Abstract
Populations of the toxigenic marine dinoflagellate Alexandrium are composed of multiple genotypes that display phenotypic variation for traits known to influence top-down processes, such as the ability to lyse co-occurring competitors and prospective grazers. We performed a detailed molecular analysis of species interactions to determine how different genotypes perceive and respond to other species. In a controlled laboratory culture study, we exposed two A. fundyense strains that differ in their capacity to produce lytic compounds to the dinoflagellate grazer Polykrikos kofoidii, and analyzed transcriptomic changes during this interaction. Approximately 5% of all analyzed genes were differentially expressed between the two Alexandrium strains under control conditions (without grazer presence) with fold-change differences that were proportionally higher than those observed in grazer treatments. Species interactions led to the genotype-specific expression of genes involved in endocytotic processes, cell cycle control and outer membrane properties, and signal transduction and gene expression regulatory processes followed similar patterns for both genotypes. The genotype-specific trait changes observed in this study exemplify the complex responses to chemically mediated species interactions within the plankton and their regulation at the gene level.
Introduction
In the classic article entitled ‘The paradox of the plankton,' Hutchinson (1961) addresses the question of how species can coexist in a relatively isotropic or unstructured environment, competing for the same resources. This apparent paradox derives from the traditional view that ‘bottom-up' processes lead to competition for abiotic resources (for example, light and nutrients) and are the main driving force in shaping the structure of plankton communities. The failure of resource competition alone to account for the observed species heterogeneity and niche differentiation in plankton has led to an increased appreciation of species interactions, considered ‘top-down' processes, as a potential driving force for annual species succession (Margalef, 1978; Smetacek et al., 2004; Murray et al., 2011). The recognition of the role of species interactions in paradigms of successional processes on seasonal and annual time scales has underscored the fundamental importance of such interactions in co-evolutionary processes determining species diversity (Smetacek, 2001; Hamm et al., 2003; Smetacek et al., 2004).
The co-evolutionary mechanisms involved in the maintenance of species diversity are even more complex when considering the coexistence of multiple genotypes that exhibit fitness differences in the laboratory under controlled environmental conditions. This phenomenon has been named ‘the second paradox of the plankton' (Hebert and Crease, 1980; Fox et al., 2010). Different genotypes within one population can differentially express traits, such as cell size, cell shape and the production of secondary metabolites. These differentially expressed traits may also include integrative biochemical mechanisms, leading to variation in intrinsic growth rates. Differences in these parameters that result from genotypic variation influence both intra- and interspecific interactions (Van Donk et al., 1999).
There are few well-defined model marine phytoplankton species for which phenotypic and genotypic diversity have been explored at the population level. In all cases to date, in-depth analyses using molecular markers have revealed a high degree of intra-population diversity within dominant phytoplankton groups, including dinoflagellates (Alpermann et al., 2010), coccolithophorids (Iglesias-Rodríguez et al., 2006) and diatoms (Rynearson and Armbrust, 2000). Among 77 clonal strains of a population of the toxigenic marine red-tide dinoflagellate Alexandrium, no repeated sampling of any genotype was observed using microsatellite and amplified fragment length polymorphism markers (Alpermann et al., 2010).
These Alexandrium genotypes also exhibit wide intra-population phenotypic variation (Alpermann et al., 2010) in the production of secondary metabolites, including paralytic shellfish toxins, a group of potent neurotoxic alkaloids responsible for toxic effects on marine organisms and human consumers of seafood (Anderson et al., 2012), and allelochemical compounds of uncertain structural affinity with lytic effects (Tillmann and John, 2002; Fistarol et al., 2004; Hattenrath-Lehmann and Gobler, 2011).
On a per cell basis, populations exhibit wide variation in lytic potency (Alpermann et al., 2010; Hattenrath-Lehmann and Gobler, 2011). Lytic allelochemicals affect both putative competitors and predators, and such genotype-specific species interactions are expected to affect community structure for both competitive and bi-trophic systems. Furthermore, mutual facilitation, whereby interspecific interactions occur between individuals with different genotypes that vary in lytic capacity, provides a benefit to one species, without negatively affecting other genotypes (John et al., 2015).
In our study, we resolved species interactions among A. fundyense clones at the genotypic level. We utilized a comparative approach whereby a lytic (Alex2) and non-lytic (Alex5) strain of A. fundyense were exposed to waterborne cues from a protistan grazer. The two strains were selected based on previously established differences in their lytic capacity, as quantified by a whole-cell bioassay using the cryptomonad Rhodomonas (Tillmann et al., 2009). For all tested cell concentrations, Alex2 showed a high lytic capacity in the Rhodomonas bioassay, whereas Alex5 showed no lytic effect (Tillmann et al., 2009). We used a functional genomics approach to (1) assess the differences between the two A. fundyense strains and (2) screen for traits that differ among genotypes during exposure to a heterotrophic grazer. Our results provide insight into traits that are selectively induced and may hence be beneficial with respect to fitness under pressure owing to biotic interactions.
Materials and methods
Alexandrium fundyense and Polykrikos kofoidii cultures
Two strains of A. fundyense, Alex2 (lytic) and Alex5 (non-lytic), previously classified as A. tamarense Group 1 (John et al., 2014), were isolated in May 2004 from the North Sea coast of Scotland (Alpermann et al., 2010). Clonal isolates were grown in natural seawater-based (salinity ~33) K-medium (Keller et al., 1987) in a temperature- and light-controlled culture chamber (18 °C, 14-h:10-h light–dark cycle, ~150 μmol m−2 s−1).
The culture of the heterotrophic dinoflagellate Polykrikos kofoidii was established from a plankton net tow (20 μm) in 2009 in coastal waters of Scotland. The stock culture was maintained in K-medium (Keller et al., 1987) in culture flasks on a plankton wheel (1 r.p.m.) at 15 °C under low light (10–20 μmol m−2 s−1). The Polykrikos cultures were fed the thecate dinoflagellate Lingulodinium polyedrum (CCMP 1738). A subculture used for experimental inoculation was starved for ~1 day so that no dinoflagellate food algae were present.
Experimental design
Axenic, mid-exponential phase cultures of A. fundyense were washed three times with sterile-filtered seawater and diluted to a concentration of ~1000 cells ml−1. The washing steps ensured the exchange of culture media containing lytic substances before the experiments. P. kofoidii cultures were diluted to a concentration of ~40 cells ml−1.
