Abstract
U2AF homology motifs (UHM) that recognize U2AF ligand motifs (ULM) are an emerging family of protein–protein interaction modules. UHM–ULM interactions recur in pre-mRNA splicing factors including U2AF1 and SF3b1, which are frequently mutated in myelodysplastic syndromes. The core topology of the UHM resembles an RNA recognition motif and is often mistakenly classified within this large family. Here, we unmask the charade and review recent discoveries of UHM–ULM modules for protein–protein interactions. Diverse polypeptide extensions and selective phosphorylation of UHM and ULM family members offer new molecular mechanisms for the assembly of specific partners in the early-stage spliceosome.
Keywords: U2AF1, SF3B1, RRM, UHM, protein–protein interaction
INTRODUCTION
Despite a multitude of interacting subunits, traditional protein–protein interaction motifs are rare among pre-mRNA splicing factors. Approximately 240 proteins are thought to contribute to the pre-mRNA splicing process (Hegele et al. 2012). Nevertheless, the spliceosome contains only five documented WW domains and lacks canonical SH2 or SH3 domains for modular protein–protein interactions. In contrast, more than 200 human proteins are annotated both to contain RNA recognition motifs (RRMs) and to function in pre-mRNA splicing (Swiss-Prot, http://www.uniprot.org). An emerging view is that the “super-sized” RRM family can be divided based on divergent sequence elements and functions of the supposed family members (e.g., Blatter et al. 2015, commentary in Loerch and Kielkopf 2015).
The canonical RRM platform centers on two ribonucleoprotein (RNP) consensus motifs and typically binds RNA through key aromatic residues in β-strands. Structures of RRM-containing ribonucleoproteins and seminal spliceosome complexes (Yan et al. 2015; Galej et al. 2016; Rauhut et al. 2016; Wan et al. 2016a,b; Yan et al. 2016) demonstrate that the RRM fold serves as a platform for multiprotein assemblies with RNA. One of the first RRM structures revealed distinct α-helical and β-sheet surfaces of the U2B′′ RRM bound to the U2A′ protein and the U2 small nuclear (sn)RNA (Price et al. 1998). Subsequent structures, including the complex of alternative splicing factors PTB with Raver1 (Rideau et al. 2006; Joshi et al. 2011), intramolecular CPEB1 contacts for polyadenylation (Afroz et al. 2014), and the Snu17p RRM bound to Bud13p in the pre-mRNA retention and splicing complex (RES) (Tripsianes et al. 2014; Yan et al. 2016), establish that separate α-helical and RNP surfaces of the RRMs often bind protein and RNA partners simultaneously.
Remarkably, other members of the RRM fold family have adapted for protein–protein interactions in lieu of the ability to bind RNA. In one class of these specialized, protein-binding “RRMs,” the aromatic RNP residues bind protein ligands in lieu of traditional RNA interfaces. The RNP motifs at such protein-interfaces remain similar to those of classical, RNA-binding RRMs and as such, can be challenging to distinguish based on primary sequences alone. Examples include Y14–Mago nashi in the exon-junction complex (Fribourg et al. 2003; Lau et al. 2003; Shi and Xu 2003; Bono et al. 2006), the nonsense-mediated decay factors Upf2–Upf3 (Kadlec et al. 2004), and inter-RRM packing of Prp24 in the U2 snRNP (Bae et al. 2007; Montemayor et al. 2014). In other cases, the “RRM” has acquired distinct sequence features for binding proteins, whereas the RNP motifs have diverged and lost the ability to bind RNA. For example, an eIF3j interface with the α-helical region of the eIF3b RRM recruits eIF3b to the 40S ribosome, yet the eIF3b RRM has acidic residues near the RNP motifs and lacks detectable association with RNAs (ElAntak et al. 2007, 2010). Primary sequence features, such as atypical RNP residues or an acidic isoelectric point, often distinguish this latter type of protein-binding “RRM” variants. Among these, the U2AF homology motif (UHM) represents a discrete “RRM-like” family, whose members have lost the ability to bind RNA and instead gained specialized features for recognizing U2AF ligand motifs (ULMs) of protein partners.
Here we focus on recent progress toward understanding and defining the UHM family. Since initial views of UHM–ULM complexes (for review, see Kielkopf et al. 2004), many new UHM–ULM relationships have been revealed, including phosphorylation and polypeptide extensions of core ULM motifs as respective means for UHMs to engage in flirtatious as opposed to stable, monogamous relationships. Recent discoveries of mutant UHM- and ULM-containing splicing factors in myelodysplastic syndromes (MDS) (for review, see Inoue et al. 2016) highlight the importance of understanding how these bona fide protein–protein interaction modules select molecular partners in the assembling spliceosome.
Establishing a “U2AF homology motif” family
The first identified UHM–ULM-mediated complex was that of the eponymous U2 small nuclear ribonucleoprotein (snRNP) auxiliary factor (U2AF) heterodimer (Figs. 1, 2). Initially, the minimal interacting domains of the U2AF heterodimer were mapped to a putative RRM within U2AF35 and a short region of U2AF65 surrounding a singular tryptophan (Zhang et al. 1992; Rudner et al. 1996). The crystal structure revealed an irregular RRM variant of human U2AF35 bound to the tryptophan-containing region of U2AF65 (Fig. 2C; Kielkopf et al. 2001), which was recently confirmed for the fission yeast homologs U2AF59 and U2AF23 (Fig. 2D; Yoshida et al. 2015). Shortly thereafter, a C-terminal “RRM” of U2AF65 was found to recognize a tryptophan-containing site of SF1 via similar core interactions (Fig. 2A,B; Selenko et al. 2003). Altogether, the U2AF35-UHM–U2AF65 ULM and SF1 ULM–U2AF65-UHM interactions mediate a SF1–U2AF65–U2AF35 ternary complex (Fig. 1), which in turn recognizes consensus pre-mRNA signals at the 3′ splice site via distinct KH and RRM domains. We coined the term “U2AF Homology Motif,” or UHM to distinguish this family from canonical RRMs. Although the RRM-like fold is conserved among these U2AF domains, separate sequence motifs have specialized for protein–protein interactions with tryptophan-containing “U2AF Ligand Motifs,” or ULMs (Tables 1–2).
FIGURE 1.

Multiple UHM–ULM interactions guide the early stages of spliceosome assembly and other pathways. Experimentally confirmed phosphorylation sites of the ULM-containing regions are marked by pink circles. “W” represents ULM tryptophans. Thick gray arrow represents the transition during spliceosome assembly. Thin black arrows represent known UHM–ULM interactions. Arrows toward SF3b155 indicate the general ULM-containing region rather than specific binding sites.
FIGURE 2.
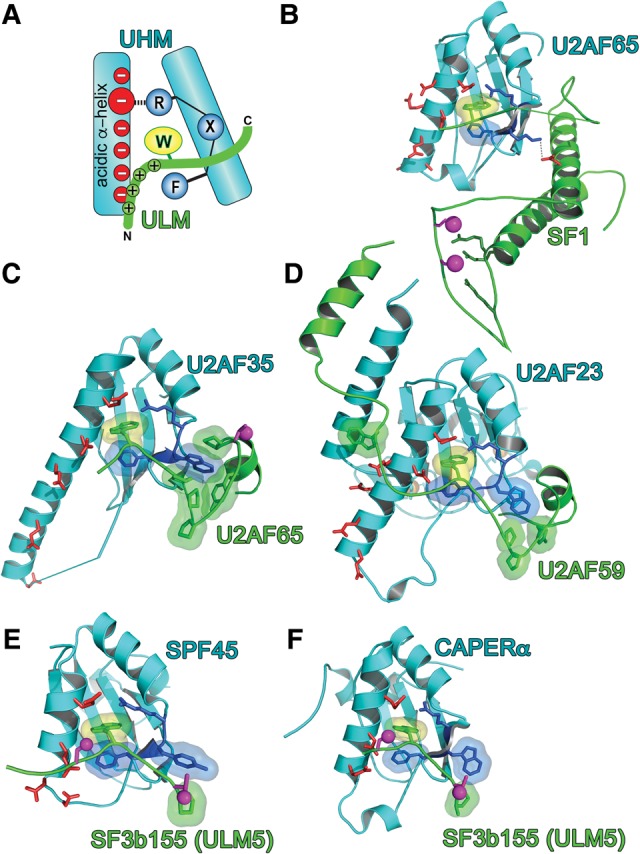
UHM–ULM recognition. (A) Schematic representation of consensus UHM (cyan)–ULM (green) interactions. (B–F) Structures of UHM–ULM complexes, including (B) U2AF65 UHM–SF1 ULM (PDB ID: 4FXW); (C) U2AF35 UHM–U2AF65 ULM (PDB ID: 1JMT); (D) U2AF23 UHM–U2AF59 ULM (PDB ID: 4YH8); (E) SPF45 UHM–SF3b155 ULM5 (PDB ID 2PEH); and (F) CAPERα UHM–SF3b155 ULM5 (PDB ID: 4OZ1). Magenta spheres mark phosphorylation sites: the SF1 and SF3b155 sites in B,E,F have been confirmed experimentally, whereas the human U2AF65 site in C is the predicted phosphorylation of a TP motif. The surface of the central ULM tryptophan is colored yellow, UHM RXF-motifs are blue, and acidic UHM α-helical residues are red; orange spheres are zinc ions on the back surface of the U2AF23 UHM in D.
TABLE 1.
