Abstract
Determining the density and morphology of dendritic spines is of high biological significance given the role of spines in synaptic plasticity and in neurodegenerative and neuropsychiatric disorders. Precise quantification of spines in three dimensions (3D) is essential for understanding the structural determinants of normal and pathological neuronal function. However, this quantification has been restricted to time- and labor-intensive methods such as electron microscopy and manual counting, which have limited throughput and are impractical for studies of large samples. While there have been some automated software packages that quantify spine number, they are limited in terms of their characterization of spine structure. This unit presents methods for objective dendritic spine morphometric analysis by providing image acquisition parameters needed to ensure optimal data series for proper spine detection, characterization, and quantification with Neurolucida 360. These protocols will be a valuable reference for scientists working towards quantifying and characterizing spines.
Keywords: dendritic spines, neurons, confocal microscopy, automated quantification
Introduction
Because of their capacity for plasticity and their role as the location of excitatory synapses, accurately characterizing the structure of dendritic spines is of profound biological significance. Spine morphology determines the strength, stability and function of excitatory synaptic connections that subserve the neuronal networks underlying cognitive function. Developmental, aging-, and disease-related structural changes in neurons and dendritic spines, and their functional consequences, remain poorly understood. Therefore, there is value in imaging and quantifying specific spine populations, types, densities, and distribution along neuronal dendrites across brain regions or cortical layers. It is also important to assess entire neurons or dendritic trees as it permits to assess tree complexity and dendritic branching, both of which may be altered in disease states. Optimally both spines and neurons should be examined at high resolution, but frequently a compromise is necessary for throughput. The necessity to analyze data sets accurately, efficiently, and in true 3D has been a major bottleneck in deriving reliable relationships between altered neuronal function and changes in spine morphology. In this chapter, we provide protocols for both imaging and analysis scenarios, starting with imaging dendritic segments (Basic protocol 1), followed by the protocols for complete spine analysis (Basic protocol 2), and full neuron imaging and analysis (Basic protocol 3 and 4).
BASIC PROTOCOL 1
TITLE: IMAGING OF FLUORESCENTLY LABELED DENDRITIC SEGMENTS
High-resolution confocal microscopy is used to visualize spines on dendritic segments or complete neurons. Prior to performing microscopy, a number of labeling techniques can be employed which may include GFP-viral transfection, iontophoretic injection of a fluorescent dye (cell loading), DiOlistic labeling, or viral expression. We have routinely used iontophoretic injection of Lucifer Yellow in our laboratory; however, the Alexa Fluor dyes have also been used because of larger variety of choices (Boyan and Liu, 2014; Ding et al., 2009; Knafo et al., 2009a; Knafo et al., 2009b; Merino-Serrais et al., 2011; Wallace and Bear, 2004). For cell loading, it is imperative to fill a neuron sufficiently to the distal tips of all apical and basal dendrites or up to 10 minutes depending on the experimental conditions. It is also important not to overfill a neuron to avoid dye leaking out of the cell or dye-coupling with other neurons which can lead to multiple “ghost” neurons being filled. Once neurons are labeled with the fluorescent marker of choice, the tissue can be taken to the microscope. A confocal microscope with objective lenses that have a low numerical aperture for low resolution and a high numerical aperture for high resolution should be used. Imaging parameters for the confocal microscope should be selected for the particular fluorophore used, animal tissue, and confocal microscope. It is important to note that the same imaging parameters must be used for the entire study.
Materials
Charged or gelatin subbed glass slides (Fisherbrand Microscope slides; Fisher Scientific)
Glass coverslips (number 1.5; Sigma-Aldrich, St. Louis,MO)
Imaging spacers (SecureSeal, Electron Microscopy Sciences, Hatfield, PA) or nail polish to seal coverslips to the slide.
VectaShield mounting medium (Vector Laboratories, Burlingame, CA), or any other preferred anti-fade fluorescence mounting media.
Labeled tissue
Confocal microscope equipped with low and high numerical aperture objective lenses.
Immersion media; choose an immersion media that matches the specifications of the high NA objective that is on your system. Examples of immersion media and their refractive index (RI) are oil (RI = 1.53–1.54), glycerol (RI = 1.47) and water (RI = 1.34). This and the lens choice should also be based on what medium the sample is mounted in (aqueous, etc.) for optimal refractive index matching and aberration-reduced imaging.
Setting up the confocal microscope
Optimize the confocal laser scanning system for the particular fluorophore by matching the laser line to the excitation peak of the fluorophore (e.g., Argon 458 nm line for Lucifer Yellow).
Match the dichroic beam-splitter in front of the laser to the laser used.
Select an emission filter to maximize the amount of collected photons. We recommend a long-pass image filter unless multiple fluorophores are used or there is background photon emission due to autofluorescence.
Choose appropriate image resolution (e.g., 512 × 512) and bit depth (16 or 8 bit). Most confocal microscopes can image with at least 12-bits. Filled neurons with large, bright soma and fine processes could benefit for more intensity resolution (assuming the sensor itself has the dynamic range).
Taking high-resolution images of dendritic segments or sections of cells
- Sample the dendritic segment in an unbiased manner. Sampling should be random (for example, do not choose the best-looking spines). Choose a segment based on brain region, type of cells, and the type of inputs into the cell you wish to obtain. Measurements can be taken on entire neurons (see below), at specific distances from the soma, or by branch orders. In practice, it is best to choose segments that can be captured in a single field-of-view without exceeding the working distance of the highest resolution objective that will be used. In addition, we recommend avoiding bifurcations, sampling from primary dendrites (due to variation in spine density compared to higher order branches), and sampling the first and last 10% of the dendrite.
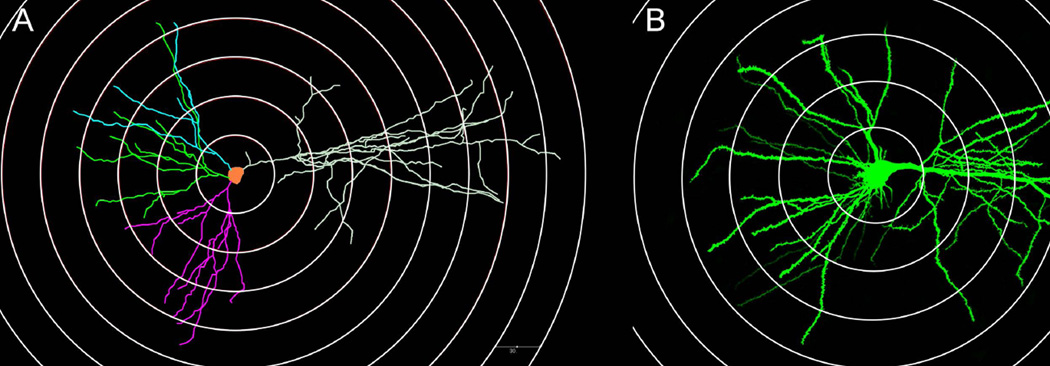
- If the neuron has been previously traced (using MBF Bioscience’s Neurolucida live on a wide-field microscope, or Neurolucida 360 on previously acquired images), use Neurolucida 360 Explorer to display the reconstructed trace with 3D Sholl rings placed at specific distances superimposed on the neuron (Figure 1A) to determine where to image along the dendritic arbor.
- Alternatively, Zen Zeiss software can be used to capture a low-resolution 3D image montage (e.g., with 10× objective) of the complete neuron and select the desired segments from this image by using the ruler tool and measuring at the desired distance, such as 50 or 100 µm from center of the soma, or desired branch order (Figure 1B). Note that this method selects branches on the basis of 2D alignment, instead of 3D distance.
Image the dendritic segment. Regardless of how you choose to sample segment, locate the neuron at low magnification (10×). Once the segment of interest is located, switch to a high numerical aperture, high magnification objective (63× or 100×), locate the segment of interest again, and place it in the center of the field of view. We recommend imaging dendritic segments at high magnification (63× or 100× with an additional digital zoom) to obtain the fine structural detail of each spine.
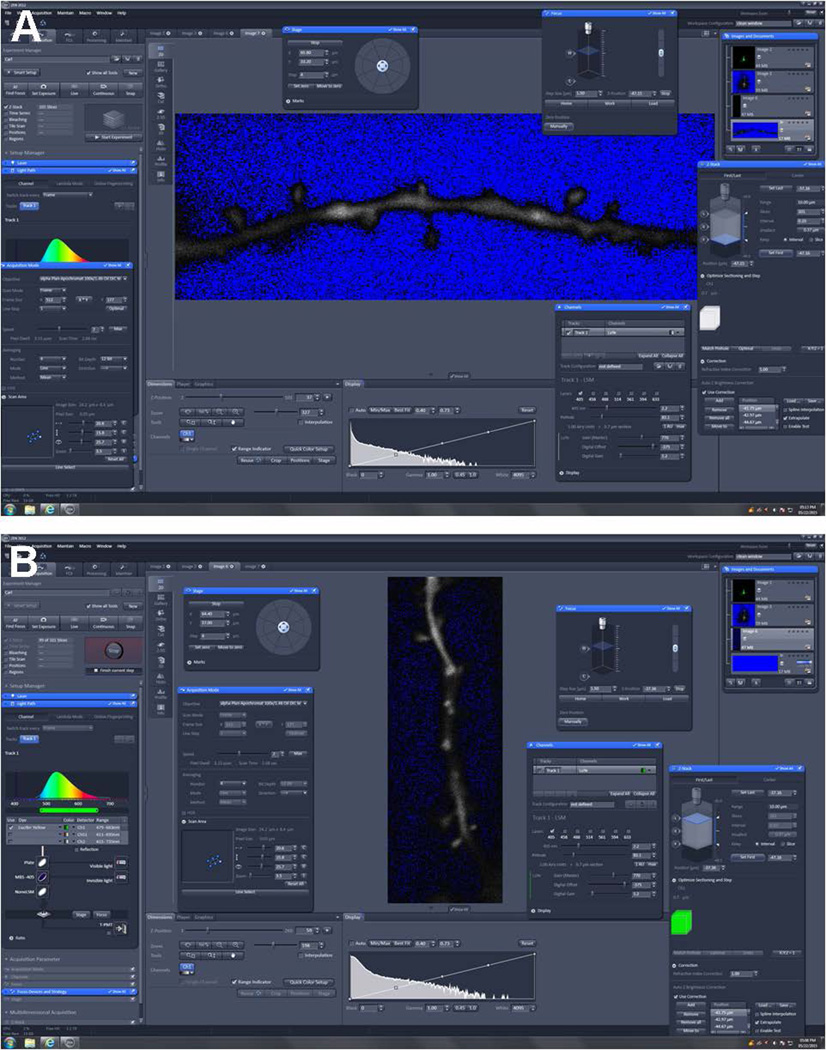
Set gain and offset to acquire the optimal image. It is very important to avoid saturated pixels. If the dendrite is saturated when imaging dendrites and spines, the size and shape may appear altered. This can lead to some spines (especially those above and below the dendrite) not being detected or not being properly characterized. Moreover, saturated pixels will result in incorrect deconvolution outcomes as described in the next section. Essentially, do not strive to get the perfect publication picture with minimized background noise. Instead, get an image with some background noise to facilitate successful post-processing deconvolution. Set the gain so that there are relatively few saturated pixels in the dendritic portion of the image and relatively zero-intensity pixels in the background of the image. A special color palette can be used on most microscopes to visualize saturated and zero intensity pixels in distinctive color (commonly red for saturated pixels and blue for pixels at zero; Figure 2).
