Abstract
Class‐switched memory B cells are key components of the “reactive” humoral immunity, which ensures a fast and massive secretion of high‐affinity antigen‐specific antibodies upon antigenic challenge. In humans, IgA class‐switched (IgA+) memory B cells and IgA antibodies are abundant in the blood. Although circulating IgA+ memory B cells and their corresponding secreted immunoglobulins likely possess major protective and/or regulatory immune roles, little is known about their specificity and function. Here, we show that IgA+ and IgG+ memory B‐cell antibodies cloned from the same healthy humans share common immunoglobulin gene features. IgA and IgG memory antibodies have comparable lack of reactivity to vaccines, common mucosa‐tropic viruses and commensal bacteria. However, the IgA+ memory B‐cell compartment contains fewer polyreactive clones and importantly, only rare self‐reactive clones compared to IgG+ memory B cells. Self‐reactivity of IgAs is acquired following B‐cell affinity maturation but not antibody class switching. Together, our data suggest the existence of different regulatory mechanisms for removing autoreactive clones from the IgG+ and IgA+ memory B‐cell repertoires, and/or different maturation pathways potentially reflecting the distinct nature and localization of the cognate antigens recognized by individual B‐cell populations.
Keywords: Autoreactivity, IgA Antibodies, Immunoglobulin genes, Memory B cells, Polyreactivity
Introduction
Activation of naïve B cells during T‐dependent immune responses induces the formation of germinal centers (GCs). In GCs, B cells undergo both class switching, which alters antibody effector functions, and somatic hypermutation, which has the potential to increase antibody affinity 1. Following antigen‐mediated selection in the GC, B cells differentiate into either memory B cells, which can react quickly to recurrent antigenic challenge, or plasma cells, that produce large quantities of antibody 1. GC‐derived memory B cells have different functional properties according to the immunoglobulin (Ig) isotype they express following Ig class switching 2, which can be geographically determined as for the induction of IgA in the mucosal immune system 3, 4. IgA antibodies primarily ensure immune protection of mucosal surfaces against invading pathogens, but also circulate and are present in large quantities in blood 5. Indeed, IgA is the most prevalent isotype of antibodies circulating in human blood after IgG. Unliganded “natural” IgA antibodies circulating in human serum are believed to exert anti‐inflammatory activities, whereas specific IgAs opsonizing pathogens or forming immune complexes would trigger immune responses by activating myeloid cells 6. In this regard, although individuals with selective IgA deficiency are usually asymptomatic, they are prone to recurrent mucosal infections but also frequently present allergic and autoimmune manifestations 7.
Most nascent B cells in the bone marrow express poly‐ and auto‐reactive B‐cell antigen receptors (BCRs), but these potentially deleterious clones are counterselected at two major tolerance checkpoints during B‐cell development in humans 8. In contrast, defective tolerance checkpoints resulting in a “leakage” of autoreactive B cells accumulating in the periphery have been documented in patients with autoimmune diseases such as systemic lupus erythematosus 9, 10, rheumatoid arthritis 11, 12, and type 1 diabetes 13, 14. Importantly, B cells can naturally re‐acquire poly‐ and self‐reactivity during the GC reaction under physiologic conditions. Nearly 40% of post‐GC IgG‐expressing memory B cells are self‐reactive, the majority of these autoreactive clones being generated as a result of antigen‐driven affinity maturation 15. Human IgA antibodies have also been shown to be polyreactive, with 25% of intestinal IgA‐expressing plasmablasts that exhibit polyreactive binding to self and foreign antigens, including commensal bacteria and rotavirus 16. However, it is still unknown whether circulating blood IgA+ memory B‐cell antibodies resemble IgG+ memory B‐cell antibodies at a molecular level, and whether the affinity maturation of IgA+ memory B‐cell antibodies can create poly‐ and self‐reactivity in healthy individuals.
In this study, we characterized the gene repertoire and reactivity of 251 recombinant monoclonal antibodies cloned from blood IgA+ memory B cells of healthy humans. Our results show that IgA+ and IgG+ antibodies cloned from circulating memory B cells exhibited very conserved immunoglobulin gene features with few subtle variations. In addition, both memory B‐cell populations showed comparable lack of reactivity to vaccines, common mucosa‐tropic viruses and commensal bacteria. However, the frequency of polyreactive clones was decreased in the IgA+ compared to the IgG+ memory B‐cell compartment. More strikingly, the frequency of self‐reactive antibodies was significantly lower for IgA+ compared to IgG+ memory B cells. Finally, we demonstrated that the self‐reactivity of IgAs was acquired following B‐cell affinity maturation, as formerly shown for IgG. Thus, although IgG+ and IgA+ memory B cells had very similar antibody gene repertoires, they differed in their reactivity to self‐antigens, suggesting different tolerance or regulatory mechanisms and/or reflecting alternative maturation pathways for their cognate antigens.
Results
Gene features of blood IgA+ memory B‐cell antibodies
To characterize the IgA antibody repertoire and reactivity of circulating blood memory B cells, we isolated single CD19+CD27+IgA+ B lymphocytes from the PBMCs of four healthy donors (Supporting Information Fig. 1, hd1 to hd4), and amplified and cloned their heavy‐ and light‐chain variable domain (IgH and IgL, respectively) genes. On the 297 unique IgH/IgL‐paired sequences analyzed (Supporting Information Table 1), we found that about two‐third of B cells expressed the IgA1 subclass (65% IgA1 vs 35% IgA2) (Fig. 1A). IgH and IgL gene features were compared with the data obtained from the single CD19+CD27+IgG+ B‐cell antibodies retrieved from donors hd2 and hd4, but also with historical data previously obtained from blood IgG memory B cells 15, 17 (Fig. 1 and Supporting Information Fig. 2). Comparative analyses of IgH and IgL variable (V) and joining (J) gene usage, and length, hydrophobicity, number of positive charges of the complementary determining region 3 (CDRH3) mostly showed conservation of gene features between IgA+ and IgG+ memory B‐cell antibodies (Fig. 1 and Supporting Information Fig. 2). Nevertheless, some significant differences were observed between the two B‐cell compartments. Compared to IgG+, IgA+ memory antibodies showed decreased frequency of VH1(DH)JH3 and VH4(DH)JH5 rearrangements (p = 0.01 and p = 0.03, respectively) (Fig. 1C). Moreover, we found an increased frequency of Jκ1 and Jκ2 gene segments for IgA+ and IgG+ B‐cell antibodies, respectively (Fig. 1G and Supporting Information Fig. 2). Finally, IgA+ memory antibodies less frequently combined VH1‐expressing IgH with Vκ3‐expressing IgL (3.4% IgA vs 11.4% IgG, p = 0.04) (Supporting Information Fig. 1D). Similarly to their IgG counterparts, blood IgA memory antibodies displayed high levels of somatic mutations in IgH variable genes (19.9±0.54 for IgA versus 19.2±0.97 for IgG, p = 0.56), independently of the IgA subclass (20.1±0.66 for IgA1 and 19.8±0.86 for IgA2) (Fig. 1E, 1I and Supporting Information Fig. 2C).
Figure 1.
