Abstract
Reliable detection of unwanted organisms is essential for meaningful health monitoring in experimental animal facilities. Currently, most rodents are housed in IVC systems, which prevent the aerogenic transmission of pathogens between cages. Typically soiled-bedding sentinels (SBS) exposed to soiled bedding collected from a population of animals within an IVC rack are tested as representatives, but infectious agents often go undetected due to inefficient transmission. Pasteurellaceae are among the most prevalent bacterial pathogens isolated from experimental mice, and the failure of SBS to detect these bacteria is well established. In this study, we investigated whether analysis of exhaust air dust (EAD) samples by using a sensitive and specific real-time PCR assay is superior to conventional SBS monitoring for the detection of Pasteurella pneumotropica (Pp) infections. In a rack with a known prevalence of Pp-positive mice, weekly EAD sampling was compared with the classic SBS method over 3 mo. In 6 rounds of testing, with a prevalence of 5 infected mice in each of 7 cages in a rack of 63 cages, EAD PCR detected Pp at every weekly time point; SBS failed to detect Pp in all cases. The minimal prevalence of Pp-infected mice required to obtain a reliable positive result by EAD PCR testing was determined to be 1 in 63 cages. Reliable detection of Pp was achieved after only 1 wk of exposure. Analysis of EAD samples by real-time PCR assay provides a sensitive, simple, and reliable approach for Pp identification in laboratory mice.
Abbreviations: EAD, exhaust-air dust; Pp, Pasteurella pneumotropica; SBS, soiled-bedding sentinel
In recent decades, the use of IVC rack systems in laboratory rodent facilities has increased. Within these microisolation units, each cage is ventilated separately and therefore represents an individual microbiologic unit. If cages are handled appropriately, the spread of infectious agents between cages is prevented. For health monitoring of IVC-housed colonies, quarterly monitoring of sentinels exposed to soiled bedding (SBS) remains the most common method of indirect testing when various factors, such as housing conditions, immunodeficiency of resident animals, or low numbers of animals prevent direct animal sampling.21 The detection of unwanted organisms by using sentinel mice relies on the sentinels becoming infected with the pathogen, regardless of the testing method used. The major disadvantage of this health monitoring strategy is that not all agents infect sentinels through soiled bedding transfer. Viruses (such as lymphocytic choriomeningitis virus and Sendai virus), bacteria (Pasteurella pneumotropica [Pp]), and murine fur mites often remain undetected.1,7,9,15,19,29 The use of contact sentinels or colony animals to improve the detection of unwanted organisms is not always feasible, and health monitoring in IVC-housed rodent colonies remains challenging.
Pp is one of the most prevalent bacterial pathogens in experimental facilities worldwide.14,27 Although the pathogenicity of most Pasteurellaceae species is low, Pp is associated with various clinical manifestations, including eye, genital tract, and respiratory infections.2,22,24 Immunodeficient animals infected with Pp develop mild to severe or even lethal disease.6,12,17,20 Even subclinical Pp infections in immunocompetent mice can represent an unwanted experimental variable that can critically influence research data.26 In Europe, the Federation of European Laboratory Animal Science Associations recommends quarterly screening for this pathogen.21 However, even when Pp was transferred to SBS, its subsequent detection and analysis can be problematic. Pasteurellaceae are usually detected through bacterial culture of swabs taken from the nasopharynx, genital tract, or large intestine, with subsequent analysis of suspect colonies by using biochemical test kits. However, the commonly used kits are optimized for human samples and frequently fail to identify rodent samples to the correct species or even family level.4,13,21 The analysis of subcultured bacterial colonies by matrix-assisted laser desorption–ionization–time-of-flight mass spectrometry has good specificity,11,32 but the equipment is very expensive, and reference databases lack sufficient murine datasets. Serological tests are inappropriate for the detection of Pp infections, because the seroconversion of infected sentinel mice is unreliable.29 PCR technology is the most reliable and sensitive method currently, and genus- and species-specific assays are available.3,5,10,18,30 Recently, we demonstrated that the combination of a real-time PCR assay that amplified a variable region in the 16S rRNA sequence with a high-resolution melting curve analysis is sufficient for the identification and differentiation of murine Pasteurellaceae species.23
Many drawbacks are associated with SBS, but an alternative health monitoring strategy has yet to be developed. Consequently PCR testing of environmental samples, such as exhaust air dust (EAD) samples from IVC racks, has been proposed.7 In a previous study, mouse hepatitis virus, Sendai virus, mouse parvovirus,7 and mouse norovirus33 were detected in the exhaust-air prefilter, and Radfordia affinis and Myobia musculi were detected in swabs of the horizontal exhaust plenum.16 Although Pp could not be detected in environmental samples by using conventional PCR,25 we investigated whether EAD samples are suitable for the detection of Pp infections in IVC-housed mice by using a specific and highly sensitive real-time PCR assay.
Here we compared real-time PCR analysis of dust samples taken from the exhaust-air prefilter of an IVC rack containing a known number of Pp-infected mice with the conventional SBS health monitoring strategy. We also investigated the minimal prevalence of infected animals necessary for reliable detection of Pp.
Materials and Methods
Mice.
