Abstract
We have previously shown that in vitro sensitivity to dexamethasone (DEX) stimulation in human endothelial cells is positively regulated by the glucocorticoid receptor (NR3C1, GR). The present study determined the role of differential GR transcriptional regulation in glucocorticoid sensitivity. We studied 25 human umbilical vein endothelial cells (HUVECs) that had been previously characterized as DEX-sensitive (n = 15), or resistant (n = 10). Real-time PCR analysis of GR 5′UTR mRNA isoforms showed that all HUVECs expressed isoforms 1B, 1C, 1D, 1F, and 1H, and isoforms 1B and 1C were predominantly expressed. DEX-resistant cells expressed higher basal levels of the 5′UTR mRNA isoforms 1C and 1D, but lower levels of the 5′UTR mRNA isoform 1F than DEX-sensitive cells. DEX treatment significantly decreased GRα and GR-1C mRNA isoform expression in DEX-resistant cells only. Reporter luciferase assays indicated that differential GR mRNA isoform expression was not due to differential promoter usage between DEX-sensitive and DEX-resistant cells. Analysis of promoter methylation, however, showed that DEX-sensitive cells have higher methylation levels of promoter 1D and lower methylation levels of promoter 1F than DEX-resistant cells. Treatment with 5-aza-2-deoxycytidine abolished the differential 5′UTR mRNA isoform expression between DEX-sensitive and DEX-resistant cells. Finally, both GRα overexpression and 5-aza-2-deoxycytidine treatment eliminated the differences between sensitivity groups to DEX-mediated downregulation of endothelial nitric oxide synthase (NOS3), and upregulation of plasminogen activator inhibitor 1 (SERPINE1). In sum, human endothelial GR 5′UTR mRNA expression is regulated by promoter methylation with DEX-sensitive and DEX-resistant cells having different GR promoter methylation patterns.
Keywords: glucocorticoids, endothelium, methylation, 5′UTR mRNA isoform
Introduction
Glucocorticoids are therapeutic agents used for reducing inflammation via targeting the glucocorticoid receptor (NR3C1, GR) of immune cells (Auphan et al. 1995, Barnes 1998). However, glucocorticoids also target the GR present in various other tissues, causing unwanted side effects. In the cardiovascular system, glucocorticoids induce short-term side effects that include hypertension, dyslipidemia and thrombosis. Chronic synthetic glucocorticoid-therapy has been associated with endothelial dysfunction and increased risk of cardiovascular events such as myocardial infarction (Yang & Zhang 2004, Walker 2007, Hadoke et al. 2009, Huang & Glass 2010, Kadmiel & Cidlowski 2013). These adverse events are mediated in part by glucocorticoid-dependent down-regulation of endothelial nitric oxide synthase (NOS3) and upregulation of plasminogen activator inhibitor 1 (SERPINE1) (Wallerath et al. 2004, Tamura et al. 2015). Of importance, human studies have revealed significant human variability in response to both endogenous (cortisol) and synthetic glucocorticoids (Ito et al. 2006, Kino 2007), but the mechanisms remain undetermined.
Glucocorticoids mediate their biological effects by the ubiquitously expressed GR (Bamberger et al. 1996, Adcock et al. 2004, Heitzer et al. 2007). Alternative splicing produces 3′UTR mRNA isoforms GRα, GRβ and GRP. GRα protein is the biologically relevant isoform, capable of binding ligand that regulates transcription and stability of target genes. GRβ is localized in the nucleus and has a dominant negative effect on GRα through the formation of GRα/GRβ heterodimers. GRP has been reported to have both synergistic and antagonistic effects on GRα (Kassel & Herrlich 2007, Biddie et al. 2012). Previously, we have shown that human endothelial sensitivity to dexamethasone (DEX) varied according to the levels of GRα protein degradation (Mata-Greenwood et al. 2013). Human umbilical vein endothelial cells (HUVECs) that were resistant to in vitro stimulation by DEX had an increased expression of the E3 ubiquitin ligase gene BCL2-athanogene 1 (BAG1) and subsequent GR protein ubiquitination and proteasomal degradation. It was found that the basal protein levels of GRα correlated with the endothelial expression of key genes, such as NOS3 and SERPINE1 (Mata-Greenwood et al. 2013). However, the transcriptional regulation of GR expression in human endothelial cells and its role in endothelial glucocorticoid sensitivity have not been well defined, and warrant further exploration.
GR mRNA expression is regulated by complex mechanisms that include transcriptional and epigenetic modification of GR promoters (Presul et al. 2007, Turner et al. 2010, Cao-Lei et al. 2011). The GR gene (NR3C1) contains at least nine 5′UTR first exons that are spliced to the common exon 2. The alternative first exons are divided into two promoter regions: a distal region located more than 30 kb upstream that contains exons 1A and 1I, and a proximal region located 5 kb upstream of the translation start site that contains exons 1B, 1C, 1D, 1E, 1F, 1H, and 1J. Each first exon is regulated by its own promoter that binds specific transcription factor complexes (Bockmuhl et al. 2011, Cao-Lei et al. 2011). Therefore, the transcription of the GR gene is regulated by complex promoter regions containing multiple transcription start sites. Furthermore, both proximal and distal GR promoters are embedded in a CpG island that is mostly methylation-free and therefore can be targeted for methylation. Recent human and rodent studies have shown tissue and disease specific expression of untranslated first exons, due in part to changes in GR promoter methylation (Turner et al. 2006, 2008, 2010, Bockmuhl et al. 2011). These studies have shown that the various GR promoters allow for tissue-specific mechanisms to control and adapt GR expression levels according to various developmental and pathophysiological stages.
Previous studies have shown that rodent heart and aortic tissue express significant levels of GR transcripts (Presul et al. 2007, Xiong & Zhang 2013), but the transcriptional regulation of GR in specific cardiovascular cells (i.e., endothelial, smooth muscle, cardiomyocytes) remains unknown. Therefore, the main objective of the present study is to uncover the mechanisms that regulate the expression of GR mRNA isoforms in human endothelial cells that have been previously characterized as DEX-sensitive or DEX-resistant. We find that DNA methylation plays an important role in the fine-tuning determination of GR mRNA isoform expression in human endothelial cells. Our study provides novel insights into the upstream mechanisms that alter GR expression in the cardiovascular system and suggest that future use of epigenetic markers will aid in identifying glucocorticoid responsive individuals.
Materials and methods
Subjects and cell culture
HUVECs isolated from 25 healthy term pregnancies were selected from a previous cohort study of 42 subjects (Mata-Greenwood et al. 2013) to further characterize the transcriptional regulation of GR in human endothelial cells. These HUVECs were previously characterized for endothelial cell purity and in vitro response to DEX (Mata-Greenwood et al. 2013). The study subject characteristics are summarized in Supplementary Table 1, see section on supplementary data given at the end of this article. The study was approved by the IRBs of the Loma Linda University and the University of California at San Diego. HUVECs were cultured using ECM media (ScienCell, Carlsbad, CA, USA). All assays were performed between passages 4 and 6. To study inter-individual differences in response to glucocorticoids, we stimulated confluent and quiescent HUVECs with DEX using DMSO as a solvent (final concentration of 0.05%). This synthetic glucocorticoid was chosen for its stability in the presence of cellular 11-β-dehydrogenase, and for its specificity for GRα binding with respect to other nuclear receptors, including the mineralocorticoid receptor (Wenting-van Wijk et al. 1999). To starve HUVECs, we used M199 (Sigma–Aldrich) supplemented with 0.95 mM HEPES, 0.1% BSA, 1% antibiotics and 1% fetal bovine serum (FBS). The acute monocytic leukemia cell line THP-1 from Sigma–Aldrich was used as a positive control as it is known to express all GR mRNA isoforms (Steer et al. 2000). THP-1 cells were grown in DMEM with 1% antibiotics and 10% FBS.
RNA extraction and real-time PCR
Total RNA was extracted with TriZOL-RNEasy kits (Invitrogen), quantified, and stored at −80 °C until analysis. The total RNA (1 µg) was reverse transcribed and real time PCR was performed for each sample in triplicate as previously described (Goyal et al. 2014). Taqman assays were obtained from Life Technologies for GR mRNA isoforms 1B (Hs01005211_m1), 1C (Hs01010775_m1), 1D (Hs03666144_m1) and the housekeeping gene 18S (Hs99999901_s1), and these were run according to the manufacturer’s recommendations using a Probe Quantitect PCR mix (Qiagen). Designed primers for SYBR green PCR analysis of the remaining GR mRNA isoforms and the housekeeping gene 18S, together with PCR conditions and accession numbers, are shown in Supplementary Table 2, see section on supplementary data given at the end of this article. SYBR green PCR was performed using Quantitect SYBR green PCR kit (Qiagen) using a 2.0 Lightcycler amplifier (Roche). PCR products were purified, sequenced, and used to obtain standard curves for each mRNA isoform. Extrapolation of unknowns from the standard curve was performed using Prism 3 (GraphPad Software, San Diego, CA, USA), predicting unknowns from the standard curve Ct values. Data is presented as fg mRNA/ng 18S. Alternatively, the percentages of each GR mRNA isoform were estimated for each subject as mRNA isoform/sum of all mRNA isoforms × 100.