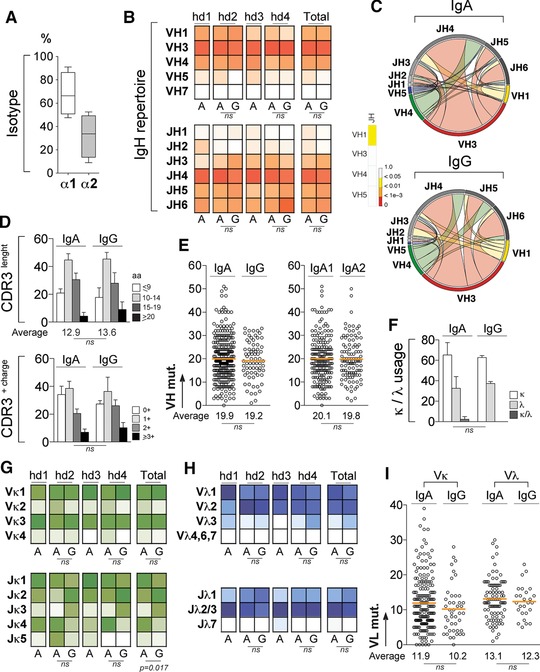
Immunoglobulin gene repertoire of IgA+ memory B‐cell antibodies. Single CD19+CD27+IgA+ and CD19+CD27+IgG+ B cells from PBMCs of healthy donors were FACS sorted and their heavy‐ and light‐chain variable domains (IgH and IgL) amplified and sequenced. All immunoglobulin gene characteristics were determined by analyzing IgH and IgL sequences of 297 single IgA+ B cells isolated from four individuals (n = 58 for hd1, n = 56 for hd2, n = 84 for hd3, and n = 99 for hd4) and 70 single IgG+ B cells from two of them (n = 31 for hd2 and n = 39 for hd4) using IgBLAST; (http://www.ncbi.nlm.nih.gov/igblast) and IMGT® (http://www.imgt.org) online tools. (A) Box plot showing the frequency of IgA antibodies expressing IgA1 (α1) or IgA2 (α2) subclass as determined using Blast® alignment tool (NIH) and Fcα1 and Fcα2 gene sequences as reference (IMGT®). (B) Heat maps comparing the VH and JH gene usages between IgA+ and IgG+ memory B‐cell antibodies. Color gradient is proportional to the frequency for gene usages with darker colors indicating high frequencies while white corresponds to a frequency of 0. Groups were compared using 2 × 5 Fisher's Exact test. ns, not significant. (C) Circos plots generated from the immunoglobulin gene analysis using “circlize” (v0.3.1) R package compare the frequency of VH(DH)JH rearrangements between IgA+ and IgG+ memory B‐cell antibodies. Heat bar shows the results of the 2 × 5 Fisher's Exact test used to compare both groups. (D) Bar graphs comparing the CDRH3 aminoacid length and number of positive charges in the CDRH3 between IgA+ and IgG+ memory B‐cell antibodies. Groups were compared using 2 × 5 Fisher's Exact test. ns, not significant. (E) Comparison of the number of mutations in VH genes (VH mut.) between IgA+ and IgG+ memory B‐cell antibodies, and between IgA1 and IgA2 subclasses. The average number of mutations is indicated below each dot plot. Groups were compared using Student's t‐test with Welch's correction. ns, not significant. (F) Bar graphs comparing the frequency of Igκ− and/or Igλ−expressing antibodies between IgA+ and IgG+ memory B‐cell compartments. Groups were compared using 2 × 5 Fisher's Exact test. ns, not significant. (G) Same as in (B) but for Vκ and Jκ gene usages. (H) Same as in (B) but for Vλ and Jλ gene usages (I) Same as in (E) but for IgA Vκ and Vλ genes.
IgA reactivity against vaccines, viral pathogens, and commensal bacteria
The nature and the spectrum of the antigens recognized by blood IgA+ memory B cells in humans remain mostly unexplored. To identify the antigen specificities of IgA+ memory B‐cell antibodies, we produced 251 recombinant IgA monoclonals from the four healthy donors, and 61 IgG monoclonals from two of them as control. All antibodies were expressed with a human IgG1 backbone, and tested for ELISA binding to a broad range of antigens including a panel of vaccines, viruses, and commensal bacteria. First, we evaluated the reactivity of both IgA and IgG memory antibodies against influenza virus (Flu), the trivalent diphtheria‐tetanus‐polio (DTP), trivalent measles‐mumps‐rubella (MMR), and hepatitis B virus (HBV) vaccines. Although, all donors were vaccinated against DTP and HBV, and two of them against MMR (Supporting Information Fig. 1A), none of the IgA or IgG antibodies specifically recognized the selected vaccines (Fig. 2A). We next assayed whether IgA memory antibodies could target common mucosal‐tropic pathogenic viruses such as adenovirus type 5, rotavirus, and rhinovirus type 1A. For comparison, we also examined their binding to the HIV‐1 envelope antigen used as negative control (all donors were HIV‐seronegative individuals). The ELISA binding experiments showed that no IgA and IgG antibodies reacted against the tested viral antigens (Fig. 2B). Finally, we tested the ELISA reactivity of the recombinant antibodies produced from IgA+ and IgG+ memory B cells against a panel of selected gut commensal bacteria; Escherichia coli, Enterobacter cloacae, Enterococcus faecalis, and Morganella morganii. Binding experiments did not allow identification of high affinity antibodies recognizing specifically a given commensal strain in any of two B‐cell memory compartments, except the 3–187 IgA antibody that bound exclusively to E. coli antigens (Fig. 2C). In fact, as observed for vaccine and virus antigens, most of the IgG and IgA antibodies exhibited a complete lack of reactivity with the exception of few IgG/IgA antibodies that reacted with all tested antigens most likely due to a polyreactive‐type of binding (Fig. 2).
Figure 2.
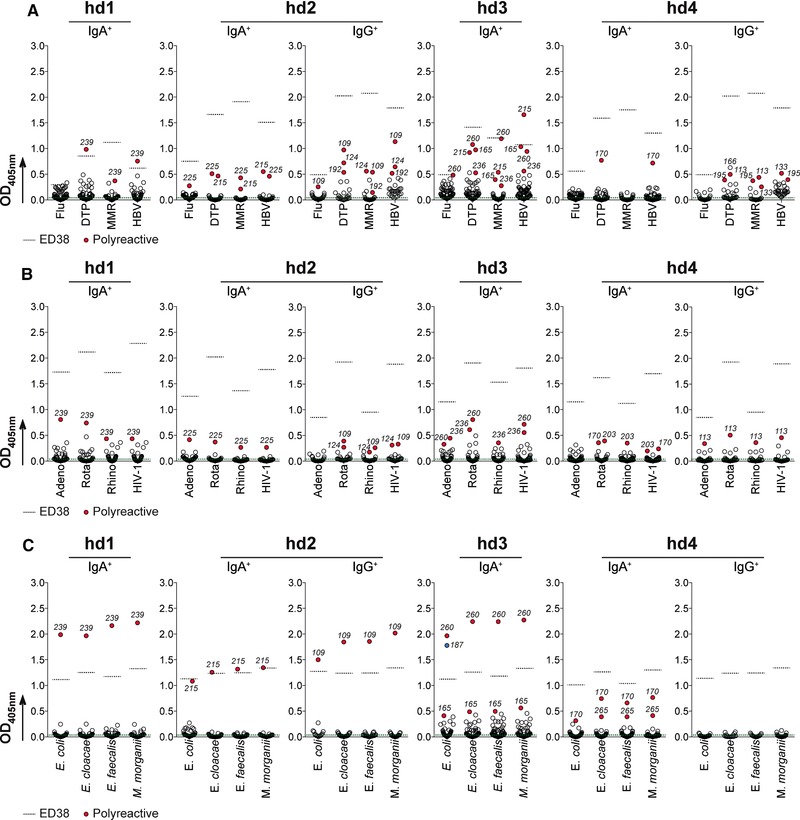
Reactivity of IgA+ memory B‐cell antibodies against vaccines, viral pathogens, and commensal bacteria. The reactivity of recombinant IgA+ and IgG+ memory antibodies against selected vaccines, mucosa‐tropic viruses, and commensal bacteria was determined by ELISA. In total, 251 recombinant IgA (from four healthy individuals; n = 59 for hd1, n = 47 for hd2, n = 67 for hd3, and n = 78 for hd4), and 61 IgG monoclonal antibodies (from hd2 and hd4 donors, n = 28 and n = 33, respectively) were produced and tested. Antibodies were tested in triplicate and in two independent experiments. Averaged data from one representative experiment are shown. (A) Flu, influenza virus; DTP, diphtheria‐tetanus‐polio; MMR, measles‐mumps‐rubella; HBV, hepatitis B virus. (B) Adeno, adenovirus type 5; Rota, rotavirus; Rhino, rhinovirus type 1A; HIV‐1. (C) Selected commensal bacteria. Dotted black lines indicate the reactivity of the polyreactive control antibody ED38 38. Dotted green line indicates the average reactivity of the negative control antibody mGO53 8 against the tested antigens. Polyreactive antibodies reacting non‐specifically with the different antigens tested are shown in red.
