Abstract
We sequenced Early Neolithic genomes from the Zagros region of Iran (eastern Fertile Crescent), where some of the earliest evidence for farming is found, and identify a previously uncharacterized population that is neither ancestral to the first European farmers nor has contributed significantly to the ancestry of modern Europeans. These people are estimated to have separated from Early Neolithic farmers in Anatolia some 46-77,000 years ago and show affinities to modern day Pakistani and Afghan populations, but particularly to Iranian Zoroastrians. We conclude that multiple, genetically differentiated hunter-gatherer populations adopted farming in SW-Asia, that components of pre-Neolithic population structure were preserved as farming spread into neighboring regions, and that the Zagros region was the cradle of eastward expansion.
The earliest evidence for cultivation and stock-keeping is found in the Neolithic core zone of the Fertile Crescent (1, 2); a region stretching north from the southern Levant through E-Anatolia and N-Mesopotamia then east into the Zagros Mountains on the border of modern-day Iran and Iraq (Fig. 1). From there farming spread into surrounding regions, including Anatolia and later Europe, southern Asia, and parts of Arabia and N-Africa. Whether the transition to agriculture was a homogenous process across the core zone, or a composite of localized domestications is unknown. Likewise, the extent to which core zone farming populations were genetically homogenous, or exhibited structure that may have been preserved as agriculture spread into surrounding regions, is undetermined.
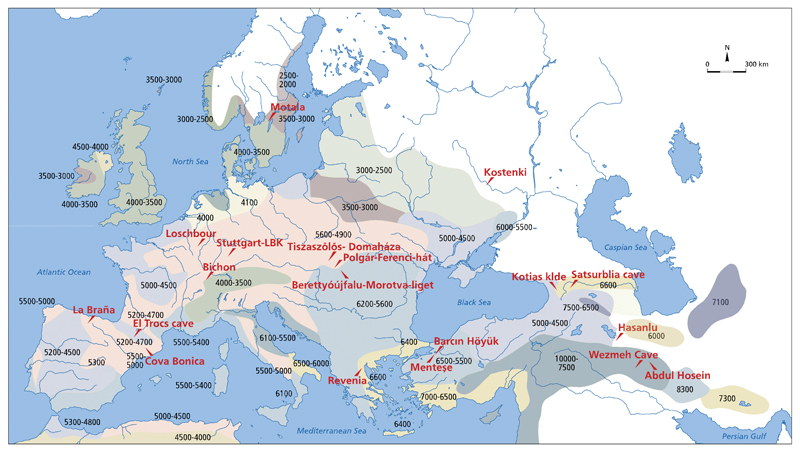
Fig 1. Map of prehistoric Neolithic and Iron Age Zagros genome locations.
Colors indicate isochrones with numbers giving approximate arrival times of the Neolithic culture in years BCE.
Ancient DNA (aDNA) studies indicate that early Aegean farmers dating to c. 6,500-6,000 BCE are the main ancestors of early European farmers (3, 4), although it is not known if they were predominantly descended from core zone farming populations. We sequenced four Early Neolithic (EN) genomes from Iran, including a well preserved sample from the central Zagros site of Wezmeh Cave (WC1, 7,455-7,082 cal BCE), the latter to a coverage of 10x. The three other individuals from Tepe Abdul Hosein were less well preserved (genome coverage between 0.6 and 1.2 x) but are around 10,000 years old, and therefore among the earliest Neolithic human remains in the world (Table S1 and S3).
Despite a lack of a clear Neolithic context, the radiocarbon inferred chronological age, and palaeodietary data support WC1 being an early farmer (Tables S1-S3, Fig. S7). WC1 bone collagen δ13C and δ15N values are indistinguishable from that of a securely assigned Neolithic individual from Abdul Hosein and consistent with a diet rich in cultivated C3 cereals rather than animal protein. Specifically, collagen from WC1 and Abdul Hosein is 13C depleted compared to those from contemporaneous wild and domestic fauna from this region (5), which consumed C4 plants. Crucially, WC1 and the Abdul Hosein farmers exhibit very similar genomic signatures.
