Abstract
Using a cell-free expression system we produced the p7 viroporin embedded into a lipid bilayer in a single-step manner. The protein quality was assessed using different methods. We examined the channel forming activity of p7 and verified its inhibition by 5-(N,N-Hexamethylene) amiloride (HMA). Fourier transformed infrared spectroscopy (FTIR) experiments further showed that when p7 was inserted into synthetic liposomes, the protein displayed a native-like conformation similar to p7 obtained from other sources. Photoactivatable amino acid analogs used for p7 protein synthesis enabled oligomerization state analysis in liposomes by cross-linking. Therefore, these findings emphasize the quality of the cell-free produced p7 proteoliposomes which can benefit the field of the hepatitis C virus (HCV) protein production and characterization and also provide tools for the development of new inhibitors to reinforce our therapeutic arsenal against HCV.
Keywords: Cell-free protein synthesis, membrane protein, viroporin, proteoliposomes, hepatitis C virus
Introduction
The hepatitis C virus (HCV) is a small enveloped, positive-sense single-stranded RNA virus of the family Flaviviridae. It is considered to be a major public health problem as 150 million people are infected currently resulting in more than 300,000 deaths annually (World Health Organization, 2013). The virus causes those chronically infected to develop liver diseases that may progress into fibrosis, cirrhosis or hepatocellular carcinoma. While there are vaccines to prevent hepatitis A and B; no vaccine for hepatitis C is yet available to prevent the spread of infection [1,2]. The current treatments’ effectiveness against HCV is genotype dependent, including therapy with pegylated interferon or recently launched interferon-free treatments using nucleoside/nucleotide NS5B polymerase inhibitors and proteases inhibitors. The HCV replication is highly error prone due to the lack of correcting mechanisms of the HCV RNA-dependent RNA polymerase [3]. As a result, this high mutation rate is consistent with the high degree of HCV diversity found across the population of infected individuals. Within genotypes and subtypes, mutations of the NS3, NS5A and NS5B proteins induce resistance to newly developed direct antiviral agents (DAAs) [4,5]. With the apparition of resistance associated variants, the high cost of the new DAAs and their scarce global availability, the discovery of complementary treatments to fight against HCV is still needed.
The viroporin p7 is located between the structural proteins: core, E1, E2 and the nonstructural protein NS2, resulting in the production of a precursor E2-p7-NS2. Activation of host signal peptidases induces cleavages in the precursor releasing p7 from the viral polyprotein [6]. The HCV p7 protein is involved in the viral life cycle including virus assembly and infectivity which makes it a promising drug target [7]. It is a 63-amino acid integral membrane protein and has different potential topologies and conformations as indicated by diverse studies including NMR [8–12]. Other studies also revealed an N-terminal α-helix and two transmembrane segments connected by a short hydrophilic cytosolic segment of 7 amino acids [13–15] (Fig. 1A). A recent review on p7 displays the different reported topologies of the protein [16]. The p7 protomer is able to oligomerize to an hexameric [15,17] and heptameric state [12] forming an ion channel. This viroporin is cation selective and a list of small molecule drugs such as amantadine, rimantadine, BIT225, hexamethylene amiloride (HMA) and long-alkyl-chain iminosugars, including N-nonyl deoxynojirimycin (NN-DNJ), have been identified as potential negative regulators of the p7 channel [17–20]. In cellulo, amantadine, NN-DNJ, and rimantadine have been shown to inhibit the release of infectious HCV and virus entry in a genotype-depent manner [21] while NN-DNJ, BIT225 and rimantadine have been reported to inhibit cell-free virus transmission [22].
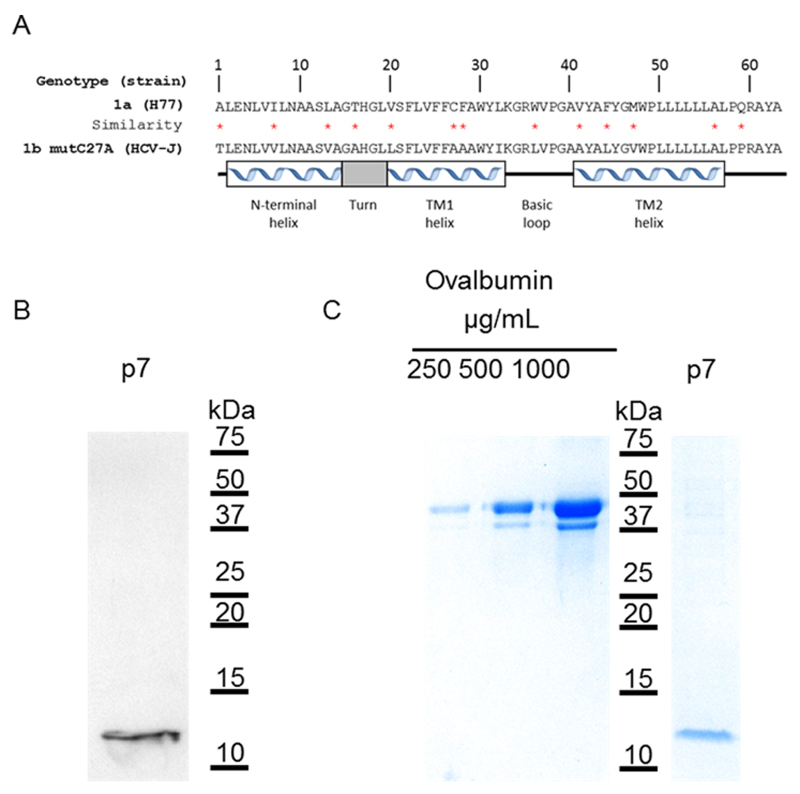
Figure 1. Sequence, structure and expression analysis of p7.
(A) The amino acid sequence of the expressed p7 from genotype 1a strain H77 is displayed and aligned with the sequence of the genotype 1b strain HCV-J. The corresponding schematic representation of helical, turn, and loop regions deduced from the NMR structure analysis of p7 genotype 1b strain HCV-J is also displayed [9] as no NMR data is available for p7 from genotype 1a strain H77. TM = transmembrane. (B) Western immunoblotting of p7 proteoliposomes; (C) Coomassie brilliant blue staining: 5 µl of ovalbumin (different concentrations were used as standards) and 1 µl of p7 proteoliposomes after sucrose gradient purification were loaded on SDS-PAGE.
Expression of the p7 membrane protein in classical overexpression systems is difficult due to its high content in hydrophobic amino acids and its small size. Several studies have been performed in order to obtain sufficient amounts of p7 for functional and structural studies, including peptide synthesis [9,18,19,23], expression in Escherichia coli (E. coli) either in inclusion bodies [12,24] or under a soluble form (i.e. when p7 is fused to the maltose binding protein (MBP) and an histidine tag [25]). However, these methods for protein production require tremendous work and display some limiting features on numerous aspects. To recover a sufficient quantity of active p7 protein embedded into a lipid bilayer, these production strategies involve several critical steps including; resolubilization, refolding and protein insertion into liposome in the presence of detergents. An attractive alternative for producing such difficult to express proteins is the use of cell-free expression systems [26,27]. Membrane proteins have been expressed using both eukaryotic and prokaryotic cell-free systems [28–30]. One of the interesting features of the cell-free expression systems resides in their capacities to be easily adapted to the production of the protein of interest because they are completely open systems, in which each reaction parameter can be modified. Production of membrane proteins in cell-free expression systems can be achieved either in a precipitated form (P-CF) without any additives or in the presence of lipids (L-CF) or detergents (D-CF) [31–35]. The detergents will shield the hydrophobic domains of the protein and help its folding and solubilization. The use of detergents can be detrimental and some of them can alter the protein structure and thus impact their activities [36,37]. They can also be incompatible with cell-free expression systems, as some can inhibit the transcription/translation machinery [38]. Addition of lipids directly in the expression reaction mix represents a more suitable alternative for producing active membrane proteins in a native-like conformation. Several studies have demonstrated that membrane proteins expressed in cell-free systems with natural or synthetic liposomes can be directly inserted into the lipid bilayer in order to form recombinant proteoliposomes [39–42]. This insertion renders the production of proteoliposomes simpler by avoiding the purification and the resolubilization of the membrane proteins using detergents before relipidation. Different lipid compositions of the vesicles mimicking the natural lipid environment of the membrane protein can be optimized for best yield and protein activity. In addition to direct addition of liposomes, small parts of lipid bilayers wrapped in a lipoprotein can form lipid disks which are able to accept membrane proteins during synthesis [29,43].
