Prenatal cell‐free DNA (cfDNA) testing, or non‐invasive prenatal testing, has been shown to be capable of detecting certain microdeletion syndromes1 and has recently been extended to genome‐wide detection of subchromosomal abnormalities, including microduplications2. Here we describe a fetus with multiple structural anomalies that was diagnosed with double segmental duplications by cfDNA testing at our laboratory (Department of Genomic Medicine, Changhua Christian Hospital, Taiwan). We confirmed the cfDNA test result by chromosomal microarray (CMA) and karyotyping, and ascertained that the fetus had an unbalanced translocation inherited from a parental carrier of a balanced translocation involving t(11;22)(q23;q11.2).
A 30‐year‐old woman, gravida 2 para 1, was referred to our center at 35 weeks of gestation due to intrauterine growth restriction (IUGR) and suspected cardiac defects. During the visit, a detailed ultrasound examination confirmed IUGR (estimated fetal weight of 1953 g, corresponding to 32 weeks of gestation) and showed oligohydramnios, an overriding aorta (Figure 1a) with a ventricular septal defect and a small pulmonary artery (Z‐score = −2.11), and renal malformations (small kidneys with poor corticomedullary differentiation; Figure 1b). Given the presence of conotruncal heart defects, we conducted invasive tests including fluorescence in‐situ hybridization (FISH), array comparative genomic hybridization (array CGH) and conventional karyotyping of amniocytes to obtain a genetic diagnosis. Prior to the invasive diagnosis, maternal blood was drawn for cfDNA testing for research purposes. cfDNA testing was performed using next‐generation sequencing with two different algorithms for aneuploidy detection: Z‐score and genome‐wide normalized score (GWNS)3. The Z‐score was based on Z statistics which quantified the deviation of the read ratio of chromosomes/segments of interest from the normal control, while GWNS normalized the read counts with the effective proportions of the corresponding chromosomes/segments based on a hypothesis that the DNA read proportion of each chromosome/segment constitutes a robust ratio among normal controls3.
Figure 1.
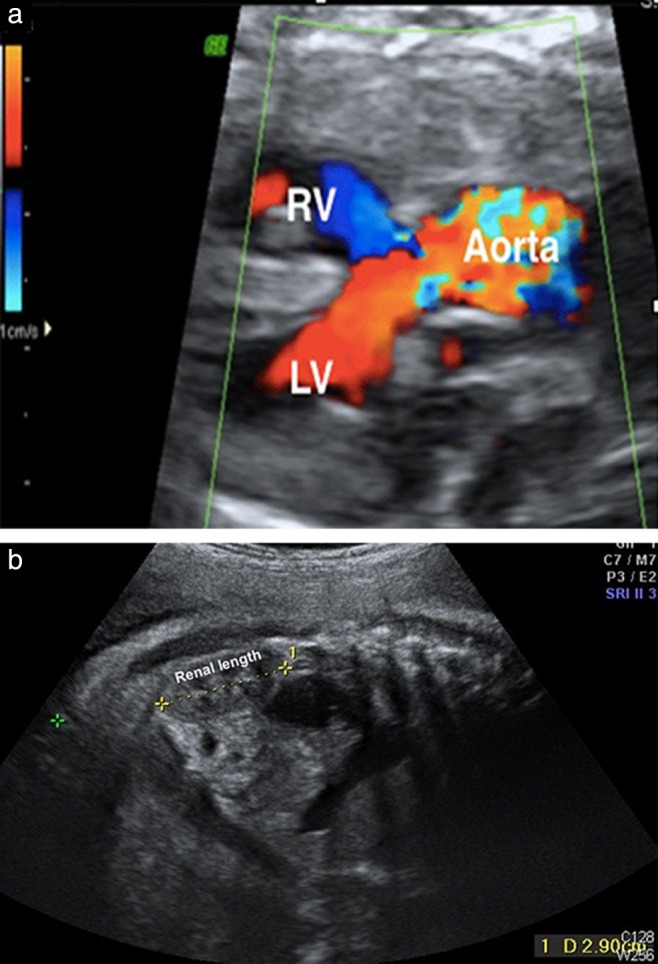
Prenatal ultrasound image in a 35‐week fetus, showing an overriding aorta (a) and a small kidney (length, 29.0 mm vs reference length at 35 weeks of 33.1 mm (3rd percentile) to 51.2 mm (97th percentile)) with poor corticomedullary differentiation (b). LV, left ventricle; RV, right ventricle.
