Abstract
ADP-ribosylation (ADPr) is a biologically and clinically important post-translational modification, but little is known about the amino acids it targets on cellular proteins. Here we present a proteomic approach for direct in vivo identification and quantification of ADPr sites on histones. We have identified 12 unique ADPr sites in human osteosarcoma cells and report serine ADPr as a new type of histone mark that responds to DNA damage.
Cells control crucial biological processes through the reversible conjugation of diverse post-translational modifications (PTMs), ranging from small chemical groups, such as phosphorylation, to complex carbohydrates and entire small proteins. ADP-ribosylation (ADPr) is the best-characterized nucleotide-based PTM, with a wealth of evidence pointing to its biomedical importance, especially in DNA damage responses and cancer therapy1. In contrast to the limited amino acid specificities of most PTMs, nearly all the chemically reactive amino acid side-chains have been reported as targets of ADPr2. However it is difficult to study most of these ADPr linkages in vivo, even to abundant targets like histones. Histone ADPr is primarily studied in DNA damage repair3, although a detailed understanding is constrained by ignorance of the exact sites in cellular contexts.
Given the biological and clinical importance of ADPr, we set out to analyze histone ADPr in cells using mass spectrometry (MS). Although endogenous histones can be easily isolated from cells, poor sequence coverage generally hampers analysis of marks. Conventional tryptic digestion of histones generates many peptides too short for productive MS analysis. Over the last decade, several sample preparation strategies have been proposed to reduce these systematic blind spots, typically integrating multiple parallel digestions based on chemical derivatizations and proteolysis with less efficient and specific proteases4, 5. Despite these sophisticated approaches, it has been practically impossible to obtain full sequence coverage for all histones.
We have developed a simple method for high-coverage sequencing of histones based on the popular FASP (“filter-aided sample preparation”) approach6, coupling partial trypsin digestion to generate longer peptides with ultrafiltration to remove undigested proteins (Fig. 1). Digestion of purified histones by this “partial FASP” strategy is reproducible according to SDS-PAGE analysis, correlation of histone peptide intensities and overlap of identified peptides (Supplementary Results, Supplementary Fig. 1). Our approach allows full sequence coverage of core histones (although the C-terminal peptide of H2A is occasionally lost) and above 95% coverage of H1 (Supplementary Fig. 2a) by generating peptides with higher mass and charge than in standard FASP protocols (Supplementary Figs. 2b-d and 3).
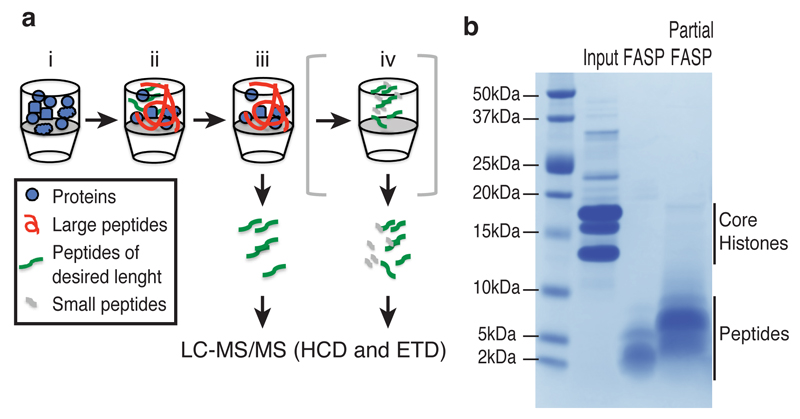
Figure 1. Partial FASP method for increased sequence coverage of histones.
(a) Workflow of the Partial FASP method. Histones are digested on the filter with limiting amount of trypsin (1:2000) for 20 minutes at 20 ºC (i, ii), the peptides amenable for MS analysis are separated from the undigested proteins by centrifugation (iii). Optionally, undigested proteins and large peptides can be further digested overnight to recover smaller peptides similarly to the FASP method (iv). See Online Methods for further details. (b) SDS-PAGE analysis of the purified histones (Input, 10 μg) and eluted peptides from the filter after a full digestion (FASP) or a partial digestion (Partial FASP) of 100 μg proteins. See Supplementary Fig. 10 for original image.
To isolate histones from cell lines, we modified a method combining sulfuric acid extraction with sulfopropyl-Sepharose (SP) chromatography7 (Supplementary Fig. 4a-b). Importantly, cell lysis in sulfuric acid preserves the physiological level of ADPr by preventing the artifactual protein ADPr observed upon lysis in non-denaturing conditions8 and the PARG-mediated degradation of polymers to free ADP-ribose, which can lead to non-enzymatic ADPr2 (Supplementary Fig. 4c-d). Although this isolation approach combined with partial FASP is sufficient for detecting ADPr by MS, to increase sensitivity we also enriched ADP-ribosylated peptides with a recently developed phosphoproteomic approach9. In order to make poly-ADPr sites detectable as well, we repeated the analyses after treating the histone peptides with a Nudix hydrolase, which converts both mono-ADP-ribose and poly-ADP-ribose to phospho-ribose10, 11. We did not use PARG, as this can lead to non-enzymatic ADPr through the generation of free ADP-ribose2.
All peptide samples were analyzed by UHPLC-MS/MS. Initial HCD (higher-energy collisional dissociation) MS/MS analysis confidently detected several peptides modified by ADPr. While low-mass ions resulting from fragmentation of ADP-ribose (Supplementary Fig. 5) unmistakably confirmed the presence of ADP-ribose12, we could not confidently localize the modification sites with HCD, since the ADPr-peptide bond appeared to be extremely labile, a behavior also observed for O-glycopeptides13. To precisely map the modified residues, we employed an alternative fragmentation mode, ETD (electron transfer dissociation), on an orbitrap Fusion (Fig. 2a). Although in theory ETD is the best method for mapping labile PTMs, its lower sensitivity makes it less suited for detecting ADPr in complex mixtures. To circumvent this limitation we exploited the lability of ADP-ribose, triggering acquisition of high-resolution ETD spectra based on the presence of adenine ions in rapidly-acquired HCD spectra12. Another drawback of ETD is that its efficiency is charge-state dependent: fully digested tryptic peptides fragment poorly, being primarily doubly charged. This limitation is solved by partial FASP, which generates highly-charged peptides (Supplementary Fig. 2b) ideal for ETD analysis.