Confirmed and candidate UHM family members
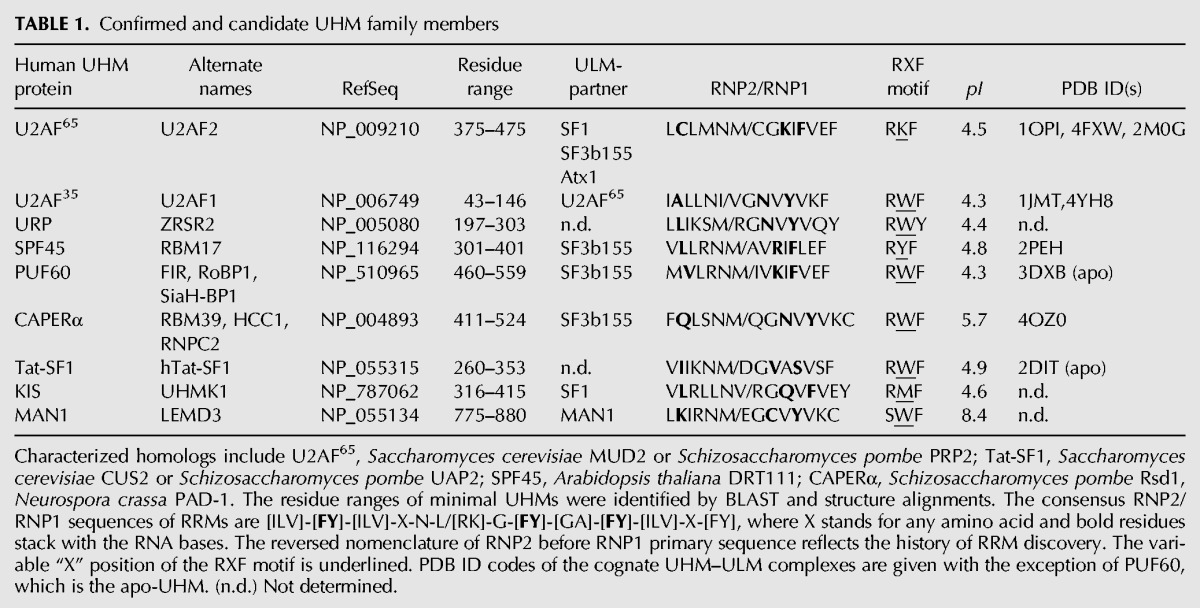
TABLE 2.
Confirmed ULM sequences
Starting from the U2AF prototypes, sequence similarity searches identified analogous “RRM” variants among diverse proteins (Table 1; Fig. 1; Kielkopf et al. 2004). Many putative UHMs have now been confirmed, either by structure determination or by functional homology (Table 1; Fig. 2E–F). Examples include SPF45 (also called RBM17 and a homolog of DRT111) (Fig. 2E; Corsini et al. 2007), PUF60 (also called FIR) (Corsini et al. 2009), and CAPERα (also called HCC1 or RBM39 and a homolog of PAD-1) (Fig. 2F; Loerch et al. 2014). These three proteins function in alternative splicing (Page-McCaw et al. 1999; Lallena et al. 2002; Dowhan et al. 2005; Hastings et al. 2007; Huang et al. 2012); PUF60 and CAPERα also are coactivators of transcription (Liu et al. 2000, 2001; Jung et al. 2002; Dowhan et al. 2005; Dutta et al. 2008). Other UHM-containing proteins participate in related pathways. A protein kinase, originally called KIS and now UHM kinase 1 (UHMK1), requires an intact UHM for site-specific phosphorylation of SF1 (Manceau et al. 2006, 2008). An inner nuclear membrane protein, MAN1, contains both a UHM and a ULM, and an intramolecular interaction between these motifs is important for downstream functions in the transforming growth factor (TGF)-β signaling pathway (Konde et al. 2010). These additions now establish UHM–ULM interactions as a recurrent protein–protein interaction mode, particularly among early-stage pre-mRNA splicing factors. Nonetheless, the Pfam database (Finn et al. 2016) continues to include UHMs as part of a super-sized RRM family.
Do UHMs bind RNA?
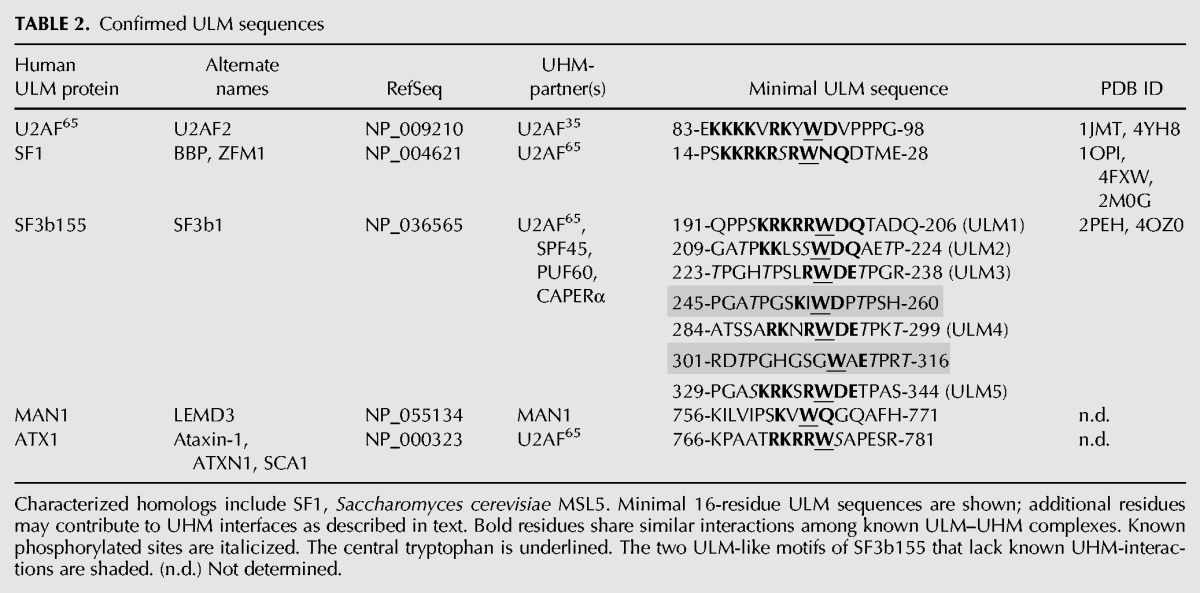
The UHM shares the core βαββαβ topology of an RRM, namely an α/β-sandwich arranged in a four-strand, antiparallel β-sheet packed against two α-helices (Fig. 3). Canonical RRMs bind RNA via two conserved RNP motifs in the central β-strands (for review, see Maris et al. 2005): RNP2 in β1 (Lahiri and Thomas 1986; Dreyfuss et al. 1988) and RNP1 in β3 (Fig. 3A; Adam et al. 1986; Sachs et al. 1986). Whereas a subset of RNP residues supports the RRM and UHM folds, the aromatic side chains at the second position of RNP2 and third/fifth positions of RNP1 are required for RRMs to bind RNA (e.g., Sickmier et al. 2006; McLaughlin et al. 2011; Bauer et al. 2012). Among confirmed UHMs, the corresponding second/third RNP2/RNP1 residues diverge from the RRM consensus (Table 1; Fig. 3B–E), consistent with dedicated UHM functions for protein–protein rather than protein–RNA interactions. Although early deletion experiments suggested that the U2AF65 UHM contributes to RNA binding (Zamore et al. 1992), the flanking RRM was inadvertently truncated. Likewise, our initial report of weak RNA binding by a minimal U2AF35 UHM U2AF65 ULM heterodimer (Kielkopf et al. 2001) may have resulted from excess U2AF65 ULM, which weakly binds RNA in the absence of UHM (C Kielkopf, unpubl.). Several groups have now established that stoichiometric complexes of U2AF65 UHM–SF1 ULM and U2AF35 UHM–U2AF65 ULM (human or fission yeast homologs) lack detectable RNA association in chemical shift and isothermal titration calorimetry (ITC) experiments (Selenko et al. 2003; Soares et al. 2006; Yoshida et al. 2015).
FIGURE 3.
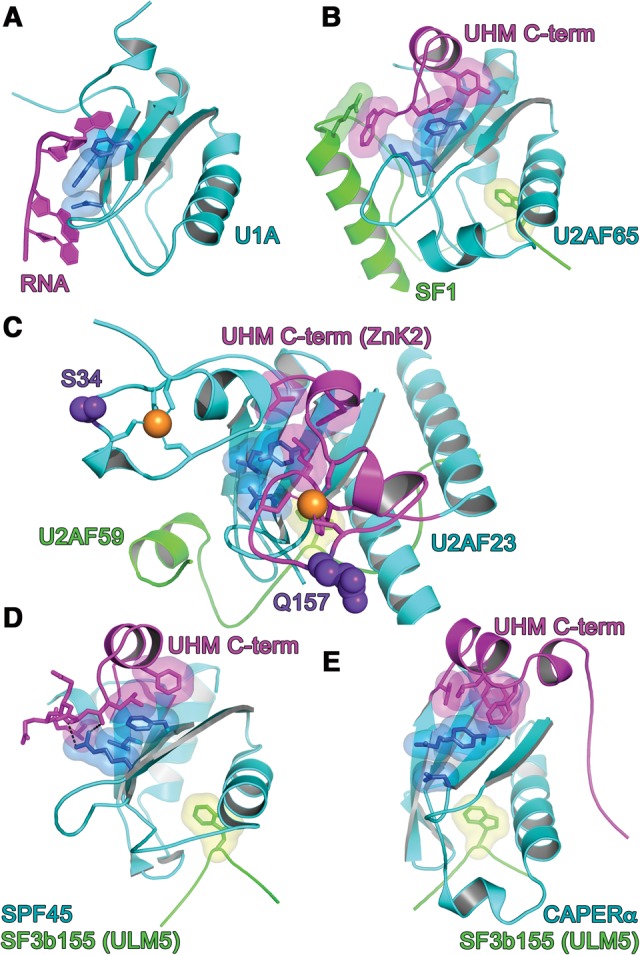
RNP interactions. (A) N-terminal RRM of U1A prototype bound to RNA ligand. The RNP surfaces of UHM (cyan)–ULM (green) complexes, including (B) U2AF65 UHM–SF1 ULM (PDB ID: 4FXW); (C) U2AF23 UHM–U2AF59 ULM (PDB ID: 4YH8); (D) SPF45 UHM–SF3b155 ULM5 (PDB ID 2PEH); and (E) CAPERα UHM–SF3b155 ULM5 (PDB ID: 4OZ1). The conserved aromatic residue at RNP1 position 5 typically is masked by a C-terminal extension (magenta) of the core UHM fold. The key residues at positions 2 of RNP2 and positions 3/5 of RNP1 are colored blue. Orange spheres represent zinc ions. Indigo, space-filling residues in C correspond to MDS-relevant mutations of the human homolog. Views are following an ∼120° rotation about the y-axis relative to Figure 2.
