Abstract
Genetic diversity is considered an important factor, stabilizing ecological functions when organisms are faced with changing environmental conditions. Although well known from terrestrial systems, documentations of this relationship from marine organisms, and particularly planktonic microorganisms, are still limited. Here we experimentally tested the effects of genotypic diversity on ecologically relevant cellular parameters (growth, primary production, particulate organic carbon, particulate organic nitrogen, particulate organic phosphorus and biogenic silica) at optimal and suboptimal salinity conditions in a marine phytoplankton species. Multiple clonal genotyped and phenotypically characterized isolates of the diatom Skeletonema marinoi from the Baltic Sea were grown in monocultures and mixes of 5 and 20 clones at native (5 psu) and reduced (3 psu) salinities and respective parameters were compared. Re-genotyping of 30 individuals from each population at five microsatellite loci at the end of the experiment confirmed maintenance of genotypic richness. Although a diversity effect on growth was not detected, primary production and particulate organic nutrients were positively affected by increased diversity independent of salinity condition. Under salinity stress, highest values of primary production and particulate organic nitrogen content were measured at the high diversity level. The observed diversity effects emphasize the importance of genetic diversity of phytoplankton populations for ecological functions.
Introduction
Globally decreasing levels of biodiversity resulting from anthropogenic perturbations are predicted to have severe effects on the ecosystem functions that are essential for the services of the marine habitats (Schläpfer and Schmidt, 1999). Theoretical and empirical research has emphasized that the stability and functioning of aquatic ecosystems may depend more than previously considered on the structural and functional variability of the system compartments (Hillebrand and Shurin, 2005; Duffy et al., 2005; Stachowicz et al., 2007). It is now well established that high diversity at different trophic levels can enhance productivity, resource use, nutrient cycling and stability of marine systems. Loss of diversity, in turn, leads to increased rates of resource collapse, and to decreased water quality and recovery potential (Worm et al., 2006).
In marine systems the work on the relationship of marine biodiversity and ecological processes have so far mainly concentrated on benthic habitats, but studies begin to emerge that demonstrate clear effects of community structure and variability on pelagic ecosystem functions such as primary productivity and resource use efficiency (Litchman and Klausmaier, 2008; Ptacnik et al., 2008). While phytoplankton species richness, species identity and trait diversity are now increasingly considered in studies on ecological functioning of marine pelagic systems, the implications of intraspecific diversity are still poorly explored.
Genetic diversity, defined as ‘any measure that quantifies the magnitude of genetic variability within a population' (Hughes et al., 2008), is a basic constituent of biodiversity which, in microorganisms, is typically reflected by the number of clones in a population (genotypic richness). Other components of genetic diversity are for instance heterozygosity and allelic richness. For example, genotypic richness or identity plays an important role in the configuration of ecosystem functions (Whitham et al., 2012; Whitlock, 2014). These include processes that affect stocks of materials (e.g. carbon, nutrients) involved in biogeochemical fluxes and cycles (Madritch et al., 2006). The most commonly documented ecological effect of increased genetic diversity involves productivity of the population, which is often found to be enhanced in mixtures of genotypes compared with monocultures (Bell, 1991; Smithson and Lenne, 1996). A number of studies have shown that genetically diverse populations can resist environmental perturbations more effectively than genetically uniform populations (Hughes and Stachowicz, 2004). Finally, genetic diversity may affect communities, trophic interactions and, when dominant species are involved, ecosystem processes (Madritch et al., 2006; Lankau and Strauss, 2007; Bangert et al., 2008). While an ecologically relevant link between genetic diversity and ecosystem functions has been extensively demonstrated for terrestrial systems (Schweitzer et al., 2005; Crutsinger et al., 2008), such studies are still rare for aquatic habitats and largely limited to benthic primary producers and invertebrates (Reusch et al., 2005; Gamfeldt and Kallström, 2007; Ehlers et al., 2008). Even less information is available for plankton microorganisms (Bell, 1991; Roger et al., 2012), despite their important role in global biogeochemical cycles.
Recent population genetic analyses have revealed high levels of genetic diversity and differentiation for a number of phytoplankton species (Godhe and Härnström, 2010; Tesson et al., 2014; Dia et al., 2014). At the same time it has become evident that individual clonal lineages of a phytoplankton population can differ quite substantially in phenotypic traits (Alpermann et al., 2010; Gsell et al., 2012). It has been proposed that such intraspecific differences in fitness-related traits should stabilize the performance of mixtures in variable environments (Bell, 1991; Roger et al., 2012), but a clear relationship between such effects and clonal richness have not been unequivocally demonstrated so far.
Here we present the results of an experiment comparing ecologically relevant eco-physiological parameters such as growth, primary production and nutrient acquisition in monoclonal and mixed cultures of S. marinoi isolated from the Baltic Sea at different salinities with the aim of substantiating a link between genetic diversity and parameters relevant for ecosystem function. Baltic low-salinity-adapted populations of the species are genotypically diverse and display considerable phenotypic diversity (Sjöqvist et al., 2014, 2015). Using microsatellite-based genotyping methods we were able to monitor and confirm true clonal diversity in experimental mixtures.
Materials and methods
Experimental setup
Batch culture experiments were conducted using genetically distinct, genotyped (five microsatellite loci) strains of Skeletonema marinoi. Five randomly chosen, genetically distinct strains (C1402, C1404, C1412, C1413 and C1426) were grown separately and in a mix. To test for potential effects of varying clonal richness, an additional experimental unit was set up mixing 20 genotypes (C1402, C1403, C1404, C1405, C1406, C1407, C1408, C1410, C1411, C1412, C1413, C1414, C1415, C1416, C1417, C1418, C1419, C1425, C1426 and C1428). By comparing monocultures against a mix of the same five strains we enabled the calculation of potential overyielding. It was not logistically feasible using our method to quantify monoclonal performance of all 20 strains. However, the high diversity level was included in order to reflect a more realistic situation, as earlier work has shown that S. marinoi populations naturally consist of hundreds of genotypes (Sjöqvist et al., 2015). All monoclonal cultures and diversity levels were incubated at a native salinity of 5 psu and at a salinity of 3 psu, the latter representing the lower end of the Baltic Sea salinity gradient and a potentially stressful condition. Both salinity treatments and each diversity level were run in three replicates. The experimental sets were inoculated from exponentially growing cells at a start concentration of 10 000 cells ml−1. Mixtures were set up to contain equal amounts of 5 or 20 genotypes. All cells were grown in 500 ml tissue culture flasks containing F/4 medium (Guillard, 1975). The experiment was run for 8 days at +10 °C at light conditions of 40 μmol m−2 s−1 and a 1410 h light:dark cycle. No acclimatization to salinity levels prior to the start of the experiment was performed since the aim was to create fluctuating conditions representing severe disturbance. All 20 experimental strains were established from a sediment sample collected in the Bothnian Sea (18.55E, 62.12N), where ambient mean salinities are 5 psu. Culture establishment and genetic characterization using microsatellites are described in detail in Sjöqvist et al. (2015).
