Abstract
Oxygenic photosynthesis evolved from anoxygenic ancestors before the rise of oxygen ~2.32 billion years ago; however, little is known about this transition. A high redox potential reaction center is a prerequisite for the evolution of the water-oxidizing complex of photosystem II. Therefore, it is likely that high-potential phototrophy originally evolved to oxidize alternative electron donors that utilized simpler redox chemistry, such as nitrite or Mn. To determine whether nitrite could have had a role in the transition to high-potential phototrophy, we sequenced and analyzed the genome of Thiocapsa KS1, a Gammaproteobacteria capable of anoxygenic phototrophic nitrite oxidation. The genome revealed a high metabolic flexibility, which likely allows Thiocapsa KS1 to colonize a great variety of habitats and to persist under fluctuating environmental conditions. We demonstrate that Thiocapsa KS1 does not utilize a high-potential reaction center for phototrophic nitrite oxidation, which suggests that this type of phototrophic nitrite oxidation did not drive the evolution of high-potential phototrophy. In addition, phylogenetic and biochemical analyses of the nitrite oxidoreductase (NXR) from Thiocapsa KS1 illuminate a complex evolutionary history of nitrite oxidation. Our results indicate that the NXR in Thiocapsa originates from a different nitrate reductase clade than the NXRs in chemolithotrophic nitrite oxidizers, suggesting that multiple evolutionary trajectories led to modern nitrite-oxidizing bacteria.
Introduction
The evolution of oxygenic photosynthesis in the ancestors of Cyanobacteria was the most important metabolic innovation in Earth's history; however, the evolutionary trajectory from ancestral anoxygenic phototrophy to oxygenic photosynthesis remains uncertain. Comparative biology of extant phototrophic microbes places important constraints on this process. There are currently seven bacterial phyla that contain members capable of chlorophyll-based phototrophy: Cyanobacteria, Alpha-, Beta- and Gammaproteobacteria, Chloroflexi, Chlorobi, Firmicutes, Acidobacteria and Gemmatimonadetes (Bryant et al., 2007; Overmann and Garcia-Pichel, 2013; Shih et al., 2013; Zeng et al., 2014). A fundamental division between phototrophs is based on the types of reaction centers that they use. In type I reaction centers (RCI and PSI) the final electron acceptor is a ferredoxin protein, whereas in type II reaction centers (RCII and PSII) the electrons are donated to quinones (Blankenship, 2014). Anoxygenic phototrophs only utilize a single reaction center, whereas oxygenic Cyanobacteria couple type I and type II reaction centers in series. Many substrates can be used as electron donors for phototrophy; however, there are important energetic limits (Supplementary Table S1). Extant anoxygenic reaction centers have redox potentials that range from +300 to +500 mV, and primarily oxidize lower potential (<+100 mV) electron donors (Supplementary Figure S1). In contrast, PSII used in oxygenic photosynthesis produces a very strong oxidant (~+1250 mV; Rappaport and Diner, 2008) that is capable of oxidizing water to molecular oxygen (+810 mV). The water-oxidizing complex in PSII could have only evolved after the origin of high-potential phototrophy (Fischer et al., 2016), suggesting that some type of high-potential anoxygenic phototrophy bridged the evolutionary gap between low-potential anoxygenic and high-potential oxygenic phototrophy (Olson, 1970; Rutherford and Faller, 2003). The highest redox potential substrate oxidized by known extant anoxygenic phototrophs is nitrite (+430 mV); therefore, phototrophic nitrite oxidizers might provide insight into the evolution of high-potential phototrophy (Griffin et al., 2007; Schott et al., 2010).
Phototrophic nitrite oxidizers are also important for understanding the evolution of chemolithotrophic nitrite-oxidizing bacteria (NOB). NOB catalyzes the second step of nitrification, which is a key process of the biogeochemical nitrogen cycle in oxic ecosystems. The highly structured intracytoplasmic membrane systems (ICMs) of the NOB Nitrobacter (Alphaproteobacteria) and Nitrococcus (Gammaproteobacteria) closely resemble the ICM of phototrophic purple bacteria. This ultrastructural similarity, and the relatively close phylogenetic affiliation of Nitrobacter and Nitrococcus with purple bacteria, led to the hypothesis that these chemolithotrophic NOB evolved from phototrophic ancestors (Teske et al., 1994). This scenario seemed to gain support from the surprising discovery of anoxygenic phototrophic nitrite oxidizers in the genera Rhodopseudomonas and Thiocapsa within the Alpha- and Gammaproteobacteria, respectively (Griffin et al., 2007; Schott et al., 2010). Recent studies revealed a second evolutionary origin of chemolithotrophic nitrite oxidation, where this metabolism evolved independently in the non-proteobacterial NOB genera Nitrospira and Nitrospina, probably through horizontal gene transfer of the nitrite-oxidizing enzyme and other functionally important proteins with anaerobic ammonium oxidizers (Lücker et al., 2010, 2013). Until now, it has remained unclear whether the extant anoxygenic phototrophic nitrite oxidizers developed along either of these two lines of NOB evolution or represent a third independent lineage.
To reveal the mechanism of phototrophic growth on nitrite, constrain the energetics of high-potential anoxygenic phototrophy, shed light on possible evolutionary links between photolithotrophic and chemolithotrophic nitrite oxidizers and elicudate the metabolic capabilities and ecophysiology of a phototrophic nitrite oxidizer, we sequenced and analyzed the genome of the nitrite-oxidizing phototroph Thiocapsa sp. strain KS1. In addition, we carried out biochemical experiments with fractionated cell-free extracts of a Thiocapsa KS1 pure culture to characterize the enzymatic nitrite-oxidizing and nitrate-reducing activities.
Materials and methods
Genome sequencing and analysis
Thiocapsa KS1 (JCM 15485) was grown as described in Supplementary Information. High-molecular-weight genomic DNA was isolated following the hexadecyltrimethylammonium bromide protocol as described elsewhere (Lücker et al., 2013). Sequencing and assembly were performed at LGC Genomics (Berlin, Germany) using GS FLX Titanium-sequencing technology. The Thiocapsa KS1 draft genome was annotated using the MicroScope annotation platform (Vallenet et al., 2013). Data from this sequencing project have been deposited at the European Nucleotide Archive under study ID PRJEB9229.
RCII and NXR sequences from genomes and metagenomes were retrieved from National Center for Biotechnology Information and DOE Joint Genome Institute. Sequence alignments were calculated using Muscle 3.8 (Edgar, 2004) or ARB (Ludwig, 2004). Phylogenetic analyses were performed on the CIPRES (Miller et al., 2010) cluster or on a desktop PC using RAxML (Stamatakis, 2014) and MrBayes (Ronquist and Huelsenbeck, 2003).