In this study, we used an E. coli cell-free expression system containing synthetic liposomes to express HCV p7 protein and obtain p7 proteoliposomes in a one step process. The liposomes enabled the correct folding of the viroporin into a native-like conformation as shown by Fourier transformed infrared spectroscopy (FTIR). The most commonly used method to analyze ion channel activity is the measurement of currents caused by the ion flow. In this context, we have demonstrated the functionality of the recombinant p7 protein by patch-clamp recordings. We show that the channel activity is inhibited by HMA as predicted. We also investigated the oligomerization states of the p7 protein embedded into a lipid bilayer by using photoactivatable amino acid analogs and a chemical agent for cross-linking studies. Our results emphasize that the use of cell-free protein synthesis in presence of liposomes is a reliable and efficient system for membrane protein expression. The obtained proteoliposomes represent a great tool for the study of the HCV p7 viroporin and the development of new treatments.
Materials and Methods
Cell-free synthesis
p7 protein sequence from HCV strain H77 genotype 1a was synthesized by DNA2.0. The gene was then cloned directly into a pIVEX2.4d (Roche Applied Science) using NdeI/XhoI restriction sites.
Cell-free reaction was carried out at 30°C for 16h with gentle agitation at 400 rpm by using an E. coli extract and energy mix provided by Synthelis SAS. Energy mix composition was prepared according to Sitaraman et al. [44] without cAMP, betaine, trehalose and with 210 mM sodium oxalate and 33 mM NAD. DNA was added at 15 µg/ml. Mg2+ and K+ ion concentrations were screened for best yield: a range from 8 mM to 30 mM Mg2+ was assayed using magnesium acetate in correlation with a range of 250 mM to 370 mM of K+ using potassium acetate.
Liposomes were prepared using a 10 mg/ml lipid mixture of (1,2-dioleoyl-sn-glycero-3-phosphocholine:1,2-dioleoyl-sn-glycero-3-phosphoethanolamine:1,2-dimyristoyl-sn-glycero-3-phosphate:cholesterol; 40:20:20:20 volume ratio) in chloroform. This composition has been previously shown to enable efficient cell-free protein synthesis [45]. All lipids were purchased from Avanti Polar lipids. Chloroform was evaporated using a univapo 150H. The thin lipid film was rehydrated with diethyl pyrocarbonate treated water to obtain a 30 mg/ml lipid slurry. This solution was sonicated using a tip sonicator (Branson Digital Sonifier 250) at 20% for 5 times 30 seconds before being filtered once with a 0.22 µm PES filter.
To purify proteoliposomes, cell-free reactions were loaded on top of 3-step discontinuous sucrose gradient (60%, 30% and 5%) prepared in 50mM Hepes pH 7.5 buffer. After centrifugation at 280,000 x g for 1hr at 4°C, fractions were collected at each interface and analyzed by Western blotting using a poly histidine antibody conjugated with a horseradish peroxidase (Sigma-Aldrich) diluted at 1:10 000 in TBS-Tween buffer, 5% nonfat milk.
To assess protein integration in the lipid membrane of liposomes, proteoliposomes were subjected to alkaline extraction. Samples were diluted 1:10 in 0.1 M sodium carbonate (pH 11.5) and incubated on ice for 30 minutes before centrifugation on discontinuous sucrose gradient and analyzed as stated above.
Protein concentration was estimated by Coomassie brilliant blue staining of an SDS-PAGE gel.
Cross-linking
Photoactivatable L-leucine and L-methionine were purchased from Thermofisher. These analogs were substituted at 2 mM final concentration to replace natural amino acids. After proteoliposome purification, the samples aliquoted in microtubes were subjected to ultra violet (UV) light at 302 nm for one minute. The reaction was quenched by incubation with 1.5 M Tris-HCl, pH 7.5, at room temperature for 15 min. Analysis of the samples was performed by SDS-PAGE.
Dithiobis(succinimidyl propionate) (DSP) was purchased from Sigma-Aldrich. Proteoliposomes were incubated for 30 minutes at room temperature with 5 mM of DSP. The reaction was quenched by incubation with 1.5 M Tris-HCl, pH 7.5, at room temperature for 15 min. SDS-PAGE analysis of cross-linked samples was performed under non-reducing conditions.
p7 secondary structure by FTIR spectroscopy
A Vertex 70 spectrophotometer (Bruker Optics GmbH, Ettlingen, Germany) equipped with a nitrogen-cooled mercury–cadmium–telluride detector was used for the acquisition of FTIR spectra. Single-channel spectra (sum of 256 spectra) were recorded between 4000 and 400 cm−1 with a 2 cm−1 resolution by means of the OPUS Software v6.5 (Bruker). The acquisition was made in transmission mode using two CaF2windows (60 µL for each sample, D2O being used as solvent). The spectrum of p7 protein was obtained by subtracting the spectrum of negative control proteoliposomes (liposomes purified from a cell-free reaction without the p7 DNA plasmid) to the one of p7 proteoliposomes.
The OPUS Software v6.5 (Bruker GmbH) was used for the treatment and deconvolution of the spectra as previously described [46,47]. Residual water and CO2 contributions were removed, and baseline correction was done manually, the same reference points were used for each group of spectra. The amide I band was fitted by using the frequency, width, and intensity as parameters. The most consistent results were obtained when all component peaks were assumed to be Gaussian. The correspondence of each component band with a given secondary structure was established by comparing the frequency of its maximum to the value given in the literature [48]. The relative contribution of each component (in %) was calculated as the ratio of the area of each peak over the area of the total amide I band [47].
Giant Unilamellar Vesicles (GUVs)
The GUVs were prepared by electroformation method in a chamber connected with the Vesicle Prep Pro setup (Nanion Technologies GmbH, Munich, Germany). The lipid stock, formed from 10 mM of 1,2-diphytanoyl-sn-glycero-3-phosphocholine (DPhPC) (Avanti Polar lipids) with 10% cholesterol, was dissolved in chloroform and then deposited on the conductive side of an electrode from the GUVs chamber. The electrodes were formed by transparent indium tin oxide slides (ITO-Slides). After total evaporation of solvent, an O-ring was placed around the dried lipid film. The lipids were assembled in a perfectly dehydrated lamellate phase. A non-ionic intracellular solution, 1 M Sorbitol was added to the lipid film without agitation. Then, the second ITO-Slide was placed on the top of the ring. The process of electroformation was controlled by the Vesicle Prep Pro setup and all parameters (amplitude, frequency, main time, rise time and fall time) of electroformation were programmed by Vesicle Control software. The parameters were an alternative tension of 3 V peak to peak with a progressive increase for the start time and a progressive decrease for the stop time to avoid an abrupt change, a frequency of 5 Hz was applied to the ITO-Slides over a period of 2 hours at 36°C.