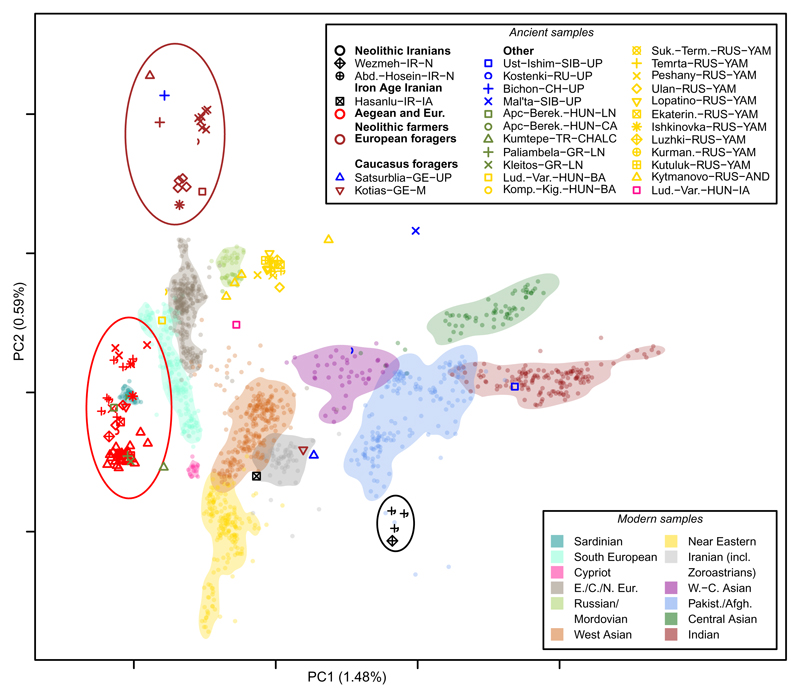
The four EN Zagros genomes form a distinct cluster in the first two dimensions of a principal components analysis (PCA; Fig. 2); they plot closest to modern-day Pakistani and Afghans and are separated from European hunter-gatherers (HG) and other Neolithic farmers. In an outgroup f3-test (6, 7) (Fig. S17-S20), all four Neolithic Iranian individuals are genetically more similar to each other than to any other prehistoric genome except a Chalcolithic genome from NW-Anatolia (see below). Despite 14C dates spanning around 1,200 years, these data are consistent with all four genomes being sampled from a single eastern Fertile Crescent EN population.
Fig 2. PCA plot of Zagros, European, and Near and Middle Eastern ancient genomes.
Comparing ancient and modern genomes, Neolithic Zagros genomes form a distinct genetic cluster close to modern Pakistani and Afghan genomes but distinct from other Neolithic farmers and European hunter-gatherers. See Animation S1 for an interactive 3D version of the PCA including the third principal component.
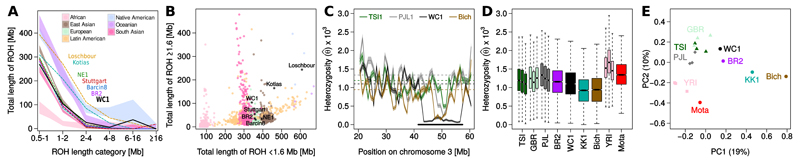
We obtained the distribution of runs of homozygosity (ROH) above 500 kb in length and compared other relevant ancient and modern genomes (Fig. 3A, B). WC1 shares a similar ROH distribution with European and Aegean Neolithics, and modern day Europeans. However, of all ancient samples considered, WC1 displays the lowest total length of short ROH suggesting he was descended from a relatively large HG population. In sharp contrast, the ROH distributions of the HG Kotias from Georgia, and Loschbour from Luxembourg indicate prolonged periods of small ancestral population size (8).
Fig 3. Level and structure of ancient genomic diversity.