Polyreactivity of blood IgA+ memory B‐cell antibodies
Antibody polyreactivity can be acquired as a result of affinity maturation, and ∼ 23% of human IgG memory B cells express BCRs capable of polyreactive binding 15. To determine the polyreactivity frequency of circulating IgA memory B‐cell antibodies, all 251 IgA monoclonals were evaluated for ELISA binding to unrelated antigens; KLH, dsDNA, Insulin and LPS (Fig. 3A). The frequency of polyreactive IgA antibodies varied moderately between individuals (ranging from 14.1 to 21.3%) (Fig. 3A), and was on average slightly but statistically lower than the ones measured for IgG from internal hd2/hd4 controls (16.7% versus 29.6%, p = 0.03) (Fig. 3B). Importantly, all IgA and IgG antibodies reacting nonspecifically against the selected vaccines, viruses and bacteria were shown to be highly polyreactive in this assay (Supporting Information Table 1). No particular IgA subclass or light chain usage was associated with more elevated polyreactivity levels (Fig. 3C). Likewise, polyreactivity was independent of the number of positive charges in the CDRH3 (Fig. 3D), VH and JH gene (Fig. 3E), number of putative N‐glycosylation sites (PNGS) (Supporting Information Fig. 3), and VL and JL gene segments used (data not shown). However, polyreactive IgA antibodies had on average higher mutation rates of their VH genes than non‐polyreactive ones (22. 98 versus 18.83, p = 0.008) (Fig. 3G), and showed a significant bias for longer CDRH3 (Fig. 3D). The association of polyreactivity with longer CDRH3 was likely resulting from the preferential recombination with JH6 genes (longest among JH families) as evidenced by the comparative analysis of VHJH link frequencies. This supports that polyreactive IgA antibodies had a significantly increased number of VH3(DH)JH6 rearrangements and reciprocally a decreased number in VH3(DH)JH4 combinations compared to nonpolyreactive IgAs (Fig. 3F).
Figure 3.
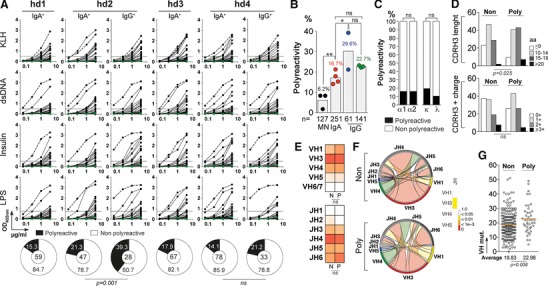
Polyreactivity of IgA+ memory B‐cell antibodies. (A) The reactivity of recombinant IgA+ and IgG+ memory antibodies against KLH, dsDNA, insulin and LPS was measured by ELISA. In total, 251 recombinant IgA (from four healthy individuals; n = 59 for hd1, n = 47 for hd2, n = 67 for hd3, and n = 78 for hd4), and 61 IgG monoclonal antibodies (from hd2 and hd4 donors, n = 28 and n = 33, respectively) were tested and analyzed. Duplicate values were obtained from at least three independent experiments. Representative data from one experiment are shown as mean values. Dotted green and black lines represent the negative control antibody mGO53 8 and positive control antibody ED38 38, respectively. Horizontal lines show cut‐off OD405 nm for positive reactivity. Pie charts summarize the frequency of polyreactive (black) and non‐polyreactive (white) IgA+ and IgG+ memory B‐cell clones. The number of tested antibodies is indicated in the pie chart center. Groups were compared using 2 × 2 Fisher's Exact test. ns, not significant. (B) Bar graph comparing the frequency of polyreactive antibodies (as indicated above bars) between IgA+ memory B cells, and other B‐cell compartments: mature naïve B cells (MN) 8, IgG+ memory B cells from donor hd2‐hd4, and from historical data 15, 17. Each symbol represents a donor. Groups were compared using 2 × 5 Fisher's Exact test.* p < 0.05; ** p < 0.01; ns, nonsignificant. The number of antibodies tested in each group is indicated below each bar. (C) Bar graph comparing the frequency of polyreactive (black) and non‐polyreactive (white) IgA+ memory B cells according to IgA sub‐class (IgA1 or IgA2), and light chains used (Igκ or Igλ). Groups were compared using 2 × 5 Fisher's Exact test.ns, nonsignificant. (D) Bar graphs comparing the frequency of polyreactive and nonpolyreactive IgA+ memory B cells according to CDRH3 aminoacid length and number of positive charges in the CDRH3. Groups were compared using 2 × 5 Fisher's Exact test. ns, nonsignificant. (E) Heat maps comparing the frequency of VH and JH gene usages between polyreactive and nonpolyreactive IgA+ memory B cells. (F) Circos plots generated from the immunoglobulin gene analysis using “circlize” (v0.3.1) R package compare the frequency of VH(DH)JH rearrangements between polyreactive and nonpolyreactive IgA+ memory B cells. Heat bar shows the results of the 2 × 5 Fisher's Exact test used to compare both groups. (G) Dot plots comparing the number of mutations in VH genes between polyreactive and non‐polyreactive IgA+ memory B cells. Groups were compared using Student's t‐test with Welch's correction. ns, not significant.