Select the averaging number, frame direction, and other parameters; they must be maintained throughout the entire experiment. We generally use a line or frame average of 4, and a pixel dwell time of approximately 6 µm per pixel. Reduced scanning speed produce cleaner images, which can lead to more accurate reconstructions.
Select a pinhole size at “1 airy unit.” It is important to resist opening the pinhole to obtain a brighter signal with less noise. Resolution along the Z (or optical) axis decreases with increasing pinhole size.
Set the X and Y parameters to 0.05 µm and the z-interval to 0.1 µm to allow the most accurate imaging (other laboratories may use different scaling to achieve a cubic voxel (Golden et al., 2013; Hao et al., 2006; Rocher et al., 2010)). We tend to spatially oversample in XYZ given the small size of spine heads and necks. Larger pixel sizes may miss these structures, and thus important information regarding the number of thin spines and length of neck will be underestimated. In addition, deconvolution can also benefit from the added information provided by oversampling.
Set the RI correction value. This is an essential step to minimize the immersion RI mismatch of your sample to reduce the spherical aberration. The best-case scenario is immersion media RI = sample RI.
Acquire the Z stack of the segment. Make sure to capture enough planes above and below the dendritic segment so that the entire segment for analysis is contained within the image stack. This will ensure complete resolution of the top and bottom of your sample and proper quantification and characterization of spine heads. A common practice is to acquire at least 0.5 µm above and below the sample.
Figure 1.
Neuronal map for determining the dendritic segments to image. (A) 3D reconstruction of a CA1 pyramidal neuron with superimposed Sholl rings created in Neurolucida Explorer (MBF Bioscience). (B) Low magnification confocal image of a CA1 pyramidal neuron with superimposed concentric circles at measured distances from the center of the cell body using the Zeiss Zen 780 software. Note: the neurons in this figure are not the same.
Figure 2. Example of optimally set gain and offset of dendritic segments.
Images were acquired on a Zeiss 780 laser scanning confocal microscope equipped with the Zen software. (A) Image demonstrates an open Zen operating window for high-resolution dendritic Z-stack imaging. Note the longitudinal dendritic segment with the presence of spines. Within this image, the gain (red) and offset (blue) are properly set to acquire the optimal finalized Z-stack image. The gain is set so there are virtually no saturated pixels (red) present prior to imaging. The offset (blue) appears somewhat mottled within a black background. This adjusted level of background noise is necessary to facilitate successful post-processing deconvolution. All other parameters (frame resolution, averaging number, frame direction, etc) are set prior to imaging. The standard ZEN operating window is open, along with some of the more advanced functions needed for high-resolution 3D imaging (e.g., Z-stacks, focus, etc). (B) A vertical dendritic representation with incorrect offset (blue) settings. While there is no over saturation of the gain (red), the offset (blue) is too high (not enough blue demonstrated). The effect on image reconstruction would be altered.
SUPPORT PROTOCOL
TITLE: POST-PROCESSING DECONVOLUTION
Deconvolution is a computational method that attempts to correct the optical distortion inherent in all light microscopic imaging systems (Holmes, 1992). This distortion, or point spread function (PSF), blurs all light passing through a microscope on its way to the detector and is heavily influenced by several system features, including the numerical aperture (NA) of the objective lens, and the refractive index of the objective lens immersion medium, specimen, and specimen mounting medium (Gibson and Lanni, 1992; Nasse and Woehl, 2010). Deconvolution of confocal image stacks results in images with an improved signal-to-noise characteristic and more easily resolved structures. Specifically, for dendritic spines, deconvolution allows better discretization of adjacent spines and well as more accurate measurements and classification (Rodriguez et al., 2008). Proper deconvolution is necessary for images with dendritic spines to reduce the optical Z-smearing of dendrites and spines in the ZY projection. Incomplete deconvolution can distort individual spines and cause problems when quantifying spine densities (as individual spines many not be discernible) and classifying spines into specific types (Rodriguez et al., 2008). Such errors can significantly impact the final data.
Materials
AutoQuant Image Deconvolution Software (Media Cybernetics; other software programs for deconvolution include those from Scientific Volume Imaging, Zeiss, FIJI, Matlab, etc., refer to their operating instructions for use).
Minimal Computer Specification: 2.8 GHz Intel® quad-core 64-bit processor (Core i7 series) or better; RAM: 16 GB memory or higher; OS: Windows® 7 (64-bit); Graphics Card: 2 GB and OpenGL® 4.2 or higher (e.g., NVIDIA GeForce® GTX series, http://www.mediacy.com/index.aspx?page=AutoQuantX3_sys_req).
Confocal images (check with the deconvolution software to ensure that native file formats from the microscope imaging software are compatible).
Deconvolving images captured with a confocal microscope
- Open images in AutoQuant and make sure that they are correctly imported.
- If licensed deconvolution software is not available, one can deconvolve using some freeware software such as ImageJ (http://imagej.net/Parallel_Iterative_Deconvolution)
Microscope settings (e.g., objective, N.A., voxel size) should be automatically imported. Ensure that these setting are correct and manually enter where necessary.
Enter the emission wavelength.
Save the output file as a 16-bit .tif file.
Record the image scaling so that it can be entered into Neurolucida 360 for analysis.
BASIC PROTOCOL 2
TITLE: DENDRITIC SPINE MODELING AND RECONSTRUCTION WITH NEUROLUCIDA 360
Morphometric analysis of image data containing dendritic branches with spines
Neurolucida 360 from MBF Bioscience is a software platform for reconstructing neuronal morphology by tracing, editing, and visualizing image data from light microscopes in 3D. It is based, in part, on the laboratory version of NeuronStudio, originally developed at the Icahn School of Medicine at Mount Sinai by Susan Wearne and her colleagues (Rodriguez et al., 2003; Rodriguez et al., 2008; Rodriguez et al., 2006; Wearne et al., 2005). Key components and algorithms implemented in NeuronStudio for process and dendritic spine reconstruction have been further developed in Neurolucida 360. Built with three algorithms for user-guided and automatic tracing, Neurolucida 360 accurately models neurons visualized with multiple methodologies and imaging techniques. Further, when the algorithms are operated in user-guided mode, the researcher can switch algorithms on-the-fly to adjust for differing conditions along a single dendrite. Automatic dendritic spine detection models the protrusions from dendrites using a mesh to capture the surface and a 5-point segment to model the spine backbone. This results in a more accurate representation of the spine length and shape for better spine classification as well as a mechanism to modify the branch assignment when spines and branches are densely packed. The companion software for analytics, Neurolucida 360 Explorer, calculates a large number of metrics, including volume, length, plane angle, surface area, and includes a notation of whether the spine was classified and how it was classified (manual or automatic).
Materials
Neurolucida 360 v2.7 or later (MBF Bioscience).
Neurolucida 360 Explorer
- Computer requirements: 2.8 GHz Intel® quad-core; RAM: 16 GB memory or higher (32 GB needed as image size increases); OS: Windows® 7, 8 or 10 (64-bit); Graphics Card: AMD Radeon series GPU with 2GB graphics memory or more (http://www.mbfbioscience.com/neurolucida360).
-
◦Performance can vary quite drastically depending on changes in hardware (for example, solid state drives (SSDs) will speed up loading/saving quite dramatically)
-
◦
Image data with known scaling either embedded within the file, or written in your laboratory notebook. File formats accepted by Neurolucida 360 include: tiff, lsm (Zeiss), czi (Zeiss), oib/oif (Olympus), VSI (Olympus), lif (Leica), jpx, ids/ics (Nikon), bigTIFF (btf, ImageJ/FIJI).
Protocol steps
Refer to the video “Automatic dendritic spine modeling with Neurolucida 360” (DendriticSpines_with_Neurolucida360.mp4) for demonstration of this protocol.
- Load the image stack into Neurolucida 360 by dragging and dropping the file into the main window.
- Critically important: confirm the image scaling parameters and immersion media for proper scaling. Adjust the values if inaccurate, or provide values if image scaling is not embedded in the image (e.g., .tiff images).
- Zoom in to the region of interest using the mouse scroll wheel. Alternatively use the pivot point icon to center the image. Click the Tree icon and select the user-guided tracing mode. In the drop-down menu select the most suitable algorithm for the image data loaded. If desired, select the “pan to window center” checkbox to have the image re-center as you trace.
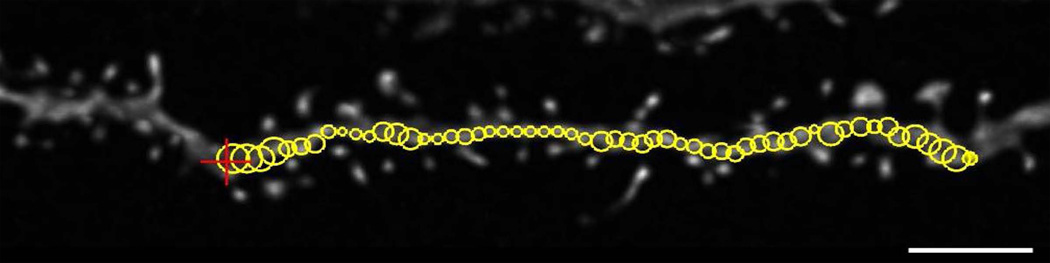
Trace the backbone of the dendritic branch. Left-click to place the initial start point, move the mouse cursor along the branch to see the path (represented by open circles) and estimated thickness provided by the tracing algorithm. Click to have the algorithm trace between points (control+Z will undo the last point). The size of the open circles provides a preview of the thickness of the dendritic branch. Ensure that large spines do not over-influence the thickness by adjusting the path of the proposed trace. To end the trace, right-click (Figure 4).
Record the tracing algorithm in the laboratory notebook, indicating which algorithm(s) was (were) used for tracing. Note: if automatic tracing is used, record all relevant parameters in addition to the tracing algorithm in the laboratory notebook.
