Abstract
The severe fever with thrombocytopenia syndrome virus (SFTSV) is an emerging, highly pathogenic bunyavirus against which neither antivirals nor vaccines are available. The SFTSV glycoproteins, Gn and Gc, facilitate viral entry into host cells. Gn and Gc are generated from a precursor protein, Gn/Gc, but it is currently unknown how the precursor is converted into the single proteins and whether this process is required for viral infectivity. Employing a rhabdoviral pseudotyping system, we demonstrate that a predicted signal sequence at the N-terminus of Gc is required for Gn/Gc processing and viral infectivity while potential proprotein convertase cleavage sites in Gc are dispensable. Moreover, we show that expression of Gn or Gc alone is not sufficient for host cell entry while particles bearing both proteins are infectious, and we provide evidence that Gn facilitates Golgi transport and virion incorporation of Gc. Collectively, these results suggest that signal peptidase liberates mature Gc from the Gn/Gc precursor and that this process is essential for viral infectivity and thus constitutes a potential target for antiviral intervention.
Introduction
Bunyaviruses constitute the largest RNA virus family and infect a wide range of hosts, including humans, arthropods and plants. Several emerging bunyaviruses pose a considerable threat to human health as exemplified by Rift Valley fever virus (RVFV) [1], Crimean Congo hemorrhagic fever virus (CCHFV) [2] and severe fever with thrombocytopenia syndrome virus (SFTSV) [3,4], which can cause severe disease in afflicted patients. SFTSV, a novel member of the phlebovirus genus, emerged in 2007 in Central and Eastern China [5,6]. The virus is transmitted from ticks to humans, with human to human transmission occurring on rare instances [7–9], and can induce a severe disease characterized by fever, gastrointestinal symptoms and thrombocytopenia. The case-fatality rate is approximately 10% in China, with the elderly being disproportionately affected, but higher rates have been observed upon SFTSV outbreaks in South Korea [10] and Japan [11]. Moreover, a virus closely related to SFTSV, Heartland virus, has been identified in the US and infection was associated with severe disease [12]. Thus, SFTSV and related viruses are important emerging agents against which at present neither specific antivirals nor vaccines are available.
Bunyaviruses contain a tripartite RNA genome consisting of three single stranded RNA segments of negative polarity termed L, M and S. The M segment encodes for the viral envelope proteins Gn and Gc, which facilitate viral entry into target cells and are the major targets for neutralizing antibodies [13,14]. In addition, the M segments of some bunyaviruses encode a non-structural protein (NSm) but no NSm open reading frame was identified in the M segment of SFTSV. The Gn and Gc proteins of SFTSV have been shown to mediate entry into several cell lines as well as primary dendritic cells and macrophages [15,16]. Entry into dendritic cells depends on the cellular lectin DC-SIGN [15], which can interact with pathogens in a glycan-dependent manner. Moreover, Gn binds to non-muscle myosin heavy chain IIA and expression of this protein is required for efficient SFTSV infection of cell lines [17]. Thus, SFTSV employs the Gn and Gc proteins as keys for entry into target cells and elucidating the processing and function of these proteins might reveal targets for antiviral intervention.
Bunyavirus Gn and Gc are generated from a precursor polyprotein, frequently termed Gn/Gc, which is proteolytically processed into the single Gn and Gc proteins and, in case of CCHFV and several other bunyaviruses, into additional viral glycoproteins and a non-structural protein, NSm [18]. For some bunyaviruses, a role of signal peptidase in Gn/Gc processing has been reported [19–21]. In addition, the activity of a proprotein convertase, subtilisin/kexin-isozyme 1 (SKI-1, also known as site-1 protease (S1P)), was shown to be required for conversion of a precursor form of CCHFV Gn into the mature Gn protein [22,23]. How the SFTSV Gn/Gc precursor is converted into Gn and Gc proteins and whether this process is required for viral infectivity is at present unclear.
Here, we show that processing of the Gn/Gc precursor into Gn and Gc is a prerequisite to infectious viral entry and we provide evidence that Gn promotes virion incorporation of Gc. Moreover, we demonstrate that potential proprotein convertase cleavage sites in Gc are dispensable for processing of the Gn/Gc precursor, while the integrity of a predicted signal sequence at the N-terminus of Gc is essential. These findings indicate that the cellular enzyme signal peptidase liberates Gc from the Gn/Gc precursor and that this process is required for viral infectivity.
Materials and Methods
Cell culture
Human embryonic kidney 293T cells (ATCC CRL-3216) and African green monkey COS-7 (ATCC CRL-1651) kidney cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; PAN Biotech) supplemented with 10% fetal bovine serum (FBS; Biochrome), 100 U/ml penicillin and 100 μg/ml streptomycin (PAN Biotech), and 1% L-glutamine (PAN Biotech). The cells were grown in a humidified atmosphere at 37°C and 5% CO2. The cells were obtained from collaborators and their identity was confirmed by STR DNA typing employing a published protocol [24].
