Abstract
At odds with their normal counterparts, hepatocellular carcinoma cells efficiently utilize ketone bodies to proliferate despite serum deprivation. These findings, which have been recently published in Cell Research, identify a novel metabolic circuitry through which tumors successfully cope with adverse microenvironmental conditions.
One of the most impressive features of malignant cells is their ability to adapt to prominent changes in the composition of the extracellular milieu1. At odds with their non-transformed counterparts, cancer cells are indeed able to proliferate in the absence of growth factors, under pronounced hypoxia, as well as when nutrients and amino acids are limited1. Throughout the past decade, work from several laboratories clarified that neoplastic cells facing adverse microenvironmental conditions can utilize a variety of metabolites to support catabolic and anabolic reactions, including (but presumably not limited to) glucose, acetate, lactate, creatine, glutamine, serine, glycine and fatty acids2. Thus, cancer cells generally rewire their metabolism, hence acquiring the capacity to utilize metabolites that are locally available to support tumor progression2. Although less universal and less specific than initially thought (meaning that different cancers can exhibit quite distinct metabolic shifts, and that at least some of the metabolic alterations that accompany malignancy are also found in highly proliferating non-transformed cells), such a rewiring process provides putative targets for the development of novel anticancer agents3. Recent work from Huafeng Zhang's group identifies a novel metabolic circuitry based on ketolysis through which hepatocellular carcinoma (HCC) cells proliferate in spite of adverse microenvironmental conditions4.
Starting from the observation that HCC cells downregulate fatty acid oxidation in response to hypoxia as a mechanism to relieve oxidative stress and support proliferation5, Zhang and collaborators initially set out to investigate the expression levels of genes involved in lipid metabolism in human HCC HepG2 cells deprived of glucose, glutamine or serum by quantitative real-time PCR. In response to all these microenvironmental challenges, HepG2 cells upregulated multiple enzymes involved in ketogenesis6, including BDH1, HMGCL, HMGCS1 and HMGCS2, consistent with the prominent role played by the liver in the provision of ketone bodies to other tissues in response to starvation. Surprisingly, HepG2 cells subjected to serum (but not glucose or glutamine) deprivation also exhibited increased levels of 3-oxoacid CoA-transferase 1 (OXCT1), which catalyzes the rate-limiting reaction of ketolysis and is normally not expressed in the adult liver7. Similar results were obtained with other human HCC cell lines, including Hep3B and PLC cells, and OXCT1 upregulation driven by serum withdrawal was confirmed by immunoblotting4. In line with this finding, exogenously supplied β-hydroxybutyrate (a ketone body) was converted by HCC cells subjected to starvation (but not by non-transformed human liver THLE3 cells, nor by HCC cells maintained in control conditions) into various intermediates of the tricyclic acid cycle, including citrate, succinate, fumarate, malate, glutamate and aspartate, supporting a time-dependent recovery in intracellular ATP levels and proliferation rates4. Importantly, the stable downregulation of OXCT1 by two distinct shRNA-coding constructs virtually abolished the ability of HepG2 cells undergoing serum deprivation to catabolize exogenous β-hydroxybutyrate. Moreover, OXCT1-depleted HepG2 cells exhibited exacerbated ATP loss in response to serum deprivation as compared to HepG2 cells expressing a control shRNA, and were completely unable to proliferate. Such defects could be rescued by the transgene-driven overexpression of a non-interferable OXCT1 variant. Of note, β-hydroxybutyrate was not exogenously supplied in this last set of experiments, suggesting that HCC can employ endogenously produced ketone bodies to proliferate despite serum deprivation. Corroborating this hypothesis, the depletion of HMGCS2 virtually abolished the proliferative advantage conferred to serum-depleted HepG2 cells by OXCT1 overexpression, unless exogenous β-hydroxybutyrate was added to the culture medium4.