Self‐reactivity of blood IgA+ memory B‐cell antibodies
Self‐reactivity is a common characteristic of circulating IgG+ memory B‐cell antibodies in healthy humans, with a third to about half of them being reactive against human cell antigens 15, 18. To determine whether IgA+ memory B‐cell antibodies in healthy individuals are also frequently autoreactive, we measured by ELISA the reactivity of 251 IgA and 61 IgG antibodies against self‐antigens contained in whole protein lysates of the human laryngeal carcinoma cell line HEp‐2 (Fig. 4A). Moreover, we also examined the reactivity of recombinant IgA and IgG antibodies against HEp‐2 cells by indirect immunofluorescence assay (IFA), and classified the various staining patterns as either cytoplasmic, nuclear or nucleo‐cytoplamic (Fig. 4B and C). When combined with the ELISA data, memory IgAs were found to be less frequently self‐reactive on average than memory IgG antibodies cloned from hd2/hd4 or from previously characterized donors (27.9 versus 45.9% for hd2/hd4 IgGs, p = 0.009) (Fig. 4D). Indeed, when the analysis was restricted to the HEp‐2 reactivities detected only by IFA, IgA antibodies had a frequency of self‐reactivity considerably decreased compared to IgGs (13.1 versus 31.1% for hd2/hd4 IgGs) (Fig. 4E). This statistical difference was also evident when considering IgA and IgG antibody groups from hd2 and hd4 donors individually (Fig. 4E). Importantly, memory IgG antibodies (n = 61) more frequently displayed autoantibody‐like specificities such anti‐nuclear (ANA; 2–136 and 2–149), anti‐vimentin‐like (2–125 and 4–139) and anti‐anchoring proteins (2–186) than IgAs for which, only one ANA (2–239) was identified amongst the 251 molecules tested. Apart from an increased number of VH mutations on average for HEp‐2‐binding IgA antibodies (21.5 versus 18.8% for nonreactive IgAs, p = 0.039), we found no association between specific IgH gene features and self‐reactivity (Supporting Information Fig. 3 and data not shown).
Figure 4.
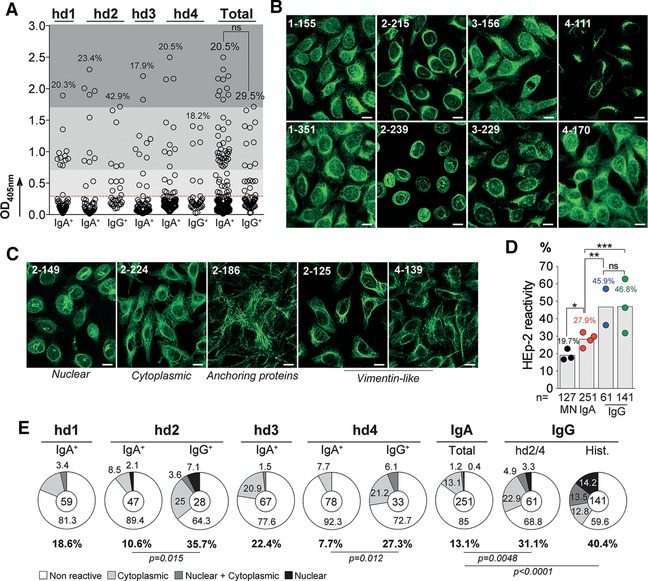
Self‐reactivity of IgA+ memory B‐cell antibodies. The reactivity of the recombinant IgA+ and IgG+ memory antibodies against HEp‐2 cell antigens was evaluated by ELISA and indirect immunofluorescence assay (IFA). In total, 251 recombinant IgA (from four healthy individuals; n = 59 for hd1, n = 47 for hd2, n = 67 for hd3, and n = 78 for hd4), and 61 IgG monoclonal antibodies (from hd2 and hd4 donors, n = 28 and n = 33, respectively) were tested and analyzed. (A) Dot plots comparing the frequency of HEp‐2 reactive antibodies by ELISA (as indicated above dots) between IgA+ and IgG+ memory B cells. The data shown correspond to the mean values of triplicate obtained in two independent experiments. (B) and (C) show representative HEp‐2 cell IFA staining patterns of antibodies cloned from IgA+ (B), and IgG+ (C) memory B cells, respectively, tested in two experiments (Magnification ×40). White scale bar indicates 12 μm. (D) Bar graph comparing the frequency of combined ELISA/IFA HEp‐2 reactive antibodies (as indicated above bars) between IgA+ memory B cells, and other B‐cell compartments: mature naïve B cells (MN) 8, IgG+ memory B cells from donor hd2‐hd4 (blue dots), and from historical data (green dots) 15, 17. Each symbol represents a donor. Groups were compared using 2 × 5 Fisher's Exact test.* p < 0.05; ** p < 0.01; *** p < 0.001; ns, nonsignificant. The number of antibodies tested in each group is indicated below each bar. (E) Pie charts comparing the frequency (as indicated below and on charts) of non HEp‐2 reactive (white) and HEp‐2 reactive IgA+ and IgG+ memory B‐cell antibodies according to the different IFA reactivity patterns observed: cytoplasmic (light grey), nuclear plus cytoplamic (dark gray), and nuclear (dark). The number of tested antibodies is indicated in the pie chart center. Groups were compared using 2 × 5 Fisher's Exact test.
The roles of class‐switching and somatic mutations for IgA poly‐ and self‐reactivities
The Fc region has been previously reported to contribute to the fine specificity of the antigen‐binding site of immunoglobulins including autoantibodies 19, 20, 21. Therefore, we next determined whether the expression of all memory IgAs as recombinant IgG monoclonals could have affected their reactivities; this would suggest a potential role of the Fc region for poly‐ and self‐reactivity of human IgA antibodies. We selected a total of 14 antibodies from all four donors, cloned their IgH variable domain fragments into IgA1 or IgA2 expression vectors 22, and purified the respective recombinant IgA1 and IgA2 monoclonals (Supporting Information Fig. 3E and F). We next evaluated by ELISA the poly‐ and self‐reactivity of the native IgA molecules in parallel with their IgG homologs. Despite some variations between individual antibodies, binding to the various antigens used in our polyreactivity assay was comparable between IgA and IgG except for 1–170 and 3–212, which showed a decreased reactivity when tested as IgA (Fig. 5A). Nonetheless, the polyreactivity frequency remained globally unaffected by isotype switching to the proper IgA sub‐class since regardless of the Fc region, all of them were shown to be polyreactive (Fig. 5A). Similarly, although the native IgAs were generally slightly less self‐reactive in comparison to their IgG homologs, all of them were reactive against HEp‐2 antigens (Fig. 5A).
Figure 5.
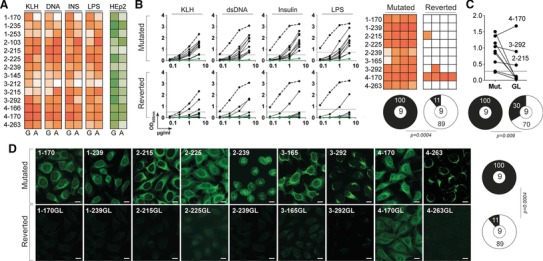
Poly‐ and self‐reactivity of class‐switched IgA+ and germline‐encoded antibodies. The reactivity of native IgA+ memory (n = 14) and reverted (n = 9) antibodies, which were selected from the four donors, were tested by ELISA against polyreactive ligands, and by ELISA and IFA against HEp‐2 cell antigens. (A) Heat map comparing the poly‐ and HEp‐2 reactivity of recombinant IgA+ memory antibodies expressed with an IgG1 or with their original IgA Fc portion. The data shown correspond to the mean values of triplicate obtained in two independent experiments. Color intensity is proportional to the reactivity level with darker colors indicating high binding while light colors show moderate binding (white = no binding). (B) ELISA graphs comparing the reactivity of mutated and reverted IgA antibodies against KLH, dsDNA, insulin, and LPS. Dotted lines represent the positive control antibody ED38 38. Horizontal lines show cut‐off OD405 nm for positive reactivity. Green line shows the negative control antibody mGO53 8. One representative experiment is shown. Heat maps summarizing the reactivity of the mutated and reverted IgA antibodies to the various antigens tested expressed as a mean of triplicate values obtained from two experiments. Color gradient is proportional to the reactivity level with darker colors indicating high binding while light colors show moderate binding (white = no binding). Pie charts summarize the frequency of polyreactive (black) and non‐polyreactive (white) antibodies. The number of tested antibodies is indicated in the pie chart center. Groups were compared using 2 × 2 Fisher's Exact test. (C) Dot plot comparing the HEp‐2 reactivity detected by ELISA of mutated (Mut.) versus reverted to germline (GL) IgA antibodies. Pie charts summarize the frequency of HEp‐2 (black) and non‐HEp‐2 (white) reactive antibodies. The number of tested antibodies is indicated in the pie chart center. The data shown correspond to the mean values of triplicate obtained in two independent experiments. Groups were compared using 2 × 2 Fisher's Exact test. (D) HEp‐2 cell IFA staining patterns obtained with the original mutated IgA antibodies and their germline counterparts (Reverted), tested in two experiments (magnification ×40). White scale bar indicates 12 μm. Pie charts summarize the frequency of HEp‐2 (black) and non‐HEp‐2 (white) reactive antibodies. The number of tested antibodies is indicated in the pie chart center. Groups were compared using 2 × 2 Fisher's Exact test.