Cell-free extracts and protein analyses
Cell-free extracts from Thiocapsa KS1 were prepared and fractionated as described in Supplementary Information. The enzymatic activities of NXR were measured continuously with spectrophotometric assays (Supplementary Information). SDS-PAGE was performed according to Muller et al. (2009). Protein bands were sent to TopLab (Martinsried, Germany) for tryptic digestion and peptide mass fingerprinting without destaining. The fingerprints were matched (Mascot search engine) against the NCBI protein database.
Results
Genome analysis
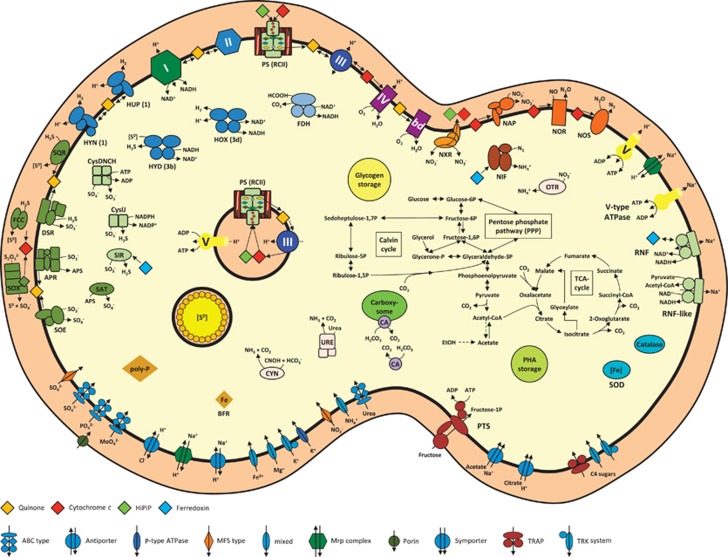
Thiocapsa KS1 is a gammaproteobacterium isolated from a sewage treatment facility in Konstanz, Germany, by selecting for photoautotrophic growth on nitrite (Griffin et al., 2007; Schott et al., 2010). This versatile phototroph can utilize a range of simple organic carbon molecules, sulfur compounds, H2 and nitrite as electron donors. We briefly describe the genes associated with respiration, phototrophy, nitrogen metabolism and other functions that confer Thiocapsa KS1 with exceptionally high ecophysiological flexibility, which makes it the most metabolically versatile nitrite-oxidizing microorganism known (Figure 1 and Supplementary Table S2).
Figure 1.
Metabolic diversity of Thiocapsa KS1. For details see main and supplemental text. APR, adenylylsulphate reductase complex; bd, cytochrome bd quinol oxidase; BFR, bacterioferritin; CA, carbonic anhydrase; CYN, cyanate hydratase; Cys, assimilatory sulfate reduction complexes; DSR, reverse dissimilatory sulfite reductase; FCC, sulfide dehydrogenase; FDH, formate dehydrogenase; HOX, HUP, HYD, HYN, hydrogenases (with enzyme classification indicated in brackets); NAP, periplasmic nitrate reductase; NIF, nitrogenase; NOR, nitric oxide reductase; NOS, nitrous oxide reductase; NXR, nitrite oxidoreductase; OTR, octaheme tetrathionate reductase; PHA, polyhydroxyalkanoate; PS, photosystem (type II reaction center); PTS, phosphotransferase system; RNF, H+/Na+-translocating NAD-ferredoxin reductase; SAT, sulfate adenylyltransferase; SIR, sulfite reductase; SOD, superoxide dismutase; SOE, sulfite-oxidizing enzyme; SOX, sulfur/thiosulfate oxidation protein complex; SQR, sulfide-quinone reductase; TCA cycle, tricarboxylic acid cycle; URE, urease. Enzyme complexes of the electron transport chain are labeled by Roman numerals: I, NADH dehydrogenase; II, succinate dehydrogenase/fumarate reductase; III, cytochrome bc1 complex; IV, cbb3-type cytochrome c oxidase; Orange, red, green and blue diamonds represent quinones, cytochrome c proteins, HiPIPs and ferredoxins, respectively.
General respiration
The genome of Thiocapsa KS1 contained one copy of a 14-subunit NADH dehydrogenase (complex I) that allows organotrophic respiration. Under photolithoautotrophic growth conditions, complex I will operate in reverse to produce NADH required for carbon fixation (Elbehti et al., 2000; Supplementary Figure S2). Biosynthetic pathways for both menaquinone and ubiquinone were present, consistent with the detection of both quinone types in the close relative T. roseopersicina (Imhoff, 1984). Thiocapsa KS1 had one cytochrome (cyt.) bc1 complex (complex III) that conserves energy during phototrophic growth, aerobic respiration and denitrification. Three high-affinity oxygen reductases were present; two C-family heme–copper oxidoreductases (cyt. cbb3 oxidase, complex IV) and a quinol-oxidizing cyt. bd oxidase. Low-affinity A-family oxygen reductases were absent. Details on ATP production and reverse electron transport are provided in Supplementary Information.
Phototrophy
Thiocapsa KS1 encoded one set of RCII genes: PufL, PufM, PufC and PuhA. These had very high sequence identities (98% for PufL and PufM) to the closest sequenced strain, T. marina DSM 5653, which was confirmed using PCR. Thiocapsa KS1 and T. marina contained a second copy of the RCII cyt. c subunit PufC, which appeared to be fused to an outer membrane protein. Multiple copies of genes for the light-harvesting complexes LH1 and LH2 were also present. Thiocapsa KS1 utilizes bacteriochlorophyll a for phototrophic growth, with genes responsible for its biosynthesis distributed throughout the genome. The pathway required for the biosynthesis of spirilloxanthin, the major pigment in T. roseopersicina (Kovács et al., 2003), was complete. Several cyt. c4-like di-heme proteins, along with two copies of high-potential iron–sulfur proteins (HiPIPs), were available to act as diffusible periplasmic electron carriers.
Sulfur metabolism
Thiocapsa KS1 encoded a wide diversity of enzymes involved in oxidative sulfur metabolism (Gregersen et al., 2011; Figure 1, Supplementary Information). Systems for the oxidation of sulfur, thiosulfate and sulfide were identified, which allow for the utilization of these sulfur compounds as electron donors for anoxygenic photosynthesis. During phototrophic growth with sulfide or thiosulfate, elemental sulfur (S0) is stored in extracytoplasmic sulfur globules within chromatophores (Pattaragulwanit et al., 1998), a property shared with other purple sulfur bacteria. These stores can later be used as electron donors when environmental sulfide and thiosulfate concentrations decrease. The complete assimilatory pathway for sulfate reduction via adenosinephosphosulfate and 3'-phosphoadenosinephosphosulfate was also present in the genome.