Genotyping of mixed cultures
At the end of the experiment, 30 cells were re-isolated from each mixture and grown to cell concentrations required for DNA extraction. We amplified five polymorphic microsatellite loci by PCR according to the previously described protocol in Godhe and Härnström (2010). The PCR products were analyzed in an ABI 3730 (Applied Biosystems Inc., Foster City, CA, USA) using an internal standard (GS500LIZ). GeneMapper v.3.0 (Applied Biosystems, Inc.) was used to determine the allele sizes for the individual loci. In total, we genotyped the 20 strains initially added to the experiment and 30 isolates per replicate at the end of the experiment to verify that the difference in clonal richness was maintained throughout the experiment and to detect changes in genotype composition.
Sampling and analyses
Samples for cell counts and chlorophyll a (chl a) were taken once per day. Cell abundance was assessed under an inverted microscope (Leica DMI 3000B, Leica Microsystems Inc., Mannheim, Germany) by counting a minimum of 400 cells using a gridded Sedgewick-Rafter chamber (1 ml). Samples for chl a were filtered onto GF/F filters, extracted in 10 ml, 96% ethanol (Jespersen and Christoffersen, 1987) and stored in −20 °C until fluorometric determination (Varian, Varian Australia Pty Ltd, Mulgrave, VIC, Australia). Growth rates (intrinsic rate of increase) were calculated based on the longest period of exponential growth as in Wood et al. (2005). On day 6, which typically coincided with the onset of the stationary phase, subsamples were filtered on acid-washed, pre-combusted GF/F filters for analyses of particulate organic carbon (POC), particulate organic nitrogen (PON) and particulate organic phosphorus (POP). POC and PON were measured on a mass spectrometer (Europa Scientific, Crewe, UK) and POP determined according to Solorzano and Sharp (1980). Samples for biogenic silica (BSi) were filtered on 0.8 μm polycarbonate filters and analyzed as described by Krause and August (1983). Samples for primary production measurements were taken on days 1, 3 and 5. In order to estimate the carbon uptake we added 20 μl of C14 (20 μCi ml−1) to 4 ml of culture in glass scintillation vials that were incubated for 2 h in the same conditions as described above. The carbon that was not taken up by the diatoms was removed by adding 100 μl of 1 m HCl. Disintegrations per minute were counted using a scintillation counter (Wallac, Turku, Finland) after addition of 7 ml of InstaGel. Concentration of dissolved inorganic carbon was measured on days 1, 3 and 5 with a Unicarbo carbon analyzer (Elektro Dynamo, Helsinki, Finland) from the experimental bottles in order to determine the ambient amount of dissolved carbon. Values for primary production, POC, PON, POP and BSi were normalized by dividing the value of each parameter by the respective chl a value.
Statistical analyses
Prior to all variance analyses normality and homoscedasticity of residuals were checked for each response variable. To study phenotypic differences between strains we conducted linear mixed effects (LME) models using the package ‘nlme' in R (Pinheiro et al., 2013). These were fitted with salinity as a main factor and strains as a random factor. The significant effect of the random factor ‘strain' was tested by comparing a generalized least squares model without the random effect, with the best LME model where salinity and strain were included. This assessment was conducted using a likelihood ratio test. The best LME for each response variable was selected based on the model's Akaike information criterion. Tukey's pairwise post hoc comparisons (Lenth, 2013) (R package ‘lsmeans') were used to determine which of the effects of explanatory factors and their interactions significantly differed from each other. To study differences in functional parameters between diversity levels and salinity conditions, general linear models (GLM) were fitted to each response variable with diversity level, salinity and their interaction as explanatory variables.
Clonal richness and Pielou's evenness index (J) (Pielou, 1966) were calculated for the multiclonal experimental units. The evenness index was calculated by dividing Shannon's diversity index (Shannon, 1948) with the natural logarithm of clonal richness. Expected heterozygosity (HE) was calculated in Microsatellite Tools for Excel (Park, 2001). Differences in clonal richness, evenness and HE within salinity conditions were assessed by one-way analysis of variance (ANOVA) and Bonferroni corrections in GraphPad Prism 6 (GraphPad Software, Inc., San Diego, CA, USA). A potential difference in diversity indices between salinity conditions at the high diversity level was tested by Student's t-test in Excel (Microsoft). Two-way ANOVAs with Bonferroni corrections were conducted to test differences of clonal proportions within and between salinity conditions.
Potential transgressive overyielding in ecological performance was analyzed in R following the script provided in Schmid et al. (2008). This was only calculated for the mix with five strains and where we found an initial positive effect of increasing diversity and a statistically significant difference in question of performance between monoclonally growing strains. The only response variable that fulfilled our requirements was C uptake. Transgressive overyielding was assumed when the mix (five genotypes) had a higher C uptake than the best sampled or simulated monoculture. By using the average performance and interclonal standard deviations of monocultures we simulated the theoretical maximum C uptake of a best monoculture. As the simulated value may be skewed the most likely value would be between the inverse of the normal cumulative distribution for the specified mean and standard deviation (function ‘NORMINV' in Excel) and the value derived from the simulations. The C uptake value of the mix was also compared with the average performance of the monocultures.
The unbiased effect size (Hedges' g), taken differing sample sizes into account, was calculated according to Hedges and Olkin (2014) in Excel by adopting the following equation:
The standardized mean difference (d) was calculated according to the equation in Friedman (1968),
The effect size was used to interpret the relative effect of salinity reduction on different diversity levels. Significantly different effect sizes were determined by calculating the 95% confidence interval for each diversity level and response variable. A significantly smaller effect size was interpreted as increased stability with respect to performance.
Results
Phenotypic differences between strains
Table 1 shows response parameters of monoclonal cultures showing differences among five random strains. A significant strain effect on growth (defined here as the intrinsic rate of increase) was identified by the LME (strain P<0.001) (Supplementary Information 1) at both salinities. Strain C1413 had a significantly lower rate of increase in 3 psu compared with C1402 (Tukey, P<0.001). In 5 psu strain C1413 had a significantly lower rate of increase compared with C1402 and C1404 (Tukey, P=0.011–0.018). All strains, except for C1426, had significantly reduced growth in 3 psu (Tukey, P<0.001). We also observed statistically significant differences in C uptake in both salinity conditions. Strain C1426 displayed a significantly higher value compared with all other strains in both salinity conditions (Tukey, P=0.001–0.007), except when compared with C1412 in 3 psu. Strains C1402, C1413 and C1426 had significantly reduced C uptake in 3 psu (Tukey, P <0.001–0.03). No differences were found between strains regarding POC, PON and POP content at any salinity condition (LME, P=0.138–0.530), nor between salinity conditions (Tukey, P=0.237–0.999). There was a significant interaction effect on BSi content (LME, strain × salinity P=0.001). Differences were observed in 5 psu where strain C1413 had significantly lower levels compared with strain C1426 (Tukey, P=0.031). In 3 psu, C1412 had significantly lower BSi content compared with C1402 (Tukey, P=0.049).