Once traced, inspect the tracing to ensure that the dendritic branch is accurately modeled for thickness in all 3 dimensions. Edit points as necessary (Figure 5). To edit, click the Edit button. Select the point mode to visualize the trace points of the branch. Each point can be moved individually by clicking the point with the left mouse button, or move points as a group (control+click to select multiple points) to adjust the position (control+Z to undo the last change). The dendritic diameter can be modified at each point by adjusting the thickness slider or by typing in a new value.
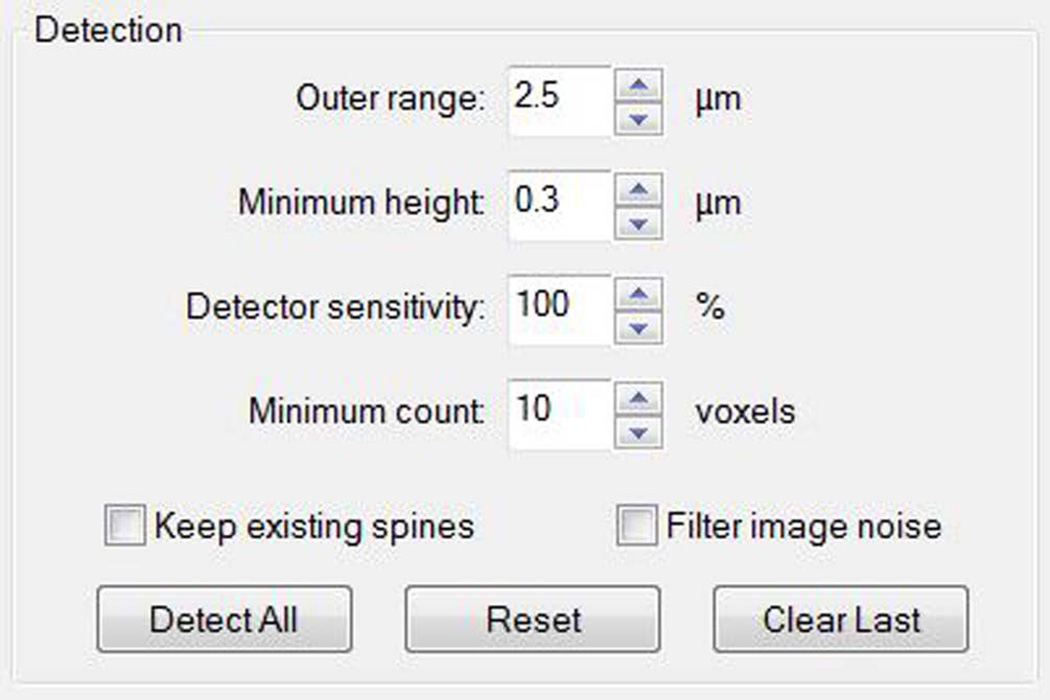
- Model dendritic spines. Click the Spine button to select the Spine detection mode. Detect all, and inspect the results. This will help parametrize the following settings (Figure 6):
- Outer range – this value defines the maximum distance from the dendritic surface that is used to search for spines.
- Minimum height – this value defines the minimum distance from the dendritic surface for a surface protrusion to be considered a spine. It helps prevent false positives due to surface irregularities. Reduce this value if the spine base is too large.
- Minimum count – this value is used to exclude individual objects that are too small to be considered a spine. This can be useful when trying to prevent image noise from being detected as spines.
- Alternative: Use Filter image noise, an image pre-processing filter that can be used with non-deconvolved images.
- Adjust parameters until the spine modeling algorithm detects the spines as desired.
- Alternatively, click a spine in the image to detect it.
- Alternatively, detect spines on a single branch of a complex dendritic arbor by selecting “use click to detect all spines on branch.”
- It is important to use the same parameters throughout the study; do not choose parameters arbitrarily since this will introduce bias and may affect the data and interpretation.
Record the detection parameters in the laboratory notebook (Figure 6).
- Edit dendritic spines. Use the Edit mode to adjust the detection by splitting and merging spines. Each detected object is displayed in a different color. You may toggle the visibility of all reconstructed objects by using the +/− tool bar button. This allows you to see the underlying image data to determine if adjacent meshes need to be combined to make one spine, or if multiple spines are encompassed by one mesh that needs to be split (Figure 7).
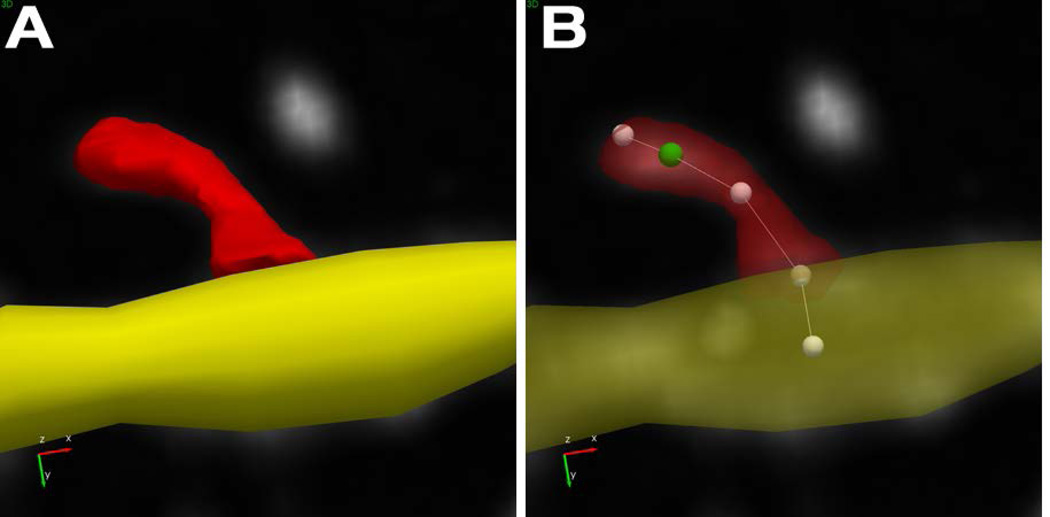
- Advanced editing: When detecting spines on multiple branches, make sure to confirm that the spines are properly assigned to the correct branch. To view and change the branch assignment, select the Points button. The spine backbone is displayed as a series of 5 points. Move the point of attachment to the new branch by selecting and dragging it to the new location. The spine will be re-detected and assigned to the specified branch (Figure 8).
Save the data file.
Perform morphometric analyses. Open the data file in the Neurolucida 360 companion program, Neurolucida 360 Explorer.
- From Neurolucida 360 Explorer’s main menu, select Branched Structure Analyses from the Analysis menu. Click the Spines tab and select reports available with spines, spine details, and dendrites.
- The Dendrite Spine report includes the total number (and type, if classified), spine density per micrometer of dendritic length.
- The spine details report includes the many metrics, including total extent (the measure of the shortest-path distance from the dendritic surface to the furthest voxel of the spine) and spine backbone length (the length of the spine from the furthest included voxel along the backbone to the insertion onto the dendritic branch.), head diameter, head:neck ratio.
- Note: other analyses may be of interest, depending on your scientific question.
Figure 4. Tracing the backbone of the dendritic segment.
Dendritic segment seen here is from a mouse pyramidal CA1 neuron filled with Lucifer Yellow. A cursor (red +) is moved along the dendritic segment to see the path and estimated thickness provided by the tracing algorithm. The open yellow circles provide a preview of dendritic branch thickness. It is important to confirm that large spines do not over-influence the thickness of the dendrite. Scale bar = 2 µm.
Figure 5. Inspection of dendritic segment thickness.
(A) Neurolucida 360 showing inspection and correction of points (green) from the dendritic branch (yellow) that were drawn off center by the large spine (arrow). (B) To edit, view the branch in point mode, select the point, and move it to the correct, centered location on the dendritic branch. Scale bar = 0.5 µm.
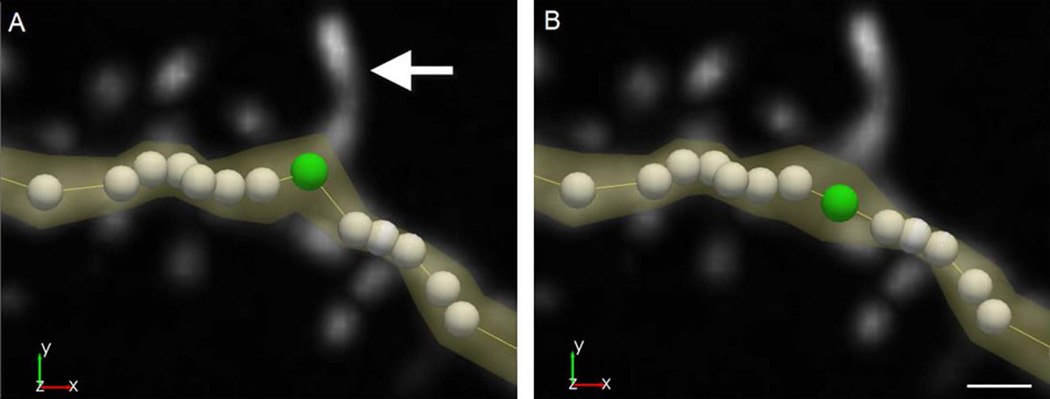
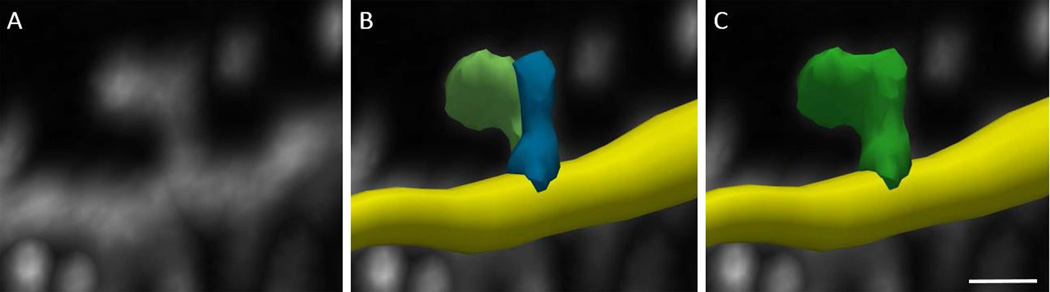
Figure 6. Merging dendritic spines.
Individual spine detections can be merged to create a single model for a dendritic spine. Inspecting the underlying image data (A) can show that a single spine was inaccurately modeled as two discrete objects (B). To correct, select each spine object in edit mode and select merge. The software remodels the spine to include all voxels previously split between the two objects as a single spine (C). Scale bar = 1 µm.
Figure 7. Dendritic spine detection parameters.