The cfDNA test used could detect aneuploidies across 22 autosomes and 16 chromosomal regions associated with 16 microdeletion diseases, including 22q11.21 (a region associated with 22q11.2 deletion syndrome; located at chr22:19,009,792‐21,452,445 [hg19]). The cfDNA test results provided the first line of evidence for fetal aneuploidy involving chromosome 11q (Z‐score = 11.84; GWNS P < 0.0001) in addition to 22q11.2 (Z‐score = 3.18; GWNS P < 0.0001). The fetal DNA concentration was estimated as 12.5% and the total mapped sequencing reads was c. 20 million after trimming 3.7% which were polymerase chain reaction duplicates and 13.1% which were unperfected reads. Interphase FISH analysis of 50 amniocytes with commercially available DNA probes (DiGeorge/VCFS TUPLE1 22q13.2 and DiGeorge TBX1/22q13.3 combinations, Cytocell, Inc., Cambridge, England) further revealed a TBX1 duplication in 90% (45/50) of the cells (nuc ish 22q11.2(TBX1×3)) (Figure 2a). Finally, array CGH with CytoScan gene chip (Agilent customer design ID 040427, Changhua Christian Hospital, Changhua, Taiwan) identified two segmental duplications involving 11q23.3q25 (18.14 megabases (Mb)) and 22q11.1q11.21 (3.21Mb) (Figure 2b) and, in addition, conventional karyotyping identified 47,XX,t(11;22)(q23;q11.2),+der(22)t(11;22)(q23;q11.2) (Figure 2c). After non‐directive genetic counseling, the couple opted for late termination of pregnancy at 36 + 3 weeks of gestation following the recommendations of the local government4. A dead 2120‐g female fetus was delivered and gross examination revealed a normal appearance except for a bulbous nose (Figure 2d). The family declined autopsy. Parental follow‐up revealed that this was a case of 22q11.2 duplication inherited from the father who carried a reciprocal balanced translocation t(11;22)(q23;q11.2).
Figure 2.
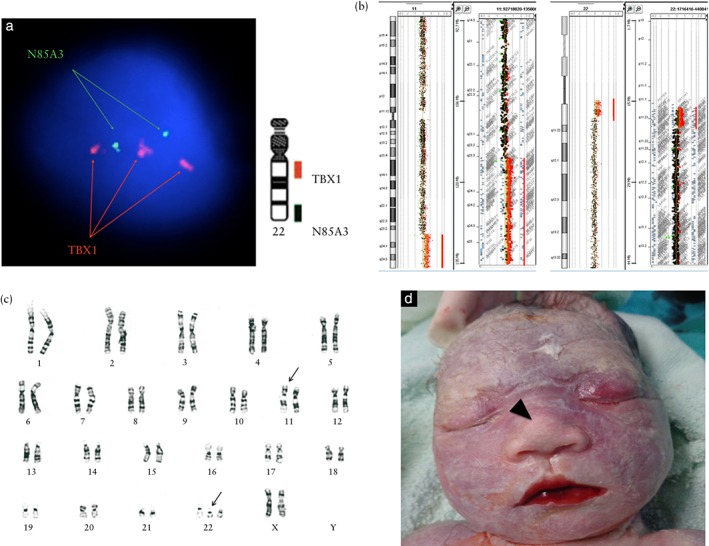
Genetic analysis of 35‐week fetus with multiple structural anomalies. (a) Interphase fluorescence in‐situ hybridization using DiGeorge TBX1/22q13.3 DNA probe (Cytocell, Inc., Cambridge, England) revealed TBX1 duplication in 90% (45/50) of amniocytes examined (nuc ish 22q11.2 (TBX1×3)). (b) Array comparative genomic hybridization using CytoScan gene chip (Agilent customer design ID 040427, Changhua Christian Hospital, Changhua, Taiwan) demonstrated two segmental duplications involving 11q23.3q25 (18.14 megabases (Mb)) (arr[hg19] 11q23.3q25(116,723,438‐134,868,407)×3) and 22q11.1q11.21 (3.21 Mb) (arr[hg19] 22q11.1q11.21(17,096,855‐20,311,763)×3). (c) Karyotype analysis of amniocytes revealed a suspect reciprocal translocation (47,XX,t(11;22)(q23;q11.2),+der(22)t(11;22)(q23;11.2)) (arrows). (d) The appearance of the terminated fetus was grossly normal except for a bulbous nose (arrowhead).
The 22q11.2 duplication can display similar features to those of 22q11.2 deletion syndrome5, in which TBX1 is thought to be the critical region responsible for conotruncal heart defects. FISH and CMA are the standard methods for genetic diagnosis6. With the development of massively parallel sequencing of cfDNA in maternal plasma, non‐invasive testing has been applied successfully to detect certain microdeletion syndromes1. However, it is still challenging to regularly detect microduplications as there is only a 1.5‐fold change in copy number (3:2) instead of a 2‐fold change (1:2) that occurs during microdeletions. In our case, we identified successfully two microduplications involving chromosomes 11q and 22q11.2 using the Z‐score algorithm and our own GWNS algorithm3, and the results were further confirmed by CMA. Based on the results of parental and fetal karyotyping, we delineated a causal interpretation of reciprocal translocation involving 11q23 and 22q11.2. This case demonstrated that cfDNA screening can detect not only microdeletions but also microduplications on some occasions, and even those as small as 3.21 Mb. Therefore, non‐invasive testing is valuable because it can provide additional genetic information which may be overlooked by targeted invasive tests, such as FISH and multiplex ligation‐dependent probe amplification. The expectant parents would therefore be informed of their reproductive choices before the baby is born. However, we believe invasive tests remain the gold standard of genetic diagnosis as multiple factors such as confined placental mosaicism, maternal mosaicism, cotwin demise or maternal malignancy may affect the accuracy of cfDNA screening. Consequently, for accurate genetic diagnosis of a fetal anomaly, clinicians should offer integrated genetic counseling after utilizing various prenatal diagnostic modalities, as shown in this case.