Table 1. Values of six different cellular parameters in five monoclonally grown S. marinoi strains in 3 and 5 psu.
| C1402 | C1404 | C1412 | C1413 | C1426 | |
|---|---|---|---|---|---|
| 3 psu | |||||
| Growth | 0.44 (0.03) | 0.41 (0.01) | 0.32 (0.04) | 0.27 (0.05) | 0.40 (>0.01) |
| C uptake | 0.09 (0.03) | 0.10 (0.02) | 0.14 (0.01) | 0.08 (0.01) | 0.21 (0.03) |
| POC | 62.15 (17.75) | 49.91 (3.59) | 49.51 (4.65) | 56.14 (15.90) | 50.56 (3.69) |
| PON | 7.96 (3.20) | 5.29 (1.00) | 3.94 (0.75) | 4.21 (2.56) | 6.11 (2.84) |
| POP | 0.61 (0.21) | 0.75 (0.07) | 0.69 (<0.01) | No data | 0.87 (0.29) |
| BSi | 15.19 (6.81) | 9.53 (2.03) | 7.47 (0.35) | 9.27 (1.11) | 8.50 (0.92) |
| 5 psu | |||||
| Growth | 0.74 (0.03) | 0.70 (0.01) | 0.58 (0.08) | 0.50 (>0.01) | 0.40 (0.16) |
| C uptake | 0.18 (0.04) | 0.17 (0.03) | 0.17 (0.06) | 0.19 (0.01) | 0.36 (0.05) |
| POC | 79.74 (19.21) | 75.60 (9.50) | 73.63 (6.30) | 60.31 (10.28) | 73.98 (19.39) |
| PON | 11.88 (3.69) | 11.67 (2.12) | 10.53 (1.05) | 8.39 (2.38) | 10.60 (3.31) |
| POP | 0.72 (0.11) | 0.61 (0.10) | 0.52 (0.05) | No data | 0.66 (0.04) |
| BSi | 10.63 (2.12) | 4.31 (0.02) | 6.54 (0.70) | 5.47 (1.11) | 13.68 (1.31) |
Standard error (s.d.) in brackets.
Clonal composition and richness of mixed cultures
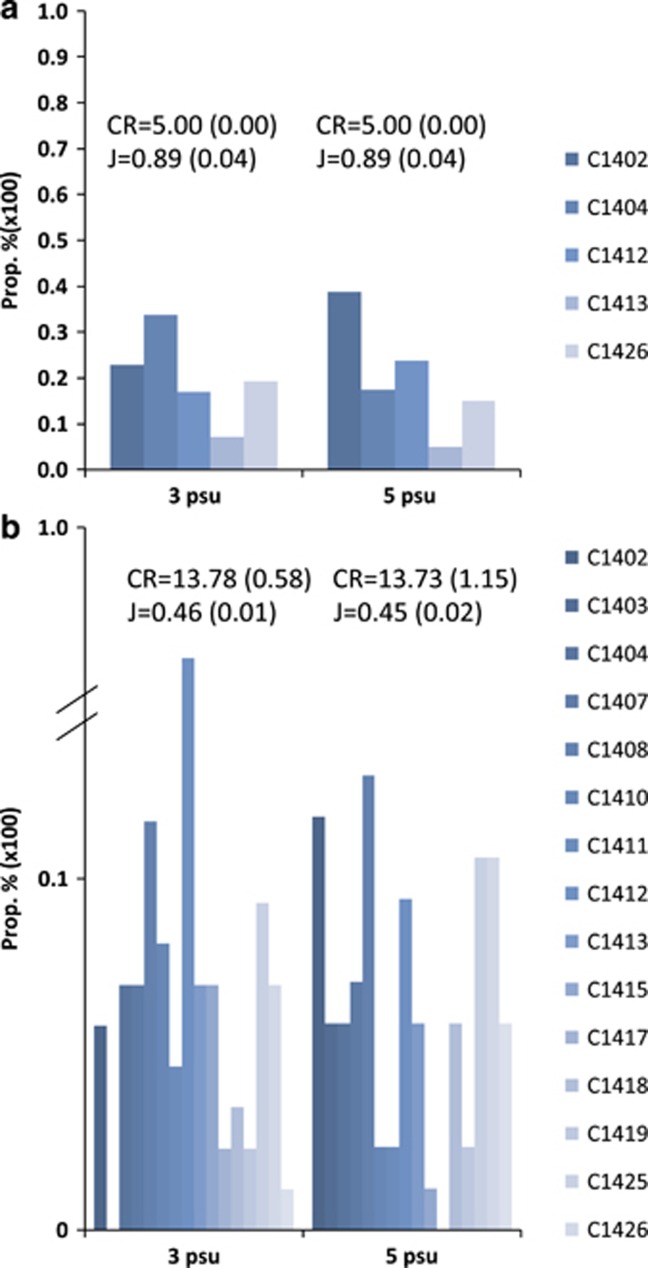
Re-genotyping at five microsatellite loci of individuals isolated from the mixed cultures at day 6 confirmed significant differences in genotypic richness among the three diversity levels (one-way ANOVA, Bonferroni, 3 psu low vs monoclonal, P<0.0001, low vs high, P<0.0001, 5 psu low vs monoclonal, P<0.0001, low vs high, P<0.0001) (Figures 1a and b). The low diversity level with an initial genotypic richness of five maintained its diversity until day 6 at both salinities. We observed a correlation of growth measured in monoclonal incubations compared with the relative proportions of respective strains in the mix (3 psu R2=0.63, 5 psu R2=0.47) (Supplementary Information 2). The genotypic richness in the high diversity level was reduced from 20 to an average of 13.17 at 3 psu and 13.73 at 5 psu when corrected for sample size (two-tailed t-test, 3 vs 5 psu, P=0.423).
Figure 1.
Clonal proportions on day 6 in the low and high diversity levels in control and stress conditions. (a) Genotypic richness (CR) was maintained until the end of the experiment. The evenness index (J) was close to 1, indicating similar relative proportions of respective genotypes. (b) Genotypic richness dropped to an average of ~13 genotypes (corrected for sample size) in both salinity conditions at the end of the experiment. As reflected by J, no genotype(s) was able to clearly dominate the populations.
Genotype composition (Figure 1a) was unaffected by salinity conditions in the low diversity level. However, a significant difference of clonal proportions existed at 3 psu (two-way ANOVA, salinity P>0.999, strain P=0.012) that were independent of salinity (Figure 1a). Multiple comparisons (Bonferroni) showed that clone C1413 had a significantly lower proportion compared with C1404 in 3 psu (P=0.012). In 5 psu, the proportions of clones C1413 and C1426 were marginally different from C1402 (Bonferroni, C1413 P=0.070, C1426 P=0.060). The evenness index (J) was similar in both salinity conditions, 0.89 in 3 psu and 0.92 in 5 psu (two-tailed t-test, P=0.382). In the high diversity level we observed a similar salinity independent strain effect on clonal proportions (two-way ANOVA, salinity P=0.952, clone P=0.017) (Figure 1b). However, none of the differences between individual clonal proportions were significant after Bonferroni correction. Evenness indexes in 3 and 5 psu (0.46 and 0.45, respectively) were not significantly different (two-tailed t-test, P=0.256). Clone C1403 was not present in 3 psu and C1417 was not present in 5 psu. Clones C1405, C1406, C1414 and C1416 were absent in both 3 and 5 psu at the end of the experiment.