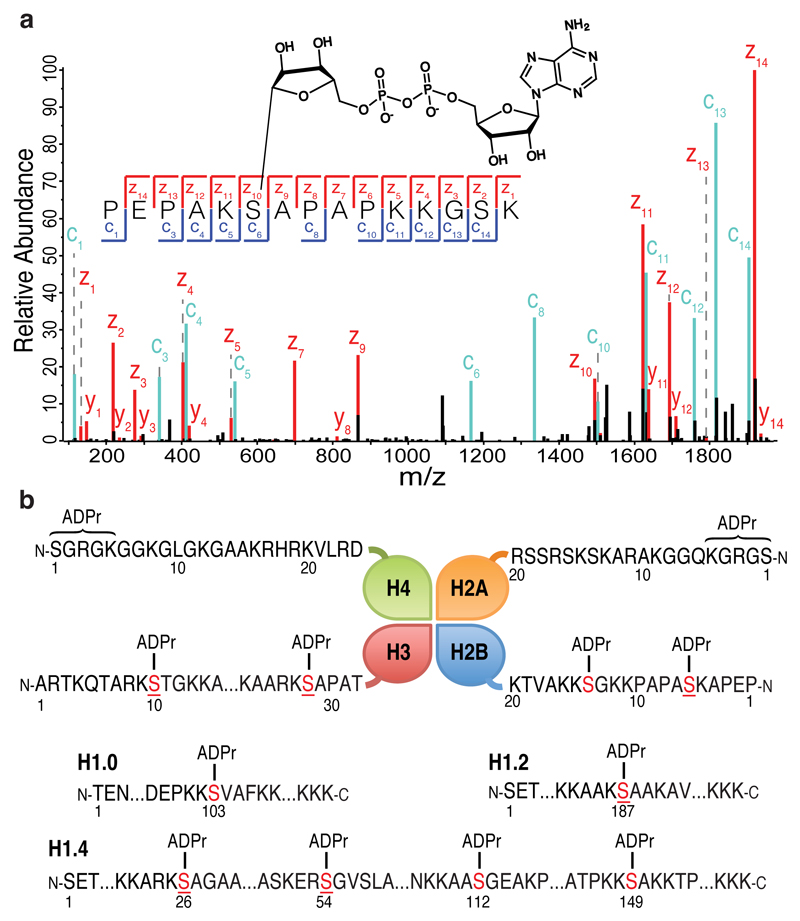
Figure 2. Identification of histone ADP-ribosylation marks.
(a) High-resolution ETD fragmentation spectra of an H2B peptide modified by ADP-ribose on Serine 6. The chemical structure of ADP-ribose is depicted. See Supplementary Fig. 7a for further details on the linkage between Serine and ADPr. (b) Schematic representation of core histones (top panel) and different H1 variants (bottom panel). The twelve novel unique ADPr marks are indicated. ADPr serine residues (in red) are located on the N-terminal tails of the four core histones. Underlined serines are reported phosphorylation sites.
All MS/MS data were processed with MaxQuant14, allowing ADPr on any chemically-reactive amino acid, with stringent criteria for peptide identification and PTM localization, followed by extensive manual validation of fragmentation spectra. We have identified twelve previously unknown, unique ADPr marks, six on the N-terminal tails of the four core histones and six on different H1 variants (Fig. 2b, Supplementary Table 1, Supplementary Note). Unexpectedly, we discovered ADPr on serine. For ten sites, high-resolution ETD spectra unambiguously pinpoint serine as the modified residue. Two modification sites near the N-termini of H2A and H4 have proven difficult to localize exactly; although some spectra suggest ADPr of Ser1, our data cannot rule out modification on Arg3 or Lys5. Spectra of longer histone peptides produced by partial FASP revealed the H2BS6 ADPr site on six distinct H2B variants. Interestingly, the sequences surrounding this H2B site are similar to the sequences of six other histone ADPr sites. Although we cannot perform rigorous sequence motif analysis on so few sites, a pattern appears to emerge with lysine before the modified serine, usually followed by alanine (Supplementary Fig. 6a).
A large number of aspartates, glutamates, arginines and lysines have been identified as ADPr acceptors on histones modified in vitro4, 12, 15. Since we did not observe these sites in our in vivo samples, we performed a series of experiments to ensure that our preparation is not biased towards serine. First, to be sure that acidic purification did not eliminate ADPr sites, we analyzed histones purified from cells without acid extraction, namely commercial HeLa histones and histones obtained by anti-FLAG purification. These experiments confirmed the modification of serine and did not conclusively identify ADPr on any other amino acids (Supplementary Fig. 6b). Although serine-linked ADPr has been described as acid labile at 37°C16, it survives our low-temperature acidic steps (Supplementary Fig. 7). Second, to ensure that the slightly alkaline pH of digestion did not introduce bias, we lowered the digestion pH to 7.39 but found no additional ADPr sites (Supplementary Fig 8). Finally, by performing in vitro ADPr reactions on histones, we found that our method can detect ADPr on amino acids other than serine (Supplementary Table 2). While these experiments do not exclude the existence of histone ADPr on other amino acids under different conditions, they only support the modification of serine in the in vivo samples examined here, even though other sites are detectable by our method.