Plasmids
Expression plasmids encoding severe fever and thrombocytopenia syndrome virus glycoprotein (SFTSV-Gn/Gc) [15], Rift valley fever virus glycoprotein (RVFV-Gn/Gc) [15], La Crosse virus glycoprotein (LACV-Gn/Gc) [15], Lassa virus glycoprotein (LASV-GPC) [25], Ebola virus GP (EBOV-GP) and Middle East respiratory syndrome coronavirus spike protein (MERS-S) with and without C-terminal V5 tag [26] have been described previously. A plasmid encoding the vesicular stomatitis virus glycoprotein (VSV-G) has also been described previously [25]. The SFTSV-Gn/Gc-V5 sequence inserted into plasmid pCAGGS served as basis for the generation of further constructs. Expression plasmids for SFTSV-Gn (aa 1–543) and SFTSV-Gc (aa 536–1073) with and without C-terminal V5- or myc-tag were generated by polymerase chain reaction (PCR) and cloning of PCR products into plasmid pCAGGS using Asp718 and XhoI restriction sites. The start codon of Gc within the Gn/Gc precursor sequence was mutated by PCR using the primers 5‘-GATCGGTACCACCATGATGAAAGT GATCTGGTTCAGCAGCC-3‘ and 5‘-CAGTGCCGCACGCGTCTAGGTCTGGCTGCCTCGATCCG-3‘ (CAT to TGC mutation underlined). The PCR product was inserted into plasmid pCAGGS using Asp718 and MluI restriction sites, resulting in mutant mutATG-Gc. To mutate RRxR motifs within Gc, overlap extension PCR was employed. Specifically, primers 5‘-CCAGTGCACCACGGCTGCAGGTCTGGCCATCTCGATCC-3‘ and 5‘-GCCAGACCTGCAGCCGTGGTGCACTGGATGTACAGCCCAGTGATCC-3‘ were used as inner primers to generate mutant mutRRxR-1 (540-AAVV-543) while 5‘-CCAGCGTGGCGGCATGTGCATGGGCAGGCGATTGCCAGAGCGG-3‘ and 5‘-TGCACATGCCGCCACGCTGGTACATCTGGACCGGGCGTCGGGC-3‘ served as inner primers for generation of mutant mutRRxR-2 (648-AACA-651). To generate mutant mutATG-Gc-RRxR-1, the inner primers 5‘-GCCAGACCTGCAGCCGTGGTGCACTGGATGTACAGCCCAGTGATCC-3’ and 5‘-CCAGTGCACCACGGCTGCAGGTCTGGCTGCCTCGATCCGCACG-3‘ were used. The PCR product encoding mutRRxR-2 was directly inserted into pCAGGS using MluI and XhoI restriction sites. In contrast, the PCR products encoding mutRRxR-1 and mutATG-Gc-RRxR-1 were first inserted into pcDNA3.1zeo containing a partial SFTSV fragment followed by insertion of the entire coding sequence into pCAGGS via Asp718 and XhoI restriction sites. The PCR and cloning strategy used for generation of mutants mutRRxR-1 and mutATG-Gc-RRxR-1 was also employed to generate mutants ΔSigP-V5 (Δ536–560) and ΔLAIGLAEG-V5 (Δ555–562). Specifically, primers 5‘-GGCAACCAGGACGACGTGCGGATCGAG GGCTGTGACGAAATGGTGCACGCCGAC-3’ and 5‘-GTCGGCGTGCACCATTTCGTCACAGCCCTCGATCCGCACGTCGTCCTGGTTGCC-3‘ were used as inner primers to generate SFTSVΔSigP-V5 (Δ536–560) while inner primers 5’-CCAGTGATCCTGACCATCTGTGACGAAATGGTGCAC-3’ and 5’-GTGCACCATTTCGTCACAGATGGTCAGGATCACTGG-3’ were employed for constructing SFTSVΔLAIGLAEG-V5 (Δ555–562). The integrity of all PCR-amplified sequences was confirmed by in house (German Primate Center) automated sequence analysis (Sanger method). Plasmids pE-GFP-ER, which encodes calreticulin fused to GFP [27], and pE-GFP-Golgi, which encodes beta-1,4-galactosyltransferase fused to GFP [28], were used to visualize the endoplasmic reticulum and the Golgi apparatus, respectively.
Analysis of glycoprotein expression and cleavage
293T cells were seeded in 6-well plates at 250,000 cells per well, incubated for 24 h and then calcium-phosphate-transfected with 2 μg of the respective glycoprotein expression plasmids. After incubation for 6 h, the cells were washed twice with warm phosphate buffered saline (PBS) and new culture medium was added. At 48 h post transfection, cells were harvested and lysed in 2 x sodium dodecyl sulfate (SDS)-lysis buffer (1.5 ml 1M Tris pH 6.8, 6 ml 80% Glycerol, 10 ml 10 x SDS, 2.5 ml β-mercaptoethanol, 5 ml 1% bromphenol blue, 50 μl 1M EDTA). For analysis of N-glycosylation, PNGase F digest was employed, using commercially available reagents (New England BioLabs, Ipswich, MA, USA). For this, transfected cells were resuspended in denaturing buffer, incubated at 100°C for 10 minutes and then exposed to 500 units PNGase F at 37°C for 1 hour.
For Western blot analysis, cell lysates were separated by SDS polyacrylamide gel electrophoresis (PAGE) and blotted onto nitrocellulose membranes (GE Healthcare, Life Sciences, Freiburg, Germany). Prior to antibody staining, membranes were blocked with 5% milk in PBS with 0.1% Tween. Expression of viral glycoproteins was analyzed with mouse monoclonal antibodies directed against the V5 tag (Invitrogen, Karlsruhe, Germany) or the myc tag [29]. As loading control, β-actin expression was detected using a mouse monoclonal antibody (Sigma-Aldrich, Taufkirchen, Germany). A horseradish peroxidase-(HRP)-conjugated secondary antibody (Dianova, Hamburg, Germany) and a chemiluminescence-based commercially available kit (GE Healthcare Life Sciences, Freiburg, Germany) were used for visualization of bound antibodies. Signals were detected with Intas ChemoCam Image 3.2. Quantification of signal intensities was performed using Fiji/ImageJ 1.51g software.
