Abstract
A lack of empirical evidence for the microbial regulation of ecosystem processes, including carbon (C) degradation, hinders our ability to develop a framework to directly incorporate the genetic composition of microbial communities in the enzyme-driven Earth system models. Herein we evaluated the linkage between microbial functional genes and extracellular enzyme activity in soil samples collected across three geographical regions of Australia. We found a strong relationship between different functional genes and their corresponding enzyme activities. This relationship was maintained after considering microbial community structure, total C and soil pH using structural equation modelling. Results showed that the variations in the activity of enzymes involved in C degradation were predicted by the functional gene abundance of the soil microbial community (R2>0.90 in all cases). Our findings provide a strong framework for improved predictions on soil C dynamics that could be achieved by adopting a gene-centric approach incorporating the abundance of functional genes into process models.
Introduction
Soil carbon (C) has a vital role in regulating climate, nutrient cycling and biodiversity and therefore in providing the ecosystem services that are essential to human well-being (Schmidt et al., 2011; Victoria et al., 2012). Managing soils to obtain multiple economic, societal and environmental benefits requires integrated policies and incentives that maintain and enhance soil C (Singh et al., 2010; Victoria et al., 2012; Trivedi et al., 2013). Soil microorganisms contribute greatly to ecosystem C budgets through their roles as decomposers, plant symbionts or pathogens, thereby modifying nutrient availability and influencing C turnover and retention in soil (Bardgett et al., 2008; Singh et al., 2010; Bardgett and van der Putten, 2014). The incredible numbers and enormous diversity of soil microbes creates huge challenge to establish the links between diversity and functions related to soil organic matter decomposition and stabilization (Hubbell, 2005). Despite the valid assumption that soil microbes influence the way in which ecosystems function, there is a very limited evidence on whether there is a direct link between microbial community structure and function in the global biogeochemical cycles of terrestrial ecosystems (Rocca et al., 2014; Kubartová et al., 2015; van der Wal et al., 2015). These gaps have kept soil microbes outside of the ongoing debates about global biodiversity loss, conservation and sustainable management policies (Bardgett and van der Putten, 2014) and have precluded the inclusion of microbial communities in global biogeochemical models such as Earth system models (ESMs) that inform citizens and policy makers of C dynamics and exchange between the biosphere and the atmosphere (Wieder et al., 2013, 2015). Understanding the extent to which soil microbial communities control ecosystem processes is thus critical to establish effective policies to preserve microbial diversity hotspots and the key ecosystem functions and services that soil microbes provide (Singh et al., 2010; Bardgett and van der Putten, 2014).
Extensive bodies of work have provided detailed insights into the mechanism(s) on the transformation of soil C pools by extracellular enzymes excreted into the soil by microbes, thus allowing researchers to develop enzyme-driven ESMs that provide a better fit to observations, especially in changing environments (Allison et al., 2010; Treseder et al., 2012; Wieder et al., 2013; Hararuk et al., 2015). Process-level analyses such as respiration and enzymatic transformation of added substrate are used as a proxy of microbial function, which gives valuable insight into overall microbial-mediated transformations in soils. However, such measurements do not provide concrete links on mechanisms, microbial functional composition and diversity that underpins process-level differences (Reeve et al., 2010; Talbot et al., 2014; You et al., 2014). Few studies conducted at local scale have observed relationships between community composition and biochemical function of microbes in soils (Fierer et al., 2012; Talbot et al., 2014; Su et al., 2015), suggesting that the composition of microbial communities, per se, may be important. However, empirical evidence of microbial regulation of ecosystem processes is lacking that limits the coupling between microbial community functional traits, the environment context and the ecosystem processes. Because of the lack of this evidence, it is generally assumed that the link between community composition and the functional and metabolic responses (in our case enzyme production) is indirect (Comte et al., 2013). This is a major constraint in our ability to develop a framework to directly incorporate microbial data into ESMs and conservation and management policy decisions.
The aim of this study was to quantify the relative contribution to, and the level of regulation of, the production of extracellular enzymes involved in C turnover by the soil microbial community. We hypothesized that the soil microbial community controls enzyme activities linked to C degradation through a linkage between structure and function and that the role of abiotic factors affecting the microbial mediated processes is via regulation of the structural attributes (composition) of the soil microbial community. Accordingly, we expected to find that: (1) microbial community composition will be delineated strongly by the sampling regions; (2) enzyme activities across regions will be likely to be driven by the selection of common active members of microbial community; and (3) the relationship between traits (production of extracellular enzymes) will be strongly correlated with functional gene structure as compared with taxonomy. To test our hypothesis, we collected soil samples (top 10 cm) across three geographical regions of Australia and multiple managed ecosystems (Supplementary Table S1) varying in a wide range of soil properties, climate and environmental parameters. We characterized bacterial and fungal communities with pyrosequencing. GeoChip 4.0 analysis provided information on the functional structure of microbial communities, including abundance of known protein-coding genes related to the production of four key enzymes involved in C degradation (Supplementary Table S2). These enzymes were selected based on their general occurrence in different soil types, key role in soil C degradation (Nannipieri et al., 2012; Burns et al., 2013; Trivedi et al., 2015) and the availability of well-established methods for determining their activity (Bell et al., 2013). Structural equation modelling was used to explicitly evaluate the relative importance of abiotic factors (soil C and pH), microbial community structure and functional genes on soil function (measured as the activity of soil enzymes related to C degradation; see Material and Methods section).
Materials and methods
Soil collection and analysis
We collected 51 soil samples in March 2013 from three key grain-producing regions in Australia (Table 1; Supplementary Table S1). The sampled regions comprised the states of New South Wales (Narrabri; 30.31°S 149.76°E; n=12), Western Australia (Cunderdin; 31.38°S; 117.14°E; n=21) and South Australia, (Karoonda; 35.08°S 139.88°E; n=18) and were 5 × 5 km2 in size. Within each sampling area, we randomly selected individual fields that were at least 500 m away from each other. In all the selected fields, wheat was grown in the previous growing season. Four soil cores (distance between two cores was at least 5 m) from each field were collected (0–10 cm depth) using a 3-cm diameter auger, thoroughly homogenized, composited in a Ziploc bag and shipped in a cooler on ice to the laboratory. Soil pH was assessed using a fresh soil-to-water ratio of 2.5 using a Delta pH-meter (Mettler-Toledo Instruments Co., Columbus, OH, USA). Total C and total N was measured on a LECO macro-CN analyser (LECO, St Joseph, MI, USA).