The software has four detection parameters to set the conditions for modeling dendritic spines. The parameters, which will vary based on the imaging settings and experimental paradigm being tested, should be chosen based on empirical data, and remain constant during the study. Do not choose parameters arbitrarily since this will introduce bias and may affect the data and interpretation.
Figure 8. Representation of dendritic spines in Neurolucida 360.
The dendritic spine is modeled with a mesh to represent the surface and volume of the spine (A). The spine backbone (B) is represented with five points. The most distal point in the backbone indicates the furthest voxel from the dendritic surface, the centroid of the spine head (green) is the second point, and the last point represents where the spine connects with the dendrite. The shape of the dendritic spine is more accurately modeled, leading to better metrics and more complex spine classes. Spines can be re-assigned to nearby branches by dragging the last point from the original dendrite location to the desired location on the alternate branch.
ALTERNATE PROTOCOL 2
TITLE: SPINE CLASSIFICATION
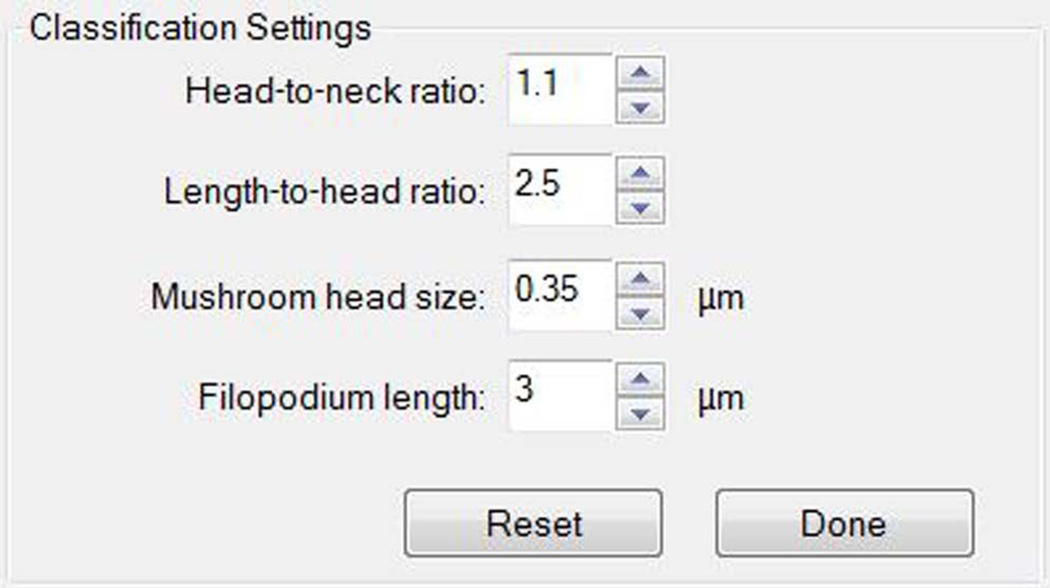
Accurate and effective dendritic spine classification remains a fundamental challenge for the neuroimaging research community. Dendritic spine morphology is thought to be crucial in synaptic plasticity and strength due to its compartmentalization of biochemical and electrical signals. A dendritic spine is a micron-sized protrusion, comprised of a spine head, where the excitatory synapse is located, and a spine neck that connects the spine to the dendritic shaft. Spines come in multiple shapes and sizes, with the more common subclasses being thin, mushroom and stubby (Harris et al., 1992; Jones and Powell, 1969; Nimchinsky et al., 2002; Peters and Kaiserman-Abramof, 1970; Spacek and Hartmann, 1983). Digital representation of spines using light microscopy has traditionally relied on manual counting from a computer screen and is prone to subjective errors. Despite the recent introduction of semi-automated tracing methods (e.g., NeuronStudio, Vaa3D, and Imaris), the problem of detecting and characterizing spine shapes automatically, in 3D, remains unsolved. Automatic classification with Neurolucida 360 greatly reduces human subjectivity and intra-operator variability. Dendritic spines from different brain areas, developmental stages, pathological conditions, or species may require different classification settings. Neurolucida 360 permits simple adjustment of the classification settings, however it is important that the classification settings are chosen on the basis of empirical research and remain unchanged throughout the study.
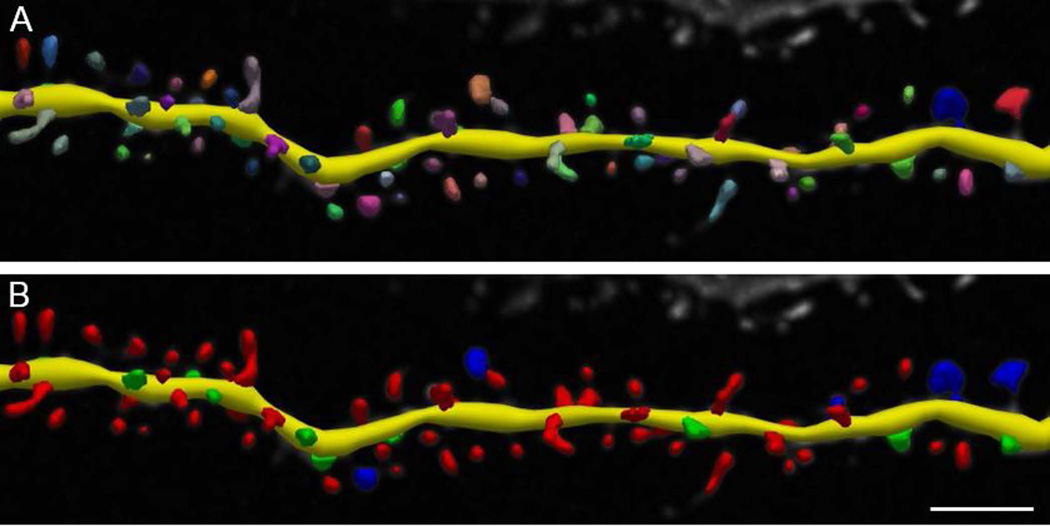
After automatic detection with Neurolucida 360, dendritic spines can be automatically classified with a simple one-button operation into one of the following types: stubby, mushroom, thin, or filopodia (Figure 9). Alternate complex types (e.g., double-headed) can be assigned manually after detection. Each spine type is color-coded for easy visualization, and can be interactively re-classified to a different canonical or complex type. By default, spine classification is assigned according to parameters determined by Rodriguez et al. (2008), which were empirically determined for mouse hippocampal neurons as the consensus from multiple experienced researchers. Default settings for detection and classification should be considered starting points and not mandates. Both detection and parameters and classification settings will be influenced by imaging methodology as well as neuron characteristics (Benavides-Piccione et al., 2002; Harris et al., 1992; Nimchinsky et al., 2002; Rodriguez et al., 2008). Report quantification parameters in your manuscripts so that others can interpret, replicate, and build upon your work. When selecting parameters based on previously published peer-reviewed papers, note that papers that use manual spine measurements are typically measuring spine head sizes in the lateral XY dimension only, which may not be directly comparable to true 3D detection and measurement.
Figure 9. Spine classification using default parameters.
Dendritic spines are first modeled on the basis of a number of detection parameters, including distance from dendritic surface and apparent size. After detection, spines are colored to differentiate each modeled spine in close proximity (A). If desired, the detected spines can be classified using classification parameters as established by Rodriguez et al., 2008 (B) or through custom specifications. It is important that the same detection parameters and classification settings (if chosen) are used for all images in the experimental study. Scale bar = 2 µm.
Materials
Neurolucida 360 v2.7 or later (MBF Bioscience)
Neurolucida 360 Explorer (MBF Bioscience)
- Computer requirements: 2.8 GHz Intel® quad-core; RAM: 16 GB memory or higher (32 GB or more needed as image size increases); OS: Windows® 7, 8 or 10 (64-bit); Graphics Card: AMD Radeon series GPU with 2GB graphics memory or more (http://www.mbfbioscience.com/neurolucida360).
-
◦Performance can vary drastically depending on changes in hardware (for example, solid state drives (SSDs) will dramatically speed up loading/saving image files).
-
◦
Data file from Neurolucida 360 with modeled dendritic spines.
Protocol steps
Step annotations
- If desired, classify the dendritic spines. Click the Classify button to instantaneously re-color the detected spines according to spine class. By default, the spines are colored red (thin), blue (mushroom), green (stubby), and yellow (filopodia) according to the metrics determined by Rodriquez et al., (2008).
- Alternatively: different metrics can be entered to define the spine classes according to values specific to the species or condition under study. Click Settings to enter the values appropriate for your experimental paradigm.
Record the classification parameters in your laboratory notebook (Figure 10).
Save the data file.
Open the data file in Neurolucida 360 Explorer and select the spine details report from Branched Structure Analysis. Spine type and assigned type will be included in the spine details report.
Figure 10. Setting spine classification parameters.
Different metrics can be entered to define the spine classes according to values specific to the species or condition under study. Parameters should not be changed during the study, and should be chosen on the basis of empirical evidence.
BASIC PROTOCOL 3
IMAGING COMPLETE NEURONS
In some instances, complete neuronal reconstructions from high-resolution images (e.g., 1024×1024) are desired in order to analyze both neuronal complexity and spine density on entire neurons. Depending on the microscope and software capabilities, the entire neuron can be captured using an automated tiling function, with an appropriate amount of overlap between tiles (e.g., 10%), along with the Z-step distance. If there is no such tiling option, individual stacks need to be separately acquired at high resolution and magnification, and then stitched together using specific software programs such as Neurolucida 360 or Volume Integration and Alignment System (VIAS; http://research.mssm.edu/cnic/tools-vias.html). As in automated tiling performed by the acquisition software, there must be an appropriate amount of overlap (e.g., 10%) to accurately stitch the segments together as a post-acquisition operation.
Find the neuron of interest on the microscope using the lowest magnification objective (e.g., 10×). An initial rapid image acquisition of the overall Z-depth and area is needed to set up for the high-resolution high magnification acquisition that follows this step.
In order to determine both the area and maximum Z-depth using rapid acquisition methods, first set the system up as follows: change image resolution to a lowered value (e.g., 256×256), change pixel dwell time to its maximum speed (fastest pixel dwell time), and open the pinhole aperture to its maximum size (largest optical slice thickness). Using these conditions allows for rapid determination of the neurons area and Z-depth. Start image acquisition and adjust for gain/offset signal/background). Since the Airy Unit aperture setting is opened to its maximum, the image adjustments are used to optimize observation of the entire neurons. If the neurons dendritic branches extend beyond the field of view, change the digital magnification from Z=1 to Z=0.9. Continue this process until you have the entire neuron in the field of view (e.g., Z=0.8, etc).
Start the Z-step function by clicking on the Z-stack window. Using the Z-step function, determine the upper and lower limits to include both the material, along with an additional Z buffer distance both above and below the tissue. It is important to capture all of the neurons terminal dendritic branches. Record both the Z-depth and the total area of the neuron.