SDS–PAGE and immunoblotting
Western blotting was performed as previously described (Mata-Greenwood et al. 2010). Briefly, protein extracts (30 µg) were prepared in a cold lysis buffer, heat denatured in a Laemmli buffer, separated on SDS-PAGE, and transferred to PVDF membranes. Membranes were blocked in 5% non-fat dried milk in 0.05% TBST for 1 h, and then probed in primary rabbit anti-GR (Santa Cruz Biotechnologies), monoclonal anti-BAG1 (Santa Cruz Biotechnologies), monoclonal anti-HSP90 (BD Biosciences, San Jose, CA, USA), and rabbit anti-FKBP51 (Stressmarq, Victoria, BC, Canada), diluted in blocking buffer (1 µg/ml) overnight at 4 °C. After three 10 min washes with TBST, the membranes were incubated with secondary antibodies that were diluted at 1:2000. Bound antibodies were visualized using the chemiluminscence substrate (ThermoFisher Scientific, Carlsbad, CA, USA). The membranes were then probed with monoclonal anti-β-Actin (Ambion, Austin, TX, USA). Data is presented as protein levels relative to β-Actin levels.
Cloning of GR promoters
GR promoters B, C, D, F, and H were cloned into the firefly luciferase-pGL3 reporter vectors (Promega), as previously described (Mata-Greenwood et al. 2010). GR promoter regions were selected to include putative essential DNA elements such as initiator (Inr) and downstream promoters (DPE). Briefly, 500 ng of HUVEC DNA (from specific donors) was used to amplify GR promoter regions using primers that contained specific restriction enzyme sites (Supplementary Table 3, see section on supplementary data given at the end of this article), followed by ligation into the basic luciferase pGL3 vector. The non-methylated GR promoter inserts were confirmed by DNA sequencing. Vector clones containing WT GR promoters were used.
Cell transfection and luciferase assays
Transfection of plasmid DNA was performed with the aid of HD Xtreme transfection reagent (Roche) as previously described (Mata-Greenwood et al. 2013). Briefly, confluent cells were transfected with the GR promoter/pGL3-luciferase constructs, using a 1:1 complex of DNA:Xtreme reagent according to the manufacturer’s protocol. TK-Renilla luciferase vector (Promega Corp.) was used as the internal control. The transfection was carried out at 37 °C for 6 h. The cells were allowed to recover in a complete culture medium for 18–20 h, and then treated with starvation media with or without DEX (0.2 and 1 µM) for another 24 h, before adding passive lysis. Firefly and Renilla luciferase activities were measured using a dual-reporter assay kit (Promega) according to the manufacturer’s protocol. Relative luciferase values were calculated as a ratio of firefly/renilla luciferase activities. Each treatment was tested in quadruplicates and averages per sample were used to calculate group averages and standard errors.
Methyl-DNA immunoprecipitation
Genomic DNA was isolated from confluent and quiescent HUVECs using the Wizard genomic DNA isolation kit (Promega). Methyl-DNA immunoprecipitation (MeDIP) was performed with the aid of a commercial kit according to the manufacturer’s protocol (Active Motif, Carlsbad, CA, USA). In brief, 20 µg of DNA was sheared into ~ 200–400 bp fragments and immunoprecipitated with a 5-methylcytosine-specific antibody at 4 °C overnight. Immunoprecipitated DNA was bound to magnetic beads, washed and eluted. Input (sheared DNA) and immunoprecipitated DNA (1 µl) was used to amplify specific promoter regions from the GR gene using SYBR green PCR, as previously described. MeDIP real-time PCR primers are shown in Supplementary Table 4, see section on supplementary data given at the end of this article. Promoter methylation levels were estimated as immunoprecipitated DNA/total (input) DNA × 100.
Bisulfite-converted DNA sequencing
To determine the methylation status of GR promoters, DNA samples were treated with bisulfite using the epiTect Bisulfite kit (Qiagen), according to the manufacturer’s instructions. Converted DNA was amplified using primers directed to bisulfite modified genomic DNA (Supplementary Table 4). Amplification conditions were as follows: 94 °C for 30 s, 40 °C for 30 s, and 72 °C for 40 sX40 cycles. Resulting amplicons were cloned into pCR4-TOPO sequencing vectors (Invitrogen) for automated fluorescent sequencing. Data were analyzed using the CLC Software, and clones showing <90% conversion or identified as clonal were not included in further analysis. The methylation levels for each subject were estimated as the methylated clones/#total clones sequenced × 100. A minimum of ten clones per amplicon were analyzed by sequencing. Averages and standard errors were then calculated for each sensitivity group.
Overexpression of GRα
Mammalian expressing vectors for the promoter-less full length GR-1Cα (NM_000176.2) were a gift from Dr John A. Cidlowski (NIEHS, Research Triangle, NC, USA). GR over-expression was achieved by transfecting log-phase proliferating HUVECs with 1 µg DNA/six well using the Xtreme transfection reagent as previously described. After 18 h of recovery, HUVECs were treated with DEX (1 µM) or not for an additional 24 h before analysis. Overexpression of GRα was confirmed by real-time PCR and immunoblotting.
Global demethylation assays
To induce global demethylation, we utilized a 5 µM dose of 5′-aza-2′-deoxycitidine (AZA) in 50% confluent cells. After 24 h of AZA treatment, cells were treated with both AZA and DEX (1 µM) or solvent for another 24 h before harvesting total RNA. DNA demethylation was confirmed by GR promoter D methylation levels of < 1%.
Procoagulant activity assay
The procoagulant activity of HUVECs was tested by a one-step recalcification time test, also known as the activated partial thromboplastin time (aPTT) as previously described (Mata-Greenwood et al. 2013). As discussed, 70% confluent HUVECs were transfected with GRα-expressing vectors, and 18 h after were treated with or without DEX (1 µM) for an additional 24 h. Cells were then washed, trypsinized, and resuspended in PBS at a concentration of one million cells per ml PBS. A 1:1 solution was prepared with cells and fresh plasma (collected from one single donor and anticoagulated with 3.8% sodium citrate, 1:9, v/v) and incubated for 180 s at 37 °C. After the addition of preheated CaCl2, the time to fibrin strand generation was recorded by a BCS XP hemostatic analyzer (Siemens, Munich, Germany).
Statistical analysis
Data are presented as means±s.e.m. Each individual variable was analyzed via ANOVA followed by post-hoc least significant difference (LSD) analysis to determine differences between sensitivity groups and between treatment groups. Homogeneity of variances was confirmed (P>0.05) using the Levene’s Test of Equality of Error Variance. For data sets with unequal variances (P<0.05), the data were log transformed and all further statistics were performed on the transformed data. For graphical presentation, the anti-logs of the mean log values were calculated and plotted. In all cases, statistical power was >0.08. A P value of <0.05 was regarded as significant. All statistical analysis was performed using SPSS, version 22.