All mice used were excess mice obtained from breeding and research colonies at our facility. The Pp-positive colony consisted of immunocompetent female AVM:ICR mice housed in groups of 5 animals per cage; they were approximately 3 mo old at the beginning of the study. The Pp-negative colony consisted of immunocompetent mice of both sexes and various strains, ages, and genetic backgrounds, to simulate a typical research mouse colony. Mice were housed in sex-matched groups of 2 to 5 animals per cage; in exceptional cases, single animals were housed separately for a short time. Female AVM:ICR mice (age, 3 mo) housed in groups of 3 animals per cage were used as Pp-negative bedding sentinels. The Pp-positive colony and the sentinel mice were obtained from a breeding colony in which no FELASA-listed agents21 were detected during routine health monitoring surveillances. The Pp-negative colony was obtained from a research colony with a known history for Helicobacter spp. and Pseudomonas aeruginosa. All experiments and animal housing were performed in strict accordance with the directive 2010/63/EU.
Animal housing.
All animals were housed in an IVC rack (SealSafe Plus, Tecniplast, Buggugiate, Italy) under SPF conditions in IVC cages (GM 500, Tecniplast), with a maximal cage density of 5 adult mice per cage (socially housed in stable groups of compatible animals; 100 cm2 floor area per mouse) and environmental conditions (12:12-h light:dark cycle, 20 to 24 °C, 45% to 65% humidity) according to directive 2010/63/EU. Routine health monitoring surveillance was performed for all agents listed in the FELASA recommendations21 with the recommended quarterly and annual tests via SBS.21 Air-handling units were run with 60 air changes per hour in positive pressure mode (15 to 22 Pa). Autoclaved wood fiber (Lignocel 3/4 S, J Rettenmaier and Söhne, Rosenberg, Germany) and nesting material (Crinkle Nature, J Rettenmaier and Söhne) were used as bedding and environmental enrichment. Sterile-filtered tap water and an irradiated standard diet for rodents (Altromin 1314, Altromin Spezialfutter, Lage, Germany) were available without restriction. Access to the animal room was gained through an air shower, and all personnel entering the room performed a complete change of clothes and shoes, and wore gloves, surgical masks, and bonnets the entire time. All cage changes and sample collections were performed in a HEPA-filtered cage-changing station. All cages and racks were cleaned and autoclaved prior to use.
Sample types and collection.
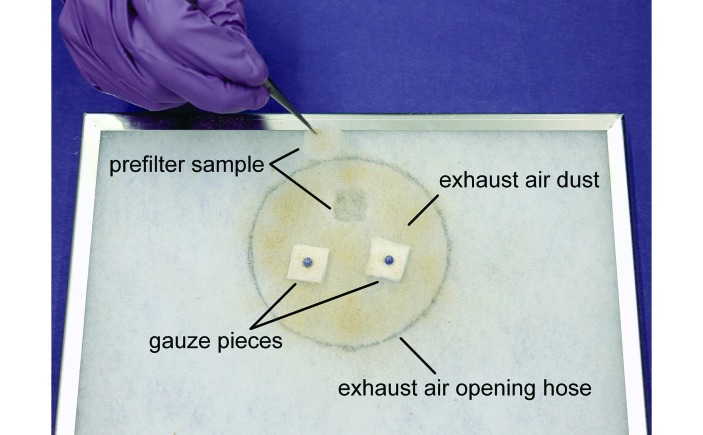
Several sample collection methods were used to monitor the Pp-positive and -negative colonies, the sentinels, and the EAD. For detection of Pp by using oral swabs, the mice were scruffed at the neck, and a sterile flocked swab (FLOQSwabs Mini Tip 80 mm, Mast Diagnostica, Reinfeld, Germany) was gently inserted approximately 1 cm into the oral cavity, rotated, and quickly removed. The flocked tip of the swab was inserted into a 2-mL microcentrifuge tube for further analysis. For detection of Pp in feces, pooled samples of 10 fecal pellets per cage or single fecal pellets from individual sentinel mice were collected by using disposable forceps and processed as described later. Once each week, approximately 10 mL soiled bedding was collected from every cage in use (49 cages for the Pp-negative colony; 7 cages for the Pp-positive colony) during routine cage changing. The sentinel cage was filled with the soiled bedding mixed with an equal amount of fresh bedding. Two sample types were used for analysis of EAD: (1) autoclaved gauze pieces (2 × 2 cm; ES Kompressen, Paul Hartmann, Heidenheim, Germany) attached to the ‘dirty side’ of the exhaust air prefilter directly above the exhaust-air hose opening and (2) 2 × 2-cm samples cut from the prefilter by using clean and disinfected scissors and forceps (Figure 1).
Figure 1.
The 2 methods for exhaust air dust (EAD) sampling of the IVC exhaust-air prefilter. The black circle indicates the position of the exhaust-air hose relative to the prefilter.
SBS were tested by using real-time PCR analysis of fecal samples and oral swabs, as well as by bacteriological culture. For bacteriologic examination, a Columbia blood agar plate containing 5% sheep blood (VWR International, Darmstadt, Germany) was inoculated with an oral swab immediately after sampling. Plates were incubated for 24 h at 37 °C. Pasteurella-like colonies were picked and analyzed by using a multiplex PCR assay based on the 16S-23S rRNA internal transcribed spacer region that can be used to differentiate between the Pp biotypes Jawetz and Heyl as well as Actinobacillus muris and other Pasteurellaceae.3 For serologic examination, serum samples of Pp-positive mice and sentinels were analyzed by an external diagnostic laboratory. Two sentinel animals were examined for all FELASA-listed organisms21 at the end of the exposure time.
DNA extraction.