Hydrogenases
Thiocapsa KS1 encoded at least five Ni–Fe hydrogenases that can oxidize H2 as an electron source for photosynthesis, recycle H2 formed during diazotrophic growth or produce H2 during phototrophic growth on reduced sulfur or carbon compounds (Maróti et al., 2010; Supplementary Information).
Carbon metabolism
Thiocapsa KS1 assimilates CO2 via the Benson–Bassham cycle, which was complete in the genome. Genes for the ribulose-bisphosphate carboxylase (type I RuBisCO) large and small subunits were duplicated and a type IV RuBisCO, which is not involved in carbon fixation (Tabita et al., 2007), was also present. Carboxysome shell proteins and carbonic anhydrases indicated the presence of carboxysomes for concentrating CO2 (Yeates et al., 2008). Phosphoglycolate formed by the oxygenase activity of RuBisCO may be fed into the tricarboxylic acid cycle via the glycolate salvage pathway and glyoxylate shunt. The complete C4-dicarboxylic acid cycle allows additional CO2 fixation by phosphoenolpyruvate carboxylation. Thiocapsa KS1 also possessed a full gene inventory for photo- or chemoorganoheterotrophic growth (Supplementary Information).
Nitrogen metabolism
As a diazotroph, Thiocapsa KS1 encoded a complete set of nif genes for nitrogen fixation including molybdenum-iron nitrogenase (NifDK) and nitrogenase reductase (NifH). Thiocapsa KS1 can assimilate ammonium and also utilize nitrite and nitrate as nitrogen sources in culture (Schott et al., 2010), although the organism did not possess genes for canonical assimilatory nitrate or nitrite reductases. Interestingly, the genome encoded several alternative mechanisms (Supplementary Information). In addition, the presence of genes encoding nickel-dependent urease, cyanate hydratase, thiocyanate hydrolase and ethanolamine ammonia-lyase indicated a broad spectrum of reduced nitrogen sources for Thiocapsa KS1.
Nitrite oxidation in Thiocapsa KS1 was mediated by a Mo-bis-MGD-binding nitrite oxidoreductase (NXR), an enzyme that can catalyze nitrite oxidation and nitrate reduction (NO2−+H2O ↔ NO3−+2e− +2H+; Tanaka et al., 1983; Sundermeyer-Klinger et al., 1984). The NXR of Thiocapsa KS1 was similar to the NXR forms found in the chemolithotrophic NOB Nitrobacter, Nitrococcus and Nitrolancea (Starkenburg et al., 2008; Sorokin et al., 2012) and to the dissimilatory membrane-bound nitrate reductase (NAR) system found in many nitrate-reducing organisms. The NXR complex consisted of the α subunit (NxrA), which contains the catalytic site, the electron-channeling β subunit (NxrB) with four cysteine-rich binding motifs for [Fe-S] clusters and the γ subunit (NxrC), a membrane protein that putatively binds two heme b groups. Electrons derived from nitrite flow from NxrA through NxrB to NxrC, which anchors NXR in the membrane and transfers the electrons to the downstream electron carriers. Like in Nitrobacter (Kirstein and Bock, 1993; Spieck et al., 1996) the NxrA and NxrB subunits were oriented toward the cytoplasm and resembled the NarGH subunits of bacterial NARs. A TorD-like chaperone, which probably inserts the Mo-bis-MGD cofactor into NxrA (Blasco et al., 1998; Ilbert et al., 2003), was encoded between the nxrB and nxrC genes.
Although Thiocapsa KS1 has candidate genes for a complete denitrification pathway (Figure 1) and can use a range of organic and inorganic low-potential electron donors for energy conservation, no growth was observed under anoxic conditions in the presence of these electron donors and nitrate as electron acceptor (Griffin et al., 2007; Schott et al., 2010). Nitrate reduction to nitrite could be performed by a periplasmic NAR (NapDAGHB). This complex is missing NapC, similar to the periplasmic NARs found in many Epsilonproteobacteria (Simon et al., 2003), suggesting that an alternative electron transfer pathway to NapA is utilized. In addition, NXR could function as a membrane-bound NAR (see below). Iron- (NirS) or copper- (NirK) dependent nitrite reductases are missing. Instead, the Thiocapsa KS1 genome contained two copies of hydroxylamine dehydrogenase-related proteins, which have been implied in nitrite reduction to NO in Methylococcus capsulatus strain Bath (Campbell et al., 2011). In addition, strain KS1 had both a nitric oxide reductase and a nitrous oxide reductase that would enable the sequential reduction of NO to N2.
Enzymatic activities of NXR
Cell-free extracts were prepared from Thiocapsa KS1 cultures to test the nitrite-oxidizing and nitrate-reducing enzyme activities. Although cultures grown photolithoautotrophically with nitrite as the sole electron donor were analyzed, no nitrite-oxidizing enzyme activity was detected by the two assays applied (Meincke et al., 1992). In contrast, a pronounced nitrate-reducing activity was measurable (Supplementary Table S3). The highest specific NAR activity (up to 1700 mU·(mg protein)−1) was found in the membrane fraction, whereas the cytosolic fraction showed less than half of this activity (Supplementary Table S3). The protein contents of the membrane and cytosolic fractions were roughly equal. No activity was detected with NAD(P)H as an alternative electron donor and dithiothreitol as reducing agent. Cells grown with the alternative electron donor H2 had no NAR activity when grown with ammonium as nitrogen source (Supplementary Table S3), and grew only poorly with nitrate as nitrogen source so that no cell extracts could be prepared from these cultures. Extracts from cells grown on fructose and nitrate had a very low NAR activity that was found exclusively in the cytosolic fraction (Supplementary Table S3), indicating the involvement of a distinct enzyme system for assimilatory nitrate reduction.
Cell fractions were also analyzed using one-dimensional SDS-PAGE (Supplementary Figure S3). The protein patterns from nitrite-grown cells contained two strong bands that were absent from H2-grown cells and thus assumed to be involved in nitrite oxidation (Supplementary Figure S3). These bands had estimated sizes of 130–150 and 55–60 kDa, respectively, which resemble the sizes of NxrA and NxrB from Nitrobacter (Meincke et al., 1992). Consistently, mass spectrometric fingerprint analysis and in silico comparison of the obtained peaks to publicly available protein sequences confirmed that the large bands (130–150 kDa) in the cytosolic and membrane fractions represented the NXR α subunit, whereas the smaller bands (55–60 kDa) were identified as the NXR β subunit (Supplementary Figure S3).