The aromatic residue at the fifth RNP1 position remains conserved among confirmed UHMs (Table 1; Fig. 3)—apart from Tat-SF1, for which only an apo-structure is available (PDB ID: 2DIT). As opposed to RNA contacts, α-helical extensions at the UHM C terminus consistently shield this conserved RNP1 residue from RNA binding (Fig. 3B–D). For the U1A RRM prototype, a similar α-helix occludes the RNP1 in the absence of RNA and rearranges in the RNA-bound conformation (Fig. 3A; Oubridge et al. 1994; Avis et al. 1996). However, this RRM–helix association is likely to be relatively weak; for example, crystallization additives displace the α-helix from the RNP1 surface (Rupert et al. 2003). By comparison, an extensive hydrophobic interface typically packs the amphipathic UHM α-helix against the RNP motifs (Selenko et al. 2003; Corsini et al. 2007, 2009; Loerch et al. 2014). As discussed below, a structure of the U2AF65 UHM bound to the N-terminal domain of SF1 further revealed that regions beyond the minimal ULM stabilize this masked UHM conformation (Fig. 3B; Wang et al. 2013; Zhang et al. 2013). It is possible that analogous, ligand-mediated reinforcements of the α-helical block on the UHM “RNA binding surface” will emerge as more UHM-containing structures are determined bound to intact, ULM-containing proteins.
Zinc knuckles mask the U2AF35 and potentially URP ribonucleoprotein motifs
The U2AF35 UHM represents a singular variation of the masked RNP1 (Fig. 3C). Rather than a C-terminal α-helix, the recent structure of the fission yeast homolog, U2AF23 reveals that N- and C-terminal zinc knuckles (ZnK1 and ZnK2) fold against the “RNP” surface of the UHM (Yoshida et al. 2015). In particular, the C-terminal ZnK2 masks the conserved aromatic residue (Y) at the fifth position of RNP1. Although these conserved ZnK motifs mask the putative RNA binding surface of the U2AF23, and by analogy, U2AF35 UHMs, the ZnK family typically interacts with RNA (e.g., Hudson et al. 2004; Teplova and Patel 2008). Approximately 10% of MDS-patients harbor acquired mutations of the U2AF1 gene that encodes the U2AF35 splicing factor (for review, see Inoue et al. 2016). These MDS-relevant U2AF1 mutations consistently modify the S34 and Q157 residues on the expected RNA binding surfaces of the respective ZnK (Fig. 3C). Indeed, the common S34F mutation alters the RNA binding characteristics of mutant U2AF35-S34F–splicing factor complexes (Okeyo-Owuor et al. 2015; Yoshida et al. 2015). As such, the U2AF35 UHM serves as a scaffold for both the U2AF heterodimer and adjoining ZnK motifs, which in turn restores RNA binding functions to the U2AF35 UHM.
The most closely related UHM-containing protein to U2AF35 is URP (38% overall sequence identity), which shares a similar domain organization of a central UHM flanked by zinc knuckles (ZnK). Both U2AF35 and URP are the products of frequently mutated genes in MDS patients (U2AF1 and ZRSR2) (for review, see Inoue et al. 2016). However, the U2AF35 mutations alter specific ZnK residues, whereas the URP mutations are randomly distributed throughout the protein sequence and often null. This difference most likely reflects the distinct functions of the two proteins: U2AF35 recognizes the intron-exon junction of the major class of splice sites in conjunction with U2AF65 (Merendino et al. 1999; Wu et al. 1999; Zorio and Blumenthal 1999), as opposed to a selective role for URP in a “minor,” stress-associated class of intron splicing (Shen et al. 2010; Madan et al. 2015). Yet, no U2AF65 paralog has been identified as a ULM partner for the URP UHM to date.
Shared structural features of UHM–ULM interactions
The core interactions of bona fide UHM–ULM complexes are shared among known structures (Fig. 2A), despite moderate primary sequence conservation (∼23% average pairwise sequence identity between matching UHM residues). A critical ULM tryptophan inserts within a hydrophobic pocket between the two α-helices of the UHM “backside” (opposite the RNP β-sheet). A phenylalanine side chain of the UHM caps the base of this pocket and stacks in a “T-type” aromatic interaction (Burley and Petsko 1985) with the tryptophan side chain. This phenylalanine is the third residue of a conserved “RXF” motif located in a loop connecting the second α-helix and terminal β-strand of the RRM-like fold. The arginine of this loop further encloses the opposite surface of the ULM tryptophan by a salt bridge with the glutamate side chain of the other UHM α-helix. The “X” residue often is an aromatic residue that either faces solvent or interacts with a C-terminal extension of the ULM, as further described below. Preceding the ULM tryptophan, a stretch of basic ULM residues interacts with an acidic UHM α-helix, which also displays a conserved glutamate residue to form the salt bridge abutting the tryptophan. Coupled with the divergence of formerly basic residues for RNA binding by an RRM ancestor, the isoelectric points of UHMs are usually acidic (Table 1). Altogether, the degenerate RNP motifs, RXF motif, acidic α-helix and low isoelectric point, are consensus features that set UHMs apart from canonical, RNA-binding RRMs.
A limited cohort of established “U2AF ligand motifs”
The ULM is highly degenerate and as such, bona fide UHM-partners are challenging to distinguish (Table 2). The consensus elements of the ULM are limited to a central tryptophan and preceding basic residues, which as described above, interact with the respective RXF motif and acidic α-helix of the UHM. A search of the Swiss-Prot database using ScanProsite (de Castro et al. 2006) and the ULM pattern [RK]-X(0,3)-W-[DN]-[EQ] matches more than 9500 protein sequences (out of 551,000 entries), which approaches the expected number of random hits (∼14,000) (Nicodeme 2001). Indeed, ULM-like mimics can bind RRM-like domains that lack the consensus UHM features in unexpected ways; for example, a ULM-like region of Bud13p forms an unrelated interface with a Snu17p RRM in the “retention and splicing” (RES) complex (Tripsianes et al. 2014).
Notwithstanding the overwhelming abundance of candidate ULMs, only five ULM-containing proteins have been confirmed to bind UHMs (Table 2). Two well-characterized ULM prototypes include U2AF65 and SF1, which respectively partner with the UHMs of U2AF35 and U2AF65 for 3′ splice site recognition (Figs. 1, 2B–D; Kielkopf et al. 2001; Selenko et al. 2003). Alternatively, the SF1 ULM is recognized by the UHM of the KIS/UHMK1, which directs phosphorylation of an SF1 SPSP motif (Manceau et al. 2006, 2008). Soon thereafter, we and others noticed seven ULM-like repeats in the N-terminal region of the SF3b155 subunit of the U2 snRNP, and confirmed that five of these putative SF3b155 ULMs detectably associate with UHMs (Thickman et al. 2006; Corsini et al. 2007, 2009; Loerch et al. 2014). The SF3b155 ULM–U2AF65 UHM complex is likely to assist SF1 displacement and stable association of the U2 snRNP during spliceosome assembly, whereas other UHM-containing splicing factors may regulate this process. Nevertheless, the functional interplays among these multiple SF3b155 ULMs and their UHM partners remain a mystery, as discussed below.
In addition to these early-stage splicing factors, UHMs bind ULM-like sequences of two other nuclear proteins (Fig. 1): the inner nuclear membrane protein MAN1 (also called LEMD3) (Konde et al. 2010) and the neurodegenerative disease-related Ataxin-1 (ATX1) (de Chiara et al. 2009). An intramolecular ULM–UHM interaction within MAN1 is thought to regulate its association with SMAD2 (Konde et al. 2010), which in turn represses TGF-β signaling for bone development (Hellemans et al. 2004; Lin et al. 2005; Pan et al. 2005). A ULM sequence near the C terminus of ATX1 binds the UHMs of U2AF65 (Lim et al. 2006; de Chiara et al. 2009) or SPF45 (Lim et al. 2008; de Chiara et al. 2009). Further, U2AF65 colocalizes in large inclusions of polyglutamine-expanded ATX1, which are characteristic of triplet-repeat expansion in spinocerebellar ataxia type 1 (de Chiara et al. 2009). As such, sequestration of UHM-containing proteins by ATX1 aggregates may contribute to the pathogenesis of this disease.
Potential functions of multiple functions of multiple SF3b155 ULMs
The presence of five confirmed ULMs in human SF3b155, rather than an isolated ULM, is unusual among ULM-containing proteins. Notably, the SF3b155 subunit is produced by the SF3B1 gene, which frequently harbors mutations of a distinct, HEAT repeat domain in MDS patients (for review, see Inoue et al. 2016). Considering that recurrent splicing factor mutations in MDS are heterozygous, rarely null, and typically affect specific accessory interactions, even the absence of MDS-relevant mutations among SF3b155 ULMs suggests essential functions for this protein region. The SF3b155 ULMs are embedded within an intrinsically disordered region (IDR) of this protein, as reflected by low sequence complexity and the absence of well-defined structure in the absence of bound UHM (Thickman et al. 2006). As such, the SF3b155 ULMs are likely to share the typical function of IDRs as dynamic signaling hubs for protein-interaction networks (for review, see Wright and Dyson 2015). The structures of individual SF3b155 ULMs bound to UHMs, including those of SPF45 and CAPERα (Fig. 2E,F; Corsini et al. 2007; Loerch et al. 2014), show the typical core UHM–ULM interactions described above. Yet, several lines of evidence indicate that these structures may be only headlines of an epic story.
The question of how many of SF3b155 ULMs can be occupied concurrently has been addressed separately for the UHMs of U2AF65 and CAPERα. The apparent stoichiometry of ITC experiments suggests that up to three U2AF65 UHMs can associate with the ULM-containing region of SF3b155 (Thickman et al. 2006). Consistent with findings for the U2AF65 UHM, a battery of methods including size exclusion chromatography, amino acid analysis, and ITC show that two CAPERα UHMs concurrently bind the SF3b155 ULM-containing region under saturating conditions (Loerch et al. 2014). The Hill coefficient of pull-downs from human cell extract further suggests that endogenous, full length CAPERα binds a GST-fused ULM-containing region of SF3b155 with positive cooperativity (Loerch et al. 2014). Despite the ability of multiple UHMs to concurrently bind the ULM-containing region of SF3b155, a one-to-one complex with U2AF65 is thought to recruit the SF3b155-tethered U2 snRNP to the pre-mRNA splice site during the early stages of spliceosome assembly (Gozani et al. 1998). Instead, other UHM-containing alternative splicing factors, such as CAPERα, may co-assemble with the U2AF65–SF3b155 complex.
