloid Interface Sci. 2009;14:349–355. [Google Scholar]
- 68.Chen Z., Rand R.P. The influence of cholesterol on phospholipid membrane curvature and bending elasticity. Biophys. J. 1997;73:267–276. doi: 10.1016/S0006-3495(97)78067-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Churchward M.A., Rogasevskaia T., Coorssen J.R. Specific lipids supply critical negative spontaneous curvature—an essential component of native Ca2+-triggered membrane fusion. Biophys. J. 2008;94:3976–3986. doi: 10.1529/biophysj.107.123984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ohta-Iino S., Pasenkiewicz-Gierula M., Kusumi A. Fast lipid disorientation at the onset of membrane fusion revealed by molecular dynamics simulations. Biophys. J. 2001;81:217–224. doi: 10.1016/S0006-3495(01)75693-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kinnunen P.K.J. Fusion of lipid bilayers: a model involving mechanistic connection to HII phase forming lipids. Chem. Phys. Lipids. 1992;63:251–258. doi: 10.1016/0009-3084(92)90041-m. [DOI] [PubMed] [Google Scholar]
- 72.Holopainen J.M., Lehtonen J.Y.A., Kinnunen P.K.J. Evidence for the extended phospholipid conformation in membrane fusion and hemifusion. Biophys. J. 1999;76:2111–2120. doi: 10.1016/S0006-3495(99)77367-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kasson P.M., Lindahl E., Pande V.S. Atomic-resolution simulations predict a transition state for vesicle fusion defined by contact of a few lipid tails. PLoS Comp. Biol. 2010;6:1000829. doi: 10.1371/journal.pcbi.1000829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Smirnova Y.G., Marrink S.-J., Knecht V. Solvent-exposed tails as prestalk transition states for membrane fusion at low hydration. J. Am. Chem. Soc. 2010;132:6710–6718. doi: 10.1021/ja910050x. [DOI] [PubMed] [Google Scholar]
- 75.Larsson P., Kasson P.M. Lipid tail protrusion in simulations predicts fusogenic activity of influenza fusion peptide mutants and conformational models. PLoS Comput. Biol. 2013;9:e1002950. doi: 10.1371/journal.pcbi.1002950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Han J., Pluhackova K., Böckmann R.A. Synaptobrevin transmembrane domain determines the structure and dynamics of the SNARE motif and the linker region. Biochim. Biophys. Acta. 2016;1858:855–865. doi: 10.1016/j.bbamem.2016.01.030. [DOI] [PubMed] [Google Scholar]
- 77.Südhof T.C., Rothman J.E. Membrane fusion: grappling with SNARE and SM proteins. Science. 2009;323:474–477. doi: 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Apellániz B., Huarte N., Nieva J.L. The three lives of viral fusion peptides. Chem. Phys. Lipids. 2014;181:40–55. doi: 10.1016/j.chemphyslip.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Martens S., Kozlov M.M., McMahon H.T. How synaptotagmin promotes membrane fusion. Science. 2007;316:1205–1208. doi: 10.1126/science.1142614. [DOI] [PubMed] [Google Scholar]
- 80.McMahon H.T., Kozlov M.M., Martens S. Membrane curvature in synaptic vesicle fusion and beyond. Cell. 2010;140:601–605. doi: 10.1016/j.cell.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 81.Kozlovsky Y., Kozlov M.M. Stalk model of membrane fusion: solution of energy crisis. Biophys. J. 2002;82:882–895. doi: 10.1016/S0006-3495(02)75450-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chernomordik L., Chanturiya A., Zimmerberg J. The hemifusion intermediate and its conversion to complete fusion: regulation by membrane composition. Biophys. J. 1995;69:922–929. doi: 10.1016/S0006-3495(95)79966-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Marrink S.-J., Mark A.E. Molecular view of hexagonal phase formation in phospholipid membranes. Biophys. J. 2004;87:3894–3900. doi: 10.1529/biophysj.104.048710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Greenfield N.J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 2006;1:2876–2890. doi: 10.1038/nprot.2006.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Litowski J.R., Hodges R.S. Designing heterodimeric two-stranded alpha-helical coiled-coils. Effects of hydrophobicity and alpha-helical propensity on protein folding, stability, and specificity. J. Biol. Chem. 2002;277:37272–37279. doi: 10.1074/jbc.M204257200. [DOI] [PubMed] [Google Scholar]
- 86.Dragan A.I., Privalov P.L. Unfolding of a leucine zipper is not a simple two-state transition. J. Mol. Biol. 2002;321:891–908. doi: 10.1016/s0022-2836(02)00699-x. [DOI] [PubMed] [Google Scholar]
- 87.Fesinmeyer R.M., Peterson E.S., Andersen N.H. Studies of helix fraying and solvation using 13C′ isotopomers. Protein Sci. 2005;14:2324–2332. doi: 10.1110/ps.051510705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.McIntosh T.J., Holloway P.W. Determination of the depth of bromine atoms in bilayers formed from bromolipid probes. Biochemistry. 1987;26:1783–1788. doi: 10.1021/bi00380a042. [DOI] [PubMed] [Google Scholar]
- 89.Hung W.-C., Lee M.-T., Huang H.W. The condensing effect of cholesterol in lipid bilayers. Biophys. J. 2007;92:3960–3967. doi: 10.1529/biophysj.106.099234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Berendsen H.J.C., Postma J.P.M., Haak J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 91.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 92.Darden T., York D., Pedersen L. Particle mesh Ewald: an N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.