Table 1. Soil type, soil properties and enzymatic activities of soil samples collected from three major grain-producing regions of Australia.
| Site | Number of samples | Soil type | pH (range) | Total C (%, range) | Total N (%, range) |
Enzymatic activities (nmol h−1
g−1
soil, range) |
|||
|---|---|---|---|---|---|---|---|---|---|
| α-Glucosidase | β-D- Xylosidase | N-acetyl-β-D-glucosaminidase | β-D- cellulosidase | ||||||
| Narrabri (New South Wales) | 12 | Vertisols | 7.8–8.4 | 1.1–1.6 | 0.07–0.14 | 1.3–2.9 | 3.7–7.7 | 5.3–15.0 | 26.4–41.9 |
| Karoonda (South Australia) | 18 | Kandosol and Calcarosol | 5.9–7.0 | 0.43–0.81 | 0.03–0.06 | 1.2–2.9 | 22.3–33.7 | 23.9–42.4 | 2.2–6.3 |
| Cunderdin (Western Australia) | 21 | Xanthic Ferralsol | 6.7–8.0 | 0.74–1.8 | 0.04–0.12 | 1.2–3.7 | 13.8–20.1 | 13.2–24.0 | 0.4–1.9 |
Soil enzymatic activities
β-D-cellulosidase (CB), β-Xylosidase (XYL), α-Glucosidase (AG) and N-acetyl-β-Glucosaminidase (NAG) activities were measured using 4-methylumbelliferyl (MUB) substrate yielding the highly fluorescent cleavage products MUB upon hydrolysis (Supplementary Table S2). All the enzyme assays were set up in 96-well microplates as described by Bell et al. (2013). Twelve replicate wells were set up for each sample and each standard concentration. The assay plate was incubated in the dark at 25 °C for 3 h to mimic the average soil temperature. Enzyme activities were corrected using a quench control. Fluorescence was measured using a microplate fluorometer (EnSpire 2300 Multilabel Reader, Perkin Elmer, Waltham, MA, USA) with 365-nm excitation and 460-nm emission filters. The activities were expressed as nmol h−1 g−1dry soil.
Molecular analysis
Soil DNA extraction
Freeze-dried soil (0.3 g) was used for DNA extraction with the FastDNA SPIN Kit for soil (MP Biomedicals, Heidelberg, Germany) according to the manufacturer's protocol. Extracted DNA was stored in a −80 °C freezer prior to molecular analysis. DNA concentrations were determined using the Qubit quantification platform with Quant-iT dsDNA BR Assay Kit (Invitrogen, Carlsbad, CA, USA).
Barcoded pyrosequencing and analysis
Fusion primers 341F-806R (Mori et al., 2013) and LR3-LR0R (Liu et al., 2012) were used to amplify multiplexed bar-coded 16S rRNA and large subunit rRNA gene sequences to profile bacterial and fungal communities, respectively. PCR products were purified, pooled and sequenced on a 454 GS FLX Titanium sequencer (Roche 454 Life Sciences, Branford, CT, USA). Detailed methodology, downstream processing and bioinformatics analysis were described previously by Singh et al. (2014) and Barnard et al. (2013). Operational taxonomic unit tables used to determine the microbial abundances were rarefied to 733 and 784 sequences for bacteria and fungi, respectively, to ensure even sampling depth. The number of sequences used for the analysis is comparable to other studies, which used a similar sequencing approach (Fierer et al., 2013).
GeoChip 4.0 analysis
For GeoChip analysis, 200 ng of total DNA was dried in a SpeedVac (ThermoSavant, Milford, MA, USA) at 45 °C for 45 min and shipped to The University of Oklahoma, Norman, OK, USA. GeoChip 4.0 analysis was performed as described previously (He et al., 2010; Tu et al., 2014). Briefly, DNA samples were labelled with the fluorescent dye Cy-5 by a random priming method (Tu et al., 2014; Su et al., 2015), followed by purification with a QIA Quick Purification Kit (Qiagen, Valencia, CA, USA). Dye incorporation was measured by a Nano-Drop ND-1000 spectrophotometer (NanoDrop Technologies Inc., Wilmington, DE, USA), and DNA was dried using a SpeedVac (ThermoSavant) at 45 °C for 45 min. Thereafter, DNA was hybridized with GeoChip 4.0 at 42 °C for 16 h in a MAUI hybridization station (BioMicro, Salt-Lake City, UT, USA) and scanned by a NimbleGen MS200 scanner (Roche, Madison, WI, USA) at 633 nm, using 100% and 75% laser power and photomultiplier tube gain, respectively. Data processing was performed as previously described (He et al., 2010) using Microarray Data Manager (http:/ieg.ou.edu/microarray/).
Quantitative PCR (qPCR)
qPCR was performed to determine gene copy numbers for bacteria and fungi, including α-Proteobacteria, β-Proteobacteria, Fimiricutes, Actinobacteria, Acidobacteria and Basidiomycota, using the primers and conditions described previously by Trivedi et al. (2012). Spearman correlation analyses were performed by using the XLSTAT software (Addinsoft SARL, Paris, France) to test the relationship between relative abundance determined by qPCR and the relative abundance determined by pyrosequencing.
Numerical and statistical analyses
We first explored whether enough spatial variability was obtained in our soil samples in terms of microbial community structure and functionality (production of enzymes involved in C degradation) to test our hypothesis. We conducted separate principal coordinates analyses in PRIMER (Clarke and Gorley, 2006) using as input the pairwise distances between microbial community composition (pyrosequencing data for bacteria and fungi), enzymatic activities and the abundance of functional genes (GeoChip data). Heat maps used to depict the variability in the abundance of major soil bacterial and fungal groups were constructed using R (http://www.r-project.org/).
We then elucidated the relationship between the microbial community and functions by linear regression analysis using the activity of enzymes involved in C degradation (measured biochemical assays) and the abundance of genes responsible for the production of each enzyme (determined by GeoChip). Because of the significant number of microbial predictors (that is, relative abundance of bacteria and fungi and functional genes), we used a classification Random Forest (RF) analysis (Breiman, 2001) as explained in Delgado-Baquerizo et al. (2015) to identify the main microbial predictors of extracellular enzyme activities. The main goal with this analysis is to reduce the number of predictors for further modelling (structural equation modelling). In our RFs, the different soil properties, bacterial and fungal relative abundances and GeoChip data were included as predictors of enzyme activities. These analyses were conducted using the RF package (Liaw and Wiener, 2002) for the R statistical software, version 3.0.2 (http://cran.r-project.org/). The significances of the model and the cross-validated R2 were assessed with 5000 permutations of the response variable using the A3 (Fortmann-Roe, 2013) R package. Similarly, the significance of the importance measures of each predictor (here soil parameters, microbial community structure, functional genes) on the response variable (enzymatic activities) was assessed by using the rfPermute (http://cran.rproject.org/web/packages/rfPermute/rfPermute.pdf) package for R.