Whether each of the multiple SF3b155 ULMs has “promiscuous” or “monogamous” relationships with UHM-containing protein partners also remains in question. Although a large number of UHM–ULM binding experiments are now complete (Table 3), the binding affinities for distinct ULM-like SF3b155 sites have rarely been comprehensively quantified for a given UHM-containing protein. To date, available evidence suggests that the constitutive splicing factor U2AF65 prefers to bind a distinct SF3b155 ULM (ULM5) (Table 2; Thickman et al. 2006) rather than compete with alternative splicing factors, although excess U2AF65 can associate with more than one ULM in vitro. Pull-down assays of full-length CAPERα by a series of SF3b155 IDR variants, each containing a single unmodified ULM, followed by ITC experiments using the three ULM peptides that detectably bound, show that CAPERα UHM has highest affinity for the first SF3b155 ULM (ULM1) (Loerch et al. 2014). Likewise, ITC and NMR data implicated SF3b155 ULM1 as the preferred binding site of the PUF60 UHM, compared to ULM2 or ULM5 (Corsini et al. 2009). These five- to sevenfold preferences of the CAPERα and PUF60 UHMs for binding the ULM1- over ULM5-containing SF3b155 variants or fragments, compared to the converse preference of the U2AF65 UHM for the distally located ULM5, suggests that the CAPERα or PUF60 splicing factors can concurrently bind SF3b155 alongside U2AF65. Accordingly, U2AF65 and PUF60 have synergistic effects on splicing in vitro (Hastings et al. 2007).
TABLE 3.
Apparent equilibrium dissociation constants (KD) of ULM–ULM complexes determined by ITC
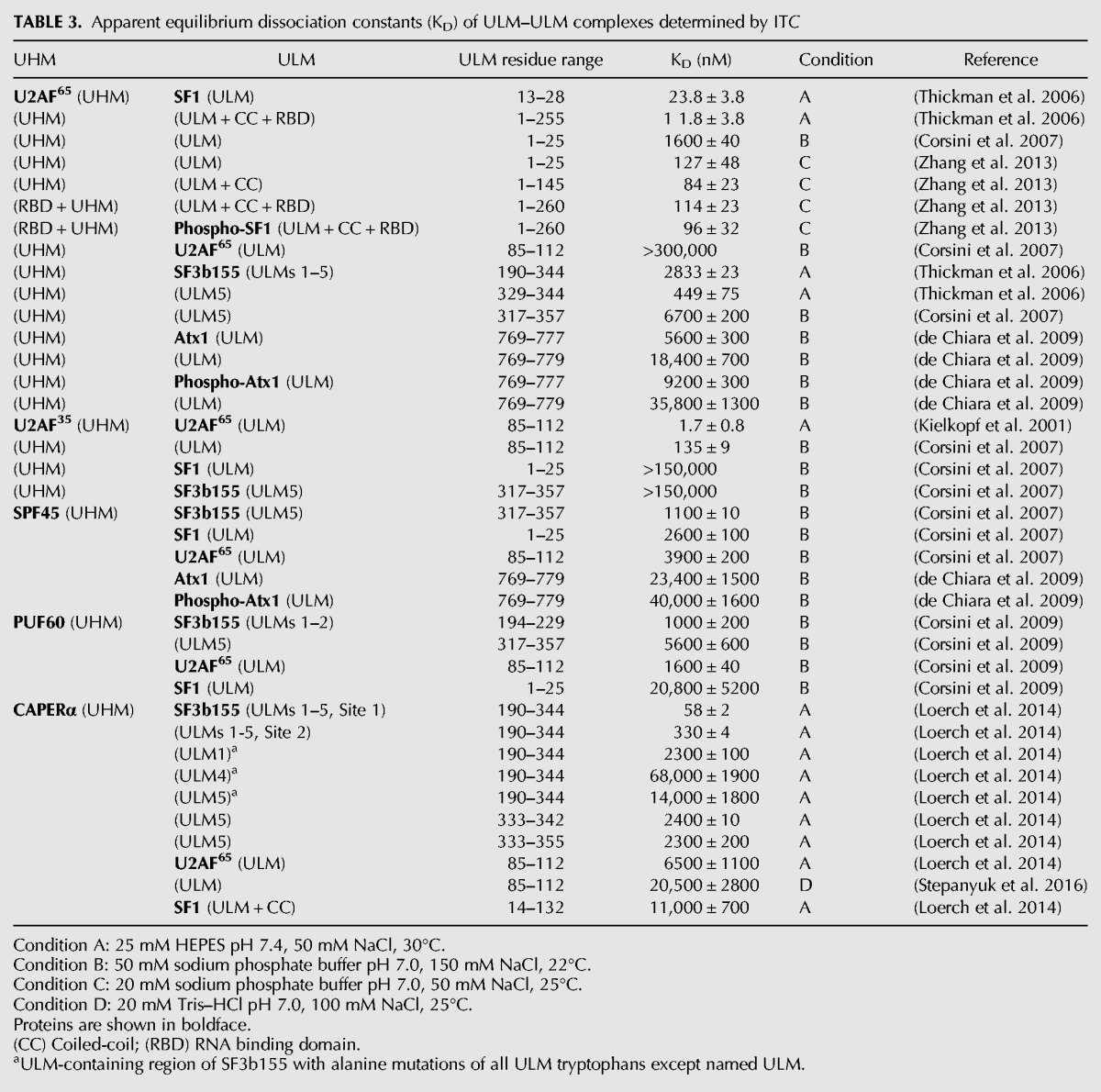
IDRs such as the SF3b155 ULM-containing region often coordinate the synergistic action of multiple weak binding sites to modulate positive or negative allostery, as recently exemplified for coordination of host CBP or pRB proteins by the adenoviral protein E1A (Ferreon et al. 2013). The ULM-containing region of SF3b155 adjoins its binding site for p14, an RRM-like U2 snRNP subunit that ultimately contacts the branch point sequence of the pre-mRNA (Will et al. 2001; Cass and Berglund 2006; Schellenberg et al. 2006; Spadaccini et al. 2006). Considering the above data, multiple UHM-containing proteins are likely to simultaneously associate with the SF3b155 IDR alongside the p14 subunit, possibly with positive cooperativity (Fig. 1). A potentially analogous precedent for cooperative assembly of an RRM-mediated spliceosome subcomplex has been set by the RES hetero-trimer of the RRM-like Snu17p, ULM-like Bud13p, and Pml1p (Tripsianes et al. 2014; Wysoczanski et al. 2014).
Emerging roles of ULM extensions for UHM specificity
The core interactions of UHM–ULM complexes are conserved, namely the electrostatic interactions between the acidic UHM α-helix and the basic ULM tail as well as the UHM RXF-mediated pocket for the ULM-tryptophan. This raises the question of how different UHMs distinguish the locally similar, yet functionally diverse, ULMs of SF1, U2AF65, SF3b155, MAN1, or ATX1 partners from numerous ULM-like sequences in the proteome. Indeed, UHM-containing proteins exhibit binding preferences for distinct ULM-containing proteins (Table 3). The U2AF35 UHM–U2AF65 ULM complex is more than 1000-fold preferred over off-target U2AF35 UHM–SF1 ULM or CAPERα– U2AF65 ULM interactions (Corsini et al. 2007; Stepanyuk et al. 2016). Likewise, the U2AF65 UHM–SF1 ULM complex is at least 200-fold preferred over an erroneous U2AF65 UHM–U2AF65 ULM homodimer (Corsini et al. 2007). Other U2AF65 UHM–ULM complexes that serve alternative functions more closely approach the affinity of its SF1 complex. For example, the U2AF65 UHM prefers binding the SF1 ULM over SF3b155 ULM5 peptides by 18-fold (Thickman et al. 2006); the U2AF65–SF3b155 complex may either be strengthened in the context of full length proteins or assist U2AF displacement during the sequential stages of pre-mRNA splicing. The affinity of the U2AF65 UHM for unphosphorylated ATX1 ULM approaches its affinity for SF1, but decreases following ATX1 phosphorylation; accordingly, ATX1-phosphorylation is thought to regulate competitive binding to U2AF65 versus a 14-3-3 protein signaling scaffold (de Chiara et al. 2009). The splicing and transcription regulator CAPERα strongly prefers to bind the intact SF3b155 ULM-containing region (e.g., by nearly 200-fold compared with SF1) (Loerch et al. 2014). The SPF45 and PUF60 UHMs also show subtle preferences for binding a representative SF3b155 ULM peptide (ULM5) over SF1 or U2AF65 ULMs, which may be enhanced in the context of the intact SF3b155 domain, as observed for CAPERα. These CAPERα, SPF45 and PUF60 associations with SF3b155 may regulate the second, U2 snRNP-dependent step of splicing, which has been shown for SPF45 (Lallena et al. 2002). Although the structural basis of SF3b155 ULM–UHM specificity remains limited to core interactions, structures of U2AF65 UHM and U2AF35 UHM complexes bound to larger SF1 or U2AF35 fragments reveal unexpected roles for ULM extensions in conferring the UHM–ULM specificity of these two complexes.
An early clue to the underpinnings of UHM–ULM-specificity was obtained from the crystal structure of the human U2AF35 UHM–U2AF65 ULM complex (Kielkopf et al. 2001). At the C terminus of the U2AF65 ULM, a short proline/aromatic-rich sequence encloses an exposed U2AF35 UHM tryptophan at the X-position of the RXF motif (Fig. 2C) and is required for high affinity interactions. This hydrophobic ULM sandwich is conserved for the fission yeast U2AF heterodimer (Fig. 2D) and encloses the UHM RWF-motif in an analogous manner as the human homolog. The “X” residues of several other UHM RXF motifs also are aromatic, including tryptophan residues for the PUF60, Tat-SF1, and MAN1 UHMs. Accordingly, a TP motif in the C-terminal region of the SF3b155 ULM is similarly positioned adjacent aromatic side chains in the RXF motifs of the CAPERα and SPF45 UHMs (Corsini et al. 2007; Loerch et al. 2014). In contrast, structurally distinct lysine and methionine residues mark the RXF motifs of the U2AF65 and KIS UHMs, which may be related to the preference of these UHMs to target SF1 over SF3b155. Although C-terminal extensions of an SF3b155 ULM5 peptide only subtly increase its binding affinity for the CAPERα UHM (Loerch et al. 2014), it is conceivable that analogous interactions between the proline-rich inter-ULM linkers of SF3b155 and aromatic residues in UHM RXF motifs contribute to UHM specificity in the context of the intact SF3b155 ULM-containing region.