A cage culture technique was used to separate Alexandrium strains from mixed Polykrikos/Alexandrium cultures to ensure RNA isolation from only the target Alexandrium and to avoid cross-hybridization on the microarray. The cages (50 ml polypropylene tubes containing a 10 μm plankton mesh at the bottom) were placed in 250 ml glass flasks. For both Alexandrium strains, 24 flasks were set-up, including three replicates of controls and three replicates of species interaction treatments for each harvesting time point (24, 48, 72 and 96 h). Each flask received 125 ml of the Alex2 or Alex5 culture diluted with 125 ml of K-medium to a concentration of ~500 cells ml−1. Cages were submerged in the flasks, and each treatment cage was inoculated with 1 ml of Polykrikos culture (~40 cells) and 1 ml of Alex5 culture (~1000 cells). Control flasks received 1 ml of Alex5 cultures within the cage. Cages were slowly moved up and down three times a day to promote the exchange of medium and dissolved compounds (including allelochemicals) between the two compartments. Experiments were terminated after 24, 48, 72 and 96 h; the first 24 h was considered to be an acclimation phase because dinoflagellates are sensitive to mixing (Berdalet et al., 2007) and often show a prolonged lag-phase after cell culture transfer.
Cell harvesting
Cages were removed from the experimental flasks and submerged in 50 ml glass flasks filled with sterile-filtered seawater to 30 ml. The contents of the cages were mixed, and 10 ml of culture suspension was preserved in acid Lugol's iodine solution for cell counts.
The flasks containing the surrounding Alexandrium cultures were also mixed, and 10 ml of the culture was preserved in acid Lugol's iodine solution for cell counts. Afterward, the culture was poured over a 10 μm nylon mesh sieve to collect the Alexandrium cells. The 10 μm nylon mesh was back-washed with sterile seawater into a 50 ml collection tube and immediately centrifuged at 4 °C for 5 min to obtain cell pellets for RNA isolation.
Cell pellets for RNA samples were mixed with 1 ml of hot (60 °C) TriReagent (Sigma-Aldrich, Steinheim, Germany), transferred to a 2 ml cryovial containing acid-washed glass beads, vortex-mixed for 1 min to shear the cells and subsequently frozen in liquid nitrogen for storage at –80 °C until RNA extraction.
Total RNA isolation
Frozen cell pellets fixed with TriReagent were lysed using a Bio101 FastPrep instrument (Thermo Savant, Illkirch, France) at maximum speed (6.5 m s−1) for 2 × 45 s. Total RNA was isolated following the Sigma-Aldrich TriReagent protocol recommendations, with linear polyacrylamide (Ambion, Life Technologies, Carlsbad, CA, USA) as a co-precipitant. Protocol details are described by Lu et al. (2014). RNA quality and integrity were verified with a RNA Nano-Chip Assay on a 2100 Bioanalyzer device (Agilent Technologies, Santa Clara, CA, USA). The RNA yield and concentration were checked with a NanoDrop ND-100 spectrometer (PeqLab, Erlangen, Germany). Only high-quality RNAs with minimum absorption ratios at 260/280 nm ⩾2 and 260/230 nm ⩾1.8 with corresponding intact ribosomal peaks were used for subsequent analyses.
454 Sequencing and complementary DNA library construction
Alex2 and Alex5 RNA samples for each treatment were pooled, and 10 μg of total RNA was used to prepare a normalized complementary DNA library for GS FLX Titanium sequencing. Complementary DNA library construction was carried out by Vertis Biotechnology AG (Freising-Weihenstephan, Germany). In total, 521 578 reads were assembled into 19 107 contigs longer than 500 bp, with an average coverage of ~27 reads per contig. The contigs were compared with an existing gene library from A. fundyense by a reciprocal best-BLAST hit search. This resulted in a final selection of 29 224 unique contigs for the microarray design.
Microarray design and hybridizations
A standardized in-house working pipeline for microarrays was used to assure reproducible sample processing and robust data analysis. Microarray probes (60-mers) were designed with the Agilent eArray online platform (https://earray.chem.agilent.com/earray/). Microarray hybridizations were designed to compare RNA from treatments and controls with respect to a reference pool of RNAs. The RNA reference pool consisted of equal volumes of subsamples of all RNAs obtained from the control treatments, and was used as a common control baseline to minimize hybridization biases (Novoradovskaya et al., 2004).
Labeling reactions were prepared from 200 ng of total RNA with the Agilent two-color Low Input Quick Amp Labeling Kit and the RNA Spike-In Mix (Agilent Technologies). In brief, RNA was reverse transcribed into complementary DNA with T(7) promoter primers and labeled fluorescently by linear amplification into complementary RNA with cyanine-3-CTPs or cyanine-5-CTPs according to the manufacturer's protocol.
Dye incorporation rates and complementary RNA concentrations were determined with a NanoDrop ND-100 spectrometer (PeqLab). Hybridizations were carried out on 8 × 60 K microarray slides with 300 ng of cyanine-3- and cyanine-5-labeled complementary RNA at 65 °C for 17 h and were washed according to the manufacturer's protocol. Microarrays were scanned with an Agilent G2565AA Scanner (Agilent Technologies).
Raw data processing was performed using Agilent Feature Extraction Software version 9.1.3.1 (FE); quality monitoring was performed using the Agilent QC Tool (v1.0) with the metric set GE2_QCMT_Feb07. Differentially expressed genes were detected using the Agilent GeneSpring GX software platform version 12. Hybridization results for biological triplicates were subjected to multiple comparison tests with a two-way analysis of variance (ANOVA) to determine the effects of treatment and time. Screening for differentially expressed genes for both genotypes under control conditions was performed in the same way. Genes were considered differentially expressed when P-values were <0.05. The data set was then reduced to include only genes for which expression changes exceeded 1.5-fold. The microarray design and hybridizations were MIAME compliant, and the experimental raw data were deposited in a MIAME-compliant database (ArrayExpress accession: E-MTAB-4060).
Gene set enrichment analysis
All sequences represented on the microarray were subjected to a hidden Markov model-based search for Pfam protein families (Finn et al., 2008). Sequences were first translated in silico (Wernersson, 2006) and subsequently online batch-processed to search for Pfam matches (http://pfam.sanger.ac.uk/). Upregulated Pfam families were tested for significant enrichment by calculating P-values from a hypergeometric distribution (Subramanian et al., 2005). Pfam families were considered significantly enriched at a given experimental time point when the P-value was <0.05.