Results
Differential GR mRNA isoform expression in HUVECs according to DEX-sensitivity
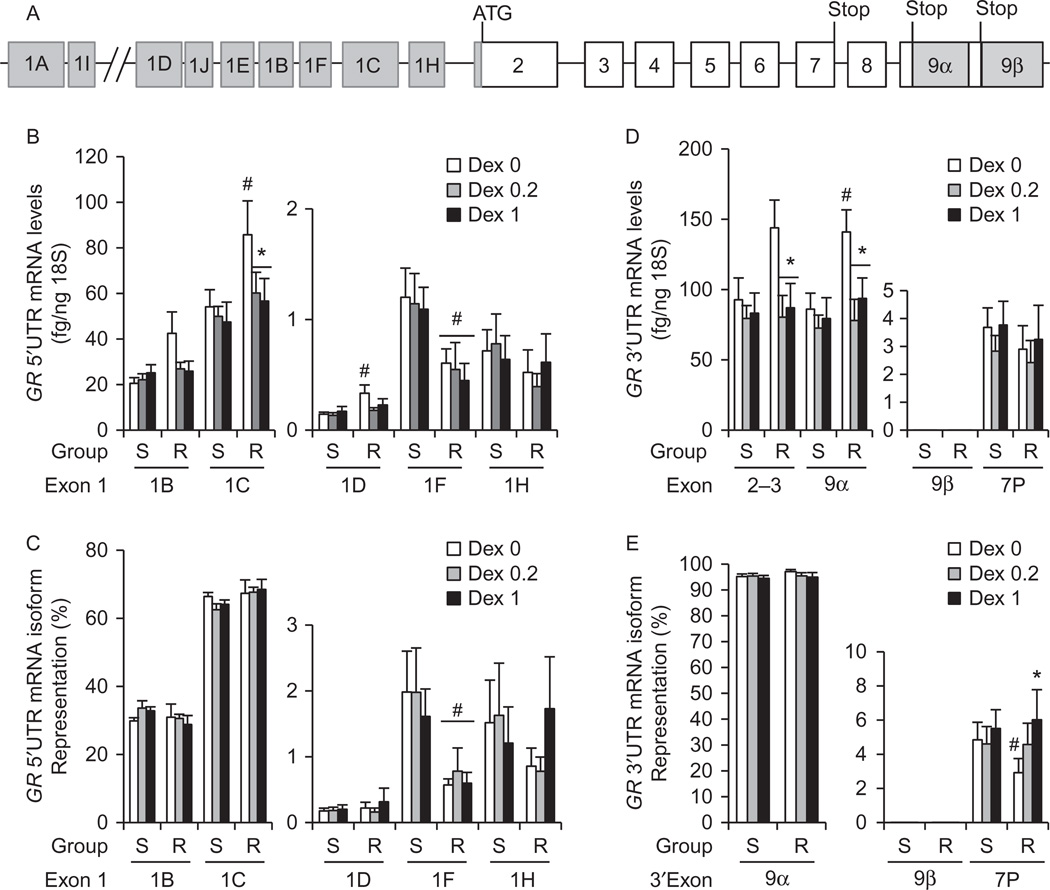
We have previously characterized a small cohort of 42 HUVECs obtained from healthy pregnancies for DEX-sensitivity. DEX-sensitive HUVECs expressed significantly higher levels of GRα protein than DEX-resistant HUVECs (Mata-Greenwood et al. 2013). Analysis of study group characteristics indicated that the pre-pregnancy maternal BMI and average systolic blood pressure were significantly higher in DEX-sensitive HUVECs compared to those of DEX-resistant cells (Supplementary Table 1), but the remaining parameters were not significantly different. To further understand the transcriptional regulation of GR in endothelial cells, and the mechanisms leading to the differences in GR expression between DEX-sensitive and DEX-resistant HUVECs, we analyzed the expression of 5′UTR mRNA isoform transcripts in 25 HUVECs (15 DEX-sensitive and 10 DEX-resistant). HUVECs expressed GR proximal mRNA isoforms 1B, 1C, 1D, 1F, and 1H, but did not express the distal isoforms 1A and 1I, or the proximal isoforms 1E and 1J (Fig. 1A and B). DEX-resistant HUVECs expressed significantly higher basal, but similar DEX-stimulated, GR mRNA isoform 1C levels (Fig. 1B); and lower percentages of mRNA isoform 1F levels (Fig. 1C) than DEX-sensitive cells. DEX treatment significantly decreased the expression of mRNA isoform 1C in DEX-resistant cells only (Fig. 1B). The percentages of GR 5′UTR mRNA isoforms in all HUVECs were ~67% (1C), ~30% (1B), ~2% (1F), ~1% (1H), and ~0.2% (1D) (Fig. 1). DEX treatment did not significantly alter the percentages of GR 5′UTR mRNA isoforms in either DEX-sensitive or DEX-resistant cells (Fig. 1C).
Figure 1.
Basal and DEX-stimulated expression of GR mRNA transcripts in DEX-sensitive (S) and DEX-resistant (R) HUVECs. (A) Structure of the human glucocorticoid receptor gene (NR3C1) that includes 9 exons 1 and 3 stop signals that result in both 5′UTR and 3′UTR mRNA isoforms. (B, C, D and E) Confluent and quiescent HUVECs were treated with solvent, DEX 0.2, and DEX1 µM for 24 h and GR mRNA isoforms were quantified by real-time PCR as described under methods. (B) Basal and DEX-stimulated expression levels of GR 5′UTR mRNA isoforms (1B, 1C, 1D, 1F, and 1H) expressed as fg GR isoform/ng 18S RNA. (C) Expression of GR 5′UTR mRNA isoforms as percent of total GR mRNA. (D) Basal and DEX-stimulated expression of GR 3′UTR mRNA isoforms (GRα, GRP and GRβ) as fg GR isoform/ng 18S RNA; (E) Expression of GR 3′UTR mRNA isoforms as percent of total GR mRNA. THP1 monocytoid cells were used as a positive control to verify that HUVECs do not express isoforms 1A, 1E, 1I and 1J. Bars represent the average±s.e.m. (n = 15 DEX-sensitive and 10 DEX-resistant). *P<0.05 DEX-treated vs untreated; #P<0.05 DEX-resistant cells vs DEX-sensitive cells.
We also analyzed the expression profile of GR 3′UTR mRNA isoforms GRα, GRβ and GRP. GRα represented ~ 96% of all 3′UTR mRNA isoforms, followed by GRP (4%), and GRβ expression was slight (<0.0006%) in all HUVECs (Fig. 1E). DEX-resistant HUVECs expressed significantly higher basal levels of GRα than DEX-sensitive HUVECs (Fig. 1D) that correlate with higher basal levels of the predominant 5′UTR mRNA isoform 1C (Fig. 1B). Importantly, DEX treatment decreased the expression of GRα (Fig. 1D) and increased the percentage of GRP (Fig. 1E) in DEX-resistant cells only. Lastly, there were no significant differences in the levels of total GR mRNA (measured by exons 2–3) between the sensitivity groups. However, DEX downregulated total GR mRNA in DEX-resistant cells only (Fig. 1D).
Role of GRα overexpression on its own transcriptional regulation and endothelial cell phenotype
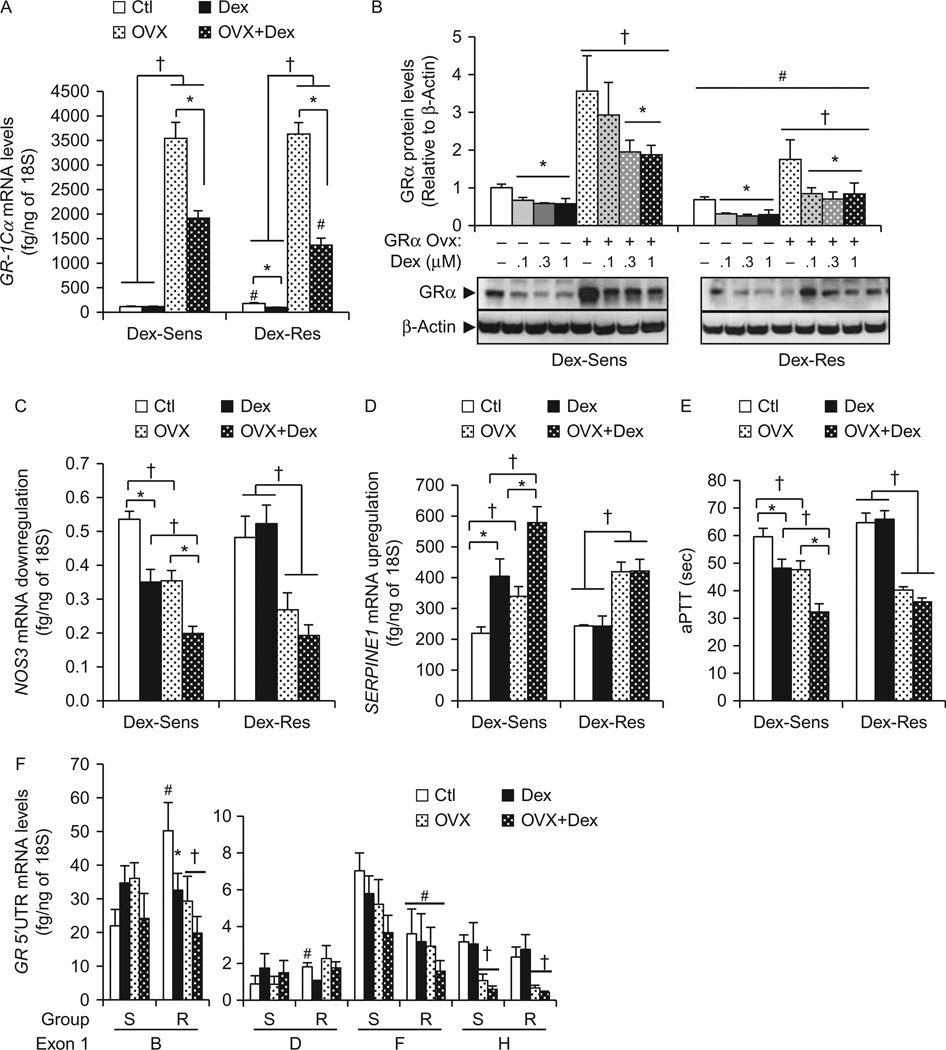
To clarify the role of differential GR 5′UTR mRNA isoform levels between sensitivity groups on endothelial phenotype, we overexpressed GRα in both DEX-sensitive and DEX-resistant HUVECs, using a promoter-less GR-1Cα-expressing vector. We hypothesized that GRα overexpression would increase sensitivity to DEX in any HUVEC tested. To validate our methods, we first showed our protocol significantly increased GR-1Cα mRNA levels by 31-fold and 20-fold, and GRα protein levels by 3.5-fold and 2.4-fold in DEX-sensitive and DEX-resistant cells respectively (Fig. 2A and B). DEX-treatment significantly decreased the levels of overexpressed GRα mRNA and protein in both sensitivity groups (Fig. 2A), suggesting DEX-dependent post-transcriptional and post-translational mechanisms of GR regulation (Pratt et al. 2006, Turner et al. 2010). However, our overexpression system had a smaller effect on GR protein levels (two- to fourfold increase) compared to GR mRNA (20- to 31-fold increase) levels that is most likely due to the reported GR protein degradation by the proteasome (Wallace et al. 2010, Mata-Greenwood et al. 2013). Of interest, overexpression of similar amount of GR-1Cα vectors yielded significantly lower levels of both GRα mRNA and protein in DEX-resistant cells compared to DEX-sensitive cells (Fig. 2B). This finding suggests that DEX-resistant cells have stronger negative autoregulatory mechanisms on GR expression compared to DEX-sensitive cells.
