Abstract
Several functional and morphological brain measures are partly under genetic control. The identification of direct links between neuroimaging signals and corresponding genetic factors can reveal cellular‐level mechanisms behind the measured macroscopic signals and contribute to the use of imaging signals as probes of genetic function. To uncover possible genetic determinants of the most prominent brain signal oscillation, the parieto‐occipital 10‐Hz alpha rhythm, we measured spontaneous brain activity with magnetoencephalography in 210 healthy siblings while the subjects were resting, with eyes closed and open. The reactivity of the alpha rhythm was quantified from the difference spectra between the two conditions. We focused on three measures: peak frequency, peak amplitude and the width of the main spectral peak. In accordance with earlier electroencephalography studies, spectral peak amplitude was highly heritable (h 2 > 0.75). Variance component‐based analysis of 28 000 single‐nucleotide polymorphism markers revealed linkage for both the width and the amplitude of the spectral peak. The strongest linkage was detected for the width of the spectral peak over the left parieto‐occipital cortex on chromosome 10 (LOD = 2.814, nominal P < 0.03). This genomic region contains several functionally plausible genes, including GRID1 and ATAD1 that regulate glutamate receptor channels mediating synaptic transmission, NRG3 with functions in brain development and HRT7 involved in the serotonergic system and circadian rhythm. Our data suggest that the alpha oscillation is in part genetically regulated, and that it may be possible to identify its regulators by genetic analyses on a realistically modest number of samples.
Keywords: alpha rhythm, heritability, linkage study, magnetoencephalography
Introduction
The constitution of individuals is under strict genetic control, readily illustrated by the morphological similarity of monozygotic twins, and such similarities extend to several functional properties regulated by polymorphic genes. Genetic‐functional correlations are expected also in system‐level functional measures with large variation between – but relatively constant appearance within – individuals. Measures of many neural processes are known to fulfill these criteria, and they can be quantified with brain imaging (van Beijsterveldt et al., 1996; Smit et al., 2006; Renvall et al., 2012). Such results suggest that it is feasible to search for genetic influences on neuronal processes and to attribute their genetic regulation to specific loci in the genome by combining neuroimaging and genetic mapping techniques.
The human cerebral cortex shows several intrinsic rhythms that can be characterized with non‐invasive neuroimaging methods such as magnetoencephalography (MEG) and electroencephalography (EEG). The most prominent of them is the ~ 10‐Hz ‘alpha’ rhythm recorded over the parieto‐occipital cortices (Berger, 1929; Adrian & Matthews, 1934; Adrian, 1944). In humans, the cortical sources of alpha activity have been located at the bilateral visual cortices and the parieto‐occipital sulcus (Salmelin & Hari, 1994; Manshanden et al., 2002). Intracortical recordings in dogs (Lopes da Silva et al., 1980) and monkeys (Bollimunta et al., 2011) have revealed simultaneous activity in the thalamic nuclei, suggesting involvement of both brain regions in the rhythm generation. Indeed, human patients with thalamic infarction show abnormal spectral spreading of alpha activity to a wide frequency range (Mäkelä et al., 1998).
The alpha rhythm is strongly attenuated by opening of the eyes, and it is modulated by tasks requiring visual attention (Worden et al., 2000; Jensen et al., 2002; Palva & Palva, 2007; Kelly et al., 2009; Saalmann et al., 2012; Gray et al., 2015), visual imagery (Salenius et al., 1995) and working memory also outside of the visual modality (Jensen et al., 2002; Jokisch & Jensen, 2007; Tuladhar et al., 2007; Haegens et al., 2010; Poch et al., 2014). Recent electrocortical recordings in monkeys have demonstrated that cortical alpha oscillations depend on the attentional state of the animal (Saalmann et al., 2012). In addition, alpha oscillations have been suggested to modulate the thalamo‐cortical (Amzica & Lopes da Silva, 2011) and cortico‐cortical information transfer (Klimesch et al., 2007; Jensen & Mazaheri, 2010), and to facilitate sensory perception (Klimesch et al., 2007). The alpha rhythm thus most likely does not reflect mere cortical idling, but seems to have important functional roles (for a review, see e.g., Lopes da Silva, 2013). The reactivity of the alpha rhythm has also been used to probe cortical functions in several neuropsychiatric diseases (for review, see e.g., Yener & Başar, 2013).
EEG studies (van Beijsterveldt et al., 1996; van Beijsterveldt & van Baal, 2002; Smit et al., 2006; Zietsch et al., 2007) have demonstrated high heritability of the alpha rhythm, but little is known about its underlying genetic determinants. Here, we adopted a sib‐pair design that allowed us to estimate the heritability of 10‐Hz parieto‐occipital rhythmic activity and attempt genetic mapping of neural features to specific genetic loci.
Materials and methods
Subjects
Altogether 210 Finnish‐speaking adults participated in the study. The subject group consisted of 91 full‐sibling pairs, eight families with three siblings and one family with four siblings (mean ± SEM age 30 ± 1 years; 148 females, 62 males; 206 right‐handed, three ambidextrous and one left‐handed; no monozygotic twins). None of the subjects had a history of neurological, psychiatric, or hearing disorders. All participants gave their written informed consent. The study conforms with the Declaration of Helsinki, and it had a prior approval from the Ethics Committee of the Hospital District of Helsinki and Uusimaa.
MEG experiment
Spontaneous cortical activity was recorded in a magnetically shielded room while the subject was seated with the head supported against the helmet‐shaped bottom of the 306‐channel Vectorview™ (Elekta Oy, Helsinki, Finland) neuromagnetometer. The device contains 102 identical triple sensors, with two orthogonal planar first‐order gradiometers that measure the tangential derivatives and of the magnetic field component normal to the helmet surface at the sensor location, and one magnetometer, each of them coupled to a SQUID (Superconducting QUantum Interference Device). The planar gradiometers detect the maximum signal directly above the active cortical area. Four head‐position‐indicator coils were attached to the scalp, and their positions were measured with a three‐dimensional digitizer; the head coordinate frame was anchored to the two periauricular points and the nasion. The head position with respect to the sensor array was determined by briefly feeding current to the marker coils before the actual measurement.
Three minutes of resting‐state data in both eyes‐closed and eyes‐open conditions were recorded. The MEG signals were band‐pass filtered to 0.03–200 Hz and sampled at 600 Hz. For external artifact suppression, the signal space separation method (Taulu & Kajola, 2005) was applied, and each individual MEG measurement was transferred to the same head position using a signal space separation ‐based head transformation algorithm (Taulu et al., 2004), implemented in MaxFilter™ software (Elekta Oy).
The power spectra in both eyes‐closed and eyes‐open conditions were estimated using Welch's method, with 4096‐point Fast Fourier Transformation resulting in frequency resolution of 0.15 Hz, eight data segments overlapping by 50% and Hamming windowing. We focused on the spectral peak at ~ 10 Hz. The relevant measures were its maximum amplitude, the frequency at which the maximum amplitude was reached, and the spectral width (full‐width at half maximum, determined based on the first frequency bin where the ascending slope reached 50% point of the maximum and the last bin where the descending slope remained above the 50% of the maximum; see Fig. 1B). Given that the sources of alpha activity are spatially distributed with typical source clusters bilaterally at the visuo‐occipital cortices and at the parieto‐occipital sulcus (Salmelin & Hari, 1994; Hari & Salmelin, 1997; Mäkelä et al., 1998; Manshanden et al., 2002), three separate subsets of planar gradiometers covering the parieto‐occipital cortex were selected for the analyses (left, middle, right; see Fig. 1A). In each of the three regions, at the channel pair showing the maximum ~ 10‐Hz amplitude, the spectral peak was quantified from the difference waveform (eyes closed – eyes open) as vector sum of the power spectra at the two gradiometer channels. Such sensor‐level measurements are well replicable and in accordance with results obtained by source modeling approaches (Virtanen et al., 1998). Importantly, these measures can be obtained with minimal subjective decisions on the data. Subsequently, the measured values were submitted to statistical testing, including heritability and linkage analyses (see below).
Figure 1.