(A) Total length of the genome in different ROH classes; shades indicate the range observed among modern samples from different populations and lines indicate the distributions for ancient samples. (B) The total length of short (<1.6Mb) vs long (≥1.6Mb) ROH. (C) Distribution of heterozygosity (θ) inferred in 1Mb windows along a portion of chromosome 3 showing the longest ROH segment in WC1. Solid lines represent the MLE estimate, shades indicate the 95% confidence intervals and dashed lines the genome-wide median for each sample. (D) Distribution of heterozygosity (θ) estimated in 1Mb windows across the autosomes for modern and ancient samples. (E) Similarity in the pattern of heterozygosity (θ) along the genome as obtained by a PCA on centered Spearman correlations. Ancient - Bich: Bichon, Upper Palaeolithic forager from Switzerland; KK1: Kotias, Mesolithic forager from Georgia; WC1: Wezmeh Cave, Early Neolithic farmer from Zagros; Mota: 4,500 year old individual from Ethiopia; BR2: Ludas-Varjú-dúló, Late Bronze Age individual from Hungary. Modern - YRI: Yoruban, W-Africa; TSI: Tuscans, Italy; PJL: Punjabi, Pakistan; GBR: British
We developed an unbiased method to estimate heterozygosity () in 1Mb windows that takes post mortem damage fully into account, even at low coverages (9; Fig. 3C, D). The mean WC1 diversity was higher than for HG individuals (Bichon and Kotias), but similar to Bronze Age individuals from Hungary and modern Europeans, and lower than for ancient (10) and modern Africans. Multidimensional scaling on a matrix of centered Spearman correlations of local heterozygosity estimates across the whole genome again puts WC1 closer to modern populations than to ancient foragers, indicating that the mean diversity and pattern of diversity distribution over the genome is similar to modern populations (Fig. 3E). However, WC1 does have an excess of long ROH segments (>1.6 Mb), relative to Aegean and European Neolithics (Fig. 3B). This includes several substantially long (7-16 Mb) ROH segments (Fig. 3A), consistent with low estimates in those regions (Fig. 3C). These regions do not show reduced coverage in WC1 nor a reduction in diversity in other samples, except for the longest such segment where we find a similar, but less extended region of reduced diversity in modern and HG individuals (7; Fig. 3B). Such long segments of reduced heterozygosity can result from cultural practices, such as consanguinity and endogamy, or demographic constraints such as a recent or ongoing bottleneck (11).
The extent of population structure in Neolithic SW-Asia has important implications for the origins of farming. High structuring would be expected under a scenario of localized independent domestication processes by distinct populations, whereas low structure would be more consistent with a single population origin of farming or a diffuse homogeneous domestication process, perhaps involving high rates of gene flow across the Neolithic core zone. The ancient Zagros individuals show stronger affinities to Caucasus HGs (Table S17.1) whereas Neolithic Aegeans showed affinities to other European HGs (Tables S17.2 and S17.3). Formal tests of admixture of the form f3(Neo_Iranian, HG; Anatolia_Neolithic) were all positive with Z-scores above 15.78 (Table S17.6), indicating that Neolithic NW-Anatolians did not descend from a population formed by the mixing of Zagros Neolithics and known HG groups. These results suggest that Neolithic populations from NW-Anatolia and the Zagros descended from distinct ancestral populations. Furthermore, while the Caucasus HGs are genetically closest to EN Zagros individuals, they also share unique drift with eastern, western, and Scandinavian European HGs (Table S16.1), indicating that they are not the direct ancestors of Zagros Neolithics.
The significant differences between ancient Iranians, Anatolian/European farmers and European HGs suggest a separation before the Neolithic. Assuming a mutation rate of 5 x 10-10 per site per year (12), the inferred mean split time for Anatolian/European farmers (as represented by Bar8) and European hunter-gatherers (LOS; Loschbour) ranged from 33-39 kya (combined 95% CI 15-61), and for the Neolithic Iranians (WC1) 46-77 kya (combined 95% CI 38-104) (13; Fig S48, Tables S34 and S35). Furthermore, the European hunter-gatherers were inferred to have an effective population size (Ne) that was ~10-20% of either Neolithic farming group.
Levels of inferred Neanderthal ancestry in WC1 are low (Fig. S22, Table S21), but fall within the general trend described in Fu et al. (14). Fu et al. (14) also inferred a basal Eurasian ancestry component in the Caucasus HG sample Satsurblia when examined within the context of a “base model” for various ancient Eurasian genomes dated from ~45,000-7,000 years ago. We examined this base model using ADMIXTUREGRAPH (6) and inferred almost twice as much basal Eurasian ancestry for WC1 as for Satsurblia (62% versus 32%) (Fig. S52), with the remaining derived from Ancient north Eurasians such as Mal`ta1 (15). Thus Neolithic Iranians appear to derive predominantly from the earliest known Eurasian population branching event (7).