If needed, a low-resolution 3D capture can be taken at this time. Otherwise, set up for high-resolution full neuron capture. Do not move the XY position. Change to an oil-immersion objective (e.g., 63× oil/N.A. 1.4 or 100× oil/N.A. 1.4). Change the pinhole value to 1 Airy Unit and optimize for offset/gain.
Set and optimize the parameters for high-resolution image capture, (e.g., 1024×1024 resolution, increased pixel dwell time, frame average of 2). Open the image information tab and record the x/y pixel resolution (e.g., 0.5×0.5×0.5 µm).
Disengage the Z-step function, and open tiling by clicking open the tiling function window. Open the function and setup (if available) for on-line stitching. Determine the number of overlapping (8–10%) tiles needed to acquire the entire neuron using the area value from the low-resolution image.
Engage the Z-step function. Using the Z-depth values from the low-resolution image, set the interval for voxel dimensions (e.g., 0.5×0.5×0.5 µm). Turn “online stitching” on. Set image overlap at 8–12% (Figure 11).
Ensure both tiling and Z-stack functions are engaged, and that all image parameters are set. Image the neuron. Alternatively, image Z-stacks can be saved independently and stitched using off-line programs (Figure 12).
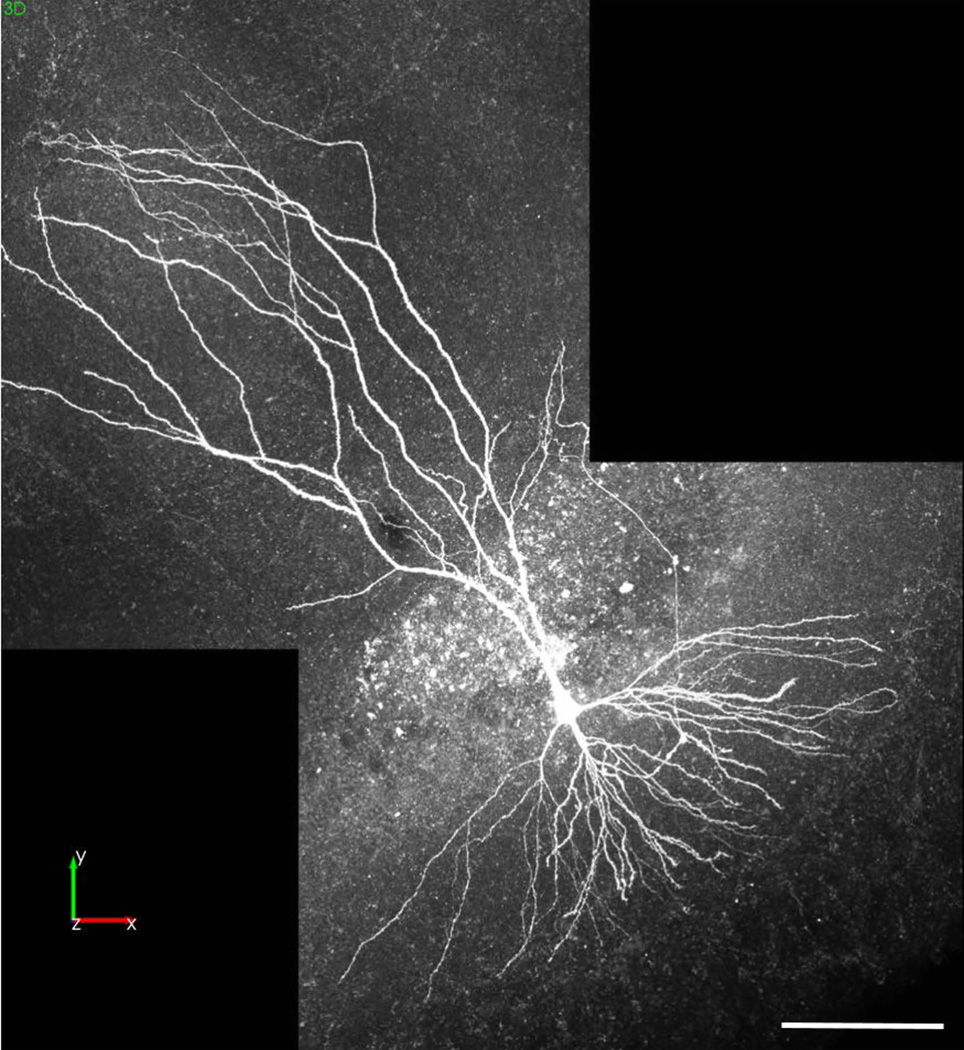
Figure 11. Complete neuron image montage of a hippocampal pyramidal neuron acquired after whole-cell recording.
A single image was created using Neurolucida 360 to montage multiple 3D images of a mouse hippocampal pyramidal neuron labeled with biocytin. Multiple 2-channel z-series images were collected with a Zeiss LSM 710 confocal microscope, mounted on an AxioImager Z2, with a 20× Plan-apochromat objective, and a 25 mW multi-wavelength (458/488/514) argon laser and a 20 mW 561 nm diode DPSS laser (Alexa-549 displayed here). Dr. Piskorowski provided the image data from an experiment performed by Vincent Robert and Ludivine Therreau in accordance with European guidelines for the care and use of laboratory animals at the Université Paris Descartes. Note: this neuron was not imaged at a resolution high enough for concurrent spine analysis. Scale bar = 100 µm.
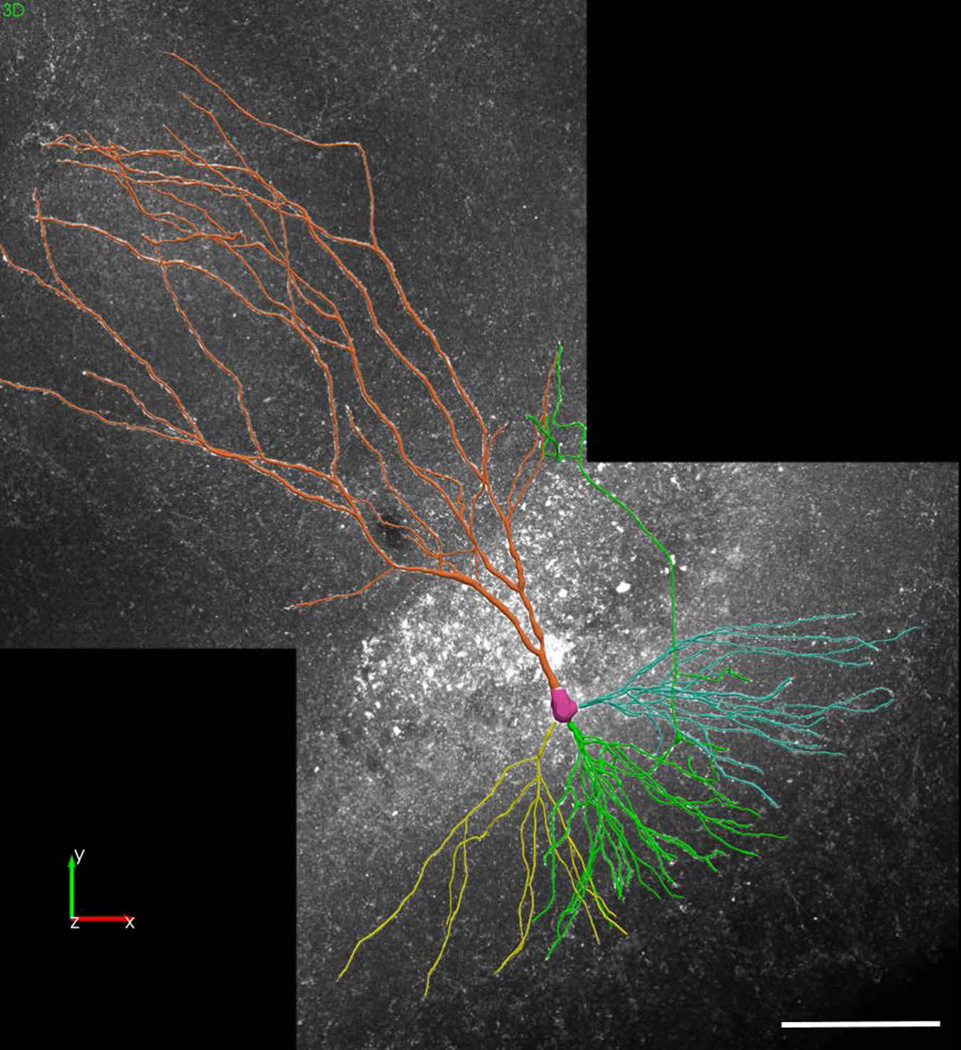
Figure 12. Complete neuron reconstruction created with Neurolucida 360.
Using Neurolucida 360, the pyramidal cell shown in Figure 11 was reconstructed using user-guided tracing and soma modeling. Scale bar = 100 µm.
BASIC PROTOCOL 4 (optional)
TITLE: NEURON RECONSTRUCTION USING NEUROLUCIDA 360
Instructions for reconstructing the entire neuronal structure follow the basic protocol for reconstructing the single dendritic segment described previously, with a few additional steps to model the soma and confirm the origin of the trees at the base of the soma.
Materials
Neurolucida 360 v2.7 or later (MBF Bioscience)
- Computer requirements: 2.8 GHz Intel® quad-core; RAM: 16 GB memory or higher (32 GB or more needed as image size increases); OS: Windows® 7, 8 or 10 (64-bit); Graphics Card: AMD Radeon series GPU with 2GB graphics memory or more (http://www.mbfbioscience.com/neurolucida360).
-
◦Performance can vary drastically depending on changes in hardware (for example, solid state drives (SSDs) will dramatically speed up loading/saving image files)
-
◦
Image data with known scaling either embedded within the file, or written in your laboratory notebook. File formats accepted by Neurolucida 360 include: tiff, lsm (Zeiss), czi (Zeiss), oib/oif (Olympus), VSI (Olympus), lif (Leica), jpx, ids/ics (Nikon), bigTIFF (btf, ImageJ/FIJI).
The width, height, number of focal planes, and depth of each pixel all contribute to the size of image files. Compression algorithms reduce the impact of image dimensions on the storage system. Though compression has no direct impact on the memory required to navigate an image, a good rule of thumb we have found is to have two times the on-disk file size in available memory when using Neurolucida 360.
Protocol steps
- Load the image stack into Neurolucida 360 by dragging and dropping the stack into the main window.
- Critically important: confirm the image scaling parameters and immersion media for proper scaling. Correct the values if inaccurate, or provide values if image scaling is not embedded in the image (e.g., tiff images).
- Zoom in to the region of interest using the mouse scroll wheel. Alternatively use the pivot point icon to center the image.