The role of C-terminal ULM extensions in cognate UHM specificity is dramatically illustrated by a striking coiled-coil extension of the SF1 ULM (Figs. 1B, 2B; Wang et al. 2013; Zhang et al. 2013). The N-terminal α-helix of the SF1 coiled-coil mediates the primary contacts between this domain and the U2AF65 UHM (Fig. 1B). Following a disordered linker to the ULM, the N-terminal section of this SF1 α-helix packs against both the U2AF65 RNP2 and a C-terminal α-helical tail of the UHM, which in turn masks the RNP1 (Fig. 2B). Thereby, the SF1 coiled-coil converts the putative RNA binding surface of the RRM-like fold into a protein–protein interface. Although a similar α-helix extends the RRM-like fold of most UHMs (Fig. 2D–E), a unique tryptophan residue of U2AF65 mediates a cation–π interaction with an arginine residue in the SF1 coiled-coil. A lysine in the U2AF65 RXF loop, which differs from the typical aromatic X-residues of other UHMs, further recognizes the C-terminal region of the same SF1 α-helix via a specific salt bridge. The SF1 coiled-coil confers only a few fold higher affinity for the U2AF65 UHM compared to the minimal ULM, likely due to an entropic cost of local folding-on-binding. Nevertheless, the specific interactions among the C-terminal U2AF65 tryptophan, lysine in the RXF loop, and the SF1 coiled-coil residues, are likely to specify this cognate UHM–ULM complex.
Remarkably, the N terminus of the fission yeast U2AF59 ULM is extended at by a distinct, anti-parallel coiled-coil with the C-terminal, α-helical extension of the U2AF23 UHM and second ZnK (Fig. 1D). The appended U2AF59 and U2AF23 α-helices, which were absent from the original U2AF65–U2AF35 structure (Kielkopf et al. 2001), are weakly conserved in the human primary sequences and instead appear to have been partly replaced by arginine-rich residues. With the exception of crystal contacts by SPF45-bound SF3b155 (Corsini et al. 2007), the N-terminal ULM residues are disordered in all UHM–ULM structures other than the U2AF59–U2AF23 heterodimer. Together with the observation that multiple mutations of these typically basic residues are needed to abolish detectable SF1–U2AF65 interactions (Selenko et al. 2003), this N-terminal ULM region is likely to contribute avidity, whereas the ordered U2AF59 α-helix confers specificity for the U2AF23 heterodimer. Given these new precedents for extended U2AF65–SF1 and U2AF59–U2AF23 heterodimers, it now remains to be seen whether regions adjoining the SF3b155, MAN1, and ATX1 ULMs likewise guide specific complexes with UHM-containing proteins.
ULM phosphorylation regulates partnerships with UHMs
Dynamic phosphorylation and dephosphorylation of splicing factors regulate spliceosome assembly and selection of splice sites (for review, see Naro and Sette 2013). For example, region-specific phosphorylation of serine–arginine rich proteins is well-known to regulate splicing factor localization and spliceosome assembly (for review, see Zhou and Fu 2013). For several ULM-containing proteins, including SF1, ATX1, and potentially U2AF65 and SF3b155, phosphorylation has emerged as a means for regulating their UHM partnerships and downstream functions.
The first example of phosphorylation regulating ULM–UHM interactions was noted for a serine in the SF1 ULM (S20, Fig. 2B; Wang et al. 1999). Phosphorylation of this site by cGMP-dependent protein kinase-I reduces SF1 association with U2AF65 and thereby inhibits spliceosome assembly. Structurally, the phosphorylated SF1 S20 would disrupt a hydrogen bond and electrostatically repel a conserved aspartate residue (D401) in the acidic α-helix of the U2AF65 UHM (Wang et al. 2013). Two of the SF3b155 ULMs display serine residues at analogous positions as SF1 S20, including SF3b155 S216 and S336 in ULM2 and ULM5; phosphorylation of SF3b155 S216 has been confirmed in an acute myeloid leukemia cell line by mass spectrometry (Weber et al. 2012). The similar positions of serines among these ULMs suggest that phosphorylation could coordinately dissociate U2AF65, and possibly other SF3b155-associated UHM-containing proteins, from SF3b155 and SF1.
A loop between the two α-helices of the SF1 coiled-coil domain harbors a highly conserved “SPSP” sequence motif (Fig. 2B), which is primarily in the phosphorylated state for SF1 isolated from human embryonic kidney cells (Manceau et al. 2006). This SPSP-phosphorylation event subtly increases the binding affinity of SF1 for U2AF65 (Manceau et al. 2006; Zhang et al. 2013), most likely by stabilizing the coiled-coil fold of the phosphorylated SF1 domain. Structures of the SPSP-phosphorylated SF1 bound to the U2AF65 UHM shows that the phosphorylated serines coordinate three arginine residues and induce folding of an otherwise disordered linker between the two α-helices of the coiled-coil (Wang et al. 2013; Zhang et al. 2013). The UHM-containing KIS kinase specifically phosphorylates the SPSP motif of SF1 (Manceau et al. 2006), and KIS-knockout mice showed subtle changes in gene expression and specific behaviors, including spontaneous activity and contextual fear conditioning (Manceau et al. 2012). However, 2D electrophoresis showed that the majority of endogenous SF1 remains in the SPSP-phosphorylated form following KIS-knockout. This result implicates other kinases in regulating the SF1 SPSP site, and leaves the potentially pluripotent functions of SF1 SPSP-phosphorylation an open question.
The most recent member of the ULM-containing family, ATX1, is a phosphorylation-sensitive molecular switch that discriminates between two ligands, U2AF65 and the multifunctional regulatory protein 14-3-3 (de Chiara et al. 2009). An S776 residue immediately adjacent the ULM tryptophan of ATX1 is phosphorylated by Akt kinase (Chen et al. 2003). The 14-3-3 protein was shown to effectively outcompete the U2AF65 UHM for binding at the phosphorylated ATX1 site, which overlaps the ATX1 ULM. Initial investigations with phospho-mimetic S776D mutations of ATX1 in mouse models suggested that phosphorylation of S776 also recruits SPF45 to ATX1 complexes (Lim et al. 2008). Although the apparent enhancement of ATX1–SPF45 association by S776 phosphorylation could not be recapitulated by ULM-containing peptides binding the isolated UHM in vitro (de Chiara et al. 2009), it remains possible that the phosphorylation-sensitivity depends on the context of the intact proteins. Notably, the abnormal expansion of an ATX1 polyglutamine-repeat in the neurodegenerative disease spinocerebellar ataxia type 1 requires the S776 residue of ATX1 and enhances ATX1–SPF45 association in vivo (Lim et al. 2008), which sets a precedent for the contribution of ULM phosphorylation to pathogenicity.
These well-characterized examples of phosphorylation-sensitive UHM–ULM interactions are only the tip of the iceberg. Numerous ULM-associated phosphorylation sites remain to be investigated. In a striking example, abundant phosphorylated threonine–proline (TP) motifs riddle the N-terminal IDR of SF3b155, including 29 TP sequences of which 18 are experimentally verified, phosphorylation sites in the Phospho.ELM database (Dinkel et al. 2011). The U2AF65 and MAN1 ULMs also contain single TP motifs, whereas the SF1 and ATX1 ULMs have none. In known structures, including the U2AF65 ULM–U2AF35 UHM heterodimer and SF3b155-ULM complexes with SPF45 or CAPERα UHMs, a TP motif located a few residues C terminal to the central ULM tryptophan packs near an exposed aromatic residue at the X-position of the UHM RXF-motif (Fig. 2B–F). Although the structural effects of ULM TP phosphorylation are currently unknown, one can envision that the proximity of the phosphorylated TP to the UHM RXF motif could result in steric clashes, anion–π repulsion, or conformational changes following peptidyl–prolyl-isomerization, which are well-known to switch cis/trans propensity following TP phosphorylation (for review, see Wulf et al. 2005).
Whether phosphorylation of these ULM-associated TP motifs controls selective ULM–UHM associations, or acts as a general “off” switch, remains a topic for speculation. While it has been shown that SF3b155 is successively phosphorylated and dephosphorylated during spliceosome assembly (Wang et al. 1998; Boudrez et al. 2002; Shi et al. 2006), a direct link to the ULM-proximal TP motifs has yet to be established. The SF3b155 subunit is phosphorylated in vitro by cyclin B–cdk1 and cyclin E–cdk2 (Seghezzi et al. 1998; Boudrez et al. 2002), which also co-immunoprecipitate with SF3b155 from cell lysates, and SF3b155 phosphorylation significantly increases during mitosis (Seghezzi et al. 1998). However, various TP motifs in the SF3b155 IDR are predicted by NetPhosK (Blom et al. 2004) to be phosphorylated by at least ten different kinases. Altogether, these observations point to selective, as well as coordinated, regulation of ULM-associated TP motifs by phosphorylation.
