Abstract
Deep-sea hypersaline anoxic basins (DHABs) in the Eastern Mediterranean Sea are considered some of the most polyextreme habitats on Earth. In comparison to microbial activities occurring within the haloclines and brines of these unusual water column habitats near the Mediterranean seafloor, relatively little is known about microbial metabolic activities in the underlying sediments. In addition, it is not known whether activities are shaped by the unique chemistries of the different DHAB brines and whether evidence exists for active microbial eukaryotes in those sediments. Metatranscriptome analysis was applied to sediment samples collected using ROV Jason from underneath the haloclines of Urania, Discovery and L'Atalante DHABs and a control site. We report on expression of genes associated with sulfur and nitrogen cycling, putative osmolyte biosynthetic pathways and ion transporters, trace metal detoxification, selected eukaryotic activities (particularly of fungi), microbe–microbe interactions, and motility in sediments underlying the haloclines of three DHABs. Relative to our control sediment sample collected outside of Urania Basin, microbial communities (including eukaryotes) in the Urania and Discovery DHAB sediments showed upregulation of expressed genes associated with nitrogen transformations, osmolyte biosynthesis, heavy metals resistance and metabolism, eukaryotic organelle functions, and cell–cell interactions. Sediments underlying DHAB haloclines that have cumulative physico-chemical stressors within the limits of tolerance for microoorganisms can therefore be hotspots of activity in the deep Mediterranean Sea.
Introduction
Hypersaline waters (generally >10% NaCl) have salinities that exceed those of most oceans (~3.5% total salt) and are therefore characterized by low water activity (aw), presenting challenges for organisms living in these habitats (Litchfield, 1998). To prevent loss of cellular water to the environment, halotolerant and halophilic organisms require ways to balance the osmotic pressure created by their hypersaline habitat. Extensive literature exists describing the adaptive mechanisms for Bacteria and Archaea and to a lesser extent Eukarya, including high GC content in DNA, high concentrations of acidic residues on exteriors of proteins, and unique lipids, cellular architectures, pigments, physiologies and metabolisms (for example, Litchfield, 1998; Oren, 2003). There is increasing evidence of extremely halophilic heterotrophic protists in solar salterns and nearly saturated brines (salinity ⩾29%) that are distinct from marine or freshwater forms, and do not grow at salinities <7.5% (Park et al., 2006, 2007, 2009; Cho et al., 2008).
Deep-sea hypersaline anoxic basins (DHABs) are considered some of the most extreme environments on Earth owing to their combination of nearly saturated salt and corresponding low aw, high hydrostatic pressure, anoxia and (sometimes) high sulfide. At least eight DHABs have been described (and more suspected) on the seafloor of the Eastern Mediterranean Sea at more than 3000 m water depth (Yakimov et al., 2015). Their unique chemistries reflect their origins from dissolved evaporites or trapped brines associated with the Miocene Messinian salinity crisis 5.59–5.33 million years ago. Basin ages range from 2000 to 176 000 years (Camerlenghi, 1990; Cita, 2006 and references therein). Their dense brines (typically ranging from 1.13 to 1.35 × 103 kg m−3 relative to Mediterranean seawater, 1.03 × 103 kg m−3) act as an impediment to mixing with overlying seawater. The halocline is the interface between normal salinity deep seawater and the brine, and can be as thin as ~2 m, depending on the basin. Haloclines have sharp oxyclines and redoxclines, with salinity increasing to ~10 times normal seawater and undetectable oxygen at the base of the halocline (Van der Wielen et al., 2005). DHAB chemistries are distinct. Some brines are thalassohaline (dominated by NaCl), while others, are athalassohaline, formed by the dissolution of bischofite (MgCl2·6 H2O), with Mg2+ concentrations up to 5000 mM compared with ~60 mM in typical seawater (Van der Wielen et al., 2005). DHABs have provided exciting new insights into the environmental factors that define the limits of life (Eder et al, 1999, 2001; Hallsworth et al, 2007) and novel planktic microbial diversity, including numerous novel candidate divisions (for example, Sass et al, 2001; Van der Wielen et al., 2005; Yakimov et al, 2007; Alexander et al., 2009; Edgcomb et al, 2009, 2011; Stock et al., 2012; Pachiadaki et al., 2014a). Less is known regarding DHAB benthos, especially microbial eukaryotes (Bernhard et al., 2014).
Here, we investigated the active prokaryotic and eukaryotic microbiota in sediments of thalassohaline and sulfidic L'Atalante and Urania DHABs, as well as the Mg2+ saturated Discovery DHAB based on metatranscriptomics (for details of brine chemistries, see Table 1; Van der Wielen et al., 2005). Metatranscriptomes provide information on the most abundant gene transcripts (a proxy for active metabolic processes) within a community at the time of preservation and is useful for casting a broad net in unusual habitats such as DHABs where little is known about in situ activities. As such, metatranscriptomic analysis is a useful tool for hypothesis generation for future follow-up investigations. Information on taxonomic diversity or on functional groups can provide insights about a community, however, information on traits (inferred from mRNA) allows a better assessment of functional attributes of communities in different habitats. Mlambo (2014) points out that ‘large-scale historical and environmental factors determine the species diversity, whereas, functional diversity is more responsive to biotic dynamics, which are structured by local habitat types.' For this reason, we include discussion not only of biological traits (for example, motility and central metabolism) but also regarding functional traits that affect and are affected by the environment (for example, sulfur and nitrogen metabolism, cell–cell interactions). To our knowledge, this is the first metatranscriptome study of sediments underlying DHAB haloclines. Our overall objective was to determine whether microbial processes and overall activities appear to be linked to the distinct chemistry (total salinity, oxygen and specific concentrations of Mg2+ and Na+, and HS−) of these three DHABs.
Table 1. Physicochemical data for sediment cores collected underneath haloclines of Urania, Discovery and L'Atalante Basins and for the brines of each DHAB.
| Sediment sample | Approximate coordinates | Depth (m) | Total salinity (PSU) | Oxygen (μM)a | Mg2+(mM)b | Na+(mM)b | HS−(mM)b | Cl−(mM)b |
|---|---|---|---|---|---|---|---|---|
| L'Atalante Lower Halocline | 35°18'N 21°23'E | 3501 | 100a | 0 | 410 (brine) | 4674 (brine) | 2.9 (brine) | 5289 (brine) |
| Urania control | 35°13'N 21°28'E | 3460 | 38b | 250–260 | 60 | 528 | 2.6 × 10–6 | 616 |
| Urania Middle Halocline | 35°13'N 21°28'E | 3470 | 172c | 0 | 316 (brine) | 3503 (brine) | 16 (brine) | 3729 (brine) |
| Discovery Upper Halocline | 35°19'N 21°41'E | 3583 | 70d | 10–75 | ||||
| Discovery Lower Halocline | 35°19'N 21°41'E | 3586 | 90d | 0–0.6 | 4995 (brine) | 68 (brine) | 0.7 (brine) | 9491 (brine) |
Data from bottom water above sediment cores (Bernhard et al., 2014).
Datum from a High Range CTD (Neil Brown Sensors) on a water column sampler deployed prior to sediment collections.
Data from halocline water column measurements (Edgcomb et al., 2011) of same depth. Note: standard conductivity sensors are not reliable for athalassohaline brines enriched in divalent cations.
Materials and methods
Sample collection
Samples were collected during R/V Atlantis cruise AT18-14 between 24 November and 6 December 2011 using the ROV Jason. Sediments were collected with Deep Submergence Lab (www.whoi.edu/groups/DSL/) Alvin-style pushcores (6.35-cm diameter) with seals to prevent contamination during ascent. Coring targeted sediments below haloclines of Urania, Discovery and L'Atalante basins as well as control samples collected from nearby (within 10 m) aerated sediments, upslope from Urania (DHAB images Figure 1) and Discovery basins. Control sediments at both DHABs (which are within ~10 nm of one another) appeared homogeneously light brown and fine grained, with no obvious megafauna. Of available core samples that were adequately preserved during recovery from the Urania control site, one was available for molecular analysis. Resource limitations prohibited analysis of more than one control sample. Predictions of microbial activities in sediments underlying the brines proper cannot be made, as the buoyancy of ROV Jason prevented us from sampling deeper than the lower haloclines. Further details of sampling are provided in Supplementary Document 1.
Figure 1.
Underwater images taken by ROV Jason of (a) Discovery DHAB and (b) Urania DHAB haloclines. Lightest color sediments delineate the upper boundary of the halocline as it impinges on the seafloor. Note that all sampling for this study was from areas that were much flatter than that shown in panel b.
Upon ROV Jason recovery onto R/V Atlantis (within 4–6 h of seafloor sampling), sealed cores were taken quickly (within 5 min) to the RV Atlantis environmental room set at 9.5 °C (±0.5 °C), where they were inspected for adequate sediment content, lack of disturbance, texture and color. On the basis of these criteria, and demands for other types of analyses, five cores of sufficient quality in total were available for metatransriptome analyses: one from L'Atalante (under the lower halocline, LLH), two from Urania (control sediment underlying the normal salinity water column outside the DHAB, UC and from under the middle halocline, UMH) and two from Discovery (one from under the upper halocline, DUH, and one from under the lower halocline, DLH). Microelectrode profiling of dissolved oxygen for replicate cores was performed in an N2-flushed glove bag as described in Bernhard et al. (2014). Salinities of overlying water were measured using a refractometer (details in Supplementary Document 1). Salinity and oxygen profiles of the water bodies of each DHAB were conducted using a high-range CTD (Neil Brown Ocean Sensors, Inc., Falmouth MA, USA) on a separately deployed water column sampler.