To reconstruct the soma. Click the Soma button to enter the Soma mode. Adjust the circular cursor using Control+scroll wheel to create a discrete search region for the algorithm. Click the soma in the image to model it. If the soma is rendered as a cube, reduce the sensitivity, clear the soma, and re-detect.
Trace the backbone of the dendritic branch. Click the Tree button. Select the user-guided tracing mode. In the drop-down menu, select the most suitable algorithm for the image data loaded. Select the “pan to window center” checkbox to have the image re-center as you trace. Click to place the initial start point, move the mouse cursor along the branch to see the path and estimated thickness provided by the tracing algorithm. Click to have the algorithm trace between points (control+Z will undo the last point). Ensure that large spines do not over-influence the thickness by adjusting the path of the proposed trace. To end the trace, right-click (Figure 4). Continue until all branches are traced. When desired, switch algorithms while tracing to match the algorithm to the image data presented. This will reduce the amount of editing needed. Directional kernels works well for punctuate label at distal branches, Rayburst Crawl and Voxel Scooping are helpful in areas of high complexity.
Record the tracing algorithm in the laboratory notebook.
- Once traced, inspect the tracing to ensure that the dendritic branch is accurately modeled for thickness in all 3 dimensions. Edit points as necessary (Figure 5).
- Click the Edit button and select the point mode to visualize the trace points of the branch. You can move points individually by clicking a point with the left mouse button, or move points as a group (control+click to select multiple points) to adjust the position (control+Z to undo the last change). You can modify thickness at each point by using the thickness slider or typing in a new value.
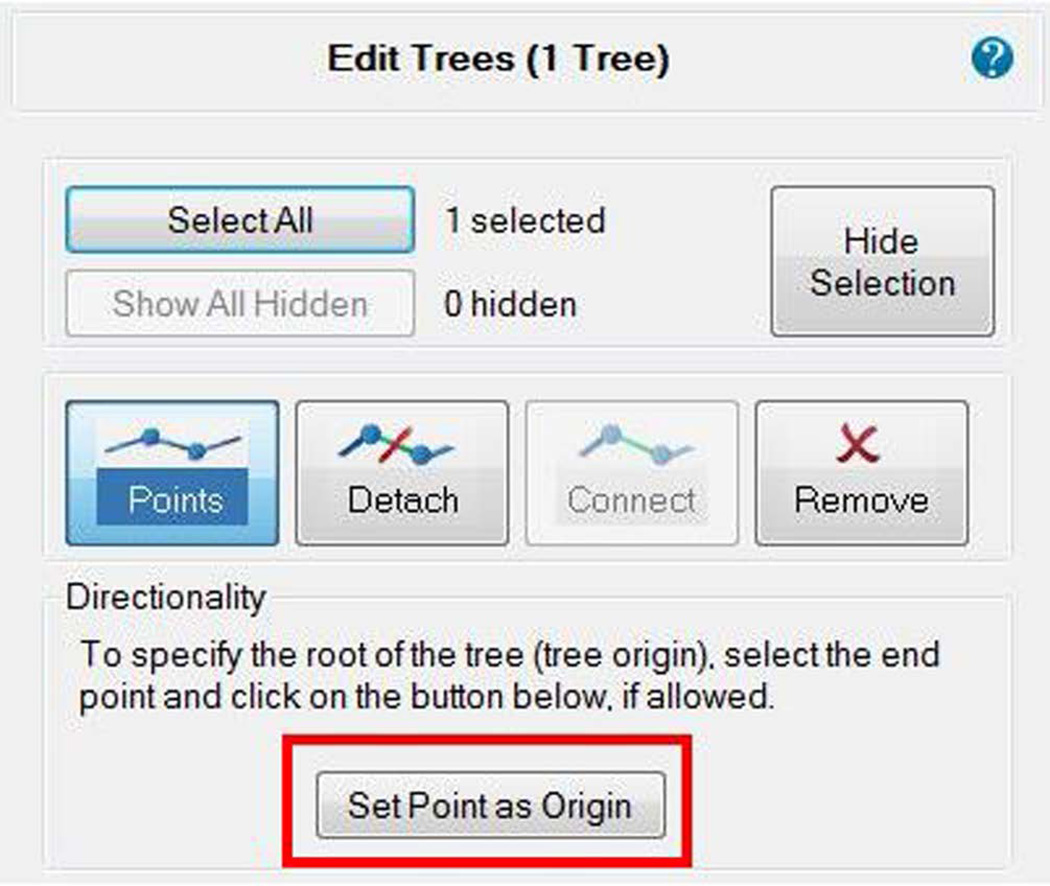
- Confirm the origin of the dendritic trees. While it is important for branch analyses to have each tree begin at the soma, it is not required to trace in any particular direction. Return to the Edit panel and select the point mode to visualize the trace points of the branch. Draw a marquee around the soma small enough so that it does not contain full dendritic trees, but large enough to contain all the points closest to the soma. Once selected, the option to set all endings to “origins” becomes available only if some trees are initiated at a different location. Select the button to reset all trees to have their origin closest to the soma (Figure 13).
Save the data file.
Proceed to Basic Protocol 2 to model dendritic spines.
Figure 13. Setting the origin of branches as the closest point to the soma.
While it is important for branch analyses to have each tree begin at the soma, it is not required to trace in any particular direction. Confirm the root of each tree in edit mode, by drawing a marquee around the soma small enough so that it does not contain full dendritic trees, but large enough to contain all the points closest to the soma. Once selected, the option to set all endings to “origins” becomes available only if some trees are initiated at a different location. Select the button to reset all trees to have their origin closest to the soma.
COMMENTARY
Background Information
Dendritic spine morphology has become a central focus in research in the fields of learning and memory, aging, and neurodegenerative diseases (for review see (Dickstein et al., 2007; Dickstein et al., 2013; Hara et al., 2012) as well as other neuropsychiatric disorders such as autism (Durand et al., 2012; Hung et al., 2008; Hutsler and Zhang, 2010; Phillips and Pozzo-Miller, 2015), schizophrenia (Hayashi-Takagi et al., 2011; Ramos-Miguel et al., 2015), and addiction (Maze et al., 2010; Selvas et al., 2015) as they are an integral component of excitatory synapses. Excitatory synapses comprise the majority of connections in the central nervous system and play a vital role in learning, memory, and cognition. Abnormal development or regulation of these synapses has been implicated in many neurodevelopmental, psychiatric, and neurodegenerative disorders. Most excitatory synapses occur at specialized postsynaptic compartments known as dendritic spines, which are tiny protrusions from the dendrites of neurons (Carlisle and Kennedy, 2005; Ethell and Pasquale, 2005; Harris and Stevens, 1989; Nimchinsky et al., 2002). Based on their morphology, spines can be divided into “canonical” types (stubby, mushroom, thin) and complex types (cup-shaped, multi-headed or branched, and filopodia) (Ethell and Pasquale, 2005; Harris et al., 1992; Kasai et al., 2010; Nimchinsky et al., 2002). The variable structure of spines determines the strength, stability, and function of the synaptic connections that facilitate the neuronal networks in the brain. Understanding the dynamics of spine morphology will help researchers to address how the brain is able to process a continuous flow of sensory information and simultaneously store and consolidate memories, sometimes for a lifetime. However, precise quantification of spines parameters has been restricted to time and labor-intensive electron microscopy, which has limited throughput and is impractical for large-scale studies.
Accordingly, there is growing interest in automating quantitative analysis of dendritic spine morphology at the light microscopic level. Traditionally, performing manual spine analysis has been, and in many situations still is, the approach of choice. This usually involves tracing dendrites and marking spines from confocal stacks as they appear in the XY plane with software packages such as Neurolucida (Brennan et al., 2009; Hao et al., 2006; Knafo et al., 2009a; Knafo et al., 2009b), Imaris (Vecellio et al., 2000), Metamorph (Wallace and Bear, 2004) and Arivis. Such counting is then followed by manual measurements of spine heads and necks using programs such as Photoshop (Hao et al., 2006). These methods, in addition to being very time-consuming, introduce much error based on observer bias and high inter-observer variability (Donohue and Ascoli, 2011). Moreover, underestimating the number of spines is a concern in these conditions since spines, which project on the Z plane of the dendrite, are often overlooked as they are obscured by the brighter dendritic shafts. Automatic algorithmic analysis of spine morphology at the light microscopic level provides for higher throughput by substantially increasing analysis speed, accuracy, and reproducibility compared to existing manual methods. In addition, it provides for observer independence and for quantitative analyses that are virtually impossible without automation (such as spine volume, surface, head diameter, neck diameter, etc.). NeuronStudio was our first semi-automated quantitative software based on the Rayburst Sampling algorithm. This software was used in multiple studies from our group and others on various animal models, such as mouse, rat and monkey, and in many brain areas including the prefrontal cortex, hippocampus, and nucleus accumbens (Bloss et al., 2011; Golden et al., 2013; Price et al., 2014; Radley et al., 2008; Rocher et al., 2010; Shansky et al., 2009; Steele et al., 2014). While programs such as Imaris (Bitplane), Amira, and NeuronStudio may make quantification easier, there remain issues regarding accuracy, in particular in defining the spine head volume and surface area (Donohue and Ascoli, 2011; Dumitriu et al., 2011).
Here, we introduce a new computational approach for detection and shape analysis of dendritic spines that incorporate the algorithms of our previous software (NeuronStudio; http://research.mssm.edu/cnic/tools-ns.html) as well as numerous improvements to make the software more broadly usable. Neurolucida 360 can read multichannel, high-bit depth images, with file sizes that exceed 50 GB. The algorithms have been tested with various labeling techniques on neurons and spines from different mammalian species (mouse, rat, monkey) and brain regions. It is important to note that successful data acquisition always relies on the quality of the materials used, and of the labeling of the cells analyzed, independent of the software performance. It is essential that imaging be performed in optimal conditions, such as these detailed in this unit. We believe that the new quantitative software package, Neurolucida 360, provides the neuroscience research community with the ability to perform higher throughput automated 3D quantitative light microscopy spine analysis under standardized conditions to accelerate the characterization of dendritic spines with greater objectivity and reliability.
Critical Parameters
Insufficient labeling of cell – the saying “garbage in, garbage out” holds true here. The best imaging systems and most sophisticated algorithms cannot correct for inconsistent or improper methodology at the bench.
Image scaling – For the most accurate estimates of quantitative measures (e.g., volume, extent, etc.) the correct image resolution and axial step size must be provided.