DNA isolation from oral swabs, gauze pieces, and prefilter material was performed by using the phenol–chloroform extraction method28 with minor modifications: sample material was incubated with lysis buffer (10 mM EDTA, 10 mM Tris-HCl pH 7.6, 0.5% SDS, 10 mM NaCl, and 0.3 mg/mL Proteinase K) for 30 min at 55 °C and shaken at 600 rpm by using a thermomixer. For oral swabs, gauze pieces, and prefilter materials, 400 µL, 500 µL, and 650 µL, respectively, of lysis buffer was used. After incubation, 300 µL of lysis mixture was transferred into a 1.5-mL microcentrifuge tube. An equal volume of phenol–chloroform–isoamyl alcohol solution was added, and the mixture was vortexed. After being centrifuged for 6 min at 15,300 × g, 200 µL of the upper aqueous phase was transferred to a new tube containing 500 µL of NaCl-saturated 100% ethanol and centrifuged for 10 min at 15,300 × g and 4 °C for precipitation. The DNA pellet was washed twice with 70% ethanol and subsequently dissolved in 35 µL of ultrapure water. DNA isolation of fecal samples was performed by using the QIAamp Fast DNA Stool Mini Kit (Qiagen, Hilden, Germany) with the following amendments: after pipetting 1 mL of InhibitEX buffer onto the fecal pellets, a 7-mm stainless steel bead (Qiagen) was added. The contents were ground for 5 min at 50 Hz by using a TissueLyser LT (Qiagen). After heating the suspension for 5 min at 70 °C and centrifuging it for 1 min at 15,300 × g, 200 µL of supernatant was collected and processed according to the manufacturer's instructions.
Real-time PCR.
A Pp Jawetz-specific real-time PCR assay10 for amplification of a sequence within the 16S rRNA gene (forward primer, 5′ CGG GTT GTA AAG TTC TTT CGG T 3′; reverse primer, 5′ GGA GTT AGC CGG TGC TTC TTC 3′) was performed by using a 6-FAM-BHQ-1 dual-labeled fluorogenic probe (5′ AAT AAG GGT ATT AAC CTT ATC ACC TTC CTC ATC 3′) in a RotorGene Q instrument (Qiagen). DNA template (2 µL) was added to a reaction mixture consisting of 5 µL of 5× HOT FIREPol Probe qPCR Mix Plus (Solis BioDyne, Tartu, Estonia), 300 nM of each primer, 200 nM probe, and ultrapure water in a 25-µL total reaction volume. The DNA extracted was not quantified prior to application. The thermocycling parameters were: initial denaturation at 95 °C for 15 min, and 40 cycles with denaturation at 95 °C for 15 s and annealing–extension at 58 °C for 60 s. For quantitative analysis, a plasmid was constructed by cloning the 93-bp real-time PCR amplicon into the pCRII-TOPO vector (Invitrogen Life Technologies, Waltham, MA) according to the manufacturer's instructions. A triplicate plasmid DNA dilution series, ranging from 1 × 107 to 1 × 102 copies/µL, was used as a standard in each run. To identify contaminants in the reaction mixture, no-template, positive, and negative controls were included. Samples that yielded at least 10 copies/µL were judged to be Pp-positive; samples with fewer than 10 copies/µL were considered equivocal and retested; these samples were considered positive when the copy number was greater than 0 after retesting. Other samples were considered negative. Data were analyzed by using RotorGene Q Software (version 2.1, Qiagen).
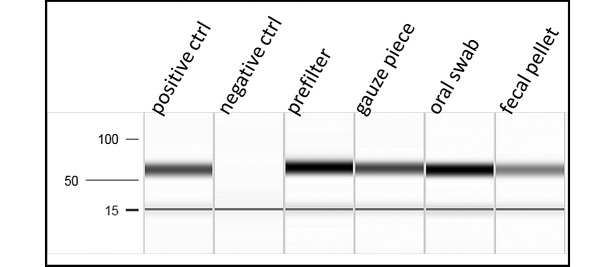
Samples of the 4 different matrix types (fecal pellet, prefilter material, gauze piece, and oral swab) were spiked with Pp DNA isolated directly from a Pp Jawetz bacterial colony to confirm that the matrix did not inhibit the analysis (Figure 2). Different DNA extraction protocols were used for each matrix as previously described. The initial amount of Pp Jawetz DNA for the spike was individually calculated for the different sample types to obtain a final DNA concentration of 1 ng/μL after DNA extraction. PCR analysis was performed by including a positive control (2 μL of 1 ng/μl DNA isolated from the Pp Jawetz bacterial colony), a no-template control, and the 4 spiked sample types. PCR products were analyzed by using the QIAxcel Advanced System (Qiagen), which is based on capillary electrophoresis, and the QIAxcel DNA Fast Analysis Kit (Qiagen). Pp DNA was PCR-amplified from all sample types after spiking negative samples (fecal pellet, prefilter material, gauze piece, and oral swab) with DNA isolated from a Pp Jawetz bacterial colony. Positive (DNA isolated from the Pp colony) and negative controls were included.
Figure 2.
Capillary electrophoresis of PCR products from different types of sampling matrix spiked with Pasteurella pneumotropica Jawetz DNA prior to DNA extraction.
Study designs.
To determine the time needed for Pp to spread from an infected to a noninfected mouse within the same cage (that is, contact infection), we set up 4 cages each containing one Pp-positive mouse and 2 Pp-negative mice. Mice were tested by using oral swabs immediately before grouping and every 3 d for a total of 21 d.