Comparison of parameters at different diversity levels
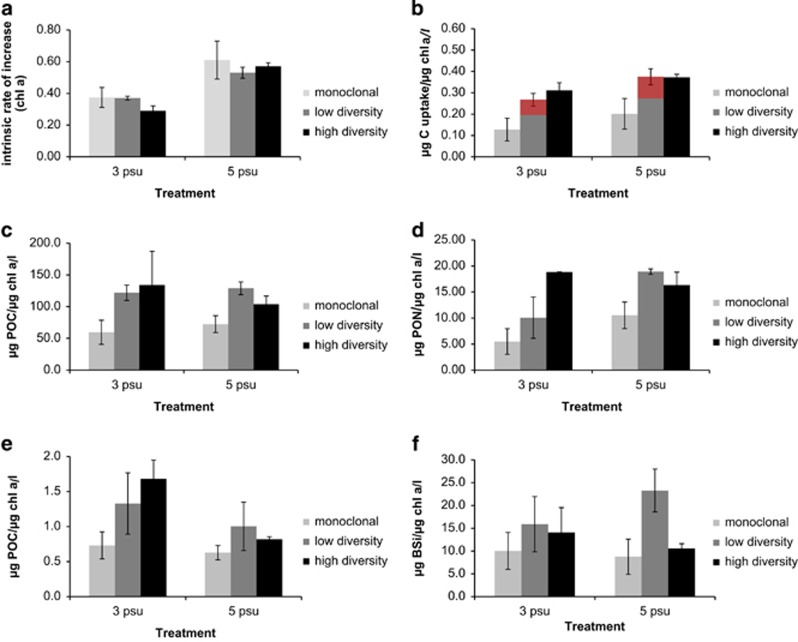
Effects of salinity and diversity level on six response variables (Figures 2a–f) were tested by GLM (Table 2). Salinity had a significant effect on μ, C uptake, PON, POP and BSi (GLM, P<0.001 to <0.01). The growth was significantly reduced in all diversity levels in 3 psu compared with 5 psu (Tukey, P<0.001). Particulate organic carbon was significantly reduced in the low and high diversity levels in 3 psu compared with 5 psu (Tukey, P<0.001). The level of PON was significantly reduced in the monoclonal cultures and the low diversity levels in 3 psu compared with 5 psu (Tukey, P<0.001). Particulate organic phosphorus was significantly higher in 3 psu compared with 5 psu with respect to both the low and the high diversity levels (Tukey, P<0.001). There was a significantly higher level of POP in the high diversity level at 3 psu compared with 5 psu (Tukey, P<0.001). Genotypic richness had no effect on growth in any salinity condition (Tukey, P=0.711–0.894). However, we observed a significant effect of diversity on C uptake, POC, PON, POP and BSi (GLM, P<0.01). The C uptake was significantly lower in the monoclonal cultures compared with both diversity levels in 3 and 5 psu (Tukey, P<0.001). No differences were observed between the low and high diversity levels (Tukey, P=0.897–0.954) for this parameter. Overyielding with respect to C uptake was detected for the mix of five clones (Figure 2b, red area). POC content was significantly lower in the monoclonal cultures compared with the low and high diversity levels in 3 psu (Tukey, P<0.001). There was also a significant difference between the monoclonal cultures and the high diversity level at 5 psu (Tukey, <0.001). The PON content was significantly lower in monoclonal cultures compared with both diversity levels at 3 and 5 psu (Tukey, P<0.001). A significant difference was observed between the low and high diversity levels at 3 psu (Tukey, P<0.001). POP content was significantly lower in monoclonal cultures compared with both diversity levels in 3 psu (Tukey, P<0.001). BSi was significantly higher in the low diversity level compared with the high diversity level at 5 psu (Tukey, P<0.001).
Figure 2.
Ecological performance of monoclonal cultures, a mix of five strains (low diversity) and a mix of (initially) 20 strains (high diversity) in 3 and 5 psu. (a) Intrinsic rate of increase based on chl a. (b) Carbon uptake. Red area represents overyield compared with the average of the monocultures. (c) POC. (d) PON. (e) POP. (f) BSi. (b–f) Normalized by chl a. See Table 2 for significant effects of diversity and salinity.
Table 2. Results of general linear models (GLM) for each response variable.
| Model type | Response variable | Explanatory variable | F | P |
|---|---|---|---|---|
| GLM | μ | Salinity | 19.17 | <0.001 |
| GLM | C uptake | Diversity | 26.67 | <0.001 |
| GLM | C uptake | Salinity | 14.69 | <0.01 |
| GLM | POC | Diversity | 35.88 | <0.001 |
| GLM | POC | Salinity | 0.04 | 0.838 |
| GLM | POC | Diversity*salinity | 3.76 | <0.05 |
| GLM | PON | Diversity | 104.03 | <0.001 |
| GLM | PON | Salinity | 32.90 | <0.001 |
| GLM | PON | Diversity*salinity | 18.62 | <0.001 |
| GLM | POP | Diversity | 37.55 | <0.001 |
| GLM | POP | Salinity | 38.16 | <0.001 |
| GLM | POP | Diversity*salinity | 10.82 | <0.01 |
| GLM | BSi | Diversity | 12.80 | <0.001 |
| GLM | BSi | Salinity | 0.16 | 0.686 |
| GLM | BSi | Diversity*salinity | 0.30 | 0.073 |
Significant effects of diversity indicate differences between diversity levels within the same salinity condition. Significant effects of salinity indicate differences in the response variables of different diversity levels between salinity conditions. Significant interaction effects suggest that the response of diversity levels were different depending on the salinity condition.
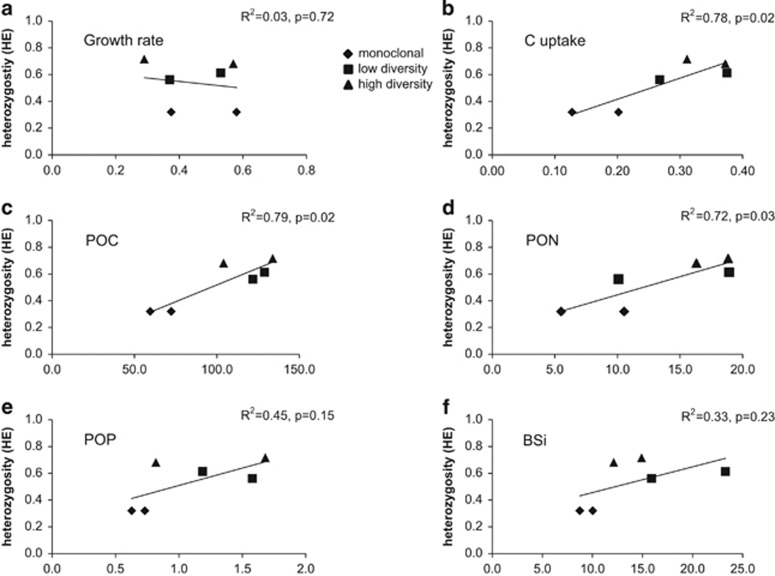
An effect of genotypic richness was indicated by a significant interaction between salinity and diversity for POC, PON and POP (GLM, P<0.001). Carbon uptake (R2=0.78, P=0.02), POC (R2=0.79, P=0.02) and PON (R2=0.72, P=0.03) correlated significantly with HE (Figures 3b–d), but no significant correlations were found between HE and growth rates, POP nor BSi (Figures 3a). Evenness and ecological function did not correlate either.
Figure 3.
Expected heterozygosity (HE) vs ecological performance. Same parameters as in Figures 2a–f. R2, Pearson's correlation coefficient.