After this, and owing to the correlative nature of our data, structural equation modelling (SEM) was used to identify the relative importance and effects of functional genes vs abiotic factors (total C and pH) and microbial composition on soil function (that is, enzyme activities). Unlike regression or analysis of variance, SEM offers the ability to separate multiple pathways of influence and view them as a system (Shipley, 2002; Grace, 2006; Delgado-Baquerizo et al., 2015). Another important capability of SEM is its ability to partition direct and indirect effects that one variable may have on another and estimate the strengths of these multiple effects (Shipley, 2002; Grace, 2006; Delgado-Baquerizo et al., 2015). SEM was generated based on the known effects and relationships among key drivers of microbial community composition and function (Supplementary Figure S1). In this study, we were interested in linking microbial composition (that is, relative abundance of main microbial taxa) to functional genes and soil functioning. This allows us to identify the main microbial groups driving functional genes and soil functioning, which would not have been possible by including a global diversity metric such as evenness or Shannon diversity. In these analyses, we chose soil C and pH as the explanatory variables because both have been demonstrated previously as primary factors determining not only the structure of soil microbial community (Rousk et al., 2010; You et al., 2014) but also the production of soil enzymes involved in C degradation (Talbot et al., 2014; You et al., 2014). In addition, we included the relative abundance of particular microbial groups that were previously identified to be important predictors of enzyme activities in our soil samples by RF analysis. Microbial community and soil properties data were normalized prior to analyses (that is, log-transformed) when needed. When these data manipulations were complete, we parameterized our model using our data set and tested its overall goodness of fit. The overall goodness of fit in our models was tested, as explained in Schermelleh-Engel et al. (2003). There is no single universally accepted test of overall goodness of fit for structural equation models applicable in all situations regardless of sample size or data distribution. Most modellers circumvent this problem by using multiple goodness of fit criteria. We used the Chi-square test (χ2; the model has a good fit when 0⩽χ2⩽2 and 0.05<P⩽1.00 and acceptable fit when 2⩽χ2⩽3 and 0.01<P⩽0.05) and the root mean square error of approximation (RMSEA; the model has a good fit when RMSEA 0⩽RMSEA⩽0.05 and 0.10<P⩽1.00 and acceptable fit when RMSEA 0.05⩽RMSEA⩽0.08 and 0.05<P⩽1.00). Additionally, and because some variables were not normal, we confirmed the fit of the model using the Bollen–Stine bootstrap test (the model has a good fit when 0.10<bootstrap P⩽1.00 and acceptable fit when 0.05<bootstrap P⩽0.10). Because our SEM was saturated (the number of degrees of freedom was zero), no probability level could be assigned to the chi-square statistic, making the model untestable. To solve this problem, the free covariance weight between pH and enzyme activity was fixed, and the best solution was chosen through maximization of the maximum likelihood function releasing a degree of freedom (see Delgado-Baquerizo et al., 2013 and García-Palacios et al., 2013 for examples). After attaining a satisfactory model fit, we introduced composite variables into our model. The use of composite variables does not alter the underlying SEM model but collapses the effects of multiple conceptually related variables into a single composite effect, aiding interpretation of model results (Grace, 2006). Microbial community composition (that is, relative abundance of main microbial phyla/classes) was included in our model as a composite variable. Finally, we calculated the standardized total effects of total C, pH, microbial community composition and functional gene on the enzyme activities. The net influence that one variable has upon another is calculated by summing all direct and indirect pathways between the two variables. If the model fits the data well, the total effect should approximately be the bivariate correlation coefficient for that pair of variables (Grace, 2006).
Results
Soil physicochemical properties
Soil samples differed significantly in a range of soil properties (Table 1; Supplementary Table S1). Soil pH ranged from 5.95 to 8.34, total C from 0.43% to 1.76% and total N from 0.031% to 0.14%. pH ranged from 7.85 to 8.34, from 5.95 to 7.02 and from 6.83 to 8.01 in samples collected from Narrabri, Karoonda and Cunderdin regions, respectively. Similarly, total C ranged from 1.1% to 1.4%, from 0.43% to 0.81% and from 1.0% to 1.76% in samples collected from Narrabri, Karoonda and Cunderdin regions, respectively. We also observed variability in the activity of enzymes involved in C degradation, which ranged from 5.3 to 42.2 (NAG), from 0.4 to 41.9 (CB), from 1.1 to 3.7 (AG) and from 3.7 to 33.7 (XYL) nmol h−1 g−1 soil (Table 1; Supplementary Table S1). Principal coordinate analysis (PCoA) of soil enzymatic data indicated strong regional differences (Supplementary Figure S2a).
Structure and function of soil microbial communities
In accordance with our initial assumption, community structure (β-diversity) for bacteria and fungi was significantly different between regions (Supplementary Figures S2c and d). PCoA analysis revealed clear separation between samples from different regions for fungal (Supplementary Figure S2c) and bacterial (Supplementary Figure S2d) communities. The heat maps showed significant differences in the relative abundance of major bacterial and fungal groups between samples from different regions (Supplementary Figures S3 and S4). The differences in community composition were primarily driven by the relative abundance of Proteobacteria (alpha, beta, delta and gamma), Acidobacteria and Actinobacteria (Supplementary Figure S4). Differences in the fungal community were linked to variation in dominant families, including Dothideomycetes, Eurotiomycetes, Sordariomycetes and Agaricomycetes (Supplementary Figure S5). Taxon-specific qPCR analysis showed similar trends as the pyrosequencing data, and we found a strong and significant correlation (P<0.0001) between the relative abundance data from pyrosequencing and taxon-specific qPCR (Supplementary Table S3). Similar to the microbial community structure observations, PCoA analysis of all detected genes (GeoChip analysis) showed that the sampling regions were well separated on first two axis, suggesting that the soil microbial functional gene structure is significantly different between different regions (Supplementary Figure S2b).
We observed variability in the abundance (measured as normalized signal intensity from GeoChip) of genes encoding the enzymes studied that ranged from 5.2 to 19.9, from 0.84 to 10.01, from 17.1 to 31.2 and from 4.02 to 15.4 for Acetylglucosaminidase (encoding NAG); Exoglucanase (encoding CB), α-amylase (encoding AG) and Xylanase (encoding XYL), respectively (Supplementary Table S1).