Reaction center analyses
A key step in the transition from anoxygenic phototrophy to oxygenic photosynthesis was the evolution of a high-potential reaction center. RCIIs using menaquinones as electron acceptors (Chloroflexi (Collins et al., 2011) and the gammaproteobacterium Halorhodospira halophila (Schoepp-Cothenet et al., 2009)) have special pair P/P+ redox potentials ranging from +270 to +390 mV, whereas those utilizing ubiquinone have redox potentials in the range of +400 to +505 mV.
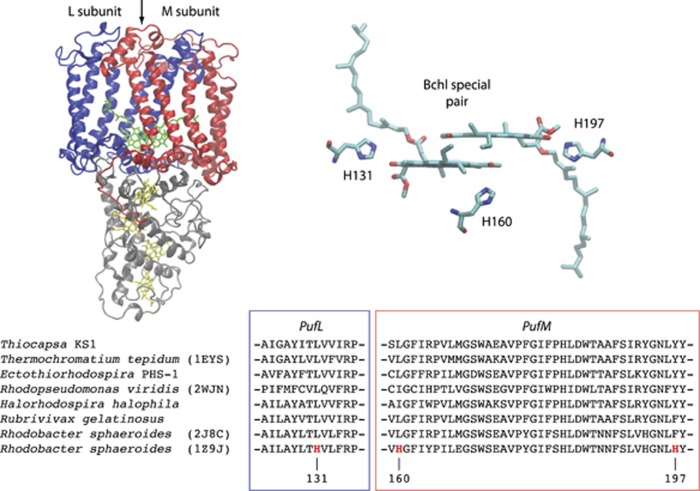
The redox potential of the P/P+ transition in RCII is determined by intrinsic chemical properties of bacteriochlorophyll, the electronic coupling of the special pair and their interactions with the surrounding protein (Allen and Williams, 2014). All known RCIIs use bacteriochlorophylls coupled in a special pair, with the redox potential predominately tuned by the protein environment surrounding the special pair. Experimental studies have shown that it is possible to increase the P/P+ redox potential of RCII by (1) forming hydrogen bonds with the 2-acetyl and 9-keto groups of the special pair of bacteriochlorophylls, (2) converting one of the special pair bacteriochlorophylls into a bacteriopheophytin by altering its Mg2+ ligand or (3) adding positive charges that electrostatically interact with the special pair. In Rhodobacter sphaeroides the effects of adding hydrogen bonds to the special pair were shown to be additive, with three additional hydrogen bonds increasing the redox potential from +505 to +765 mV (Lin et al., 1994; Allen and Williams, 2011; Figure 2). In Blastochloris viridis the replacement of a special pair bacteriochlorophyll histidine ligand with leucine resulted in the loss of Mg2+ (converting it to a bacteriopheophytin), shifting the redox potential from +517 to +772 mV (Ponomarenko et al., 2009). Furthermore, extensive mutations in R. sphaeroides also showed that the addition of a positive charge within 10 Å of the special pair increased the redox potential by 50 mV. Protein modifications such as these can be easily identified in multiple sequence alignments.
Figure 2.
High-potential RCIIs? Shown are known mutations that produce high-potential RCIIs. Sequence analysis detected no natural variants capable of forming hydrogen bonds at these positions (Supplementary Table S4). PufL is blue, PufM is red and PufC is gray.
The RCII from Thiocapsa KS1 has none of the modifications described above that could raise its redox potential, and has the exact same residues interacting with the special pair as Thermochromatium tepidum (Ivancich et al., 1996), implying a redox potential of ~+500 mV (Figure 2, Supplementary Table S4).
To determine whether RCIIs found in nature exhibited any modifications that would enable high-potential phototrophy, we analyzed the RCII genes from all sequenced genomes and publically available metagenomes. Sequence alignments of >3000 RCII proteins (Supplementary Table S5) identified no variants in positions that could form additional hydrogen bonds with either the 2-acetyl and 9-keto groups of the special pair (Supplementary Table S4). In addition, no sequences were found that had modified bacteriochlorophyll ligands, and no variants were identified that would modify electrostatic interactions with the special pair (Krammer et al., 2009). Together, this implies that extant RCIIs are unable to achieve redox potentials greater than ~+500 mV.
Discussion
Ecophysiology of Thiocapsa KS1
Although chemolithoautotrophic NOB have been studied for decades, we are just beginning to understand the physiology of phototrophic nitrite oxidizers. Traditionally, the chemolithoautotrophic NOBs were assumed to be highly specialized and metabolically restricted organisms. This picture changed with discoveries such as the chemoorganoheterotrophic Nitrobacter (Bock, 1976) and the aerobically H2-oxidizing, or anaerobically formate-consuming and nitrate-reducing, Nitrospira (Koch et al., 2014, 2015). However, the Thiocapsa KS1 genome has revealed an exceptionally high degree of metabolic versatility not found in other NOB. This flexibility likely allows Thiocapsa KS1 to colonize a wide variety of ecological niches and to persist under changing environmental conditions, using photolithotrophic nitrite oxidation, as only one of several alternative lifestyles. Further studies on the ecophysiology of phototrophic NOB will be crucial for assessing the relative contribution of photolithotrophic nitrite oxidation to overall nitrification in the environment. The whole-genome analysis of Thiocapsa KS1 presented here provides a number of hypotheses on the ecophysiology of phototrophic NOB that can be tested in targeted experiments in either pure cultures of Thiocapsa KS1 or in microbial communities.
Evolution of nitrite oxidation in Thiocapsa KS1 and chemolithotrophic nitrite oxidizers
Thiocapsa KS1 is the first identified (Griffin et al., 2007) and genomically characterized (this study) nitrite-oxidizing phototroph. As several genomes of chemolithotrophic NOB had been sequenced earlier, we were able to compare the nitrite-oxidizing systems of these organisms to elucidate whether and how the evolutionary pathways of photo- and chemolithotrophic nitrite oxidation are related.