To determine whether hypermutations are also involved in the acquisition of poly‐ and self‐reactivities of IgA+ memory B cells as shown for IgG memory antibodies 15, we reverted the IgH and IgL chain genes of 9 IgA memory antibodies to their germline form and compared their reactivity to the original mutated antibodies (Supporting Information Table 2). The removal of somatic hypermutations induced a loss of polyreactivity for eight out of nine antibodies tested (Fig. 5B). Most germline antibodies lacked reactivity with HEp‐2 antigens by ELISA (6 out of 9) (Fig. 5C). But more importantly, with the exception of 4–170GL, which was positive by IFA, none of the germline versions could recapitulate the HEp‐2 immunofluorescence staining observed with their respective mutated forms (Fig. 5D).
Discussion
Despite the fact that IgA+ memory B cells are numerous in human blood where they act as sentinels of the memory cell pool for IgA‐mediated immune response, very little is known about their repertoire specifically with regard to antibody gene features, specificity and function. In this study, we have characterized the immunoglobulin gene repertoire and reactivity of recombinant monoclonal antibodies cloned from single blood IgA+ memory B cells from healthy individuals. Overall, IgA memory antibodies displayed very similar gene features as compared to their IgG antibody counterparts. As previously reported for intestinal IgA+ plasmablasts 16, IgA+ memory B cells showed an increased Jκ1 usage suggesting frequent, productive primary immunoglobulin gene rearrangements. Importantly, IgA immunoglobulin genes carried high levels of somatic mutations indicating that circulating CD27+IgA+ B cells are shaped following an antigen‐driven affinity maturation process. Thus, the vast majority of IgA+ memory B cells circulating in the blood derive from T cell‐dependent immune responses.
What are the antigens specifically recognized by blood IgA+ memory B cells in humans yet remained largely unknown. Considering the enormous diversity of antibody specificities, we were unable to identify the cognate antigens bound by IgA+ memory B cells, but we did not observe any differences in the reactivity of IgA and IgG antibodies against vaccines, mucosa‐tropic viruses and gut commensal bacteria. In mice, maintenance of immune homeostasis and mucosal integrity relies on T‐cell‐independent and to lesser extend T‐cell‐dependent IgAs that “coat” commensals and thus shape microbiota composition and diversity 23, 24. Human IgA and IgG antibodies produced by intestinal plasmablasts react with strains of the commensal flora 16, and blood CD27−IgA+ B cells have been described to express polyreactive antibodies binding to commensal and pathogenic bacteria 25. Our results, however, suggest that IgA+ memory B cells circulating in human blood under physiological conditions rarely react and with low affinity, with commensal bacteria colonizing the gut.
In mice, naïve but also memory B cells in Peyer's patches can migrate to the spleen and also other mucosal tissues such as mammary glands where they differentiate into plasma cells 26. Memory B‐cell recirculation between different compartments might also occur in humans, as suggested by the egress of transitional B cells from gut associated lymphoid tissues (GALT) to the splenic marginal zone 27, and the presence of mucosa‐derived B‐cell subset in healthy humans’ blood 28. However, the proportion of IgA+ memory B‐cell clones generated in GALT that can enter blood and lymphatic circulation to reach other compartments is still unknown. Interestingly, only one third of peripheral IgA+ memory B cells analyzed here express IgA2 sub‐class, which is the main immunoglobulin subtype of ileum‐derived antibodies 16. This is in agreement with the predominance of serum IgA1 over IgA2, and with the distribution of their corresponding plasma cell populations in the bone marrow 29. In addition, germ‐free and alymphoblastic mice are typically characterized by a profound mucosal IgA deficiency still maintain half of normal serum IgA levels 30, 31. Therefore, although a non‐neglectable fraction of blood IgA+ memory B cells could originate from the GALT, we propose that the majority of blood CD27+IgA+ B cells may derive from systemic or other mucosal compartments.
Antibody polyreactivity is a common property of IgG+ memory B cells 15, 18, but that is also found in IgA+ memory B cells with 17% of IgA+ memory antibodies exhibiting polyreactive binding. This frequency is very comparable to the one recently reported for human blood CD27+IgA+ B lymphocytes (16%) 25, and relatively lower than the frequency of polyreactive IgG+ memory antibodies from the same healthy donors (30%). Polyreactive clones were enriched with IgA displaying longer CDRH3 and had on average more mutated VH genes as described previously 25. IgA polyreactivity was not related to class switching but was dependent on the presence of somatic hypermutations since 90% of the mutated IgA antibodies lost polyreactive binding upon reversion to their germline state. As opposed to IgG+ memory B cells, IgA+ memory B cells were rarely self‐reactive, with less than 15% being HEp‐2 reactive by IFA as shown recently 25, and only 0.4% of IgA antibodies presenting an ANA‐type of staining pattern. As for polyreactivity, self‐reactivity of IgA memory antibodies mainly resulted from the inclusion of somatic mutations in Ig genes as already demonstrated for IgG+ memory B lymphocytes 15. Of note, although antibodies have been recently cloned from human blood CD27+ and CD27− IgA+ B cells and compared, their relationship to IgG molecules expressed by memory B cells has never been determined directly 25. Considering that IgA+ and IgG+ memory B cells have comparable gene features including equivalent hypermutation loads likely involved for both populations in the acquisition of self‐reactivity, it is unclear why IgA+ memory antibodies showed a considerable reduction of poly‐ and self‐reactivity. This difference could be explained by the presence of distinct tolerance checkpoints between the two compartments or alternatively, by a more stringent tolerance threshold for counterselecting autoreactive IgA+ memory B‐cell clones. In this regard, it has been proposed that activation in the GALT of transitional B cells arriving from the bone marrow through blood could constitute a selective tolerance checkpoint before their entry into the circulating mature naïve B‐cell pool 27. It is interesting to highlight that only few autoimmune diseases are characterized by a predominant production of autoantigen‐specific IgA antibodies such as coeliac disease 32, and IgA autoimmune blistering dermatoses 33, whereas pathogenic and non‐pathogenic IgG autoantibodies are a hallmark for most autoimmune disorders. On the other hand, the rarity of self‐reactive IgA+ memory B cells could originate from the nature of the cognate antigen recognized and the maturation process involved. Finally, we cannot exclude that IgA class‐switching could disfavor the persistence in the circulating memory B‐cell pool of clones with polyspecific reactivities 34.