Inhibition of glycoprotein cleavage
For inhibition of cellular proteases, the following compounds were used: Proprotein convertase inhibitor (Merck Millipore, Darmstadt, Germany), AEBSF (4-(2-aminoethyl)-benzenesulfonylfluoride hydrochloride) and PF-429242 dihydrochloride (4-[(Diethylamino)methyl]-N-[2-(2-methoxyphenyl)ethyl]-N-(3R)-3-pyrrolidinyl-benzamide dihydrochloride) (both Sigma-Aldrich, Taufkirchen, Germany). All inhibitors were diluted in solvent as recommended by the manufacturers and applied in the indicated concentrations. Target cells were treated with the inhibitors from 6 h post transfection until cells were harvested.
Analysis of SFTSV-Gn/Gc-driven host cell entry
For analysis of viral entry a previously described rhabdoviral pseudotyping system was used [15]. In brief, 293T cells were seeded in T-25 cell culture flasks at 500,000 cells/flask and calcium-phosphate-transfected with 6 μg of the glycoprotein expression plasmid. After a 6 h incubation period, the cells were washed twice with warm PBS and fresh culture medium was added. At 30 h post transfection, the cells were transduced at an MOI of 0.2 with a replication-defective vesicular stomatitis virus (VSVΔG) pseudotyped with VSV-G and encoding luciferase and green fluorescent protein (GFP). After a 1 h incubation step, the cells were washed five times with PBS and fresh culture medium was added. Twenty-four hours post transduction rhabdoviral pseudotypes were harvested from the culture supernatants via sterile-filtration through 0.45 μm filters and stored at -80°C. For analysis of infectivity, 293T cells were seeded in 96-well plates at 30,000 cells/well. At 24 h post seeding, cells were incubated with 50 μl pseudotyped virus for 1 h followed by replacement of the medium by fresh culture medium. Transduction efficiency was quantified at 24 h post inoculation by determining luciferase activity in cell lysates using a commercially available beetle-juice luciferase assay kit (p.j.k., Kleinblittersdorf, Germany). For analysis of particle incorporation of viral glycoproteins, sterile-filtered virus stocks were concentrated by centrifugation through a 20% sucrose cushion (17,000 g, 2 h), lysed and analyzed by SDS-PAGE and Western blotting, as described above. A monoclonal anti-VSV-M antibody (Kerafast, Boston, MA, USA) was used to detect the presence of M-protein in viral particles.
Immunofluorescence and confocal microscopy
COS-7 cells were seeded on glass coverslips in 24-well plates at 40,000 cells per well and incubated for 24 h at 37°C. Subsequently, the cells were calcium-phosphate cotransfected with 0.5 μg of plasmids encoding viral glycoproteins (SFTSV-Gn-myc and/or SFTSV-Gc-V5 or SFTSV-Gn/Gc-V5) and marker proteins localized to the endoplasmic reticulum (calreticulin fused to GFP, [27]) or Golgi apparatus (β-1,4-galactosyltransferase fused to GFP, [28]). Transfection of empty pCAGGS plasmid served as negative control. After 6 h of incubation at 37°C, cells were washed twice with PBS and fresh culture medium was added. At 48 h post transfection, the glass coverslips were washed once with PBS followed by fixation in 4% paraformaldehyde for 10 min at RT. After washing three times with PBS, the glass coverslips were incubated in 0.2% Triton X-100 in PBS for 10 min at RT and washed three times with PBS. Prior to antibody staining, samples were blocked for 30 min at 37°C with PBS containing 0.5% Tween and 1% bovine serum albumin (BSA). The primary antibodies mouse monoclonal IgG2a anti-V5 (Invitrogen, Karlsruhe, Germany) and rabbit polyclonal anti-c-Myc (Sigma-Aldrich, Taufkirchen, Germany) were diluted 1:500 in PBS containing 0.5% Tween and 1% BSA and glass coverslips were placed upside down on 25 μl droplets of the antibody dilutions and incubated at RT for 1 h. Glass coverslips were washed three times with PBS and again incubated upside down on 25 μL droplets containing secondary antibodies, Alexa fluor 647 donkey anti-mouse IgG (Life Technologies, Eugene, OR, USA) and Alexa fluor 546 donkey anti-rabbit IgG (Life Technologies, Eugene, OR, USA) diluted 1:1,000 in PBS supplemented with 0.5% Tween and 1% BSA in PBS. After 1 h incubation at RT in the dark, the glass coverslips were washed three times with PBS followed by two washes with distilled H2O prior to mounting onto a microscope slide with mowiol. Fluorescence images were acquired with the confocal laser scanning microscope LSM 5 Pascal Axioskop 2 MOT plus (Carl Zeiss Microimaging GmbH, Göttingen, Germany) using a 63x, NA1.4 Zeiss Plan Apochromat objective and LSM Pascal 5 software version 3. Confocal images were colored and converted to JPEG using the software AxioVision Rel. 4.8 (Carl Zeiss Microimaging GmbH). All images were cropped to 70% of the original image for presentation purposes. No further image modifications were performed.
Statistical analysis
Statistical analysis was performed by Welch’s t-test [30].
Results
The SFTSV-Gn/Gc precursor is efficiently processed into Gn and Gc
We first asked whether expression of the SFTSV-Gn/Gc precursor protein results in production of mature Gn and Gc proteins in 293T cells, a highly transfectable cell line previously used to generate infectious SFTSV-Gn/Gc-bearing vectors [15]. For this, cells were transfected with plasmid encoding the Gn/Gc precursor protein of SFTSV. For comparison, Gn/Gc of RVFV and LACV, other members of the bunyavirus family, were also studied. In addition, the glycoproteins of Ebola virus (EBOV-GP) and Lassa virus (LASV-GPC) were analyzed as positive controls for glycoprotein processing, since they are known to be cleaved by proprotein convertases [31,32]. All glycoproteins studied contained a C-terminal V5 antigenic tag for convenient detection by Western blot.