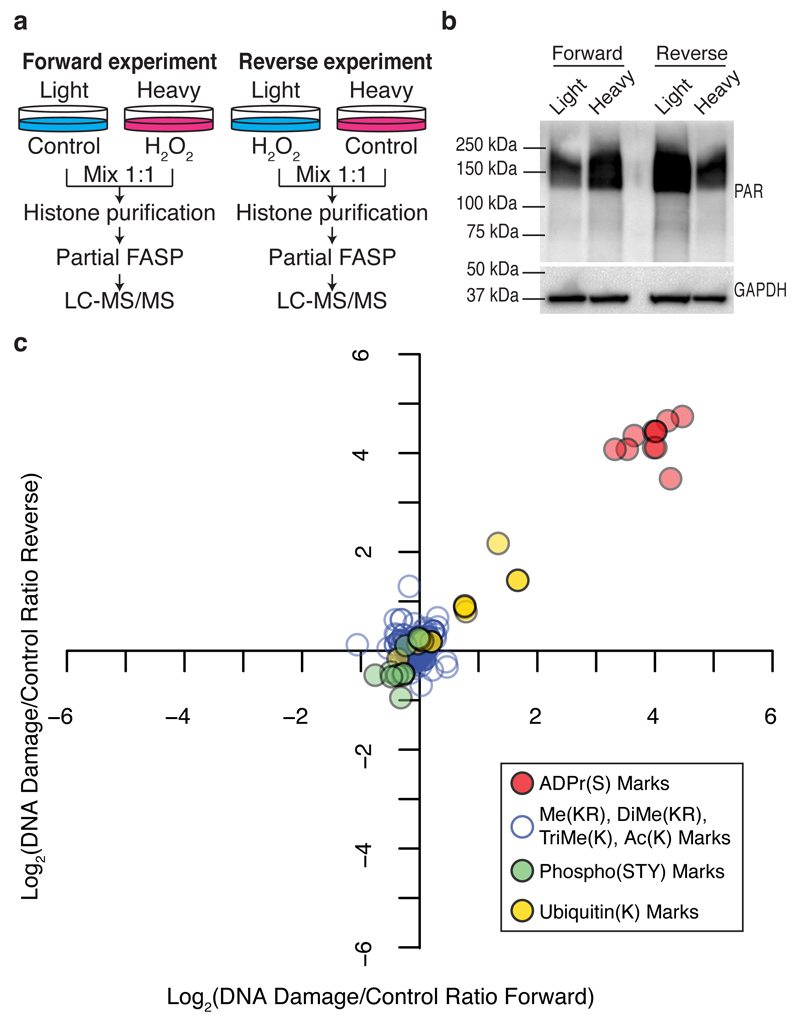
Histones have long been known to be acceptors of ADPr in response to DNA damage17. To investigate whether our newly identified serine ADPr marks respond to DNA damage, we performed quantitative proteomics using SILAC (stable isotope labeling by amino acids in cell culture)18. All the detected ADPr marks increased 10- to 27-fold in human osteosarcoma cells exposed to oxidative DNA damage by hydrogen peroxide (H2O2) for 10 minutes. Meanwhile, the canonical histone PTMs remained largely unchanged, except for two ubiquitination marks that increased slightly (Fig. 3, Supplementary Dataset). Histone phosphorylation marks, including gamma-H2AX, a known responder to DNA damage, did not change after only 10 minutes of exposure to H2O2, an early time point with maximum ADPr (Supplementary Fig. 9a). The suppression of DNA damage-induced serine ADPr in cells treated with the inhibitor olaparib suggests that this modification is enzymatic (Supplementary Fig. 9b-d).
Figure 3. Modulation of serine ADP-ribosylation marks on core histones upon DNA damage.
(a) Schematic representation of the SILAC-based strategy to quantify core histone marks upon 10 minutes of 2 mM H2O2-induced DNA damage. (b) Western blot analysis of total protein poly-ADP-ribosylation levels prior to mixing light and heavy lysates from each SILAC experiment. Anti-GAPDH was used as a loading control. See Supplementary Fig. 10 for original images. (c) Log transformed Heavy/Light SILAC ratios from the forward and Light/Heavy SILAC ratios from the reverse experiment were plotted against each other. Each point represents a unique histone mark.
Our study not only establishes serine ADPr as a novel histone mark but thereby identifies the first proteins modified by ADPr on serine. An early report based on indirect characterization of chemical stability of ADPr-protein bonds suggested serine as a possible acceptor of ADPr16. Here we provide the first direct evidence of proteins linked to ADP-ribose through the hydroxyl group of serine, a linkage chemically identical to the o-glycosidic bond linking ADP-ribose monomers in poly-ADP-ribose19 and the bond between carbohydrates and o-glycosylated proteins20 (Supplementary Fig. 7a). Six out of the twelve reported ADPr sites have also been reported to be phosphorylated (Fig. 2b), suggesting possible interplay between these two PTMs. ADPr on serines 10 and 28 of H3 is particularly interesting, since these residues undergo crucial phosphorylation during mitosis21.
Owing to the chemical instability of ADPr, studying this PTM at the molecular level is not trivial. We have developed an MS-based approach for mapping and quantifying biologically relevant histone ADPr marks in cells, and in the process identified serine ADPr as a new histone mark. Given the large number of proteins ADP-ribosylated after DNA damage8, we expect serine ADPr not to be restricted to histones. This work provides both a new direction of enquiry in the role of ADPr in cellular function, and a new tool with which to explore it.
Online Methods
Cell culture and SILAC labeling
Human U2OS osteosarcoma cells and HEK293T cells (293T) were acquired from ATCC. Chicken bursal lymphoma DT40 cells were a generous gift from Prof. William Earnshaw (University of Edinburgh). U2OS and 293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, penicillin/streptomycin (100U/ml) at 37 °C, 5% CO2. DT40 cells were cultured in RPMI 1640 Medium supplemented with 10% fetal bovine serum, 1% chicken serum, penicillin/streptomycin (100U/ml) at 37 °C, 5% CO2. All cell lines were regularly tested for Mycoplasma by PCR-based detection analysis and discarded if positive.
For SILAC labeling18, U2OS cells were grown in medium containing unlabeled L-lysine (#L8662, Sigma-Aldrich) as the light condition, or isotopically labeled L-lysine (13C6,15N2, # 608041, Sigma-Aldrich) as the heavy condition. Both light and heavy DMEM were supplemented with 10% dialyzed FBS (Thermo Scientific). Cells were cultured for more than 7 generations to achieve complete labeling. Incorporation efficiency (>99%) was determined by MS.
Histone purification
Histones were purified with a method that combines sulfuric acid extraction with ion exchange chromatography (modified from 7).