Relationships between microbial community structure and function
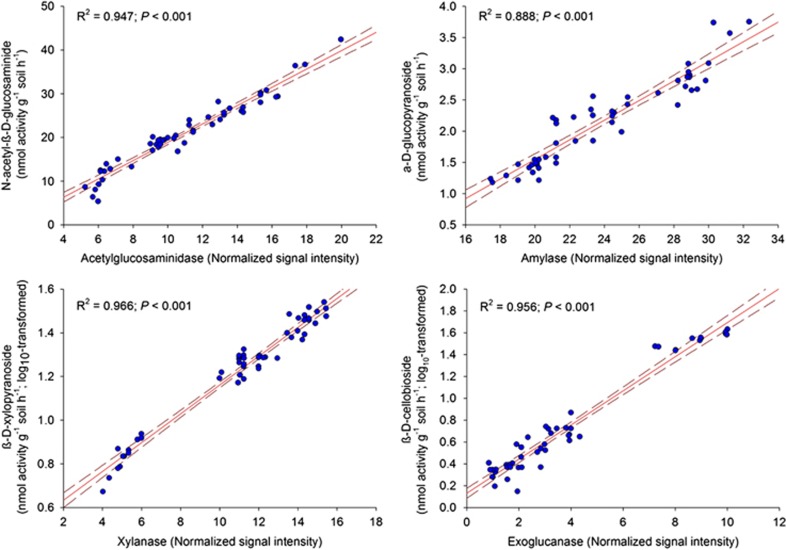
We observed a strong correlation between the relative abundance of microbial functional genes and the activity of NAG (R2=0.947), AG (R2=0.888), XYL (R2=0.966) and CB (R2=0.956), which were highly significant in all cases (P<0.001; Figure 1). Similarly, we observed a strong correlation (R2=0.896 and 0.949 for bacteria and fungi, respectively; P<0.005 in both cases) between taxonomic and functional diversity in our studied sites (Supplementary Figure S5).
Figure 1.
Relationships between GeoChip data and their corresponding enzyme activities (n=51). Solid lines represent the fitted linear regressions and dashed lines represent 95% confidence intervals.
Identifying drivers of activity of soil enzymes linked to soil C degradation
RF analysis
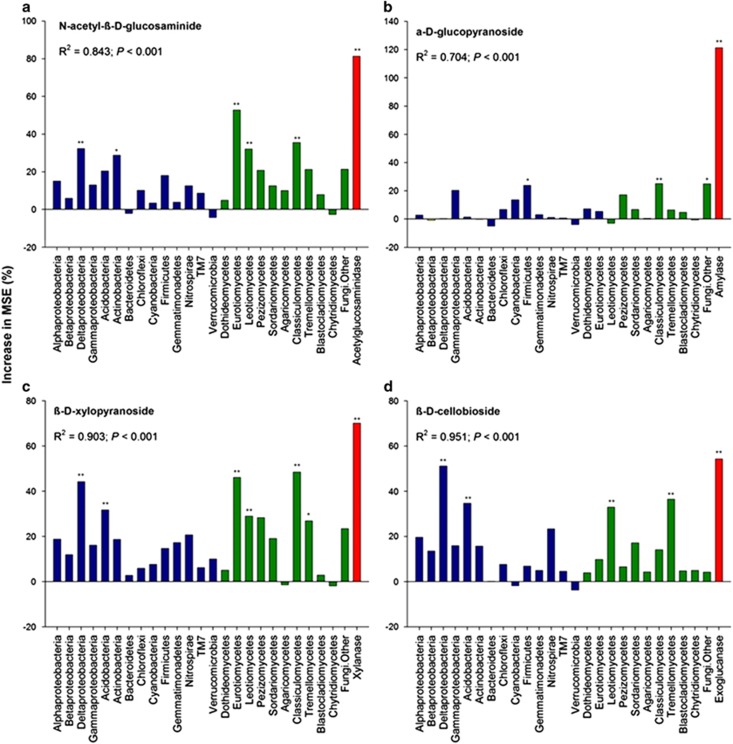
The abundance of functional genes was the single most important variable for the activity of all four studied enzymes (P<0.01; Figure 2). Among bacteria, δ-Proteobacteria was an important variable for predicting NAG (P<0.05), XYL (P<0.01) and CB (P<0.01) (Figure 2). Actinobacteria (for NAG; P<0.05), Firmicutes (for AG, P<0.05) and Acidobacteria (for XYL and CB; both P<0.01) were other important phyla predicting the activities of different enzymes. Among fungal families, Eurotiomycetes (for NAG and XYL; both P<0.01), Leotiomycetes (for NAG (P<0.01), CB (P<0.01), and XYL (P<0.05)), Classiculomycetes (for NAG and XYL (both P<0.01) and AG (P<0.05)) and Tremellomucetes (for XYL (P<0.05) and CB (P<0.01)) were also important variables for predicting activities of different enzymes.
Figure 2.
RF mean predictor importance (percentage of increase of mean square error) of bacterial and fungal relative abundances and GeoChip data as drivers of the different enzyme activities (a: NAG; b: AG; c: XYL; and d: CB). This accuracy importance measure was computed for each tree and averaged over the forest (5000 trees). Significance levels are as follows: *P<0.05 and **P<0.01.