Planar lipid bilayer formation and reconstitution of p7
In order to form a planar lipid bilayer, 5 µl of the GUVs solution was pipetted onto the patch clamp chip. The microstructured chip had an aperture of approximately 1 micron in diameter. The GUVs were positioned onto the aperture in the chip by application of a slight negative pressure, typically (−) 10 to (−) 40 mbars. When the GUVs touched the glass surface of the chip, they burst and formed planar bilayers with a seal resistance of tens to hundreds of giga-ohms.
Electrophysiological recordings
All lipid bilayer recordings were done using the Port-a-Patch planar-patch-clamp system (Nanion Technologies, Munich, Germany). Proteins were reconstituted into planar bilayers using p7 proteoliposome fusion. Signals were acquired with an EPC 10 amplifier and the data acquisition software PatchMaster (both from HEKA, Lambrecht, Germany) at a sampling rate of 50 kHz. The recorded data were digitally filtered at 3 kHz. In bilayer experiments, the following symmetrical solution was employed: 200 mM KCl, 10 mM HEPES/KOH pH 7.2. All patch-clamp experiments were performed at room temperature. For inhibition assays, HMA was used at 10 µM, 100 µM and 1 mM.
Results and discussion
The study of membrane proteins is of high interest as they represent almost 30% of the genes from a genome and play essential biological roles. However, reaching the high expression level required for structural and functional studies remains a challenging task.
Different approaches have been taken to overcome problems related to membrane protein expression. Previous reports have used high molecular weight expression tags such as maltose MBP, GST or TrpΔLE to express p7 in inclusion bodies [8,24,25,49]. However, this method requires additional steps such as resolubilization of inclusion bodies, cleavage of the expression tag and refolding steps. Many problems can be encountered during the cleavage step alone such as low yield, precipitation of the target protein, high cost of proteases and failure to recover active or structurally intact protein [50]. It has also been reported that adding a tag could negatively affect the target protein by altering its biological activity [51], adding undesired flexibility in structural studies [52] and also changing protein conformation [53]. Several groups have used p7 from chemical peptide synthesis [9], but folding problems can also occur after synthesis.
Today, cell-free expression systems provide a real alternative for membrane protein expression, enabling the study of their structure and function [54–56]. Cell-free expression systems are open systems, which allow the fine-tuning of each condition to improve the quality and quantity of proteins expression. In the production of membrane proteins, membrane mimicking additives help expression, folding and stability.
In order to gain insight into the function of the viroporin p7, we first tested the expression of the recombinant full-length p7 protein from HCV strain H77 genotype 1a using an E. coli cell-free expression system. Synthetic liposomes were directly added to the cell-free reaction. The quality of cell-free expression depends on a variety of factors such as ion concentration, type and liposome concentrations [39,42,57]. Mg2+ and K+ ion concentrations were optimized for best yield. Mg2+ concentration was found to be at 16 mM, while K+ was found to be at 310 mM. The expression efficiency was detected by Western blotting using an anti-histidine antibody (Fig. 1B) and quantified by Coomassie brilliant blue staining (Fig. 1C). The batch expression yield of the partially purified proteoliposomes ranged from 0.5 to 0.8 mg/ml of reaction. In comparison, this yield was up to four hundred times larger than the yield reported in an earlier study for a FLAG tagged p7, which was evaluated at 2.5 mg/L [12] and p7 obtained from inclusion bodies, which was estimated at 2 mg/L [24]. Although the cost of cell-free expression is still higher than conventional E. coli protein expression, independently of the progress made throughout the last decade, cell-free expression of p7 is less labor intensive, which in turn lowers the final cost of total protein expression. To our knowledge, this is the first time that a full-length p7 protein from HCV was expressed using a cell-free system in a recombinant proteoliposome form in a one-step manner. The theoretical protein molecular weight was expected to be 9.9 kDa. However, the protein migrated above the 10 kDa molecular weight marker at 11 kDa, this in part due to the presence of residual lipids. Migration of membrane proteins in SDS-PAGE gel is often anomalous compared to soluble proteins [58].
p7 protein relipidation appears to be a hard task with product loss and giving different outcomes depending on the method used to carry out this process. Indeed, Gan et al. reported inconsistent p7 structure and function results depending on the reconstitution protocol chosen [25]. Moreover, a recent molecular simulation study on the M2 protein from the influenza B virus, a viroporin analogue to p7, showed that the zwitterionic detergent dihexanoylphosphatidylcholine (DHPC) molecules form “nonphysiological” interactions with the M2 protein termini [59]; underlining the tedious and careful selection of detergents and to a certain extent the need to avoid them when possible.
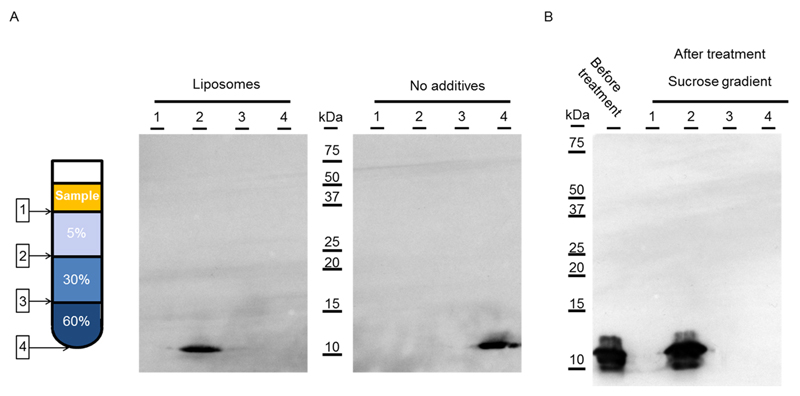
In this study, we used two methods to demonstrate the direct integration of the neo-synthesized p7 into synthetic liposomes when expressed in an E. coli cell-free system. First, the cell-free reactions expressing p7 with or without liposomes were loaded onto the top of a discontinuous sucrose gradient, were centrifuged, and the interfaces were analyzed by Western blotting from top to bottom (Fig. 2A). We observed that the entire p7 protein expressed with liposomes was retained at the interface between 5% and 30% sucrose. In the absence of liposomes, p7 was found in the pellet in the form of aggregates. These results suggest that the p7 protein was interacting with liposomes, while p7 expressed without additives was precipitated and therefore found in the pellet. To assess whether the p7 protein was only associated with the lipid bilayer of liposomes or if it was integrated within the bilayer, we performed an alkaline extraction. This method has been widely used to assess if an integral membrane protein has achieved stable insertion into a lipid bilayer [55,60]. Liposomes are disrupted into sheets and protein-protein interactions are disturbed while protein-lipid interactions are maintained. After alkaline extraction, the sample was centrifuged on a discontinuous sucrose gradient as previously described. The p7 protein was found at the same interface as prior to treatment and no protein was found in the pellet (Fig. 2B). These experiments confirmed the proper insertion of the p7 protein into the bilayer of liposomes during the cell-free expression.
Figure 2. p7 integration into liposomes.
Analysis of the interfaces of a sucrose gradient after ultracentrifugation (280 000 x g for 1hr at 4°C) by Western immunoblotting. Interfaces are represented as such: 1 (Sample buffer-5% sucrose), 2 (5%-30% sucrose), 3 (30%-60% sucrose) and 4 (bottom of the tube). (A) comparison between p7 expressed with liposomes (left) and without additives (right). After ultracentrifugation, p7 is found at the 5%-30% sucrose interface with lipids while without additive, p7 is pelleted at the bottom of the tube; (B) Ultracentrifugation of purified p7 proteoliposomes after alkaline extraction. The sample is found at the 5%-30% sucrose interface while no protein is found in the pellet.