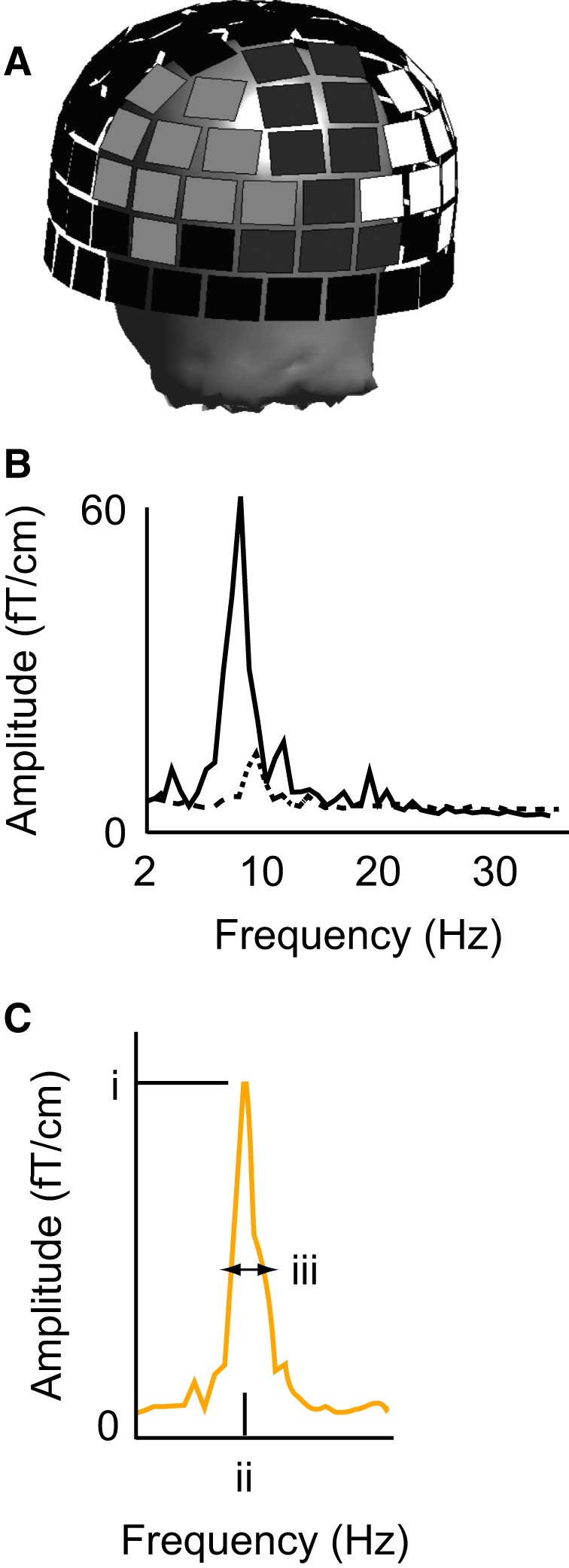
MEG signals. (A) Difference spectra between eyes‐closed and eyes‐open conditions were measured at the maximum channels over three sensor regions (marked with light gray, dark gray and white over the helmet). (B) Example responses from one subject at a representative channel in the eyes‐closed (solid line) and eyes‐open (dotted line) conditions. (C) Vector sum of each channel pair was used for estimating (i) the amplitude of the ~ 10‐Hz spectral peak, (ii) the exact frequency at which the maximum amplitude was reached, and (iii) the spectral width (difference between the 50% point of the ascending and descending slopes).
Genotyping
Deoxyribonucleic acid (DNA) was extracted from blood samples with FlexiGene DNA Kit (Qiagen, Hilden, Germany) and genotyped on Affymetrix 250K StyI Single‐Nucleotide Polymorphism (SNP) arrays (Affymetrix, Santa Clara, CA, USA). For all samples with an initial success rate > 92.9%, genotypes were called with Affymetrix Genotyping Console software using the BRLLM algorithm. Additional quality control filtering was done using Plink version 1.07 (Purcell et al., 2007; http://pngu.mgh.harvard.edu/purcell/plink/). The exclusion criteria for SNPs were as follows: (i) genotyping success rate < 98%; (ii) minor allele frequency < 5% and (iii) deviation from Hardy–Weinberg equilibrium with P < 0.0001 in two subsets of 98 unrelated samples. Individuals with genotyping success rate < 95% were excluded. The relatedness of siblings and the unrelatedness of other pairs of individuals was then checked based on levels of allele sharing. In addition, individuals with no remaining siblings were removed, resulting in a dataset of 203 individuals and 153 640 SNPs. For linkage analysis, SNPs were further pruned based on linkage disequilibrium (LD; r 2 > 0.1) to eliminate the inflation of linkage measures. After the pruning, 203 individuals and 28 184 autosomal SNPs remained (average distance between SNPs ≈ 0.1 cm). The lowest genotyping success per individual was 97.9% in the pruned and 98.1% in the non‐pruned dataset, and the overall genotyping success rate in each dataset was 99.8%.
Heritability, linkage and association analyses and phenotypic correlations
Phenotype heritabilities were calculated based on all successfully phenotyped and genotyped individuals (n = 203) using the variance component option of Merlin 1.1.2 (Abecasis et al., 2002). The genotypes were checked using the error detection and correction option of Merlin, and the likely erroneous genotypes were coded as missing. A linkage analysis (n = 203) was conducted with the variance component option in Merlin, using the built‐in LD correction feature to account for the density of SNPs (Abecasis & Wigginton, 2005) and the built‐in normality correction feature (InverseNormal), because most of the phenotypes were not normally distributed and did not yield well to standard normality corrections.
The genome‐wide empirical significance of the observed logarithm of odds (LOD) scores was determined by simulations. Datasets (n = 1001) without any genetic linkages but with phenotypes, relatedness between individuals, and missing genotype patterns identical to the real data were simulated in Merlin. For each simulation and phenotype, the highest simulated LOD score per chromosome was recorded, resulting in an empirical distribution of LOD scores. The significance of the observed LOD scores was evaluated by counting from the distribution of simulated LODs the expected number of linkage peaks (n exp) with a LOD score equal to or higher than the observed LOD score. Thus, we estimated how many times per simulation, on average, a LOD score exceeding the observed one was detected. The lower the n exp, the less likely it is to obtain as high a LOD score by chance, without the involvement of any genes. According to the standard guidelines by Lander & Kruglyak (1995), observed LOD scores with n exp < 0.05 were declared significant and those with n exp < 1 suggestive.
Because linkage scans were performed for several phenotypes, the overall significance level was corrected for multiple comparisons. First, the best simulated LOD scores per chromosome and across simulations (i.e., n = 22*1001 LOD scores) were ranked for a given phenotype. Then the observed LOD scores for that phenotype were compared to the simulated ones, and the rank(s) of the observed nominally significant/suggestive LOD(s) were recorded. The ranks for the k observed significant or suggestive linkage peaks were then compared to the k best ranks (across phenotypes) obtained on each simulation round. The probability of having observed the k significant or suggestive LODs by chance was calculated as the proportion of the simulation rounds where the observed LODs were exceeded, so that the best simulated LOD exceeded the best observed LOD in rank, the second‐best simulated LOD exceeded the second‐best observed LOD in rank, etc., to the ranks of the kth simulated vs. observed LOD.
Association analysis was performed with r package genabel (Aulchenko et al., 2007), using a specific mixed linear model called GRAMMAR‐gamma (Svischeva et al., 2012). This method produces a correct distribution of the test statistics and unbiased estimates of regression coefficients. It corrects for the relatedness of the individuals by estimating a relatedness matrix from the genetic data and using this as a random effect in the mixed model.
Phenotype correlations between siblings were calculated using Spearman's correlation in r version 3.0.1. Within each family, siblings were randomly designated as Sibling 1 and Sibling 2 (and Sibling 3 in the families represented by three individuals), and the correlation between Sibling 1 and Sibling 2 was calculated for each phenotype. Across 100 such random designations, the median rho and P value of the correlation (rho > 0) were recorded. Phenotype correlation between unrelated individuals was calculated by similar random assignment of 100 sets of 98 pairs of unrelated individuals. Correlations between the nine phenotypes were calculated using all phenotyped individuals (n = 210).
Results
We first considered the within‐individual replicability and inter‐individual variability in different measures of the signal patterns. For individual participants, the peak width and peak amplitude of the 10‐Hz spectral peak (eyes‐closed vs. eyes‐open conditions) over the middle parieto‐occipital region remained fairly consistent even across measurements separated by 16 months, whereas the peak frequencies varied (Fig. 2A). The spectral peak amplitudes were also very similar between siblings (Fig. 2B, Table 1; Spearman's rho = 0.42, P < 0.001) but varied by more than tenfold between pairs of unrelated individuals (Fig. 2C; Spearman's rho = −0.013, P = 0.55). At individual level, correlations between the phenotypes from the left, middle and right parieto‐occipital region were relatively high for each of the three measures (Spearman's rho 0.54–0.62 for the spectral peak width, 0.73–0.78 for the peak frequency and 0.83–0.93 for the peak amplitude), whereas correlations between the three measures were generally lower (Spearman's rho from −0.055 to 0.12 between spectral peak width and frequency, from −0.036 to −0.082 between peak amplitude and frequency and from −0.33 to −0.54 between spectral peak width and amplitude).