Structural equation modelling
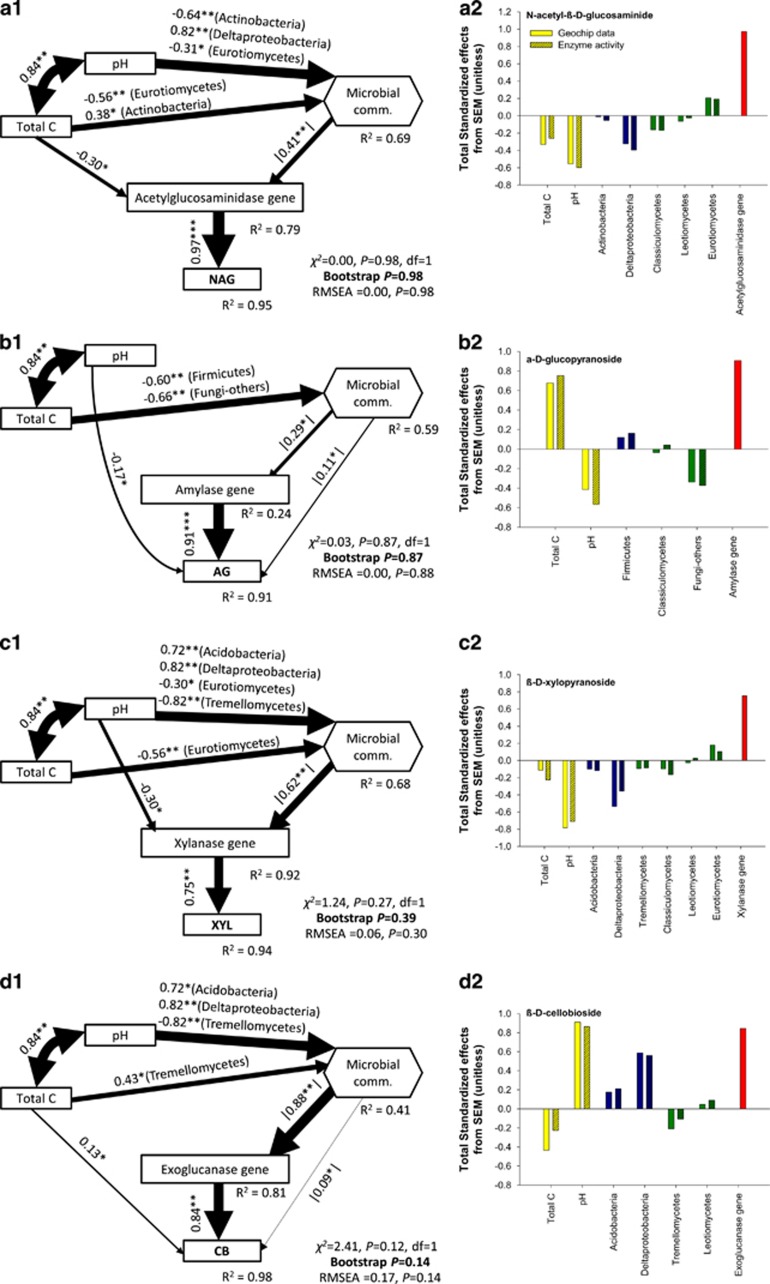
SEM explained 91.0–97.0% of the variation in enzyme activities and provided a good fit using χ2 test, RMSEA and Bollen–Stine bootstrap metrics (Schermelleh-Engel et al., 2003; Grace, 2006) (Figures 3a–d). Most importantly, our SEM analysis provided evidence that the direct effect of functional genes on enzyme activities was maintained even when considering key abiotic and biotic factors, such as total C, pH and microbial community composition (Figures 3a–d). Interestingly, our SEM analysis further suggested that the effects of soil properties on enzyme activities were indirectly driven via microbial community composition and functional gene abundance (P<0.01; Figures 3a–d). Further, the abundance of the genes involved in driving the enzymatic activity of the four studied enzymes was directly linked with microbial community composition (P<0.05 for AG; P<0.01 for NAG, XYL and CB, respectively). However, the structure of the soil microbial community had no direct effect on the enzymatic activity of NAG and XYL and a very low direct effect on AG and CB. This interesting result further indicated that the structure of the soil microbial community indirectly regulated the activity of extracellular enzymes via functional genes.
Figure 3.
Structural equation models based on the effects of soil properties (total C and pH), bacterial and fungal relative abundances and Geochip data on enzyme activities. Numbers adjacent to arrows are standardized path coefficients, analogous to partial regression weights and indicative of the effect size of the relationship (panels a1–d1). The sign of the the microbial community composition (microbial comm.) composite is not interpretable; thus absolute values are presented. Arrow width is proportional to the strength of path coefficients. As in other linear models, R2 indicates the proportion of variance explained and appears above every response variable in the model. Model fitness details (χ2 vs RMSEA and non-parametric Bootstrap parameters are close by each figure) are close to each figure. Significance levels are as follows: *P<0.05 and **P<0.01. Panels (a2–d2) represent standardized total effects (direct plus indirect effects) derived from the structural equation model used.
Discussion
Overall the motivation of this study was not to provide a comprehensive census of taxonomic or functional diversity but to determine the regulation of functions by the soil microbial community. Taxonomic and functional profiling in the present study was used as a means to evaluate the differences in the structure and functional potential of the soil microbial community. Altogether, our sampling regions varied considerably in their soil chemical and physical characteristics (Table 1; Supplementary Table S1; Supplementary Figure S2a), climatic conditions, microbial community structure and composition (Supplementary Figures S2c and d, S3 and S4) and gene functionality (Supplementary Figure S2b) and thus provide an excellent framework to test our hypothesis (see below).
Strong relationship between functional community composition and enzymatic activities
Our study provides novel evidence of a strong relationship between the structure of the soil microbial community and the abundance of genes encoding four different enzymes involved in C degradation. In particular, our results indicated a strong statistical correlation between the GeoChip data that measures the abundance of genes related to the production of four key soil enzymes involved in C mineralization processes and biochemical data that measures the activity of the corresponding enzymes. Several studies have suggested that coarse measures of microbial communities based on DNA (whether taxonomic or functional) may be insufficient to understand the changes in the functional contributions of these communities (Rocca et al., 2014; Wood et al., 2015). As an example, Wood et al. (2015) found no relationship between C mineralization and gene abundance in farms in Africa under a tropical agricultural system. The authors suggested that the process rate should be controlled by the expression of related genes, rather than the overall abundance. We argue that an ecosystem process that relies on a cascade of other reactions involving a variety of enzymes will not represent an accurate measure to relate gene abundance with function. However, the measurement of activity of a particular enzyme, rather than the process that it catalyses, will be a realistic scenario to relate gene abundance with function. In support of our argument, Reeve et al. (2010) have stated that correlations between traditional techniques and soil DNA might be stronger than with soil mRNA because DNA may better represent the potential functional capability of the microbial biomass rather than its current and presumably transient state represented by mRNA. Our results emphasize that to make valid assumptions on the biodiversity–functional relationships in future, parameters selected to measure ecosystem multifunctionality (multiple functions and services as in Byrnes et al., 2014) need to carefully consider their component parts, what drives these processes, how they relate to one another and also how the individual functions that they comprise should be weighted and measured.
The extremely strong correlation between all enzyme activities with functional genes further suggests that soil microbes are a good proxy for soil functionality. This can provide accurate information that can then be used for ecosystem and global change modelling and conservation and management policies (Wieder et al., 2013, 2015). In addition, this strong correlation also indicates that functional genes can be used to develop a gene-centric approach to integrate environmental genomics into simulation models in order to improve their predictive power and accuracy of ESMs (Reed et al., 2014).