For EBOV-GP and LASV-GPC, the expected C-terminal processing products of 20 kDa and 35 kDa were detected in addition to the precursor proteins (Fig 1A), indicating that these glycoproteins were processed by host cell proteases. Similarly, signals corresponding to the molecular weight expected for mature Gc were detected in cells expressing SFTSV, RVFV and LACV Gn/Gc proteins (Fig 1A). In addition, a weak signal of the molecular weight expected for the SFTSV-Gn/Gc precursor (116 kDa) was detected. These results indicate that SFTSV-Gn/Gc and other bunyavirus Gn/Gc precursor proteins were efficiently processed into mature proteins under the conditions studied.
Fig 1. Gn and Gc proteins are efficiently expressed from separate plasmids.
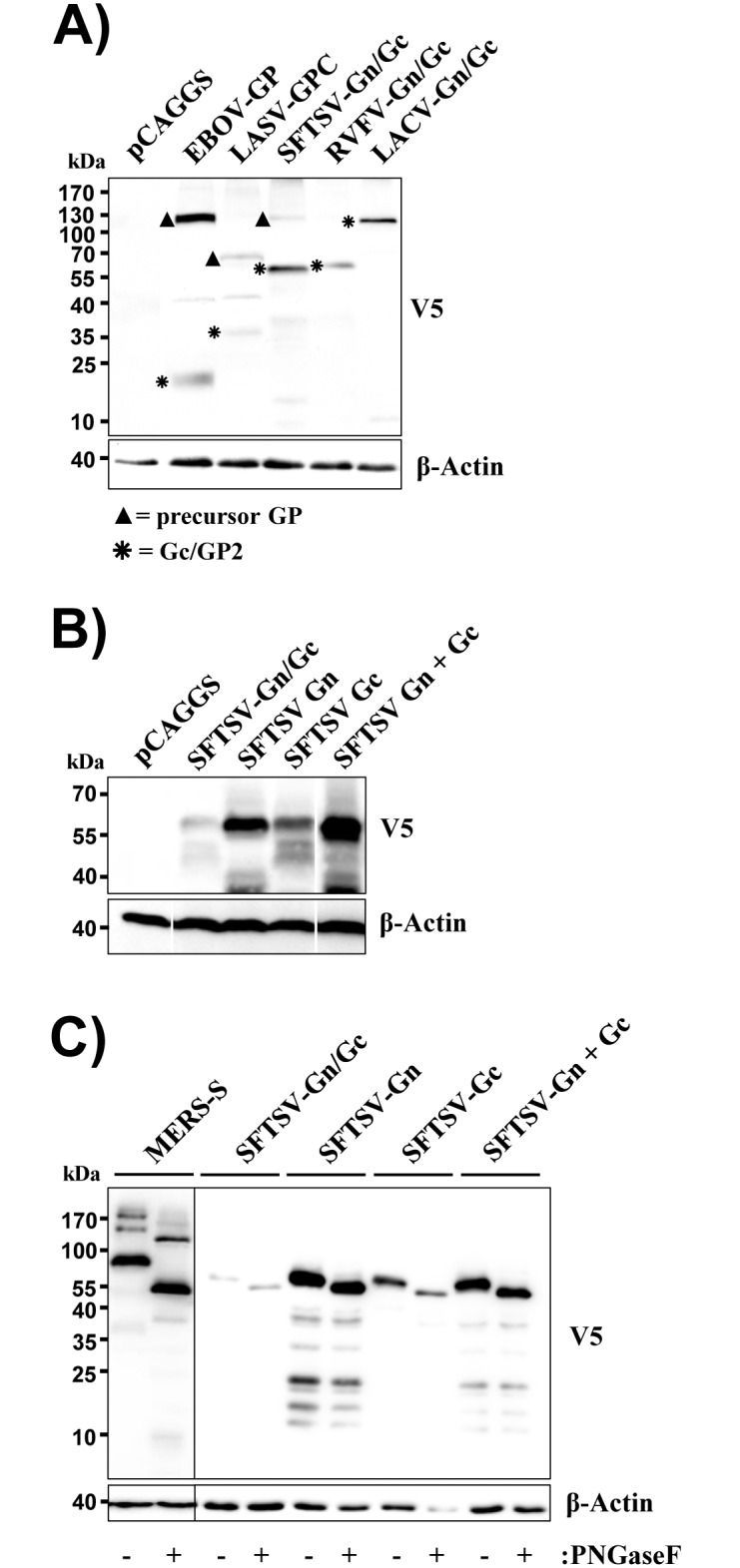
(A) 293T cells were transfected with empty pCAGGS plasmid or pCAGGS encoding the indicated viral glycoproteins with a C-terminal V5 antigenic tag. Glycoprotein expression was analyzed by Western blotting using a V5-specific monoclonal antibody (top panel). Detection of β-actin served as loading control (bottom). Unprocessed glycoprotein precursor proteins are marked with filled triangles while asterisks indicate processed Gc (SFTSV, RVFV, LACV) or GP2 (EBOV, LASV). A single representative blot is shown from which irrelevant lanes were excised. Similar results were obtained in three independent experiments. (B) The experiment was conducted as described for panel A but plasmids encoding both the Gn/Gc precursor and single proteins (all containing a V5 tag) were transfected. A single representative blot is shown from which irrelevant lanes were excised. Similar results were obtained in three independent experiments. (C) The experiment was conducted as described for panel B but cell lysates were treated with PNGaseF to remove N-glycans or Mock treated. As a control for successful removal of N-glycans, cells expressing the spike protein of MERS-CoV were PNGase F treated. The results were confirmed in three independent experiments.