In summary, blood CD27+IgG+ and CD27+IgA+ B cells derive from the GC‐dependent pathway, and are often considered as a relatively homogenous memory B‐cell population in terms of immunoglobulin gene features and gene expression profiles 25, 35. However, we showed that they differ considerably regarding their level of poly‐ and self‐reactivity, with a drastically decreased frequency of self‐reactive antibodies in the IgA+ memory B‐cell compartment in comparison to its IgG+ counterpart. Therefore, the observations described herein have important implications in understanding the development of IgA+ memory B‐cell response, and its potential dysregulation during certain autoimmune diseases.
Materials and methods
Human samples
Samples were obtained as part of the BHUANTIVIH research protocol performed in accordance with and after ethical approval from all the French legislation and regulation authorities. Peripheral blood mononuclear cells (PBMC) and sera were collected from a cohort of healthy donors in the ICAReB biobanking platform (Institut Pasteur) under the CoSimmGen protocol approved by the French Agence nationale de sécurité du médicament (ANSM) on May 24th 2012, and the Comité de Protection des Personnes (CPP) on January 17th 2014. The BHUANTIVIH protocol received approval from the Comité Consultatif pour le Traitement de l'Information en matière de Recherche dans le domaine de la Santé (CCTIRS) on December 12th 2013, and the Commission Nationale de l'Informatique et des Libertés (CNIL) on August 8th 2014. All donors gave written consent to participate in this study, and data were collected under pseudo‐anonymized conditions using subject coding. Vaccine and HIV serology status of the donors was provided by the ICAReB platform, and the absence of antinuclear (ANA) and anti‐neutrophil cytoplasmic (ANCA) autoantibodies was confirmed at the time of blood collection with routine diagnostic tests performed in a licensed biomedical lab (CBCV lab, Institut Pasteur) (Supporting Information Fig. 1A).
Single B‐cell FACS sorting and expression‐cloning of antibodies
Peripheral B cells were isolated from donors’ PBMCs by magnetic B‐cell enrichment using human CD19 or CD20 microbeads according to the manufacturer's instructions (Miltenyi). Single CD19+CD27+IgA+ B cells were identified and sorted into 96‐well PCR plates using a FACS Aria III sorter (Becton Dickinson) as previously described 8, 36. Single‐cell cDNA synthesis using SuperScript III reverse transcriptase (Fisher Scientific) followed by nested‐PCR amplifications of IgH, Igκ and Igλ genes, and sequences analyses for Ig gene features were performed as previously described 8, 16, 36. For the reversion to germline (GL) of the selected IgA antibodies, sequences were constructed by replacing the mature VH‐JH and VL‐JL gene segments with their GL counterparts as previously described 37. Purified digested PCR products and synthetic GL genes were cloned into human Igγ1‐, Igκ‐, or Igλ‐expressing vectors as previously described 15, and for some selected ones, into Igα1‐, or Igα2‐expressing vectors as previously described 22. Recombinant antibodies were produced by transient co‐transfection of exponentially growing Freestyle™ 293‐F suspension cells (Fisher Scientific) using polyethylenimine (PEI)‐precipitation method as previously described 22. Recombinant human antibodies were purified by batch/gravity‐flow affinity chromatography using peptide M‐coupled agarose (Invivogen) for IgAs, and protein G sepharose 4 fast flow beads (GE Healthcare) for IgGs.
Immunoglobulin polymerization of selected IgA and IgG antibodies was evaluated by SDS‐PAGE using 3–8% Tris‐Acetate Novex gels (Fisher Scientific) in non‐reducing conditions followed by a silver staining (Fisher Scientific), and quantified by size‐exclusion chromatography (SEC). Briefly, after column equilibration with PBS, antibodies (0.4‐0.5 mg) were injected in a Superdex 200 SEC column (Increase 10/300GL) at a flow rate of 0.3 ml/min, and separated using the AKTA pure liquid chromatography system (GE Healthcare) as previously described 22. UV absorbance and conductivity measurements were recorded and analyzed using Unicorn 6.3 software (GE Healthcare). The proportion of multimeric immunoglobulins was calculated using area under the curve values of the specific SEC pics (compared to the total signal). N‐glycans content in selected IgG antibodies was estimated using infrared immunoblot analysis following protein separation by SDS‐PAGE (using 250 ng of IgG/IgA) and electrotransfer onto nitrocellulose membranes. Briefly, after a 2 h saturation step in 3% BSA, 0.05% Tween 20‐PBS (PBST), filters were washed with PBST and incubated 2 h with biotynilated Tritilum vulgaris lectin (Sigma) (20 μg/mL in 3% BSA‐PBST). After washings, filters were incubated 1 h with 1:10,000‐diluted AF690‐conjugated donkey anti‐human IgG antibodies and AF790‐conjugated streptavidin (Jackson Immunoresearch), washed and examined with the Odyssey Infrared Imaging system (LI‐COR Biosciences).
Polyreactivity ELISA
Polyreactivity ELISA was performed as followed. Briefly, high‐binding 96‐well ELISA plates (Costar) were coated overnight with 0.5 μg/well of purified double stranded (ds)‐DNA, KLH, LPS, and 0.25 μg/well of purified insulin (Sigma) in PBS. After washings with 0.001% Tween‐PBS, plates were blocked 2 h with 2% BSA, 1 mM EDTA, PBST (Blocking buffer). After washings, coated plates were incubated 2 h with IgG or IgA antibodies diluted at 26.67 nM and three consecutive 1:4 dilutions in PBS. After washings, the plates were revealed by incubation for 1 h with goat HRP‐conjugated anti‐human IgG (Jackson ImmunoReseach, 0.8 μg/mL final), or anti‐human IgG/IgM/IgA antibodies (Immunology Jackson ImmunoReseach, 0.8 μg/mL final), and by adding 100 μL of HRP chromogenic substrate (ABTS solution, Euromedex) after washing steps. After 1‐h incubation, optical densities were measured at 405 nm (OD405nm), and background values given by incubation of PBS alone in coated wells were subtracted. High positive (ED38) 38 and negative (mGO53) 8 antibody controls were included in each experiment, and threshold values for reactivity were determined as previously described 39. All experiments were performed using HydroSpeed™ microplate washer and Sunrise™ microplate absorbance reader (Tecan).
HEp‐2 ELISA and IFA
Protein extract of human laryngeal carcinoma cells (ATCC® CCL‐23™) was prepared from trypsinized HEp‐2 cell monolayers using hypotonic lysis buffer (20 mM Tris‐HCl, pH 7.5, 5 mM MgCl2, 5 mM CaCl2, 1 mM DTT, 1 mM EDTA, plus proteases inhibitors (Pierce) as previously described 40. High‐binding 96‐well ELISA plates (Costar) were coated overnight with 0.5 μg/well of HEp‐2 whole cell lysate in PBS. After washings with PBST, plates were blocked 2 h with the ELISA blocking buffer. After washings with PBST, coated plates were incubated 2 h with IgG or IgA antibodies diluted at 6.67 nM in PBS (in triplicate), and revealed as described above. Recombinant IgG and IgA antibodies (150 μg/mL), and control antibodies (mGO53 8, ED38 38, and internal controls in the kit) were analyzed by indirect immunofluorescence assay (IFA) on HEp‐2 cells (ANA AeskruSlides, Ingen) sections using FITC‐conjugated anti‐human IgG antibodies as the tracer according to the manufacturer’ instructions. Sections were examined using the fluorescence microscope Axio Imager 2 (Zeiss) with ApoTome.2 system, and pictures were taken at magnification ×40 with 7000 ms‐acquisition using ZEN imaging software (Zen 2.0 blue version, Zeiss) at the Imagopole platform (Institut Pasteur).