Briefly, cells were stimulated with 2 mM H2O2 for 10 minutes, washed twice with ice-cold PBS and lysed by rotation in 0.1 M H2SO4 at 4ºC for 2 hours. The lysate was centrifuged at 2200 g at 4ºC for 20 min. The pellet with non-soluble proteins and cell debris was discarded. Perchloric acid (PCA) was added to the supernatant to a final concentration of 4% v/v. After overnight incubation at 4ºC, this fraction was centrifuged at 21,000 g at 4ºC for 45 min. The supernatant containing PCA-soluble proteins was concentrated using 10 kDa cut-off centrifugal concentrators (Vivacon 500, Sartorius Stedim). The pellet (PCA-insoluble proteins) was washed subsequently with 4% PCA (2 x 1 ml), 0.2% HCl in acetone (2 x 1 ml) and acetone (2 x 1 ml). After drying, the pellet was resuspended in Binding Buffer (50 mM Tris-HCl pH: 8.0, 0.5 M NaCl, 2 mM EDTA, 1 mM DTT). For ion exchange chromatography, sulfopropyl(SP)-Sepharose resin (#S1799, Sigma-Aldrich) was packed into a column and pre-equilibrated with 10 volumes of Binding Buffer. The resuspended pellet containing PCA-insoluble proteins was passed through the column. The resin was washed with 10 volumes of Binding Buffer and 30 volumes of Washing Buffer (50 mM Tris-HCl pH 8.0, 0.6 M NaCl, 2 mM EDTA, 1 mM DTT). Proteins were eluted with Elution Buffer (50 mM Tris-HCl pH 8.0, 2 M NaCl, 2 mM EDTA, 1 mM DTT) in 10 fractions.
Eluted proteins were precipitated overnight in 4% (v/v) PCA at 4ºC. The fractions were then centrifuged at 21,000 g at 4ºC for 45 min and the resulting pellets were washed with 4% PCA (2 x 1 ml), 0.2% HCl in acetone (2 x 1 ml) and acetone (2 x 1 ml). This acidic precipitation appears to cause some degree of modification loss (Supplementary Fig. 7b).
To avoid this loss we also implemented a variation of the protocol for purifying the core histones without protein precipitation with PCA. Briefly, sulfuric acid-soluble proteins were neutralized with 1M Tris-HCl pH 8.0. NaCl, EDTA and DTT were added to a final concentration of 0.5 M, 2 mM, 1 mM, respectively and core histones were purified using SP-Sepharose resin as described above. Proteins eluted with Elution Buffer were concentrated using a C18 cartridge (hydrophobicity interaction; Sep-Pak SPE Waters). We would propose this approach as a viable lower-loss alternative for histone recovery.
SILAC experiments
Untreated cells and cells pretreated for 40 min with 2 μM olaparib (#10621, Cayman Chemical) were either stimulated with 2 mM H2O2 for 10 minutes or left unstimulated (Figure 3 and Supplementary Fig. 9). Cells were washed twice with ice-cold PBS and lysed by rotation in 0.1 M H2SO4 at 4ºC for 2 hours. Prior to centrifugation at 4ºC, aliquots from the light and heavy lysates were retained for Western blot analysis. Supernatants containing sulfuric acid-soluble fractions from light and heavy lysates were mixed 1:1 and histones were purified as described above. Each SILAC experiment was composed of at least two technical replicate analyses.
Immuno-slot blot and Western blot analysis
Immuno-slot blot analysis of PAR was assayed as previously described22. Briefly, Hybond N+ membranes (GE Healthcare) were prewashed with ultrapure water and samples were loaded using the Bio-Dot SF apparatus (Bio-Rad) following manufacturer instructions. Membranes were then washed once with ultrapure water and with 0.4 M NaOH and kept in water until further processing.
For Western blot analysis, samples were subjected to a standard SDS-PAGE method. Proteins were transferred to PVDF membranes (Merck Millipore) and kept in water until further processing.
Immuno-slot blot and Western blot membranes were blocked with TBS-T buffer (25 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.05% Tween 20 and 5% non-fat dried milk) and probed overnight with primary antibodies at 4 °C, followed by a one hour incubation with peroxidase-conjugated secondary antibodies (Amersham) at room-temperature. Heat-inactivated TBS-T buffer (1h at 60ºC) was used for Western blot analysis of phosphorylated histones. Blots were developed using ECL Select (Amersham) and signals were captured using a ChemiDoc MP System (Bio-Rad). Anti-poly-ADP-ribose polyclonal antibody was purchased from Trevigen (#4336-BPC-100, diluted at 1:2000). Anti-phospho-Histone H2A.X (Ser139) monoclonal antibody (clone JBW301) was purchased from Merck Millipore (#05-636-I, diluted at 1:500). Anti-phospho-Histone H3 (Ser10) polyclonal antibody and anti-phospho-Histone H3 (Ser28) polyclonal antibody were purchased from Cell Signaling Technology (#9701 and #9713 respectively, diluted at 1:1000). Anti-phospho-Histone H3.3 (Ser31) monoclonal antibody (clone EPR1873) was purchased from Abcam (#ab92628, diluted at 1:1000). Anti-GAPDH monoclonal antibody (clone 6C5) was purchased from Calbiochem (#CB1001, diluted at 1:2000). Loading controls (GAPDH) were run on the same blot.
For the analysis of histone phosphorylation marks (Supplementary Fig. 9a), U2OS cells were either stimulated with 2 mM H2O2 for the indicated time points or left unstimulated. Cells were washed twice with ice-cold PBS, lysed in Laemmli buffer and analyzed by Western blotting as described above.
Analysis of PAR stability
The original paper describing the loss of serine ADPr in acidic conditions16 shows that temperature is an important parameter for acid lability of serine ADPr. We have used PAR chains as a model conjugate to test our incubation conditions, since the attachment of ADPr to serine and the bond between ADP-ribose monomers in poly-ADP-ribose (PAR) are both acetal linkages (see Supplementary Fig. 7a). To investigate the extent of the potential loss of serine ADPr during our acid incubations, we monitored the degradation of PAR under different acidic conditions by anti-PAR slot blot (Supplementary Fig. 7b). 6 pmol of poly-ADP-ribose (PAR) standard (#ALX-202-043-C001, Enzo Biochem, Inc) were incubated in a final volume of 20 µl in: 200 mM Tris pH 7.3 for 1 hour at 4 ºC, or in 200 mM Tris pH: 7.3 for 1 hour at 37ºC, or in 0.1M H2SO4 for 2 hours at 4 ºC, or in 4% PCA for 16 hours at 4 ºC, or in 44% formic acid (FA) for 1 hour at 37 ºC, or in 44% FA for 1 hour at 4 ºC, or in 200 mM Tris pH: 7.3 in the presence of 1 µM human PARG for 20 minutes at 25ºC. After incubation, samples were neutralized with 80 µl of Tris (0.5g/ml). NaOH and EDTA were added to a final concentration of 0.4 M and 10 mM respectively. Samples were split into two different fractions containing 1/6 and 5/6 of the total volume and analyzed by immuno-slot blotting.