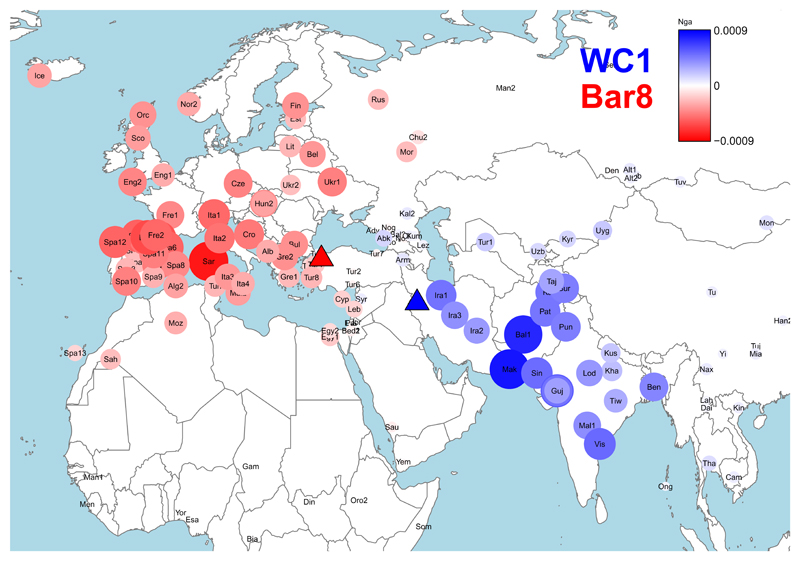
We applied ‘chromosome painting’ and a Bayesian mixture model (7) to infer which individuals from 160-220 modern groups are most recently related to WC1 on the basis of shared haplotypes. WC1 was inferred to share a high proportion (>95%) of recent ancestry with groups sampled from the Middle East, Caucasus and India. We compared WC1's profile of haplotype sharing with modern groups to that of three high coverage Neolithic genomes from NW-Anatolia (Barcın, Fig 4), Germany (LBK, Stuttgart) and Hungary (NE1, Polgár-Ferenci-hát). In stark contrast to WC1, these Anatolian and European Neolithics shared ~60-90% of recent ancestry with modern groups sampled from South Europe (Figs. S24, S30, S32-S37, Table S22).
Fig 4. Modern-day peoples with affinity to WC1.
Modern groups with an increasingly higher (respectively lower) inferred proportion of haplotype sharing with the Iranian Neolithic Wezmeh Cave (WC1, 7,455-7,082 cal BCE, blue triangle) compared to the Anatolian Neolithic Barcın genome (Bar8; 6,212–6,030 cal BCE, red triangle) are depicted with an increasingly stronger blue color (respectively red color). Circle sizes illustrate the relative absolute proportion of this difference between WC1 versus Bar8. The key for the modern group labels is provided in Table S24.
We examined recent haplotype sharing between each modern group and ancient Neolithic genomes from Iran (WC1) and Europe (LBK, NE1), two ancient HG genomes sampled from Luxembourg (LOS) and the Caucasus (KK1; Kotias), a 4.5k-year old genome from Ethiopia (Mota) and Ust’-Ishim, a 45k-year old genome from Siberia. Modern groups from S-, C and NW-Europe shared haplotypes predominantly with European Neolithic samples LBK and NE1, and European HGs, while modern Near and Middle Eastern, and Indian samples had higher sharing with WC1 (Fig. S28-29). Modern Pakistani, Iranian, Armenian, Tajikistani, Uzbekistani and Yemeni samples were inferred to share >10% of haplotypes most recently with WC1; even when we include modern groups from neighboring geographic regions as ancestry surrogates (Fig. S26-27, Table S23). This includes Iranian Zoroastrians, having the highest inferred sharing with WC1 out of all modern groups (Table S23). Consistent with this, f3 statistics indicate Iranian Zoroastrians are the most genetically similar to all four Neolithic Iranians, followed by other modern Iranians (Fars), Balochi (SE Iran, Pakistan and Afghanistan), Brahui (Pakistan and Afghanistan), Kalash (Pakistan) and Georgians (Figs. S12-15). Interestingly, WC1 most likely had brown eyes, relatively dark skin, and black hair, although Neolithic Iranians do carry reduced pigmentation-associated alleles in several genes, and display derived alleles at 7 of the 12 loci showing the strongest signatures of selection in ancient Eurasians (3) (Tables S29-S33). While there is a strong Neolithic component in these modern S-Asian populations, simulation of allele sharing rejected full population continuity under plausible ancestral population sizes, indicating some population turnover in Iran since the Neolithic (7).