The stabilizing effect of clonal richness
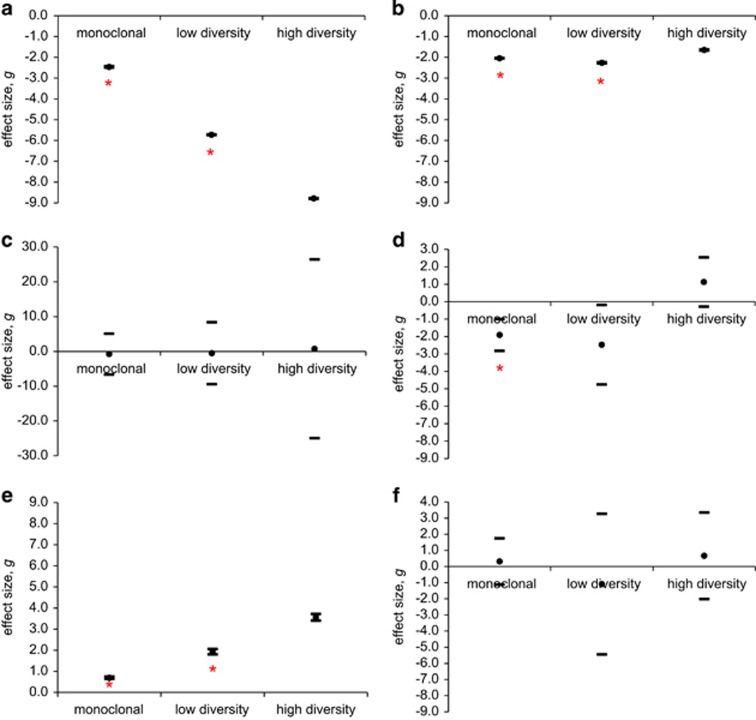
Hedges' g estimating the extent of salinity effects on different diversity levels to assess the stabilizing role of diversity on ecological performance revealed a significant positive effect size of diversity on C uptake, POC and PON. A negative effect size was determined for growth whereas effects on POP and BSi were neutral (Figures 4a–f).
Figure 4.
Effect size (g) of salinity reduction across diversity levels. Same parameters as in Figures 2a–f. The x-axis represents a neutral effect. Lower performance under reduced salinity conditions results in a negative effect size (below x-axis). Positive effect size above x-axis. Red asterisk indicate statistically different effect sizes (P<0.05) between diversity levels. A full color version of this figure is available online at the ISME Journal website.
Discussion
The experiments presented here revealed significant phenotypic differences in ecologically relevant cellular parameters, such as growth rates, primary production, POP and BSi content, among genetically distinct strains of S. marinoi, which were, furthermore, specifically affected by salinity stress. When clones were grown in mixtures, clonal diversity was maintained, though slight shifts in genotype composition occurred through time and at different salinities. Although a significant effect of diversity on growth was not observed, other functions such as primary production and particulate organic nutrient contents increased compared with monocultures, indicating that diversity effects may not primarily be manifested as increase of growth. Under salinity stress, primary production was least affected in the high diversity treatment, suggesting a stabilizing impact of increased genetic diversity.
Phenotypic differences among strains
It has become increasingly evident that clonal lineages of phytoplankton species and populations can be phenotypically very different. For example, blooms of S. marinoi consist of genetically distinct clones that differ largely in growth characteristics and cell chain morphology (Sjöqvist et al., 2014). Other phytoplankton species vary intraspecifically in environmental tolerances (Gsell et al., 2012; Ribeiro et al., 2013; Skjelbred et al., 2013) and cellular toxicity (Suikkanen et al., 2013). Such intraspecific differences in phenotypic traits form the basis for variability in respective ecosystem functions. Our results confirm strain-specific growth rates and growth performance under salinity stress and provide evidence for significant phenotypic variation in primary production rates. The strains also responded differently to salinity stress since the growth rate of C1426 seemed unaffected by salinity changes compared with all other strains, which had reduced growth rates in the lower salinity condition. Interestingly, cellular nutrient quotas were relatively stable among strains. The lack of significant variation in particulate nutrients in our data may be attributed to growth phase-dependent aspects. The sampling for POC, PON, POP and BSi was conducted in the late exponential/early stationary phase where other studies have observed decreasing variation of intracellular C, N and P among clonal isolates of the same species (Vrede et al., 2002). Salinity stress consistently affected several cellular parameters in the five monoclonal experimental cultures. Growth rates were significantly reduced as expected at suboptimal conditions (Supplementary Information 3), which is in accordance with earlier observations for Skeletonema (Rijstenbil et al., 1989). Slower growth at 3 psu likely explains the increased BSi levels—an effect reported for other marine diatoms (Tuchmann et al., 1984; Vrieling et al., 1999).
Diversity and performance
Most commonly, diversity-function studies report a positive effect on fitness of the species in question (Williams, 2001; Reed and Frankham, 2001). In phytoplankton, fitness is most commonly defined by growth rates (Klausmeier et al., 2004; Lohbeck et al., 2012). The two studies that have so far addressed the relevance of genetic diversity for ecological performance in phytoplankton showed that growth rates were in fact higher in mixtures compared with monoclonal cultures (Bell, 1991; Roger et al., 2012). Our experiment did not show such an effect. However, the studies are not directly comparable, since the genotype frequencies differed considerably: The before-mentioned studies demonstrated or considered dominance of one strain in their diversity treatments that may have been facilitated by strong intraspecific competition, whereas genotypic richness was maintained throughout the experiments reported here. The evenness numbers (low diversity 0.89, high diversity 0.45) suggest that intraspecific competition was generally moderate in our mixtures, though slightly more intense at the high diversity level. It is, however, likely that the high evenness values observed in our short-term experiment would decrease over longer time scales, where even subtle fitness differences would lead to dominance of one or only a few strains, provided that the environment remains stable. Slight differences in strain frequencies were observed in the two salinity treatments. Although after the short exposure period, these were (with one exception) not significant, altered strain distributions could indicate the beginning of selection processes by an altered salinity environment. However, frequency changes did not directly relate to salinity preferences of the respective strains and thus the indicated short-term selection seems to be a result of complex competitive interactions, rather than strain-specific adaptation to low salinity. The indicated selection processes suggest that population structure in marine phytoplankton may change under the influence of climate-related salinity changes.
Besides growth rate, other quantifiable ecologically relevant cellular or population parameters are considered as components representing fitness of an organism (Reed and Frankham, 2003). In our study, some assimilation processes were significantly enhanced by genotypic diversity. As revealed by primary production data, carbon uptake per chl a was significantly higher in mixtures compared with monocultures. In the 5-clone mixes at both salinity treatments we were able to even detect transgressive overyielding, that is, better performance by the mixture compared with the best monoculture (Harper, 1977), which confirms a robust link between genotypic richness and C assimilation in Baltic S. marinoi. Significantly higher C assimilation of the mix compared with the best performing monoclonal average can be a result of either niche partitioning or some other non-competitive interclonal interaction that resulted in better C assimilation of the entire population (Roscher et al., 2005). Also, growth rates measured in monoclonal incubations correlated only weakly with the relative proportions of strains in the genotypic mixture, suggesting that other factors than the pace of monoclonal growth governed dominance patterns in the population.