PARP1 activity assay
Automodification activity of PARP1 was assayed as previously described23. Briefly, PARP1 (#4668, Trevigen) was incubated for 20 minutes at room-temperature in reaction buffer (50 mM Tris-HCl pH 7.5, 50 mM NaCl, 1 mM MgCl2) containing 130 ng of activated DNA (#4671-096-06, Trevigen) in the presence or absence of 200 µM NAD (#4684-096-02, Trevigen) and in the presence or absence of 0.1 M H2SO4. Reactions were stopped with 2 µM Olaparib and analyzed by Western blotting.
PARP10 expression and in vitro ADP-ribosylation of histones
GST-PARP10cd was expressed from a pGEX-4T1 GST-PARP10cd (amino acids 818–1025) plasmid that was a kind gift from Bernhard Lüscher (RWTH Aachen University)24. Recombinant protein was purified from transformed Rosetta2 (DE) competent cells. Briefly, transformed bacteria were grown overnight in LB supplemented with 100 µg/ml of Ampicillin and 34 µg/ml chloramphenicol at 37°C. Overnight culture was diluted in 4 liters of media and grown at 37°C until the absorbance measured at 600 nm reached 0.8. Temperature was then cooled down to 18°C, bacteria induced with 0.2 mM IPTG and culture prolonged for 16 hours. Bacteria were then lysed using BugBuster protein extraction reagent (Novagen) and Benzonase (Sigma) in PBS buffer supplemented with 10% glycerol, 1mM DTT and Complete Protease Inhibitor (Roche). After a 1-hour incubation, the lysate was clarified by centrifugation and supernatant applied on glutathione sepharose beads (GE Healthcare). Beads were incubated with the lysate for 50 minutes at 4°C and then washed in 20 column volumes of lysis buffer. GST-tagged protein was eluted in lysis buffer supplemented with 20 mM reduced glutathione (Sigma, readjusted pH to 7.4). Fractions were then collected, assayed by SDS-PAGE and coomassie blue staining (Instant Blue, Expedeon). Best fractions were pooled and dialysed in 25 mM Tris-HCl pH 7.5, 150 mM NaCl, 10% glycerol, 1 mM DTT.
In vitro histone ADP-ribosylation was performed as described in Rosenthal et al12. Briefly, 600 µg of histones were incubated with 2 nmol of PARP10 for 15 min at 30°C. The reaction buffer consisted of 50 mM Tris-HCl pH 8.0, 4 mM MgCl2, 250 µM DTT and 160 µM NAD+. The reaction was stopped by the addition of 0.1 M H2SO4 and histones were purified again exactly as described above.
PARG activity assay
PARG activity was assayed as previously described23. Briefly, PARP1 was automodified as described above. The reaction was stopped with 2 µM Olaparib and split into equal volume fractions. The fractions were incubated for another 20 minutes at room temperature in the presence or absence of 1 µM human PARG and in the presence or absence of 0.1 M H2SO4. Reactions were stopped by the addition of Laemmli loading buffer and analyzed by Western blotting.
Analysis of protein poly-ADP-ribosylation during cell lysis
U2OS cells were washed twice with ice-cold PBS, harvested and split into equal volume fractions. Half of the fractions were lysed in modified RIPA buffer (50 mM Tris-HCl pH 7.5, 400 mM NaCl, 1 mM EDTA, 1% NP-40, 0.1% Na-deoxycholate) supplemented with protease inhibitor cocktail (#04693116001, Roche), 2 mM Na-orthovanadate, 5 mM NaF, 5 mM Glycero-2-phosphate and 1 µM ADP-HPD (#118415, Calbiochem) for 2 hours in the presence or absence of 2 μM olaparib and 10 mM 3-aminobenzamide (3-ABA, #165350, Calbiochem). The remaining fractions were lysed in 0.1 M H2SO4 in the presence or absence of 2 μM olaparib and 10 mM 3-aminobenzamide for 2 hours. All the fractions were analyzed by Western blotting (Supplementary Fig. 4d).
Jungmichel et al. (Mol. Cell 2013; Fig S2C) showed that mechanical post-lysis shearing of DNA triggers PARP-1 activity leads to an artificially high level of PARylation. Supplementary Fig. 4d demonstrates that acid extraction prevents this undesirable post-lysis increase in PARylation and therefore preserves the endogenous level of PAR, which is low without induction of DNA damage. With non-denaturing lysis buffers, such as the modified RIPA buffer used by Jungmichel et al., this phenomenon can be prevented by the addition of PARP-1 inhibitors, although other enzymes, such as PARG, cannot be easily inactivated. The important advantage of our method is that virtually all the conjugation/deconjugation enzymes are inactive during lysis. This is crucial to prevent both non-physiological (but enzymatic) ADPr and non-enzymatic ADPr. With acid extraction PARG is prevented from generating free ADPr via post-lysis degradation of PAR.
Protein digestion
Pelleted histones (~50 μg) were resuspended in 200 μl 8 M urea in 0.1 M Tris-HCl pH 8.0, 10 mM tris(2-carboxyethyl)phosphine (TCEP), 20 mM chloroacetamide and transferred to 10 kDa cut-off Vivacon® 500 flat filters (Sartorius Stedim), unless otherwise stated. Samples were centrifuged at 14,000g at 20 ºC for 20 min, followed by three washes with 200 μl of 50 mM ammonium bicarbonate (ABC). Digestions were performed with trypsin Gold (Promega) on the filters in 50 μl 50 mM ABC.
For 'Partial FASP’ digestion, 1:2000 trypsin to protein ratio was used for 20 minutes at 20 ºC. The digestion was stopped by the addition of formic acid to lower the pH below 3. Peptides were collected by centrifugation at 14,000g at 4 ºC for 10 min, followed by a wash with 50 μl 50 mM ABC.
The retentate containing undigested proteins was further digested in 50 μl 50 mM ABC overnight at 37 ºC, with 1:50 trypsin to protein ratio.