Figure 2.
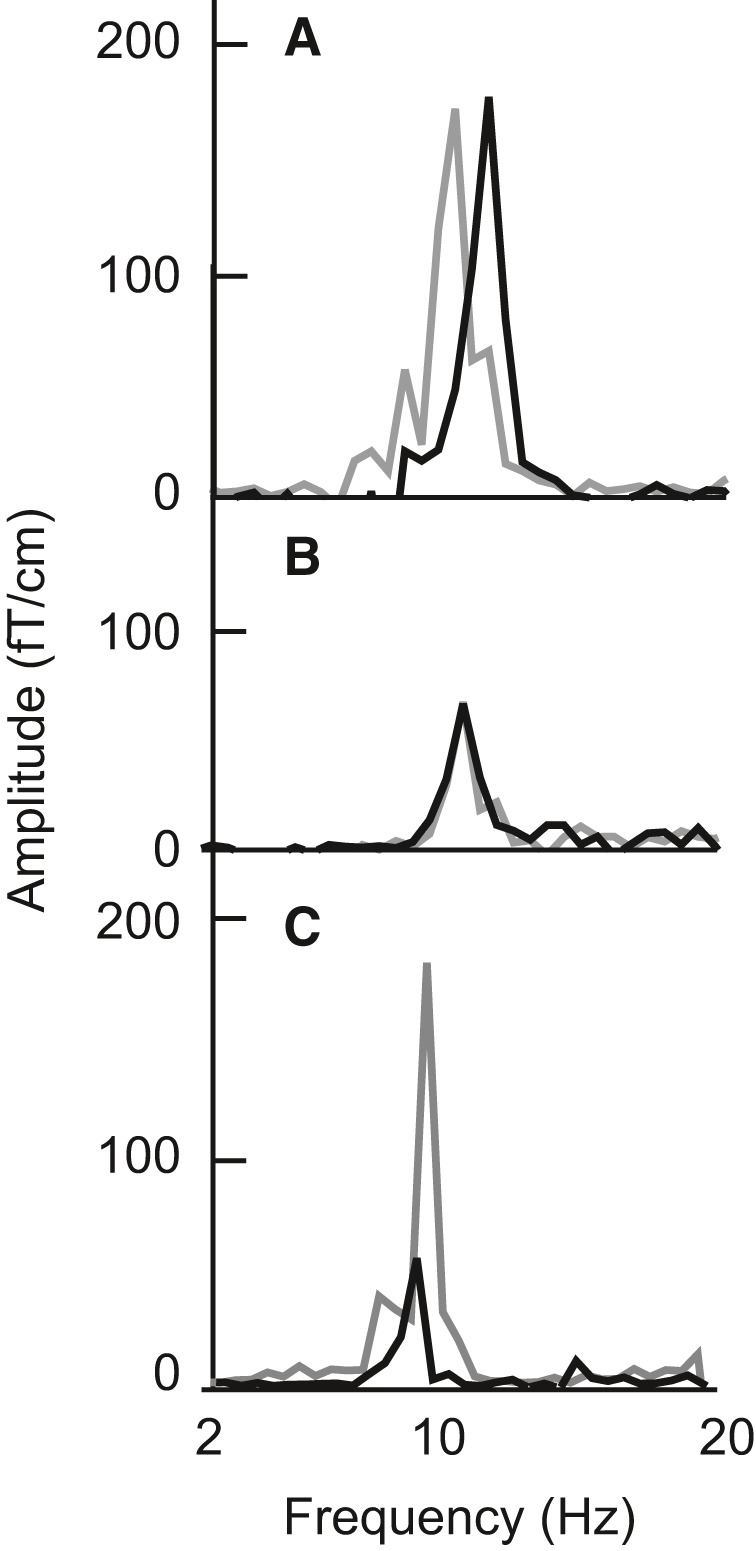
Examples of the difference spectra (eyes closed vs. eyes open) over the middle parieto‐occipital region in one subject at two measurements separated by 16 months (A), in two siblings (B) and in two unrelated subjects (C).
Table 1.
Phenotype distributions and heritabilities
| Phenotype | h 2 value | Median | Q1 | Q3 | rho | P value |
|---|---|---|---|---|---|---|
| L peak amplitude | 0.83 | 126.96 | 41.15 | 254.00 | 0.40 | 0.000030 |
| M peak amplitude | 0.87 | 143.42 | 46.30 | 289.24 | 0.42 | 0.000010 |
| R peak amplitude | 0.75 | 59.30 | 20.87 | 118.75 | 0.36 | 0.00018 |
| L peak width | 0.36 | 0.88 | 0.44 | 1.25 | 0.17 | 0.046 |
| M peak width | 0.12 | 0.73 | 0.44 | 1.32 | 0.055 | 0.30 |
| R peak width | 0.36 | 0.88 | 0.59 | 1.54 | 0.14 | 0.091 |
| L peak frequency | 0.17 | 10.03 | 9.30 | 10.55 | 0.073 | 0.24 |
| M peak frequency | 0.48 | 10.11 | 9.67 | 10.62 | 0.23 | 0.011 |
| R peak frequency | 0.44 | 9.96 | 9.38 | 10.55 | 0.23 | 0.011 |
The heritability estimates (h 2) of the measured 10‐Hz spectral peak phenotypes (from Merlin; n = 203), their median, first quartile (Q1) and third quartile (Q3) values calculated from 100 unrelated subjects, and their Spearman correlation between siblings (rho, P value). L, left; M, middle; R, right.
We calculated heritability (h 2) values for phenotypes with large variation between subjects (spectral peak width and amplitude; Table 1, Fig. S1) as well as for spectral peak frequency that had been reported heritable in previous EEG studies (van Beijsterveldt & van Baal, 2002; Smit et al., 2006). The measured spectral peak amplitudes showed high heritability (h 2 > 0.75), also in accordance with earlier EEG results (van Beijsterveldt et al., 1996; van Beijsterveldt & van Baal, 2002; Smit et al., 2006; Zietsch et al., 2007), whereas the heritability did not exceed 0.5 for frequency or spectral peak width (Table 1).
We proceeded to genetic linkage analysis using 28 184 pruned autosomal SNPs and the variance component option of Merlin. The linkage analysis of the nine phenotypes revealed one nominally significant and 11 suggestive linkage peaks; altogether seven linkage peaks were seen for the spectral peak width phenotypes, four for the amplitudes, and one for the frequencies (Figs 3 and 4; Table 2). The overall probability of observing an equally significant result by chance in nine linkage scans was P = 0.046. The significant linkage was observed on chromosome 10q23.2 for the phenotype of left hemisphere spectral peak width. The effect seemed fairly localized to the left side, as no linkage at this locus was found for the middle or right‐sided signals (Fig. 3).
Figure 3.

Linkage analysis for ‘spectral peak width’ phenotype. LOD scores of variance component‐based linkage analysis between the three width‐value phenotypes and ∼28 000 autosomal SNPs, plotted against genomic location in centimorgan units. The threshold LOD scores for significant and suggestive results are indicated by continuous and dashed lines, respectively. The gray‐and‐white shading denotes the succession of even‐ and odd‐numbered chromosomes.
Figure 4.