Troubleshooting
| Problem | Possible Cause | Solution |
|---|---|---|
| Poor images (low signal-to-noise) |
Poor/incomplete labeling of cells | Only use tissue with strong fluorescent signal and complete arborization |
| Z-smear | Identified cell beyond working distance of objective |
Image only cells that lie within 80 µm of the tissue surface |
| Low Z resolution | Microscope type (e.g., confocal, spinning disk, 2 photon), pinhole size too large, objective is not sufficient to resolve the needed features |
Adjust imaging parameters to obtain the optimal image |
| Dendrites appear distorted |
Air bubble in mounting media | Remount the tissue |
| Dendrite appears to be moving across the screen during confocal imaging |
Insufficient seal of coverslip | If you used spacers when mounting the tissue, make sure there are enough spacers used for the thickness of the tissue. Each spacer is 120 µm If you used nail polish, either there is not enough to create a proper seal or it has not hardened enough. It is always best to wait a couple of days between mounting the tissue and imaging Make sure the objective does not compress the coverslip. This happens when the working distance is mismatched with the tissue |
| Dendrite appears to be compressed while imaging |
The segment is too deep in the tissue (>80 µm) |
Image segments more parallel to the tissue section |
| Dendrites appear to be flat once reconstructed |
Objective unable to fully adjust to the Z-step depth |
Image segments more parallel to the tissue section |
| Software does not target the image |
Loading tiff images into Neurolucida 360 without correct scaling |
Correct size of voxels |
| Dendrites appear beaded |
Poor perfusion/fixation leads to poorly loaded neurons |
Cells are not usable for analysis |
| Cell loses fluores- cence intensity during imaging |
Poor perfusion/fixation leads to poorly loaded neurons |
Cells are not usable for analysis |
| Overlapping den- dritic segments from adjacent cells/same cell in same imaging plane |
Certain techniques (e.g., viral expression using GFP, DiOlistic, cells loaded too closely) |
In certain cases this is unavoidable (e.g., GFP). If dendritic segments are separated in the Z-plane they can still be used. |
Anticipated Results
After reconstruction in 3D with Neurolucida 360, a number of metrics are calculated. Using the companion software, Neurolucida 360 Explorer, spine analyses include number, total extent, plane angle, volume, surface area, contact area, XYZ coordinates, head diameter and length, neck length, head diameter to neck diameter ratio, as well as notations of attachment, type, and method for classification. Several analyses are also available for dendrites, including but not limited to: Sholl analysis, branch order, dendritic length, number of branch points, spine density, spine density by type, convex hull, polar histogram. These analyses can be exported directly to Microsoft Excel. The data file can also be exported in a format suitable for third-party 3D rendering software (e.g., Blender), or further utilized with programming software (such as MatLab) to perform additional computations.
Time Considerations
Image acquisition
Acquisitions of dendritic segments or full neurons are the most time-consuming aspects of dendritic spine analysis. The time it takes to image dendritic segments can vary depending on the depth of the segment, the Z-step, scan time, and averaging. It is important to avoid shortcuts when acquiring confocal images to preserve the fine structural details of dendritic spines. Moreover, it is important to remain unbiased when choosing dendritic segments to image (e.g., avoid only imaging shallow dendritic segments to save time).
The time to image a complete neuron will vary depending on the type of data to be collected from the cell. For dendritic spine density only, neurons can be imaged at a lower magnification. For more detailed information about spines (e.g., spine type, and estimates of surface area, volume, and neck length), higher magnifications are needed but this will increase the imaging duration as more image stacks need to be obtained and stitched together. A complete neuron at 100× can take up to 10 hours to image, depending on neuron size, complexity, and microscope configuration.
Dendritic spine analysis
Automatic dendritic spine analysis with Neurolucida 360 requires less than 1 minute for branch reconstruction. Parameterization of spine detection can take 3–10 minutes, with automatic detection occurring in less than 1 minute. Spine classification is instantaneous. Manual editing of branch and dendritic spines will vary depending on the complexity of the image data. For short dendritic segments similar to those described here, editing typically requires less than 5 minutes.
Complete neuronal reconstruction
The time required for automatic montaging of a complete neuron from discrete, overlapping image stacks will depend on the number of stacks. Typically, image montaging requires approximately 5 minutes with a powerful computer. Once the image has been stitched, the reconstruction of the entire neuronal structure will also depend on the complexity. Typically, user-guided reconstruction can take between 10 minutes and 3 hours.
Supplementary Material
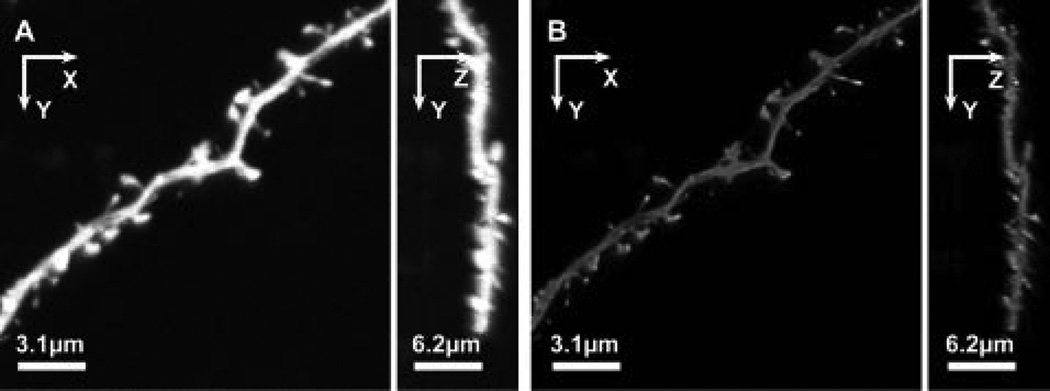
Figure 3. Deconvolution of dendritic segments.
XY and ZY maximal projections of a typical image stack before (A) and after (B) deconvolution with AutoDeblur. Compared to the raw data (A), the deconvolved data exhibit good relative intensity equalization of spines and dendrites, and significantly reduced Z-axis “stretching” from optical smear, in the ZY projection (B). Adapted from (Rodriguez et al., 2008).XY and ZY maximal projections of a typical image stack before (A) and after (B) deconvolution with AutoDeblur. Compared to the raw data (A), the deconvolved data exhibit good relative intensity equalization of spines and dendrites, and significantly reduced Z-axis “stretching” from optical smear, in the ZY projection (B). (Adapted from (Rodriguez et al., 2008).
Significance Statement.
Understanding the role of dendritic spines is an important area of neuroscience research. We introduce a methodology for performing morphometric dendritic spine analysis from 3D confocal images of dendritic segments. The protocol describes the process of selecting segments for analysis, confocal image acquisition guidelines, deconvolution, and analysis with Neurolucida 360. Neurolucida 360 improves the reliability and accuracy of spine morphometrics while providing an objective means to rapidly analyze spines in 3D. Quantitative neuron and dendritic spine analyses could accelerate the understanding of the relationship between brain structure and function under physiological and pathological conditions and thereby improve the development of novel treatment strategies for complex CNS diseases.
Acknowledgments
The development of Neurolucida 360 was funded by a NIMH Lab-to-Marketplace grant (R44 MH093011) to Paul J. Angstman and Patrick R. Hof. Videos were created by Pasang Sherpa. We appreciate the thoughtful edits by our colleague, Dr. Sandrine Dincki.
Footnotes
KEY REFERENCE
Drs. Dickstein and Tappan hosted a webinar on this topic based on a previous version of Neurolucida 360. The recorded version of the webinar is available for viewing at this link: https://youtu.be/HczuQjeNcR4
INTERNET RESOURCES
Software described in the protocol is available from the following companies. Free versions or free trials are available from each vendor.
Zen Black image acquisition software (Zeiss) http://www.zeiss.com/microscopy/en_us/products/microscope-software/zen-lite.html
AutoQuant image deconvolution software (Media Cybernetics) http://www.mediacy.com/index.aspx?page=AutoQuant
Neurolucida 360 automated neuron reconstruction software (MBF Bioscience) http://www.mbfbioscience.com/neurolucida360
VIDEOS
Automatic dendritic spine modeling with Neurolucida 360 (DendriticSpines_with_Neurolucida360.mp4)
This video demonstrates all steps in Neurolucida 360 for modeling dendritic spines on dendritic segments from loading the image data to classification.