For the ‘FASP’ digestion, 1:50 trypsin to protein ratio was used and the digestion was performed in 50 mM ABC overnight at 37 ºC, as described in the original protocol6.
Variations of the partial digestion method
H1-containing fractions were partially digested in a more ‘continuous’ way: trypsin was added in 50 μl 50 mM ABC to 1:2000 ratio to the filters and peptide elution was started immediately by centrifugation at 14,000g at 20 ºC for 10 min. After that, 50 μl 50 mM ABC was added to the filter and centrifuged for another 10 min. This step was repeated once more. The eluted peptides were dried down in Speedvac concentrator. The retentate was further digested in 50 μl 50mM ABC for 1 hr at 20 ºC, with 1:50 trypsin: protein ratio.
To generate even longer peptides using the principle of the Partial FASP, we also digested histones in a progressive mode using the Amicon Pro system (Millipore) with a V-shaped 10-kDa cut-off filter. 100 μg of histones were diluted with 200 μl 8 M urea in 0.1M Tris-HCl pH 8.0, 10 mM tris(2-carboxyethyl)phosphine (TCEP), 20 mM chloroacetamide and transferred into the V-shaped filter. From the upper reservoir, a highly-diluted trypsin solution (2 μg trypsin in 5 ml 1 M urea, 50 mM ABC) was slowly introduced into the histone-containing filter device during 40 minutes of centrifugation at 4,000 rpm in a swing-bucket centrifuge. The steady flow of buffer removed early cleavage products (containing multiple miscleavages) from further tryptic digestion as soon as they were short enough to pass through the 10-kDa cut-off filter. The retained mixture of longer partial digest products was thus subject to a steadily increasing concentration of trypsin to facilitate cleavage at less accessible sites. The flow-through was acidified with formic acid until the pH was below 3 and desalted on C18 cartridges (3M Empore).
In all cases eluted peptides were dried down in Speedvac concentrator and resuspended in 0.1% formic acid (FA) prior to LC-MS/MS analysis.
Digestion at pH 7.3
In order to test whether we lose ADPr linked to serine or other residues due to basic pH, we performed partial FASP digestion at a lower pH, at 7.3 9. Purified endogenous histones were denatured in 8M Urea pH 7.3 and then digested in Tris-HCl pH 7.3. A comparison of peptide intensities from this experiment to one digested at pH 8 (Supplementary Fig. 8a) revealed no negative effect of using pH 8 on the analysis of serine ADPr, nor did we detect new sites when the digestion pH was lowered to 7.3.
Next, to thoroughly address the effect of the digestion pH on glutamate and aspartate ADPr, we performed pH 7.3 partial FASP on histones ADP-ribosylated in vitro by PARP1024. Upon comparison to the rest of the peptide population, ADPr on arginine and acidic residues appears to be only marginally favored by pH 7.3 (Supplementary Fig. 8b).
Commercial HeLa core histones were purchased from Cayman Chemicals and digested the same way as described above (Partial FASP). According to the manufacturer, these histones are isolated via hydroxyapatite chromatography from HeLa S3 nuclear pellet, and the purification method does not involve acid extraction.
FLAG-PARP1 immunoprecipitation
HEK293T cells were transfected with a pLenti-CMV construct expressing FLAG-PARP1. Transient DNA transfection was performed with Fugene6 (Promega), according to the manufacturer’s instructions.
Cells were lysed in TX100 buffer (50 mM Tris-HCl pH 8, 100 mM NaCl, 1% Triton X-100, 1 mM DTT, protease and phosphatase inhibitors, 1 μM Olaparib, 1 μM PARG inhibitor (ADP-HDP) and Benzonase nuclease (Sigma) by rotation at 4 ºC for 15 min and centrifuged at 14,000 g for 10 min at 4 ºC. Lysates were added to FLAG-M2 affinity gel and incubated for 25 min whilst rotating at 4 ºC. The gel was then washed several times with lysis buffer without any Triton X-100. Proteins were eluted using 100 μg/ml 3X FLAG peptide (Sigma) in TBS. Immunoprecipitated complexes (including histones) were digested with the Partial FASP method and analyzed by mass spectrometry, as described.
Hydrolysis of ADP-ribosylated peptides by Nudix16
In order to render poly-ADP-ribosylation sites amenable to MS analysis without the risk of non-enzymatic ADPr due to high levels of free ADP-ribose, peptides were processed with Nudix 16 hydrolase, which converts both ADPr and poly-ADPr to phosphoribose and free AMP, which cannot lead to re-elongation2. Peptides were resuspended in 50 μl reaction buffer (50 mM Tris-HCl pH 8.0, 15 mM MgCl2, 50 mM NaCl, 0.1 mM TCEP) and incubated with 2 mM recombinant Nudix1610, 11 (purified as in Palazzo et al, 2015) at 30 ºC overnight. Peptides were then desalted on either C18 cartridges (3M Empore) or using in-house manufactured StageTips25, depending on the peptide amounts. Eluted peptides were dried down in Speedvac concentrator and either resuspended in 40% acetonitrile (ACN), 0.1% FA prior to phosphoenrichment, or in 0.1% FA for direct analysis by LC-MS/MS.
Enrichment of ADPr/or phosphoribosylated peptides with immobilized metal affinity chromatography (IMAC)
The enrichment was performed as in Daniels et al., 20149, with some modifications. Peptides were resuspended in 40% ACN, 0.1% FA (binding buffer) and incubated with 10 μl PHOS-select beads (Sigma) for 1 hour, with end-to-end rotation at 25 ºC. The beads were then spun down and the supernatant was further incubated with 10 μl of beads. The bead-bound peptides were washed once with binding buffer and transferred to a pre-equilibrated StageTip. There the beads were washed twice with binding buffer and acidified with 1% FA. Peptides were eluted onto the StageTip with 0.5 M potassium phosphate pH 7, acidified again with 1 % FA and washed with 0.1% FA. They were eluted with 40% ACN, 0.1% FA and dried down in the SpeedVac concentrator.