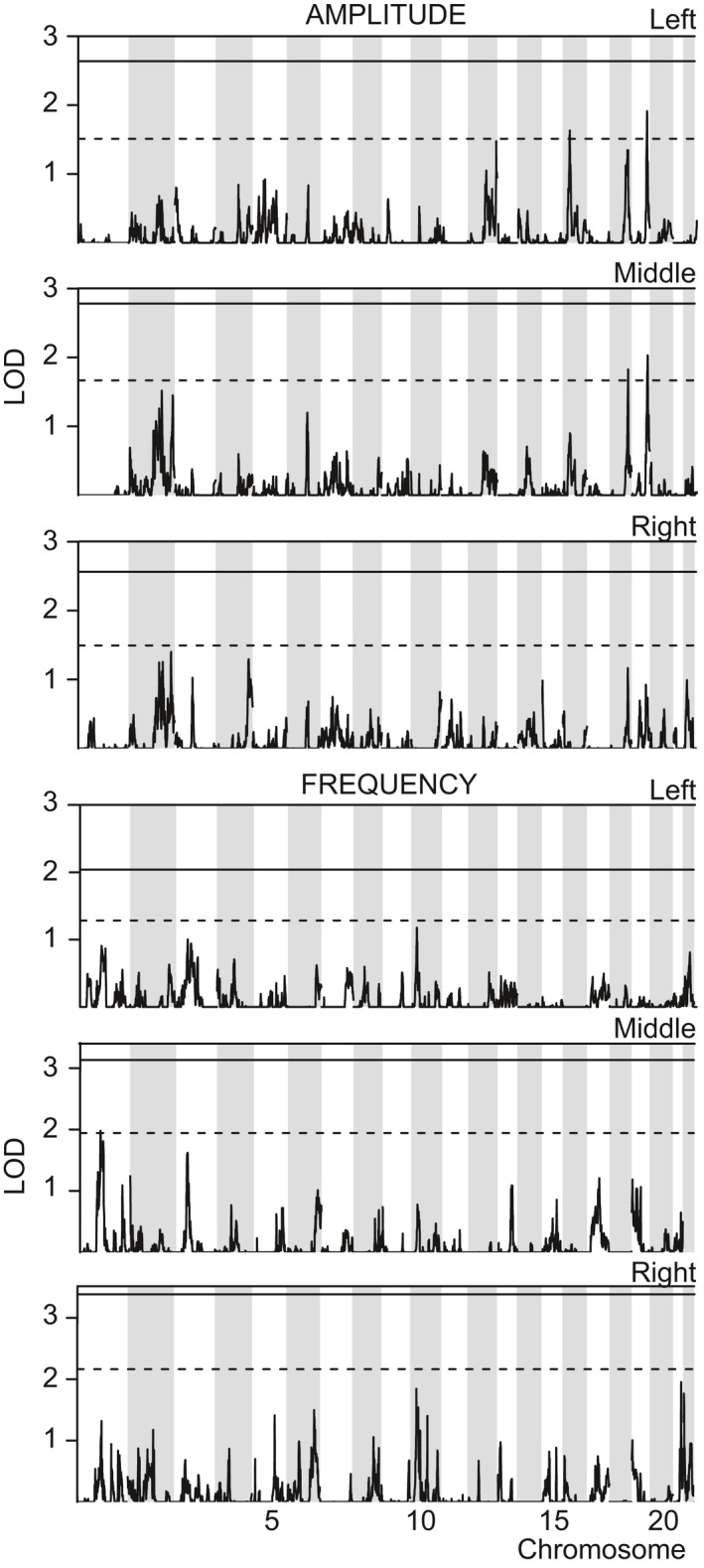
Linkage analysis for ‘spectral peak amplitude’ and ‘spectral peak frequency’ phenotypes. LOD scores of variance component‐based linkage analysis between the phenotypes and ∼28 000 autosomal SNPs, plotted against genomic location in centimorgan units. The threshold LOD scores for significant and suggestive results are indicated by continuous and dashed lines, respectively. The gray‐and‐white shading denotes the succession of even‐ and odd‐numbered chromosomes.
Table 2.
Linkage analysis results
| Phenotype | chr | LOD | n exp | Best SNP | Location |
|---|---|---|---|---|---|
| L peak width | 10 | 2.814 | 0.034 | rs10509410 | Intronic in PAPSS2 |
| R peak width | 16 | 2.339 | 0.146 | rs11861062 | Intergenic |
| M peak width | 2 | 1.118 | 0.208 | rs9288662 | Intronic in SP140 |
| L peak amplitude | 19 | 1.912 | 0.373 | rs1293703 | 3′in DPRX |
| M peak amplitude | 19 | 2.031 | 0.390 | rs269940 | Intronic in NLRP7 |
| R peak width | 8 | 1.996 | 0.414 | rs7015861 | Intronic in FAM91A1 |
| M peak amplitude | 18 | 1.827 | 0.661 | rs9945030 | Intronic in RP11‐47G4.2 |
| L peak amplitude | 16 | 1.631 | 0.738 | rs226042 | Intronic in CRYM |
| R peak width | 13 | 1.775 | 0.804 | rs7333894 | Intergenic |
| M peak frequency | 1 | 1.980 | 0.902 | rs11161750 | Intronic in COL24A1 |
| M peak width | 7 | 0.849 | 0.941 | rs10486813 | Intronic in SUGCT |
| L peak width | 18 | 1.802 | 0.949 | rs676603 | Intronic in AQP4‐AS1 |
Suggestive and significant linkages for the nine phenotypes, along with their chromosomal location (chr), significance based on 1001 permutations (n exp), name of best‐linked SNP and its location relative to genes. PAPSS2, 3′‐phosphoadenosine 5′‐phosphosulfate synthase 2; SP140, SP140 nuclear body protein; DPRX, divergent‐paired related homeobox; NLRP7, NLR family, pyrin domain‐containing 7; FAM91A1, family with sequence similarity 91 member A1; RP11‐47G4.2, gene RP11‐47G4.2; CRYM, crystallin mu; COL24A1, collagen, type XXIV, alpha 1; SUGCT, succinyl‐CoA,glutarate‐CoA transferase; AQP4‐AS1, AQP4 antisense RNA 1. L, left; M, middle; R, right parieto‐occipital region.
On chromosome 10, the region with strongest evidence of linkage (defined as the area with at least suggestive linkage, i.e., LOD > 1.778) spanned approximately 12.7 Mb (from 79.8 Mb to 92.4 Mb on the hg38 map assembly; Fig. 5A). The region contained 144 annotated loci (Table S1), including 65 protein‐coding genes (Fig. 5B). Many of the genes in the region are expressed in the brain, either specifically or ubiquitously, but not all are well annotated for function. Genes with suggestively interesting functions relevant for this study include GRID1 (glutamate receptor, ionotropic, delta 1), ATAD1 (ATPase family, AAA domain‐containing, member 1), NRG3 (neuregulin‐3) and HTR7 (5‐hydroxytryptamine receptor 7).
Figure 5.
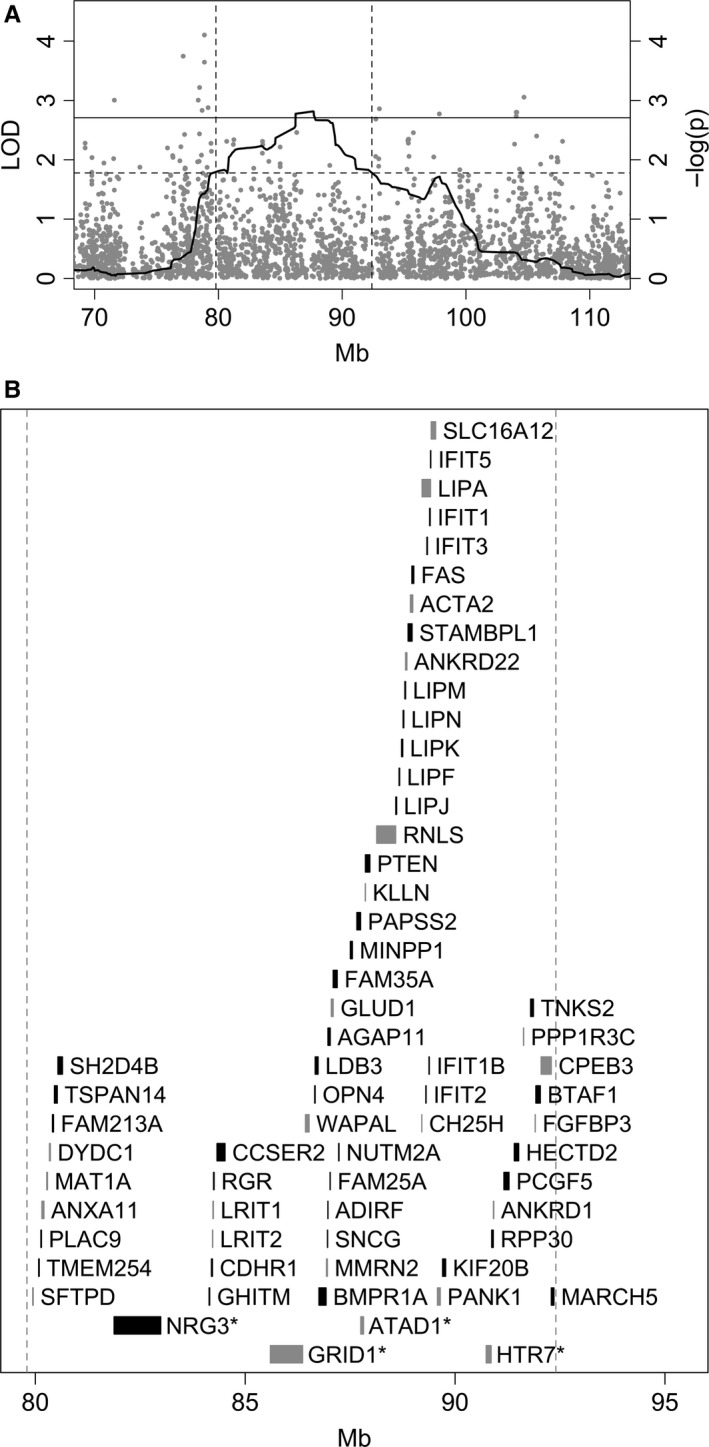
The linkage region on chromosome 10. (A) Association results for left‐hemisphere spectral peak width in and adjacent to the linkage peak. The SNPs tested for association are depicted as gray dots, plotted according to their genomic location on the x axis and –log(P) value from the association test on the y axis. The linkage analysis LOD score across the region is shown as bold black curve, with horizontal lines depicting the empirical thresholds for significant linkage (solid line) and suggestive linkage (dashed line). Vertical lines mark the region of suggestive linkage. (B) The location of all known protein‐coding genes in the linkage region from A, with color denoting their direction of transcription (black, from centromeric to telomeric; gray, from telomeric to centromeric). The four brain‐related genes treated in Discussion are on the two bottom lines, denoted by an asterisk at the end of their name.