Early Neolithic samples from eastern and western SW-Asia differ conspicuously, but comparisons to genomes from Chalcolithic Anatolia and Iron Age Iran indicate a degree of subsequent homogenization. Kumtepe6, a ~6,750 year old genome from NW-Anatolia (16), was more similar to Neolithic Iranians than any other non-Iranian ancient genome (Fig. S17-20; Table S18.1). Furthermore, our Iron Age genome (F38; 971-832 BCE; sequenced to 1.9x) from Tepe Hasanlu in NW-Iran shares greatest similarity with Kumtepe6 (Fig. S21) even compared to Neolithic Iranians (Table S20). We inferred additional non-Iranian or non-Anatolian ancestry in F38 from sources such as European Neolithics and even post-Neolithic Steppe populations (Table S20). Consistent with this, the F38 individual carried a N1a sub-clade mtDNA, which is common in early European and NW-Anatolian farmers (3). In contrast, his Y-chromosome belongs to sub-haplogroup R1b1a2a2, also found in five Yamnaya individuals (17) and in two individuals from the Poltavka culture (3). These patterns indicate that post-Neolithic homogenization in SW-Asia involved substantial bidirectional gene flow between the East and West of the region, as well as possible gene flow from the Steppe.
Migration of people associated with the Yamnaya culture has been implicated in the spread of Indo-European languages (17, 18) and some level of Near Eastern ancestry was inferred in southern Russian pre-Yamnaya populations (3). Our tests suggest that Neolithic Iranians were unlikely to be the main source of Near Eastern ancestry in the Steppe population (Table S20), and that ancestry in pre-Yamnaya populations originated primarily in the west of SW-Asia.
We also inferred shared ancestry between Steppe and Hasanlu Iron Age genomes, distinct from that in EN Iranians (Table S20, (7)). In addition, modern Middle Easterners and South Asians appear to possess mixed ancestry from ancient Iranian and Steppe populations (Table S19 and S20). However, Steppe-related ancestry may also have been acquired indirectly from other sources (7) and it is not clear if this is sufficient to explain the spread of Indo-European languages from a hypothesized Steppe homeland to the region where Indo-Iranian languages are spoken today. On the other hand, the affinities of Zagros Neolithic individuals to modern populations of Pakistan, Afghanistan, Iran, and India is consistent with a spread of Indo-Iranian languages, or of Dravidian languages (which includes Brahui), from the Zagros into southern Asia, in association with farming (19).
The Neolithic transition in SW-Asia involved the appearance of different domestic species, particularly crops, in different parts of the Neolithic core zone, with no single center (20). Early evidence of plant cultivation and goat management between the 10th and the 8th millennium BCE highlight the Zagros as a key region in the Neolithisation process (1). Given evidence of domestic species movement from East to West across SW-Asia (21), it is surprising that EN human genomes from the Zagros are not closely related to those from NW-Anatolia and Europe. Instead they represent a previously undescribed Neolithic population. Our data show that the chain of Neolithic migration into Europe does not reach back to the eastern Fertile Crescent, also raising questions about whether intermediate populations in southeastern and Central Anatolia form part of this expansion. On the other hand, it seems probable that the Zagros region was the source of an eastern expansion of the SW-Asian domestic plant and animal economy. Our inferred persistence of ancient Zagros genetic components in modern day S-Asians lends weight to a strong demic component to this expansion.
Supplementary Materials
Materials and Methods
Figs. S1 - S52
Tables S1 - S37
References (22 - 162)
Animation S1 (3D animation of PCA)
One Sentence Summary.
Neolithic genomes from Zagros region of Iran are ancestral to modern South Asians but distinct from early NW-Anatolian and European farmers.
Acknowledgments
This paper is a product of the Palaeogenome Analysis Team (PAT). FB was supported by funds of Johannes Gutenberg-University Mainz given to JB. ZH and RM are supported by a Marie Curie Initial Training Network (BEAN / Bridging the European and Anatolian Neolithic, GA No: 289966). CS was supported by the EU: SYNTHESYS / Synthesis of Systematic Resources, GA No: 226506-CP-CSA-INFRA, and DFG: (BO 4119/1). AS was supported by the EU: CodeX Project No: 295729. MC was supported by Swiss NSF grant 31003A_156853. AK, DW were supported by Swiss NSF grant 31003A_149920. SL is supported by BBSRC (Grant Number BB/L009382/1). LvD is supported by CoMPLEX via EPSRC (Grant Number EP/F500351/1). GH is supported by a Sir Henry Dale Fellowship jointly funded by the Wellcome Trust and the Royal Society (Grant Number 098386/Z/12/Z) and supported by the National Institute for Health Research University College London Hospitals Biomedical Research Centre. MGT and YD are supported by a Wellcome Trust Senior Research Fellowship awarded to MGT. JB is grateful for support by the HPC cluster MOGON (funded by DFG; INST 247/602-1 FUGG). AH . MM was supported by the UMR 7209, CNRS/ MNHN/SU and ANR-14-CE03-0008-01- CNRS ANR Kharman. OM was supported by Institut Français de Recherche en Iran (October 2015). FBi, MM, OM and HD thank the National Museum of Iran and especially Dr. Jebrael Nokandeh, director of National Museum of Iran. We thank Nick Patterson for early access to the latest version of qpgraph.