To enable photolithotrophic growth on nitrite, Thiocapsa KS1 utilizes NXR. Notably, in the known aerobic chemolithotrophic NOB two different forms of NXR have been identified. They differ in the localization of their α and β subunits on either the cytoplasmic or periplasmic side of the membrane, and thus in the bioenergetics of nitrite oxidation. The NXR of Nitrospira (Lücker et al., 2010) and Nitrospina (Lücker et al., 2013) is located on the periplasmic side of the membrane, indicating that nitrite oxidation occurs in the periplasm of these organisms. This is energetically ideal, given that protons generated by the reaction directly contribute to the pmf. In contrast, NOB from the Proteobacteria, such as Nitrobacter (Starkenburg et al., 2006), and from the Chloroflexi (Sorokin et al., 2012), oxidizes nitrite on the cytoplasmic side of the membrane. In this case protons from nitrite oxidation are released into the cytoplasm, with no contribution to pmf, and electrons are transferred across the membrane to a soluble cyt. c550 on the periplasmic side (Figure 1 and Supplementary Figure S4). The redox potential of this cyt. c550 appears to be low (~+280 mV in Nitrobacter; Ketchum et al., 1969), and this step of the electron transport chain might require energy from the electrochemical membrane potential (Cobley, 1976; Ferguson, 1982). Assuming a membrane potential of ~150 mV, the electrons from the nitrite/nitrate couple may reach a potential of ~+280 mV when transferred along the electrochemical gradient to the positive side of the membrane.
The NXR of Thiocapsa KS1 faces similar constraints as the Nitrobacter system because its α subunit with the active site is also located on the cytoplasmic side of the membrane. In phototrophic bacteria, for example, R. capsulatus, there are at least two electron transport pathways to the reaction center that involve c-type cytochromes, one via the soluble cyt. c2 and one via the membrane-bound cyt. cy (Jenney and Daldal, 1993; Jenney et al., 1994). With a range of +345 to +395 mV (Pettigrew et al., 1978), the redox potential of various cyt. c2 from different phototrophic purple bacteria would be in the right range to allow electron transfer from nitrite to the reaction center. Alternatively, a HiPIP protein could be employed as soluble electron carrier instead of cyt. c. For example, the purple sulfur bacterium Allochromatium vinosum preferentially uses HiPIP instead of c-type cytochromes during photo-organotrophic growth (Van Driessche et al., 2003). HiPIP potentials range from +50 to +500 mV (Heering et al., 1995), and the HiPIP protein of T. roseopersicina has a redox potential of +342 mV (Przysiecki et al., 1985). Hence, electron transfer from nitrite via NXR to a HiPIP or cyt. c should be possible and might be facilitated by the membrane potential as outlined above for the chemolithotrophic NOB and cyt. c550.
Although the NXRs from Thiocapsa KS1 and Nitrobacter are both membrane-associated and oriented toward the cytoplasm, we identified the key differences in these enzymes. In Thiocapsa KS1, the transfer of electrons from NXR across the cell membrane and onto the soluble electron carrier most likely involves a unique di-heme cyt. c that is fused onto the γ subunit of NXR (NxrC; Supplementary Figure S4). This fusion is unique among the known nitrite oxidizers, and its functional analog in all known chemolithotrophic NOBs with a cytoplasmic NXR, such as Nitrobacter, is a di-heme cyt. c encoded by a separate gene upstream of the nxrA gene (Sorokin et al., 2012). Protein sequence analyses indicate different evolutionary origins for these cyt. c moieties. The separate cyt. c occurs also in several dissimilatory membrane-bound NARs from heterotrophic denitrifiers such as Thermus thermophilus, where it belongs to a unique electron transport chain from a special NADH oxidase via NAR to nitrite-, NO- and N2O reductases (Cava et al., 2008).
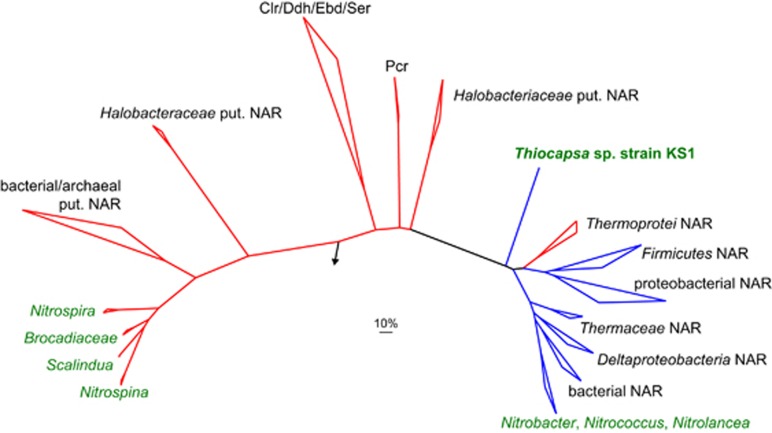
Phylogenetic analyses of NxrA and NxrB protein sequences showed that the periplasmic NXR forms of Nitrospira and Nitrospina are phylogenetically distinct from the cytoplasmic forms of Nitrobacter, Nitrococcus and Nitrolancea (Lücker et al., 2010, 2013; Sorokin et al., 2012; Figure 3, Supplementary Figure S5). Interestingly, the NxrA and NxrB sequences from Thiocapsa KS1 are not closely related to either of these two NXR groups in phylogenetic trees, but instead form a separate deep-branching lineage, which is only distantly related to the cytoplasmic NXRs from chemolithotrophic NOB (Supplementary Figure S5). Both cytoplasmic NXR lineages are related to NARs from phylogenetically diverse bacteria and archaea (Figure 3, Supplementary Figure S5). Consistent with this phylogenetic placement, the NXRs of Nitrobacter (Sundermeyer-Klinger et al., 1984) and of Thiocapsa KS1 (this study) have strong nitrate-reducing activity. The lack of detectable nitrite-oxidizing activity in the whole-cell extracts from strain KS1 might reflect the experimental design, for example, the absence of a membrane potential in the cell-free extracts that could facilitate the electron transfer from nitrite to the soluble acceptor. Thus, the topologies of the phylogenetic trees (Figure 3, Supplementary Figure S5) and the nitrate-reducing activities strongly suggest that the cytoplasmic NXR of Thiocapsa KS1 originated from a different NAR ancestor than the NXRs found in chemolithotrophic NOB. Similar conversions of reductases into oxidases have been described previously in other nitrogen cycle enzymes (Klotz and Stein, 2008).
Figure 3.
Phylogenetic analyses of Thiocapsa KS1 NXR and related enzymes of the dimethyl sulfoxide reductase type II family. Bayesian inference tree of the large (NxrA) subunit. Names of validated enzymes are indicated (Clr, chlorate reductase; Ddh, dimethylsulfide dehydrogenase; Ebd, ethylbenzene dehydrogenase; NAR, nitrate reductase; Pcr, perchlorate reductase; Ser, selenatereductase). The arrow indicates the outgroup; the scale bar represents 10% estimated sequence divergence. Enzyme complexes with the active center located on the cytoplasmic side of the membrane are indicated in blue, complexes oriented toward the periplasm in red. Names of organisms containing a NXR are shown in green. Note the three independent origins of NXR.