Subsamples of pushcores were taken from the core center using a sterile 20-ml syringe barrel (inner diameter of ~1.4 cm, length of ~10 cm), sectioned in 2-cm increments, and stored immediately at −80 °C until RNA extraction. Sediment samples were designated according to DHAB (L, U, D) and position under the halocline (depth, refractometer data): upper halocline (UH), mid-halocline (MH) and lower halocline (LH).
RNA isolation and cDNA synthesis
RNA from~8 g from each core was extracted from homogenized sediments from the top 2 cm using an optimized protocol and the RNA Power Soil kit (MoBio, Carlsbad, CA, USA) after testing and assessing RNA extraction efficiencies of two other widely used extraction kits for sediment nucleic acids. Minor modifications included introduction of three cycles of freeze–thaw (−80 °C, 5 min, 65 °C, 5 min), bead beating with 2 × 5 min intervals on a horizontal vortexer, an overnight nucleic acid precipitation and a 1-h centrifugation during the precipitation step. In addition, we introduced two DNAase treatments using TurboDNAase (Ambion, Foster City, CA, USA). Removal of DNA was confirmed by PCR using as the forward primer the oligonucleotide (5′-AYTGGGYDTAAAGNG-3′) and a mix of reverse primers (5′-GCCTTGCCAGCCCGCTCAG, TACCRGGGTHTCTAATCC, TACCAGAGTATCTAATTC, CTACDSRGGTMTCTAATC and TACNVGGGTATCTAATCC-3′ in a 6:1:2:12 ratio, respectively), designed to cover most of the Bacteria domain (Cole et al., 2009). The Ovation Single Cell RNA-seq kit (Nugen, San Carlos, CA, USA) was used to create barcoded cDNA libraries. We replaced the First Strand Primer Mix, A1 version 15 (S01882) with the First Strand Primer Mix, A1 version 13 (S01858) to avoid biases against prokaryotic transcripts. Libraries were sequenced at the DNA Sequencing & Genotyping Center, University of Delaware using three lanes of 150-bp paired-end Illumina HiSeq (Illumina, Inc., San Diego, CA, USA).
Metatranscriptome data analysis
Forward and reverse reads were filtered using Trimmomatic (Bolger et al., 2014), which performed a ‘sliding window' trimming, cutting once the average quality within the window (8-nt) fell below a threshold score (12). Reads of ⩾50 nucleotides that were retained were assembled using Trinity (Grabherr et al., 2011) release r20140717. Metapathways 2.5 (Konwar et al., 2015) was used for ORF predictions using Prodigal (Hyatt et al., 2010). Functional annotation of the contigs used LAST (Kiełbasa et al., 2011) against the RefSeq non-redundant database (Pruitt et al., 2005) update 2014-01-18, and visualization of metabolic pathways used Pathway Tools (Karp et al., 2010). The paired-end reads were mapped to the assembled transcripts and normalized using a bwa-based version of the RPKM (RPKM=[# of fragments]/[length of transcript in kilobase]/[million mapped reads]) (Li and Durbin, 2010) with default values, as implemented in Metapathways 2.5 (Konwar et al., 2015).
The RPKM reads were imported into the DEGseq package v. 1.12.1 (Wang et al., 2010), as implemented in R version 3.0.2 (http://www.r-project.org) for identification of differentially expressed genes in un-replicated datasets. The DEGexp function applies a MA-plot with Random Sampling model (Wang et al., 2010).
Results and discussion
Chemistries of the overlying waters and O2 profiles are reported in Table 1 and Bernhard et al. (2014). Oxygen was undetectable in UMH, LLH and DLH sediments, and was 10–75 and 250–260 μM in the top 2 cm in DUH and UC, respectively. The basic sequencing output metrics for each of our metatranscriptome libraries are presented in Supplementary Table 1. Annotations of our assembled reads for all libraries produced far superior results than annotations of raw reads, so only annotations of assembled reads are discussed. Pairwise comparisons of expressed genes that are present in at least one of between libraries using DEGseq provide some general insight into the degree to which expression of particular genes varies between habitats, although we interpret these single replicate comparisons cautiously. The analysis includes a total of 6950 assembled, and annotated transcripts were present in at least one sample. Results indicate the presence of transcripts that were differentially (P-value<0.001) expressed in all comparisons (Supplementary Table 2 and Supplementary Figure 1). Sequences are deposited in datasets separated by habitat in the GenBank Short Reads Archive under study accession SRP061621.
Our previous study of 16S rRNA diversity in sediments from the same cores analyzed here (Kormas et al., 2015) indicated that DHAB environmental factors significantly shaped microbial communities. In that contribution, archaeal operational taxonomic units were detected only in few halocline samples. Archaeal signatures were dominated by taxa associated with methane-rich deep-sea sediments, but might not be directly involved in methanogenesis/anaerobic oxidation of methane because we did not detect transcripts of methyl coenzyme M-reductase. Kormas et al. (2015) found each site harbored only a few site-specific dominant bacterial operational taxonomic units that included members of α-, β- and γ-Proteobacteria, Firmicutes, Actinobacteria and the Bacteroidetes/Chlorobi group, with most operational taxonomic units in lower haloclines related to halotolerant taxa or phylotypes described from high-salt habitats. We interpret taxonomic distributions of rRNA transcripts within our data sets with caution, as library preps can introduce biases, and preservation of rRNA of inactive/dead organisms under anoxic conditions has been reported (for example, Orsi et al., 2013). Consistent with this is our observation of recovery of some rRNA sequences affiliated to chloroplasts in our metatranscriptomes. We therefore avoid discussing percentages of particular taxonomic groups within the rRNA transcript pool for each sample, and restrict discussion to the groups that make up >90% of rRNA reads in each dataset. In our UC sample, rRNA reads came almost exclusively from Gammaproteobacteria affiliated with Pseudoalteromonas sp., with contributions from Deltaproteobacteria affiliated to Desulfobacteraceae and Desulfoarculaceae, as well as Firmicutes affiliated to Clostridiaceae. Pseudoalteromonas sp. and Deltaproteobacteria were detected in the Kormas et al. (2015) UC iTAG libraries. Our DUH dataset was comprised primarily of rRNA sequences of Pseudoalteromonas sp., Clostridia and the family Flavobacteraceae. Consistent with this, Kormas et al. (2015) found a Clostridium-like phylotype to dominate DUH sediments. In the DLH sample, sequences affiliating to Pseudoalteromonas sp. again were the most prevalent, along with Clostridium sp. and Mycoplasma sp., the first two of which dominated the DLI sample in Kormas et al. (2015). In UMH, rRNA transcripts affiliating to the genera Pseudomonas, Rhodobacter and Clostridia dominated. In agreement with Kormas et al. (2015), we speculate that the rRNA signatures of Pseudomonas sp. may not be active in the UMH. The LLH sample was dominated by rRNA signatures affiliated with the genus Clostridia, various Deltaproteobacteria and the phylum Chloroflexi. We note that we recovered some rRNA signatures of previously described novel groups (MSBL9, MSBL6) associated with DHABs (Yakimov et al., 2014) from our LLH, DLH and DUH samples. Given the lack of published data on sediment chemistry for the DHABs we investigated, interpretations of links between gene expression and DHAB sediment chemistry must rely on available information for the brines of these DHABs.
Microorganisms may share genetic resources through mutual ‘cheating and cooperation' (Fullmer et al., 2015). Analysis of the metatranscriptomes in our core samples therefore provides insights into overall community metabolism. We acknowledge that analysis of single cores per habitat may underestimate the breadth of metabolic activities, however, the relatively isolated DHAB sediments may be less heterogeneous than normoxic marine sediments. By normalizing each sample's read count by taking into account read number as a percentage of total reads and read length using RPKM values (fragments per kilobase per million), we can evaluate the relative importance of specific activities in contrasting habitats. Of course this must be interpreted with caution because metatranscriptomes can harbor biases from preparation steps and because they reflect primarily the most abundant transcripts, particularly for high biodiversity habitats.
Our working hypothesis was that the unique chemistry of each basin's halocline would select not only for different microbiota in the underlying sediments, but also different functional traits. We focus discussion on several categories of biological and functional activities, including transcripts attributed to microbial eukaryotes.