Next, Zhang and colleagues investigated the signal transduction pathways responsible for the upregulation of OXCT1 in serum-deprived HCC cells by interrogating a panel of chemical inhibitors. Amongst a few molecules with distinct specificities, LY-294002 (an inhibitor of phosphoinositide-3-kinases and AKT1) was the sole agent capable of avoiding OXCT1 upregulation by serum withdrawal. Corroborating a direct implication of AKT1 in this process, HepG2 cells expressing two distinct AKT1-specific shRNAs lost their ability to upregulate OXCT1 upon serum deprivation, as did HepG2 cells depleted of an essential component of the AKT1 activator mechanistic target of rapamycin (MTOR) complex 2 (mTORC2), namely, RPTOR independent companion of MTOR, complex 2 (RICTOR). These findings were further confirmed with rapamycin, a macrolide that activates mTORC2 as it inhibits mTORC18. Moreover, the ability of mTORC2-activated AKT1 to drive the upregulation of OXCT1 was mechanistically ascribed to the transcription factor SP1. Thus, similar to their AKT1-deprived counterparts, SP1-depleted HepG2 cells were unable to catabolize exogenous β-hydroxybutyrate4. Interestingly, the ability of HepG2 cells to upregulate OXCT1 was associated with limited activation of the energy sensor AMP-activated protein kinase (AMPK) upon serum withdrawal9, presumably reflecting an improved capacity to preserve bioenergetic homeostasis via ketolysis. Accordingly, depletion of OXCT1 or AKT1 aggravated, while OXCT1 overexpression quenched, the autophagic response of HepG2 cells to serum deprivation, knowing that autophagy is an evolutionarily ancient mechanism of adaptation to adverse microenvironmental conditions10,11. Exogenous β-hydroxybutyrate also limited autophagy in serum-starved HepG2 cells, provided that OXCT1 was normally expressed. Moreover, the overexpression of OXCT1 abolished the ability of an AKT1-specific shRNA to exacerbate serum starvation-induced autophagy4.
Finally, Zhang and collaborators validated the relevance of their findings in vivo. OXCT1-overexpressing HepG2 cells xenografted into athymic nu/nu mice manifested increased proliferative potential and decreased markers of autophagy as compared to their control counterparts. Conversely, HepG2 cell xenografts stably expressing an OXCT1-targeting shRNA displayed autophagy activation and reduced proliferation rates. Moreover, the intraperitoneal administration of β-hydroxybutyrate accelerated tumor progression in vivo, but only when HepG2 cells overexpressed OXCT14. Circulating β-hydroxybutyrate was increased in a cohort of 35 HCC patients as compared to 29 healthy subjects. Moreover, 20 HCC lesions displayed increased levels of OXCT1 (mRNA), OXCT1 (protein), SP1 (protein) and phosphorylated SP1 (active protein), as compared to adjacent normal tissues. Finally, in a cohort of 148 HCC patients, high expression levels of OXCT1 in malignant cells positively correlated with clinical stage and indicated dismal prognosis in monovariate analysis4.