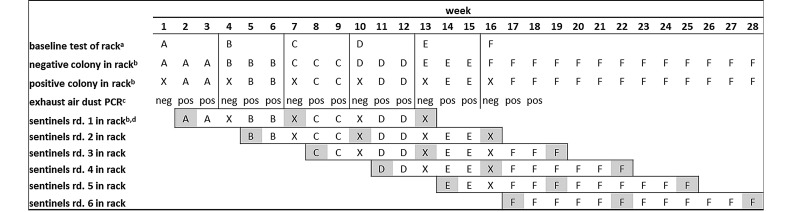
To compare EAD analysis and SBS monitoring, a cleaned and autoclaved IVC rack connected to a clean and disinfected (Pursept-FD, Merz Hygiene, Frankfurt am Main, Germany) air-handling unit was populated with the Pp-negative colony. The Pp-negative status of the colony was confirmed by testing one mouse per cage during the baseline testing week at the beginning of each experimental round. Single mice were considered sufficient, given that our contact infection study revealed that a Pp infection spreads rapidly within a single cage. All mice were tested in rotation in subsequent rounds to verify the status of each individual mouse at least once during the experiment. Old or sick mice of the Pp-negative colony underwent veterinary examination and were replaced with new Pp-negative mice as necessary. All sick mice were confirmed by oral swab testing to be Pp-negative to exclude that clinical signs are caused by Pp infection. One horizontal row of the IVC rack was equipped with empty cages to provide space for sentinel mice. Three sentinel mice (2 for health monitoring surveillance and one backup) were determined to be free of Pp by testing of oral swabs and single fecal samples during the baseline week. After the IVC rack system was populated with the Pp-negative colony, a gauze piece was attached directly above the opening of the exhaust-air hose on a new exhaust air prefilter in the air-handling unit (Figure 1). This gauze piece and the prefilter were removed and analyzed after the baseline testing week, to exclude any remaining DNA contamination within the IVC rack system and the air-handling unit and to confirm the negative infectious status of the negative colony. Afterward, a new prefilter containing 2 new gauze pieces was applied. At the beginning of the test period, a Pp-positive colony consisting of 7 cages, each containing 5 positive mice, and the sentinel mice were placed in the rack. To confirm infection of the positive mice, the entire positive colony was tested every second week by using individual oral swabs and pooled fecal samples from each cage. Six experimental rounds were determined to be necessary to produce statistically relevant results. An optimal hygienic monitoring period lasts 10 to 12 wk to provide sufficient time for infection and seroconversion to various agents, such as Mycoplasma pulmonis and Pp 21. Therefore, sentinel mice were retested by using individual oral swabs and individual fecal samples after 6 and 12 wk of exposure to soiled bedding; after 12 wk of exposure, serologic, bacteriologic, and complete health monitoring surveillance according to FELASA recommendations was performed.
For EAD analysis, a gauze piece was collected each week and a prefilter sample was collected every 2 wk. EAD testing was considered positive and the experimental round was stopped when dust samples yielded positive results 2 wk in a row. This experiment was repeated 5 times by using cleaned and autoclaved IVC racks (Figure 3). Each experimental round consisted of an initial baseline testing period, to confirm that the negative colony and the IVC were negative for Pp, and a testing period, during which the rack was repopulated with the positive colony. All sentinels were housed for 12 wk on soiled bedding derived from all cages, including the positive colony, even when the positive colony was located in another rack during the baseline test weeks. At the end of the 6 rounds, blood samples for serologic examination were collected from 12 mice from the Pp-positive colony, which had tested positive by repeated oral swab and fecal analysis over a 6-mo period.
Figure 3.
Experimental design for comparing EAD analysis with sentinel monitoring. Letters A through F indicate different IVC racks. Each new round of experimentation began with a clean and autoclaved IVC rack, after detection of 2 positive EAD samples in the previous round; X indicates an IVC rack where Pp-positive and sentinel mice were held during the baseline test week. a, Samples for baseline testing were collected after the rack (containing the Pp-negative colony) operated for 1 wk; the Pp-positive colony and the sentinel mice were then placed in experimental IVC rack. b, Localization of Pp-positive and - negative as well as sentinel mice. c, Real-time PCR results of EAD sample. d, Sentinel mice in all 6 rounds were exposed to soiled bedding over a period of 12 wk. During baseline testing weeks, the sentinel mice were transferred to and held in rack X and further exposed to soiled bedding from the Pp-positive and -negative colonies; highlighted letters indicate sentinel testing (immediately before placement in the experimental rack, as well as after 6 and 12 wk of exposure time). Pp, Pasteurella pneumotropica; EAD, exhaust air dust; rd, round; pos, positive EAD real-time PCR result; neg, negative EAD real-time PCR result; bar, rack change.
To investigate the minimal prevalence of infected animals that is necessary for reliable detection of Pp in the EAD, we used decreasing numbers of Pp-positive mice within a rack of 63 cages otherwise completely filled with Pp-negative cages. Three rounds of dust testing with 4, 2, and 1 cage, each occupied by 5 Pp-positive mice, were performed as described previously but without sentinel mice. As previously, the experimental round was terminated when we obtained 2 consecutive positive dust samples. At the end of the experiment, the entire negative colony was retested by using oral swabs to exclude Pp transmission within the rack during the experiment.
Results
Rapidity and duration of P. pneumotropica transmission by direct contact.
To better understand the speed of transmission from infected to uninfected mice, the duration and intensity of the infection, and the duration and intensity of bacterial colonization of the nasopharynx, we cohoused Pp-infected mouse with 2 Pp-negative mice in a single cage, in a total of 4 cages (Table 1). At the beginning of the experiment, the infection status of all mice was confirmed by PCR analysis of oral swabs. Additional oral swabs were PCR-analyzed every 3 d for a total of 21 d. One animal with no detectable Pp infection had to be euthanized after 3 d due to emaciation and poor general condition. All initially negative mice tested Pp-positive between days 3 and 12; additional testing showed that Pp was continuously detectable in all mice until day 21 (Table 1).