Nitrogen cycling
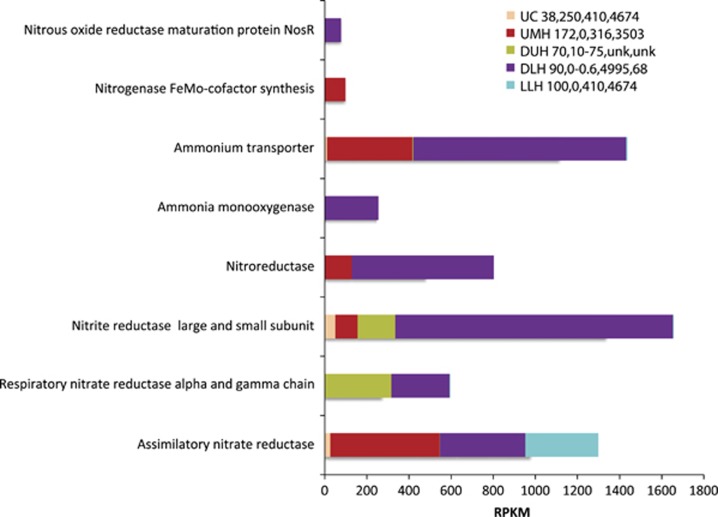
Aside from supporting normal biosynthetic pathways such as amino acid synthesis, assimilatory nitrogen metabolism may have a role in supporting the production of nitrogen-hungry osmolytes (see discussion in Pachiadaki et al. (2014a) for Thetis DHAB). We note expression of genes for assimilatory nitrogen metabolism in all samples (for example, assimilatory nitrate and nitrite reductases, nitroreductases, and ammonium transporters, as well as all genes in the pathway required for assimilatory nitrate reduction to ammonium and incorporation into L-glutamine and L-glutamate). Ammonium transporter and nitrite reductase genes were significantly upregulated in the DLH sample (P-value<0.001) in comparison with all other samples. We also note evidence for dissimilatory nitrogen metabolism, particularly in UMH and DLH (Figure 2). Transcripts of nitrite reductase (large and small subunit), respiratory nitrate reductase, nitric oxide reductase and nitrous oxide reductase maturation proteins suggest that denitrification is active in sediments below Urania and Discovery haloclines, and to a lesser extent in the control and LLH sediments. Interestingly, transcripts of ammonia monooxygenase, a key enzyme associated with aerobic ammonia oxidation, were detected in anoxic DLH sediments. Evidence for persistence of aerobic respiration in anoxic marine subsurface (D'Hondt et al. 2015) and sapropel (Süß et al., 2004) sediments has been reported. A possible explanation would be the presence of a pathway that can produce oxygen, although transcripts of chlorite dismutase (associated with perchlorate-respiring bacteria capable of producing oxygen; Coates et al., 1999; de Geus et al., 2009) were not detected in DLH. Ettwig et al. (2010, 2012) proposed a microbiologically mediated process that could produce oxygen by converting two nitric oxide molecules to dinitrogen and oxygen, although the enzyme catalyzing this reaction is still not characterized.
Figure 2.
Relative expression (presented as RPKM values) of genes associated with nitrogen cycling in the Urania Control (UC), Urania Middle Halocline (UMH), Discovery Upper Halocline (DUH), Discovery Lower Halocline (DLH) and L'Atalante Lower Halocline (LLH) sediment samples. Numbers following legend label refer to total salinity (PSU), oxygen (μM), and Mg2+ (mM) and Na+ (mM) concentrations in sediments as explained in Table 1. Unk=not measured.
There are interesting links between nitrogen metabolism and production of enzymes associated with reactive oxygen species. In UMH and DLH sediments, we observed relatively high expression of genes associated with reactive oxygen species inactivation, such as glutathione peroxidase and reductase, as well as superoxide dismutases and thioredoxin reductases (Supplementary Figure 2; Carmel-Harel and Storz, 2000). Production of glutathione peroxidase is highly dependent on nitrogen for precursor biosynthesis (Fahey and Sundquist, 1991). Expression of reactive oxygen species-associated enzyme genes appears higher in the same habitats where assimilatory nitrogen and ammonia monooxygenase transcripts are highest, suggesting that UMH and DLH sediments are fairly active relative to sediments from our control and LLH sites.
Sulfur cycling
As reported previously, the steep interfaces between normal salinity seawater and DHAB brines result in rapid biological oxygen depletion and decreases in redox potential to values near 100 mV in the upper halocline, and to −400 mV in the lower halocline (Van der Wielen et al., 2005; Daffonchio et al., 2006; Yakimov et al., 2007, 2013; Hallsworth et al., 2007; Borin et al., 2009). The steep density gradients are known to trap both organic detritus from the overlying water column and organics produced de novo (Yakimov et al., 2007). This pool supports a spectrum of aerobic and anaerobic metabolisms along the transition. The brines of DHABs we examined contain varied sulfide concentrations (Table 1), most likely produced in each case by bacterial sulfate reduction (La Cono et al. 2011). Evidence for active sulfur cycling (oxidation reduction) has been reported from DHAB halocline water layers on the basis of 16S rRNA genes, successful isolations of δ- and ɛ-Proteobacteria, activity measurements (for example, Borin et al., 2009), metagenomes (Ferrer et al., 2012) and metatranscriptomes (Pachiadaki et al., 2014a).
In our dataset, sulfur transformations were evident from transcripts of sulfide-quinone reductases, molybdopterine synthase sulfurylases, rhodanese-related sulfurtranferases, sulfite reductase hemoprotein beta-component, phosphoadenosine phosphosulfate reductases, sulfate adenyltransferases (the last three comprise 75% of the enzymes required for assimilatory sulfate reduction) and other transcripts associated with general sulfur metabolism (Supplementary Figure 3). Some highly expressed transcripts in UC, DLH and UMH samples may be associated with dissimilatory sulfate reduction (for example, D-lactate dehydrogenase, adenylsulfate reductase, sulfate adenyltransferase) or sulfur oxidation (sulfite oxidation III and Fe+3-dependent sulfur oxidation II pathway genes expressed in DLH). These genes were upregulated (P-value<0.001) in DLH and UMH relative to other samples. However, their participation in assimilatory metabolic pathways is also possible (Keller et al., 2014). For example, sulfate reduction is performed by sulfate adenyltransferase, an enzyme also participating in purine metabolism, selenoamino acid metabolism and sulfur metabolism. Overall, sulfur cycling appeared most active in the UMH and DLH sediments.
Osmolyte biosynthesis and ion transporters
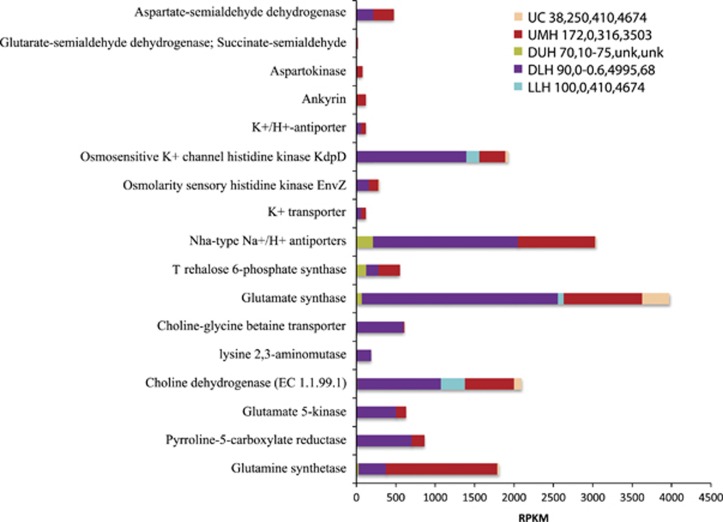
Microorganisms in hypersaline environments use two general strategies for coping with osmotic stresses: they can regulate concentrations of inorganic ions such as KCl (‘salt-in' approach) or they can produce and accumulate low-molecular-weight organic compounds that protect enzymes and macromolecular structures (‘salt-out' approach) (Grant, 2004; Stan-Lotter and Fendrihan, 2013). Expression of genes associated with osmotic stress was higher in all halocline sediment samples relative to the control (Figure 3). Expression of putative osmotic protectants and ion transporters in LLH was low, consistent with overall low recovery of transcripts from that sample. The highest expression of putative osmolytes, transporters and other putative genes involved in osmotic stress responses was in sediments from athalassohaline Discovery, particularly Nha-type Na+/H+ antiporters, osmosensitive K+ channel histidine kinase (KdpD sensor kinase proteins are involved in potassium transport and sensing turgor pressure), glutamate and glutamine synthetases, choline-glycine betaine transporters, choline dehydrogenase (possibly involved in betaine biosynthesis) and pyrroline-5-carboxylate reductase, which can be involved in proline biosynthesis during osmotic stress (Perez-Arellano et al., 2010). These genes were all significantly upregulated in the DLH (P-value<0.001) relative to our other samples. Higher expression of these genes in DLH vs DUH sediments supports their involvement in adaptation to hypersalinity, particularly to high MgCl2 concentrations. For further discussion, see Supplementary Document 2.
Figure 3.
Relative expression (presented as RPKM values) of genes associated with osmotic stress in the Urania Control (UC), Urania Middle Halocline (UMH), Discovery Upper Halocline (DUH), Discovery Lower Halocline (DLH) and L'Atalante Lower Halocline (LLH) sediment samples. Numbers following legend label refer to total salinity (PSU), oxygen (μM), and Mg2+ (mM) and Na+ (mM) concentrations in sediments as explained in Table 1. Unk=not measured.