SFTSV-Gn and Gc are both required for infectious entry
We next asked whether Gn and Gc proteins can be expressed separately and whether expression of both proteins is required for host cell entry. Mature Gn and Gc are predicted to exhibit almost identical molecular weights of 60 kDa and 58 kDa, respectively, and the corresponding bands were indeed detected in cells transfected with separate expression plasmids encoding these proteins (Fig 1B). For at present unclear reasons, expression of Gc from a plasmid encoding only the Gc ORF was more efficient than expression from a Gn/Gc encoding plasmid, and Gn was expressed to higher levels than Gc (Fig 1B). Digestion with PNGase F revealed that Gn and Gc expressed from separate plasmids were N-glycosylated (Gn harbors two consensus sequences for N-glycosylation while three consensus sequences are present in Gc), indicating that these proteins enter the secretory pathway (Fig 1C), as expected.
Based on our finding that separate expression of Gn and Gc is feasible, we next asked whether both proteins are required for infectious entry. For this, we employed a vesicular stomatitis virus (VSV)-based pseudotyping system previously shown to be suitable for analysis of SFTSV-Gn/Gc-driven entry [15]. Particles produced in cells transfected with Gn/Gc encoding plasmid incorporated mature Gc (we did not have the means to analyse Gn incorporation) and were readily able to transduce cells (Fig 2A and 2B), although with roughly 100-fold reduced efficiency compared to VSV-G bearing particles, in line with our previous findings [15]. In contrast, expression of Gn alone was not sufficient to render particles infectious, despite efficient particle incorporation of the protein (Fig 2A and 2B). Moreover, particles produced in the presence of Gc did not harbor Gc and were not infectious. Finally, particles produced in cells cotransfected with separate Gn and Gc plasmids were able to transduce cells, although with slightly reduced efficiency as compared to particles from Gn/Gc expressing cells (Fig 2A and 2B). These results suggest that both Gn and Gc are required for infectious SFTSV entry and demonstrate that simultaneous expression of these proteins from separate plasmids is compatible with infectivity.
Fig 2. Gn facilitates virion incorporation of Gc and both proteins are required for host cell entry.
(A-B) Rhabdoviral vectors encoding luciferase and pseudotyped with the indicated glycoproteins with V5 tag were analyzed for incorporation of glycoprotein and VSV matrix (M) protein (panel A) and for transduction of 293T cells (panel B). Transduction efficiency was analyzed by determining luciferase activities in lysates of transduced cells. Transduction measured with particles generated in Gn/Gc expressing cells was set as 1. Particles harboring no glycoprotein (pCAGGS) were used as a control. (C-D) The experiment was carried out as described for panels A-B but the indicated V5- and myc-tagged glycoproteins were used. For infectivity assays (panels B and D), the average of three independent experiments performed with triplicate samples are shown. Error bars indicate standard error of the mean (SEM). Asterisks indicate a statistical significant increase (p<0.05) of relative infectivity with respect to the control (pCAGGS). The virion incorporation data were confirmed in at least four independent experiments. Representative blots are shown in panel C from which irrelevant lanes were excised.
Gn is required for particle incorporation of Gc
Mature Gc was incorporated into particles upon expression of the Gn/Gc precursor protein but not upon expression of Gc alone, as discussed above. These findings posed the question whether Gn facilitates virion incorporation of Gc. To investigate this possibility, particle incorporation of the viral glycoproteins and infectivity were analyzed upon coexpression of vector with combinations of Gn and Gc proteins harboring either the V5 or the myc antigenic tag. These studies confirmed that expression of Gc alone is not compatible with particle incorporation and infectivity (Fig 2C and 2D). In addition, they demonstrated that Gc is incorporated into particles upon coexpression of Gn and showed that simultaneous particle incorporation of both Gn and Gc is associated with infectivity (Fig 2C and 2D).
Gn facilitates transport of Gc from the endoplasmic reticulum into the Golgi apparatus
The finding that Gn is required for virion incorporation of Gc raised the question whether Gn might facilitate the transport of Gc to the site of viral budding, the Golgi apparatus. To address this question, we analyzed the cellular localization of epitope tagged Gn, Gc and Gn/Gc via confocal laser scanning microscopy. Cellular proteins fused to GFP and known to localize to the endoplasmic reticulum (calreticulin) and the Golgi apparatus (β-1,4-galactosyltransferase), respectively, were employed as organelle markers. We found that Gn and Gn/Gc were predominantly localized to the Golgi apparatus while Gc alone was mainly found in the ER (Fig 3A). Moreover, coexpression of Gn with Gc resulted in accumulation of Gc in the Golgi apparatus (Fig 3B), indicating that Gn facilitates transport of Gc into this organelle.
Fig 3. Gn facilitates Gc transport into the Golgi apparatus.