Vaccines, viruses, and commensal bacteria ELISAs
High‐binding 96‐well ELISA plates (Costar) were coated overnight with 50 μL/well of vaccines and viral lysates diluted in PBS as follows: Vaxigrip (2014‐2015, Sanofi Pasteur), and GenHevac B (Sanofi Pasteur) at 1 μg/mL, and Revaxis (Sanofi Pasteur) and Priorix (GSK) at a 1:50 dilution. ELISA plates were also coated with protein lysates of adenovirus type 5, rhinovirus type 1A, and rotavirus (Zepto Metrix Corporation) at 5 μg/mL and purified trimeric YU‐2 gp140 protein at 2.5 μg/mL 22. M. morganii (CIP 231T), E. faecalis (CIP 103241), E. cloacae (CIP 60.85T) were obtained from the biological resource center of Institut Pasteur (CRBIP), and E. coli used correspond to DH10β cells (NEB). Bacteria were grown as single colony, fixed with 0.2% paraformaldehyde (Sigma) for 20 min, washed, and used to coat Poly‐L‐Lysine‐treated high‐binding 96‐well ELISA plates (Costar) as previously described 16. After washings with 0.05% Tween‐20‐PBS (PBST), plates were blocked 2 h with the ELISA blocking buffer. After washings with PBST, coated plates were incubated 2 h with recombinant IgG antibodies diluted to 1 μg/mL in PBS, and revealed as described above.
Statistics
P values for Ig gene repertoire analyses, analysis of lengths, positive charges, of IgH CDR3, PNGS, and antibody reactivity were calculated by two‐sided 2 × 2 or 2 × 5 Fisher's Exact test. The numbers of VH, Vκ, and Vλ mutations were compared across groups of antibodies using unpaired Student's t‐test with Welch's correction. Statistical analyses were performed using GraphPad Prism sofware (v6.0a), and SISA online tools for 2 × 5 Fisher test (http://www.quantitativeskills.com/sisa).
Conflict of Interest
The authors declare no commercial or financial conflict of interest.
Abbreviations
- BCRs
B‐cell antigen receptors
- CDRH3
complementary determining region 3
- DTP
trivalent diphtheria‐tetanus‐polio
- dsDNA
double‐stranded DNA
- Flu
influenza virus
- GALT
gut associated lymphoid tissues
- GC(s)
germinal center(s)
- GL
germline
- IFA
indirect immunofluorescence assay
- Ig
immunoglobulin
- IgA+ and IgG+
IgA and IgG class‐switched
- IgH and IgL
heavy‐ and light‐chain variable domain
- HBV
hepatitis B virus
- KLH
keyhole limpet hemocyanin
- LPS
lipopolysaccharide
- MMR
trivalent measles‐mumps‐rubella
- PBMC
Peripheral blood mononuclear cells
- PNGS
putative N‐glycosylation sites
- SEC
size‐exclusion chromatography
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Peer review correspondence
Supporting Information Fig. 1
Supporting Information Fig. 2
Supporting Information Fig. 3
Acknowledgments
We are grateful to all participants who consented to be part of this study. We thank Olivia Chény, Nathalie Jolly and Servane Warrot from the Pôle Intégré de Recherche Clinique (PIRC, Institut Pasteur) for their assistance on the preparation and submissions of the BHUANTIVIH protocol, Marie‐Noëlle Ungeheuer and her team from the ICAReB platform for providing us the human samples, Valentina Libri, Sandrine Schmutz, and Sophie Novault for their assistance with single cell sorting at the Center for Human Immunology (CIH, Institut Pasteur), and members of the Imagopole platform for their help with microscopy experiments. We are grateful to Caroline Eden (Icahn School of Medicine at Mount Sinai) for helpful comments and manuscript editing. J. P. was supported by a postdoctoral fellowship from the ANRS (Agence Nationale de Recherche sur le Sida et les hépatites virales). This work was supported by the European Research Council (ERC) – Seventh Frame‐work Program (ERC‐2013‐StG 337146). H.M. was supported by the G5 Institut Pasteur Program and the Milieu Intérieur Program (ANR‐10‐LABX‐69‐01).
References
- 1. Victora, G. D. and Nussenzweig, M. C. , Germinal centers. Annu. Rev. Immunol. 2012. 30: 429–457. [DOI] [PubMed] [Google Scholar]
- 2. Tarlinton, D. and Good‐Jacobson, K. , Diversity among memory B cells: origin, consequences, and utility. Science 2013. 341: 1205–1211. [DOI] [PubMed] [Google Scholar]
- 3. Macpherson, A. J. , McCoy, K. D. , Johansen, F. E. and Brandtzaeg, P. , The immune geography of IgA induction and function. Mucosal Immunol 2008. 1: 11–22. [DOI] [PubMed] [Google Scholar]
- 4. Cerutti, A. , The regulation of IgA class switching. Nat. Rev. Immunol. 2008. 8: 421–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kaetzel, C. S. , Mucosal Immune Defense : Immunoglobulin A. Springer, New York: 2007. [Google Scholar]
- 6. Mkaddem, S. B. , Christou, I. , Rossato, E. , Berthelot, L. , Lehuen, A. and Monteiro, R. C., IgA, IgA receptors, and their anti‐inflammatory properties. Curr. Top. Microbiol. Immunol. 2014. 382: 221–235. [DOI] [PubMed] [Google Scholar]
- 7. Mestecky, J. and Hammarström, L. , IgA‐associated diseases Mucosal Immune Defense: Immunoglobulin A. Springer, New York: 2007, 321–344. [Google Scholar]
- 8. Wardemann, H. , Yurasov, S. , Schaefer, A. , Young, J. W. , Meffre, E. and Nussenzweig, M. C. , Predominant autoantibody production by early human B cell precursors. Science 2003. 301: 1374–1377. [DOI] [PubMed] [Google Scholar]
- 9. Yurasov, S. , Tiller, T. , Tsuiji, M. , Velinzon, K. , Pascual, V. , Wardemann, H. and Nussenzweig, M. C. , Persistent expression of autoantibodies in SLE patients in remission. J. Exp. Med. 2006. 203: 2255–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yurasov, S. , Wardemann, H. , Hammersen, J. , Tsuiji, M. , Meffre, E. , Pascual, V. and Nussenzweig, M. C. , Defective B cell tolerance checkpoints in systemic lupus erythematosus. J. Exp. Med. 2005. 201: 703–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Samuels, J. , Ng, Y. S. , Coupillaud, C. , Paget, D. and Meffre, E. , Impaired early B cell tolerance in patients with rheumatoid arthritis. J. Exp. Med. 2005. 201: 1659–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Menard, L. , Samuels, J. , Ng, Y. S. and Meffre, E. , Inflammation‐independent defective early B cell tolerance checkpoints in rheumatoid arthritis. Arthritis. Rheum. 2011. 63: 1237–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chamberlain, N. , Massad, C. , Oe, T. , Cantaert, T. , Herold, K. C. and Meffre, E. , Rituximab does not reset defective early B cell tolerance checkpoints. J. Clin. Invest. 2016. 126: 282–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Menard, L. , Saadoun, D. , Isnardi, I. , Ng, Y. S. , Meyers, G. , Massad, C. , Price, C. et al, The PTPN22 allele encoding an R620W variant interferes with the removal of developing autoreactive B cells in humans. J. Clin. Invest. 2011. 121: 3635–3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tiller, T. , Tsuiji, M. , Yurasov, S. , Velinzon, K. , Nussenzweig, M. C. and Wardemann, H. , Autoreactivity in human IgG+ memory B cells. Immunity 2007. 