W.‐J. Wu†‡#, G.‐C. Ma†§¶#, Y.‐S. Lin**, C.‐H. Yeang††, Y.‐H. Ni‡‡, W.‐C. Li§§, H.‐D. Tsai‡, S. Shur‐Fen Gau¶¶*** and M. Chen*‡***†††‡‡‡# †Department of Genomic Medicine and Center for Medical Genetics, Changhua Christian Hospital; and Department of Genomic Science and Technology, Changhua Christian Hospital Healthcare System, Changhua, Taiwan; ‡Department of Obstetrics and Gynecology, Changhua Christian Hospital, Changhua, Taiwan; §Institute of Biochemistry, Microbiology and Immunology, Chung Shan Medical University, Taichung, Taiwan; ¶Department of Medical Laboratory Science and Biotechnology, Central Taiwan University of Science and Technology, Taichung, Taiwan; **Welgene Biotechnology Company, Nangang Business Park, Taipei, Taiwan; ††Institute of Statistical Science, Academia Sinica, Taipei, Taiwan; ‡‡Department of Pediatrics, National Taiwan University Hospital and College of Medicine, Taipei, Taiwan; §§Department of Obstetrics and Gynecology, Puli Christian Hospital, Nantou, Taiwan; ¶¶Department of Psychiatry, National Taiwan University Hospital and College of Medicine, Taipei, Taiwan; ***Department of Medical Genetics, National Taiwan University Hospital, Taipei, Taiwan; †††Department of Obstetrics and Gynecology, National Taiwan University Hospital and College of Medicine, Taipei, Taiwan; ‡‡‡Department of Life Science, Tunghai University, Taichung, Taiwan *Correspondence. (e‐mail: mingchenmd@gmail.com; mchen_cch@yahoo.com) #W.‐J.W., G.‐C.M. and M.C. contributed equally to the management and documentation of this case.
Acknowledgments
This study was kindly supported by research grants from the Ministry of Science and Technology (MOST 104‐2314‐B‐371‐009‐MY3 to M.C.) and Changhua Christian Hospital (101‐CCH‐IRP‐40 to G.‐C.M. and 102‐CCH‐IRP‐034 to M.C.), Taiwan.
References
- 1. Peters D, Chu T, Yatsenko SA, Hendrix N, Hogge WA, Surti U, Bunce K, Dunkel M, Shaw P, Rajkovic A. Noninvasive prenatal diagnosis of a fetal microdeletion syndrome. N Engl J Med 2011; 365: 1847–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yin AH, Peng CF, Zhao X, Caughey BA, Yang JX, Liu J, Huang WW, Liu C, Luo DH, Liu HL, Chen YY, Wu J, Hou R, Zhang M, Ai M, Zheng L, Xue RQ, Mai MQ, Guo FF, Qi YM, Wang DM, Krawczyk M, Zhang D, Wang YN, Huang QF, Karin M, Zhang K. Noninvasive detection of fetal subchromosomal abnormalities by semiconductor sequencing of maternal plasma DNA. Proc Natl Acad Sci USA 2015; 112: 14670‐14675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yeang CH, Ma GC, Hsu HW, Lin YS, Chang SM, Cheng PJ, Chen CA, Ni YH, Chen M. Genome‐wide normalized score: a novel algorithm to detect fetal trisomy 21 during non‐invasive prenatal testing. Ultrasound Obstet Gynecol 2014; 44: 25–30. [DOI] [PubMed] [Google Scholar]
- 4. Chiang S. Late Abortion: A Comprehensive Review. Taiwan J Obstet Gynecol 2005; 44: 318–326. [Google Scholar]
- 5. Welsfeld‐Adams JD, Edelmann L, Gadi K, Mehta L. Phenotypic heterogeneity in a family with a small atypical microduplicationof chromosome 22q11.2 involving TBX1. Eur J Med Genet 2012; 55: 732–736. [DOI] [PubMed] [Google Scholar]
- 6. Chen M, Yang YS, Shih JC, Lin WH, Lee DJ, Lin YS, Chou CH, Cameron AD, Ginsberg NA, Chen CA, Lee ML, Ma GC. Microdeletions/duplications involving TBX1 gene in fetuses with conotruncal heart defects which are negative for 22q11.2 deletion on fluorescence in‐situ hybridization. Ultrasound Obstet Gynecol 2014; 43: 396–403. [DOI] [PubMed] [Google Scholar]