Results
Differentially expressed genes between genotypes
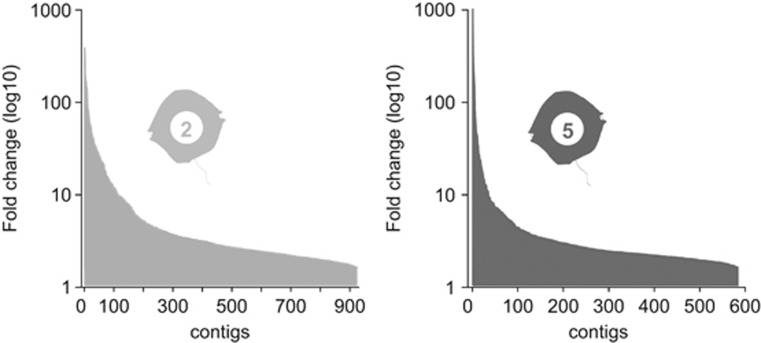
In an analysis of the variability in gene expression patterns between the two Alexandrium genotypes under control conditions, we found that ~5% of all genes represented on the microarray were differentially expressed at all time points. Among these, 923 genes were always more highly expressed in Alex2 cultures, and 584 were more highly expressed in Alex5 cultures. For these differentially expressed genes, we observed maximum fold-change differences between control cultures of 385 for Alex2 and 977 for Alex5 (Figure 1). The top 10 differentially expressed genes for each strain and their assigned Pfam protein families are listed in Table 1 (Alex2) and Table 2 (Alex5).
Figure 1.
Differentially expressed genes between control cultures of the two Alexandrium fundyense genotypes. Fold-changes and total number of differentially expressed genes for Alex2 (left) and Alex5 (right).
Table 1. Ten most highly overexpressed genes in Alex2 control cultures (in comparison with Alex5) and overexpressed genes coding for proteins potentially involved in secondary metabolism.
| Contig | Mean FC value | Pfam protein family name | Attributed function |
|---|---|---|---|
| Top 10 overexpressed genes in Alex2 | |||
| contig11508 | 385.70 | — | — |
| contig47146 | 232.12 | — | — |
| Atam01000 | 199.42 | — | — |
| contig01736 | 188.40 | Ion channel | Ion flux control |
| Atam27888 | 172.62 | — | — |
| Atam09090 | 164.73 | — | — |
| contig48538 | 144.33 | — | — |
| contig14000 | 141.05 | — | — |
| contig54109 | 140.65 | — | — |
| Atam19896 | 133.97 | — | — |
| Prominent secondary metabolite protein coding genes overexpressed in Alex2 | |||
| contig46061 | 28.56 | Glycosyl transferase family 8 | Transfer of glycosyl groups |
| contig27559 | 9.43 | ABC transporter transmembrane region | Transport |
| Atam22087 | 5.87 | FAD binding domain | Redox reactions |
| contig50076 | 5.44 | ABC-2 type transporter | Transport |
| contig53666 | 5.41 | Glycosyl transferase family 8 | Transfer of glycosyl groups |
| contig03340 | 4.17 | Glycosyl transferase family 8 | Transfer of glycosyl groups |
| contig30821 | 3.81 | Major facilitator superfamily | Transport |
| contig33504 | 3.79 | Major facilitator superfamily | Transport |
| Atam04504 | 2.93 | Aldo/keto reductase family | Redox reactions |
| contig29413 | 2.90 | Aldo/keto reductase family | Redox reactions |
| contig47324 | 2.82 | Taurine catabolism dioxygenase TauD, TfdA family | Redox reactions |
| contig51994 | 2.69 | Aminotransferase class-III | Special, transaminase activity |
| contig05529 | 2.68 | Zinc-binding dehydrogenase | Redox reactions |
| contig06697 | 2.66 | Acyl-CoA dehydrogenase, middle domain | Redox reactions |
| Atam18919 | 2.55 | Glycosyl transferase family 8 | Transfer of glycosyl groups |
| contig08880 | 2.52 | ABC-2 type transporter | Transport |
| Atam25893 | 2.49 | ABC transporter | Transport |
| contig02047 | 2.47 | SnoaL-like polyketide cyclase | Polyketide synthase |
| contig18051 | 2.22 | ABC transporter transmembrane region | Transport |
| Atam05470 | 2.19 | Cytochrome P450 | Redox reactions |
| contig21528 | 2.17 | Methyltransferase domain (2) | Methyl group transfer |
| contig14651 | 2.09 | Beta-ketoacyl synthase, C- and N-terminal domain | Polyketide synthase |
| contig40268 | 2.09 | Oxidoreductase family, C-terminal alpha/beta domain | Redox reactions |
| contig41426 | 1.82 | Glycosyl transferase family 8 | Transfer of glycosyl groups |
The table includes fold-change (FC) values and the assigned Pfam families for each gene, as well as the attributed function of each annotatable gene.
Table 2. Ten most highly overexpressed genes in Alex5 control cultures (compared with Alex2) and overexpressed genes coding for proteins potentially involved in secondary metabolism.
| Contig | Mean FC value | Pfam family name | Assessed function |
|---|---|---|---|
| Top 10 overexpressed genes in Alex5 | |||
| contig52451 | 977.12 | Fatty acid desaturase | Unsaturated fatty acid synthesis |
| contig10253 | 318.80 | — | — |
| contig22596 | 205.62 | — | — |
| contig22292 | 156.37 | — | — |
| contig03283 | 143.74 | — | — |
| contig14894 | 78.45 | — | — |
| 58180097 | 59.28 | — | — |
| contig05597 | 51.09 | EF-hand domain pair (2) | Calcium-ion binding |
| contig22152 | 45.17 | — | — |
| Atam02515 | 42.39 | — | — |
| Prominent secondary metabolite protein coding genes overexpressed in Alex5 | |||
| contig48765 | 10.39 | Methyltransferase domain | Methyl group transfer |
| Atam13914 | 5.12 | Methyltransferase domain | Methyl group transfer |
| Atam35211 | 4.65 | EamA-like transporter family | Transport |
| Atam25791 | 3.31 | Acetyltransferase (GNAT) family | Acetyl group transfer |
| Atam23762 | 2.94 | Cytochrome P450 | Redox reactions |
| contig33556 | 2.86 | Aldo/keto reductase family | Redox reactions |
| contig34741 | 2.04 | ABC-2 type transporter | Transport |
| contig01330 | 1.99 | Taurine catabolism dioxygenase TauD, TfdA family | Redox reactions |
| Atam01694 | 1.88 | NAD-dependent epimerase/dehydratase family | Redox reactions |
| contig06638 | 1.84 | EamA-like transporter family | Transport |
| contig16321 | 1.80 | ABC-2 type transporter | Transport |
| contig37655 | 1.79 | Periplasmic binding protein | Transport |
| Atam12516 | 1.76 | Acyltransferase family | Acyl group transfer |
The table includes fold-change (FC) values and the assigned Pfam families for each gene, as well as the attributed function of each annotatable gene.