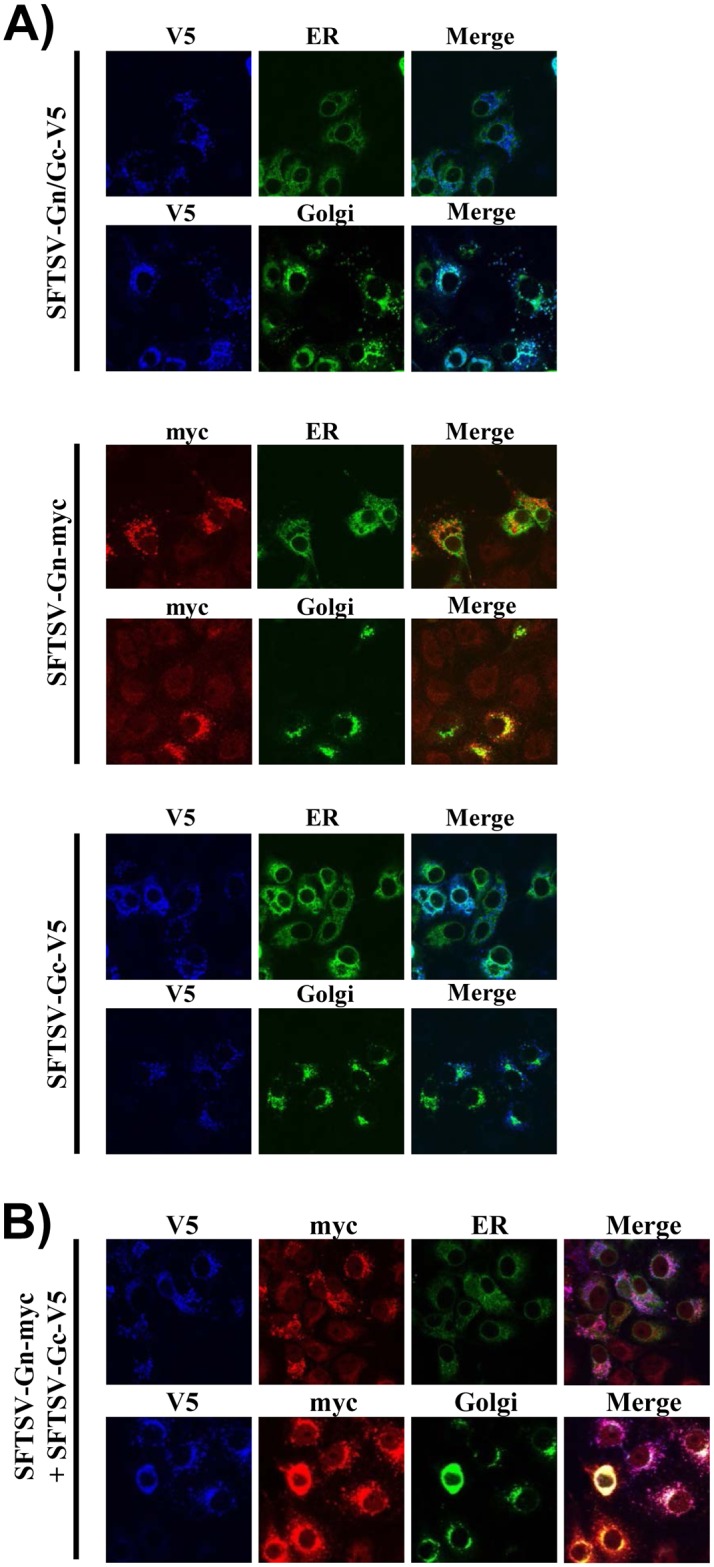
COS-7 cells seeded on glass coverslips were cotransfected with plasmids encoding the indicated glycoproteins (panel A: SFTSV-Gn-myc, SFTSV-Gc-V5 or SFTSV-Gn/Gc-V5; panel B: SFTSV-Gn-myc and SFTSV-Gc-V5) and markers for the endoplasmic reticulum and the Golgi apparatus, respectively. At 48 h post transfection, immunofluorescence staining was performed using antibodies directed against the antigenic tags as primary antibodies and Alexa fluor 647- and Alexa fluor 546-labelled antibodies as secondary antibodies. Fluorescence images were taken at 63x resolution and cropped to 70% of the original image for presentation purposes. No further image modifications were performed. Similar results were obtained in a separate experiment.
RRxR motifs in Gc are dispensable for Gn/Gc processing
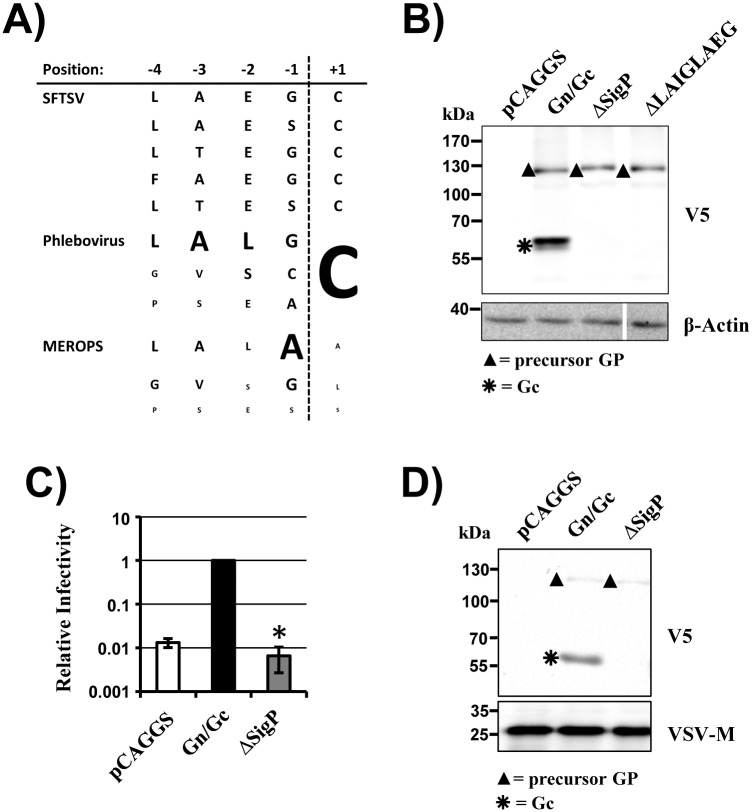
We next asked which protease is responsible for Gn/Gc processing into mature Gn and Gc. In silico analyses predicted signal peptides at the N-termini of Gn and Gc (Fig 4A). Moreover, an RRxR motif, a potential cleavage site for proprotein convertases or related enzymes, was detected at position 5 within Gc (Fig 4A). Cleavage at this motif would be compatible with the observed molecular weight of Gc (Fig 1B). Therefore, we determined whether mutation of this motif impacts Gn/Gc cleavage (mutant RRxR-1). Mutation of an internal RRxR motif (position 113) within Gc served as control (mutant RRxR-2). Moreover, we addressed whether the N-terminal methionine of Gc is required for Gc expression (mutant mutATG-Gc), which would be expected if Gc was translated separately from Gn, potentially due to an unidentified IRES element. Finally, the effect of combined mutation of the first RRxR motif and the AUG was studied (mutant mutATG-Gc-RRxR-1).