Heavy metals
During formation of the brines and/or evaporites that became the sources of DHAB brines, different metal-containing compounds in seawater precipitated onto the seafloor according to their solubility, concentrating different metals at different stages in this process (Cita, 2006 and references therein). Additional focusing of these deposits can occur by subsurface brine flows (for example, discussed in Warren, 2000). Unfortunately, little is currently known about heavy metal concentrations in brines and sediments of most DHABs; however, the few existing studies (primarily of Bannock Basin) indicate that DHABs may be trace metal-rich. Van der Sloot et al. (1990) reported extremely high concentrations of cobalt (0.015%), copper (1.35%) and Zn (0.28%) in suspended matter collected at the seawater/brine interface of Tyro and Bannock basins (E. Mediterranean). In anoxic waters such as DHAB brines, concentrations of some dissolved metals may be low owing to the formation of sulfide precipitates and/or adsorption to sediment particles (Saager et al., 1992; Schuf et al., 1995). The composition and concentration of heavy metals such as cadmium, mercury, lead, arsenic, cobalt and copper are known to shape the structure and composition of microbial communities because of their toxicity (for example, Oliveira and Pampulha, 2006; Ravikumar et al., 2007). They can bind to vital cellular structural proteins, enzymes and nucleic acids, interfering with their function (for example, Srivastava and Goyal, 2010; Olaniran et al., 2013). Heavy metal-tolerant halophiles are known (Nieto et al., 1989; Oren, 2006; Sowmya et al., 2014), hence it is not surprising we find expression of genes associated with metal detoxification in our DHAB metatranscriptomes.
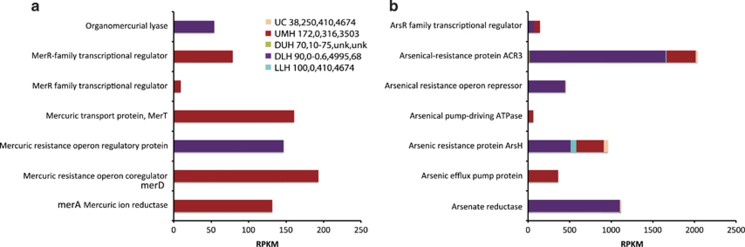
Transcripts associated with mercury (Hg) detoxification (Figure 4a) were not detected in the DUH, LLH or UC samples, but were expressed in the DLH and UMH samples, suggesting that Urania and Discovery lower haloclines may be relatively enriched in mercury. Expression of organomercurial lyase and mercuric resistance operon genes was detected in DLH. In UMH, we detected the expression of mercuric ion reductase (merA), a mercuric resistance operon co-regulator, a mercuric transport protein (merT) and transcriptional regulators (MerR) that are known to be involved in responses to environmental stimuli including recognition of heavy metals (Brown et al., 2003).
Figure 4.
Relative expression (presented as RPKM values) of genes associated with processing (a) mercury and (b) arsenic in the Urania Control (UC), Urania Middle Halocline (UMH), Discovery Upper Halocline (DUH), Discovery Lower Halocline (DLH) and L'Atalante Lower Halocline (LLH) sediment samples. Numbers following legend label refer to total salinity (PSU), oxygen (μM), and Mg2+ (mM) and Na+ (mM) concentrations in sediments as explained in Table 1. Unk=not measured.
Transcripts associated with arsenic detoxification appear to be more highly expressed (Figure 4b) than for mercury detoxification, particularly in the DLH sample, suggesting that Discovery brine may be rich in both mercury and arsenic. Significantly higher expression (P<0.001) of the gene for arsenical resistance protein (ACR3) was observed in DLH relative to other samples. Microorganisms in anoxic sediments are known to be capable of respiring through the coupled reduction of iron or sulfur and toxic metals such as arsenic (Reyes et al., 2008), and to use chaperone proteins to counter heavy metal-induced protein aggregation (Tamas et al., 2014). We found evidence for expression of genes that may facilitate the binding and movement of arsenate or arsenite ions across the cell membrane, and therefore, may support the ability of DHAB microorganisms to cope with in situ metal concentrations. These included the phosphate transport system (Pst) complex (pstA, pstB, pstC) responsible for phosphate-specific transport, and the glycerol uptake facilitator (GlpF) aquaglyceroporin protein responsible for osmoregulation and potentially, movement of arsenite (Gomes et al., 2009). The phosphate transport system complex genes were significantly upregulated (P-value<0.001) in the DLH relative to other samples. The pattern of expression of these genes in terms of habitat mirrored the expression of As detoxification genes noted above. For discussion of transcripts associated with additional heavy metals, see Supplementary Document 2.
Eukaryotic activities
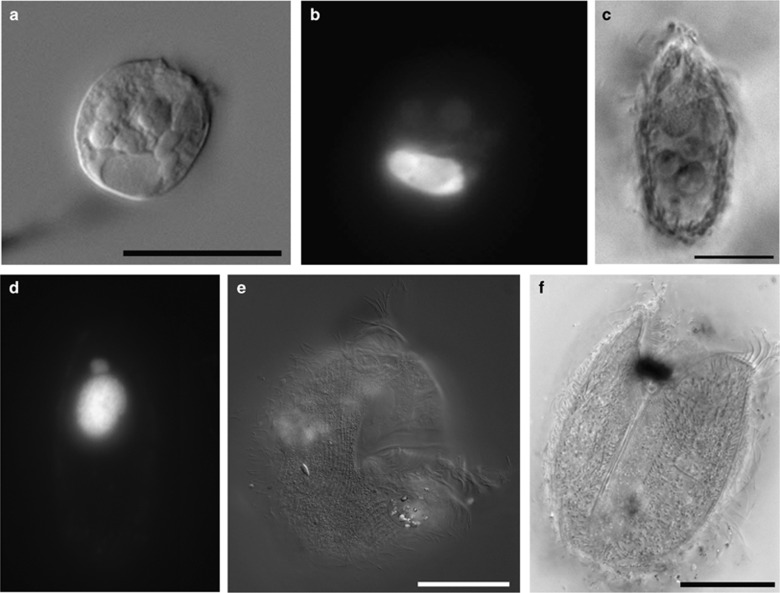
The extent of diversity of microbial eukaryotes in extremely hypersaline (over 30%) environments has been debated (Edgcomb and Bernhard, 2013 and references therein). Evidence for active eukaryotes in DHABs came initially from ribosomal DNA- and RNA-based studies (Alexander et al., 2009; Edgcomb et al., 2009). Microscopic images of intact and presumably living (based on intact nuclei) planktonic microbial eukaryotes provides an important second line of evidence (Edgcomb et al., 2011; Edgcomb and Bernhard, 2013; Edgcomb and Orsi, 2013; Edgcomb and Pachiadaki, 2014) (Figure 5). Actin and tubulin transcripts in the Thetis DHAB upper halocline waters were >4 times as abundant than in the normal salinity deep water column (~2200 m depth) (Pachiadaki et al., 2014a). Higher numbers of particular unicellular eukaryotes have been reported in the upper haloclines relative to control waters (Edgcomb et al., 2011).
Figure 5.
Light micrographs of protists from DHAB haloclines. (a, b) Paired DIC (a) and epifluorescence (b; DAPI labeling) micrographs of unknown protist from Urania halocline (core 607-608 cE). (c, d) Paired phase contrast (c) and epifluorescence (d; DAPI labeling) micrographs of ciliate from Urania upper halocline (core 607 c10). (e) DIC image of ciliate from L'Atalante mid-halocline (core 611 c10). (f) Phase contrast image of ciliate from L'Atalante mid-halocline (core 611 c10). e and f are specimens also illustrated in Bernhard et al., 2014. a=20 μm, c=10 μm, e=50 μm; f=25 μm.
Similar to our discussion of bacterial and archaeal rRNA transcripts above, we refrain from interpretation of the relative abundances of eukaryotic rRNA transcripts, but we compare the composition of the most abundant rRNA reads (>90% of each library) broadly to the results of Bernhard et al. (2014) who report on eukaryotic iTAG diversity from the same core samples. Consistent with Bernhard et al. (2014), our UC sample was dominated by rRNA signatures of fungi related to Malasseziomycetes within Basidiomycota. In contrast, we also observed abundant signatures of Ascomycota affiliated with the genera Penicillium and Aspergillus. In DUH, and consistent with Bernhard et al. (2014), our library was also comprised primarily of similar rRNA signatures. In our DLH sample, the most abundant rRNA sequences came from the marine protist rhizarian genus Acantharia, and the fungal genera Penicillium and Aspergillus. This habitat was not sampled by Bernhard et al. (2014), so a comparison with that study cannot be made. Our UMH sample contained rRNA signatures primarily of the fungal genus Aspergillus, as well as sequences of Acantharia, Penicillium and the protist amoebozoan genus Acanthamoeba, the first three of which were also detected by Bernhard et al. (2014). The LLH sample was comprised primarily of sequences affiliated with the genus Aspergillus, along with various other ascomycetes and basidiomycetes. The Bernhard et al. (2014) study found this sample to be dominated by the Basidiomycota related to Malasseziomycetes. Overall, these results suggest that the taxonomic composition of the eukaryotic fraction of our metatranscriptomes is roughly consistent with previously published iTAG diversity profiles.