We analyzed differentially expressed genes between representative lytic (Alex2) and non-lytic (Alex5) genotypes, specifically for the occurrence of Pfam protein family annotations with known involvement in secondary metabolite production (Medema et al., 2011), and found that 24 genes in these protein families were more highly expressed in the lytic Alex2 than in the non-lytic Alex5 (Table 1). By comparison, 13 genes coding for proteins involved in secondary metabolism were more highly expressed in Alex5 than in Alex2 (Table 2). However, none of these genes were differentially expressed in response to grazers (Supplementary File S1).
Cage experiment results
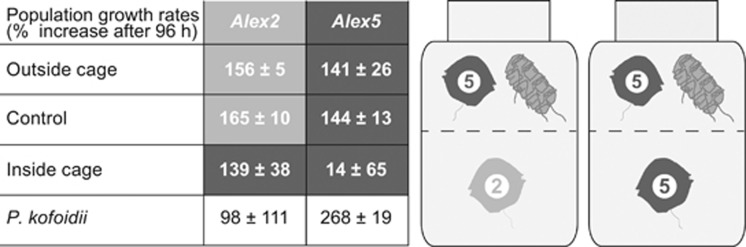
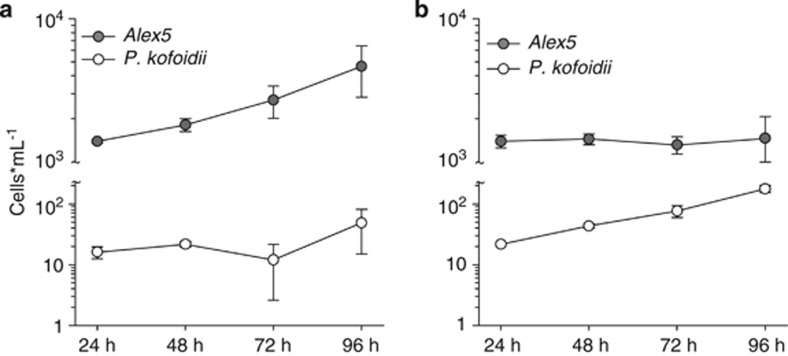
Exposure of both Alexandrium genotypes to Polykrikos grazing had different effects on conspecifics (Figures 2 and 3). When the mixture of Polykrikos and non-lytic Alex5 cultures was surrounded by lytic Alex2 cultures, the number of Alexandrium cells inside the cage increased significantly over time (Figures 2 and 3a; ANOVA, P=0.015). The Polykrikos cell concentration inside the cage in this treatment did not increase over time (ANOVA, P=0.196).
Figure 2.
Growth rates for Alexandrium fundyense and Polykrikos kofoidii and schematic representation of the experimental set-up. The relative growth rates of Alex2 and Alex5 treatment versus control cultures, as well as for P. kofoidii cultures, are given as percentage cell population increases over 4 days. The experimental set-up shows the partition of A. fundyense and P. kofoidii cells in the cage and a surrounding compartment.
Figure 3.
Cell concentration of Alex5 and Polykrikos kofoidii inside the cage. Panel (a) shows the cell concentrations and standard deviations (n=3) for Alex5 and P. kofoidii when surrounded by Alex2 cultures; panel (b) shows the cell numbers and s.d. (n=3) for Alex5 and P. kofoidii when surrounded by Alex5 cultures.
By contrast, exposure of the Polykrikos/Alex5 mixture to non-lytic Alexandrium cultures resulted in a significant increase in Polykrikos cells (Figures 2 and 3b; ANOVA, P<0.001). The abundance of Alexandrium cells inside the cage did not change significantly over time (ANOVA, P=0.793).
Differentially expressed genes with respect to grazer presence
The number of upregulated genes increased by >9-fold for the lytic Alex2 during the exposure period (Table 3). In the non-lytic Alex5 cultures, the number of upregulated genes doubled over time (Table 3).
Table 3. Gene expression analysis for the combined treatment with both Alexandrium fundyense strains.
| Genotype | 24 vs 48 h | 24 vs 72 h | 24 vs 96 h | Common upregulated at all time points |
|---|---|---|---|---|
| Alex2 | 604 | 959 | 5724 | 244 |
| Alex5 | 496 | 1177 | 909 | 61 |
The table displays the numbers of upregulated genes for comparisons at various time points after exposure for each A. fundyense strain to the cage containing interacting P. kofoidii cells and conspecific A. fundyense. The last column shows the number of genes that were upregulated at all points.
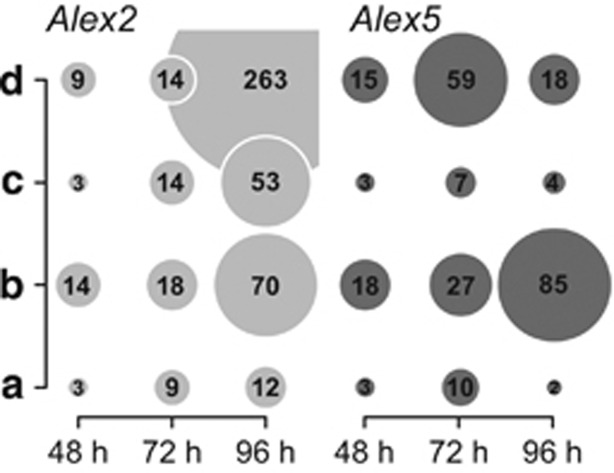
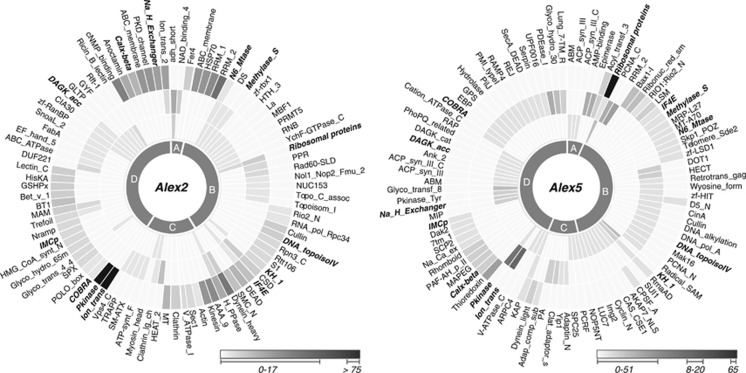
The genes that code for enriched Pfam family proteins were assigned to categories (A: secondary metabolic processes; B: cell cycle control and genetic information storage and processing; C: intracellular vesicle-associated processes; and D: signal transduction processes) and are summarized in Figure 4. Genes assigned to category B showed a similar increase over time in transcript abundance for both strains, but differed with respect to their Pfam family annotations (Figures 4 and 5). Categories A, C and D showed differences in frequency over time in both strains, for transcript numbers and assigned gene products (Figures 4 and 5).
Figure 4.