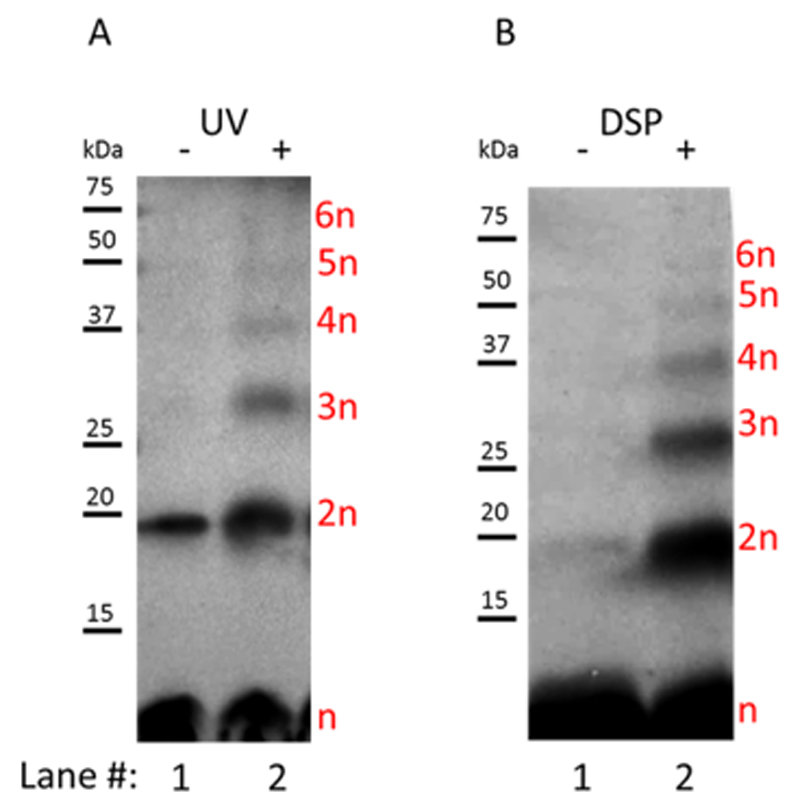
Homo-oligomerization of p7 being required to form a functional viroporin, we studied the oligomerization state of p7 by in vitro cross-linking assays. This method enabled cross-linking between specific protein-protein interacting domains in their natural environment, in this case the lipid membrane. One of the major benefits of cell-free expression is the possibility to add modified amino acids easily. Here, L-leucine and L-methionine usually added to the cell-free reaction were substituted by L-photo-leucine and L-photo-methionine. This approach avoids the use of cross-linking agents that could alter natural behavior of the target. The 3 methionine and 15 leucine residues of our p7 construct allowed cross-linking reactions. These analogs were incorporated during protein synthesis and then activated by ultraviolet (UV) light to covalently cross-link interacting proteins [61]. The p7 proteoliposomes were purified and then subjected to UV light from a Biorad Imager before analysis by Western blotting. A sample that had not been subjected to UV light was used as a control (Fig. 3A, lane 1). Upon UV exposure, diazirine molecules were activated leading to the cross-linking of interacting proteins. In accordance with previous studies p7 [15,17], p7 oligomers were detected (Fig. 3A, Lane 2) and up to 6 protomers assembled to create an hexamer. The cross-linking experiment was repeated using the lipid-soluble cross-linking reagent DSP. This compound has been previously used to demonstrate oligomerization of p7 from genotype 1b [17]. We observed, as reported [17], the formation of a p7 hexamer when using DSP as a cross-linker (Fig. 3B, lane2).
Figure 3. Cross-linking assays of p7 in liposome membranes. Immunoblotting analysis of p7 resolved by SDS-PAGE.
(A) Using photoactivatable amino acid analogs. Samples were exposed to UV light to trigger cross-linking reaction. (B) Using DSP. SDS-PAGE was conducted in non-reducing conditions for the cross-linking analysis of p7 using DSP. n = protomer, 2n = dimer, 3n = trimer, 4n = tetramer, 5n = pentamer and 6n = hexamer
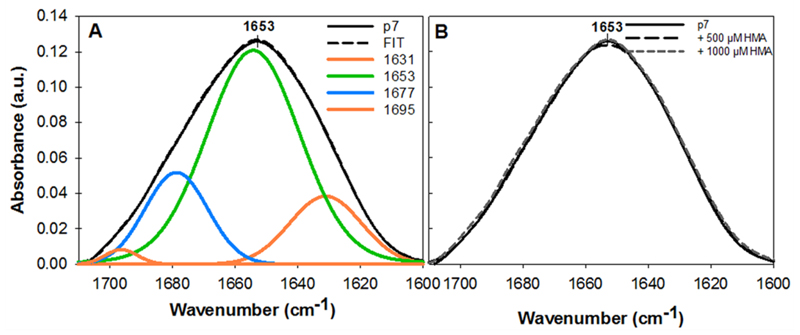
FTIR spectroscopy is an established tool for structural characterization of proteins and suitable for studies in lipid bilayers. We used this method to show that liposomes provided an environment that enabled the proper folding of the p7 protein. The analysis of the amide I band (1700 – 1600 cm-1 region) is widely used to investigate the secondary structure of proteins [62–64]. By analyzing this band, centered at ~1653 cm-1, we showed that most of the p7 protein is represented by α-helix structures (Fig. 4A). Two other minor contributions were identified and based on FTIR data on proteins [65] can be attributed to β-sheets (1631 and 1695 cm-1) and to β-turns structures (1677 cm-1). The result of the deconvolution of the band was a percentage of 66% of α-helix structures and 16% of β-sheets. This data is consistent with the 69% predicted structure using Chou and Fasman secondary structure prediction server and consistent with previously published data obtained by circular dichroism or attenuated total reflectance FTIR [9,23,25]. In addition, our measurements showed the contribution β-structures in the spectrum. This observation was also recently reported by Gan et al.[25], independently of the protein source (i.e. recombinant and from peptide synthesis). While α-helix structures constitute the transmembrane domains of the protein, the role of β-structures is still unclear. It has been suggested that β-structures likely account for the permeability of large molecules [25]. No studies of the p7 protomer using NMR have reported β-structures, however, not all of them where performed on the p7 oligomer. It is possible that the β-structures are induced upon oligomerisation. Moreover, the discrepancy between structures could be explained by the genetic diversity of the amino acid sequences and the experimental conditions adopted in each study. The effect of HMA, a compound that had been previously reported as a p7 inhibitor [18], on the secondary structure was also assessed. Addition of HMA to p7 proteoliposomes did not change the recorded spectrum, even at high concentration such as 1 mM (Fig. 4B). The secondary structure stability of p7 was also investigated after one week storage at 4°C. The spectrum did not differ significantly from the fresh sample, thus indicating p7 protein stability within a lipid bilayer.
Figure 4. FTIR spectra of p7 proteoliposomes.
(A) spectrum and fit of cell-free expressed p7 proteoliposomes (Fit: orange: β-sheets, green: α-helix, blue: β-turns); (B) spectra of p7 proteoliposomes without HMA (solid line), with 500 µM HMA (large dashed line) and with 1000 µM HMA (small dashed line). The mean spectrum of three spectra is represented.