Two T. roseopersicina strains are also capable of phototrophic nitrite oxidation (Schott et al., 2010); however, NXR is not found in T. marina DSM 5653 or any other sequenced member of the Chromatiaceae. This suggests that the common ancestor of Thiocapsa KS1 and T. roseopersicina acquired NXR via lateral gene transfer after divergence from T. marina. The donor of the nxr genes in this lateral gene transfer event remains unknown, but cannot be a member of the known chemolithotrophic NOB clades because the NXR of Thiocapsa is not closely affiliated with the NXR forms of any of these organisms (Figure 3; Supplementary Figure S5).
Hence, our analyses of Thiocapsa KS1 do not support the earlier hypothesis that nitrite oxidation in Nitrobacter and other chemolithotrophic NOB evolved from a phototrophic ancestor. Instead, the evolutionary history of nitrite oxidation seems to be more complex and comprises at least three independent origins: two for the cytoplasmic NXRs and one for the periplasmic forms (Figure 3, Supplementary Figure S5).
The limits of high-potential anoxygenic phototrophy
The high redox potential of nitrite (+430 mV) poses a challenge for anoxygenic phototrophy. Currently, studied RCIIs from Proteobacteria and Chloroflexi have redox potentials in the range of +270 to +505 mV (Supplementary Table S1). Expectedly, RCIIs that reduce menaquinol, such as those found in Chloroflexi (Collins et al., 2011) and Halorhodospira halophila SL1 (Schoepp-Cothenet et al., 2009), have redox potentials at the lower end of this range. The RCIIs from Chloroflexi, Gemmatimonadetes and the majority of Proteobacteria contain a tetra-heme cyt. c protein (PufC) that is bound on the periplasmic side of the RCII complex and serves as a wire connecting the soluble electron donor with the special pair. The redox potentials of these hemes vary, with the direct donor to the special pair (heme c559 in Figure 2) having a potential of ~+380 mV (Nogi et al., 2005). The majority of electron donors for anoxygenic phototrophy have redox potentials ~0 mV or lower, providing a significant thermodynamic driving force for the overall electron transfer reaction (Supplementary Figure S1). Nitrite has a much higher redox potential—higher than PufC and very close to the redox potential of the RCII special pair in Thiocapsa (+490 mV in T. pfennigii; Prince, 1978). This raises a key question: how do anoxygenic phototrophs use nitrite as an electron donor for photoautotrophic growth?
To drive nitrite oxidation, anoxygenic phototrophs have two options: either transfer electrons from nitrite to the electron carrier pool utilized by other electron donors, possibly by expending energy, or use a high-potential cyt. c (similar to those found in acidophilic iron oxidizers) and a modified reaction center that is able to generate a higher redox potential that can produce the overpotential needed to drive the reaction (Supplementary Figure S2). Thiocapsa KS1 employs the first scenario. This shows that a high-potential reaction center is not required for nitrite oxidation. However, it remains possible that other anoxygenic phototrophs might utilize a high-potential reaction center (Supplementary Figure S2) to oxidize nitrite or other high-potential substrates. In the analyses of ~3000 RCII sequences from genomic and environmental metagenomic data sets, we observed no mutations that would confer a higher potential on any RCII found to date (Supplementary Table S4). This implies that RCII can oxidize only substrates with redox potentials lower than ~+500 mV, and that high-potential anoxygenic phototrophy using RCII is either uncommon or absent in modern environments. Thus, extant RCIIs offer a limited understanding of the evolution of the high-potential phototrophy required for the evolution of oxygenic photosynthesis.
Acknowledgments
We are grateful to LABGeM and the National Infrastructure ‘France Genomique' for annotation support within the MicroScope platform. Support for this work was provided by the Caltech Center for Environmental Microbial Interactions (WWF), the David and Lucile Packard Foundation (WWF), the National Science Foundation Graduate Research Fellowship program (JEJ), the Austrian Science Fund (FWF, grant P24101-B22), the Radboud Excellence Initiative and the Netherlands Organization for Scientific Research (NWO, VENI grand 863.14.019 to SL), the Deutsche Forschungsgemeinschaft, Bonn, Germany, grant Schi 180/12 (BS) and the Agouron Institute (JH and WWF). JH is an Agouron Postdoctoral Scholar.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Allen JP, Williams JC. (2014). Energetics of cofactors in photosynthetic complexes: relationship between protein–cofactor interactions and midpoint potentials. In: Golbeck JH, Van der Est A (eds) The Biophysics of Photosynthesis. Springer: New York, NY, USA, pp 275–295. [Google Scholar]
- Allen JP, Williams JC. (2011). The evolutionary pathway from anoxygenic to oxygenic photosynthesis examined by comparison of the properties of photosystem II and bacterial reaction centers. Photosyn Res 107: 59–69. [DOI] [PubMed] [Google Scholar]
- Blankenship RE. (2014) Molecular Mechanisms of Photosynthesis, 2nd edn. Wiley-Blackwell. [Google Scholar]
- Blasco F, Santos Dos JP, Magalon A, Frixon C, Guigliarelli B, Santini CL et al. (1998). NarJ is a specific chaperone required for molybdenum cofactor assembly in nitrate reductase A of Escherichia coli. Mol Microbiol 28: 435–447. [DOI] [PubMed] [Google Scholar]
- Bock E. (1976). Growth of Nitrobacter in the presence of organic matter. Arch Microbiol 108: 305–312. [DOI] [PubMed] [Google Scholar]
- Bryant DA, Costas AMG, Maresca JA, Chew AGM, Klatt CG, Bateson MM et al. (2007). Candidatus Chloracidobacterium thermophilum: an aerobic phototrophic Acidobacterium. Science 317: 523–526. [DOI] [PubMed] [Google Scholar]
- Campbell MA, Nyerges G, Kozlowski JA, Poret-Peterson AT, Stein LY, Klotz MG. (2011). Model of the molecular basis for hydroxylamine oxidation and nitrous oxide production in methanotrophic bacteria. FEMS Microbiol Lett 322: 82–89. [DOI] [PubMed] [Google Scholar]