An association analysis of the non‐LD‐pruned dataset (153 640 SNPs) revealed a suggestive association with left‐hemisphere spectral peak width close to the corresponding linkage peak on chromosome 10 (Fig. 5A). The region was located directly adjacent to the suggestive linkage peak region and contained five SNPs with nominal P value smaller than 0.001; the most significantly associated SNP was rs529442 with P = 7.91e‐05. The second‐best associated SNP (rs17391134, P = 1.80e‐04) was situated intronically in the KCNMA1 gene (Homo sapiens potassium channel, calcium activated large conductance subfamily M alpha, member 1). However, considering the number of SNPs tested (n > 1600 within the linkage peak region), the observed significances do not survive correction for multiple testing.
Discussion
In this study, we focused on the genetic basis of the human 10‐Hz rhythm, measured with MEG, by studying the heritability of selected phenotypic measures and performing a genetic linkage mapping study in a sib‐pair setup. Our present MEG data and heritability analysis confirm earlier observations of high heritability for peak amplitude of the 10‐Hz rhythm (van Beijsterveldt et al., 1996; van Beijsterveldt & van Baal, 2002; Smit et al., 2006; Zietsch et al., 2007). Furthermore, we observed significant genetic linkage for left‐hemisphere spectral peak width emerging in the genome‐wide analysis for one locus, and suggestive linkage for 11 loci across phenotypes, which would be an empirically unlikely finding in randomized data (P = 0.046).
The heritability estimates for alpha peak amplitudes (0.75–0.87) resembled closely those reported in the EEG literature (e.g., van Beijsterveldt & van Baal, 2002). However, the heritability estimates for peak frequencies were lower than reported in the earlier EEG studies. Part of this discrepancy may be explained by our estimates being based on the difference spectra between eyes‐closed and eyes‐open conditions, whereas the EEG studies have typically reported peak frequencies while subjects have their eyes closed. Importantly, EEG and MEG measurements reflect partially different neuronal populations, and the alpha peak frequency has been demonstrated to vary between EEG and MEG recordings within the same individuals (Srinivasan et al., 2006). This is most likely related to EEG picking up both frontal and occipital 10‐Hz activity and being sensitive to radial and tangential distributed neuronal layers, while MEG is rather restricted to tangentially oriented activity in the occipital region with very little frontal contribution to the measurements (Srinivasan et al., 2006).
The spectral width of the alpha peak probably reflects variability in the subjects’ alpha frequency range during the measurement. Alpha wave bandwidth is known to differ between individuals (Niedermeyer, 2004), and its morphology has been related for example to subjects’ memory performance (Klimesch et al., 2000). For migraine patients, alpha bandwidth increased during preattack period (Bjørk et al., 2009), and patients with thalamic infarctions showed spreading of alpha rhythm to lower frequencies (Mäkelä et al., 1998). The cellular‐level mechanisms behind the variations in alpha peak width have earlier been related to desynchronization in the cortical, or thalamocortical, neural generators (Bjørk et al., 2009). Alpha peak frequency has been shown to increase together with cognitive load, possibly related to, e.g., engagement of different neuronal networks (Haegens et al., 2014).
Our results are in line with earlier studies in suggesting that the variability in the alpha rhythm is partly genetically regulated; in our study the finding was strongest for the spectral peak width. Obviously, we cannot exclude weak genetic effects for any of the suggestively linked loci or loci undetected here, but focus here only on the one significant locus. The significantly linked region contains four genes with known relevance to brain function. Several other genes in the region are also expressed in the brain according to the comprehensive FANTOM5 database (FANTOM, 2014), but as their functions and relevance to brain physiology are poorly known, we here exclude them from discussion.
Glutamate receptor channels are responsible for the major part of the rapid, excitatory synaptic transmission in the brain, and they have been implicated also in synaptic plasticity (Yamazaki et al., 1992; Debanne et al., 2003). GRID1, located nearest to the linkage peak along with ATAD1, is a subunit in the glutamate receptor channels, and it is highly expressed in different parts of the brain with almost no expression in peripheral tissues (Nagase et al., 1999). GRID1 has been associated with susceptibility to schizophrenia in several independent studies (Fallin et al., 2005; Guo et al., 2007; Zhu et al., 2009), and in healthy subjects to gray matter variation in prefrontal and anterior thalamic regions (Nenadic et al., 2012). ATAD1 is involved in the regulation of cell surface expression of neuronal glutamate receptor complex proteins, specifically GRIP1 (glutamate receptor‐interacting protein 1) and GLUR2 (glutamate receptor 2) with which it interacts and which bind and regulate AMPA (alpha‐amino‐3‐hydroxy‐5‐methyl‐4‐isoxazole propionate) type receptors postsynaptically (Dai et al., 2010; Zhang et al., 2011). Atad1 knock‐out mice have severe seizures, and deficits in short‐term memory and in spatial working memory tasks (Zhang et al., 2011). Thus, either GRID1 or ATAD1 might have a specific functional effect in the regulation of brain rhythmic activity through genetic variation affecting glutamate signaling pathways.
Neuregulins, including NRG3 located in the present linkage region on chromosome 10, are a family of proteins that are expressed specifically in neurons (Zhang et al., 1997; Carteron et al., 2006). NRG3 binds specifically the ERBB4 receptor tyrosine kinase and stimulates its phosphorylation (Zhang et al., 1997). Developmental NRG3 overexposure results in deficits in social behavior in adult mice (Paterson & Law, 2014), and, in humans, variations in NRG3 have been associated with several neuropsychiatric and neurocognitive disorders, such as schizophrenia (Fallin et al., 2003; Faraone et al., 2006; Wang et al., 2008; Kao et al., 2010) and autism (Balciuniene et al., 2007).
Serotonin, or 5‐hydroxytryptamine, can bind to a number of distinct receptors, one of which is HTR7 in the present linkage region on chromosome 10. The serotonin receptors are G‐protein coupled receptors and can use alternative downstream signaling pathways; HTR7 signals through adenylate cyclase activation. In addition to neurotransmission, HTR7 has been shown to structurally regulate brain reward pathways postnatally (Adriani et al., 2006; Leo et al., 2009), modulate hippocampal neuronal morphology (Kobe et al., 2012) and enhance neurite outgrowth (Speranza et al., 2013). Furthermore, HTR7 has been associated with frontal brain theta oscillations (Zlojutro et al., 2011), circadian rhythms and the sleep‐wake cycle (Leopoldo et al., 2011; Adriani et al., 2012).