Table 1.
Pasteurella pneumotropica infection status of initially negative mice during 21 d of contact with infected cage mates
| Infection statusa | Real-time PCR results on day |
||||||||
| Cage | Mouse | 3 | 6 | 9 | 12 | 15 | 18 | 21 | |
| 1 | 1 | – | – | nt | nt | nt | nt | nt | nt |
| 2 | – | – | – | – | + | + | + | + | |
| 3 | + | + | + | + | + | + | + | + | |
| 2 | 1 | – | – | + | + | + | + | + | + |
| 2 | – | – | – | – | + | + | + | + | |
| 3 | + | + | + | + | + | + | + | + | |
| 3 | 1 | – | + | + | + | + | + | + | + |
| 2 | – | + | + | + | + | + | + | + | |
| 3 | + | + | + | + | + | + | + | + | |
| 4 | 1 | – | – | – | + | + | + | + | + |
| 2 | – | + | + | + | + | + | + | + | |
| 3 | + | + | + | + | + | + | + | + | |
–, negative real-time PCR result; +, positive real-time PCR result; nt, not tested (mouse euthanized)
Infection status, as determined immediately before grouping
Comparison of PCR analysis of EAD with SBS for detecting Pp infections in IVC-housed mouse colonies.
Sentinels were exposed to soiled bedding from a colony with known Pp prevalence for a total of 12 wk. To minimize the time required, each experimental round was terminated and a new round started when 2 consecutive positive EAD results were obtained. However, the sentinels from each experimental round were retained until they had been exposed to soiled bedding for a total of 12 wk (Figure 3). After baseline EAD testing of rack A and the negative colony during week 1, the positive colony (35 mice in 7 cages) and the SBS (3 mice in one cage) for round 1 were transferred from the holding rack (rack X) to rack A at the beginning of week 2 (start of round 1). After positive EAD results were obtained at the end of weeks 2 and 3, the negative colony (49 cages containing the Pp-negative mice) was transferred to a fresh rack (rack B). Baseline testing for round 2 was performed in the new rack at the end of week 4. The positive colony and the sentinels were held in rack X and returned to rack B during week 5, together with the sentinels for round 2, after baseline testing. Therefore, during weeks 5 to 6, sentinels from rounds 1 and round 2 were placed in rack B. After 2 positive EAD results were obtained during weeks 5 and 6, EAD testing of round 2 was terminated and round 3 was prepared, and so on. This pattern was completed a total of 6 times. EAD testing for round 6 was terminated in rack F during week 18. The SBS for round 1 were taken from rack D during week 13 for terminal testing, and the SBS from experimental round 6 remained in rack F until week 28. No cross-contamination occurred between the infected and noninfected colonies. The negative mice were confirmed to be free of Pp infection by the results of real-time PCR of all oral swabs tested throughout the experimental period.
The positive mice showed continuous colonization of the oral cavity, as determined by biweekly oral swab analysis of 33 of the 35 infected mice over 1 y. Only 2 infected mice tested negative, at one time point for 1 or 3 biweekly tests. These spurious results might reflect technical problems during DNA extraction (for example, loss of the DNA pellet during DNA isolation procedure) or intermittent shedding of Pasteurellaceae. All pooled fecal samples from cages containing infected mice (10 pellets per cage) tested every 2 wk yielded positive PCR results at all time points. Prior to the beginning of each experimental round, the experimental racks (A through F) containing the negative colony were operated for 1 wk for baseline testing. The absence of residual Pp DNA in the washed and autoclaved IVC racks, their pipes, and fresh IVC prefilters as well as the negative infection status of the negative colony was verified by testing gauze pieces and prefilter material at the end of the baseline week. All samples taken for baseline testing were negative at all times. After the baseline tests, the experimental rack was populated with the positive colony and the sentinels. Gauze pieces attached to the prefilter were tested weekly and prefilter samples were tested biweekly, to detect Pp DNA in the EAD. In all 6 rounds, Pp could be detected after 1 wk; positive results were confirmed during the subsequent week in all 6 rounds. For SBS monitoring, a cage occupied by 3 sentinel mice was added to racks A through F after baseline testing in each experimental round. Care was taken that all sentinels were continuously exposed to soiled bedding from the experimental colony with a known prevalence of Pp over 12 wk. In addition, all sentinels were exposed to Pp-positive soiled bedding when kept in rack X during baseline testing. All 3 SBS in a cage were tested after 6 and 12 wk of exposure by using oral swabs and single-animal fecal samples. All sentinel mice tested negative at all times. To confirm these results, Pp serology, bacterial culture of an oral swab, and complete FELASA health monitoring were performed for 2 SBS in each round at the end of the exposure time. In total, 3 animals tested positive for Helicobacter hepaticus, but all other animals were negative for all FELASA-listed organisms. PCR analysis of oral swabs and feces from the Pp-positive colony showed continuous colonization of the oral cavity and excretion in the feces, respectively, for 6 mo. The blood samples taken at the end of the study showed that all 12 of these mice were seronegative.
Minimal prevalence of Pp-infected mice for reliable detection by EAD PCR analysis.