The discrepancy between the responses of primary production and growth to clonal diversity remains somewhat puzzling since generally these two parameters are directly related (Cloern et al., 1995). An explanation may be found in the way of how growth was determination here. The growth rates calculations were based on chl a measurements, which may be highly sensitive to environmental conditions and not always represent cell growth realistically. Carbon assimilation per chl a unit is prone to significant inherent variation and is not only a function of, for example, light intensity (Ryther and Yentsch, 1957), but also reflects the nutritional status of surrounding waters (Curl and Small, 1965). Field studies have shown that phytoplankton primary production may be uncoupled from chl a values (Marañón et al., 2000, 2003), meaning a population may grow slowly but produce unexpectedly high amounts of carbon. It is also worth mentioning that the chl a growth rates determined here closely reflected growth rates calculated based on cell numbers (data not shown r2 =0.89). Finally, the higher POC values in multiclonal cultures reflected enhanced primary production, describing the capacity for carbon fixation in another way. Our results thus emphasize that diversity effects may not necessarily show in fitness as expressed by growth, but in other cellular parameters with ecological significance.
Carbon uptake rates, POC and PON also correlated with increasing heterozygosity, indicating that not only the number of genotypes favored ecological performance but also increasing diversity of genes a strain harbors seem to be linked to improved performance. The heterozygosity of the neutral markers used here are most likely not related to any functional genes and are thus not directly linked to any fitness measures. However, Reed and Frankham (2003) showed that heterozygosity of neutral markers was positively and significantly correlated with population fitness and may be used as a proxy for heterozygosity throughout the genome. The explanatory power of heterozygosity in neutral molecular markers for fitness measures is commonly low, about 15–20% (Reed and Frankham, 2003). Our significant R2 coefficients of 0.72–0.79 are exceptionally high and may be explained by the highly significant linkage disequilibrium that was found in 8 out of 10 loci pairs (data not shown). Significant linkage disequilibrium is known to generate significant heterozygosity-fitness-correlations (HFCs) (Hansson et al., 2004) through linkage between the markers and the fitness loci. Linkage disequilibrium is a probable feature in monoclonal laboratory strains as they have gone through the strongest genetic bottleneck and are kept inbred. Thus, the HFCs in this study should be interpreted with caution and are most likely lower in natural populations. However, the significant HFCs support the hypothesis of a link between genetic diversity and ecological performance in this species.
C:chl a ratios are often used for global or regional comparisons of aquatic C sequestration and used in models of biogeochemical processes (Cloern et al., 1995). The values for parameterization in such models are usually derived from single clone cultures. Our results add even another layer of complexity, and emphasize that clonal diversity and effects of clonal interactions in diverse populations may need to be incorporated into models aiming at realistic assessments of global C fixation. This being the first study showing a clear relationship between genetic diversity and primary production in a phytoplankton species, it is obvious that more data, and data over longer time scales on several other species, are needed before any reliable equations relating to genetic diversity and primary production can be built.
Diversity and response to salinity stress
The predicted reduction in sea surface salinity in the Northern Baltic Sea during the next century lies between 1.5 and 2.0 psu (Meier et al., 2012). The response of phytoplankton populations to such stress, with respect to their ecological functions, may in part depend on their level of genetic diversity. Stability aspects may be investigated by examining the extent of variation in experimental studies, an interesting aspect that is commonly overlooked (Benedetti-Cecchi, 2003). Stability in ecological functions is expected to increase with higher genetic diversity (Worm et al., 2006), suggesting that increased diversity leads to smaller changes in ecological performance in contrasting environmental conditions. Therefore, the magnitude and direction of ecological effects on populations with varying diversity levels in contrasting environmental conditions may be inspected by effect sizes (Osenberg et al., 1994). The effect size takes into consideration the ratio between the size of an effect and the variability among replicates (Cohen, 1988), which increases the statistical power and the ecological interpretability of experimental data. When effect sizes of salinity reduction were calculated for different diversity levels, our results indicated significantly smaller effects in the multiclonal cultures with respect to C uptake and PON. These results suggest a stabilizing effect with increasing genotypic richness for these specific parameters. In marine systems, it has been shown that increased genotypic diversity of, for example, Zostera marina populations increases their resistance to perturbations (Hughes and Stachowicz, 2004; Ehlers et al., 2008). The explanation for increased stability with increasing diversity may be ascribed to overyielding as we have shown and explained above. Overyielding may result from plasticity in resource use, where genotypes upon competition can switch their preferences for, for example, different chemical forms of nitrogen (Ashton et al., 2010). In theory, this may lead to a total production that exceeds the expectation based on yields from monocultures. Overyielding may also emerge because of facilitation, where diatom-associated bacteria that are genotype specific may chemically mobilize otherwise unavailable forms of micronutrients. This has not been shown in diatom–bacteria studies, but similar mechanisms are known from terrestrial plant systems (Li et al., 2014). Alternatively, the ‘portfolio effect' involving a suite of species (suite of genotypes in our case) that exhibit differential responses to environmental perturbations creates stability that preserves the integrity of a physiological trait (Tilman, 1999, 2006). Theory predicts that the portfolio effect will be reduced at low evenness as the vulnerability of a community (or population) increases when fewer species (or genotypes) are dominating (Doak et al. 1998, Hillebrand et al., 2008). The high evenness of our multiclonal populations indicates a high potential for an active role of the portfolio effect in stabilizing ecologically relevant parameters in contrasting salinity conditions in this study.
Conclusions
Our results imply that the simple measurement of cell numbers or chl a may be a poor proxy for estimating diversity effects on ecologically relevant parameters in phytoplankton. The uptake of C and the amount of particulate nutrients were significantly higher in multiclonal cultures, confirming a link between genotypic richness and cellular parameters. Transgressive overyielding in question of C uptake supports this conclusion and indicates an active role of intraspecific interactions. Also, multiple significant HFCs strengthen the concept of link between sub-species diversity and ecological performance. The evidence for a stabilizing effect of genotypic richness with respect to primary productivity implies a good capacity of diverse S. marinoi populations to resist climate change-related salinity reduction. We expect our results to highlight the importance of sub-species level variation and its link to ecosystem functions in marine phytoplankton species. Modeling studies aiming at describing the effects of environmental change on ecosystem level functions in marine phytoplankton would benefit from more knowledge about the role of genetic diversity. Future attempts to quantify the global aquatic primary productivity would benefit from including effects of intraspecific interactions.