Conceivably, any of these genes, with highly specific brain expression patterns, relevant functions in synaptic signaling, and association with brain oscillations, might be candidates to control part of the variation in the 10‐Hz rhythmic activity. The cellular‐level mechanisms underlying the thalamo‐cortico‐thalamic loop of 10‐Hz oscillations remain under debate, but they seem to involve the GABAergic connections of the reticular nucleus (Steriade, 1999). In addition, the activation of glutamate receptors (mGluR) in the thalamus has been shown to induce rhythmic activity resembling alpha oscillations (Hughes et al., 2004), with a suggested role in blocking unwanted stimuli from reaching the cortex (Vijayan & Kopell, 2012). The role of glutamergic functions in the generation of alpha oscillation is especially interesting in light of the potential genetic link to GRID1 and ATAD1 proposed by our present findings. Another plausible link in this study points to HTR7 – serotonergic HTR7 receptors are highly abundant in the thalamus (Ruat et al., 1993), and serotonergic hallucinogen psilocybin was recently shown to modulate cortical alpha activity in humans (Kometer et al., 2013).
Left‐lateralized modulation of alpha activity has been recently described in relation to suppression of task‐irrelevant information (Okazaki et al., 2014), and genetic correlates for functional differences in the brain have been reported (e.g., Darki et al., 2012). Hemispheric differences in the functionality of alpha activity would thus be in reasonable agreement with our finding that significant genetic linkage for 10‐Hz spectral peak width was observed exclusively in the left hemisphere. On a more general level, our finding might point to a genetic architecture of the phenotype where the regulatory loci and/or their effect sizes differ between the hemispheres. The observed lateralized genetic linkage could thus stem from an influence of a larger number of genes on the right than left hemispheric activity, with individual genes having too small effects to be detected in this study, or it may suggest a more profound stochastic or environmental component on the right hemispheric activity.
Similarly, genetic architecture may explain the observed differences in heritability (highest for peak amplitude) vs. linkage results (significant for spectral peak width) – peak amplitude may be governed by a combination of small effects of many genes, which this study has limited power to detect, whereas the regulation of spectral peak width may be more oligogenic, and its large‐effect determinant(s) therefore easier to locate. However, we also wish to point out that a part of the difference – as well as of the lateralization discussed above – may result from the limited power of this study, leading to imprecise estimates of the heritabilities and linkage strength and to suggestive rather than significant linkage signals; obviously, replication studies will be needed to determine the significance of the suggestive linkages detected here. Further increases in resolution and power could be achieved by larger sample sizes, still within practical limits. Importantly, our previous results (Renvall et al., 2012) and those reported here emphasize the applicability and promise of the present genetic mapping scheme for selected functional brain measures, even at modest sample sizes. Given that not only the alpha rhythm but also all other prominent spectral bands of the human EEG/MEG, i.e., delta, theta, beta and gamma rhythms have been demonstrated to be highly heritable (van Beijsterveldt et al., 1996; van Beijsterveldt & van Baal, 2002; Zietsch et al., 2007; van Pelt et al., 2012), studies on their genetic determinants would be of high interest for further uncovering their functional roles.
Conflict of interest
The authors declare no conflicting interests.
Supporting information
Fig. S1. Distributions of the phenotype measurements for spectral peak width (A), peak frequency (B) and peak amplitude (C). The distributions depict two sets of 98 unrelated individuals (Set1 and Set2) and measurements from the left (L), middle (M) and right (R) parieto‐occipital region.
Table S1. List of annotated loci in the linkage peak region on chromosome 10.
Acknowledgements
This study was supported by Biocentrum Helsinki (ES), Sigrid Juselius Foundation (JKe, RS), Jenny and Antti Wihuri Foundation (ES), Jane and Aatos Erkko Foundation (JKe), Academy of Finland (grant #127401 and #277655 for HR, #257576 for JKu and #255349, #256459 and #283071 for RS) and Finnish Cultural Foundation (OH, MV). The funding sources had no role in study design, analysis, manuscript writing or submitting decision. We thank Riitta Lehtinen and Ingegerd Fransson for expert technical assistance.
Abbreviations
- DNA
deoxyribonucleic acid
- EEG
electroencephalography
- LD
linkage disequilibrium
- LOD
logarithm of odds
- MEG
magnetoencephalography
- SNP
single‐nucleotide polymorphism
References
- Abecasis, G.R. & Wigginton, J.E. (2005) Handling marker‐marker linkage disequilibrium: pedigree analysis with clustered markers. Am. J. Hum. Genet., 77, 754–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abecasis, G.R. , Cherny, S.S. , Cookson, W.O. & Cardon, L.R. (2002) Merlin–rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet., 30, 97–101. [DOI] [PubMed] [Google Scholar]
- Adrian, E.D. (1944) Brain rhythms. Nature, 153, 360–362. [Google Scholar]
- Adrian, E.D. & Matthews, B.H.C. (1934) The Berger rhythm: potential changes from the occipital lobes in man. Brain, 57, 355–384. [DOI] [PubMed] [Google Scholar]
- Adriani, W. , Leo, D. , Greco, D. , Rea, M. , di Porzio, U. , Laviola, G. & Perrone‐Capano, C. (2006) Methylphenidate administration to adolescent rats determines plastic changes on reward‐related behavior and striatal gene expression. Neuropsychopharmacology, 31, 1946–1956. [DOI] [PubMed] [Google Scholar]
- Adriani, W. , Travaglini, D. , Lacivita, E. , Saso, L. , Leopoldo, M. & Laviola, G. (2012) Modulatory effects of two novel agonists for serotonin receptor 7 on emotion, motivation and circadian rhythm profiles in mice. Neuropharmacology, 62, 833–842. [DOI] [PubMed] [Google Scholar]
- Amzica, F. & Lopes da Silva, F.H. (2011) Cellular substrates of brain rhythms In Schomer D.L. & Lopes da Silva F.H. (Eds), Niedermeyer's Electroencephalography: Basic Principles, Clinical Applications and Related Fields, 6th Edn Lippincott Williams & Wilkins, Philadelphia, pp. 33–64. [Google Scholar]
- Aulchenko, Y.S. , Ripke, S. , Isaacs, A. & van Duijn, C.M. (2007) GenABEL: an R library for genome‐wide association analysis. Bioinformatics, 23, 1294–1296. [DOI] [PubMed] [Google Scholar]
- Balciuniene, J. , Feng, N. , Iyadurai, K. , Hirsch, B. , Charnas, L. , Bill, B.R. , Easterday, M.C. , Staaf, J. et al (2007) Recurrent 10q22–q23 deletions: a genomic disorder on 10q associated with cognitive and behavioral abnormalities. Am. J. Hum. Genet., 80, 938–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Beijsterveldt, C.E.M. & van Baal, G.C.M. (2002) Twin and family studies of the human electroencephalogram: a review and a meta‐analysis. Biol. Psychol., 61, 111–138. [DOI] [PubMed] [Google Scholar]
- van Beijsterveldt, C.E. , Molenaar, P.C. , de Geus, E.J. & Boomsma, D.I. (1996) Heritability of human brain functioning as assessed by electroencephalography. Am. J. Hum. Genet., 58, 562–573. [PMC free article] [PubMed] [Google Scholar]
- Berger, H. (1929) Über das Elektrenkephalogramm des Menschen. Archiv für Psychiatrie und Nervenkrankheiten, 87, 527–570. [Google Scholar]
- Bjørk, M.H. , Stovner, L.J. , Nilsen, B.M. , Stjern, M. , Hagen, M. & Sand, T. (2009) The occipital alpha rhythm related to the “migraine cycle” and headache burden: A blinded, controlled longitudinal study. Clin. Neurophysiol., 120, 464–471. [DOI] [PubMed] [Google Scholar]