Fig 4. RRxR motifs and methionine at position 1 within Gc are dispensable for Gn/Gc processing.
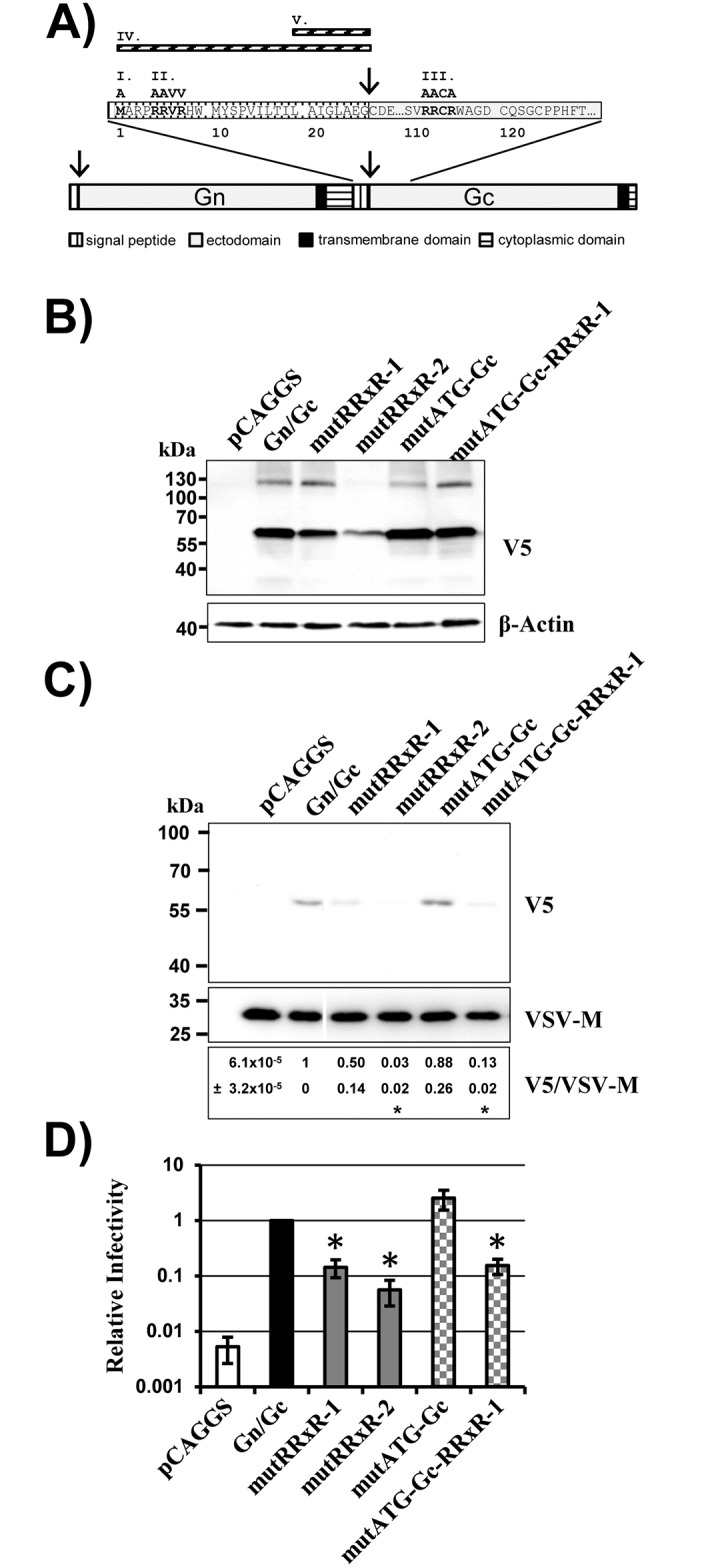
(A) Schematic overview of the SFTSV-Gn/Gc glycoprotein. Domains within Gn and Gc are indicated, the sequences of the putative signal peptide of Gc and internal sequences in Gc surrounding a RRxR motif are shown. Mutations introduced into the Gn/Gc precursor are shown in roman numerals. I., mutant mutATG-Gc, in which the methionine at position 1 was changed to an alanine. II. and III, mutants mutRRxR-1 and mutRRxR-2, in which RRxR motifs within the Gc signal peptide and in the Gc ectodomain were mutated as indicated. IV, mutant ΔSigP, in which the signal peptide of Gc was deleted. V. ΔLAIGLAEG, deletion of the eight amino acids upstream of the internal signal peptidase cleavage site. Numbering of the sequence cutout starts at the first amino acid of the internal Gc signal peptide. (B) 293T cells were transfected with empty plasmid (pCAGGS) or pCAGGS encoding the indicated glycoproteins. Glycoprotein expression was analyzed by Western blotting using a V5-specific antibody (top) or a β-actin antibody (bottom) as loading control. Similar results were obtained in at least three independent experiments. (C) Equal volumes of rhabdoviral vectors pseudotyped with SFTSV Gn/Gc wt or the indicated Gn/Gc mutants were pelleted through a sucrose cushion and pellets were analyzed by Western blot for incorporation of glycoproteins and matrix protein M. A single representative blot is shown from which irrelevant lanes were excised. The results were confirmed in three independent experiments and relative quantification of signal intensities was performed. Numbers represent the mean values of relative V5/VSV-M signal intensity ratios ± SEM. Asterisks denote a statistically significant decrease (p<0.05) of V5/VSV-M ratios with respect to the V5/VSV-M ratio of Gn/Gc wt. (D) The rhabdoviral pseudotypes analyzed in panel C were used for transduction of 293T cells. The average of three independent experiments performed with triplicate samples is shown. Transduction measured with particles generated in Gn/Gc expressing cells was set as 1. Error bars indicate SEM. Asterisks denote a statistical significant decrease (p<0.05) of relative infectivity with respect to particles bearing Gn/Gc.