Acknowledgments
This project was funded by the Academy of Finland Grants #251564, 256074 and 283061. We would like to thank Tvärminne Zoological Station for providing excellent research facilities for a pilot study. Fragment analyses were performed at KIGene, Neurogenetics, Karolinska University Hospital, Stockholm by Annika Eriksson. Jessica Titocci and Jani Strengell are acknowledged for assistance in the laboratory.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Alpermann TJ, Tillmann U, Beszteri B, Cembella AD, John U. (2010). Phenotypic variation and genotypic diversity in a planktonic population of the toxigenic marine dnoflagellate Alexandrium tamarense (Dinophyceae). Phycology 46: 18–32. [Google Scholar]
- Ashton IW, Miller AE, Bowman WD, Suding KN. (2010). Niche complementarity due to plasticity in resource use: plant partitioning of chemical N forms. Ecology 91: 3252–3260. [DOI] [PubMed] [Google Scholar]
- Bangert RK, Lonsdorf EV, Wimp GM, Shuster SM, Fischer D, Schweitzer JA et al. (2008). Genetic structure of a foundation species: scaling community phenotypes from the individual to the region. Heredity 100: 121–131. [DOI] [PubMed] [Google Scholar]
- Bell G. (1991). The ecology and genetics of fitness in Chlamydomonas. IV. The properties of mixtures of genotypes of the same species. Evolution 17: 1036–1046. [DOI] [PubMed] [Google Scholar]
- Benedetti-Cecchi L. (2003). The importance of the variance around the mean effect size of ecological processes. Ecology 84: 2335–2346. [Google Scholar]
- Cloern JE, Grenz C, Vidergar-Lucas L. (1995). An empirical model of the phytoplankton chlorophyll: carbon ratio—the conversion factor between productivity and growth rate. Limnol Oceanogr 40: 1313–1321. [Google Scholar]
- Cohen J. (1988) Statistical Power Analysis for the Behavioral Sciences. 2nd edn. Lawrence Erlbaum: Hillsdale, NJ, USA. [Google Scholar]
- Crutsinger GM, Souza L, Sanders NJ. (2008). Intraspecific diversity and dominant genotypes resist plant invasions. Ecol Lett 11: 16–23. [DOI] [PubMed] [Google Scholar]
- Curl H, Small LF. (1965). Variations in photosynthetic assimilation rates in natural, marine phytoplankton communities. Limnol Oceanogr 10: R67–R73. [Google Scholar]
- Dia A, Guillou L, Mauger S, Bigeard E, Marie D, Valero M et al. (2014). Spatiotemporal changes in the genetic diversity of harmful algal blooms caused by the toxic dinoflagellate Alexandrium minutum. Mol Ecol 23: 549–560. [DOI] [PubMed] [Google Scholar]
- Doak DF, Bigger D, Harding E, Marvier M, O'Malley R, Thomson D. (1998). The statistical inevitability of stability-diversity relationships in community ecology. Am Nat 151: 264–276. [DOI] [PubMed] [Google Scholar]
- Duffy JE, Richardson JP, France KE. (2005). Ecosystem consequences of diversity depend on food chain length in estuarine vegetation. Ecol Lett 8: 301–309. [Google Scholar]
- Ehlers A, Worm B, Reusch TBH. (2008). Importance of genetic diversity in eelgrass Zostera marina for its resilience to global warming. Mar Ecol Prog Ser. 355: 1–7. [Google Scholar]
- Friedman H. (1968). Magnitude of experimental effect and a table for its rapid estimation. Psychol Bull 70: 245–251. [Google Scholar]
- Gamfeldt L, Kallström B. (2007). Increasing intraspecific diversity increases predictability in population survival in the face of perturbations. Oikos 116: 700–705. [Google Scholar]
- Godhe A, Härnström K. (2010). Linking the planktonic and benthic habitat: genetic structure of the marine diatom Skeletonema marinoi. Mol Ecol 19: 4478–4490. [DOI] [PubMed] [Google Scholar]
- Gsell AS, Senerpont Domis LN, Pryztulska-Bartosiewicz A, Mooij WM, Donk E, Ibelings BW. (2012). Genotype-by-temperature interactions may help to maintain clonal diversity in Asterionella formosa (Bacillariophyceae). J Phycol 48: 1197–1208. [DOI] [PubMed] [Google Scholar]
- Guillard RRL. (1975). Culture of phytoplankton for feeding marine invertebrates. In: Smith WL, Chanley MH (eds). Culture of Marine Invertebrate Animals. Plenum Press: New York, pp 29–60. [Google Scholar]
- Hansson B, Westerdahl H, Hasselquist D, Åkesson M, Bensch S. (2004). Does linkage disequilibrium generate heterozygosity-fitness correlations in Great reed warblers? Evolution 58: 870–879. [DOI] [PubMed] [Google Scholar]
- Harper JL. (1977) Population Biology of Plants. Academic Press: London, UK. [Google Scholar]
- Hedges LV, Olkin I. (2014) Statistical Method for Meta-analysis. Academic press: New York. [Google Scholar]
- Hillebrand H, Bennett DM, Cadotte MW. (2008). Consequences of dominance: a review of evenness effects on local and regional ecosystem processes. Ecology 89: 1510–1520. [DOI] [PubMed] [Google Scholar]
- Hillebrand H, Shurin J (2005). In: Belgrano A, Scharler UM, Dunne J, Ulanowicz RE (eds). Aquatic Food Webs. Oxford University Press: Oxford.
- Hughes AR, Stachowicz JJ. (2004). Genetic diversity enhances the resistance of a seagrass ecosystem to disturbance. Proc Natl Acad Sci USA 101: 8998–9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AR, Inouye BD, Johnson MTJ, Underwood N, Vellend M. (2008). Ecological consequences of genetic diversity. Ecol Lett 11: 609–623. [DOI] [PubMed] [Google Scholar]
- Jespersen A, Christoffersen K. (1987). Measurements of chlorophyll-a from phytoplankton using ethanol as extraction solvent. Archiv für Hydrobiologie 109: 445–454. [Google Scholar]
- Klausmeier CA, Litchman E, Daufresne T, Levin SA. (2004). Optimal nitrogen-to-phosphorus stoichiometry of phytoplankton. Nature 429: 171–174. [DOI] [PubMed] [Google Scholar]
- Krause RT, August EM. (1983). Applicability of a multiresidue method and high performance liquid chromatography for determining quinomethionate in apples and oranges. J Assoc Off Anal Chem 66: 1018–1022. [PubMed] [Google Scholar]
- Lankau RA, Strauss SY. (2007). Mutual feedbacks maintain both genetic and species diversity in a plant community. Science 317: 1561–1563. [DOI] [PubMed] [Google Scholar]
- Lenth RV. (2013). lsmeans: Least-squares means. R package version, 2.
- Li L, Tilman D, Lambers H, Zhang FS. (2014). Plant diversity and overyielding: insights from belowground facilitation of intercropping in agriculture. New Phytologist 203: 63–69. [DOI] [PubMed] [Google Scholar]
- Litchman E, Klausmaier CA. (2008). Trait-based community ecology of phytoplankton. Ann Rev Ecol Evol Syst 39: 615–639. [Google Scholar]
- Lohbeck KT, Riebesell U, Reusch TBH. (2012). Adaptive evolution of a key phytoplankton species to ocean acidification. Nat Geosci 5: 346–351. [Google Scholar]
- Madritch M, Donaldson JR, Lindroth RL. (2006). Genetic identity of Populus tremuloides litter influences decomposition and nutrient release in a mixed forest stand. Ecosystems 9: 528–537. [Google Scholar]
- Marañón E, Behrenfeld MJ, Gonzáles N, Mouriño B, Zubkov MV. (2003). High variability of primary production in oligotrophic waters of the Atlantic Ocean: uncoupling from phytoplankton biomass and size structure. Mar Ecol Prog Ser 257: 1–11. [Google Scholar]
- Marañón E, Holligan PM, Varela M, Mouriño B, Bale AJ. (2000). Basin-scale variability of phytoplankton biomass, production and growth in the Atlantic Ocean. Deep Sea Res Part 1 Oceanogr Res Pap 47: 825–857. [Google Scholar]
- Meier H, Eilola K, Gustafsson B, Kuznetsov I, Neumann T, Savchuk O. (2012) Uncertainty Assessment of Projected Ecological Quality Indicators in Future Climate. Rapport Oceanografi No. 112, SMHI: Norrköping, Sweden. [Google Scholar]
- Osenberg CW, Schmitt RJ, Holbrook SJ, Abu-Saba KE, Flegal AR. (1994). Detection of environmental impacts: natural variability, effect size, and power analysis. Ecol Appl 4: 16–30. [Google Scholar]
- Park SDE. (2001). Trypanotolerance in West African cattle and the population genetic effects of selection. Ph.D. thesis, University of Dublin.