- Bollimunta, A. , Mo, J. , Schroeder, C.E. & Ding, M. (2011) Neuronal mechanisms and attentional modulation of corticothalamic α oscillations. J. Neurosci., 31, 4935–4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carteron, C. , Ferrer‐Montiel, A. & Cabedo, H. (2006) Characterization of a neural‐specific splicing form of the human neuregulin 3 gene involved in oligodendrocyte survival. J. Cell Sci., 119, 898–909. [DOI] [PubMed] [Google Scholar]
- Dai, C. , Liang, D. , Li, H. , Sasaki, M. , Dawson, T.M. & Dawson, V.L. (2010) Functional identification of neuroprotective molecules. PLoS One, 5, e15008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darki, F. , Peyrard‐Janvid, M. , Matsson, H. , Kere, J. & Klingberg, T. (2012) Three dyslexia susceptibility genes, DYX1C1, DCDC2, and KIAA0319, affect temporo‐parietal white matter structure. Biol. Psychiat., 72, 671–676. [DOI] [PubMed] [Google Scholar]
- Debanne, D. , Daoudal, G. , Sourdet, V. & Russier, M. (2003) Brain plasticity and ion channels. J. Physiology‐Paris, 97, 403–414. [DOI] [PubMed] [Google Scholar]
- Fallin, M.D. , Lasseter, V.K. , Wolyniec, P.S. , McGrath, J.A. , Nestadt, G. , Valle, D. , Liang, K.Y. & Pulver, A.E. (2003) Genomewide linkage scan for schizophrenia susceptibility loci among Ashkenazi Jewish families shows evidence of linkage on chromosome 10q22. Am. J. Hum. Genet., 73, 601–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallin, M.D. , Lasseter, V.K. , Avramopoulos, D. , Nicodemus, K.K. , Wolyniec, P.S. , McGrath, J.A. , Steel, G. , Nestadt, G. et al (2005) Bipolar I disorder and schizophrenia: a 440‐single‐nucleotide polymorphism screen of 64 candidate genes among Ashkenazi Jewish case‐parent trios. Am. J. Hum. Genet., 77, 918–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FANTOM Consortium and the RIKEN PMI and CLST (DGT) (2014) A promoter‐level mammalian expression atlas. Nature, 507, 462–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraone, S.V. , Hwu, H.G. , Liu, C.M. , Chen, W.J. , Tsuang, M.M. , Liu, S.K. , Shieh, M.H. , Hwang, T.J. et al (2006) Genome scan of Han Chinese schizophrenia families from Taiwan: confirmation of linkage to 10q22.3. Am. J. Psychiat., 163, 1760–1766. [DOI] [PubMed] [Google Scholar]
- Gray, M.J. , Frey, H.P. , Wilson, T.J. & Foxe, J.J. (2015) Oscillatory recruitment of bilateral visual cortex during spatial attention to competing rhythmic inputs. J. Neurosci., 35, 5489–5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, S.Z. , Huang, K. , Shi, Y.Y. , Tang, W. , Zhou, J. , Feng, G.Y. , Zhu, S.M. , Liu, H.J. et al (2007) A case‐control association study between the GRID1 gene and schizophrenia in the Chinese Northern Han population. Schizophr. Res., 93, 385–390. [DOI] [PubMed] [Google Scholar]
- Haegens, S. , Osipova, D. , Oostenveld, R. & Jensen, O. (2010) Somatosensory working memory performance in humans depends on both engagement and disengagement of regions in a distributed network. Hum. Brain Mapp., 31, 26–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haegens, S. , Cousijn, H. , Wallis, G. , Harrison, P.J. & Nobre, A.C. (2014) Inter‐ and intra‐individual variability in alpha peak frequency. NeuroImage, 92, 46–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hari, R. & Salmelin, R. (1997) Human cortical oscillations: a neuromagnetic view through the skull. Trends Neurosci., 20, 44–49. [DOI] [PubMed] [Google Scholar]
- Hughes, S.W. , Lorincz, M. , Cope, D.W. , Blethyn, K.L. , Kekesi, K.A. , Parri, H.R. , Juhász, G. & Crunelli, V. (2004) Synchronized oscillations at alpha and theta frequencies in the lateral geniculate nucleus. Neuron, 42, 253–268. [DOI] [PubMed] [Google Scholar]
- Jensen, O. & Mazaheri, A. (2010) Shaping functional architecture by oscillatory alpha activity, gating by inhibition. Front. Hum. Neurosci., 4, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, O. , Gelfand, J. , Kounios, J. & Lisman, J.E. (2002) Oscillations in the alpha band (9‐12 Hz) increase with memory load during retention in a short‐term memory task. Cereb. Cortex, 12, 877–882. [DOI] [PubMed] [Google Scholar]
- Jokisch, D. & Jensen, O. (2007) Modulation of gamma and alpha activity during a working memory task engaging the dorsal or ventral stream. J. Neurosci., 27, 3244–3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao, W.T. , Wang, Y. , Kleinman, J.E. , Lipska, B.K. , Hyde, T.M. , Weinberger, D.R. & Law, A.J. (2010) Common genetic variation in Neuregulin 3 (NRG3) influences risk for schizophrenia and impacts NRG3 expression in human brain. Proc. Natl. Acad. Sci. USA, 107, 15619–15624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, S.P. , Gomez‐Ramirez, M. & Foxe, J.J. (2009) The strength of anticipatory spatial biasing predicts target discrimination at attended locations: a high‐density EEG study. Eur. J. Neurosci., 30, 2224–2234. [DOI] [PubMed] [Google Scholar]
- Klimesch, W. , Vogt, F. & Doppelmayr, M. (2000) Individual differences in alpha and theta power reflect memory performance. Intelligence, 27, 347–362. [Google Scholar]
- Klimesch, W. , Sauseng, P. & Hanslmayr, S. (2007) EEG alpha oscillations: The inhibition‐timing hypothesis. Brain Res. Rev., 53, 63–88. [DOI] [PubMed] [Google Scholar]
- Kobe, F. , Guseva, D. , Jensen, T.P. , Wirth, A. , Renner, U. , Hess, D. , Müller, M. , Medrihan, L. et al (2012) 5‐HT7R/G12 signaling regulates neuronal morphology and function in an age‐dependent manner. J. Neurosci., 32, 2915–2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kometer, M. , Schmidt, A. , Jäncke, L. & Vollenweider, F.X. (2013) Activation of serotonin 2A receptors underlies the psilocybin‐induced effects on α oscillations, N170 visual‐evoked potentials, and visual hallucinations. J. Neurosci., 33, 10544–10551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, E. & Kruglyak, L. (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat. Genet., 11, 241–247. [DOI] [PubMed] [Google Scholar]
- Leo, D. , Adriani, W. , Cavaliere, C. , Cirillo, G. , Marco, E.M. , Romano, E. , di Porzio, U. , Papa, M. et al (2009) Methylphenidate to adolescent rats drives enduring changes of accumbal Htr7 expression: implications for impulsive behavior and neuronal morphology. Genes Brain Behav., 8, 356–368. [DOI] [PubMed] [Google Scholar]
- Leopoldo, M. , Lacivita, E. , Berardi, F. , Perrone, R. & Hedlund, P.B. (2011) Serotonin 5‐HT7 receptor agents: structure‐activity relationships and potential therapeutic applications in central nervous system disorders. Pharmacol. Ther., 129, 120–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes da Silva, F. (2013) EEG and MEG: relevance to neuroscience. Neuron, 80, 1112–1128. [DOI] [PubMed] [Google Scholar]
- Lopes da Silva, F.H. , Vos, J.E. , Mooibroek, J. & Van Rotterdam, A. (1980) Relative contributions of intracortical and thalamo‐cortical processes in the generation of alpha rhythms, revealed by partial coherence analysis. Electroen. Clin. Neuro., 50, 449–456. [DOI] [PubMed] [Google Scholar]
- Mäkelä, J.P. , Salmelin, R. , Kotila, M. , Salonen, O. , Laaksonen, R. , Hokkanen, L. & Hari, R. (1998) Modification of neuromagnetic cortical signals by thalamic infarctions. Electroen. Clin. Neuro., 106, 433–443. [DOI] [PubMed] [Google Scholar]
- Manshanden, I. , De Munck, J.C. , Simon, N.R. & Lopes da Silva, F.H. (2002) Source localization of MEG sleep spindles and the relation to sources of alpha band rhythms. Clin. Neurophysiol., 113, 1937–1947. [DOI] [PubMed] [Google Scholar]
- Nagase, T. , Ishikawa, K. , Kikuno, R. , Hirosawa, M. , Nomura, N. & Ohara, O. (1999) Prediction of the coding sequences of unidentified human genes. XV. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res., 6, 337–345. [DOI] [PubMed] [Google Scholar]
- Nenadic, I. , Maitra, R. , Scherpiet, S. , Gaser, C. , Schultz, C.C. , Schachtzabel, C. , Smesny, S. , Reichenbach, J.R. et al (2012) Glutamate receptor delta 1 (GRID1) genetic variation and brain structure in schizophrenia. J. Psychiat. Res., 46, 1531–1539. [DOI] [PubMed] [Google Scholar]