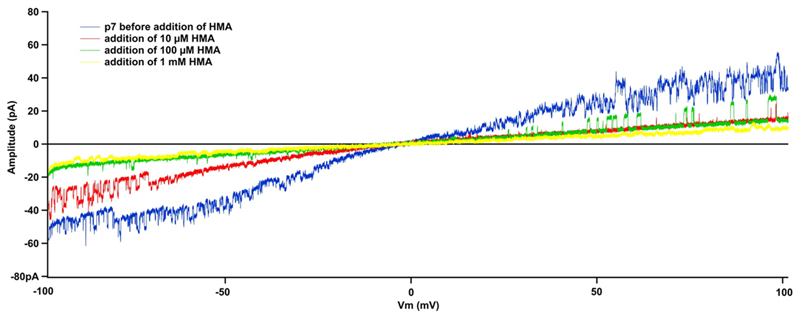
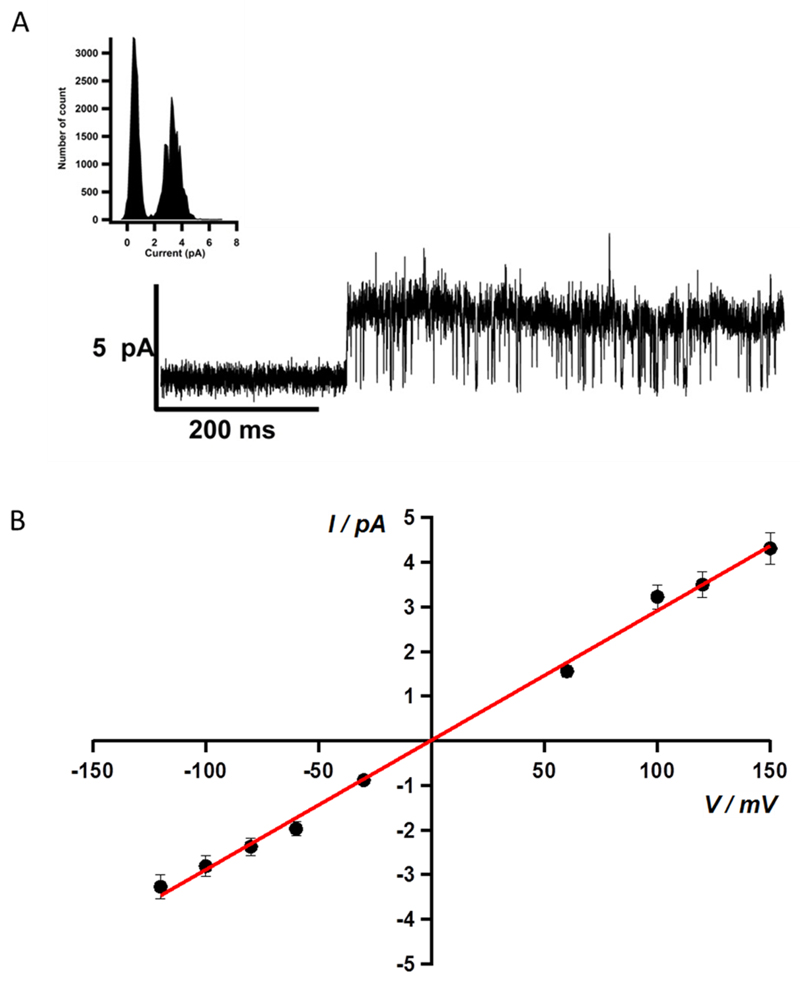
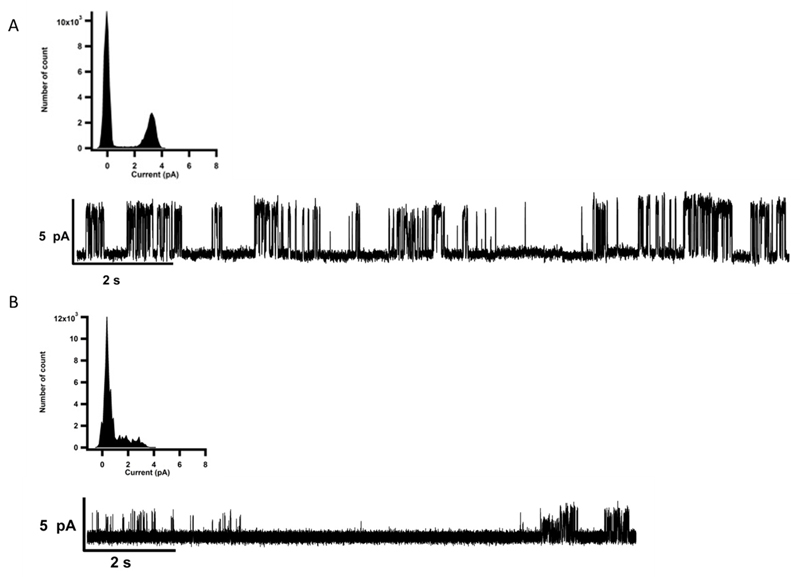
Furthermore, the correct folding and oligomerisation was also confirmed by testing the function of the recombinant p7 viroporin. Proteoliposomes have the ability to fuse with membranes allowing patch-clamp recordings. The p7 proteoliposomes were added on top of a planar lipid bilayer. Proteoliposome fusion to the underlying bilayer exhibited a typical capacitive spike in the current as previously reported [25]. Directly after addition, currents were measured in response to a voltage ramp applied to a DPhPC and cholesterol lipid bilayer from - 100 mV to +100 mV in 2 seconds (Fig. 5). The channel activity of p7 was confirmed by measuring single channel currents in continuous recording at +100 mV (Fig. 6A) holding potential (inset: the all point histogram). The channel conductance value observed at + 100 mV was 32.27 ± 1.42 pS in 200 mM KCl, 2mM EDTA, 10 mM HEPES, pH 7.4. Control measurements were taken using E2 proteoliposomes, after fusion, and no pore formation and channel activity was observed (Supplementary Fig. S1A). The current voltage relationships of all p7 conductance states observed exhibited ideal ohmic behaviour throughout the voltage range tested, from -100 mV to +100 mV (Fig. 6B), confirming that the ion channel and related states are not voltage-gated and that there was no difference in channel behaviour at positive or negative voltages as reported in previous work [18,23,66]. In the IV curve presented in figure 6B, a conductance level of 29 ± 0.6 pS was obtained from the slope. The single channel IV curve was extracted from the all point histograms at each voltage, with recordings of at least 180 s (n=5). HMA was added to the recording chamber to block channel activity and further characterize the viroporin. In the presence of 10 µM HMA, the amplitude of the recorded currents of the voltage ramps was reduced by 40–50% (Fig. 5 red trace). At 1 mM HMA concentration (Fig. 5 yellow trace), we noticed a full blockage of the channel. These results indicated that the ion channel activity was inhibited by HMA in a concentration dependent manner. HMA was able to inhibit p7 channel activity in a concentration dependent fashion in our in vitro assay which is in agreement with previously reported results [18]. p7 single channel activity was fully blocked in the presence of 500 µM HMA (Fig. 7A). The channel conductance value was decreased to 9.26 ± 1.25 pS with the drug and could be restored after washing out the HMA to a value of 32.8 ± 2.1 pS (Fig. 7B). Addition of 500 µM HMA did not affect the control recordings (Supplementary Fig. S1B). Amantadine did not show inhibition of the channel activity. This compound has been described as having a genotype dependent efficiency both in vitro and in vivo [21,67,68]. In contrast, when amantadine was used at high concentration a rise in conductivity could be detected. This observation had already been noticed by Montserret et al. [9]. It had also been reported that due to its high hydrophobicity, amantadine is able to interact with the lipid bilayer and disturb its integrity [69]. Further, we noticed that at high concentration, amantadine was able to break giga-ohm seals obtained during the patch clamp recording suggesting that the compound can alter the lipid membrane causing its rupture. Since HMA inhibits p7 ion conductivity, the compound is likely to bind to the protein. Using FTIR, we noted that HMA did not alter the secondary structure of p7. A study of p7 from the J4 genotype using NMR reported that HMA did bind the protein, which did not seem to alter the secondary structure [8]. Therefore, HMA binding might prevent ion conductivity by steric hindrance, clogging the channel, or changing the conformation of the pore without altering the secondary structure of the complex as modeled by Ouyang et al. for amantadine [10]. Another interesting hypothesis is that HMA could destabilise protomer interaction and oligomer formation preventing the creation of the pore necessary for ion channel activity. HMA could therefore inhibited p7 ion conductivity in a similar manner than the one of NN-DNJ as reported previously [8].