Differentially expressed genes and their time-dependent abundances for Alex2 and Alex5 in response to grazer presence. Genes presented here only exhibited changes in expression in the grazer treatment and not in the respective control. Selected differentially expressed genes are grouped into categories (a–d). The area of a circle represents the number of genes per category and per time point. (a) Secondary metabolic processes; (b) cell cycle control and genetic information storage and processing; (c) intracellular vesicle-associated processes; (d) signal processing and modification.
Figure 5.
Abundance of selected significantly enriched Pfam protein families and domains at various time points. The Pfam families and domains are grouped into categories, as described for Figure 4. The varying shades of grey represent the number of genes within the respective Pfam domain/family. Scales are shown below each diagram. The three circles represent different analysis times: inner circle, 48 h; middle, 72 h; outer circle, 96 h. Pfam domains/families in bold italic font were significantly enriched in both genotypes.
Discussion
We observed that species interactions induce gene expression changes in a genotype-specific manner. These genotype-specific responses may be influenced by the differences in the allelochemical properties of both strains as only one is lytic. At least under the experimental conditions, this suggests that the lytic strain Alex2 constructs an ecological niche that is distinct from that of the non-lytic strain Alex5. This suggests that genotype-specific interactions and associated trait variation within a population maintain disequilibrium among genotypes and can, therefore, among other mechanisms, explain the ‘paradox of the plankton'. The presence of paralytic shellfish poisoning toxins appears to have a negligible or neutral effect with respect to the species interactions investigated here (Senft-Batoh et al., 2015).
Inherent genotype-specific transcription patterns
In our transcriptome analysis, ~5% of all tested genes differed between the two Alexandrium strains under control conditions and we detected unusually high fold-change values for dinoflagellates (Figure 1). Among treatments, we detected maximal gene expression differences of ~15-fold (Alex2) and ~77-fold (Alex5), and in the strain comparison between controls, we observed maximal differences of ~385-fold (Alex2) and ~977-fold (Alex5). Dinoflagellates have a tendency to expand particular genes via tandem DNA repeats, and high transcript abundances are correlated with the genomic abundance of such genes (Bachvaroff and Place, 2008). Further studies are needed to determine whether the observed differences in transcriptional activity are caused by genome expansions. If indeed this is the case, different traits could arise by the selective duplication of particular genes in dinoflagellates. mRNA recycling mechanisms proposed for dinoflagellates (Slamovits and Keeling, 2008) could contribute to the manifestation and diversification of traits via gene duplication.
The distinct genetic programs that are apparently active in the two strains may also be related to differences in cues originating from conspecific strains in the cage, that is, Alex2 is surrounded by the conspecific Alex5 and in the experimental set-up with Alex5, no other strain is present. Indeed, complex strain-specific interactions between co-cultured genotypes have been observed (Van Gremberghe et al., 2009), and these interactions may be facilitated and/or initiated by differential gene expression. Both mechanisms (cues from conspecifics and potential strain-specific gene expansions in the genome) could have caused the observed differences in gene expression between the controls. At least for the transcripts with high fold-change values (Tables 1 and 2), genomic expansion may indeed have contributed to the differences in fixed phenotypic traits, that is, the allelochemical properties of the strains. Differences in the expression of genes involved in secondary metabolism (Tables 1 and 2) should be further investigated to determine their role in the production of lytic compounds in Alexandrium strains because none of these genes were upregulated in the presence of grazers (Supplementary File S1). Lytic compound production appears to be a stable trait in Alexandrium that, if present, is constitutively expressed (Zhu and Tillmann, 2012). The existence of an inducible genetic basis for this trait, quantifiable as differential gene expression, is thus questionable.
Allelochemical effects of Alexandrium on grazer growth rates
Differences in allelochemical properties, specifically lytic activity, between the Alexandrium strains Alex2 and Alex5 influence the survival rate of co-occurring unicellular heterotrophs (Tillmann et al., 2008). These observations are consistent with our findings for multispecies interactions, whereby the lytic Alex2 strain facilitated the growth of its conspecific Alexandrium strain inside the cage by constraining an increase in the abundance of the putative predator Polykrikos. By contrast, the presence of the non-lytic Alex5 strain allowed an increase in actively grazing Polykrikos cells, which prevented a further increase in grazing-controlled conspecifics within the cage (Figure 3).
Secondary metabolic processes
Protein families that are potentially involved in secondary metabolic processes increased in frequency over time in the lytic Alex2 and peaked in abundance after 72 h in the non-lytic Alex5 (Figure 4). However, the overall abundance of genes in this category was comparably low. The significantly enriched protein families in Alex2 were mainly broadly acting (involved in redox-reactions (3), transmembrane transport (1) and glycosyl group transfer (1); see Figure 5), making it difficult to interpret their potential roles in biotic interactions. The non-lytic Alex5 showed an enrichment for proteins involved in terpenoid and potential polyketide synthesis (ABM and ACP_synt_III) after 48 h, which could indicate that further unknown secondary metabolites are involved in the response to grazer cues. However, their precise role remains elusive. In addition, it is not clear whether they are part of a specific response or display a ‘moving target strategy,' where species switch phenotypes stochastically as a defense response (Adler and Karban, 1994).
Endocytotic processes and cell recognition features
In the lytic Alex2 strain, gene expression linked to intracellular vesicle-associated processes increased over time, whereas these genes were only slightly enriched in the non-lytic Alex5 strain and did not exhibit changes in expression over time (Figure 4, category C). Changes in dissolved and particulate organic matter may have triggered the expression of these genes. The lytic effects of Alex2 increase the levels of dissolved organic matter and particulate organic matter in the growth medium, which may be used as an additional nutritional source. The molecular genetic basis of dissolved organic matter/particulate organic matter uptake in marine protists has only been described in a metatranscriptomic study (Lin et al., 2010; Lin, 2011). In our data set, we found several genes associated with vesicle transport processes that were significantly enriched in treatments where cells were lysed. Genes necessary for the formation of clathrin-coated vesicles were significantly enriched at every experimental time point for the lytic Alex2 strain (Figure 5). Clathrin-coated vesicles sequester solutes or receptor-bound molecules from the surrounding medium and subsequently develop into lysosomes, which are digestive compartments with an acidic pH (Alberts et al., 2008). Proton-translocating inorganic pyrophosphatases may have an important role in acidifying these digestive compartments. We found a significant enrichment and an increase over time in genes encoding such inorganic pyrophosphatases for the lytic Alex2 (Figure 5). Inorganic pyrophosphatases are proton pumps distinct from F-, P- and V-ATPases that utilize inorganic pyrophosphate hydrolysis as a driving force for H+-ion movement across the membrane (Rea and Poole, 1993). In general, inorganic pyrophosphate is considered a waste product of anabolism (Pérez-Castiñeira et al., 2001), and the use of inorganic pyrophosphate, rather than ATP, to acidify lysosomes would therefore be an efficient and energetically favorable recycling process in Alexandrium cells to sequester nutrients derived from endocytosis.