Regulatory pathways of the activity of enzymes involved in C degradation
Identifying the structural–functional relationships for microbial organisms is particularly critical to determine the importance of the soil microbial community in regulating ecosystem processes, and thus there is keen interest in developing theoretical and experimental approaches to disentangle the microbial regulation of soil functions from other biotic and abiotic drivers (for example, Strickland et al., 2009; Wallenstein and Hall, 2012; Talbot et al., 2014; You et al., 2014; Wood et al., 2015). Albeit we found that functional genes were strongly related to enzyme activities, these results are correlative in nature and hence potentially non-causative. Therefore, we used SEM to identify the relative importance of functional genes vs other important abiotic (total C and pH) and biotic (microbial composition) drivers on enzyme activities. Interestingly, our results indicated that the direct effects of functional genes on soil functions were maintained after considering multiple biotic and abiotic drivers simultaneously. In fact, most effects of soil properties and microbial composition on soil function were indirectly driven via functional genes. In this respect, we found that soil chemical variables had a direct impact on the structure (measured in terms of the relative abundance of major Phyla (and also different classes within Proteobacteria) and families for bacteria and fungi, respectively) of the soil microbial community.
Our SEM analysis further showed that soil C and pH have either no (for Amylase and Exoglucanase) or very weak (for Acetylglucosaminidase and Xylanase) direct impact on the abundance of functional genes for enzyme production. Similarly, soil C and pH either do not (for XYL and NAG) or had a weak (CB and AG) direct influence on the enzyme activity (Figures 3a–d) and that most effects from soil properties on functional genes and enzyme activities were indirectly mediated by the composition of microbial community. These results are supported by You et al. (2014) who showed that soil nutrient status imposes a significantly higher effect on the composition of the soil microbial community in comparison to soil enzymatic activities. Overall, SEM analysis explained most of the variation in the activity of enzymes involved in C degradation that is mainly predicted by the functional structure of the soil microbial community. These results support our initial hypothesis and provide direct evidence for the microbial regulation of soil processes linked to C degradation in terrestrial ecosystems. Our results suggest environmental filters are primary responsible for the assembly of soil microbial community and determine the strong differences in the community composition between different geographic regions (Fierer et al., 2013; Talbot et al., 2014). However, the functional potential of the microbial community will be driven by the selection of active taxa within different lineages. Functional attributes are embedded in the genomic blueprint and the capacity to produce extracellular enzymes varies at relatively fine-scale phylogenetic resolution (Philippot et al., 2010; Trivedi et al., 2013; Zimmerman et al., 2013). Because the composition of microbial community does not correlate strongly with the enzyme activity, functional genes and processes may offer a better predictive framework for investigating the ecological consequences of microbial traits conserved at higher phylogenetic resolutions than inferring function based on phylogenetic marker genes. Overall, our results suggest that the activity and composition of soil microbial communities (particularly functional gene abundance) can serve as a predictor for overall C dynamics (retention, release, storage). This is important as this illustrates the need of incorporating the microbial community contributions to ESMs.
Different microbial groups explained the abundance of genes encoding different enzymes (Figures 2 and 3a2–d2). There is a difference in the number of genes involved in the degradation of various C sources among different microbial groups and in many instances the genes involved in the degradation of moderately labile and recalcitrant forms of C are phylogenetically conserved (Trivedi et al., 2013; Zhao et al., 2013). The production of NAG and XYL was significantly linked to δ-Proteobacteria, Actinobacteria and Acidobacteria in bacteria and Eurotiomycetes (subphylum Pezizomycotina) and Leotiomycetes (very closely related to subphylum Pezizomycotina) in fungi (Figures 3a2 and c2). Genomic analysis of these groups has shown that they have a higher potential to produce these enzymes as compared with other groups within bacteria and fungi (Karlsson and Stenlid, 2008; Trivedi et al., 2013; Zhao et al., 2013). SEM showed that bacterial phyla Acidobacteria, Deltaproteobacteria and Actinobacteria are important predictors of activity of enzymes CB, XYL and NAG. These groups are classified as oligotrophs and thrive on moderately labile and recalcitrant forms on C (Fierer et al., 2007; Trivedi et al., 2013). We observed a small but significant (P<0.05) direct control of microbial structure on AG and CB activity. This is not surprising as AG and CB are involved in degradation of labile forms of C and the gene(s) encoding the production of AG and CB are present in a number of soil microbial groups (Trivedi et al., 2013; Zhao et al., 2013). The activity of all four studied enzymes was directly and significantly regulated by the abundance of genes encoding the respective enzymes (Figure 3). These results suggest that microbial community structure influences enzymatic activities through the relative abundance of functional genes. Future studies should expand to include a wide array of possible enzymes involved in C dynamics. Nonetheless, our findings suggest that future studies on structure–function relationship should explicitly include functional genes. This is now possible given the rapid advancement in sequencing and probe-based technologies over recent years.
The lack of explicit evidence that soil microbes regulate the enzymatic activities has been the major bottleneck in incorporating their structural and functional composition for predictions. Recently, Wieder et al. (2015) have incorporated microbial functional types that exhibit copiotrophic and oligotrophic growth strategies in a process-based model to predict soil C turnover. Even at very course level of microbial representation, the model predictions were better than conventional biogeochemical models. Previous studies have shown that trophic strategy is strongly reflected in genomic content and genomic signatures can be used as a proxy for determining the ecological characteristics of soil microorganisms (Trivedi et al., 2013). Our study provides evidence for explicit links between microbial communities and enzymatic activities. We also noticed that inclusion of different microbial groups provide better predictions for different enzymatic activities. Based on the results of our study (microbes having a direct control on the production of C degrading enzymes) and others (incorporation of microbial data set improve model predictions; Wieder et al., 2015), we argue that grouping of microbes in simplified functional groups will allow to parameterize and accurately simulate soil biogeochemical functions in ESMs.
Conclusions
Increased interest in microbial responses to soil processes such as C cycling is largely based on the uncertainties surrounding belowground responses to global climate change (Hawkes and Keitt, 2015). Our study provides empirical evidence that variation in microbial community composition leads to differences in functional gene abundance, which in turn has consequences for the activity of enzymes directly linked to C degradation at field to regional scales. Further work should include range of land use types, environmental conditions and soil properties to confirm global applications of these findings. By directly linking enzyme activities and edaphic soil parameters to the genetic composition of microbial communities, our study provides a framework for achieving mechanistic insights into patterns and biogeochemical consequences of soil microbes. This framework can be extended to identify the consequences of changes in microbial diversity on other ecosystem functions and services. Such an approach is critical for informing our understanding of the key role microbes have in modulating Earth's biogeochemistry.
Data deposition
The raw sequence data have been deposited in the NCBI Sequence Read Archive (BioProject accession no. PRJNA308378).