Summary and perspectives
Significant progress toward establishing a network of UHM–ULM interactions (Fig. 1) has been made since the discovery of these RRM-like protein–protein interaction domains fifteen years ago. Most suspected members of the UHM family have now been structurally confirmed and implicated in UHM–ULM partnerships. Since the first U2AF65 ULM–U2AF35 UHM and SF1 ULM–U2AF65 UHM prototypes (Kielkopf et al. 2001; Selenko et al. 2003), structures of SPF45 and CAPERα UHMs reveal similar core interactions with the individual SF3b155 ULMs (Corsini et al. 2007; Loerch et al. 2014). The apo-PUF60 UHM structure also has been determined and the PUF60 UHM binds an SF3b155 ULM (Corsini et al. 2009). The UHM-containing KIS kinase has been shown to selectively phosphorylate SF1 in a ULM-dependent manner (Manceau et al. 2006). These UHM–ULM partnerships are likely to contribute to spliceosome assembly and alternative splicing, yet new functional roles for UHM–ULM protein interaction modules continue to emerge. An intramolecular UHM–ULM switch in the inner nuclear membrane protein MAN1 regulates SMAD2 binding for TGF-β signaling (Konde et al. 2010). Similarly, a ULM in the glutamine-repeat expanded ataxin protein competes for UHM-mediated association with U2AF65 versus a phosphorylation-sensitive, 14-3-3 protein signaling hub (de Chiara et al. 2009). Altogether, these recent findings demarcate a bona fide UHM family of ULM-interaction modules (Fig. 3), underlying a deceptively RRM-like charade.
An outstanding question is whether new members of the UHM family will be discovered in the future. To date, nine UHM family members (consolidating U2AF65/MUD2, SPF45/DRT111, Tat-SF1/CUS2/UAP2, CAPERα/PAD-1/Rds1 homologs) were first identified based on primary sequence searches (Kielkopf et al. 2004) and now have been shown to partner with ULM-containing proteins (Table 1). Seven atypical RRMs with sequence features that appeared distantly related to UHMs also were flagged previously (Kielkopf et al. 2004). Among these candidates, new structures already rule out UHM-type interactions for the N-terminal RRMs of Prp24 and SAP49. Even so, these Prp24 and SAP49 RRMs do function in protein–protein interactions. The N-terminal RRM of Prp24 utilizes the RNP1/RNP2 surface to bind a neighboring RRM rather than RNA (Bae et al. 2007; Montemayor et al. 2014). The N-terminal SAP49 RRM engages a neighboring Cus1 subunit in the activated yeast spliceosome (Yan et al. 2016) via a tryptophan-mediated interaction that is qualitatively similar to a UHM. In addition to the SAP49–Cus1 complex (Yan et al. 2016), new examples of tryptophan-mediated interactions with RRM-like domains have emerged outside the UHM family, including eIF3b–eIF3j in the translation initiation complex (ElAntak et al. 2010), Snu17p– Bud13p in the RES complex (Tripsianes et al. 2014), mRNA export complexes of viral proteins with AlyREF (Tunnicliffe et al. 2011; Tunnicliffe et al. 2014), and an intramolecular interface of CPEB1 (Afroz et al. 2014). The exact conformations of the tryptophan-containing ligands differ among these atypical RRMs, which also lack the RXF motif, acidic α1, and C-terminal α-helix of UHMs. Also unlike UHMs, SAP49, Snu17p and CPEB1 simultaneously bind RNA and protein partners. As such, tryptophan-mediated protein-interactions with atypical, RRM-like domains represent a recurring theme among ribonucleoprotein complexes that is likely to expand in the future.
Hitherto, a skeleton in the UHM family closet has been the remarkably similar core interactions with ULMs, which raise the conundrum of how specific UHM–ULM partnerships are arranged. Several answers to this puzzle have begun to fall into place. New structures of larger U2AF and SF1 complexes implicate regions adjoining the ULMs as accessories for UHM engagement (Wang et al. 2013; Zhang et al. 2013; Yoshida et al. 2015). Phosphorylation sites within and flanking the ULMs can regulate the marriage of phosphorylated SF1 with the U2AF65 UHM (Wang et al. 2013; Zhang et al. 2013) or exacerbate the divorce of the phosphorylated ATX1 ULM from the U2AF65 UHM in lieu of a 14-3-3 binding partner (Lim et al. 2008; de Chiara et al. 2009). Still questionable are the potential polygamous versus monogamous relationships of the multiple SF3b155 ULMs with different UHM-binding partners. Despite many exposés, whether new confessions of ULM extensions, phosphorylation-sensitivity, and cooperative binding will emerge as recurring dramas among UHM–ULM partners for now remain tantalizing family secrets. In the longer term, understanding the interplay among UHMs and ULMs may offer new therapeutic avenues for targeting the spliceosome (for review, see Bonnal et al. 2012), which has been highlighted recently for MYC-driven malignancies (Hirsch et al. 2015; Hsu et al. 2015; Koh et al. 2015) and already is promising for compounds that target the ULM-containing protein SF3b155 (for review, see Salton and Misteli 2016).
ACKNOWLEDGMENTS
This review was supported by National Institutes of Health grants GM070503 and GM117005 and an EvansMDS award to C.L.K.
Footnotes
Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.057950.116.
REFERENCES
- Adam SA, Nakagawa T, Swanson MS, Woodruff TK, Dreyfuss G. 1986. mRNA polyadenylate-binding protein: gene isolation and sequencing and identification of a ribonucleoprotein consensus sequence. Mol Cell Biol 6: 2932–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afroz T, Skrisovska L, Belloc E, Guillen-Boixet J, Mendez R, Allain FH. 2014. A fly trap mechanism provides sequence-specific RNA recognition by CPEB proteins. Genes Dev 28: 1498–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avis JM, Allain FH, Howe PW, Varani G, Nagai K, Neuhaus D. 1996. Solution structure of the N-terminal RNP domain of U1A protein: the role of C-terminal residues in structure stability and RNA binding. J Mol Biol 257: 398–411. [DOI] [PubMed] [Google Scholar]
- Bae E, Reiter NJ, Bingman CA, Kwan SS, Lee D, Phillips GN Jr, Butcher SE, Brow DA. 2007. Structure and interactions of the first three RNA recognition motifs of splicing factor prp24. J Mol Biol 367: 1447–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer WJ, Heath J, Jenkins JL, Kielkopf CL. 2012. Three RNA recognition motifs participate in RNA recognition and structural organization by the pro-apoptotic factor TIA-1. J Mol Biol 415: 727–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatter M, Dunin-Horkawicz S, Grishina I, Maris C, Thore S, Maier T, Bindereif A, Bujnicki JM, Allain FH. 2015. The signature of the five-stranded vRRM fold defined by functional, structural and computational analysis of the hnRNP L protein. J Mol Biol 427: 3001–3022. [DOI] [PubMed] [Google Scholar]
- Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S, Brunak S. 2004. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 4: 1633–1649. [DOI] [PubMed] [Google Scholar]
- Bonnal S, Vigevani L, Valcarcel J. 2012. The spliceosome as a target of novel antitumour drugs. Nat Rev Drug Discov 11: 847–859. [DOI] [PubMed] [Google Scholar]
- Bono F, Ebert J, Lorentzen E, Conti E. 2006. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell 126: 713–725. [DOI] [PubMed] [Google Scholar]
- Boudrez A, Beullens M, Waelkens E, Stalmans W, Bollen M. 2002. Phosphorylation-dependent interaction between the splicing factors SAP155 and NIPP1. J Biol Chem 277: 31834–31841. [DOI] [PubMed] [Google Scholar]
- Burley SK, Petsko GA. 1985. Aromatic-aromatic interaction: a mechanism of protein structure stabilization. Science 229: 23–28. [DOI] [PubMed] [Google Scholar]
- Cass DM, Berglund JA. 2006. The SF3b155 N-terminal domain is a scaffold important for splicing. Biochemistry 45: 10092–10101. [DOI] [PubMed] [Google Scholar]
- Chen HK, Fernandez-Funez P, Acevedo SF, Lam YC, Kaytor MD, Fernandez MH, Aitken A, Skoulakis EM, Orr HT, Botas J, et al. 2003. Interaction of Akt-phosphorylated ataxin-1 with 14–3–3 mediates neurodegeneration in spinocerebellar ataxia type 1. Cell 113: 457–468. [DOI] [PubMed] [Google Scholar]
- Corsini L, Bonnal S, Basquin J, Hothorn M, Scheffzek K, Valcarcel J, Sattler M. 2007. U2AF-homology motif interactions are required for alternative splicing regulation by SPF45. Nat Struct Mol Biol 14: 620–629. [DOI] [PubMed] [Google Scholar]
- Corsini L, Hothorn M, Stier G, Rybin V, Scheffzek K, Gibson TJ, Sattler M. 2009. Dimerization and protein binding specificity of the U2AF homology motif of the splicing factor Puf60. J Biol Chem 284: 630–639. [DOI] [PubMed] [Google Scholar]
- de Castro E, Sigrist CJ, Gattiker A, Bulliard V, Langendijk-Genevaux PS, Gasteiger E, Bairoch A, Hulo N. 2006. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res 34: W362–W365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Chiara C, Menon RP, Strom M, Gibson TJ, Pastore A. 2009. Phosphorylation of S776 and 14-3-3 binding modulate ataxin-1 interaction with splicing factors. PLoS One 4: e8372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkel H, Chica C, Via A, Gould CM, Jensen LJ, Gibson TJ, Diella F. 2011. Phospho.ELM: a database of phosphorylation sites—update 2011. Nucleic Acids Res 39: D261–D267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowhan DH, Hong EP, Auboeuf D, Dennis AP, Wilson MM, Berget SM, O'Malley BW. 2005. Steroid hormone receptor coactivation and alternative RNA splicing by U2AF65-related proteins CAPERα and CAPERβ. Mol Cell 17: 429–439. [DOI] [PubMed] [Google Scholar]
- Dreyfuss G, Swanson MS, Pinol-Roma S. 1988. Heterogeneous nuclear ribonucleoprotein particles and the pathway of mRNA formation. Trends Biochem Sci 13: 86–91. [DOI] [PubMed] [Google Scholar]