Prior investigations of endocytosis in marine heterotrophic protists revealed that protein kinases (Hartz et al., 2008) and lectins (Wootton et al., 2007) can be directly linked to phagocytosis. Genes that clustered into category D in Alex2 included those for lectins (C-type and Ricin-B-type), which were significantly enriched in the expression analysis after 48 and 96 h (Figure 5). In addition to phagocytosis, lectins have an important role in cell–cell recognition as well as receptor-mediated and clathrin-dependent endocytosis (Roberts et al., 2006; Varki et al., 2009). In addition, Alex2 exhibited a significant enrichment for genes associated with other adhesive (MAM) receptors after 48 and 96 h (Figure 5), and these loci may contribute to endocytotic processes. Genes for ion channels and protein kinases accounted for the majority of genes assigned to category D at 96 h in the lytic Alex2. Such an increase in the expression of ion channels can be directly connected to endocytosis, as a necessary mechanism to maintain intracellular ion homeostasis. Owing to the increased expression of genes for ion channels and protein kinases, we observed an increased abundance of genes assigned to category D (72 h) for the non-lytic Alex5 strain (Figure 5), although the enrichment of protein families involved in endocytotic processes was not as strong as it was for the lytic Alex2 strain (Figure 4). Alex5 cultures may, however, still utilize organic compounds excreted from the heterotrophic grazer.
In Alex2, we detected increased expression of motor proteins, kinesins, and, predominately, dyneins, including associated motor units (Figure 5). This pattern strongly indicates the establishment of intracellular trafficking routes and could be directly associated with vesicular transport and endosome and lysosome organization and maintenance. Furthermore, after 96 h, we observed increased expression of acidifying V-ATPases and inorganic pyrophosphatases. These observations provide evidence that endocytotic processes were strongly induced and maintained over time in the lytic Alex2 strain (Figures 4 and 5).
Protein families that clustered into category D for Alex2 and, to a lesser extent, for Alex5 included several types of ligand-binding receptors (Figure 5). These proteins are associated with receptor-mediated endocytosis and glycoprotein and glycolipid modifications, which can alter the properties of outer glycosylated membrane surfaces. The biochemical compositional change of the glycosylated membrane surface determines important traits, such as self-self recognition, non-self discrimination and ‘cell taste' (Gahmberg, 1981; Wolfe, 2000), and thus could have a strong cascading effect on further interactions with conspecifics, competitors and/or predators.
Signal transduction processes
Significantly enriched protein families involved in signaling processes provide molecular genetic evidence for signal transduction via G-protein-coupled receptors, as proposed by Hartz et al. (2008). Cascades induced by ligand binding to G-protein-coupled receptors lead to increased levels of diacylglycerol (DAG), an important intracellular secondary messenger (Alberts et al., 2008). Our data show that DAG kinase accessory domains are significantly enriched in both Alexandrium strains after 48 h and additionally after 72 h in the lytic Alex2 strain (Figure 5). DAG kinase drives the conversion of DAG to phosphatidic acid (PA), which is involved in a wide range of biotic (for example, pathogen related) and abiotic stress responses in plants (Arisz et al., 2009). In higher plants, PA has therefore emerged as an important lipid secondary messenger with direct effects on vesicular trafficking and membrane surface charge. Our transcriptome data suggest that similar mechanisms may operate in less well-studied marine protists. The involvement of PA signaling in biotic interactions between marine protists may therefore be an important link between the activation of G-protein-coupled receptors and subsequent responses. Our expression data indicate that the conversion of DAG into PA takes place in both Alexandrium strains, underpinning the importance of G-protein-coupled receptors and DAG/PA signaling as an intracellular response inducer in biotic interactions.
Transcriptional regulation
The number of significantly enriched genes associated with protein families involved in information storage and processing and/or cell cycle control increased over time for both Alexandrium strains (Figure 4, category B). For Alex2, this category primarily comprised protein families that act on several levels of transcriptional and translational control. The increased expression after 96 h was predominantly explained by an increase in RNA-binding proteins, which act on post-transcriptional regulation of gene expression. In addition, several genes linked to protein families with transcription factor properties were enriched. We observed a co-occurring expression pattern at all time points in Alex2, and at 96 h in the Alex5 treatment in which there was an upregulation of genes associated with protein families involved in DNA m6adenine methylation and the primary translation initiation factor If4e, which binds the cap structure of mRNAs (Figure 5). M6adenine methylation is an epigenetic regulation process in prokaryotes and lower eukaryotes that differs from cytosine methylation because it activates transcription, whereas cytosine methylation suppresses transcription (Hattman, 2005). Given the low abundance of histones in the dinoflagellate genome (Lin, 2011) and their importance in transcriptional control by opening the chromatin structure (Zhang and Reinberg, 2001), epigenetic processes like m6adenine methylation may be a convergent mechanism (with respect to histone acetylation) in dinoflagellates and could potentially have profound effects on the observed changes in transcriptional regulation in response to species interactions.
Cell division and growth-related processes
Protein families clustered in category B for the non-lytic Alex5 indicated an influence on cell cycle progression (Figures 4 and 5). Protein families involved in cell cycle control were significantly enriched, followed by an upregulation of ribosomal protein (RP) genes after 96 h (Figure 5). The synthesis of ribosomal proteins in lower eukaryotes, such as yeasts, is often linked to nutrient availability and stress-related signals (Powers et al., 2004). Studies in yeast reveal that the RP-gene regulon is activated via the protein kinase A pathway (Jorgensen et al., 2004), which regulates a range of growth-related processes. Our transcriptomic enrichment data provide evidence that a protein kinase A-activated transcription regulator (AKAP7_NLS) is expressed after 48 h, concurrent with the expression of proliferation cell nuclear antigen (PCNA) and a cyclin domain-containing gene (Figure 5). This pattern is followed by the expression of additional genes encoding proliferation cell nuclear antigens after 72 h, along with several genes that encode DNA replication, modification and repair proteins. Finally, after 96 h, the transcriptional accumulation of RP-genes was apparent (Figure 5). RP-gene expression also increased in the first 48 h in the lytic strain Alex2, followed by an enhanced expression of genes coding for proteins involved in transcriptional and translational control (Figure 5). Investment in new ribosomes provides a platform for faster growth, if all other essential components are available (Warner, 1999). Protein kinase A-mediated transcriptional regulation along with RP-gene transcription and cell cycle gene expression exemplifies how cellular growth may be internally regulated on a molecular basis in Alexandrium. However, external cues derived from biotic interactions also have the potential to restructure the internal growth rhythm and cellular processes. This influence of chemical cues derived from species interactions on phytoplankton growth was recently shown for a diatom by Amin et al. (2015).