Figure 2.
Role of GRα overexpression in endothelial phenotype, expression autoregulation, and response to DEX. HUVECs were transfected with a promoter-less GR-1Cα expressing vector, then treated for 24 h with or without DEX (1 µM). Total RNA was extracted, reverse-transcribed and quantified by real-time PCR. (A and B) GRα overexpression in HUVECs upregulated both mRNA (A) and protein (B) levels in both DEX-sensitive (n = 5) and DEX-resistant (n = 5) HUVECs. (C, D and E) Effect of GRα overexpression on NOS3 downregulation (C), SERPINE1 upregulation (D) and activated partial thromboplastin time, aPTT (E). (F) Effect of GRα overexpression on GR 5′UTR mRNA isoform levels. Bars represent the average±s.e.m. (n = 5 DEX-sensitive and 5 DEX-resistant). *P<0.05 DEX-treated vs untreated; #P<0.05 DEX-resistant vs DEX-sensitive cells; †P<0.05 basal vs GRα overexpression.
We then investigated the role of increased expression of GRα in endothelial phenotype of both DEX-sensitive and DEX-resistant cells (Fig. 2C, D and E). We chose to study NOS3 and SERPINE1 expression, as these genes are key endothelial genes regulated by glucocorticoids, and their expression changes also regulate endothelial phenotype (Lou et al. 2001, Muzaffar et al. 2005, Kimura et al. 2009). GRα overexpression correlated with significant decreases in NOS3 (Fig. 2C) and increases in SERPINE1 (Fig. 2D) basal mRNA expression in both DEX-sensitive and DEX-resistant cells. Overexpression of GRα also increased the basal procoagulant activity of HUVECs as shown by the aPTT assay (Fig. 2E). However, there were significant differences between the sensitivity groups in response to DEX. DEX-treatment led to NOS3 down-regulation, SERPINE1 upregulation and aPTT increases in GRα-overexpressing DEX-sensitive cells, but not in GRα-overexpressing DEX-resistant cells (Fig. 2D and E). Therefore, overexpression of GR-1Cα alone did not correct the lack of response to DEX in resistant cells. However, GR-1Cα overexpression did alter basal endothelial phenotype in both sensitivity groups suggesting important non-ligand dependent effects of GR in human endothelial cells.
We also investigated the effect of GR-1Cα over-expression on its own expression. We found that GR-1Cα overexpression (alone or in combination with DEX treatment) decreased the expression of GR mRNA isoform 1B in DEX-resistant cells, and that of isoform 1H in both sensitivity groups (Fig. 2F). GR-1Cα overexpression did not alter DEX effects on GR mRNA isoform expression (Fig. 2F).
Finally, we investigated the expression of GR chaperones FKBP51, BAG1 and HSP90 in HUVECs overexpressing GRα. We found that resistant cells that overexpress GRα have significantly higher expression of the GRα inhibitor FKBP51 and higher levels of HSP90 than sensitive cells (Supplementary Figure 1, see section on supplementary data given at the end of this article), and could be a reason for the lack of DEX response in GRα overexpressing resistant cells. Therefore, differences in DEX-sensitivity in our HUVECs can also be accounted by differential regulation of GR chaperones. In summary, these results demonstrate that exogenous upregulation of GRα mRNA/protein levels can significantly alter both basal and glucocorticoid-stimulated endothelial phenotype in HUVECs, but does not eliminate the differences in GR expression and function regulation between the sensitivity groups.
Correlation of GR 5′UTR mRNA isoform expression with promoter usage
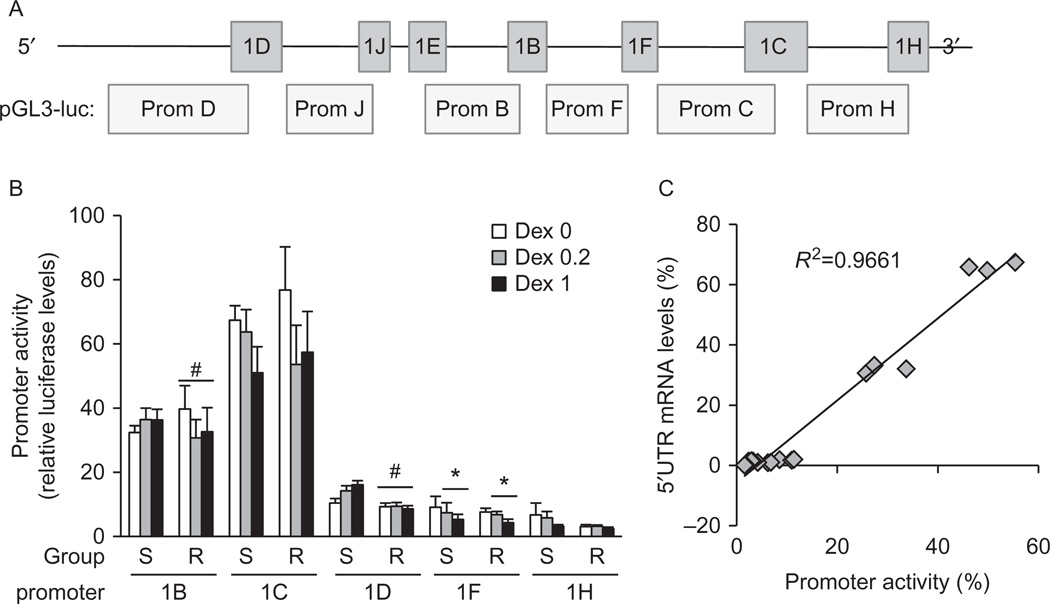
To further characterize the differences observed in GR mRNA isoform expression, we cloned five of the proximal TATA-less GR promoters to the pGL3 luciferase reporter vector to study promoter activity in HUVECs (Fig. 3A). Clones were sequenced and analyzed for putative internal elements (Inr), DPE, and transcription factor binding sites using the MattInspector software (Supplementary Figure 2, see section on supplementary data given at the end of this article). The consensus Inr element sequence (YYANWYY) located within the first 5 bp of the transcriptional starting site (+1) was found in promoter 1C only. The consensus TATA-less DPE elements (DPE=RGWYV) were found in promoters 1B and 1C, while partial DPEs were found in the remaining GR promoters (Supplementary Figure 2). Putative promoter elements for transcription factors were found for each promoter: those reported to be highly expressed in the cardiovascular system are shown in green.
Figure 3.
Basal and DEX-stimulated transcriptional activity of proximal GR promoters. (A) Structure of the proximal CpG island of the human GR gene. (B and C) HUVECs (12 DEX-sensitive and 8 DEX-resistant) were transfected with GR promoter-luciferase vectors and then treated with solvent, DEX 0.2, or DEX 1 µM for 24 h. Promoter activity was estimated by the levels of firefly luciferase activity, normalized to Renilla luciferase activity and expressed in fold activity of the empty pGL3 firefly basic reporter vector. Transfections were performed in quadruples. (B) Basal and DEX-dependent GR promoter usage in HUVECs. (C) Linear regression analysis of promoter usage (percent luciferase activity) against 5′UTR mRNA isoform expression (percent isoform levels). Averages for all HUVECs for each treatment group (DEX 0, 0.2 and 1 µM) were used. Bars represent the average±s.e.m.. *P<0.05 DEX-treated vs untreated; #P<0.05 DEX-resistant cells vs DEX-sensitive cells.