Searching metatranscriptomes for mRNA transcripts that can be unambiguously annotated to eukaryotes is challenging, even with current binning approaches, owing to the relative paucity of public genomic data for marine protists, common evolutionary origins of some genes with prokaryotes and lateral gene transfer events. We used BLAST (NCBI RefSeq-NR database 2014-01-18) to sort transcripts within categories that could be assigned to eukaryotes with less ambiguity; including eukaryotic central metabolism, organelle functions, intra-organelle trafficking and structural proteins. The greatest activity identified within these categories was recorded from UMH sediments, followed by the DLH, with most transcripts affiliating with Fungi (Figure 6 and Supplementary Figures 4 and 5). This is consistent with Bernhard et al. (2014) who found microscopic and rRNA pyrotag evidence for protists and fungi in these same core samples, with most pyrotags affiliating with fungal classes, particularly Malasseziomycetes, Dothideomycetes and Microbotryomycetes.
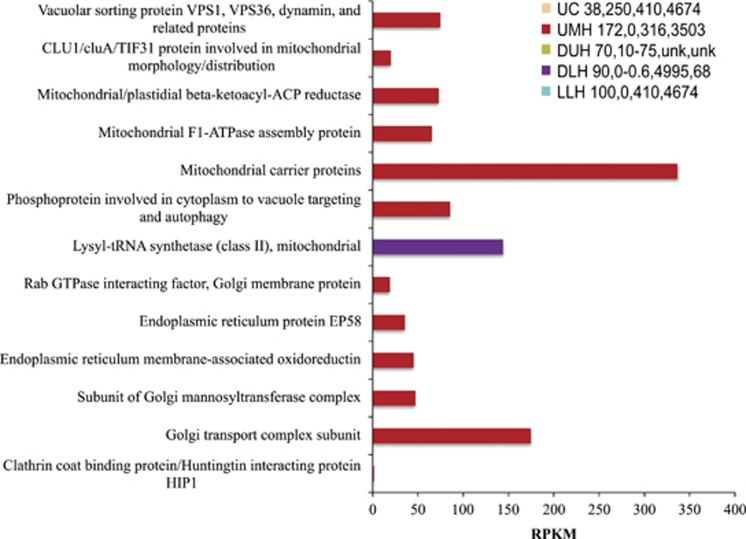
Figure 6.
Relative expression (presented as RPKM values) of genes associated with eukaryotic intracellular transport in the Urania Control (UC), Urania Middle Halocline (UMH), Discovery Upper Halocline (DUH), Discovery Lower Halocline (DLH) and L'Atalante Lower Halocline (LLH) sediment samples. Numbers following legend label refer to total salinity (PSU), oxygen (μM), and Mg2+ (mM) and Na+ (mM) concentrations in sediments as explained in Table 1. Unk=not measured.
All transcripts within the category of intracellular transport came from the UMH sample with the exception of a mitochondrial lysyl-tRNA synthetase (class II) identified from the DLH sample. The most abundant transcripts were mitochondrial carrier proteins and Golgi transport proteins (Figure 6) affiliated with relatives of the fungal genera Aspergillus and Penicillium. Transcripts for other mitochondrial proteins were detected in the UMH sample, including mitochondrial F1-ATPase assembly proteins, mitochondrial/plastidial beta-ketoacyl-ACP reductases and CLU1 proteins involved in mitochondrial morphology and distribution, mostly affiliating with genes described from various yeasts (Fields et al., 1998; Venkatesan et al., 2014). Expression of a clathrin coat-binding protein of Aspergillus involved in clathrin-mediated endocytosis (Carter et al., 1993) was detected in the UMH sample, suggesting that saprophytic fungi are active in UMH. Fungal transcripts for Golgi function were detected in the same sample, including a Golgi transport complex, a subunit of the Golgi mannosyltransferase complex and membrane Rab GTPase that contains a filamin domain and a KDEL motif (Hutagalung and Novick, 2011). Expression of genes associated with endoplasmic reticulum was detected in the UMH sample; including membrane-associated oxidoreductin and the protein EP58 (involved in disulfide bond formation, Kimata et al., 2000; Kettner et al., 2004). Other transcripts included the vacuolar sorting proteins VPS1, VPS36, dynamin and related proteins, and phosphoproteins involved in cytoplasm to vacuole targeting and autophagy (Klionsky et al., 1990). These were affiliated mostly to sequences of fungal relatives of Aspergillus and Penicillium and a few protist taxa (Acanthamoeba and unidentified protists). Stress proteins (heat shock protein 60 and 90, as well as chaperonin complex component TCP-1 subunits) putatively affiliated with eukaryotes were detected in all halocline sediment samples. Future culture-based studies of DHAB isolates may be able to shed light on the role of these proteins.
Expression of several structural proteins of eukaryotes was detected in all our sediment samples, including our control sample (Supplementary Figure 5). Because of their low amino acid similarity compared with prokaryotic structural proteins and unique posttranslational modifications, actin and tubulin transcripts can be used to detect evidence for eukaryotic activity. These included expression of α-actinin, a Ca2+-regulatory actin-binding and bundling protein (Sjöblom et al., 2008), and actin-related proteins in UMH and DLH sediments. The regulatory protein transcript was annotated to Aspergillus, codes for a protein that interacts with microtubules and is in some cases associated with phagocytosis (Alberts et al., 2002). In our UMH, DUH and DLH samples, transcripts of arginyltransferase, involved in tRNA-dependent posttranslational addition of arginine in the N-terminus of various eukaryotic substrates including actin (Karakozova et al., 2006), were detected.
Alpha tubulin transcripts were detected in DUH, DLH and LLH sediment samples. According to the ‘tubulin code,' tubulin can undergo unique posttranslational modifications in the carboxy-terminal tail (Sirajuddin et al., 2014), including tyrosination/detyrosination, polyglutamination and polyglycylation processes. Polyglutamination and tyrosination are performed by tubulin-tyrosin ligase domain proteins (Janke et al., 2005). Polyglutamination is required for the stability and motility of cilia/flagella, and controlling the length of flagella (Kubo et al., 2015). Expression of predicted and putative tubulin-tyrosin ligase was detected in our UC, UMH and DLH samples, and were taxonomically annotated to diverse fungi as well as a few amoebae, ciliates and flagellates. Expression of eukaryotic structural proteins was not significantly upregulated in the lower salinity DUH sample relative to the DLH sample.
Eukaryotic transcripts associated with central metabolism were also detected in all samples except for our control and LLH samples (Supplementary Figure 4). This is consistent with more abundant recovery of microbial eukaryote cells and SSU rRNA signatures in sediments under DHAB haloclines relative to normal saline deep Mediterranean Sea sediments (Bernhard et al., 2014). This is likely made possible by prokaryotic abundances and activities that are higher in halocline waters than in control deep-sea waters above DHABs (for example, Sass et al., 2001; Pachiadaki et al., 2014b). We interpret the lack of detection of eukaryotic transcripts associated with intracellular transport and organelle trafficking in our UC and LLH sediment samples as indicating extremely low eukaryotic activities in those sediments. Sodium concentrations in the LLH may exceed the tolerance of many eukaryotes. For details of central metabolism transcripts, see Supplementary Document 2.
Collectively, these results suggest active eukaryotic populations in sediments underlying the haloclines of Discovery and Urania DHABs relative to control sediments outside the influence of the haloclines, and that communities are likely dominated by Fungi. The chemistry (high NaCl) of LLH may present a barrier to all but the most halotolerant eukaryotes. Recovery of transcripts for only a low number of genes in the categories discussed suggests a greater sequencing depth is needed to gain a more comprehensive indication of DHAB sediment eukaryotic metabolic activities.
Microbe–microbe interactions and motility
As discussed by Klein (2015), modern molecular ecology approaches relying on bulk extraction of nucleic acids risk capturing not only the molecules of microorganisms that were active in situ, but the molecules of inactive and dormant organisms, and extracellular nucleic acids. By examining mRNA, we minimize contributions from these confounding pools. As defined by Pielou (1975), a community is defined as ‘any assemblage of plants and animals living together in one place and to a greater or lesser degree, interacting with one another—in a word, an ecological community.' The community of interacting microorganisms is what is relevant to the ecology of DHAB habitats. We found evidence of transcripts associated with activities that indirectly imply cell–cell interactions in these polyextreme habitats.
Cell–cell connections and motility
In the UMH, DUH and DLH sediments, we detected expression of genes associated with pili formation, including Gram-negative pili assembly chaperones, prepilin peptidases, mannose-sensitive haemagglutinin pilin proteins, pilin-like competence factors and type IV pilin (Supplementary Table 3). The genes for pili assembly chaperones, competence factors and type IV pilin were significantly upregulated (P-value<0.001) in the DLH vs DUH samples. Absence of these transcripts in the UC and LLH datasets implies they are either absent or expressed at levels below detection. While the hair-like pili can be the attachment site of bacteriophages, some can generate motile forces, and some can be used for bacterial conjugation. We also detected the expression of flagellar proteins, including basal-body rod proteins, flagellar biosynthesis proteins, hook proteins, motor rotation proteins, motor switch proteins, P-ring proteins and flagellin, primarily in UMH, DUH and DLH sediments. Adhesin proteins, which are found on bacterial cell surfaces or appendages, facilitate attachment to other cells or surfaces and are, therefore, another sign of interactions between cells and their environment. In DHAB sediments, adhesins possibly have a role in colonization of organic and inorganic surfaces. Adhesion-related transcripts were also primarily detected in the three DHAB environments where flagellar- and pili-related transcripts were detected (Supplementary Table 3).