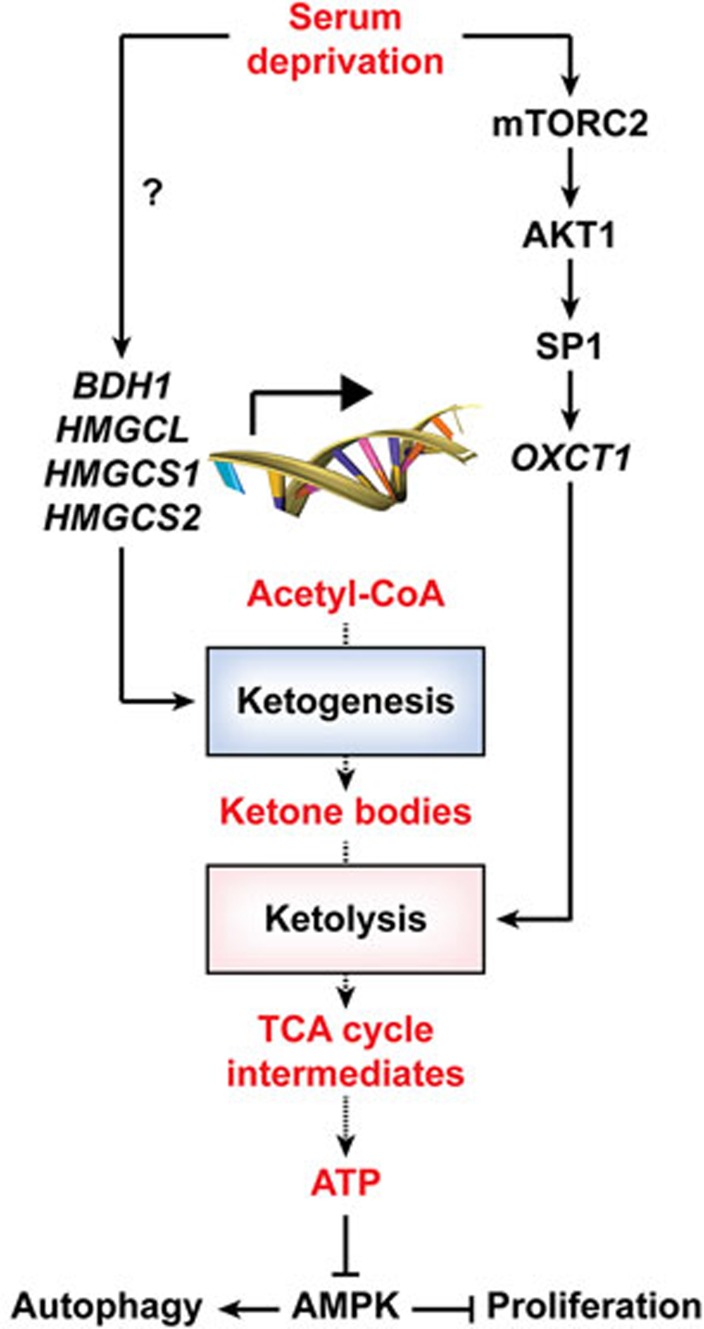
In essence, Zhang and co-authors demonstrated that HCC cells acquire the capacity to consume ketone bodies to cope with the nutritional stress imposed by serum starvation, in thus far contrasting with normal hepatocytes, which are only capable of ketogenesis. Moreover, they delineated the signal transduction cascade involving mTORC2, AKT1, SP1 and OXCT1 through which HCC cells activate ketolysis in response to adverse microenvironmental conditions (Figure 1). It will be interesting to determine whether this mechanism can be targeted pharmacologically for the development of new drugs against HCC.
Figure 1.
Molecular mechanisms supporting the proliferation of HCC cells despite serum withdrawal. In response to serum deprivation, HCC cells upregulate multiple enzymes involved in ketogenesis, resembling normal hepatocytes. In addition, serum-deprived HCC cells also upregulate OXCT1 expression, via a signal transduction pathway involving the sequential activation of mTORC2, AKT1 and SP1. Thus, contrary to normal hepatocytes, HCC cells also respond to serum deprivation by acquiring the ability to employ endogenous and exogenous ketone bodies for ATP synthesis. Such a capacity allows HCC cells to proliferate despite the nutritional stress imposed by serum withdrawal. TCA, tricyclic acid.
References
- Hanahan D, Weinberg RA. Cell 2011; 144:646–674. [DOI] [PubMed]
- Martinez-Outschoorn UE, Peiris-Pages M, Pestell RG, et al. Nat Rev Clin Oncol 2016. May 4. doi: 10.1038/nrclinonc.2016.60
- Galluzzi L, Kepp O, Vander Heiden MG, et al. Nat Rev Drug Discov 2013; 12:829–846. [DOI] [PubMed]
- Huang D, Li T, Wang L, et al. Cell Res 20 September 2016; 26:1112–1130. [DOI] [PMC free article] [PubMed]
- Huang D, Li T, Li X, et al. Cell Rep 2014; 8:1930–1942. [DOI] [PubMed]
- Pietrocola F, Galluzzi L, Bravo-San Pedro JM, et al. Cell Metab 2015; 21:805–821. [DOI] [PubMed]
- Llovet JM, Zucman-Rossi J, Pikarsky E, et al. Nat Rev Dis Primers 2016; 2:16018. [DOI] [PubMed]
- Shimobayashi M, Hall MN. Nat Rev Mol Cell Biol 2014; 15:155–162. [DOI] [PubMed]
- Hardie DG, Ross FA, Hawley SA. Nat Rev Mol Cell Biol 2012; 13:251–262. [DOI] [PMC free article] [PubMed]
- Galluzzi L, Pietrocola F, Bravo-San Pedro JM, et al. EMBO J 2015; 34:856–880. [DOI] [PMC free article] [PubMed]
- Sica V, Galluzzi L, Bravo-San Pedro JM, et al. Mol Cell 2015; 59:522–539. [DOI] [PubMed]