Figure 5. Effect of the HMA on p7.
Different concentrations of HMA were applied to the bilayer containing p7. The recordings were done using a ramp protocol from -100 mV to +100 mV during 2 s (100 mV/s ramp).
Figure 6. Functional characterization of p7.
(A) Single-channel recording of p7 reconstituted into a DPhPC and cholesterol bilayer examined by the patch clamp technique in 200mM KCl, 2 mM EDTA, 10mM HEPES, pH 7.4. The corresponding point amplitude histogram shows a conductance of 32.27 ± 1.42 pS; (B) p7 current-voltage (I-Vm) relationship. The main conductance, calculated from the slope of the linear regression, was determined to be 29.02 ± 0.6 pS. Coefficient values ± one standard deviation: a = 0.015147 ± 0.0627; b = 0.029017 ± 0.000642
Figure 7. Effect of the HMA on p7 single channel recorging.
(A) p7 in presence of 500 µM HMA, the all point histogram shows a reduced activity and the p7 single channel conductance in presence of HMA was 9.26 ± 1.25 pS (n= 5); (B) p7 after wash out of HMA, the conductance of p7 is recovered after washing with a conductance of 32.8 ± 2.1 pS.
In conclusion, we demonstrated the efficiency of cell-free protein synthesis to express the p7 viroporin at high yields (up to 0.8 mg/mL). Addition of liposomes in the synthesis reaction enabled the production of proteoliposomes in a one-step manner. The obtained p7 proteoliposomes were validated and fully characterized on a structural level, as well as a functional level. We believe that p7 proteoliposomes produced using cell-free synthesis can be great tools for the study of the protein and the development of novel therapies against HCV. We also believe that this strategy could be extended to other viroporins of interest.
Supplementary Material
Aknowledgments
We would like to thank Marie-Anne Petit and Emmanuel Drouet for their guidance and advice throughout this project. C.P. wishes to thank the European Commission for support in the framework of FP7 via an ERC Starting grant 2010 (GA 259370, BIOMIM). This work was supported by a CIFRE (Conventions Industrielles de Formation par la Recherche) convention number 2011/0251 of the ANRT (Association Nationale de la Recherche et l'innovation Technologique) and Synthelis SAS.
References
- [1].Liang TJ. Current progress in development of hepatitis C virus vaccines. Nat Med. 2013;19:869–78. doi: 10.1038/nm.3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Xue J, Zhu H, Chen Z. Therapeutic vaccines against hepatitis C virus. Infect Genet Evol. 2014;22:120–129. doi: 10.1016/j.meegid.2014.01.008. [DOI] [PubMed] [Google Scholar]
- [3].Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat Rev Microbiol. 2007;5:453–463. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- [4].Dietz J, Susser S, Berkowski C, Perner D, Zeuzem S, Sarrazin C. Consideration of Viral Resistance for Optimization of Direct Antiviral Therapy of Hepatitis C Virus Genotype 1-Infected Patients. PLoS One. 2015;10:e0134395. doi: 10.1371/journal.pone.0134395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Aloia AL, Locarnini S, Beard MR. Antiviral resistance and direct-acting antiviral agents for HCV. Antivir Ther. 2012;17:1147–1162. doi: 10.3851/IMP2426. [DOI] [PubMed] [Google Scholar]
- [6].Lin C, Lindenbach BD, Pragat BM, Mccourt DW, Rice CM. Processing in the Hepatitis C Virus E2-NS2 Region: Identification of p7 and Two Distinct E2-Specific Products with Different C Termini. J Virol. 1994;68:5063–5073. doi: 10.1128/jvi.68.8.5063-5073.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Jones CT, Murray CL, Eastman DK, Tassello J, Rice CM. Hepatitis C Virus p7 and NS2 Proteins Are Essential for Production of Infectious Virus. J Virol. 2007;81:8374–8383. doi: 10.1128/JVI.00690-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Cook Ga, Dawson LA, Tian Y, Opella SJ. Three-Dimensional Structure and Interaction Studies of Hepatitis C Virus p7 in 1,2-Dihexanoyl- sn -glycero-3-phosphocholine by Solution Nuclear Magnetic Resonance. Biochemistry. 2013;52:5295–5303. doi: 10.1021/bi4006623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Montserret R, Saint N, Vanbelle C, Salvay G, Simorre J, Ebel C, et al. NMR Structure and Ion Channel Activity of the p7 Protein from Hepatitis C Virus. J Biol Chem. 2010;285:31446–31461. doi: 10.1074/jbc.M110.122895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].OuYang B, Xie S, Berardi MJ, Zhao X, Dev J, Yu W, et al. Unusual architecture of the p7 channel from hepatitis C virus. Nature. 2013;498:521–5. doi: 10.1038/nature12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Patargias G, Zitzmann N, Dwek R, Fischer WB. Protein - Protein Interactions: Modeling the Hepatitis C Virus Ion Channel p7. J Med Virol. 2006;49:648–655. doi: 10.1021/jm050721e. [DOI] [PubMed] [Google Scholar]
- [12].Clarke D, Griffin S, Beales L, Gelais CSt, Burgess S, Harris M, et al. Evidence for the Formation of a Heptameric Ion Channel Complex by the Hepatitis C Virus P7 Protein in Vitro *. J Biol Chem. 2006;281:37057–37068. doi: 10.1074/jbc.M602434200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Carrère-kremer S, Montpellier-pala C, Cocquerel L, Wychowski C, Penin F, Dubuisson J. Subcellular Localization and Topology of the p7 Polypeptide of Hepatitis C Virus. J Virol. 2002;76:3720–3730. doi: 10.1128/JVI.76.8.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Griffin SDC, Harvey R, Clarke DS, Barclay WS, Harris M, Rowlands DJ. A conserved basic loop in hepatitis C virus p7 protein is required for amantadine-sensitive ion channel activity in mammalian cells but is dispensable for localization to mitochondria. J Gen Virol. 2004;85:451–461. doi: 10.1099/vir.0.19634-0. [DOI] [PubMed] [Google Scholar]
- [15].Luik P, Chew C, Aittoniemi J, Chang J, Wentworth P, Dwek RA, et al. The 3-dimensional structure of a hepatitis C virus p7 ion channel by electron microscopy. PNAS. 2009;106:12712–12713. doi: 10.1073/pnas.0905966106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Madan V, Bartenschlager R. Structural and Functional Properties of the Hepatitis C Virus p7 Viroporin. Viruses. 2015;7:4461–4481. doi: 10.3390/v7082826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Griffin S. The p7 protein of hepatitis C virus forms an ion channel that is blocked by the antiviral drug, Amantadine. FEBS Lett. 2003;535:34–38. doi: 10.1016/S0014-5793(02)03851-6. [DOI] [PubMed] [Google Scholar]
- [18].Premkumar A, Wilson L, Ewart GD, Gage PW. Cation-selective ion channels formed by p7 of hepatitis C virus are blocked by hexamethylene amiloride. FEBS Lett. 2004;557:99–103. doi: 10.1016/S0014-5793(03)01453-4. [DOI] [PubMed] [Google Scholar]