- Niedermeyer, E. (2004) The normal EEG of the waking adult In Niedermeyer, E. & Lopes da Silva, F. (Eds), Electroencephalography. Basic Principles, Clinical Applications, and Related Fields, 5th Edn Lippincott Williams & Wilkins, Philadelphia, pp. 167–192. [Google Scholar]
- Okazaki, Y.O. , De Weerd, P. , Haegens, S. & Jensen, O. (2014) Hemispheric lateralization of posterior alpha reduces distracter interference during face matching. Brain Res., 1590, 56–64. [DOI] [PubMed] [Google Scholar]
- Palva, S. & Palva, J.M. (2007) New vistas for α‐frequency band oscillations. Trends Neurosci., 30, 150–158. [DOI] [PubMed] [Google Scholar]
- Paterson, C. & Law, A.J. (2014) Transient overexposure of neuregulin 3 during early postnatal development impacts selective behaviors in adulthood. PLoS One, 9, e104172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Pelt, S. , Boomsma, D.I. & Fries, P. (2012) Magnetoencephalography in twins reveals a strong genetic determination of the peak frequency of visually induced γ‐band synchronization. J. Neurosci., 32, 3388–3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poch, C. , Campo, P. & Barnes, G.R. (2014) Modulation of alpha and gamma oscillations related to retrospectively orienting attention within working memory. Eur. J. Neurosci., 40, 2399–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell, S. , Neale, B. , Todd‐Brown, K. , Thomas, L. , Ferreira, M.A. , Bender, D. , Maller, J. , Sklar, P. et al (2007) PLINK: a tool set for whole‐genome association and population‐based linkage analyses. Am. J. Hum. Genet., 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renvall, H. , Salmela, E. , Vihla, M. , Illman, M. , Leinonen, E. , Kere, J. & Salmelin, R. (2012) Genome‐wide linkage analysis of human auditory cortical activation suggests distinct loci on chromosomes 2, 3, and 8. J. Neurosci., 17, 14511–14518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruat, M. , Traiffort, E. , Leurs, R. , Tardivel‐Lacombe, J. , Diaz, J. , Arrang, J.M. & Schwartz, J.C. (1993) Molecular cloning, characterization, and localization of a high‐affinity serotonin receptor (5‐HT7) activating cAMP formation. Proc. Natl. Acad. Sci. USA, 90, 8547–8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saalmann, Y.B. , Pinsk, M.A. , Wang, L. , Li, X. & Kastner, S. (2012) The pulvinar regulates information transmission between cortical areas based on attention demands. Science, 337, 753–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salenius, S. , Kajola, M. , Thompson, W.L. , Kosslyn, S. & Hari, R. (1995) Reactivity of magnetic parieto‐occipital alpha rhythm during visual imagery. Electroen. Clin. Neuro., 95, 453–462. [DOI] [PubMed] [Google Scholar]
- Salmelin, R. & Hari, R. (1994) Characterization of spontaneous MEG rhythms in healthy adults. Electroen. Clin. Neuro., 91, 237–248. [DOI] [PubMed] [Google Scholar]
- Smit, C.M. , Wright, M.J. , Hansell, N.K. , Geffen, G.M. & Martin, N.G. (2006) Genetic variation of individual alpha frequency (IAF) and alpha power in a large adolescent twin sample. Int. J. Psychophysiol., 61, 235–243. [DOI] [PubMed] [Google Scholar]
- Speranza, L. , Chambery, A. , Di Domenico, M. , Crispino, M. , Severino, V. , Volpicelli, F. , Leopoldo, M. , Bellenchi, G.C. et al (2013) The serotonin receptor 7 promotes neurite outgrowth via ERK and Cdk5 signaling pathways. Neuropharmacology, 67, 155–167. [DOI] [PubMed] [Google Scholar]
- Srinivasan, R. , Winter, W.R. & Nunez, P.L. (2006) Source analysis of EEG oscillations using high‐resolution EEG and MEG. Prog. Brain Res., 159, 29–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steriade, M. (1999) Coherent oscillations and short‐term plasticity in corticothalamic networks. Trends Neurosci., 22, 337–345. [DOI] [PubMed] [Google Scholar]
- Svischeva, G. , Axenovich, T.I. , Belonogova, N.M. , van Duijn, C.M. & Aulchenko, Y.S. (2012) Rapid variance components‐based method for whole‐genome association analysis. Nat. Genet., 44, 1166–1170. [DOI] [PubMed] [Google Scholar]
- Taulu, S. & Kajola, M. (2005) Presentation of electromagnetic multichannel data: the signal space separation method. J. Appl. Phys., 97, 1–10. [Google Scholar]
- Taulu, S. , Kajola, M. & Simola, J. (2004) Suppression of interference and artefacts by the Signal Space Separation Method. Brain Topogr., 16, 269–275. [DOI] [PubMed] [Google Scholar]
- Tuladhar, A.M. , ter Huurne, N. , Schoffelen, J.M. , Maris, E. , Oostenveld, R. & Jensen, O. (2007) Parieto‐occipital sources account for the increase in alpha activity with working memory load. Hum. Brain Mapp., 28, 785–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayan, S. & Kopell, N.J. (2012) Thalamic model of awake alpha oscillations and implications for stimulus processing. Proc. Natl. Acad. Sci. USA, 109, 18553–18558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtanen, J. , Ahveninen, J. , Ilmoniemi, R. , Näätänen, R. & Pekkonen, E. (1998) Replicability of MEG and EEG measures of the auditory N1/N1 m‐response. Electroen. Clin. Neuro., 108, 291–298. [DOI] [PubMed] [Google Scholar]
- Wang, Y.C. , Chen, J.Y. , Chen, M.L. , Chen, C.H. , Lai, I.C. , Chen, T.T. , Hong, C.J. , Tsai, S.J. et al (2008) Neuregulin 3 genetic variations and susceptibility to schizophrenia in a Chinese population. Biol. Psychiat., 64, 1093–1096. [DOI] [PubMed] [Google Scholar]
- Worden, M.S. , Foxe, J.J. , Wang, N. & Simpson, G.V. (2000) Anticipatory biasing of visuospatial attention indexed by retinotopically specific alpha‐band electroencephalography increases over occipital cortex. J. Neurosci., 20, RC63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki, M. , Araki, K. , Shibata, A. & Mishina, M. (1992) Molecular cloning of a cDNA encoding a novel member of the mouse glutamate receptor channel family. Biochem. Biophys. Res. Commun., 183, 886–892. [DOI] [PubMed] [Google Scholar]
- Yener, G.G. & Başar, E. (2013) Brain oscillations as biomarkers in neuropsychiatric disorders: following an interactive panel discussion and synopsis. Suppl. Clin. Neurophys., 62, 343–363. [DOI] [PubMed] [Google Scholar]
- Zhang, D. , Sliwkowski, M.X. , Mark, M. , Frantz, G. , Akita, R. , Sun, Y. , Hillan, K. , Crowley, C. et al (1997) Neuregulin‐3 (NRG3): a novel neural tissue‐enriched protein that binds and activates ErbB4. Proc. Natl. Acad. Sci. USA, 94, 9562–9657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , Wang, Y. , Chi, Z. , Keuss, M.J. , Pai, Y.M. , Kang, H.C. , Shin, J.H. , Bugayenko, A. et al (2011) The AAA+ ATPase Thorase regulates AMPA receptor‐dependent synaptic plasticity and behavior. Cell, 145, 284–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y. , Kalbfleisch, T. , Brennan, M.D. & Li, Y. (2009) A MicroRNA gene is hosted in an intron of a schizophrenia‐susceptibility gene. Schizophr. Res., 109, 86–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zietsch, B.P. , Hansen, J.L. , Hansell, N.K. , Geffen, G.M. , Martin, N.G. & Wright, M.J. (2007) Common and specific genetic influences on EEG power bands delta, theta, alpha, and beta. Biol. Psychol., 75, 154–164. [DOI] [PubMed] [Google Scholar]
- Zlojutro, M. , Manz, N. , Rangaswamy, M. , Xuei, X. , Flury‐Wetherill, L. , Koller, D. , Bierut, L.J. , Goate, A. et al (2011) Genome‐wide association study of theta band event‐related oscillations identifies serotonin receptor gene HTR7 influencing risk of alcohol dependence. Am. J. Med. Genet. B, 156B, 44–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Distributions of the phenotype measurements for spectral peak width (A), peak frequency (B) and peak amplitude (C). The distributions depict two sets of 98 unrelated individuals (Set1 and Set2) and measurements from the left (L), middle (M) and right (R) parieto‐occipital region.
Table S1. List of annotated loci in the linkage peak region on chromosome 10.