Cell–cell competition
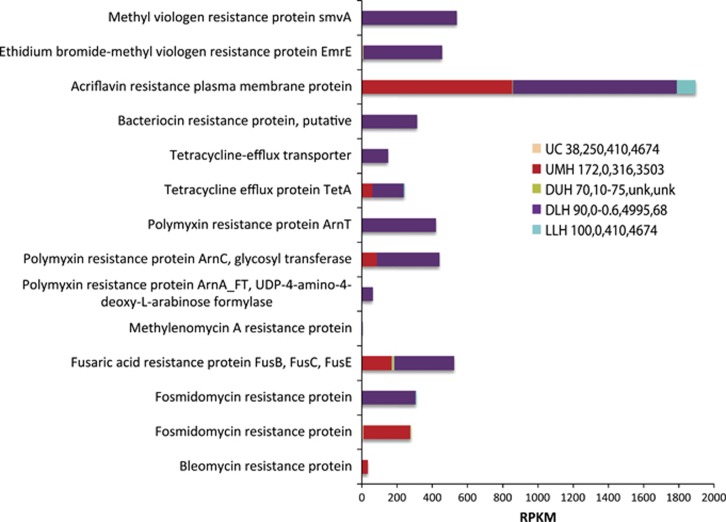
We found evidence for active competition between members of the microbial community in the form of toxin and antibiotic production. Toxin transcripts were almost exclusively detected in UMH, DUH and DLH sediments. These included transcripts for toxin-antitoxin, HigA and HigB toxins, K1 killer toxin, zeta toxins, RTX toxins (produced by Gram-negative bacteria) and YoeB toxin (Supplementary Table 1), the latter two of which were significantly upregulated (P-value<0.001) in DLH relative to the DUH sample. Cell–cell competition was even more evident when looking at transcripts associated with antibiotic and antimicrobials production (Figure 7). In DUH, DLH and UMH samples, expression of antibiotic production or resistance genes was the greatest, including tetracycline efflux proteins, and resistance proteins to polymyxin (produced by Gram-positive bacteria), fusaric acid (produced by fungal species, such as Fusarium), and fosmidomycin, methylenomycin and bleomycin produced by bacteria affiliated to Streptomyces (Mahajan and Balachandran, 2015). In addition, we detected the expression of genes for bacteriocins, which bacteria use to inhibit the growth of closely related strains (Cotter et al., 2013), as well as bacterial acriflavine resistance proteins and ethidium bromide-methyl viologen resistance proteins (Bay et al., 2008). All these antibiotic and antimicrobial production or resistance genes were significantly upregulated (P-value<0.001) in the DLH relative to the DUH sample. Active fungal-bacterial competition for resources is suggested by detection of transcripts associated with mechanisms to resist these antimicrobials in all samples, including drug resistance transporters, efflux pumps and multidrug resistance proteins (Supplementary Table 1). Transcripts associated with polyketide synthase and, specifically, mitomycin antibiotics typically produced by Streptomyces were detected in all samples except LLH sediments (Supplementary Table 1). Polyketide synthases are involved in secondary metabolite synthesis, and are important sources of common antibiotics (Koehn and Carter, 2005).
Figure 7.
Relative expression (presented as RPKM values) of genes associated with antibiotic/antimicrobial production in the Urania Control (UC), Urania Middle Halocline (UMH), Discovery Upper Halocline (DUH), Discovery Lower Halocline (DLH) and L'Atalante Lower Halocline (LLH) sediment samples. Numbers following legend label refer to total salinity (PSU), oxygen (μM), and Mg2+ (mM) and Na+ (mM) concentrations in sediments as explained in Table 1. Unk=not measured.
Evidence of phage activity
An active phage population implies an active target host population. Our datasets revealed phage transcripts associated with glycoproteins typically involved in capsid formation (Sae-Ueng et al., 2014), including head-tail pre-connector proteins, bacteriophage glycoproteins and glycoprotein baseplate subunit and tail lysozyme. Interestingly, these glycoprotein transcripts were not detected in the control sample. This is consistent with a recent study of extracellular DNA in sediments of two DHABs that indicates DHABs are hotspots of viral infections, releasing high amounts of extracellular DNA that may be an important trophic resource for benthic prokaryotes (Corinaldesi et al., 2014).
Conclusion
Relative to our control sediment sample collected outside of Urania Basin and to the LLH sediments, microbial communities in the UMH, DUH and DLH sediments appeared more active. This is consistent with previous halocline water column studies and the Bernhard et al. (2014) benthos study that showed DHAB habitats can be microbial hotspots in the deep ocean. The almost saturated NaCl concentrations of L'Atalante brine appear to suppress the activities of most prokaryotic and eukaryotic microorganisms based on analysis of our LLH sample. Eukaryotic transcripts, particularly under Urania and Discovery haloclines appear to be predominantly, but not exclusively, fungal. In Urania and Discovery DHAB samples, we found evidence of active nitrogen and sulfur cycling, trace metal detoxification, cell motility and numerous forms of cell–cell interactions (for example, antibiotic and antimicrobial production and defense as well as cell-to-cell connections). Diverse transcripts associated with production of potential osmolytes were expressed in greater numbers in samples from DLH than DUH, strong evidence for their role in adaptation to the athalassohaline Discovery DHAB.
Acknowledgments
We would like to thank the captain and crew of the R/V Atlantis as well as the ROV Jason team for assuring the success of our sampling objectives. We also thank Colin Morrison for sampling assistance, Dave Beaudoin for lab assistance, Andrew Newman for graphics assistance, and Dave Beaudoin and Colin Morrison for contributing to microscopic photo-documentation. This research was funded by NSF OCE-0849578 to VE and JMB and OCE-1061391 to JMB and VE, WHOI scholarship award for MP and the Investment in Science Fund at WHOI.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. (2002). How cells regulate their cytoskeletal filaments. In: Molecular Biology of the Cell 4th edition Garland Science: New York, USA. [Google Scholar]
- Alexander E, Stock A, Breiner HW, Behnke A, Bunge J, Yakimov MM et al. (2009). Microbial eukaryotes in the hypersaline anoxic L'Atalante deep-sea basin. Env Microbiol 11: 360–381. [DOI] [PubMed] [Google Scholar]
- Bay DC, Rommens KL, Turner RJ. (2008). Small multidrug resistance proteins: a multidrug transporter family that continues to grow. Biochim Biophys Acta 1778: 1814–1838. [DOI] [PubMed] [Google Scholar]
- Bernhard JM, Kormas KA, Pachiadaki MG, Rocke E, Beaudoin DJ, Morrison C et al. (2014). Benthic protists and fungi of Mediterranean deep hypersaline anoxic basin redoxcline sediments. Front Microbiol 5: 605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. (2014). Trimmomatic: A flexible trimmer for Illumina Sequence Data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borin S, Brusetti L, Mapelli F, D'Auria G, Brusa T, Marzorati M et al. (2009). Sulfur cycling and methanogenesis primarily drive microbial colonization of the highly sulfidic Urania deep hypersaline basin. Proc Natl Acad Sci USA 106: 9151–9156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NL, Stoyanov JV, Kidd SP, Hobman JL. (2003). The MerR family of transcriptional regulators. FEMS Microbiol Rev 27: 145–163. [DOI] [PubMed] [Google Scholar]
- Camerlenghi A. (1990). Anoxic basins of the eastern Mediterranean: geological framework. Mar Chem 31: 1–19. [Google Scholar]
- Carmel-Harel O, Storz G. (2000). Roles of the glutathione- and thioredoxin-dependent reduction systems in the Escherichia coli and Saccharomyces cerevisiae responses to oxidative stress. Annu Rev Microbiol 54: 439–461. [DOI] [PubMed] [Google Scholar]
- Carter LL, Redelmeier TE, Woolenweber LA, Schmid SL. (1993). Multiple GTP-binding proteins participate in clathrin-coated vesicle-mediated endocytosis. J Cell Biol 120: 37–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho BC, Park JS, Xu K, Choi JK. (2008). Morphology and molecular phylogeny of Trimyema koreanum n. sp., a ciliate from the hypersaline water of a solar saltern. J Eukaryot Microbiol 55: 417–426. [DOI] [PubMed] [Google Scholar]
- Cita MB. (2006). Exhumation of Messinian evaporites in the deep-sea and creation of deep anoxic brine-filled collapsed basins. Sediment Geol 188-189: 357–378. [Google Scholar]
- Coates JD, Michaelidou U, Bruce RA, O'Connor SM, Crespi JN, Achenbach LA. (1999). Ubiquity and diversity of dissimilatory (per)chlorate-reducing bacteria. Appl Environ Microbiol 65: 5234–5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ et al. (2009). The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37: 141–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corinaldesi C, Tangherlini M, Luna GM, Dell'Anno A. (2014). Extracellular DNAcan preserve the genetic signatures of present and past viral infection events in deep hypersaline anoxic basins. Proc R Soc 281: 20133299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter PD, Ross RP, Hill C. (2013). Bacteriocins – a viable alternative to antibiotics? Nat Rev Microbiol 11: 95–105. [DOI] [PubMed] [Google Scholar]
- Daffonchio D, Borin S, Brusa T, Brusetti L, van der Wielen PW, Bolhuis H et al. (2006). Stratified prokaryote network in the oxic-anoxic transition of a deep-sea halocline. Nature 440: 203–207. [DOI] [PubMed] [Google Scholar]
- de Geus DC, Thomassen EAJ, Hagedoorn P-L, Pannu NS, Van Duijn E, Abrahams JP. (2009). Crystal structure of chlorite dismutase, a detoxifying enzyme producing molecular oxygen. J Mol Biol 387: 192–206. [DOI] [PubMed] [Google Scholar]
- D'Hondt S, Inagaki F, Zarikian CA, Abrams LJ, Dubois N, Engelhardt T et al. (2015). Presence of oxygen and aerobic communities from sea floor to basement in deep-sea sediments. Nat Geosci 8: 299–304. [Google Scholar]