Luciferase assays revealed significant differences in GR promoter usage in all HUVECs, with promoter 1C having the highest activity (55%), followed by promoter 1B (30%), promoter 1D (10%), promoter 1F (8%), and promoter 1H (3%) (Fig. 3B). There were no significant differences in basal promoter usage between the sensitivity groups to account for the observed differences in GR 5′UTR mRNA isoforms 1C, 1D, and 1F (Fig. 1B). There were only small significant differences in DEX-stimulated promoter usage; sensitive cells responded to DEX with a slight increase in promoter 1B and 1D usage compared to DEX-resistant cells (Fig. 3B). DEX-treatment also significantly decreased promoter 1F usage in both DEX-sensitive and DEX-resistant cells, with non-significant decreases in promoter 1C usage in both groups as well (Fig. 3B). Therefore, our promoter assays suggest that the observed differences in GR mRNA isoforms 1C, 1D, and 1F between sensitivity groups (Fig. 1B and C) are not the result of differential transcriptional activation of GR promoters.
We analyzed the correlation of promoter usage with 5′UTR mRNA isoform expression by plotting the average % mRNA levels with average % promoter usage in all HUVECs at basal, low DEX (0.2 µM) and high DEX (1 µM) dose (Fig. 3C). We found a significant positive correlation between these two parameters, with only promoter 1D usage being disproportionally higher than the expression of the 1D mRNA isoform (Fig. 3C), suggesting epigenetic regulation of promoter 1D.
Role of promoter methylation in GR 5′UTR mRNA isoform expression
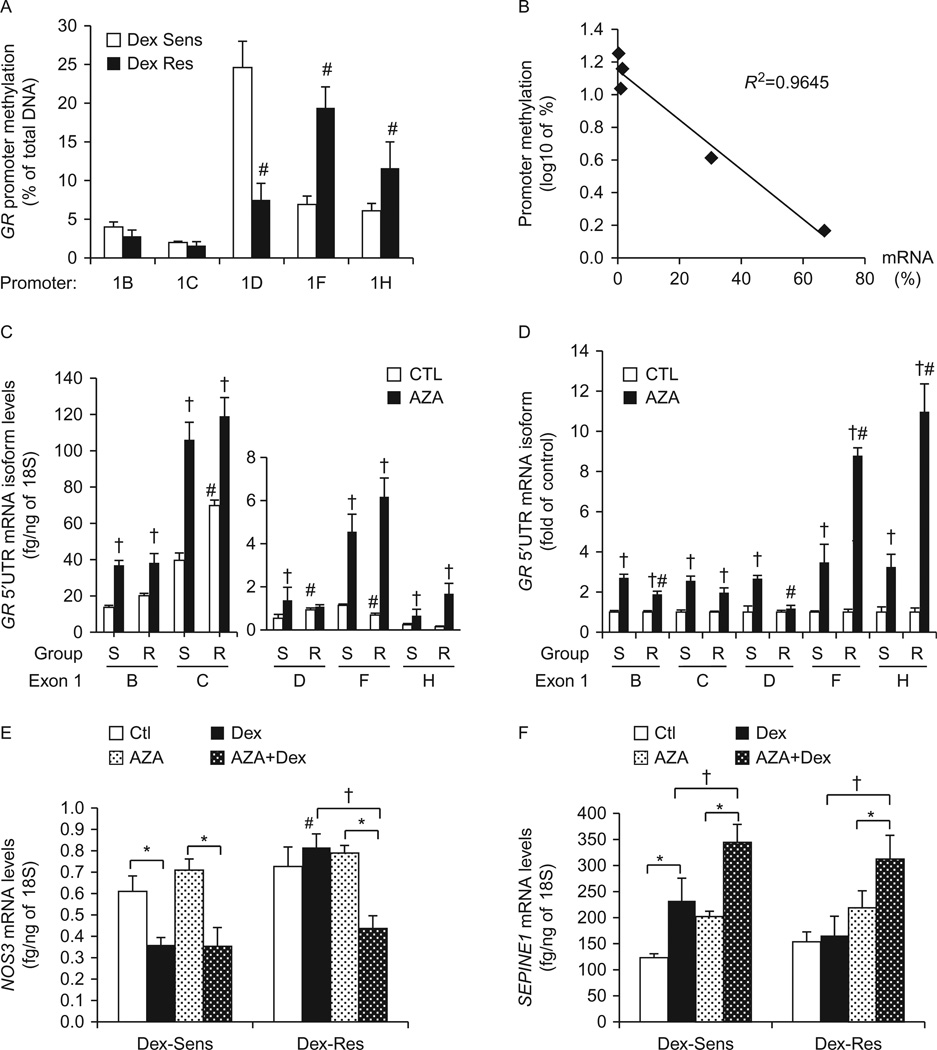
Because our reporter assays failed to reveal transcriptional mechanisms that would account for the observed differences between DEX-sensitive and DEX-resistant cells in GRα expression regulation (particularly of mRNA isoforms 1C, 1D, and 1F), we hypothesized that there could be differential methylation patterns of GR promoters. We first studied promoter methylation levels by immunoprecipitation of 5-methylcytosine (Fig. 4A). As expected, promoters 1B and 1C showed very low methylation levels of <4 and <1.7% respectively (Fig. 4A). Promoter 1D showed the highest methylation levels of ~20%, followed by promoter 1F (~14%), and promoter 1H (~10%). There were significant differences in promoter methylation levels between DEX-sensitive and DEX-resistant HUVECs (Fig. 4A). DEX-resistant cells had significantly lower methylation levels of promoter 1D and higher methylation levels of promoter 1F, compared to DEX-sensitive cells (Fig. 4A). The basal promoter methylation levels (i.e. averages for all cells: sensitive and resistant) correlated inversely with the expression of GR 5′UTR mRNA isoforms, with promoter 1C having the lowest methylation levels and higher mRNA isoform 1C levels, and promoter 1D having the highest methylation levels together with lower mRNA isoform 1D expression levels (Fig. 4B). These results explain why the expression of the 5′UTR mRNA isoform 1D is lower than that of isoform 1F, although the cloned promoter 1D-driven luciferase activity was higher than that of promoter 1F (Figs 1B and 3B), as cloned promoter-vectors do not have any cytosine methylations.
Figure 4.
Role of GR promoter methylation on GR expression and response to DEX. (A) Methylation of proximal promoters as determined by immunoprecipitation of 5-methylcytosine in DEX-sensitive (S) and DEX-resistant (R) cells (n=8/group). Results are shown as average percentage methylation. (B) The correlation between basal HUVEC GR promoter methylation and GR 5′UTR mRNA isoform expression was determined by linear regression analysis. Each data point represents the average levels for each sensitivity group. Promoter methylation percentages are shown as Iog10 values. (C and D) Global demethylation was achieved with 48 h of AZA treatment, and the expression of GR 5′UTR mRNA isoform was determined by real-time PCR (n = 5/sensitivity group). Results are shown as fg GR isoform/ng 18S RNA (C), and fold expression of untreated cells (D). (E and F) HUVECs were treated with AZA as described for (C) and (D) in the presence or absence of DEX 1µM, and then analyzed for NOS3 (E) and SERPINE1 (F) mRNA expression by real-time PCR. Bars represent the average±s.e.m. *P<0.05 DEX-treated vs untreated; #P<0.05 DEX-resistant cells vs DEX-sensitive cells; †P<0.05 non-AZA vs AZA treatment.
To confirm our MeDIP results, we then investigated the specific methylation sites in promoters 1D and 1F via bisulfite sequencing. Promoters 1D and 1F were chosen because they exhibited the highest methylation levels and because of differences in the expression of mRNA isoforms 1D and 1F between sensitivity groups. Bisulfite sequence analysis confirmed the differences observed in our MeDIP assays (Supplementary Figure 3, see section on supplementary data given at the end of this article and Fig. 4A). We found five methylation sites in promoter 1D with higher methylation percentages in DEX-sensitive cells than DEX-resistant cells (Supplementary Figure 3). DEX-resistant cells showed significant promoter 1F methylation of five different cytosines in comparison with none found in DEX-sensitive cells. In sum, bisulfite analysis confirmed that DEX-sensitive cells have higher methylation levels for promoter 1D but lower methylation levels for promoter 1F, compared to DEX-resistant cells. It is important to note that the MeDIP assay and bisulfite sequencing showed methylation levels for promoter 1F of ~7 and 0%, respectively, in DEX-sensitive cells. This discrepancy is likely due to the presence of other methylation sites further upstream or downstream from the selected DNA segment analyzed via bisulfite sequencing.