- Cava F, Zafra O, Berenguer J. (2008). A cytochrome c containing nitrate reductase plays a role in electron transport for denitrification in Thermus thermophilus without involvement of the bc respiratory complex. Mol Microbiol 70: 507–518. [DOI] [PubMed] [Google Scholar]
- Cobley JG. (1976). Energy-conserving reactions in phosphorylating electron-transport particles from Nitrobacter winogradskyi. Activation of nitrite oxidation by the electrical component of the protonmotive force. Biochem J 156: 481–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins AM, Kirmaier C, Holten D, Blankenship RE. (2011). Kinetics and energetics of electron transfer in reaction centers of the photosynthetic bacterium Roseiflexus castenholzii. Biochim Biophys Acta 1807: 262–269. [DOI] [PubMed] [Google Scholar]
- Edgar RC. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbehti A, Brasseur G, Lemesle-Meunier D. (2000). First evidence for existence of an uphill electron transfer through the bc(1) and NADH-Q oxidoreductase complexes of the acidophilic obligate chemolithotrophic ferrous ion-oxidizing bacterium Thiobacillus ferrooxidans. J Bacteriol 182: 3602–3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson SJ. (1982). Is a proton-pumping cytochrome oxidase essential for energy conservation in Nitrobacter? FEBS Lett 146: 239–243. [Google Scholar]
- Fischer WW, Hemp J, Johnson JE. (2016). Evolution of oxygenic photosynthesis. Annu Rev Earth Planet Sci 44 doi:10.1146/annurev-earth-060313-054810. [Google Scholar]
- Gregersen LH, Bryant DA, Frigaard N-U. (2011). Mechanisms and evolution of oxidative sulfur metabolism in green sulfur bacteria. Front Microbiol 2: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin BM, Schott J, Schink B. (2007). Nitrite, an electron donor for anoxygenic photosynthesis. Science 316: 1870. [DOI] [PubMed] [Google Scholar]
- Heering HA, Bulsink BM, Hagen WR, Meyer TE. (1995). Influence of charge and polarity on the redox potentials of high-potential iron-sulfur proteins: evidence for the existence of two groups. Biochemistry 34: 14675–14686. [DOI] [PubMed] [Google Scholar]
- Ilbert M, Méjean V, Giudici-Orticoni M-T, Samama J-P, Iobbi-Nivol C. (2003). Involvement of a mate chaperone (TorD) in the maturation pathway of molybdoenzyme TorA. J Biol Chem 278: 28787–28792. [DOI] [PubMed] [Google Scholar]
- Imhoff JF. (1984). Quinones of phototrophic purple bacteria. FEMS Microbiol Lett 25: 85–89. [Google Scholar]
- Ivancich A, Kobayashi M, Drepper F, Fathir I, Saito T, Nozawa T et al. (1996). Hydrogen-bond interactions of the primary donor of the photosynthetic purple sulfur bacterium Chromatium tepidum†. Biochemistry 35: 10529–10538. [DOI] [PubMed] [Google Scholar]
- Jenney FE, Daldal F. (1993). A novel membrane-associated c-type cytochrome, cyt cy, can mediate the photosynthetic growth of Rhodobacter capsulatus and Rhodobacter sphaeroides. EMBO J 12: 1283–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenney FE, Prince RC, Daldal F. (1994). Roles of the soluble cytochrome c2 and membrane-associated cytochrome cy of Rhodobacter capsulatus in photosynthetic electron transfer. Biochemistry 33: 2496–2502. [DOI] [PubMed] [Google Scholar]
- Ketchum PA, Sanders HK, Gryder JW, Nason A. (1969). Characterization of cytochrome c from Nitrobacter agilis. Biochim Biophys Acta 189: 360–365. [DOI] [PubMed] [Google Scholar]
- Kirstein K, Bock E. (1993). Close genetic relationship between Nitrobacter hamburgensis nitrite oxidoreductase and Escherichia coli nitrate reductases. Arch Microbiol 160: 447–453. [DOI] [PubMed] [Google Scholar]
- Klotz MG, Stein LY. (2008). Nitrifier genomics and evolution of the nitrogen cycle. FEMS Microbiol Lett 278: 146–156. [DOI] [PubMed] [Google Scholar]
- Koch H, Galushko A, Albertsen M, Schintlmeister A, Gruber-Dorninger C, Lucker S et al. (2014). Growth of nitrite-oxidizing bacteria by aerobic hydrogen oxidation. Science 345: 1052–1054. [DOI] [PubMed] [Google Scholar]
- Koch H, Lücker S, Albertsen M, Kitzinger K, Herbold C, Spieck E et al. (2015). Expanded metabolic versatility of ubiquitous nitrite-oxidizing bacteria from the genus Nitrospira. Proc Natl Acad Sci USA 112: 11371–11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovács AT, Rákhely G, Kovács KL. (2003). Genes involved in the biosynthesis of photosynthetic pigments in the purple sulfur photosynthetic bacterium Thiocapsa roseopersicina. Appl Environ Microbiol 69: 3093–3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer E-M, Sebban P, Ullmann GM. (2009). Profile hidden Markov models for analyzing similarities and dissimilarities in the bacterial reaction center and photosystem II. Biochemistry 48: 1230–1243. [DOI] [PubMed] [Google Scholar]
- Lin X, Murchison HA, Nagarajan V, Parson WW, Allen JP, Williams JC. (1994). Specific alteration of the oxidation potential of the electron donor in reaction centers from Rhodobacter sphaeroides. Proc Natl Acad Sci USA 91: 10265–10269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W. (2004). ARB: a software environment for sequence data. Nucleic Acids Res 32: 1363–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lücker S, Nowka B, Rattei T, Spieck E, Daims H. (2013). The genome of Nitrospina gracilis illuminates the metabolism and evolution of the major marine nitrite oxidizer. Front Microbiol 4: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lücker S, Wagner M, Maixner F, Pelletier E, Koch H, Vacherie B et al. (2010). A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc Natl Acad Sci USA 107: 13479–13484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maróti J, Farkas A, Nagy IK, Maróti G, Kondorosi E, Rákhely G et al. (2010). A second soluble Hox-type NiFe enzyme completes the hydrogenase set in Thiocapsa roseopersicina BBS. Appl Environ Microbiol 76: 5113–5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meincke M, Bock E, Kastrau D, Kroneck P. (1992). Nitrite oxidoreductase from Nitrobacter hamburgensis: redox centers and their catalytic role. Arch Microbiol 158: 127–131. [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T (2010). Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees. Computing Environments Workshop (GCE); 14 November 2010; New Orleans, LA. IEEE: New Orleans, LA, USA, pp 1–8.