After successful detection of Pp by EAD PCR at a prevalence of 7 of 63 cages in a rack within 1 wk of exposure, we investigated the minimal prevalence necessary for reliable detection. The experimental setup was the same as that in the previous experiment (except without sentinel mice), but the number of cages containing Pp-positive mice was reduced from 4 to 2 to 1 during 3 rounds of testing. Again, a new round was started after 2 consecutive positive EAD results were obtained. In all instances, even with a prevalence of just 1 in 63 cages per rack, the presence of Pp-positive mice was detected during the first week and confirmed in the second week. Again, the Pp-negative colony, remained negative according to oral swab PCR testing throughout the entire experimental period.
Discussion
Accurate data on the health status of laboratory animals is crucial for their use in research projects. Undetected subclinical infection by unwanted organisms represents an uncontrolled variable that might interfere with the results of animal experiments.2,8,31 As discussed previously, the standard procedures for the detection of Pp in mouse colonies can be nonspecific or produce false-negative results. As a consequence, Pp infections are frequently overlooked or reported incorrectly.
Analysis of EAD samples repeatedly and reliably detected Pp infections even at the very low prevalence of one cage of infected mice in an IVC rack. In contrast, the use of SBS failed to detect Pp infections even at a prevalence of 7 cages in a rack. None of the sentinel mice in our study became infected with Pp at any time. None of the methods used to test sentinels—real-time PCR of oral swabs and fecal samples, bacterial culture of oral swabs, and serology—detected infection of the sentinel mice. Although mice easily become infected with Pp by direct contact and display continuous colonization of the oral cavity and gut, as shown in the current study, exposure to soiled bedding seems insufficient to infect sentinel mice. Because all pooled fecal samples from cages of infected mice showed positive results over a period of 6 mo, infected mice likely continuously shed bacterial nucleic acids with their feces. Given that the viability of Pasteurellaceae on wood bedding is limited to 30 min,29 live bacteria likely never reached the sentinel cage, because cage changing and the preparation of soiled bedding takes more than 30 min. Alternatively transferred bacteria might have lost their infectivity or an infectious dose may not have been transferred, resulting in lack of infection of the sentinel animals with Pp.
The Pp-negative colony was tested by using pooled fecal samples from each cage as well as oral swabs collected from one mouse per cage at the beginning of the experiment, between rounds 1 and 6 and after round 6 for a period of 6 mo. Testing only one animal per cage (as a representative of all cage inhabitants) by oral swab seemed reasonable given that we demonstrated that cagemates became infected within 3 to 12 d of cohabitation.
Contrary to our findings, a previous study25 failed to detect the presence of Pp on swab samples taken from a ventilated rack housing infected animals. In addition, analysis of swab samples taken from cages and laminar flow hoods, as well as analysis of the inner surface of the prefilter, failed to detect bacterial DNA.25 Several reasons might explain these differences. In contrast to the cited study, we used a specific and highly sensitive real-time PCR method that allows detection of Pp nucleic acids at a much lower copy number. No detailed information about the rack model or air handling unit and air changes per hour was provided in the previous study;25 perhaps the prefilter was not the optimal sampling site in that particular rack system. Moreover, the analysis of gauze pieces or prefilter material itself might result in a higher copy number than swab samples, due to the higher amount of dust. Differences in the preparation of DNA from dust might also be a reason for lower sensitivity in the previous study.25
One of the key factors in EAD analysis is choosing an optimal sampling site. In our experiments, the dust was collected by using gauze pieces attached to the ‘dirty side’ of the exhaust-air prefilter directly above the exhaust-air hose opening. Thus, the collection gauze was in an optimal position to collect dust particles potentially contaminated with Pp nucleic acids. In addition, we analyzed prefilter samples collected from the same position. At this location, soiled prefilters showed visible quantities of dust evenly distributed over the surface at the end of the regular exchange interval of 4 wk. For the IVC system used in our studies, we therefore regard the exhaust air prefilter as an appropriate sampling site. However, the preferred position for EAD sampling within a rack must be determined for each different IVC rack system, because airflow direction within the rack can vary. The terminal vertical exhaust plenum in another rack system, for example, was demonstrated to be an unreliable sampling site for the detection of fur mites, whereas sampling at the shelf exhaust manifold allowed detection of a single infested cage.16 When the number of infected animals is low, as is often the case in endemic infection with agents of low transmissibility, the effectiveness of EAD analysis of gauze pieces attached to the rack prefilter is likely to be highly dependent on the uniformity of airflow within the rack. Unbalanced airflow might lead to inaccurate sampling of dust from cages occupied by infected rodents.7
The presence of residual DNA from the previous experimental round within the rack, tubes, or air handling unit, which would lead to false-positive results, was excluded by baseline testing. A fresh, cleaned, and autoclaved rack was used for each round of experimentation. When using EAD PCR for hygienic monitoring, residual nucleic acids, which pose a risk of generating false-positive results, might constitute a major drawback. Discriminating between previous and current infections might not be possible. We regard EAD testing as a highly sensitive screening method that permits surveillance of agent-free colonies. Therefore, a positive result, when obtained for the first time, would cause concern and prompt further confirmation and possibly an elimination strategy. A possible strategy might include pooled samples (for example, oral swabs, fecal samples) to trace infections stepwise down to individual cages or involve removing mice from positive cages and repeatedly retesting. Once a colony has an established agent-positive status, verifying the absence of the unwanted agent after an elimination attempt requires careful sanitizing of the equipment to avoid contamination with residual living or dead infectious agents or residual nucleic acids. We recommend washing and autoclaving the contaminated rack and then running it for several days before performing a baseline test and adding any cages to the system. In our studies, the infectious status of the Pp-negative colony had already been verified, and therefore these cages were included in the baseline test. Changing the prefilter only might not ensure complete elimination of residual contamination and could lead to false-positive results. When using EAD samples for health monitoring of ongoing endemic infections, IVC racks should be washed (for example, in a rack washer) and autoclaved on a regular basis.