- [19].Pavlovic D, Neville DCA, Argaud O, Blumberg B, Dwek RA, Fischer WB, et al. The hepatitis C virus p7 protein forms an ion channel that is inhibited by long-alkyl-chain iminosugar derivatives. PNAS. 2003;100:6104–6108. doi: 10.1073/pnas.1031527100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Luscombe CA, Huang Z, Murray MG, Miller M, Wilkinson J, Ewart GD. A novel Hepatitis C virus p7 ion channel inhibitor , BIT225 , inhibits bovine viral diarrhea virus in vitro and shows synergism with recombinant interferon-α-2b and nucleoside analogues. Antiviral Res. 2010;86:144–153. doi: 10.1016/j.antiviral.2010.02.312. [DOI] [PubMed] [Google Scholar]
- [21].Griffin S, Stgelais C, Owsianka AM, Patel AH, Rowlands D, Harris M. Genotype-Dependent Sensitivity of Hepatitis C Virus to Inhibitors of the p7 Ion Channel. Hepatology. 2008;48:1779–1790. doi: 10.1002/hep.22555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Meredith LW, Zitzmann N, McKeating Ja. Differential effect of p7 inhibitors on hepatitis C virus cell-to-cell transmission. Antiviral Res. 2013;100:636–9. doi: 10.1016/j.antiviral.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Whitfield T, Miles AJ, Scheinost JC, Offer J, W P, Jr, Dwek RA, et al. The influence of different lipid environments on the structure and function of the hepatitis C virus p7 ion channel protein. Mol Membr Bol. 2011;28:254–264. doi: 10.3109/09687688.2011.581253. [DOI] [PubMed] [Google Scholar]
- [24].Cook Ga, Stefer S, Opella SJ. Expression and purification of the membrane protein p7 from hepatitis C virus. Biopolymers. 2011;96:32–40. doi: 10.1002/bip.21453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gan SW, Surya W, Vararattanavech A, Torres J. Two Different Conformations in Hepatitis C Virus p7 Protein Account for Proton Transport and Dye Release. PLoS One. 2014;9:1–14. doi: 10.1371/journal.pone.0078494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Katzen F, Peterson TC, Kudlicki W. Membrane protein expression: no cells required. Trends Biotechnol. 2009;27:455–460. doi: 10.1016/j.tibtech.2009.05.005. [DOI] [PubMed] [Google Scholar]
- [27].Schwarz D, Dötsch V, Bernhard F. Production of membrane proteins using cell-free expression systems. Proteomics. 2008;8:3933–3946. doi: 10.1002/pmic.200800171. [DOI] [PubMed] [Google Scholar]
- [28].Dondapati SK, Kreir M, Quast RB, Wüstenhagen Da, Brüggemann A, Fertig N, et al. Membrane assembly of the functional KcsA potassium channel in a vesicle-based eukaryotic cell-free translation system. Biosens Bioelectron. 2014;59C:174–183. doi: 10.1016/j.bios.2014.03.004. [DOI] [PubMed] [Google Scholar]
- [29].Cappuccio JA, Blanchette CD, Sulchek TA, Arroyo ES, Kralj JM, Hinz AK, et al. Cell-free Co-expression of Functional Membrane Proteins and Apolipoprotein , Forming Soluble Nanolipoprotein Particles. Mol Cell Proteomics. 2008;7:2246–2253. doi: 10.1074/mcp.M800191-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Periasamy A, Shadiac N, Amalraj A, Nagarajan Y, Waters S, Mertens HDT, et al. Cell-free protein synthesis of membrane ( 1 , 3 ) - β - D -glucan ( curdlan ) synthase: Co-translational insertion in liposomes and reconstitution in nanodiscs. Biochim Biophys Acta. 2013;1828:743–757. doi: 10.1016/j.bbamem.2012.10.003. [DOI] [PubMed] [Google Scholar]
- [31].Klammt C, Lo F, Scha B, Glaubitz C, Bernhard F. High level cell-free expression and specific labeling of integral membrane proteins. Eur J Biochem. 2004;580:568–580. doi: 10.1111/j.1432-1033.2003.03959.x. [DOI] [PubMed] [Google Scholar]
- [32].Keller T, Schwarz D, Bernhard F, Dötsch V, Hunte C, Gorboulev V, et al. Cell Free Expression and Functional Reconstitution of Eukaryotic Drug. Biochemistry. 2008;47:4552–4564. doi: 10.1021/bi800060w. [DOI] [PubMed] [Google Scholar]
- [33].Ishihara G, Goto M, Saeki M, Ito K, Hori T, Kigawa T. Expression of G protein coupled receptors in a cell-free translational system using detergents and thioredoxin-fusion vectors. Protein Expr Purif. 2005;41:27–37. doi: 10.1016/j.pep.2005.01.013. [DOI] [PubMed] [Google Scholar]
- [34].Klammt C, Schwarz D, Fendler K, Haase W, Do V, Bernhard F. Evaluation of detergents for the soluble expression of a -helical and b -barrel-type integral membrane proteins by a preparative scale individual cell-free expression system. FEBS J. 2005;272:6024–6038. doi: 10.1111/j.1742-4658.2005.05002.x. [DOI] [PubMed] [Google Scholar]
- [35].Park K, Berrier C, Lebaupain F, Pucci B, Popot J, Ghazi A, et al. Fluorinated and hemifluorinated surfactants as alternatives to detergents for membrane protein cell-free synthesis. Biochem J. 2007;187:183–187. doi: 10.1042/BJ20061473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Engel CK, Chen L, Prive GG. Stability of the lactose permease in detergent solutions. Biochim Biophys Acta. 2002;1564:47–56. doi: 10.1016/s0005-2736(02)00397-8. [DOI] [PubMed] [Google Scholar]
- [37].White JF, Grisshammer R. Stability of the neurotensin receptor NTS1 free in detergent solution and immobilized to affinity resin. PLoS One. 2010;5:1–8. doi: 10.1371/journal.pone.0012579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Park K, Billon-denis E, Dahmane T, Zito F, Lebaupain F, Pucci B. In the cauldron of cell-free synthesis of membrane proteins: playing with new surfactants. N Biotechnol. 2010;00:1–7. doi: 10.1016/j.nbt.2010.08.008. [DOI] [PubMed] [Google Scholar]
- [39].Marques B, Liguori L, Paclet M-H, Villegas-Mendéz A, Rothe R, Morel F, et al. Liposome-mediated cellular delivery of active gp91 (phox) PLoS One. 2007;2:e856. doi: 10.1371/journal.pone.0000856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Liguori L, Blesneac I, Madern D, Vivaudou M, Lenormand J. Single-step production of functional OEP24 proteoliposomes. Protein Expr Purif. 2010;69:106–111. doi: 10.1016/j.pep.2009.07.004. [DOI] [PubMed] [Google Scholar]
- [41].Liguori L, Marques B, Villegas-mendez A, Rothe R, Lenormand J. Liposomes-mediated delivery of pro-apoptotic therapeutic membrane proteins. J Control Release. 2008;126:217–227. doi: 10.1016/j.jconrel.2007.12.004. [DOI] [PubMed] [Google Scholar]
- [42].Kalmbach R, Chizhov I, Schumacher MC, Friedrich T, Bamberg E, Engelhard M. Functional Cell-free Synthesis of a Seven Helix Membrane Protein : In situ Insertion of Bacteriorhodopsin into Liposomes. J Mol Biol. 2007:639–648. doi: 10.1016/j.jmb.2007.05.087. [DOI] [PubMed] [Google Scholar]
- [43].Katzen F, Fletcher JE, Yang J, Kang D, Peterson C, Cappuccio JA, et al. Insertion of Membrane Proteins into Discoidal Membranes Using a Cell-Free Protein Expression Approach. J Proteome Res. 2008:3535–3542. doi: 10.1021/pr800265f. [DOI] [PubMed] [Google Scholar]