- Muller N, Schleheck D, Schink B. (2009). Involvement of NADH:acceptor oxidoreductase and butyryl coenzyme A dehydrogenase in reversed electron transport during syntrophic butyrate oxidation by Syntrophomonas wolfei. J Bacteriol 191: 6167–6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogi T, Hirano Y, Miki K. (2005). Structural and functional studies on the tetraheme cytochrome subunit and its electron donor proteins: the possible docking mechanisms during the electron transfer reaction. Photosyn Res 85: 87–99. [DOI] [PubMed] [Google Scholar]
- Olson JM. (1970). The evolution of photosynthesis. Science 168: 438–446. [DOI] [PubMed] [Google Scholar]
- Overmann J, Garcia-Pichel F. (2013). The phototrophic way of life. In: Rosenberg E, Delong EF, Lory S, Stackebrandt E, Thompson F (eds) The Prokaryotes. Springer: Berlin, Heidelberg, pp 203–257. [Google Scholar]
- Pattaragulwanit K, Brune DC, Trüper HG, Dahl C. (1998). Molecular genetic evidence for extracytoplasmic localization of sulfur globules in Chromatium vinosum. Arch Microbiol 169: 434–444. [DOI] [PubMed] [Google Scholar]
- Pettigrew GW, Bartsch RG, Meyer TE. (1978). Redox potentials of the photosynthetic bacterial cytochromes c2 and the structural bases for variability. Biochim Biophys Acta 503: 509–523. [DOI] [PubMed] [Google Scholar]
- Ponomarenko NS, Li L, Marino AR, Tereshko V, Ostafin A, Popova JA et al. (2009). Structural and spectropotentiometric analysis of Blastochloris viridis heterodimer mutant reaction center. Biochim Biophys Acta 1788: 1822–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince RC. (1978). The reaction center and associated cytochromes of Thiocapsa pfennigii: their thermodynamic and spectroscopic properties, and their possible location within the photosynthetic membrane. Biochim Biophys Acta 501: 195–207. [DOI] [PubMed] [Google Scholar]
- Przysiecki CT, Meyer TE, Cusanovich MA. (1985). Circular dichroism and redox properties of high redox potential ferredoxins. Biochemistry 24: 2542–2549. [DOI] [PubMed] [Google Scholar]
- Rappaport F, Diner BA. (2008). Primary photochemistry and energetics leading to the oxidation of the (Mn)4Ca cluster and to the evolution of molecular oxygen in Photosystem II. Coord Chem Rev 252: 259–272. [Google Scholar]
- Ronquist F, Huelsenbeck JP. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Rutherford AW, Faller P. (2003). Photosystem II: evolutionary perspectives. Philos Trans R Soc Lond B Biol Sci 358: 245–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoepp-Cothenet B, Lieutaud C, Baymann F, Verméglio A, Friedrich T, Kramer DM et al. (2009). Menaquinone as pool quinone in a purple bacterium. Proc Natl Acad Sci USA 106: 8549–8554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott J, Griffin BM, Schink B. (2010). Anaerobic phototrophic nitrite oxidation by Thiocapsa sp. strain KS1 and Rhodopseudomonas sp. strain LQ17. Microbiology 156: 2428–2437. [DOI] [PubMed] [Google Scholar]
- Shih PM, Wu D, Latifi A, Axen SD, Fewer DP, Talla E et al. (2013). Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc Natl Acad Sci USA 110: 1053–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J, Sänger M, Schuster SC, Gross R. (2003). Electron transport to periplasmic nitrate reductase (NapA) of Wolinella succinogenes is independent of a NapC protein. Mol Microbiol 49: 69–79. [DOI] [PubMed] [Google Scholar]
- Sorokin DY, Lücker S, Vejmelkova D, Kostrikina NA, Kleerebezem R, Rijpstra WIC et al. (2012). Nitrification expanded: discovery, physiology and genomics of a nitrite-oxidizing bacterium from the phylum Chloroflexi. ISME J 6: 2245–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spieck E, Aamand J, Bartosch S, Bock E. (1996). Immunocytochemical detection and location of the membrane-bound nitrite oxidoreductase in cells of Nitrobacterand Nitrospira. FEMS Microbiol Lett 139: 71–76. [Google Scholar]
- Stamatakis A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starkenburg SR, Chain PSG, Sayavedra-Soto LA, Hauser L, Land ML, Larimer FW et al. (2006). Genome sequence of the chemolithoautotrophic nitrite-oxidizing bacterium Nitrobacter winogradskyi Nb-255. Appl Environ Microbiol 72: 2050–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starkenburg SR, Larimer FW, Stein LY, Klotz MG, Chain PSG, Sayavedra-Soto LA et al. (2008). Complete genome sequence of Nitrobacter hamburgensis X14 and comparative genomic analysis of species within the genus Nitrobacter. Appl Environ Microbiol 74: 2852–2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundermeyer-Klinger H, Meyer W, Warninghoff B. (1984). Membrane-bound nitrite oxidoreductase of Nitrobacter: evidence for a nitrate reductase system. Arch Microbiol 140: 153–158. [Google Scholar]
- Tabita FR, Hanson TE, Li H, Satagopan S, Singh J, Chan S. (2007). Function, structure, and evolution of the RubisCO-like proteins and their RubisCO homologs. Microbiol Mol Biol Rev 71: 576–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Fukumori Y, Yamanaka T. (1983). Purification of cytochrome a 1 c 1 from Nitrobacter agilis and characterization of nitrite oxidation system of the bacterium. Arch Microbiol 135: 265–271. [Google Scholar]
- Teske A, Alm E, Regan JM, Toze S, Rittmann BE, Stahl DA. (1994). Evolutionary relationships among ammonia- and nitrite-oxidizing bacteria. J Bacteriol 176: 6623–6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallenet D, Belda E, Calteau A, Cruveiller S, Engelen S, Lajus A et al. (2013). MicroScope—an integrated microbial resource for the curation and comparative analysis of genomic and metabolic data. Nucleic Acids Res 41: D636–D647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Driessche G, Vandenberghe I, Devreese B, Samyn B, Meyer TE, Leigh R et al. (2003). Amino acid sequences and distribution of high-potential iron-sulfur proteins that donate electrons to the photosynthetic reaction center in phototropic proteobacteria. J Mol Evol 57: 181–199. [DOI] [PubMed] [Google Scholar]
- Yeates TO, Kerfeld CA, Heinhorst S, Cannon GC, Shively JM. (2008). Protein-based organelles in bacteria: carboxysomes and related microcompartments. Nat Rev Microbiol 6: 681–691. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Feng F, Medová H, Dean J, Koblížek M. (2014). Functional type 2 photosynthetic reaction centers found in the rare bacterial phylum Gemmatimonadetes. Proc Natl Acad Sci USA 111: 7795–7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.