Acknowledgments
We thank Dr IC Rochester, Dr G Vadakattu, Dr K Flower, Dr D Minkey and Dr M McNee for their help in sample collection. The Grains Research and Development Corporation (GRDC), Australia via Grant number UWS000008 and Australian Research Council via Grant number DP13010484 funded this work. Drs C Janitz and J King from Next Generation Sequencing Facility of WSU are acknowledged for Pyrosequencing analysis.
Author contributions
PT, MD-B, ICA and BKS designed the study. PT, MDB, CT, BKS, TCJ, HH and JZ analysed the data. PT wrote the manuscript and all the other authors contributed to revisions.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Allison SD, Wallenstein MD, Bradford MA. (2010). Soil-carbon response to warming dependent on microbial physiology. Nat Geosci 3: 336–340. [Google Scholar]
- Bardgett RD, Freeman C, Ostle NJ. (2008). Microbial contribution to climate change though carbon cycle feedbacks. ISME J 2: 805–814. [DOI] [PubMed] [Google Scholar]
- Bardgett RD, van der Putten WH. (2014). Belowground biodiversity and ecosystem functioning. Nature 515: 505–511. [DOI] [PubMed] [Google Scholar]
- Barnard RL, Osborne CA, Firestone MK. (2013). Responses of soil bacterial and fungal communities to extreme desiccation and rewetting. ISME J 7: 2229–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CW, Fricks BE, Rocca JD, Steinweg JM, McMahon SK, Wallenstein MD. (2013). High-throughput fluorometric measurement of potential soil extracellular enzyme activities. J Vis Exp, e50961. [DOI] [PMC free article] [PubMed]
- Breiman L. (2001). Random forests. Mach Learn 45: 5–32. [Google Scholar]
- Burnham KP, Anderson DR. (2002) Model Selection and Multimodel Inference. A Practical Information-Theoretical Approach. Springer-Verlag: New York, USA. [Google Scholar]
- Burns RG, DeForest JL, Marxsen J, Sinsabaugh RL, Stromberger ME, Wallenstein MD et al. (2013). Soil enzymes in a changing environment: current knowledge and future directions. Soil Biol Biochem 58: 216–234. [Google Scholar]
- Byrnes JEK, Gamfeldt L, Isbell F, Lefcheck JS, Griffin JN, Hector A et al. (2014). Investigating the relationship between biodiversity and ecosystem multifunctionality: challenges and solutions. Methods Ecol Evol 5: 111–124. [Google Scholar]
- Clarke KR, Gorley RN. (2006) PRIMER v6: User Manual/Tutorial, 6 edn. PRIMER-E: Plymouth, UK. [Google Scholar]
- Comte J, Fauteux L, Del Giorgio PA. (2013). Links between metabolic plasticity and functional redundancy in freshwater bacterioplankton communities. Front Microbiol 4: 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado-Baquerizo M, Maestre FT, Gallardo A, Quero JL, Ochoa V et al. (2013). Aridity modulates N availability in arid and semiarid Mediterranean grasslands. PLoS One 8: e59807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado-Baquerizo M, Gallardo A, Covelo F, Prado-Comesaña A, Ochoa V, Maestre FT. (2015). Differences in thallus chemistry are related to species-specific effects of biocrust-forming lichens on soil nutrients and microbial communities. Funct Ecol 8: 1087–1098. [Google Scholar]
- Fierer N, Bradford MA, Jackson RB. (2007). Toward an ecological classification of soil bacteria. Ecology 88: 1354–1364. [DOI] [PubMed] [Google Scholar]
- Fierer N, Leff JW, Adams BJ, Nielsen UN, Bates ST, Lauber CL et al. (2012). Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc Natl Acad Sci USA 109: 21390–21395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Ladau J, Clemente JC, Leff JW, Owens SM, Pollard KS et al. (2013). Reconstructing the microbial diversity and function of pre-agricultural tallgrass prairie soils in the United States. Science 342: 621–624. [DOI] [PubMed] [Google Scholar]
- Fortmann-Roe S. (2013). Accurate, Adaptable and Accessible Error Metrics for Predictive Models. R Package Version 0.9.2.
- Fotheringham AS, Brunsdon C, Charlton M. (2002) Geographically Weighted Regression: The Analysis of Spatially Varying Relationships. Wiley: Hoboken, NJ, USA. [Google Scholar]
- García-Palacios P, Maestre FT, Milla R. (2013). Community-aggregated plant traits interact with soil nutrient heterogeneity to determine ecosystem functioning. Plant Soil 364: 119–129. [Google Scholar]
- Grace JB. (2006) Structural Equation Modelling and Natural Systems. Cambridge University Press: Cambridge, UK. [Google Scholar]
- Hararuk O, Smith MJ, Luo Y. (2015). Microbial models with data-driven parameters predict stronger soil carbon responses to climate change. Global Change Biol 6: 2439–2453. [DOI] [PubMed] [Google Scholar]
- Hawkes CV, Keitt TH. (2015). Resilience vs historical contingency in microbial responses to environmental change. Ecol Lett 18: 612–625. [DOI] [PubMed] [Google Scholar]
- He Z, Deng Y, Van Nostrand JD, Tu Q, Xu M, Hemme CL et al. (2010). GeoChip 3.0 as a high-throughput tool for analyzing microbial community composition, structure and functional activity. ISME J 4: 1167–361. [DOI] [PubMed] [Google Scholar]
- Hubbell SP. (2005). Neutral theory in community ecology and the hypothesis of functional equivalence. Funct Ecol 19: 166–172. [Google Scholar]
- Karlsson M, Stenlid J. (2008). Comparative evolutionary histories of the fungal chitinase gene family reveal non-random size expansions and contractions due to adaptive natural selection. Evol Bioinform Online 4: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubartová A, Ottossen E, Stenlid J. (2015). Linking fungal communities to wood density loss after 12 years of log decay. FEMS Microbiol Ecol 91: 1–11. [DOI] [PubMed] [Google Scholar]
- Liu KL, Porras-Alfaro A, Kuske CR, Eichorst SA, Xie G. (2012). Accurate, rapid taxonomic classification of fungal large-subunit rRNA genes. Appl Environ Microbiol 78: 1523–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaw A, Wiener M. (2002). Classification and regression by random forest. R News 2: 18–22. [Google Scholar]
- Mori H, Maruyama F, Kato H, Toyoda A, Dozono A, Ohtsubo Y et al. (2013). Design and experimental application of a novel non-degenerate universal primer set that amplifies prokaryotic 16S rRNA genes with a low possibility to amplify eukaryotic rRNA genes. DNA Res 25: dst052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nannipieri PA, Giagnoni LA, Renella GI, Puglisi ED, Ceccanti BR, Masciandaro GR et al. (2012). Soil enzymology: classical and molecular approaches. Biol Fertil Soils 48: 743–762. [Google Scholar]