- Eder W, Jahnke LL, Schmidt M, Huber R. (2001). Microbial diversity of the brine-seawater interface of the Kebrit Deep, Red Sea, studied via 16S rRNA gene sequences and cultivation methods. Appl Environ Microbiol 67: 3077–3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eder W, Ludwig W, Huber R. (1999). Novel 16S rRNA gene sequences retrieved from highly saline brine sediments of Kebrit Deep, Red Sea. Arch Microbiol 172: 213–218. [DOI] [PubMed] [Google Scholar]
- Edgcomb V, Orsi W, Leslin C, Epstein SS, Bunge J, Jeon S et al. (2009). Protistan community patterns within the brine and halocline of deep hypersaline anoxic basins in the eastern Mediterranean Sea. Extremophiles 13: 151–167. [DOI] [PubMed] [Google Scholar]
- Edgcomb VP, Bernhard JM. (2013). Heterotrophic protists in hypersaline microbial mats and deep hypersaline basin water columns. Special Issue ‘Extremophiles and Extreme Environments'. Life 3: 346–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgcomb VP, Orsi W, Breiner HW, Stock A, Filker S, Yakimov MM et al. (2011). Novel active kinetoplastids associated with hypersaline anoxic basins in the Eastern Mediterranean deep-sea. Deep-Sea Res PT I 58: 1040–1048. [Google Scholar]
- Edgcomb VP, Orsi W. (2013). Microbial eukaryotes in hypersaline anoxic deep sea basins. In: Seckbach J, Oren A, Stan-Lotter H (eds). COLE Series: Polyextremophiles – Life Under Multiple Forms of Stress, Cellular Origin, Life in Extreme Habitats and Astrobiology Vol. 27 Springer Verlag: Springer Science and Business Media, Dordrecht, p 634. [Google Scholar]
- Edgcomb VP, Pachiadaki M. (2014). Ciliates along oxyclines of permanently stratified marine water columns. J Euk Microbiol 61: 434–445. [DOI] [PubMed] [Google Scholar]
- Ettwig KF, Butler MK, Le Paslier D, Pelletier E, Mangenot S, Kuypers MM et al. (2010). Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464: 543–548. [DOI] [PubMed] [Google Scholar]
- Ettwig KF, Speth DR, Reimann J, Wu ML, Jetten MSM, Keltjens JT. (2012). Bacterial oxygen production in the dark. Front Microbiol 3: 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahey RC, Sundquist AR. (1991). Evolution of glutathione metabolism. Adv Enzymol Relat Areas Mol Biol 64: 1–53. [DOI] [PubMed] [Google Scholar]
- Ferrer M, Werner J, Chernikova TN, Bargiela R, Fernandez L, La Cono V et al. (2012). Unveiling microbial life in the new deep-sea hypersaline Lake Thetis. Part II: a metagenomic study. Environ Microbiol 14: 268–281. [DOI] [PubMed] [Google Scholar]
- Fields SD, Conrad MN, Clarke M. (1998). The S. cerevisiae CLU1 and D. discoideum cluA genes are functional homologues that influence mitochondrial morphology and distribution. J Cell Science 111: 1717–1727. [DOI] [PubMed] [Google Scholar]
- Fullmer MS, Soucy SM, Gogarten JP. (2015). The pan-genome as a shared genomic resource: mutual cheating, cooperation and the black queen hypothesis. Front Microbiol 6: 728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes D, Agasse A, Thiebaud P, Delrot S, Geros H, Chaumont F. (2009). Aquaporins are multifunctional water and solute transporters highly divergent in living organisms. Biochim Biophys Acta 1788: 1213–1228. [DOI] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I et al. (2011). Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol 29: 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant WD. (2004). Life at low water activity. Phil Trans R Soc Lond B Biol Sci 359: 1249–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallsworth JE, Yakimov MM, Golyshin PN, Gillion JL, Auria D, de Lima G et al. (2007). Limits of life in MgCl2-containing environments: chaotropicity defines the window. Environ Microbiol 9: 801–813. [DOI] [PubMed] [Google Scholar]
- Hutagalung AH, Novick PJ. (2011). Role of Rab GTPases in membrane traffic and cell physiology. Physiol Rev 91: 119–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser LJ. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform 11: 119–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, Rogowski K, Wloga D, Regnard C, Kajava AV, Strub JM et al. (2005). Tubulin polyglutamylase enzymes are members of the TTL domain protein family. Science 308: 1758–1762. [DOI] [PubMed] [Google Scholar]
- Karakozova M, Kozak M, Wong CC, Bailey AO, Yates JR 3rd, Mogilner A et al. (2006). Arginylation of beta- actin regulates actin cytoskeleton and cell motility. Science 313: 192–196. [DOI] [PubMed] [Google Scholar]
- Karp PD, Paley SM, Krummenacker M, Latendresse M, Dale JM, Lee TJ et al. (2010). Pathway Tools version 13.0: integrated software for pathway/genome informatics and systems biology. Brief Bioinformatics 11: 40–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller KL, Rapp-Giles BJ, Semkiw ES, Porat I, Brown SD, Wall JD. (2014). New model for electron flow for sulfate reduction in Desulfovibrio alaskensis G20. App Environ Microbiol 80: 855–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettner K, Blomberg A, Rodel G. (2004). Schizosaccharomyces pombe ER oxidoreductin-like proteins SpEro1a p and SpEro1b p. Yeast 21: 1035–1044. [DOI] [PubMed] [Google Scholar]
- Kiełbasa SM, Wan R, Sato K, Horton P, Frith MC. (2011). Adaptive seeds tame genomic sequence comparison. Genome Res 21: 487–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimata Y, Ooboki K, Nomura-Furuwatari C, Hosoda A, Tsuru A, Kohno K. (2000). Identification of a novel mammalian endoplasmic reticulum-resident KDEL protein using an EST database motif search. Gene 261: 321–327. [DOI] [PubMed] [Google Scholar]
- Klein DA. (2015). Partial formalization: an approach for critical analysis of definitions and methods used in bulk extraction-based molecular microbial ecology. Open Journal of Ecology 5: 400–407. [Google Scholar]
- Klionsky DJ, Herman PK, EMR SD. (1990). The fungal vacuole: composition, function and biogenesis. Microbiol Rev 54: 266–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehn FE, Carter GT. (2005). The evolving role of natural products in drug discovery. Nat Rev Drug Discov 4: 206–220. [DOI] [PubMed] [Google Scholar]
- Konwar KM, Hanson NW, Bhatia MP, Kim D, Wu SJ, Hahn AS et al. (2015). MetaPathways v2.5: quantitative functional, taxonomic and usability improvements. Bioinformatics 32: 3345–3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kormas KA, Pachiadaki M, Karayanni H, Leadbetter ER, Bernhard JM, Edgcomb VP. (2015). Inter-comparison of the potentially active prokaryotic communities in the halocline sediments of Mediterranean deep-sea hypersaline basins. Extremophiles 19: 949–960. [DOI] [PubMed] [Google Scholar]
- Kubo K, Hirono M, Aikawa T, Kamiya R, Witman GB. (2015). Reduced tubulin polyglutamylation suppresses flagellar shortness in Chlamydomonas. Mol Biol Cell 26: 2810–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Cono V, Smedile F, Bortoluzzi G, Arcadi E, Maimone G et al. (2011). Unveiling microbial life in new deep-sea hypersaline Lake Thetis. Part I: Prokaryotes and environmental settings. Environ Microbiol 13: 2250–2268. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. (2010). Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26: 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litchfield CD. (1998). Survival strategies for microorganisms in hypersaline environments and their relevance to life on early Mars. Meteorit Planet Sci 33: 813–819. [DOI] [PubMed] [Google Scholar]
- Mahajan G, Balachandran L. (2015). Biodiversity in production of antibiotics and other bioactive compounds. Adv Biochem Eng Biotechnol 147: 37–58. [DOI] [PubMed] [Google Scholar]
- Mlambo MC. (2014). Not all traits are ‘functional': insights from taxonomy and biodiversity-ecosystem functioning research. Biodivers Conserv 23: 781–790. [Google Scholar]
- Nieto JJ, Fernandez-Castillo R, Marquez MC, Ventosa A, Quesada E, Ruiz-Berraquero F. (1989). Survey of metal tolerance in moderately halophilic bacteria. Appl Environ Microbiol 44: 2385–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olaniran AO, Balgobind A, Pillay B. (2013). Bioavailability of heavy metals in soil: Impact on microbial biodegradation of organic compounds and possible improvement strategies. Int J Mol Sci 14: 10197–10228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira A, Pampulha ME. (2006). Effects of long-term heavy metal contamination on soil microbial characteristics. J Biosci Bioeng 102: 157–161. [DOI] [PubMed] [Google Scholar]
- Oren A. (2003) Halophilic Microorganisms and Their Environments. Kluwer Academic: Dordrecht, p 575. [Google Scholar]
- Oren A. (2006). Halophilic microorganisms and their environments. In: Seckbach J (ed). Cellular Origin and Life in Extreme Habitats. Kluwer Academic Publishers: Boston, MA, USA, p 575. [Google Scholar]
- Orsi W, Biddle J, Edgcomb VP. (2013). Deep sequencing of subseafloor eukaryotic rRNA reveals active fungi across marine subsurface provinces. PLoS One 8: e56335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachiadaki MG, Yakimov MM, Leadbetter E, Edgcomb VP. (2014. a). Unveiling microbial activities along the halocline of Thetis, a deep hypersaline anoxic basin in the Eastern Mediterranean Sea. ISME J 8: 2478–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachiadaki M, Taylor C, Oikomomou A, Yakimov M, Stoeck T, Edgcomb VP. (2014. b). In situ grazing experiments apply new technology to gain insights into deep-sea food webs. Deep Sea Res II doi:doi:10.1016/j.dsr2.2014.10.019.