Use of EAD PCR for screening makes it possible to monitor entire experimental animal facilities efficiently by using reasonable sample numbers. Because the exhaust-air prefilter in IVC systems must be changed regularly, collection of prefilter material for PCR analysis requires only minimal additional work. Recently, 2 suppliers offer updated IVC rack systems that provide system-optimized sampling devices to enable contamination-free recovery of EAD samples from the optimal location; other suppliers likely will follow soon.
EAD sampling has the potential to constitute a key contribution to the basic 3Rs principles regarding the use of laboratory animals. The associated improvement in Pp detection leads to increased knowledge regarding animal health status, might reduce the number of experimental animals needed, and represents a technologic refinement given that mice no longer have to be manipulated for oral swabbing or disturbed for fecal collection. This simple and low-cost sampling method has great potential to become a useful tool for primary health monitoring and surveillance of rodent populations. Further experiments are needed to investigate whether EAD analysis can be adapted for use for other agents, as it has been for mouse hepatitis virus, mouse parvovirus, Helicobacter muridarum,7 mouse norovirus,33 and murine fur mites.16
Acknowledgments
We thank Natalie Humm, Michael Opitz, and the animal caretaker team for excellent support and Claudia Kiermayer for providing Pp-negative mice.
References
- 1.Artwohl JE, Cera LM, Wright MF, Medina LV, Kim LJ. 1994. The efficacy of a dirty-bedding sentinel system for detecting Sendai virus infection in mice: a comparison of clinical signs and seroconversion. Lab Anim Sci 44:73–75. [PubMed] [Google Scholar]
- 2.Baker DG. 1998. Natural pathogens of laboratory mice, rats, and rabbits and their effects on research. Clin Microbiol Rev 11:231–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benga L, Benten WP, Engelhardt E, Bleich A, Gougoula C, Sager M. 2013. Development of a multiplex PCR assay based on the 16S–23S rRNA internal transcribed spacer for the detection and identification of rodent Pasteurellaceae. J Microbiol Methods 95:256–261. [DOI] [PubMed] [Google Scholar]
- 4.Boot R, Van den Brink M, Handgraaf P, Timmermans R. 2004. The use of the API 20 NE bacteria classification procedure to identify Pasteurellaceae strains in rodents and rabbits. Scand J Lab Anim Sci 31:177–183. [Google Scholar]
- 5.Bootz F, Kirschnek S, Nicklas W, Wyss SK, Homberger FR. 1998. Detection of Pasteurellaceae in rodents by polymerase chain reaction analysis. Lab Anim Sci 48:542–546. [PubMed] [Google Scholar]
- 6.Chapes SK, Mosier DA, Wright AD, Hart ML. 2001. MHCII, TLR4, and Nramp1 genes control host pulmonary resistance against the opportunistic bacterium Pasteurella pneumotropica. J Leukoc Biol 69:381–386. [PubMed] [Google Scholar]
- 7.Compton SR, Homberger FR, Paturzo FX, Clark JM. 2004. Efficacy of 3 microbiological monitoring methods in a ventilated cage rack. Comp Med 54:382–392. [PubMed] [Google Scholar]
- 8.Connole MD, Yamaguchi H, Elad D, Hasegawa A, Segal E, Torres-Rodriguez JM. 2000. Natural pathogens of laboratory animals and their effects on research. Med Mycol 38 Suppl 1:59–65. [PubMed] [Google Scholar]
- 9.Cundiff DD, Riley LK, Franklin CL, Hook RR, Jr, Besch-Williford C. 1995. Failure of a soiled bedding sentinel system to detect cilia-associated respiratory bacillus infection in rats. Lab Anim Sci 45:219–221. [PubMed] [Google Scholar]
- 10.Dole VS, Banu LA, Fister RD, Nicklas W, Henderson KS. 2010. Assessment of rpoB and 16S rRNA genes as targets for PCR-based identification of Pasteurella pneumotropica. Comp Med 60:427–435. [PMC free article] [PubMed] [Google Scholar]
- 11.Frey J, Kuhnert P. 2014. Identification of animal Pasteurellaceae by MALDI–TOF mass spectrometry. Methods Mol Biol 1247:235–243. [DOI] [PubMed] [Google Scholar]
- 12.Hart ML, Mosier DA, Chapes SK. 2003. Toll-like receptor 4-positive macrophages protect mice from Pasteurella pneumotropica-induced pneumonia. Infect Immun 71:663–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayashimoto N, Aiba T, Itoh K, Kato M, Kawamoto E, Kiyokawa S, Morichika Y, Muraguchi T, Narita T, Okajima Y, Takakura A, Itoh T. 2005. Identification procedure for Pasteurella pneumotropica in microbiologic monitoring of laboratory animals. Exp Anim 54:123–129. [DOI] [PubMed] [Google Scholar]
- 14.Hayashimoto N, Morita H, Ishida T, Yasuda M, Kameda S, Uchida R, Tanaka M, Ozawa M, Sato A, Takakura A, Itoh T, Kagiyama N. 2013. Current microbiological status of laboratory mice and rats in experimental facilities in Japan. Exp Anim 62:41–48. [DOI] [PubMed] [Google Scholar]