- Dutta J, Fan G, Gelinas C. 2008. CAPERα is a novel Rel-TAD-interacting factor that inhibits lymphocyte transformation by the potent Rel/NF-κB oncoprotein v-Rel. J Virol 82: 10792–10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ElAntak L, Tzakos AG, Locker N, Lukavsky PJ. 2007. Structure of eIF3b RNA recognition motif and its interaction with eIF3j: structural insights into the recruitment of eIF3b to the 40 S ribosomal subunit. J Biol Chem 282: 8165–8174. [DOI] [PubMed] [Google Scholar]
- ElAntak L, Wagner S, Herrmannova A, Karaskova M, Rutkai E, Lukavsky PJ, Valasek L. 2010. The indispensable N-terminal half of eIF3j/HCR1 cooperates with its structurally conserved binding partner eIF3b/PRT1-RRM and with eIF1A in stringent AUG selection. J Mol Biol 396: 1097–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreon ACM, Ferreon JC, Wright PE, Deniz AA. 2013. Modulation of allostery by protein intrinsic disorder. Nature 498: 390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, et al. 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44: D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fribourg S, Gatfield D, Izaurralde E, Conti E. 2003. A novel mode of RBD-protein recognition in the Y14-Mago complex. Nat Struct Biol 10: 433–439. [DOI] [PubMed] [Google Scholar]
- Galej WP, Wilkinson ME, Fica SM, Oubridge C, Newman AJ, Nagai K. 2016. Cryo-EM structure of the spliceosome immediately after branching. Nature 537: 197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozani O, Potashkin J, Reed R. 1998. A potential role for U2AF-SAP 155 interactions in recruiting U2 snRNP to the branch site. Mol Cell Biol 18: 4752–4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings ML, Allemand E, Duelli DM, Myers MP, Krainer AR. 2007. Control of pre-mRNA splicing by the general splicing factors PUF60 and U2AF65. PLoS One 2: e538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegele A, Kamburov A, Grossmann A, Sourlis C, Wowro S, Weimann M, Will CL, Pena V, Luhrmann R, Stelzl U. 2012. Dynamic protein–protein interaction wiring of the human spliceosome. Mol Cell 45: 567–580. [DOI] [PubMed] [Google Scholar]
- Hellemans J, Preobrazhenska O, Willaert A, Debeer P, Verdonk PC, Costa T, Janssens K, Menten B, Van Roy N, Vermeulen SJ, et al. 2004. Loss-of-function mutations in LEMD3 result in osteopoikilosis, Buschke-Ollendorff syndrome and melorheostosis. Nat Genet 36: 1213–1218. [DOI] [PubMed] [Google Scholar]
- Hirsch CL, Coban Akdemir Z, Wang L, Jayakumaran G, Trcka D, Weiss A, Hernandez JJ, Pan Q, Han H, Xu X, et al. 2015. Myc and SAGA rewire an alternative splicing network during early somatic cell reprogramming. Genes Dev 29: 803–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu TY, Simon LM, Neill NJ, Marcotte R, Sayad A, Bland CS, Echeverria GV, Sun T, Kurley SJ, Tyagi S, et al. 2015. The spliceosome is a therapeutic vulnerability in MYC-driven cancer. Nature 525: 384–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Zhou Z, Wang H, Kleinerman ES. 2012. CAPER-α alternative splicing regulates the expression of vascular endothelial growth VEGF165 in Ewing's sarcoma cells. Cancer 118: 2106–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson BP, Martinez-Yamout MA, Dyson HJ, Wright PE. 2004. Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat Struct Mol Biol 11: 257–264. [DOI] [PubMed] [Google Scholar]
- Inoue D, Bradley RK, Abdel-Wahab O. 2016. Spliceosomal gene mutations in myelodysplasia: molecular links to clonal abnormalities of hematopoiesis. Genes Dev 30: 989–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi A, Coelho MB, Kotik-Kogan O, Simpson PJ, Matthews SJ, Smith CW, Curry S. 2011. Crystallographic analysis of polypyrimidine tract-binding protein-Raver1 interactions involved in regulation of alternative splicing. Structure 19: 1816–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung DJ, Na SY, Na DS, Lee JW. 2002. Molecular cloning and characterization of CAPER, a novel coactivator of activating protein-1 and estrogen receptors. J Biol Chem 277: 1229–1234. [DOI] [PubMed] [Google Scholar]
- Kadlec J, Izaurralde E, Cusack S. 2004. The structural basis for the interaction between nonsense-mediated mRNA decay factors UPF2 and UPF3. Nat Struct Mol Biol 11: 330–337. [DOI] [PubMed] [Google Scholar]
- Kielkopf CL, Rodionova NA, Green MR, Burley SK. 2001. A novel peptide recognition mode revealed by the X-ray structure of a core U2AF35/U2AF65 heterodimer. Cell 106: 595–605. [DOI] [PubMed] [Google Scholar]
- Kielkopf CL, Lucke S, Green MR. 2004. U2AF homology motifs: protein recognition in the RRM world. Genes Dev 18: 1513–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh CM, Bezzi M, Low DH, Ang WX, Teo SX, Gay FP, Al-Haddawi M, Tan SY, Osato M, Sabo A, et al. 2015. MYC regulates the core pre-mRNA splicing machinery as an essential step in lymphomagenesis. Nature 523: 96–100. [DOI] [PubMed] [Google Scholar]
- Konde E, Bourgeois B, Tellier-Lebegue C, Wu W, Perez J, Caputo S, Attanda W, Gasparini S, Charbonnier JB, Gilquin B, et al. 2010. Structural analysis of the Smad2-MAN1 interaction that regulates transforming growth factor-β signaling at the inner nuclear membrane. Biochemistry 49: 8020–8032. [DOI] [PubMed] [Google Scholar]
- Lahiri DK, Thomas JO. 1986. A cDNA clone of the hnRNP C proteins and its homology with the single-stranded DNA binding protein UP2. Nucleic Acids Res 14: 4077–4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallena MJ, Chalmers KJ, Llamazares S, Lamond AI, Valcarcel J. 2002. Splicing regulation at the second catalytic step by Sex-lethal involves 3′ splice site recognition by SPF45. Cell 109: 285–296. [DOI] [PubMed] [Google Scholar]
- Lau CK, Diem MD, Dreyfuss G, Van Duyne GD. 2003. Structure of the Y14-Magoh core of the exon junction complex. Curr Biol 13: 933–941. [DOI] [PubMed] [Google Scholar]
- Lim J, Hao T, Shaw C, Patel AJ, Szabo G, Rual JF, Fisk CJ, Li N, Smolyar A, Hill DE, et al. 2006. A protein–protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell 125: 801–814. [DOI] [PubMed] [Google Scholar]
- Lim J, Crespo-Barreto J, Jafar-Nejad P, Bowman AB, Richman R, Hill DE, Orr HT, Zoghbi HY. 2008. Opposing effects of polyglutamine expansion on native protein complexes contribute to SCA1. Nature 452: 713–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F, Morrison JM, Wu W, Worman HJ. 2005. MAN1, an integral protein of the inner nuclear membrane, binds Smad2 and Smad3 and antagonizes transforming growth factor-β signaling. Hum Mol Genet 14: 437–445. [DOI] [PubMed] [Google Scholar]
- Liu J, He L, Collins I, Ge H, Libutti D, Li J, Egly JM, Levens D. 2000. The FBP interacting repressor targets TFIIH to inhibit activated transcription. Mol Cell 5: 331–341. [DOI] [PubMed] [Google Scholar]
- Liu J, Akoulitchev S, Weber A, Ge H, Chuikov S, Libutti D, Wang XW, Conaway JW, Harris CC, Conaway RC, et al. 2001. Defective interplay of activators and repressors with TFIH in xeroderma pigmentosum. Cell 104: 353–363. [DOI] [PubMed] [Google Scholar]
- Loerch S, Kielkopf CL. 2015. Dividing and conquering the family of RNA recognition motifs: a representative case based on hnRNP L. J Mol Biol 427: 2997–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loerch S, Maucuer A, Manceau V, Green MR, Kielkopf CL. 2014. Cancer-relevant splicing factor CAPERα engages the essential splicing factor SF3b155 in a specific ternary complex. J Biol Chem 289: 17325–17337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madan V, Kanojia D, Li J, Okamoto R, Sato-Otsubo A, Kohlmann A, Sanada M, Grossmann V, Sundaresan J, Shiraishi Y, et al. 2015. Aberrant splicing of U12-type introns is the hallmark of ZRSR2 mutant myelodysplastic syndrome. Nat Commun 6: 6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manceau V, Swenson M, Le Caer JP, Sobel A, Kielkopf CL, Maucuer A. 2006. Major phosphorylation of SF1 on adjacent Ser-Pro motifs enhances interaction with U2AF65. FEBS J 273: 577–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manceau V, Kielkopf CL, Sobel A, Maucuer A. 2008. Different requirements of the kinase and UHM domains of KIS for its nuclear localization and binding to splicing factors. J Mol Biol 381: 748–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manceau V, Kremmer E, Nabel EG, Maucuer A. 2012. The protein kinase KIS impacts gene expression during development and fear conditioning in adult mice. PLoS One 7: e43946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris C, Dominguez C, Allain FH. 2005. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J 272: 2118–2131. [DOI] [PubMed] [Google Scholar]
- McLaughlin KJ, Jenkins JL, Kielkopf CL. 2011. Large favorable enthalpy changes drive specific RNA recognition by RNA recognition motif proteins. Biochemistry 50: 1429–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merendino L, Guth S, Bilbao D, Martinez C, Valcarcel J. 1999. Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature 402: 838–841. [DOI] [PubMed] [Google Scholar]
- Montemayor EJ, Curran EC, Liao HH, Andrews KL, Treba CN, Butcher SE, Brow DA. 2014. Core structure of the U6 small nuclear ribonucleoprotein at 1.7-Å resolution. Nat Struct Mol Biol 21: 544–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naro C, Sette C. 2013. Phosphorylation-mediated regulation of alternative splicing in cancer. Int J Cell Biol 2013: 151839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicodeme P. 2001. Fast approximate motif statistics. J Comput Biol 8: 235–248. [DOI] [PubMed] [Google Scholar]
- Okeyo-Owuor T, White BS, Chatrikhi R, Mohan DR, Kim S, Griffith M, Ding L, Ketkar-Kulkarni S, Hundal J, Laird KM, et al. 2015. U2AF1 mutations alter sequence specificity of pre-mRNA binding and splicing. Leukemia 29: 909–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oubridge C, Ito N, Evans PR, Teo CH, Nagai K. 1994. Crystal structure at 1.92 Å resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature 372: 432–438. [DOI] [PubMed] [Google Scholar]