26: 205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Benckert, J. , Schmolka, N. , Kreschel, C. , Zoller, M. J. , Sturm, A. , Wiedenmann, B. and Wardemann, H. , The majority of intestinal IgA+ and IgG+ plasmablasts in the human gut are antigen‐specific. J. Clin. Invest. 2011. 121: 1946–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mietzner, B. , Tsuiji, M. , Scheid, J. , Velinzon, K. , Tiller, T. , Abraham, K. , Gonzalez, J. B. et al, Autoreactive IgG memory antibodies in patients with systemic lupus erythematosus arise from nonreactive and polyreactive precursors. Proc. Natl Acad. Sci. USA 2008. 105: 9727–9732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Koelsch, K. , Zheng, N. Y. , Zhang, Q. , Duty, A. , Helms, C. , Mathias, M. D. , Jared, M. et al, Mature B cells class switched to IgD are autoreactive in healthy individuals. J. Clin. Invest. 2007. 117: 1558–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Torres, M. and Casadevall, A. , The immunoglobulin constant region contributes to affinity and specificity. Trends Immunol. 2008. 29: 91–97. [DOI] [PubMed] [Google Scholar]
- 20. Tudor, D. , Yu, H. , Maupetit, J. , Drillet, A. S. , Bouceba, T. , Schwartz‐Cornil, I. , Lopalco, L. et al, Isotype modulates epitope specificity, affinity, and antiviral activities of anti‐HIV‐1 human broadly neutralizing 2F5 antibody. Proc. Natl Acad. Sci. USA 2012. 109: 12680–12685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xia, Y. , Pawar, R. D. , Nakouzi, A. S. , Herlitz, L. , Broder, A. , Liu, K. , Goilav, B. et al, The constant region contributes to the antigenic specificity and renal pathogenicity of murine anti‐DNA antibodies. J. Autoimmun. 2012. 39: 398–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lorin, V. and Mouquet, H. , Efficient generation of human IgA monoclonal antibodies. J. Immunol. Methods 2015. 422: 102–110. [DOI] [PubMed] [Google Scholar]
- 23. Bunker, J. J. , Flynn, T. M. , Koval, J. C. , Shaw, D. G. , Meisel, M. , McDonald, B. D. , Ishizuka, I. E. et al, Innate and adaptive humoral responses coat distinct commensal bacteria with immunoglobulin A. Immunity 2015. 43: 541–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fransen, F. , Zagato, E. , Mazzini, E. , Fosso, B. , Manzari, C. , El Aidy, S. , Chiavelli, A. et al, BALB/c and C57BL/6 mice differ in polyreactive IgA abundance, which impacts the generation of antigen‐specific IgA and microbiota diversity. Immunity 2015. 43: 527–540. [DOI] [PubMed] [Google Scholar]
- 25. Berkowska, M. A. , Schickel, J. N. , Grosserichter‐Wagener, C. , de Ridder, D. , Ng, Y. S. , van Dongen, J. J. , Meffre, E. et al, Circulating human CD27‐IgA+ memory B cells recognize bacteria with polyreactive Igs. J. Immunol. 2015. 195: 1417–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lindner, C. , Thomsen, I. , Wahl, B. , Ugur, M. , Sethi, M. K. , Friedrichsen, M. , Smoczek, A. et al, Diversification of memory B cells drives the continuous adaptation of secretory antibodies to gut microbiota. Nat. Immunol. 2015. 16: 880–888. [DOI] [PubMed] [Google Scholar]
- 27. Vossenkamper, A. , Blair, P. A. , Safinia, N. , Fraser, L. D. , Das, L. , Sanders, T. J. , Stagg, A. J. et al, A role for gut‐associated lymphoid tissue in shaping the human B cell repertoire. J. Exp. Med. 2013. 210: 1665–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fernandes, J. R. and Snider, D. P. , Polymeric IgA‐secreting and mucosal homing pre‐plasma cells in normal human peripheral blood. Int. Immunol. 2010. 22: 527–540. [DOI] [PubMed] [Google Scholar]
- 29. Woof, J. M. and Mestecky, J., Chapter 17 ‐ Mucosal Immunoglobulins In Lambrecht J. M. S. W. R. L. K. C. N. (Ed.) Mucosal Immunology (Fourth Edition). Academic Press, Boston, 2015, pp 287–324. [Google Scholar]
- 30. Macpherson, A. J. , Gatto, D. , Sainsbury, E. , Harriman, G. R. , Hengartner, H. and Zinkernagel, R. M. , A primitive T cell‐independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science 2000. 288: 2222–2226. [DOI] [PubMed] [Google Scholar]
- 31. Macpherson, A. J. , Lamarre, A. , McCoy, K. , Harriman, G. R. , Odermatt, B. , Dougan, G. , Hengartner, H. et al, IgA production without mu or delta chain expression in developing B cells. Nat. Immunol. 2001. 2: 625–631. [DOI] [PubMed] [Google Scholar]
- 32. Husby, S. and Murray, J. A. , Diagnosing coeliac disease and the potential for serological markers. Nat Rev Gastroenterol Hepatol 2014. 11: 655–663. [DOI] [PubMed] [Google Scholar]
- 33. Baum, S. , Sakka, N. , Artsi, O. , Trau, H. and Barzilai, A. , Diagnosis and classification of autoimmune blistering diseases. Autoimmun. Rev. 2014. 13: 482–489. [DOI] [PubMed] [Google Scholar]
- 34. Gitlin, A. D. , von Boehmer, L. , Gazumyan, A. , Shulman, Z. , Oliveira, T. Y. and Nussenzweig, M. C. , Independent roles of switching and hypermutation in the development and persistence of B lymphocyte memory. Immunity 2016. 44: 769–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Berkowska, M. A. , Driessen, G. J. , Bikos, V. , Grosserichter‐Wagener, C. , Stamatopoulos, K. , Cerutti, A. , He, B. et al, Human memory B cells originate from three distinct germinal center‐dependent and ‐independent maturation pathways. Blood 2011. 118: 2150–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tiller, T. , Meffre, E. , Yurasov, S. , Tsuiji, M. , Nussenzweig, M. C. and Wardemann, H. , Efficient generation of monoclonal antibodies from single human B cells by single cell RT‐PCR and expression vector cloning. J. Immunol. Methods 2008. 329: 112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mouquet, H. , Scharf, L. , Euler, Z. , Liu, Y. , Eden, C. , Scheid, J. F. , Halper‐Stromberg, A. et al, Complex‐type N‐glycan recognition by potent broadly neutralizing HIV antibodies. Proc. Natl Acad. Sci. USA 2012. 109: E3268–E3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Meffre, E. , Schaefer, A. , Wardemann, H. , Wilson, P. , Davis, E. and Nussenzweig, M. C. , Surrogate light chain expressing human peripheral B cells produce self‐reactive antibodies. J. Exp. Med. 2004. 199: 145–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mouquet, H. , Klein, F. , Scheid, J. F. , Warncke, M. , Pietzsch, J. , Oliveira, T. Y. , Velinzon, K. et al, Memory B cell antibodies to HIV‐1 gp140 cloned from individuals infected with Clade A and B viruses. PLoS One 2011. 6: e24078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ji, H. , Lysis of cultured cells for immunoprecipitation. Cold Spring Harb Protoc 2010. 2010: pdb prot5466. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Peer review correspondence
Supporting Information Fig. 1
Supporting Information Fig. 2
Supporting Information Fig. 3