LC-MS/MS analysis
Liquid chromatography for all LC-MS/MS runs was performed on an EASY-nLC 1000 Liquid Chromatography system (Thermo Scientific) coupled to the spectrometers via modified NanoFlex sources (Thermo scientific). Peptides were loaded onto 250-mm x 75-μm PicoFrit (C18 2 μm medium) analytical columns (New Objective) at a maximum pressure of 800 bar. Solutions A and B for the UPLCs were 0.1% formic acid in water and acetonitrile, respectively. Samples were loaded in 0.1% formic acid in water to maximize retention of highly hydrophilic peptides. Gradients varied slightly in length (90 to 150 min) and mixture, and may be extracted from the respective raw files. In general they incorporated a linear gradient from very low or zero %B to 20 or 30% for 65-100 minutes, followed by a steeper phase and a wash. This length of gradient was maintained despite the relative simplicity of the protein mixture in order to improve the resolution and identification of as many modified peptide forms as possible, including those of low abundance.
HCD (Higher-energy Collision-induced Dissociation) acquisitions
Pure HCD datasets were acquired on a Q Exactive Plus mass spectrometer (Thermo Scientific). Optimal data acquisition for our low-complexity samples was achieved by increasing the sensitivity of the analysis26. MS1 spectra were acquired in the 300-1800 m/z scan range with a resolution of 120,000. AGC targets were set to 3,000,000 ions; maximum injection time was 100 ms. Up to 5 data-dependent MS2 spectra were acquired at 60,000 resolution. AGC target for MS2 was set to 1,000,000 ions. In order to reach this target, long MS2 injection times were allowed (500 ms). Unassigned or singly charged ions were rejected and the dynamic exclusion option was enabled (duration: 20 s).
For the quantification of very low abundance ADP-ribosylated peptides, a similar method with a smaller scan range (400-900 m/z) and one with an inclusion list (based on previous runs) were also used.
ETD (Electron Transfer Dissociation) acquisitions
Pure ETD and mixed HCD/ETD datasets were acquired on an Orbitrap Fusion instrument (Thermo Scientific). Our methods for targeted acquisition of ETD spectra of modified precursors fell into two essential types – offline and online. In the offline method, in-house scripts were used to generate a precursor inclusion list from previous HCD data based either directly on the presence of one or more diagnostic peaks (i.e. adenine - 136.062 Da) in a precursor’s fragmentation spectrum in the raw data or on the subsequent identification of the precursor as a modified peptide (with poor or no localization information). These selected precursors were then fragmented by ETD in a targeted fashion upon detection. The method also allowed for ETD fragmentation of additional precursors in the absence of listed targets.
The online method used the product ion trigger feature in the decision tree of a TopSpeed acquisition method. As they eluted, multiply-charged precursors were rapidly fragmented in HCD mode (low injection time: 30 ms; resolution 15,000; AGC target: 50,000) to screen a maximum number of precursors for diagnostic Adenine ions (typically among the strongest fragment ion signals). Upon detection of an intense diagnostic peak, the respective precursor was immediately isolated again and subjected to ETD12. Since the entire cycle time was held to a maximum of 3 seconds, the entire process from MS1, through screening, to ETD fragmentation took place well within the width of a chromatographic peak. The maximum MS2 injection time and AGC for ETD were set to 1000 ms and 500,000 ions in order to achieve single amino-acid resolution for particularly difficult precursors. Resolution was set to 30,000.
Data analysis
Raw files were analyzed with MaxQuant proteomics suite of algorithms (version 1.5.0.30)14, integrated with the search engine Andromeda27 (available from www.maxquant.org). The data were searched against databases containing human and chicken histone sequences with the following parameters. The initial maximum allowed mass deviation was set to 7 ppm for precursor ions; the minimum peptide length was set to 6 amino acids and the maximum number of missed cleavages was set to 8 with the maximum charge state 10. Variable modifications included oxidation (M), acetylation (Protein N-term and K), phosphorylation (STY), methylation (KR), dimethylation (K), ADP-ribosylation (DEKRSTYCMNQHM) or phosphoribose (DEKRSYTCMNQHM). The variable modification ADP-ribosylation allowed for neutral losses of adenine (m/z 136.0618); adenosine with loss of water (m/z 250.0935); AMP (m/z 348.0704); ADP (m/z 428.0367) and ADP-ribose (m/z 542.0684)28. FTMS top peaks per 100 Da were set to 20. For confident identification of ADP-ribosylation sites, we considered only ETD MS/MS spectra and required a minimum Andromeda score of 100, mass deviation smaller than 3 ppm after MaxQuant recalibration and a localization score above 0.9. In addition, we manually validated all the representative spectra by requiring extensive coverage of the peptide backbone fragment ions. For localization we required the clear presence of multiple high-intensity fragment ions pinpointing the modification site. Contrary to the HCD fragmentation, the presence ADP-ribose specific diagnostic ions, which cannot be detected in the ETD mode, was not considered as a criterion for validation of spectra.
For SILAC experiments, multiplicity was set to 2, with Lys8 as the Heavy Label. Max. labeled AAs were set to 7. Max missed cleavages were set to 6, max charge was 7. The minimum peptide length was set to 6 amino acids. Variable modifications included acetylation (Protein N-term and K), phosphorylation (STY), methylation (KR), di-methylation (KR), Tri-Methylation (K), ubiquitination (on K, both glygly and LRGG modification) ADP-ribosylation (S). The ‘Re-Quantify’ option was enabled.
For comparison of the FASP and Partial FASP methods, evidence and peptide tables from MaxQuant were combined to create a single evidence file per unique peptide (modified and unmodified), using an in-house script. This was further analyzed by the Perseus software (http://www.perseus-framework.org) to create the peptide intensity/density plots and perform statistical analyses by calculating correlation coefficients, or in Microsoft Excel to visualize the distribution of peptide mass. For charge state comparison, data were taken from the RawMeat software (Vast Scientific) and visualized in Microsoft Excel. Bar plots representing the peptides identified from a FASP or Partial FASP triplicate experiment were visualized using an in-house script.
Supplementary Material
Acknowledgements
Many thanks to Alisa Zhiteneva, Prof. William Earnshaw (University of Edinburgh) and Dr. Michael Tatham (University of Dundee) for comments on the manuscript. This work is funded by Deutsche Forschungsgemeinschaft (Cellular Stress Responses in Aging-Associated Diseases) [grant number EXC 229 (to I.M.)]. The work in the I.A. laboratory is funded by the Wellcome Trust (grant number 101794) and the European Research Council (grant number 281739). The research leading to these results has received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Sklodowska-Curie grant agreement No 657501.