Finally, we utilized AZA to induce global demethylation in order to study the role of GR promoter methylation on its own expression and function. Analysis of the GR 5′UTR mRNA isoform expression demonstrated that AZA treatment resulted in increased expression of all GR 5′UTR mRNA isoforms in DEX-sensitive cells, and in all isoforms except isoform 1D in DEX-resistant cells (Fig. 4C and D). Importantly, AZA treatment eliminated the differences in GR 5′UTR mRNA isoforms 1C, 1D and 1F expression between DEX-sensitive and DEX-resistant cells (Fig. 4C). Analysis of AZA-induced fold increases in GR mRNA expression further highlighted differences between sensitivity groups. AZA-treatment produced higher (fold of control) increases in the expression of GR mRNA isoforms 1B and 1D in DEX-sensitive cells compared to DEX resistant cells, and a higher fold increase of isoforms 1F and 1H in DEX-resistant cells compared to DEX sensitive cells, and these data correlated with GR promoter methylation levels (Fig. 4A and D).
Consistent with the effect of AZA on GR expression, we found that AZA pretreatment increased the response to DEX in the resistant group in terms of NOS3 and SERPINE1 gene expression regulation, and thereby eliminated the differential response to DEX between the sensitivity groups (Fig. 4E and F). Of note, AZA treatment significantly increased the basal expression of SERPINE1 on all cells. This result is likely due to the effect of AZA on SERPINE1 promoter demethylation and basal transcriptional upregulation. However, we did not study the effect of AZA on the promoter methylation status of other genes. Therefore, AZA effects on in vitro DEX response could be mediated by demethylation of other genes that are differentially methylated in DEX-sensitive and DEX-resistant cells. In sum, we found significant differences in GR promoter methylation that result in differential GR 5′UTR mRNA isoform expression, and these differences further characterize DEX-sensitive and DEX-resistant HUVECs.
Discussion
We uncovered novel mechanisms of GR expression regulation in human endothelial cells that include differential expression of 5′UTR mRNA isoforms due to promoter methylation We have also found significant differences in GR promoter methylation and 5′UTR mRNA isoform expression according to DEX-sensitivity. The results of our studies on GR mRNA and protein regulation in HUVECs are summarized in Table 1. First, we have determined that the primary 5′UTR mRNA isoform present in human endothelial cells is isoform 1C, followed by isoform 1B, with smaller contributions from isoforms 1F, 1H, and 1D. Previous studies have shown similar dominance of expression of isoform 1C in tissues such as the liver, heart, kidney, and lung (Presul et al. 2007, Turner et al. 2008, 2010). Our promoter analysis suggests that the presence of putative Inr elements and DPE in promoter 1C leads to transcriptional dominance of mRNA isoform 1C compared to other isoforms. These DNA elements are known to recruit RNA polymerase complexes in TATA-less promoters (Butler & Kadonaga 2002, Yang et al. 2007, Juven-Gershon & Kadonaga 2010). Interestingly, mRNA isoforms 1J, 1D, 1H, and 1F have been reported to be highly expressed in non-vascular tissues such as the hippocampus and peripheral mononuclear cells (Turner et al. 2008, Cao-Lei et al. 2013). In our studies, these exons represent < 10% of all endothelial GR transcripts. Therefore, our data indicate that regulation of isoforms 1C and 1B expression, with smaller contributions from mRNA isoforms 1D, 1F and 1H, will have a higher impact on total GR mRNA expression.
Table 1.
Summary of major findings on GR expression regulation in HUVECs
| Figure 1: GR mRNA isoform levels | Resistant cells have higher expression of GR isoforms 1C and 1D, and lower expression of isoform 1F than sensitive cells in basal conditions. |
| Dexamethasone-treatment downregulated 5′UTR mRNA isoform 1C and 3′UTR mRNA isoform α in resistant cells only. | |
|
Figure 2: Effect of GR-1Cα Overexpression |
GR-1Cα overexpression in resistant cells did not improve in vitro response to dexamethasone as determined by NOS3 downregulation, SERPINE1 upregulation, and decreases in aPTT. |
|
GR-1Cα overexpression further downregulated the expression of 5′UTR mRNA isoforms 1B (in resistant cells only) and 1H in both sensitivity groups. | |
| Figure 3: GR promoter activity | There were no significant differences in basal GR promoter (unmethylated) activity between sensitive and resistant cells. |
| Dexamethasone treatment decreased promoter 1F activity in both sensitivity groups, decreased promoter 1B activity in resistant cells only and increased promoter 1D activity in sensitive cells only. | |
| Promoter activity differences between sensitive and resistant cells did not correlate with differences in GR 5′UTR mRNA levels. | |
|
Figure 4: GR promoter methylation and AZA treatment |
Sensitive cells have higher promoter 1D methylation levels and lower promoter 1F and 1H methylation levels compared to resistant cells. |
| Promoter methylation levels correlated with GR 5′UTR mRNA levels. | |
| AZA treatment upregulated the expression of all GR mRNA isoforms except isoform 1D in resistant cells. | |
| AZA treatment improved in vitro dexamethasone response in resistant cells as determined by NOS3 downregulation and SERPINE1 upregulation. | |
Previously we reported that increased in vitro DEX sensitivity of HUVECs correlated positively with GRα protein levels (Mata-Greenwood et al. 2013). DEX-sensitive HUVECs showed lower GRα protein turnover through the proteasome system, and this was partly due to lower expression of the E3 ubiquitin ligase BAG1. In the present study, we have found that increased GRα protein levels observed in DEX-sensitive, compared to DEX-resistant, cells are not the result of increased GRα mRNA levels, as previously hypothesized. However, we did observe significant differences in GRα mRNA isoform expression among DEX-sensitive and DEX-resistant HUVECs. Resistant cells, but not sensitive cells, responded to DEX treatment with a significant down-regulation of the main GR 5′UTR mRNA isoform 1C and 3′UTR mRNA isoform α (Table 1). Because glucocorticoids downregulate GRα mRNA expression (Petersen et al. 2004, Ramamoorthy & Cidlowski 2013), we now hypothesize that DEX-resistant cells show significant DEX-dependent downregulation of GR mRNA expression in addition to increased GR protein degradation via the proteasome. Our GRα overexpression data supports this hypothesis; DEX treated-resistant cells had lower levels of overexpressed GRα mRNA and protein than DEX-treated-sensitive cells. Therefore, DEX-resistance in endothelial cells could arise from a strong negative autoregulation that includes both mRNA and protein expression decreases.
Another interesting result was that GRα overexpression did not increase in vitro response to DEX (measured as changes in NOS3, SERPINE1, and aPTT) in resistant cells (Table 1). Furthermore, GRα overexpression significantly altered basal endothelial phenotype (NOS3, SERPINE1, and aPTT levels) in both sensitivity groups with a greater effect shown in resistant cells. We currently speculate these data to be caused by differences in GR:chaperone interactions between sensitive and resistant cells. In support of this hypothesis, we found that GRα overexpression upregulated the expression of GR inhibitors BAG1 and FKBP51. Further studies on GR chaperones are therefore needed to fully understand the differential in vitro response to DEX in HUVECs. In summary, these data suggest that posttranslational regulation of GRα protein, such as GRα proteasomal degradation and GR:chaperone interactions, and not transcriptional mechanisms, are key factors in determining basal and DEX-stimulated levels of biologically active GR and, thereby, in the biological response to glucocorticoids (Table 1).
Perhaps the most novel result of this study is the inverse association between GR 5′UTR mRNA isoform expression and GR promoter methylation together with the finding of significant differences in promoter methylation between DEX-sensitive and DEX-resistant cells (Table 1). This study is the first to report that methylation plays an important role in GR mRNA expression in endothelial cells. DEX-sensitive HUVECs showed significantly higher methylation of promoter 1D, but lower methylation of promoters 1F and 1H. Methylation levels of promoter 1D and 1F correlated inversely with the expression of their corresponding 5′UTR mRNA isoforms 1D and 1F. Of importance was the finding that AZA treatment abrogated the differences in GR 5′UTR mRNA isoform expression between sensitivity groups, thereby showing that methylation differences in GR promoters are a principal reason for the observed differences in GR 5′UTR mRNA isoforms expression between DEX-sensitive and DEX-resistant cells. Unexpectedly, AZA treatment increased the expression of GR mRNA isoforms 1B and 1C, even though their proximal promoters were highly unmethylated. We hypothesize that AZA demethylation of upstream sequences, such as promoter 1D or GR enhancers, increases the expression of mRNA isoforms 1B and 1C. Our hypothesis is in agreement with previous reports on YY1, a promoter 1D binding transcription factor, and the mediated transcriptional activation of isoform1B (Cao-Lei et al. 2011). To our knowledge, this study is the first to show that the expression of GR mRNA isoforms 1B and 1C is also regulated by methylation. Numerous studies have shown a significant role of promoter 1F methylation in brain tissue, and its correlation with psychiatric diseases (Moser et al. 2007, Alt et al. 2010, Labonte et al. 2014, Van der Knaap et al. 2014). In rats, maternal care has shown to decrease methylation levels of promoter 1–7 in brain and liver tissue (Szyf et al. 2005, Witzmann et al. 2012), while hypoxia and other stressors increase it (Xiong & Zhang 2013, Gonzalez-Rodriguez et al. 2014). Recent human studies on promoter 1F methylation in umbilical cord mononuclear cells revealed increased promoter 1F methylation in gestational infants exposed to maternal stressors (Filiberto et al. 2011, Drake et al. 2012, Sinclair et al. 2012, Hompes et al. 2013). This finding is of interest because our DEX-sensitive cells originated from mothers with significantly higher pre-pregnancy BMI than those from the DEX-resistant group. Therefore, we hypothesize that maternal weight could be an important factor (stressor) in determining fetal endothelial glucocorticoid sensitivity. Altogether, our data suggest that differential methylation of GR promoters have significance in regulating GR mRNA isoform expression and are associated with glucocorticoid sensitivity.