Conclusions
Our comparative transcriptional analysis indicates that a heterotrophic protistan grazer induced changes in several traits in the here investigated Alexandrium strains that differ in their capacity to facilitate the lysis of potential grazers or competitors. This study thus provides new insight into chemically mediated processes involved in plankton species interactions and their regulation. The increased expression of genes involved in endocytotic processes, particularly in the lytic Alex2, suggests that this strain preferably switches to organic matter as an alternative food source, even under favorable laboratory conditions where ample inorganic nutrients and light are available. The production of lytic compounds thus not only provides a competitive advantage via grazing protection, but it may also have a positive feedback effect by providing a selective advantage to outcompete other prey organisms under grazing pressure via increased uptake of organic matter.
Based on the gene expression data, additional traits that changed in a strain-specific manner in response to species interactions included properties of the outer glycosylated membrane, which could influence further interactions. Such strain-specific trait alterations indicate a complex interplay within community ecological processes. Thus, depending upon ‘who interacts with whom and when,' there may be short-term differences in the outcome of species interactions, which set the stage for the survival of dominant genotypes in seasonal succession processes. Future investigations should therefore account for how specific genotypes alter the community structure and also determine the impact of phenotypic diversity of multiple genotypes on a given community composition.
Acknowledgments
A file containing Pfam domains and families associated with secondary metabolite synthesis was kindly provided by Takano and Medema (University of Groningen, The Netherlands) who constructed the antiSMASH antibiotics and Secondary Metabolite Analysis Shell. We thank Gernot Glöckner from the Medical Faculty, University of Cologne, Germany, for the assembly of the raw sequences and Stephan Frickenhaus (AWI) for the reciprocal best-BLAST search analysis. Financial support was provided by the PACES research program of the Alfred Wegener Institute Helmholtz Centre for Polar and Marine Research and by the German Research Foundation (DFG) Priority Programme DynaTrait (1704).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Adler FR, Karban R. (1994). Defended fortresses or moving targets? Another model of inducible defenses inspired by military metaphors. Am Nat 5: 813–832. [Google Scholar]
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. (2008). Molecular Biology of the Cell: Reference Edition: 1601.
- Alpermann TJ, Tillmann U, Beszteri B, Cembella AD, John U. (2010). Phenotypic variation and genotypic diversity in a planktonic population of the toxigenic marine dinoflagellate Alexandrium tamarense (Dinophyceae). J Phycol 46: 18–32. [Google Scholar]
- Amin SA, Hmelo LR, van Tol HM, Durham BP, Carlson LT, Heal KR et al. (2015). Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature 522: 98–101. [DOI] [PubMed] [Google Scholar]
- Anderson DM, Alpermann TJ, Cembella AD, Collos Y, Masseret E, Montresor M. (2012). The globally distributed genus Alexandrium: multifaceted roles in marine ecosystems and impacts on human health. Harmful Algae 14: 10–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arisz SA, Testerink C, Munnik T. (2009). Plant PA signaling via diacylglycerol kinase. Biochim Biophys Acta 1791: 869–875. [DOI] [PubMed] [Google Scholar]
- Bachvaroff TR, Place AR. (2008). From stop to start: tandem gene arrangement, copy number and trans-splicing sites in the dinoflagellate Amphidinium carterae. PLoS One 3: e2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdalet E, Peters F, Koumandou VL, Roldán C, Guadayol Ò, Estrada M. (2007). Species-specific physiological response of dinoflagellates to quantified small-scale turbulence. J Phycol 43: 965–977. [Google Scholar]
- Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz H-R et al. (2008). The Pfam protein families database. Nucleic Acids Res 36: D281–D288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fistarol GO, Legrand C, Selander E, Hummert C, Stolte W, Granéli E. (2004). Allelopathy in Alexandrium spp.: effect on a natural plankton community and on algal monocultures. Aquat Microb Ecol 35: 45–56. [Google Scholar]
- Fox JW, Nelson WA, McCauley E. (2010). Coexistence mechanisms and the paradox of the plankton: quantifying selection from noisy data. Ecology 91: 1774–1786. [DOI] [PubMed] [Google Scholar]
- Gahmberg CG. (1981). Membrane glycoproteins and glycolipids: structure, localization and function of the carbohydrate. In: Finean JB, Michell RH. (eds), Membrane Structure. Elsevier B.V.: Oxford, UK, pp 127–160. [Google Scholar]
- Hamm CE, Merkel R, Springer O, Jurkojc P, Maier C, Prechtel K et al. (2003). Architecture and material properties of diatom shells provide effective mechanical protection. Nature 421: 841–843. [DOI] [PubMed] [Google Scholar]
- Hartz AJ, Sherr BF, Sherr EB. (2008). Using inhibitors to investigate the involvement of cell signaling in predation by marine phagotrophic protists. J Eukaryot Microbiol 55: 18–21. [DOI] [PubMed] [Google Scholar]
- Hattenrath-Lehmann TK, Gobler CJ. (2011). Allelopathic inhibition of competing phytoplankton by North American strains of the toxic dinoflagellate, Alexandrium fundyense: evidence from field experiments, laboratory experiments, and bloom events. Harmful Algae 11: 106–116. [Google Scholar]
- Hattman S. (2005). DNA-[adenine] methylation in lower eukaryotes. Biochemistry 70: 550–558. [DOI] [PubMed] [Google Scholar]
- Hebert PDN, Crease TJ. (1980). Clonal coexistence in Daphnia pulex (Leydig): another planktonic paradox. Science 207: 1363–1365. [Google Scholar]
- Hutchinson GE. (1961). The paradox of the plankton. Am Nat 95: 137–145. [Google Scholar]
- Iglesias-Rodríguez MD, Schofield OM, Batley J, Medlin LK, Hayes PK. (2006). Intraspecific genetic diversity in the marine coccolithophore Emiliania huxleyi (Prymnesiophyceae): the use of microsatellite analysis in marine phytoplankton population studies1. J Phycol 42: 526–536. [Google Scholar]