- Philippot L, Andersson SG, Battin TJ, Prosser JI, Schimel JP, Whitman WB et al. (2010). The ecological coherence of high bacterial taxonomic ranks. Nat Rev Microbiol 8: 523–529. [DOI] [PubMed] [Google Scholar]
- Reed DC, Algar CK, Huber JA, Dick GJ. (2014). Gene-centric approach to integrating environmental genomics and biogeochemical models. Proc Natl Acad Sci USA 111: 1879–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve JR, Schadt CW, Carpenter-Boggs L, Kang S, Zhou J, Reganold JP. (2010). Effects of soil type and farm management on soil ecological functional genes and microbial activities. ISME J 4: 1099–1107. [DOI] [PubMed] [Google Scholar]
- Rocca JD, Hall EK, Lennon JT, Evans SE, Waldrop MP, Cotner JB et al. (2014). Relationships between protein-encoding gene abundance and corresponding process are commonly assumed yet rarely observed. ISME J 9: 1693–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousk J, Bååth E, Brookes PC, Lauber CL, Lozupone C, Caporaso JG et al. (2010). Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J 4: 1340–1351. [DOI] [PubMed] [Google Scholar]
- Schermelleh-Engel K, Moosbrugger H, Müller H. (2003). Evaluating the fit of structural equation models: tests of significance and descriptive goodness-of-fit measures. Methods Psychol Res Online 8: 23–74. [Google Scholar]
- Schmidt MW, Torn MS, Abiven S, Dittmar T, Guggenberger G, Janssens IA et al. (2011). Persistence of soil organic matter as an ecosystem property. Nature 478: 49–56. [DOI] [PubMed] [Google Scholar]
- Shipley B. (2002) Cause and Correlation in Biology: A User's Guide to Path Analysis, Structural Equations and Causal Inference. Cambridge University Press: Cambridge, UK. [Google Scholar]
- Singh BK, Bardgett RD, Smith P, Reay DS. (2010). Microorganisms and climate change: terrestrial feedbacks and mitigation options. Nat Rev Microbiol 8: 779–790. [DOI] [PubMed] [Google Scholar]
- Singh BK, Quince C, Macdonald CA, Khachane A, Thomas N, Al-Soud WA et al. (2014). Loss of microbial diversity in soils is coincident with reductions in some specialized functions. Environ Microbiol 16: 2408–2420. [DOI] [PubMed] [Google Scholar]
- Strickland MS, Lauber C, Fierer N, Bradford MA. (2009). Testing the functional significance of microbial community composition. Ecology 90: 441–451. [DOI] [PubMed] [Google Scholar]
- Su JQ, Ding LJ, Xue K, Yao HY, Quensen J, Bai S et al. (2015). Long-term balanced fertilization increases the soil microbial functional diversity in a phosphorus-limited paddy soil. Mol Ecol 24: 136–150. [DOI] [PubMed] [Google Scholar]
- Talbot JM, Bruns TD, Taylor JW, Smith DP, Branco S, Glassman SI et al. (2014). Endemism and functional convergence across the North American soil mycobiome. Proc Natl Acad Sci USA 111: 6341–6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treseder KK, Balser TC, Bradford MA, Brodie EL, Dubinsky EA, Eviner VT et al. (2012). Integrating microbial ecology into ecosystem models: challenges and priorities. Biogeochemistry 109: 7–18. [Google Scholar]
- Trivedi P, Anderson IC, Singh BK. (2013). Microbial modulators of soil carbon storage: integrating genomic and metabolic knowledge for global prediction. Trends Microbiol 21: 641–651. [DOI] [PubMed] [Google Scholar]
- Trivedi P, He Z, Van Nostrand JD, Albrigo G, Zhou J, Wang N. (2012). Huanglongbing alters the structure and functional diversity of microbial communities associated with citrus rhizosphere. ISME J 6: 363–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi P, Rochester R, Trivedi C, Anderson IC, Singh BK. (2015). Soil aggregate size mediates the impacts of cropping regimes on soil carbon and microbial communities. Soil Biol Biochem 91: 169–181. [Google Scholar]
- Tu Q, Yu H, He Z, Deng Y, Wu L, Van Nostrand JD et al. (2014). GeoChip 4: a functional gene arrays-based high throughput environmental technology for microbial community analysis. Mol Ecol Resour 14: 914–928. [DOI] [PubMed] [Google Scholar]
- van der Wal A, Ottosson E, de Boer W. (2015). Neglected role of fungal community composition in explaining variation in wood decay rates. Ecology 96: 124–133. [DOI] [PubMed] [Google Scholar]
- Victoria R, Banwart SA, Black H, Ingram H, Joosten H, Milne E et al. (2012) The benefits of soil carbon: managing soils for multiple economic, societal and environmental benefits Foresight Chapter. In: UNEP Year Book 2012: Emerging Issues in Our Global Environment. UNEP: Nairobi, Kenya, pp 19–33. [Google Scholar]
- Wallenstein MD, Hall EK. (2012). A trait-based framework for predicting when and where microbial adaptation to climate change will affect ecosystem functioning. Biogeochemistry 109: 35–47. [Google Scholar]
- Wieder WR, Bonan GB, Allison SD. (2013). Global soil carbon predictions are improved by modelling microbial processes. Nat Climate Change 3: 909–912. [Google Scholar]
- Wieder WR, Grandy AS, Kallenbach CM, Taylor PG, Bonan GB. (2015). Representing life in the Earth system with soil microbial functional traits in the MIMICS model. Geosci Model Dev 8: 2011–2052. [Google Scholar]
- Wood SA, Bradford MA, Gilbert JA, McGuire KL, Palm CA, Tully KL et al. (2015). Agricultural intensification and the functional capacity of soil microbes on smallholder African farms. J Appl Ecol 52: 744–752. [Google Scholar]
- You Y, Wang J, Huang X, Tang Z, Liu S, Sun OJ. (2014). Relating microbial community structure to functioning in forest soil organic carbon transformation and turnover. Ecol Evol 4: 633–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Liu H, Wang C, Xu JR. (2013). Comparative analysis of fungal genomes reveals different plant cell wall degrading capacity in fungi. BMC Genomics 14: 274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman AE, Martiny AC, Allison SD. (2013). Microdiversity of extracellular enzyme genes among sequenced prokaryotic genomes. ISME J 7: 1187–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.