LITERATURE CITED
- Benavides-Piccione R, Ballesteros-Yanez I, DeFelipe J, Yuste R. Cortical area and species differences in dendritic spine morphology. J Neurocytol. 2002;31:337–346. doi: 10.1023/a:1024134312173. [DOI] [PubMed] [Google Scholar]
- Bloss EB, Janssen WG, Ohm DT, Yuk FJ, Wadsworth S, Saardi KM, McEwen BS, Morrison JH. Evidence for reduced experience-dependent dendritic spine plasticity in the aging prefrontal cortex. J Neurosci. 2011;31:7831–7839. doi: 10.1523/JNEUROSCI.0839-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyan G, Liu Y. Dye coupling and immunostaining of astrocyte-like glia following intracellular injection of fluorochromes in brain slices of the grasshopper, Schistocerca gregaria. Methods Mol Biol. 2014;1082:99–113. doi: 10.1007/978-1-62703-655-9_7. [DOI] [PubMed] [Google Scholar]
- Brennan AR, Yuan P, Dickstein DL, Rocher AB, Hof PR, Manji H, Arnsten AF. Protein kinase C activity is associated with prefrontal cortical decline in aging. Neurobiol Aging. 2009;30:782–792. doi: 10.1016/j.neurobiolaging.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlisle HJ, Kennedy MB. Spine architecture and synaptic plasticity. Trends Neurosci. 2005;28:182–187. doi: 10.1016/j.tins.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Dickstein DL, Kabaso D, Rocher AB, Luebke JI, Wearne SL, Hof PR. Changes in the structural complexity of the aged brain. Aging Cell. 2007;6:275–284. doi: 10.1111/j.1474-9726.2007.00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickstein DL, Weaver CM, Luebke JI, Hof PR. Dendritic spine changes associated with normal aging. Neuroscience. 2013;251:21–32. doi: 10.1016/j.neuroscience.2012.09.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding JB, Takasaki KT, Sabatini BL. Supraresolution imaging in brain slices using stimulated-emission depletion two-photon laser scanning microscopy. Neuron. 2009;63:429–437. doi: 10.1016/j.neuron.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohue DE, Ascoli GA. Automated reconstruction of neuronal morphology: an overview. Brain Res Rev. 2011;67:94–102. doi: 10.1016/j.brainresrev.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumitriu D, Rodriguez A, Morrison JH. High-throughput, detailed, cell-specific neuroanatomy of dendritic spines using microinjection and confocal microscopy. Nat Protoc. 2011;6:1391–1411. doi: 10.1038/nprot.2011.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand CM, Perroy J, Loll F, Perrais D, Fagni L, Bourgeron T, Montcouquiol M, Sans N. SHANK3 mutations identified in autism lead to modification of dendritic spine morphology via an actin-dependent mechanism. Mol Psychiatry. 2012;17:71–84. doi: 10.1038/mp.2011.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ethell IM, Pasquale EB. Molecular mechanisms of dendritic spine development and remodeling. Prog Neurobiol. 2005;75:161–205. doi: 10.1016/j.pneurobio.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Gibson SF, Lanni F. Experimental test of an analytical model of aberration in an oil-immersion objective lens used in three-dimensional light microscopy. J Opt Soc Am A. 1992;9:154–166. doi: 10.1364/josaa.9.000154. [DOI] [PubMed] [Google Scholar]
- Golden SA, Christoffel DJ, Heshmati M, Hodes GE, Magida J, Davis K, Cahill ME, Dias C, Ribeiro E, Ables JL, Kennedy PJ, Robison AJ, Gonzalez-Maeso J, Neve RL, Turecki G, Ghose S, Tamminga CA, Russo SJ. Epigenetic regulation of RAC1 induces synaptic remodeling in stress disorders and depression. Nat Med. 2013;19:337–344. doi: 10.1038/nm.3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao J, Rapp PR, Leffler AE, Leffler SR, Janssen WG, Lou W, McKay H, Roberts JA, Wearne SL, Hof PR, Morrison JH. Estrogen alters spine number and morphology in prefrontal cortex of aged female rhesus monkeys. J Neurosci. 2006;26:2571–2578. doi: 10.1523/JNEUROSCI.3440-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara Y, Rapp PR, Morrison JH. Neuronal and morphological bases of cognitive decline in aged rhesus monkeys. Age (Dordr) 2012;34:1051–1073. doi: 10.1007/s11357-011-9278-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KM, Jensen FE, Tsao B. Three-dimensional structure of dendritic spines and synapses in rat hippocampus (CA1) at postnatal day 15 and adult ages: implications for the maturation of synaptic physiology and long-term potentiation. J Neurosci. 1992;12:2685–2705. doi: 10.1523/JNEUROSCI.12-07-02685.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KM, Stevens JK. Dendritic spines of CA 1 pyramidal cells in the rat hippocampus: serial electron microscopy with reference to their biophysical characteristics. J Neurosci. 1989;9:2982–2997. doi: 10.1523/JNEUROSCI.09-08-02982.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi-Takagi A, Barker PB, Sawa A. Readdressing synaptic pruning theory for schizophrenia: Combination of brain imaging and cell biology. Commun Integr Biol. 2011;4:211–212. doi: 10.4161/cib.4.2.14492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes TJ. Blind deconvolution of quantum-limited incoherent imagery: maximum-likelihood approach. J Opt Soc Am A. 1992;9:1052–1061. doi: 10.1364/josaa.9.001052. [DOI] [PubMed] [Google Scholar]
- Hung AY, Futai K, Sala C, Valtschanoff JG, Ryu J, Woodworth MA, Kidd FL, Sung CC, Miyakawa T, Bear MF, Weinberg RJ, Sheng M. Smaller dendritic spines, weaker synaptic transmission, but enhanced spatial learning in mice lacking Shank1. J Neurosci. 2008;28:1697–1708. doi: 10.1523/JNEUROSCI.3032-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutsler JJ, Zhang H. Increased dendritic spine densities on cortical projection neurons in autism spectrum disorders. Brain Res. 2010;1309:83–94. doi: 10.1016/j.brainres.2009.09.120. [DOI] [PubMed] [Google Scholar]
- Jones EG, Powell TP. Morphological variations in the dendritic spines of the neocortex. J Cell Sci. 1969;5:509–529. doi: 10.1242/jcs.5.2.509. [DOI] [PubMed] [Google Scholar]
- Kasai H, Fukuda M, Watanabe S, Hayashi-Takagi A, Noguchi J. Structural dynamics of dendritic spines in memory and cognition. Trends Neurosci. 2010;33:121–129. doi: 10.1016/j.tins.2010.01.001. [DOI] [PubMed] [Google Scholar]
- Knafo S, Alonso-Nanclares L, Gonzalez-Soriano J, Merino-Serrais P, Fernaud-Espinosa I, Ferrer I, DeFelipe J. Widespread changes in dendritic spines in a model of Alzheimer's disease. Cereb Cortex. 2009a;19:586–592. doi: 10.1093/cercor/bhn111. [DOI] [PubMed] [Google Scholar]
- Knafo S, Venero C, Merino-Serrais P, Fernaud-Espinosa I, Gonzalez-Soriano J, Ferrer I, Santpere G, DeFelipe J. Morphological alterations to neurons of the amygdala and impaired fear conditioning in a transgenic mouse model of Alzheimer's disease. J Pathol. 2009b;219:41–51. doi: 10.1002/path.2565. [DOI] [PubMed] [Google Scholar]
- Maze I, Covington HE, 3rd, Dietz DM, LaPlant Q, Renthal W, Russo SJ, Mechanic M, Mouzon E, Neve RL, Haggarty SJ, Ren Y, Sampath SC, Hurd YL, Greengard P, Tarakhovsky A, Schaefer A, Nestler EJ. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science. 2010;327:213–216. doi: 10.1126/science.1179438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino-Serrais P, Knafo S, Alonso-Nanclares L, Fernaud-Espinosa I, DeFelipe J. Layer-specific alterations to CA1 dendritic spines in a mouse model of Alzheimer's disease. Hippocampus. 2011;21:1037–1044. doi: 10.1002/hipo.20861. [DOI] [PubMed] [Google Scholar]
- Nasse MJ, Woehl JC. Realistic modeling of the illumination point spread function in confocal scanning optical microscopy. J Opt Soc Am A Opt Image Sci Vis. 2010;27:295–302. doi: 10.1364/JOSAA.27.000295. [DOI] [PubMed] [Google Scholar]
- Nimchinsky EA, Sabatini BL, Svoboda K. Structure and function of dendritic spines. Annu Rev Physiol. 2002;64:313–353. doi: 10.1146/annurev.physiol.64.081501.160008. [DOI] [PubMed] [Google Scholar]
- Peters A, Kaiserman-Abramof IR. The small pyramidal neuron of the rat cerebral cortex. The perikaryon, dendrites and spines. Am J Anat. 1970;127:321–355. doi: 10.1002/aja.1001270402. [DOI] [PubMed] [Google Scholar]
- Phillips M, Pozzo-Miller L. Dendritic spine dysgenesis in autism related disorders. Neurosci Lett. 2015;601:30–40. doi: 10.1016/j.neulet.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price KA, Varghese M, Sowa A, Yuk F, Brautigam H, Ehrlich ME, Dickstein DL. Altered synaptic structure in the hippocampus in a mouse model of Alzheimer's disease with soluble amyloid-beta oligomers and no plaque pathology. Mol Neurodegener. 2014;9:41. doi: 10.1186/1750-1326-9-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radley JJ, Rocher AB, Rodriguez A, Ehlenberger DB, Dammann M, McEwen BS, Morrison JH, Wearne SL, Hof PR. Repeated stress alters dendritic spine morphology in the rat medial prefrontal cortex. J Comp Neurol. 2008;507:1141–1150. doi: 10.1002/cne.21588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Miguel A, Barr AM, Honer WG. Spines, synapses, and schizophrenia. Biol Psychiatry. 2015;78:741–743. doi: 10.1016/j.biopsych.2015.08.035. [DOI] [PubMed] [Google Scholar]
- Rocher AB, Crimins JL, Amatrudo JM, Kinson MS, Todd-Brown MA, Lewis J, Luebke JI. Structural and functional changes in tau mutant mice neurons are not linked to the presence of NFTs. Exp Neurol. 2010;223:385–393. doi: 10.1016/j.expneurol.2009.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Ehlenberger D, Kelliher K, Einstein M, Henderson SC, Morrison JH, Hof PR, Wearne SL. Automated reconstruction of three-dimensional neuronal morphology from laser scanning microscopy images. Methods. 2003;30:94–105. doi: 10.1016/s1046-2023(03)00011-2. [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Ehlenberger DB, Dickstein DL, Hof PR, Wearne SL. Automated three-dimensional detection and shape classification of dendritic spines from fluorescence microscopy images. PLoS One. 2008;3:e1997. doi: 10.1371/journal.pone.0001997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Ehlenberger DB, Hof PR, Wearne SL. Rayburst sampling, an algorithm for automated three-dimensional shape analysis from laser scanning microscopy images. Nat Protoc. 2006;1:2152–2161. doi: 10.1038/nprot.2006.313. [DOI] [PubMed] [Google Scholar]
- Selvas A, Coria SM, Kastanauskaite A, Fernaud-Espinosa I, DeFelipe J, Ambrosio E, Miguens M. Rat-strain dependent changes of dendritic and spine morphology in the hippocampus after cocaine self-administration. Addict Biol. 2015 doi: 10.1111/adb.12294. [DOI] [PubMed] [Google Scholar]
- Shansky RM, Hamo C, Hof PR, McEwen BS, Morrison JH. Stress-induced dendritic remodeling in the prefrontal cortex is circuit specific. Cereb Cortex. 2009;19:2479–2484. doi: 10.1093/cercor/bhp003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spacek J, Hartmann M. Three-dimensional analysis of dendritic spines. I. Quantitative observations related to dendritic spine and synaptic morphology in cerebral and cerebellar cortices. Anat Embryol (Berl) 1983;167:289–310. doi: 10.1007/BF00298517. [DOI] [PubMed] [Google Scholar]
- Steele JW, Brautigam H, Short JA, Sowa A, Shi M, Yadav A, Weaver CM, Westaway D, Fraser PE, St George-Hyslop PH, Gandy S, Hof PR, Dickstein DL. Early fear memory defects are associated with altered synaptic plasticity and molecular architecture in the TgCRND8 Alzheimer's disease mouse model. J Comp Neurol. 2014;522:2319–2335. doi: 10.1002/cne.23536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecellio M, Schwaller B, Meyer M, Hunziker W, Celio MR. Alterations in Purkinje cell spines of calbindin D-28 k and parvalbumin knock-out mice. Eur J Neurosci. 2000;12:945–954. doi: 10.1046/j.1460-9568.2000.00986.x. [DOI] [PubMed] [Google Scholar]
- Wallace W, Bear MF. A morphological correlate of synaptic scaling in visual cortex. J Neurosci. 2004;24:6928–6938. doi: 10.1523/JNEUROSCI.1110-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wearne SL, Rodriguez A, Ehlenberger DB, Rocher AB, Henderson SC, Hof PR. New techniques for imaging, digitization and analysis of three-dimensional neural morphology on multiple scales. Neuroscience. 2005;136:661–680. doi: 10.1016/j.neuroscience.2005.05.053. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.