- John U, Litaker RW, Montresor M, Murray S, Brosnahan ML, Anderson DM. (2014). Formal revision of the Alexandrium tamarense species complex (Dinophyceae) taxonomy: the introduction of five species with emphasis on molecular-based (rDNA) classification. Protist 165: 779–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John U, Tillmann U, Hülskötter J, Alpermann TJ, Wohlrab S, Van de Waal DB. (2015). Intraspecific facilitation by allelochemical mediated grazing protection within a toxigenic dinoflagellate population. Pro R Soc Lond B Biol Sci 282: doi:10.1098/rspb.2014.1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen P, Rupes I, Sharom JR, Schneper L, Broach JR, Tyers M. (2004). A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev 18: 2491–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller MD, Selvin RC, Claus W, Guillard RRL. (1987). Media for the culture of oceanic ultraphytoplankton. J Phycol 23: 633–638. [Google Scholar]
- Lin S. (2011). Genomic understanding of dinoflagellates. Res Microbiol 162: 551–569. [DOI] [PubMed] [Google Scholar]
- Lin S, Zhang H, Zhuang Y, Tran B, Gill J. (2010). Spliced leader-based metatranscriptomic analyses lead to recognition of hidden genomic features in dinoflagellates. Proc Natl Acad Sci USA 107: 20033–20038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Wohlrab S, Glöckner G, Guillou L, John U. (2014). Genomic insights into processes driving the infection of Alexandrium tamarense by the Parasitoid Amoebophrya sp. Eukaryot Cell 13: 1439–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margalef R. (1978). Life-forms of phytoplankton as survival alternatives in an unstable environment. Oceanol Acta 1: 493–509. [Google Scholar]
- Medema MH, Blin K, Cimermancic P, de Jager V, Zakrzewski P, Fischbach MA et al. (2011). antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res 39: W339–W346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray SA, Mihali TK, Neilan BA. (2011). Extraordinary conservation, gene loss, and positive selection in the evolution of an ancient neurotoxin. Mol Biol Evol 28: 1173–1182. [DOI] [PubMed] [Google Scholar]
- Novoradovskaya N, Whitfield M, Basehore L, Novoradovsky A, Pesich R, Usary J et al. (2004). Universal Reference RNA as a standard for microarray experiments. BMC Genomics 5: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers T, Dilova I, Chen CY, Wedaman K. (2004). Yeast TOR signaling: a mechanism for metabolic regulation. Curr Top Microbiol Immunol 279: 39–51. [DOI] [PubMed] [Google Scholar]
- Pérez-Castiñeira JR, Gómez-García R, López-Marqués RL, Losada M, Serrano A. (2001). Enzymatic systems of inorganic pyrophosphate bioenergetics in photosynthetic and heterotrophic protists: remnants or metabolic cornerstones? Int Microbiol 4: 135–142. [DOI] [PubMed] [Google Scholar]
- Rea PA, Poole RJ. (1993). Vacuolar H+ -translocating pyrophosphatase. Annu Rev Plant Physiol Plant Mol Biol 44: 157–180. [Google Scholar]
- Roberts EC, Zubkov MV, Martin-Cereceda M, Novarino G, Wootton EC. (2006). Cell surface lectin-binding glycoconjugates on marine planktonic protists. FEMS Microbiol Lett 265: 202–207. [DOI] [PubMed] [Google Scholar]
- Rynearson TA, Armbrust EV. (2000). DNA fingerprinting reveals extensive genetic diversity in a field population of the centric diatom Ditylum brightwellii. Limnol Oceanogr 45: 1329–1340. [Google Scholar]
- Senft-Batoh CD, Dam HG, Shumway SE, Wikfors GH. (2015). A multi-phylum study of grazer-induced paralytic shellfish toxin production in the dinoflagellate Alexandrium fundyense: a new perspective on control of algal toxicity. Harmful Algae 44: 20–31. [Google Scholar]
- Slamovits CH, Keeling PJ. (2008). Widespread recycling of processed cDNAs in dinoflagellates. Curr Biol 18: R550–R552. [DOI] [PubMed] [Google Scholar]
- Smetacek V. (2001). A watery arms race. Nature 411: 745. [DOI] [PubMed] [Google Scholar]
- Smetacek V, Assmy P, Henjes J. (2004). The role of grazing in structuring Southern Ocean pelagic ecosystems and biogeochemical cycles. Antarct Sci 16: 541–558. [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillmann U, Alpermann T, John U, Cembella A. (2008). Allelochemical interactions and short-term effects of the dinoflagellate Alexandrium on selected photoautotrophic and heterotrophic protists. Harmful Algae 7: 52–64. [Google Scholar]
- Tillmann U, Alpermann TL, da Purificação RC, Krock B, Cembella A. (2009). Intra-population clonal variability in allelochemical potency of the toxigenic dinoflagellate Alexandrium tamarense. Harmful Algae 8: 759–769. [Google Scholar]
- Tillmann U, John U. (2002). Toxic effects of Alexandrium spp. on heterotrophic dinoflagellates: an allelochemical defence mechanism independent of PSP-toxin content. Mar Ecol Prog Ser 230: 47–58. [Google Scholar]
- Van Donk E, Lürling M, Lampert W. (1999). Consumer induced changes in phytoplankton: inducibility, costs, benefits and the impact on grazers. In: Tollrian R, Harvell DC (eds), The Ecology and Evolution of Inducible Defenses. Princeton University Press: Princeton, NJ, USA, pp 89–103. [Google Scholar]
- Van Gremberghe I, Vanormelingen P, Vanelslander B, Van der Gucht K, D'hondt S, De Meester L et al. (2009). Genotype-dependent interactions among sympatric Microcystis strains mediated by Daphnia grazing. Oikos 118: 1647–1658. [Google Scholar]
- Varki A, Cummings RD, Esko JD et al. eds (2009). Essentials of Glycobiology, 2nd edn. Cold Spring Harbor: New York, USA, http://www.ncbi.nlm.nih.gov/books/NBK1908/. [PubMed] [Google Scholar]
- Warner JR. (1999). The economics of ribosome biosynthesis in yeast. Trends Biochem Sci 24: 437–440. [DOI] [PubMed] [Google Scholar]
- Wernersson R. (2006). Virtual Ribosome—a comprehensive DNA translation tool with support for integration of sequence feature annotation. Nucleic Acids Res 34: W385–W388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe GV. (2000). The chemical defense ecology of marine unicellular plankton: constraints, mechanisms, and impacts. Biol Bull 198: 225–244. [DOI] [PubMed] [Google Scholar]
- Wootton EC, Zubkov MV, Jones HD, Jones RH, Martel CM, Thornton CA et al. (2007). Biochemical prey recognition by planktonic protozoa. Environ Microbiol 9: 216–222. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Reinberg D. (2001). Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev 15: 2343–2360. [DOI] [PubMed] [Google Scholar]
- Zhu M, Tillmann U. (2012). Nutrient starvation effects on the allelochemical potency of Alexandrium tamarense (Dinophyceae). Mar Biol 159: 1449–1459. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.