Footnotes
Author contributions
I.M., O.L. and J.J.B. designed research. O.L. and J.J.B. performed experiments, acquired and analyzed MS data. T.C. optimized MS methods and contributed to acquisition and analysis of MS data. Q.Z. performed histone purification and digestion experiments. I. At. performed bioinformatics analyses. A.S. performed immuno-slot blot experiments. L.P. and R.Z. performed protein purification and in vitro assays. I. Ah. and R.Z. contributed to the histone purification experiments and supporting studies. I.M. analyzed data and wrote the manuscript.
Competing Financial Interests Statement
The authors declare no competing financial interests.
References
- 1.Feng FY, de Bono JS, Rubin MA, Knudsen KE. Chromatin to Clinic: The Molecular Rationale for PARP1 Inhibitor Function. Mol Cell. 2015;58:925–934. doi: 10.1016/j.molcel.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Daniels CM, Ong SE, Leung AK. The Promise of Proteomics for the Study of ADP-Ribosylation. Mol Cell. 2015;58:911–924. doi: 10.1016/j.molcel.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hottiger MO. Nuclear ADP-Ribosylation and Its Role in Chromatin Plasticity, Cell Differentiation, and Epigenetics. Annu Rev Biochem. 2015;84:227–263. doi: 10.1146/annurev-biochem-060614-034506. [DOI] [PubMed] [Google Scholar]
- 4.Huang H, Lin S, Garcia BA, Zhao Y. Quantitative proteomic analysis of histone modifications. Chem Rev. 2015;115:2376–2418. doi: 10.1021/cr500491u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tan MJ, et al. Identification of 67 Histone Marks and Histone Lysine Crotonylation as a New Type of Histone Modification. Cell. 2011;146:1015–1027. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 7.Rodriguez-Collazo P, Leuba SH, Zlatanova J. Robust methods for purification of histones from cultured mammalian cells with the preservation of their native modifications. Nucleic Acids Res. 2009;37 doi: 10.1093/nar/gkp273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jungmichel S, et al. Proteome-wide identification of poly(ADP-Ribosyl)ation targets in different genotoxic stress responses. Mol Cell. 2013;52:272–285. doi: 10.1016/j.molcel.2013.08.026. [DOI] [PubMed] [Google Scholar]
- 9.Daniels CM, Ong SE, Leung AK. Phosphoproteomic approach to characterize protein mono- and poly(ADP-ribosyl)ation sites from cells. J Proteome Res. 2014;13:3510–3522. doi: 10.1021/pr401032q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palazzo L, et al. Processing of protein ADP-ribosylation by Nudix hydrolases. Biochem J. 2015;468:293–301. doi: 10.1042/BJ20141554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daniels CM, Thirawatananond P, Ong SE, Gabelli SB, Leung AK. Nudix hydrolases degrade protein-conjugated ADP-ribose. Sci Rep. 2015;5:18271. doi: 10.1038/srep18271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenthal F, Nanni P, Barkow-Oesterreicher S, Hottiger MO. Optimization of LTQ-Orbitrap Mass Spectrometer Parameters for the Identification of ADP-Ribosylation Sites. J Proteome Res. 2015;14:4072–4079. doi: 10.1021/acs.jproteome.5b00432. [DOI] [PubMed] [Google Scholar]
- 13.Steentoft C, et al. Mining the O-glycoproteome using zinc-finger nuclease-glycoengineered SimpleCell lines. Nature Methods. 2011;8:977–982. doi: 10.1038/nmeth.1731. [DOI] [PubMed] [Google Scholar]
- 14.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 15.Messner S, et al. PARP1 ADP-ribosylates lysine residues of the core histone tails. Nucleic Acids Res. 2010;38:6350–6362. doi: 10.1093/nar/gkq463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cervantes-Laurean D, Loflin PT, Minter DE, Jacobson EL, Jacobson MK. Protein modification by ADP-ribose via acid-labile linkages. J Biol Chem. 1995;270:7929–7936. doi: 10.1074/jbc.270.14.7929. [DOI] [PubMed] [Google Scholar]
- 17.Messner S, Hottiger MO. Histone ADP-ribosylation in DNA repair, replication and transcription. Trends Cell Biol. 2011;21:534–542. doi: 10.1016/j.tcb.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 18.Ong SE, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Molecular & Cellular Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 19.D'Amours D, Desnoyers S, D'Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J. 1999;342(Pt 2):249–268. [PMC free article] [PubMed] [Google Scholar]
- 20.Moremen KW, Tiemeyer M, Nairn AV. Vertebrate protein glycosylation: diversity, synthesis and function. Nat Rev Mol Cell Biol. 2012;13:448–462. doi: 10.1038/nrm3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang F, Higgins JM. Histone modifications and mitosis: countermarks, landmarks, and bookmarks. Trends Cell Biol. 2013;23:175–184. doi: 10.1016/j.tcb.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 22.Affar EB, et al. Immunodot blot method for the detection of poly(ADP-ribose) synthesized in vitro and in vivo. Anal Biochem. 1998;259:280–283. doi: 10.1006/abio.1998.2664. [DOI] [PubMed] [Google Scholar]
- 23.Slade D, et al. The structure and catalytic mechanism of a poly(ADP-ribose) glycohydrolase. Nature. 2011;477:616–620. doi: 10.1038/nature10404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kleine H, et al. Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol Cell. 2008;32:57–69. doi: 10.1016/j.molcel.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 25.Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem. 2003;75:663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 26.Kelstrup CD, Young C, Lavallee R, Nielsen ML, Olsen JV. Optimized fast and sensitive acquisition methods for shotgun proteomics on a quadrupole orbitrap mass spectrometer. J Proteome Res. 2012;11:3487–3497. doi: 10.1021/pr3000249. [DOI] [PubMed] [Google Scholar]
- 27.Cox J, et al. Andromeda: A Peptide Search Engine Integrated into the MaxQuant Environment. Journal of Proteome Research. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 28.Hengel SM, Goodlett DR. A review of tandem mass spectrometry characterization of adenosine diphosphate-ribosylated peptides. International Journal of Mass Spectrometry. 2012;312:114–121. doi: 10.1016/j.ijms.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.