- Park JS, Cho BC, Simpson AG. (2006). Halocafeteria seoinensis gen. et sp. nov. (Bicosoecida), a halophilic bacterivorous nanoflagellate isolated from a solar saltern. Extremophiles 10: 493–504. [DOI] [PubMed] [Google Scholar]
- Park JS, Simpson AG, Brown S, Cho BC. (2009). Ultrastructure and molecular phylogeny of two heterolobosean amoebae, Euplaesiobystra hypersalinica gen. et sp. nov. and Tulamoeba peronaphora gen. et sp. nov., isolated from an extremely hypersaline habitat. Protist 160: 265–283. [DOI] [PubMed] [Google Scholar]
- Park JS, Simpson AG, Lee WJ, Cho BC. (2007). Ultrastructure and phylogenetic placement within Heterolobosea of the previously unclassified, extremely halophilic heterotrophic flagellate Pleurostomum flabellatum (Ruinen 1938). Protist 158: 397–413. [DOI] [PubMed] [Google Scholar]
- Perez-Arellano I, Carmona-Alvarez F, Martinez AI, Rodriguez-Diaz J, Cervera J. (2010). Pyrroline-5-carboxylate synthase and proline biosynthesis: from osmotolerance to rare metabolic disease. Protein Sci 19: 372–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pielou EC. (1975) Ecological Diversity. John Wiley and Sons, Inc: New York, NY, USA. [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR. (2005). NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res 33: D501–D504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar S, Williams GP, Shanthy S, Gracelin NA, Babu S, Parimala PS. (2007). Effect of heavy metals (Hg and Zn) on the growth and phosphate solubilising activity in halophilic phosphobacteria isolated from Manakudi mangrove. J Environ Biol 28: 109–114. [PubMed] [Google Scholar]
- Reyes C, Lloyd JR, Saltikov CW. (2008) Geomicrobiology of iron and arsenic in anoxic sediments. In: Arsenic Contamination of Groundwater: Mechanism, Analysis and Remediation. John Wiley and Sons, Inc.: New York, NY, USA, p 387. [Google Scholar]
- Saager PM, Schuf J, De Baar HJW. (1992). Trace-metal distributions in seawater and anoxic brines in the eastern Mediterranean Sea. Geochim Cosmochim Acta 57: 1419–1432. [Google Scholar]
- Sae-Ueng U, Liu T, Catalano CE, Huffman JB, Homa FL, Evilevitch A. (2014). Major capsid reinforcement by a minor protein in herpesviruses and phage. Nucleic Acids Res 42: 9096–9107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sass AM, Sass H, Coolen MJ, Cypionka H, Overmann J. (2001). Microbial communities in the chemocline of a hypersaline deep-sea basin (Urania basin, Mediterranean Sea). Appl Environ Microbiol 67: 5392–5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuf J, De Baar HJW, Millero FJ. (1995). Vertical distributions and speciation of dissolved rare earth metals in the anoxic brines of Bannock Basin, eastern Mediterranean Sea. Geochim Cosmochim Acta 59: 3285–3299. [Google Scholar]
- Sirajuddin S, Rice LM, Vale RD. (2014). Regulation of microtubule motors by tubulin isotypes and post-translational modifications. Nat Cell Biol 16: 335–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjöblom B, Salmazo A, Djinovic-Carugo K. (2008). Alpha-actinin structure and regulation. Cell Mol Life Sci 65: 2688–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowmya M, Rejula MP, Rejith PG, Mohan M, Karuppiah M, Hatha AA. (2014). Heavy metal tolerant halophilic bacteria from Vembanad Lake as possible source for bioremediation of lead and cadmium. J Environ Biol 35: 655–660. [PubMed] [Google Scholar]
- Srivastava S, Goyal P. (2010) Novel Biomaterials: Decontamination of Toxic Metals from Wastewater. Springer-Verlag: New York, NY, USA, p 139. [Google Scholar]
- Stan-Lotter H, Fendrihan S. (2013). Survival strategies of halophilic oligotrophic and desiccation resistant prokaryotes. In: Seckbach J, Oren A, Stan-Lotter H (eds). Polyextremophiles: Life Under Multiple Forms of Stress. Springer: New York, pp 235–248. [Google Scholar]
- Stock A, Breiner H-W, Pachiadaki M, Edgcomb V, Filker S, Cono V et al. (2012). Microbial eukaryote life in the new hypersaline deep-sea basin Thetis. Extremophiles 16: 21–34. [DOI] [PubMed] [Google Scholar]
- Süß J, Engelen B, Cypionka H, Sass H. (2004). Quantitative analysis of bacterial communities from Mediterranean sapropels based on cultivation-dependent methods. FEMS Microbiol Ecol 51: 109–121. [DOI] [PubMed] [Google Scholar]
- Tamas MJ, Sharma S, Ibstedt S, Jacobson T, Christen P. (2014). Heavy metals and metalloids as a cause for protein misfolding and aggregation. Biomolecules 4: 252–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Sloot H, Hoede D, Hamburg G, Woittiez JRW, van der Weijden CH. (1990). Trace elements in suspended matter from the anoxic hypersaline Tyro and Bannock Basins (eastern Mediterranean). Marine Chemistry 31: 187–203. [Google Scholar]
- Van der Wielen PW, Bolhuis H, Borin S, Daffonchio D, Corselli C, Giuliano L et al. (2005). The Enigma of Prokaryotic Life in Deep Hypersaline Anoxic Basins. Science 307: 121–123. [DOI] [PubMed] [Google Scholar]
- Venkatsan R, Sah-Teli SK, Awoniyi LO, Jiang G, Prus P, Kastanoitis AJ et al. (2014). Insights into mitochondrial fatty acid synthesis from the structure of heterotetrameric 3-ketoacyl-ACP reductase/3R-hydroxyacyl-ACP reductase/3R-hydroxyacyl-CoA dehydrogenase. Nat Commun 5: 4805. [DOI] [PubMed] [Google Scholar]
- Wang LK, Feng Z, Wang X, Zhang X. (2010). DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26: 136–138. [DOI] [PubMed] [Google Scholar]
- Warren JK. (2000). Evaporites, brines and base metals: low temperature ore emplacement controlled by evaporite diagenesis. Australian J Earth Sci 47: 179–208. [Google Scholar]
- Yakimov MM, La Cono V, Denaro R, D'Auria G, Decembrini F, Timmis KN et al. (2007). Primary producing prokaryotic communities of brine, interface and seawater above the halocline of deep anoxic lake L'Atalante, Eastern Mediterranean Sea. ISME J 1: 743–755. [DOI] [PubMed] [Google Scholar]
- Yakimov MM, La Cono V, Ferrer M, Golyshin PN, Giuliano L. (2014). Metagenomics of deep hypersaline anoxic basins. Encyclopedia of Metagenomics doi:doi:10.1007/978-1-4614-6418-1_36-1.
- Yakimov MM, La Cono V, Slepak VZ, La Spada G, Arcadi E, Messina E et al. (2013). Microbial life in the Lake Medee, the largest deep-sea salt-saturated formation. Sci Rep 3: 3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakimov MM, La Cono V, Spada GL, Bortoluzzi G, Messina E, Smedile F et al. (2015). Microbial community of the deep-sea brine Lake Kryos seawater-brine interface is active below the chaotropicity limit of life as revealed by recovery of mRNA. Env Microbiol 17: 364–382. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.