Together, our findings show that GRα gene expression in human endothelial cells is highly complex and that GR promoter methylation patterns differ between DEX-sensitive and DEX-resistant HUVECs. Although these differences in GR promoter methylation and 5′UTR mRNA expression do not account for the differences observed in GRα protein expression, function, and further alterations of endothelial phenotype, we hypothesize that they could be exploited to develop epigenetic markers of vascular glucocorticoid sensitivity. Importantly, global demethylation studies with AZA eliminated the differences between sensitivity groups in the in vitro response to DEX, indicating an important role of methylation patterns in establishing HUVEC sensitivity to glucocorticoids. Future research into the mechanisms that lead to differential methylation of GR proximal promoters, and the potential role of maternal obesity in programming fetal endothelial glucocorticoid sensitivity, are warranted.
Supplementary Material
Acknowledgments
We thank Dr John A Cidlowski (NIEHS) for providing the full length human GR-1Cα expressing vectors, Dr Fuxia Xiong for helping in the design of cloning primers, and Dr Donna Thorpe for helping with the statistical analysis.
Funding
The present study was supported by the AHA SDG award #0630297N to Dr E M-G and by the School of Medicine, Loma Linda University.
Footnotes
Supplementary data
This is linked to the online version of the paper at http://dx.doi.org/10.1530/JME-15-0124.
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
References
- Adcock IM, Ito K, Barnes PJ. Glucocorticoids: effects on gene transcription. Proceedings of the American Thoracic Society. 2004;1:247–254. doi: 10.1513/pats.200402-001MS. [DOI] [PubMed] [Google Scholar]
- Alt SR, Turner JD, Klok M, Meijer OC, Lakke EAJF, DeRijk RH, Muller CP. Differential expression of glucocorticoid receptor transcripts in major depressive disorder is not epigenetically programmed. Psychoneuroendocrinology. 2010;35:544–556. doi: 10.1016/j.psyneuen.2009.09.001. [DOI] [PubMed] [Google Scholar]
- Auphan N, DiDonato JA, Helmberg A, Karin M. Immunosuppression by glucocorticoids: inhibition of NF-κ B activity through induction of I κ B synthesis. Science. 1995;270:232–233. doi: 10.1126/science.270.5234.286. [DOI] [PubMed] [Google Scholar]
- Bamberger CM, Schulte HM, Chrousos GP. Molecular determinants of glucocorticoid receptor function and tissue sensitivity of glucocorticoids. Endocrine Reviews. 1996;17:245–261. doi: 10.1210/edrv-17-3-245. [DOI] [PubMed] [Google Scholar]
- Barnes PJ. Anti-inflammatory actions of glucocorticoids: molecular mechanisms. Clinical Science. 1998;94:557–572. doi: 10.1042/cs0940557. [DOI] [PubMed] [Google Scholar]
- Biddie SC, Conway-Campbell BL, Lightman SL. Dynamic regulation of glucocorticoid signaling in health and disease. Rheumatology. 2012;51:403–412. doi: 10.1093/rheumatology/ker215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockmuhl Y, Murgatroyd CA, Kuczynska A, Adcock IM, Almeida OF, Spengler D. Differential regulation and function of 5′-Untranslated GR-Exon 1 transcripts. Molecular Endocrinology. 2011;25:1100–1110. doi: 10.1210/me.2010-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler J, Kadonaga JT. The RNA polymerase II core promoter: a key component in the regulation of gene expression. Genes and Development. 2002;16:2583–2592. doi: 10.1101/gad.1026202. [DOI] [PubMed] [Google Scholar]
- Cao-Lei L, Leija SC, Kumsta R, Wust S, Meyer J, Turner JD, Muller CP. Transcriptional control of the human glucocorticoid receptor: identification and analysis of alternative promoter regions. Human Genetics. 2011;129:533–543. doi: 10.1007/s00439-011-0949-1. [DOI] [PubMed] [Google Scholar]
- Cao-Lei L, Suwansirikul S, Jutavijittum P, Meriaux SB, Turner JD, Muller CP. Glucocorticoid receptor gene expression and promoter CpG modifications throughout the human brain. Journal of Psychiatric Research. 2013;47:1597–1607. doi: 10.1016/j.jpsychires.2013.07.022. [DOI] [PubMed] [Google Scholar]
- Drake AJ, McPherson RC, Godfrey KM, Cooper C, Lillycrop KA, Hanson MA, Meehan RR, Seckl JR, Reynolds RM. An unbalanced maternal diet in pregnancy associates with offspring epigenetic changes in genes controlling glucocorticoid action and foetal growth. Clinical Endocrinology. 2012;77:808–815. doi: 10.1111/j.1365-2265.2012.04453.x. [DOI] [PubMed] [Google Scholar]
- Filiberto AC, Maccani MA, Koestler D, Wilhelm-Benartzi C, Avissar-Whiting M, Banister CE, Gagne LA, Marsit CJ. Birthweight is associated with DNA promoter methylation of the glucocorticoid receptor in human placenta. Epigenetics. 2011;6:566–572. doi: 10.4161/epi.6.5.15236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Rodriguez PJ, Xiong F, Li Y, Zhou J, Zhang L. Fetal hypoxia increases vulnerability of hypoxic-ischemic brain injury in neonatal rats: role of glucocorticoid receptors. Neurobiology of Disease. 2014;65:172–179. doi: 10.1016/j.nbd.2014.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal R, Zhang L, Blood AB, Baylink DJ, Longo LD, Oshiro B, Mata-Greenwood E. Characterization of an animal model of pregnancy-induced vitamin D deficiency due to metabolic dysregulation. American Journal of Physiology. Endocrinology and Metabolism. 2014;306:256–266. doi: 10.1152/ajpendo.00528.2013. [DOI] [PubMed] [Google Scholar]
- Hadoke PWF, Iqbal J, Walker BR. Therapeutic manipulation of glucocorticoid metabolism in cardiovascular disease. British Journal of Pharmacology. 2009;156:689–712. doi: 10.1111/j.1476-5381.2008.00047.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitzer MD, Wolf IM, Sanchez ER, Witchel SF, DeFranco DB. Glucocorticoid receptor physiology. Reviews in Endocrine & Metabolic Disorders. 2007;8:321–330. doi: 10.1007/s11154-007-9059-8. [DOI] [PubMed] [Google Scholar]
- Hompes T, Izzi B, Gellens E, Morreels M, Fieuws S, Pexsters A, Schops G, Dom M, Van Bree R, Freson K, et al. Investigating the influence of maternal cortisol and emotional state during pregnancy on the DNA methylation status of the glucocorticoid receptor gene (NR3C1) promoter region in cord blood. Journal of Psychiatric Research. 2013;47:880–891. doi: 10.1016/j.jpsychires.2013.03.009. [DOI] [PubMed] [Google Scholar]
- Huang W, Glass CK. Nuclear receptors and inflammation control: molecular mechanisms and pathophysiological relevance. Atherosclerosis, Thrombosis, and Vascular Biology. 2010;30:1542–1549. doi: 10.1161/ATVBAHA.109.191189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Chung KF, Adcock IM. Update on glucocorticoid action and resistance. Journal of Allergy and Clinical Immunology. 2006;117:522–543. doi: 10.1016/j.jaci.2006.01.032. [DOI] [PubMed] [Google Scholar]
- Juven-Gershon T, Kadonaga JT. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Developmental Biology. 2010;339:225–229. doi: 10.1016/j.ydbio.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadmiel M, Cidlowski JA. Glucocorticoid receptor signaling in health and disease. Trends in Pharmacological Sciences. 2013;34:518–530. doi: 10.1016/j.tips.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassel O, Herrlich P. Crosstalk between the glucocorticoid receptor and other transcription factors: molecular aspects. Molecular and Cellular Endocrinology. 2007;275:13–29. doi: 10.1016/j.mce.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Kimura H, Li X, Torii K, Okada T, Kamiyama K, Mikami D, Takahashi N, Yoshida H. Dexamethasone enhances basal and TNF-α stimulated production of PAI-1 via the glucocorticoid receptor regardless of 11β-hydroxysteroid dehydrogenase 2 status in human proximal renal tubular cells. Nephrology, Dialysis, Transplantation. 2009;24:1759–1765. doi: 10.1093/ndt/gfn756. [DOI] [PubMed] [Google Scholar]