- [44].Sitaraman K, Chatterjee DK. Chapter 15 High-Throughput Protein Expression Using Cell-Free System. Methods Mol Biol. 2009;498:229–244. doi: 10.1007/978-1-59745-196-3. [DOI] [PubMed] [Google Scholar]
- [45].Blesneac I, Moreau C, Liguori L. A Simple Method for the Reconstitution of Membrane Proteins into Giant Unilamellar Vesicles. 2010:85–92. doi: 10.1007/s00232-010-9227-8. [DOI] [PubMed] [Google Scholar]
- [46].Schwinté P, Ball V, Szalontai B, Haikel Y, Voegel J-C, Schaaf P. Secondary structure of proteins adsorbed onto or embedded in polyelectrolyte multilayers. Biomacromolecules. 2002;3:1135–43. doi: 10.1021/bm025547f. [DOI] [PubMed] [Google Scholar]
- [47].Gilde F, Guillot R, Mano JF, Logeart-avramoglou D. Secondary Structure of rhBMP - 2 in a Protective Biopolymeric Carrier Material. Biomacromolecules. 2012;13:3620–3626. doi: 10.1021/bm3010808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Barth A, Zscherp C. What vibrations tell us about proteins. Quaterly Rev Biophys. 2002;4:369–430. doi: 10.1017/S0033583502003815. [DOI] [PubMed] [Google Scholar]
- [49].Cook GA, Zhang H, Park SH, Wang Y, Opella SJ. Comparative NMR studies demonstrate profound differences between two viroporins : p7 of HCV and Vpu of HIV-1. BBA - Biomembr. 2011;1808:554–560. doi: 10.1016/j.bbamem.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Baneyx F, Mujacic M. Recombinant protein folding and misfolding in Escherichia coli. Nat Biotechnol. 2004;22:1399–1408. doi: 10.1038/nbt1029. [DOI] [PubMed] [Google Scholar]
- [51].Fonda I, Kenig M, Gaberc-Porekar V, Pristovaek P, Menart V. Attachment of histidine tags to recombinant tumor necrosis factor-alpha drastically changes its properties. ScientificWorldJournal. 2002;2:1312–25. doi: 10.1100/tsw.2002.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Smith DB, Corcoran LM. Expression and purification of glutathione-S-transferase fusion proteins. Curr Protoc Mol Biol. 2001;Chapter 16 doi: 10.1002/0471142727.mb1607s28. Unit16.7. [DOI] [PubMed] [Google Scholar]
- [53].Amor-Mahjoub M, Suppini J-P, Gomez-Vrielyunck N, Ladjimi M. The effect of the hexahistidine-tag in the oligomerization of HSC70 constructs. J Chromatogr B Analyt Technol Biomed Life Sci. 2006;844:328–34. doi: 10.1016/j.jchromb.2006.07.031. [DOI] [PubMed] [Google Scholar]
- [54].Deniaud A, Liguori L, Blesneac I, Lenormand JL, Pebay-peyroula E. Biochimica et Biophysica Acta Crystallization of the membrane protein hVDAC1 produced in cell-free system. BBA - Biomembr. 2010;1798:1540–1546. doi: 10.1016/j.bbamem.2010.04.010. [DOI] [PubMed] [Google Scholar]
- [55].Long AR, Brien CCO, Alder NN. The Cell-Free Integration of a Polytopic Mitochondrial Membrane Protein into Liposomes Occurs Cotranslationally and in a Lipid-Dependent Manner. PLoS One. 2012;7 doi: 10.1371/journal.pone.0046332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Klammt C, Schwarz D, Lo F, Schneider B, Do V, Bernhard F. Cell-free expression as an emerging technique for the large scale production of integral membrane protein. FEBS J. 2006;273:4141–4153. doi: 10.1111/j.1742-4658.2006.05432.x. [DOI] [PubMed] [Google Scholar]
- [57].Kai L, Kaldenhoff R, Lian J, Zhu X, Do V, Bernhard F, et al. Preparative Scale Production of Functional Mouse Aquaporin 4 Using Different Cell-Free Expression Modes. PLoS One. 2010;5:2–9. doi: 10.1371/journal.pone.0012972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Rath A, Glibowicka M, Nadeau VG, Chen G, Deber CM. Detergent binding explains anomalous SDS-PAGE. PNAS. 2009;106:1760–1765. doi: 10.1073/pnas.0813167106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Rouse SL, Sansom MSP. Interactions of Lipids and Detergents with a Viral Ion Channel Protein: Molecular Dynamics Simulation Studies. J Phys Chem B. 2015;119:764–772. doi: 10.1021/jp505127y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].van der Laan M, Meinecke M, Dudek J, Hutu DP, Lind M, Perschil I, et al. Motor-free mitochondrial presequence translocase drives membrane integration of preproteins. Nat Cell Biol. 2007;9:1152–9. doi: 10.1038/ncb1635. [DOI] [PubMed] [Google Scholar]
- [61].Suchanek M, Radzikowska A, Thiele C. Photo-leucine and photo-methionine allow identification of protein-protein interactions in living cells. Nat Methods. 2005:1–7. doi: 10.1038/NMETH752. [DOI] [PubMed] [Google Scholar]
- [62].Izumrudov Va, Kharlampieva E, Sukhishvili Sa. Multilayers of a globular protein and a weak polyacid: role of polyacid ionization in growth and decomposition in salt solutions. Biomacromolecules. 2005;6:1782–8. doi: 10.1021/bm050096v. [DOI] [PubMed] [Google Scholar]
- [63].Bi X, Taneva S, Keough KMW, Mendelsohn R, Flach CR. Thermal Stability and DPPC / Ca 2 + Interactions of Pulmonary Surfactant SP-A from Bulk-Phase and Monolayer IR Spectroscopy. Biochemistry. 2001;40:13659–13669. doi: 10.1021/bi011188h. [DOI] [PubMed] [Google Scholar]
- [64].Zhang X, Ge N, Keiderling Ta. Electrostatic and hydrophobic interactions governing the interaction and binding of beta-lactoglobulin to membranes. Biochemistry. 2007;46:5252–60. doi: 10.1021/bi602483p. [DOI] [PubMed] [Google Scholar]
- [65].Schwinté P, Voegel J-C, Picart C, Haikel Y, Schaaf P, Szalontai B. Stabilizing Effects of Various Polyelectrolyte Multilayer Films on the Structure of Adsorbed/Embedded Fibrinogen Molecules: An ATR–FTIR Study. J Phys Chem B. 2001;105:11906–11916. doi: 10.1021/jp0123031. [DOI] [Google Scholar]
- [66].Chew CF, Vijayan R, Chang J, Zitzmann N, Biggin PC. Determination of Pore-Lining Residues in the Hepatitis C Virus p7 Protein. Biophysj. 2009;96:L10–L12. doi: 10.1016/j.bpj.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Stgelais C, Foster TL, Verow M, Atkins E, Fishwick CWG, Rowlands D, et al. Determinants of Hepatitis C Virus p7 Ion Channel Function and Drug Sensitivity Identified In Vitro. J Virol. 2009;83:7970–7981. doi: 10.1128/JVI.00521-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Castelain S, Bonte D, Penin F, Franc C, Capron D, Dedeurwaerder S. Hepatitis C Virus p7 Membrane Protein Quasispecies Variability in Chronically Infected Patients Treated With Interferon and Ribavirin , With or Without Amantadine. J Med Virol. 2007;154:144–154. doi: 10.1002/jmv. [DOI] [PubMed] [Google Scholar]
- [69].Tverdislov VA, Kharitonenkov IG, Glaser R, Herrmann A, Lentzsch P, Donath J. Interaction of the Antivirus Agents Remantadine and Amantadine with Lipid Membranes and the Influence on the Curvature of Human Red Cells. Gen Physiol Biophys. 1986:61–75. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.