- Pielou EC. (1966). The measurement of diversity in different types of biological collections. J Theor Biol 13: 131–144. [Google Scholar]
- Pinheiro J, Bates D, Debroy S, Sarkar D. (2013) R Development Core Team (2012) nlme: Linear and Nonlinear Mixed Effects Models. R package version 3.1. R Foundation for Statistical Computing: Vienna. [Google Scholar]
- Ptacnik R, Solimini AG, Andersen T, Tamminen T, Brettum P, Lepistö L et al. (2008). Diversity predicts stability and resource use efficiency in natural phytoplankton communities. Proc Natl Acad Sci USA 105: 5134–5138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed DH, Frankham R. (2001). How closely correlated are molecular and quantitative measures of genetic variation? A meta-analysis. Evolution 55: 1095–1103. [DOI] [PubMed] [Google Scholar]
- Reed DH, Frankham R. (2003). Correlation between fitness and genetic diversity. Conserv Biol 17: 230–237. [Google Scholar]
- Reusch TBH, Ehlers A, Hämmerli A, Worm B. (2005). Ecosystem recovery after climatic extremes enhanced by genotypic diversity. Proc Natl Acad Sci USA 102: 2826–2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro S, Berge T, Lundholm N, Ellegaard M. (2013). Hundred years of environmental change and phytoplankton ecophysiological variability archived in coastal sediments. PLoS One 8: e61184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijstenbil J, Mur L, Wijnholds J, Sinke J. (1989). Impact of a temporal salinity decrease on growth and nitrogen metabolism of the marine diatom Skeletonema costatum in continuous cultures. Mar Biol 101: 121–129. [Google Scholar]
- Roger F, Godhe A, Gamfeldt L. (2012). Genetic diversity and ecosystem functioning in the face of multiple stressors. PloS One 7: e45007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roscher C, Temperton VM, Scherer-Lorenzen M, Schmitz M, Schumacher J, Schmid B et al. (2005). Overyielding in experimental grassland communities—irrespective of species pool or spatial scale. Ecol Lett 8: 419–429. [Google Scholar]
- Ryther JH, Yentsch CS. (1957). The estimation of phytoplankton production in the ocean from chlorophyll and light data. Limnol Oceanogr 2: 281–286. [Google Scholar]
- Schmid B, Hector A, Saha P, Loreau M. (2008). Biodiversity effects and transgressive overyielding. J Plant Ecol 1: 95–102. [Google Scholar]
- Schweitzer JA, Bailey JK, Hart SC, Whitham TG. (2005). Nonadditive effects of mixing cottonwood genotypes on litter decomposition and nutrient dynamics. Ecology 86: 2834–2840. [Google Scholar]
- Shannon CE. (1948). A mathematical theory of communication. Bell Syst Tech J 27: 379–423. [Google Scholar]
- Schläpfer F, Schmidt B. (1999). Ecosystem effects of biodiversity: a classification of hypotheses and exploration of empirical results. Ecol Appl 9: 893–912. [Google Scholar]
- Sjöqvist C, Godhe A, Jonsson P, Sundqvist L, Kremp A. (2015). Local adaptation and oceanographic connectivity patterns explain genetic differentiation of a marine diatom across the North Sea-Baltic Sea salinity gradient. Mol Ecol 24: 2871–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjöqvist C, Kremp A, Lindehoff E, Båmstedt U, Egardt J, Gross S et al. (2014). Effects of grazer presence on genetic structure of a phenotypically diverse diatom population. Microb Ecol 67: 83–95. [DOI] [PubMed] [Google Scholar]
- Skjelbred B, Edvardsen B, Andersen T. (2013). Environmental optima for seven strains of Pseudochattonella (Dictyochophyceae, Heterokonta). J Phycol 49: 54–60. [DOI] [PubMed] [Google Scholar]
- Smithson JB, Lenne JM. (1996). Varietal mixtures: a viable strategy for sustainable productivity in subsistence agriculture. Ann Appl Biol 128: 127–158. [Google Scholar]
- Solorzano L, Sharp JH. (1980). Determination of total dissolved phosphorus and particulate phosphorus in natural waters. Limnol Oceanogr 25: 754–758. [Google Scholar]
- Stachowicz JJ, Bruno JF, Duffy JE. (2007). Understanding the effects of marine biodiversity on communities and ecosystems. Ann Rev Ecol Evol Syst 38: 739–766. [Google Scholar]
- Suikkanen S, Kremp A, Hautala H, Krock B. (2013). Paralytic shellfish toxins or spirolides? The role of environmental and genetic factors in toxin production of the Alexandrium ostenfeldii complex. Harmful Algae 26: 52–59. [Google Scholar]
- Tesson SVM, Montresor M, Procaccini G, Kooistra WHCF. (2014). Temporal changes in population structure of a marine planktonic diatom. PLoS One 9: e114984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilman D. (1999). The ecological consequences of changes in biodiversity: a search for general principles. Ecology 80: 1455–1474. [Google Scholar]
- Tilman D, Reich PB, Knops JM. (2006). Biodiversity and ecosystem stability in a decade-long grassland experiment. Nature 441: 629–632. [DOI] [PubMed] [Google Scholar]
- Tuchmann ML, Theriot E, Stoermer EF. (1984). Effects of low level salinity concentrations on the growth of Cyclotella meneghiniana Kütz (Bacillariophyta). Arch Protistenkd 128: 319–326. [Google Scholar]
- Vrede K, Heldal M, Norland S, Bratbak G. (2002). Elemental composition (C, N, P) and cell volume of exponentially growing and nutrient-limited bacterioplankton. Appl Environ Microbiol 68: 2965–2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrieling EG, Poort L, Beelen TPM, Gieskes WWC. (1999). Growth and silica content of the diatoms Thalassiosira weissflogii and Navicula salinarum at different salinities and enrichments with aluminium. Eur J Phycol 34: 307–316. [Google Scholar]
- Whitham TG, Gehring CA, Lamit LJ, Wojtowicz T, Evans LM, Keith AR et al. (2012). Community specificity: life and afterlife effects of genes. Trends Plant Sci 17: 271–281. [DOI] [PubMed] [Google Scholar]
- Whitlock R. (2014). Relationships between adaptive and neutral genetic diversity and ecological structure and functioning: a meta-analysis. J Ecol 102: 857–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SL. (2001). Reduced genetic diversity in eelgrass transplantations affects both population growth and individual fitness. Ecol Appl 11: 1472–1488. [Google Scholar]
- Wood AM, Everroad RC, Wingard LM. (2005). Measuring growth rates in microalgal cultures. In: Anderson RA (ed). Algal Culturing Techniques. Elsevier Academic Press: Burlington, MA, pp 269–286.
- Worm B, Barbier EB, Beaumont N, Duffy JE, Folke C, Halpern BS et al. (2006). Impacts of biodiversity loss on ocean ecosystem services. Science 314: 787–790. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.