- Kino T. Tissue glucocorticoid sensitivity: beyond stochastic regulation on the diverse actions of glucocorticoids. Hormone and Metabolic Research. 2007;39:420–424. doi: 10.1055/s-2007-980193. [DOI] [PubMed] [Google Scholar]
- Labonte B, Zaoulay N, Yerko V, Turecki G, Brunet A. Epigenetic modulation of glucocorticoid receptors in posttraumatic stress disorder. Translational Psychiatry. 2014;4:e368. doi: 10.1038/tp.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou Y, Wen C, Li M, Adams DJ, Wang M, Yang F, Morris BJ, Whitworth JA. Decreased renal expression of nitric oxide synthase isoforms in adrenocorticotropin-induced and corticosterone-induced hypertension. Hypertension. 2001;37:1164–1170. doi: 10.1161/01.hyp.37.4.1164. [DOI] [PubMed] [Google Scholar]
- Mata-Greenwood E, Liao WX, Wang W, Zheng J, Chen DB. Activation of AP-1 transcription factors differentiates FGF2 and vascular endothelial growth factor regulation of endothelial nitric oxide synthase expression in placental artery endothelial cells. Journal of Biological Chemistry. 2010;285:17348–17358. doi: 10.1074/jbc.M109.092791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata-Greenwood E, Stewart JM, Steinhorn RH, Pearce WJ. Role of BAG1 on differential sensitivity of human endothelial cells to glucocorticoids. Atherosclerosis, Thrombosis, and Vascular Biology. 2013;33:1046–1057. doi: 10.1161/ATVBAHA.113.301247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser D, Molitor A, Kumsta R, Tatschner T, Riederer P, Meyer J. The glucocorticoid receptor gene exon 1-F promoter is not methylated at the NGFI-A binding site in human hippocampus. World Journal of Biological Psychiatry. 2007;8:262–268. doi: 10.1080/15622970701429862. [DOI] [PubMed] [Google Scholar]
- Muzaffar S, Shukla N, Angelini GD, Jeremy JY. Prednisolone augments superoxide formation in porcine pulmonary artery endothelial cells through differential effects on the expression of nitric oxide synthase and NADPH oxidase. British Journal of Pharmacology. 2005;145:688–697. doi: 10.1038/sj.bjp.0706235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen KB, Geng CD, Vedeckis WV. Three mechanisms are involved in glucocorticoid receptor autoregulation in a human T-lymphoblast cell line. Biochemistry. 2004;43:10851–10858. doi: 10.1021/bi049458u. [DOI] [PubMed] [Google Scholar]
- Pratt WB, Morishima Y, Murphy M, Harrell M. Chaperoning of glucocorticoid receptors. Handbook of Experimental Pharmacology. 2006;172:111–138. doi: 10.1007/3-540-29717-0_5. [DOI] [PubMed] [Google Scholar]
- Presul E, Schmidt S, Kofler R, Helmberg A. Identification, tissue expression, and glucocorticoid responsiveness of alternative first exons of the human glucocorticoid receptor. Journal of Molecular Endocrinology. 2007;38:79–90. doi: 10.1677/jme.1.02183. [DOI] [PubMed] [Google Scholar]
- Ramamoorthy S, Cidlowski JA. Ligand-induced repression of the glucocorticoid receptor gene is mediated by an NCoR1 repression complex formed by long-range chromatin interactions with intragenic glucocorticoid response elements. Molecular and Cellular Biology. 2013;33:1711–1722. doi: 10.1128/MCB.01151-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair D, Fullerton JM, Webster MJ, Weickert CS. Glucocorticoid receptor 1B and 1C mRNA transcript alterations in schizophrenia and bipolar disorder, and their possible regulation by GR gene variants. PLoS ONE. 2012;7:e31720. doi: 10.1371/journal.pone.0031720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steer JH, Kroeger KM, Abraham LJ, Joyce DA. Glucocorticoids suppress tumor necrosis factor-α expression by human monocytic THP-1 cells by suppressing transactivation through adjacent NF-kB and c-Jun-activating transcription factor-2 binding sites in the promoter. Journal of Biological Chemistry. 2000;275:18432–18440. doi: 10.1074/jbc.M906304199. [DOI] [PubMed] [Google Scholar]
- Szyf M, Weaver ICG, Champagne FA, Diorio J, Meaney MJ. Maternal programming of steroid receptor expression and phenotype through DNA methylation in the rat. Frontiers in Neuroendocrinology. 2005;26:139–162. doi: 10.1016/j.yfrne.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Tamura Y, Kawao N, Yano M, Okada K, Okumoto K, Chiba Y, Matsuo O, Kaji H. Role of plasminogen activator inhibitor-1 in glucocorticoid-induced diabetes and osteopenia in mice. Diabetes. 2015;64:2194–2206. doi: 10.2337/db14-1192. [DOI] [PubMed] [Google Scholar]
- Turner JD, Schote AB, Macedo JA, Pelascini LPL, Muller CP. Tissue specific glucocorticoid receptor expression, a role for alternative first exon usage? Biochemical Pharmacology. 2006;72:1529–1537. doi: 10.1016/j.bcp.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Turner JD, Pelascini LPL, Macedo JA, Muller CP. Highly individual methylation patterns of alternative glucocorticoid receptor promoters suggest individualized epigenetic regulatory mechanisms. Nucleic Acid Research. 2008;36:7207–7218. doi: 10.1093/nar/gkn897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JD, Alt SR, Cao L, Vernocchi S, Trifonova S, Battello N, Muller CP. Transcriptional control of the glucocorticoid receptor: CpG islands, epigenetics and more. Biochemical Pharmacology. 2010;80:1860–1868. doi: 10.1016/j.bcp.2010.06.037. [DOI] [PubMed] [Google Scholar]
- Van der Knaap LF, Riese H, Hudziak JJ, Verbiest MMPJ, Verhulst FC, Oldehinkel AJ, van Oort FVA. Glucocorticoid receptor gene (NR3C1) methylation following stressful events between birth and adolescence. The TRAILS study. Translational Psychiatry. 2014;4:e381. doi: 10.1038/tp.2014.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BR. Glucocorticoids and Cardiovascular Disease. European Journal of Endocrinology. 2007;157:545–559. doi: 10.1530/EJE-07-0455. [DOI] [PubMed] [Google Scholar]
- Wallace AD, Cao Y, Chandramouleeswaran S, Cidlowski JA. Lysine 419 targets human glucocorticoid receptor for proteasomal degradation. Steroids. 2010;75:1016–1023. doi: 10.1016/j.steroids.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallerath T, Gödecke A, Molojawi A, Li H, Schrader J, Förstermann U. Dexamethasone lacks effect on blood pressure in mice with a disrupted endothelial NO synthase gene. Nitric Oxide. 2004;10:36–41. doi: 10.1016/j.niox.2004.01.008. [DOI] [PubMed] [Google Scholar]
- Wenting-van Wijk MJG, Blankenstein MA, Lafeber FPJG, Bijlsma JWJ. Relation of plasma dexamethasone to clinical response. Clinical and Experimental Rheumatology. 1999;17:305–312. [PubMed] [Google Scholar]
- Witzmann SR, Turner JD, Meriaux SB, Meijer OC, Muller CP. Epigenetic regulation of the glucocorticoid receptor promoter 1(7) in adult rats. Epigenetics. 2012;7:1290–1301. doi: 10.4161/epi.22363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong F, Zhang L. Role of the hypothalamic-pituitary-adrenal axis in developmental programming of health and disease. Frontiers in Neuroendocrinology. 2013;34:27–46. doi: 10.1016/j.yfrne.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Zhang L. Glucocorticoids and vascular reactivity. Current Vascular Pharmacology. 2004;2:1–12. doi: 10.2174/1570161043476483. [DOI] [PubMed] [Google Scholar]
- Yang C, Bolotin E, Jiang T, Sladek FM, Martinez E. Prevalence of the Initiator over the TATA box in human and yeast genes and identification of DNA motifs enriched in human TATA-less core promoters. Gene. 2007;389:52–65. doi: 10.1016/j.gene.2006.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.