- Page-McCaw PS, Amonlirdviman K, Sharp PA. 1999. PUF60: a novel U2AF65-related splicing activity. RNA 5: 1548–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D, Estevez-Salmeron LD, Stroschein SL, Zhu X, He J, Zhou S, Luo K. 2005. The integral inner nuclear membrane protein MAN1 physically interacts with the R-Smad proteins to repress signaling by the transforming growth factor-β superfamily of cytokines. J Biol Chem 280: 15992–16001. [DOI] [PubMed] [Google Scholar]
- Price SR, Evans PR, Nagai K. 1998. Crystal structure of the spliceosomal U2B′′-U2A′ protein complex bound to a fragment of U2 small nuclear RNA. Nature 394: 645–650. [DOI] [PubMed] [Google Scholar]
- Rauhut R, Fabrizio P, Dybkov O, Hartmuth K, Pena V, Chari A, Kumar V, Lee CT, Urlaub H, Kastner B, et al. 2016. Molecular architecture of the Saccharomyces cerevisiae activated spliceosome. Science 353: 1399–1405. [DOI] [PubMed] [Google Scholar]
- Rideau AP, Gooding C, Simpson PJ, Monie TP, Lorenz M, Huttelmaier S, Singer RH, Matthews S, Curry S, Smith CW. 2006. A peptide motif in Raver1 mediates splicing repression by interaction with the PTB RRM2 domain. Nat Struct Mol Biol 13: 839–848. [DOI] [PubMed] [Google Scholar]
- Rudner DZ, Kanaar R, Breger KS, Rio DC. 1996. Mutations in the small subunit of the Drosophila U2AF splicing factor cause lethality and developmental defects. Proc Natl Acad Sci 93: 10333–10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupert PB, Xiao H, Ferre-D'Amare AR. 2003. U1A RNA-binding domain at 1.8 A resolution. Acta Crystallogr D Biol Crystallogr 59: 1521–1524. [DOI] [PubMed] [Google Scholar]
- Sachs AB, Bond MW, Kornberg RD. 1986. A single gene from yeast for both nuclear and cytoplasmic polyadenylate-binding proteins: domain structure and expression. Cell 45: 827–835. [DOI] [PubMed] [Google Scholar]
- Salton M, Misteli T. 2016. Small molecule modulators of pre-mRNA splicing in cancer therapy. Trends Mol Med 22: 28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellenberg MJ, Edwards RA, Ritchie DB, Kent OA, Golas MM, Stark H, Luhrmann R, Glover JN, MacMillan AM. 2006. Crystal structure of a core spliceosomal protein interface. Proc Natl Acad Sci 103: 1266–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seghezzi W, Chua K, Shanahan F, Gozani O, Reed R, Lees E. 1998. Cyclin E associates with components of the pre-mRNA splicing machinery in mammalian cells. Mol Cell Biol 18: 4526–4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selenko P, Gregorovic G, Sprangers R, Stier G, Rhani Z, Kramer A, Sattler M. 2003. Structural basis for the molecular recognition between human splicing factors U2AF65 and SF1/mBBP. Mol Cell 11: 965–976. [DOI] [PubMed] [Google Scholar]
- Shen H, Zheng X, Luecke S, Green MR. 2010. The U2AF35-related protein Urp contacts the 3′ splice site to promote U12-type intron splicing and the second step of U2-type intron splicing. Genes Dev 24: 2389–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Xu RM. 2003. Crystal structure of the Drosophila Mago nashi-Y14 complex. Genes Dev 17: 971–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Reddy B, Manley JL. 2006. PP1/PP2A phosphatases are required for the second step of pre-mRNA splicing and target specific snRNP proteins. Mol Cell 23: 819–829. [DOI] [PubMed] [Google Scholar]
- Sickmier EA, Frato KE, Shen H, Paranawithana SR, Green MR, Kielkopf CL. 2006. Structural basis for polypyrimidine tract recognition by the essential pre-mRNA splicing factor U2AF65. Mol Cell 23: 49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares LM, Zanier K, Mackereth C, Sattler M, Valcarcel J. 2006. Intron removal requires proofreading of U2AF/3′ splice site recognition by DEK. Science 312: 1961–1965. [DOI] [PubMed] [Google Scholar]
- Spadaccini R, Reidt U, Dybkov O, Will C, Frank R, Stier G, Corsini L, Wahl MC, Luhrmann R, Sattler M. 2006. Biochemical and NMR analyses of an SF3b155-p14-U2AF-RNA interaction network involved in branch point definition during pre-mRNA splicing. RNA 12: 410–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanyuk GA, Serrano P, Peralta E, Farr CL, Axelrod HL, Geralt M, Das D, Chiu HJ, Jaroszewski L, Deacon AM, et al. 2016. UHM-ULM interactions in the RBM39-U2AF65 splicing-factor complex. Acta Crystallogr D Struct Biol 72: 497–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teplova M, Patel DJ. 2008. Structural insights into RNA recognition by the alternative-splicing regulator muscleblind-like MBNL1. Nat Struct Mol Biol 15: 1343–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thickman KR, Swenson MC, Kabogo JM, Gryczynski Z, Kielkopf CL. 2006. Multiple U2AF65 binding sites within SF3b155: thermodynamic and spectroscopic characterization of protein–protein interactions among pre-mRNA splicing factors. J Mol Biol 356: 664–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripsianes K, Friberg A, Barrandon C, Brooks M, van Tilbeurgh H, Seraphin B, Sattler M. 2014. A novel protein-protein interaction in the RES (REtention and Splicing) complex. J Biol Chem 289: 28640–28650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunnicliffe RB, Hautbergue GM, Kalra P, Jackson BR, Whitehouse A, Wilson SA, Golovanov AP. 2011. Structural basis for the recognition of cellular mRNA export factor REF by herpes viral proteins HSV-1 ICP27 and HVS ORF57. PLoS Pathog 7: e1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunnicliffe RB, Hautbergue GM, Wilson SA, Kalra P, Golovanov AP. 2014. Competitive and cooperative interactions mediate RNA transfer from herpesvirus saimiri ORF57 to the mammalian export adaptor ALYREF. PLoS Pathog 10: e1003907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan R, Yan C, Bai R, Huang G, Shi Y. 2016a. Structure of a yeast catalytic step I spliceosome at 3.4 Å resolution. Science 353: 895–904. [DOI] [PubMed] [Google Scholar]
- Wan R, Yan C, Bai R, Wang L, Huang M, Wong CC, Shi Y. 2016b. The 3.8 Å structure of the U4/U6.U5 tri-snRNP: insights into spliceosome assembly and catalysis. Science 351: 466–475. [DOI] [PubMed] [Google Scholar]
- Wang C, Chua K, Seghezzi W, Lees E, Gozani O, Reed R. 1998. Phosphorylation of spliceosomal protein SAP 155 coupled with splicing catalysis. Genes Dev 12: 1409–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Bruderer S, Rafi Z, Xue J, Milburn PJ, Kramer A, Robinson PJ. 1999. Phosphorylation of splicing factor SF1 on Ser20 by cGMP-dependent protein kinase regulates spliceosome assembly. EMBO J 18: 4549–4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Maucuer A, Gupta A, Manceau V, Thickman KR, Bauer WJ, Kennedy SD, Wedekind JE, Green MR, Kielkopf CL. 2013. Structure of phosphorylated SF1 bound to U2AF65 in an essential splicing factor complex. Structure 21: 197–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber C, Schreiber TB, Daub H. 2012. Dual phosphoproteomics and chemical proteomics analysis of erlotinib and gefitinib interference in acute myeloid leukemia cells. J Proteomics 75: 1343–1356. [DOI] [PubMed] [Google Scholar]
- Will CL, Schneider C, MacMillan AM, Katopodis NF, Neubauer G, Wilm M, Luhrmann R, Query CC. 2001. A novel U2 and U11/U12 snRNP protein that associates with the pre-mRNA branch site. EMBO J 20: 4536–4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright PE, Dyson HJ. 2015. Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol 16: 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Romfo CM, Nilsen TW, Green MR. 1999. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature 402: 832–835. [DOI] [PubMed] [Google Scholar]
- Wulf G, Finn G, Suizu F, Lu KP. 2005. Phosphorylation-specific prolyl isomerization: is there an underlying theme? Nat Cell Biol 7: 435–441. [DOI] [PubMed] [Google Scholar]
- Wysoczanski P, Schneider C, Xiang S, Munari F, Trowitzsch S, Wahl MC, Luhrmann R, Becker S, Zweckstetter M. 2014. Cooperative structure of the heterotrimeric pre-mRNA retention and splicing complex. Nat Struct Mol Biol 21: 911–918. [DOI] [PubMed] [Google Scholar]
- Yan C, Hang J, Wan R, Huang M, Wong CC, Shi Y. 2015. Structure of a yeast spliceosome at 3.6-angstrom resolution. Science 349: 1182–1191. [DOI] [PubMed] [Google Scholar]
- Yan C, Wan R, Bai R, Huang G, Shi Y. 2016. Structure of a yeast activated spliceosome at 3.5 Å resolution. Science 353: 904–911. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Park SY, Oda T, Akiyoshi T, Sato M, Shirouzu M, Tsuda K, Kuwasako K, Unzai S, Muto Y, et al. 2015. A novel 3′ splice site recognition by the two zinc fingers in the U2AF small subunit. Genes Dev 29: 1649–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore PD, Patton JG, Green MR. 1992. Cloning and domain structure of the mammalian splicing factor U2AF. Nature 355: 609–614. [DOI] [PubMed] [Google Scholar]
- Zhang M, Zamore PD, Carmo-Fonseca M, Lamond AI, Green MR. 1992. Cloning and intracellular localization of the U2 small nuclear ribonucleoprotein auxiliary factor small subunit. Proc Natl Acad Sci 89: 8769–8773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Madl T, Bagdiul I, Kern T, Kang HS, Zou P, Mausbacher N, Sieber SA, Kramer A, Sattler M. 2013. Structure, phosphorylation and U2AF65 binding of the N-terminal domain of splicing factor 1 during 3′-splice site recognition. Nucleic Acids Res 41: 1343–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Fu XD. 2013. Regulation of splicing by SR proteins and SR protein-specific kinases. Chromosoma 122: 191–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorio DA, Blumenthal T. 1999. Both subunits of U2AF recognize the 3′ splice site in Caenorhabditis elegans. Nature 402: 835–838. [DOI] [PubMed] [Google Scholar]