Accession numbers: Mitochondrial genome sequences are deposited in GenBank (KX353757-KX353761). Genomic data are available at ENA with the accession number PRJEB14180 in BAM format.
References
- 1.Riehl S, Zeidi M, Conard N. Emergence of Agriculture in the Foothills of the Zagros Mountains of Iran. Science. 2013;341:65–57. doi: 10.1126/science.1236743. [DOI] [PubMed] [Google Scholar]
- 2.Mattews R, Mattews W, Mohammadifar Y, editors. The Earliest Neolithic of Iran: 2008 Excavations at Sheikh-e Abad and Jani Central Zagros. Vol. 1. Oxbow Books; Oxford: 2013. [Google Scholar]
- 3.Mathieson I, et al. Genome-wide patterns of selection in 230 ancient Eurasians. Nature. 2015;528:499–503. doi: 10.1038/nature16152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hofmanová Z et al. Early farmers from across Europe directly descended from Neolithic Aegeans. PNAS. 2016 doi: 10.1073/pnas.1523951113. 1523951113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Müldner G. Isotope Analysis of Animal Bone. In: Matthews R, Matthews W, Mohammadifar Y, editors. The Earliest Neolithic of Iran: 2008 Excavations at Sheikh-e Abad and Jani. Oxbow Books; 2013. [Google Scholar]
- 6.Patterson N, et al. Ancient admixture in human history. Genetics. 2012;192:1065–1093. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Materials and methods are available as supplementary material at the Science website
- 8.Jones ER, et al. Upper Palaeolithic genomes reveal deep roots of modern Eurasians. Nat Commun. 2015;6 doi: 10.1038/ncomms9912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kousathanas A, et al. Inferring heterozygosity from ancient and low coverage genomes. bioRxiv. 2016 doi: 10.1534/genetics.116.189985. 046748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Llorente MG, et al. Ancient Ethiopian genome reveals extensive Eurasian admixture in Eastern Africa. Science. 2015;350:820–822. doi: 10.1126/science.aad2879. [DOI] [PubMed] [Google Scholar]
- 11.Pemberton TJ, et al. Genomic Patterns of Homozygosity in Worldwide Human Populations. Am J Hum Genet. 2012;91:275–292. doi: 10.1016/j.ajhg.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kong A, et al. Rate of de novo mutations, father’s age, and disease risk. Nature. 2012;488:471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nachman MW, Crowell SL. Estimate of the mutation rate per nucleotide in humans. Genetics. 2000;156:297–304. doi: 10.1093/genetics/156.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu Q, et al. The genetic history of Ice Age Europe. Nature. 2016;534:200–205. doi: 10.1038/nature17993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raghavan M, et al. Upper Palaeolithic Siberian genome reveals dual ancestry of Native Americans. Nature. 2014;505:87–91. doi: 10.1038/nature12736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Omrak A, et al. Genomic Evidence Establishes Anatolia as the Source of the European Neolithic Gene Pool. Curr Biol. 2016;26:270–275. doi: 10.1016/j.cub.2015.12.019. [DOI] [PubMed] [Google Scholar]
- 17.Haak W, et al. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature. 2015;522:207–211. doi: 10.1038/nature14317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allentoft ME, et al. Population genomics of Bronze Age Eurasia. Nature. 2015;522:167–172. doi: 10.1038/nature14507. [DOI] [PubMed] [Google Scholar]
- 19.Mallory JP, Adams DQ, editors. Encyclopedia of Indo-European Culture. Taylor & Francis; London: 1997. [Google Scholar]
- 20.Fuller DQ, Willcox G, Allaby RG. Early agricultural pathways: moving outside the 'core area' hypothesis in Southwest Asia. J Exp Bot. 2012;63:617–633. doi: 10.1093/jxb/err307. [DOI] [PubMed] [Google Scholar]
- 21.Scheu A, et al. The genetic prehistory of domesticated cattle from their origin to the spread across Europe. Bmc Genet. 2015;16 doi: 10.1186/s12863-015-0203-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Materials and Methods
Figs. S1 - S52
Tables S1 - S37
References (22 - 162)
Animation S1 (3D animation of PCA)