- 15.Ike F, Bourgade F, Ohsawa K, Sato H, Morikawa S, Saijo M, Kurane I, Takimoto K, Yamada YK, Jaubert J, Berard M, Nakata H, Hiraiwa N, Mekada K, Takakura A, Itoh T, Obata Y, Yoshiki A, Montagutelli X. 2007. Lymphocytic choriomeningitis infection undetected by dirty-bedding sentinel monitoring and revealed after embryo transfer of an inbred strain derived from wild mice. Comp Med 57:272–281. [PubMed] [Google Scholar]
- 16.Jensen ES, Allen KP, Henderson KS, Szabo A, Thulin JD. 2013. PCR testing of a ventilated caging system to detect murine fur mites. J Am Assoc Lab Anim Sci 52:28–33. [PMC free article] [PubMed] [Google Scholar]
- 17.Kawamoto E, Sasaki H, Okiyama E, Kanai T, Ueshiba H, Ohnishi N, Sawada T, Hayashimoto N, Takakura A, Itoh T. 2011. Pathogenicity of Pasteurella pneumotropica in immunodeficient NOD/ShiJic-scid/Jcl and immunocompetent Crlj:CD1 (ICR) mice. Exp Anim 60:463–470. [DOI] [PubMed] [Google Scholar]
- 18.Kodjo A, Villard L, Veillet F, Escande F, Borges E, Maurin F, Bonnod J, Richard Y. 1999. Identification by 16S rDNA fragment amplification and determination of genetic diversity by random amplified polymorphic DNA analysis of Pasteurella pneumotropica isolated from laboratory rodents. Lab Anim Sci 49:49–53. [PubMed] [Google Scholar]
- 19.Lindstrom KE, Carbone LG, Kellar DE, Mayorga MS, Wilkerson JD. 2011. Soiled bedding sentinels for the detection of fur mites in mice. J Am Assoc Lab Anim Sci 50:54–60. [PMC free article] [PubMed] [Google Scholar]
- 20.Macy JD, Jr, Weir EC, Compton SR, Shlomchik MJ, Brownstein DG. 2000. Dual infection with Pneumocystis carinii and Pasteurella pneumotropica in B-cell–deficient mice: diagnosis and therapy. Comp Med 50:49–55. [PubMed] [Google Scholar]
- 21.Mähler M, Berard M, Feinstein R, Gallagher A, Illgen-Wilcke B, Pritchett-Corning K, Raspa M. 2014. FELASA recommendations for the health monitoring of mouse, rat, hamster, guinea pig, and rabbit colonies in breeding and experimental units. Lab Anim 48:178–192. [DOI] [PubMed] [Google Scholar]
- 22.Mikazuki K, Hirasawa T, Chiba H, Takahashi K, Sakai Y, Ohhara S, Nenui H. 1994. Colonization pattern of Pasteurella pneumotropica in mice with latent pasteurellosis. Jikken Dobutsu 43:375–379. [PubMed] [Google Scholar]
- 23.Miller M, Zorn J, Brielmeier M. 2015. High-resolution melting curve analysis for identification of Pasteurellaceae species in experimental animal facilities. PLoS One 10:e0142560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.National Research Council . 1991. Infectious diseases of mice and rats. Washington (DC): The National Academies Press. [Google Scholar]
- 25.Ouellet M, Cowan M, Laporte A, Faubert S, Heon H. 2011. Implementation of a PCR assay of Pasteurella pneumotropica to accurately screen for contaminated laboratory mice. Lab Anim (NY) 40:305–312. [DOI] [PubMed] [Google Scholar]
- 26.Patten CC, Jr, Myles MH, Franklin CL, Livingston RS. 2010. Perturbations in cytokine gene expression after inoculation of C57BL/6 mice with Pasteurella pneumotropica. Comp Med 60:18–24. [PMC free article] [PubMed] [Google Scholar]
- 27.Pritchett-Corning KR, Cosentino J, Clifford CB. 2009. Contemporary prevalence of infectious agents in laboratory mice and rats. Lab Anim 43:165–173. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Russell DW. 2006. The condensed protocols from Molecular Cloning: A Laboratory Manual. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press. [Google Scholar]
- 29.Scharmann W, Heller A. 2001. Survival and transmissibility of Pasteurella pneumotropica. Lab Anim 35:163–166. [DOI] [PubMed] [Google Scholar]
- 30.Wang RF, Campbell W, Cao WW, Summage C, Steele RS, Cerniglia CE. 1996. Detection of Pasteurella pneumotropica in laboratory mice and rats by polymerase chain reaction. Lab Anim Sci 46:81–85. [PubMed] [Google Scholar]
- 31.Working Group on Hygiene of the Gesellschaft fur Versuchstierkunde—Society for Laboratory Animal Science (GV-SOLAS) 1999. Implications of infectious agents on results of animal experiments. Lab Anim 33 Suppl 1: 39–87. [PubMed] [Google Scholar]
- 32.Zangenah S, Guleryuz G, Borang S, Ullberg M, Bergman P, Ozenci V. 2013. Identification of clinical Pasteurella isolates by MALDI–TOF—a comparison with Vitek 2 and conventional microbiological methods. Diagn Microbiol Infect Dis 77:96–98. [DOI] [PubMed] [Google Scholar]
- 33.Zorn J, Ritter B, Miller M, Kraus M, Northrup E, Brielmeier M. 2016. Murine norovirus detection in the exhaust air of IVCs is more sensitive than serological analysis of soiled-bedding sentinels. Lab Anim [DOI] [PubMed] [Google Scholar]