None of the mutations tested interfered with processing of the Gn/Gc precursor protein (Fig 4B), although mutation of the second RRxR motif reduced protein expression (Fig 4B). Moreover, mutation of either of two RRxR motifs reduced virion incorporation (Fig 4C) of Gc and particle infectivity (Fig 4D). In contrast, mutation of the ATG did not interfere with expression and virion incorporation of Gc or with particle infectivity (Fig 4C and 4D). These results suggest that the RRxR motifs in Gc do not constitute cleavage sites for proprotein convertases or other cellular proteases. Indeed, blockade of proprotein convertases inhibited processing of the LASV-GPC and EBOV-GP (Fig 5A and 5B), as expected [31,32], but had no impact on processing of SFTSV-Gn/Gc (Fig 5C). Similarly, blockade of a serine protease activity previously shown to be required in target cells for efficient Gn/Gc-driven entry [15] was dispensable for Gn/Gc processing (Fig 5). Collectively, our observations indicate that Gn/Gc processing is independent of proprotein convertase activity and RRxR motifs in Gc. Moreover, they suggest that the ATG of the Gc ORF does not serve as a start codon for translation in the context of the Gn/Gc mRNA.
Fig 5. Proprotein convertase activity is dispensable for processing of SFTSV Gn/Gc.
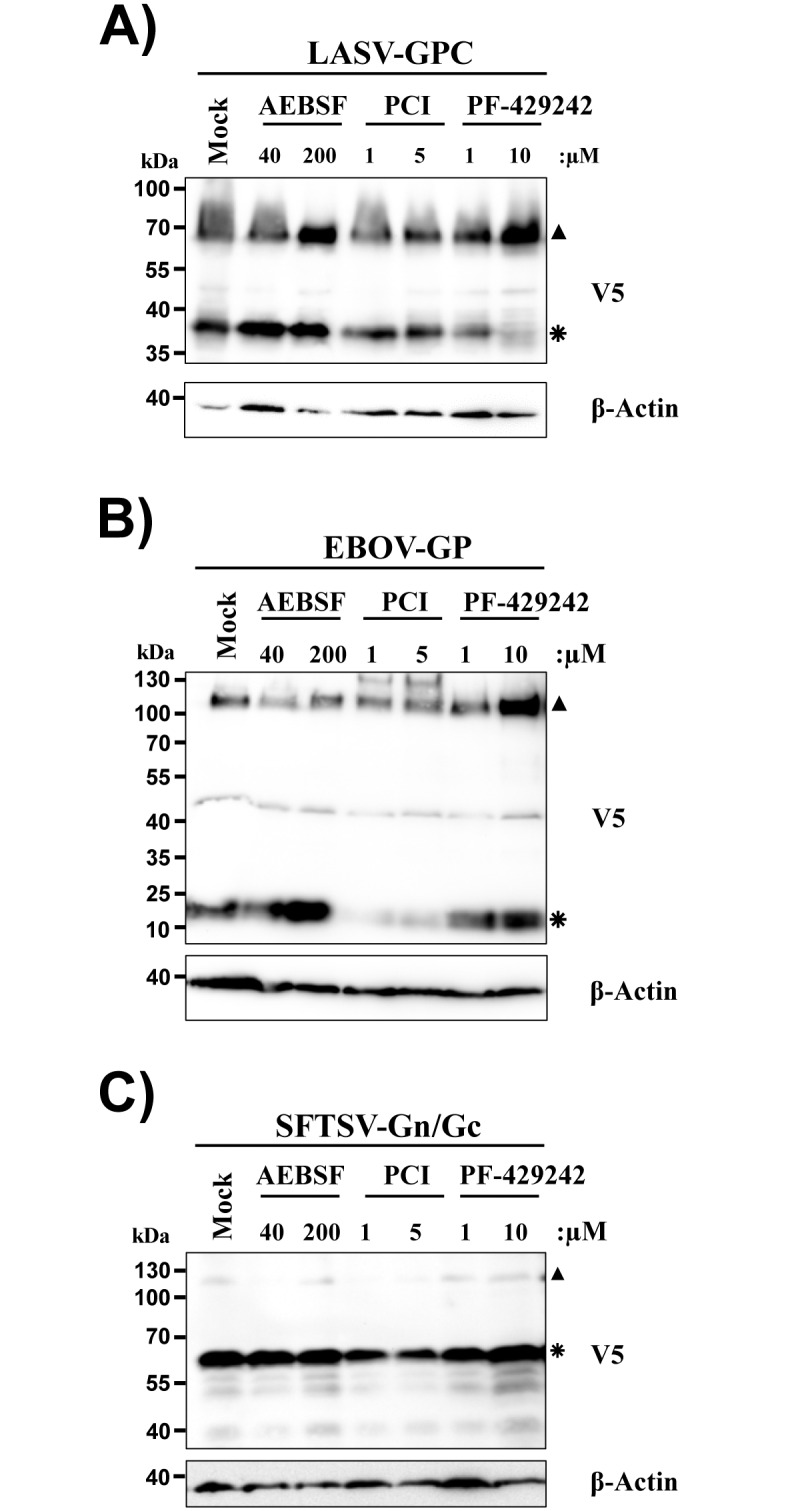
293T cells were transfected with plasmid pCAGGS encoding SFTSV-Gn/Gc, EBOV-GP or LASV-GPC. At 6 h post transfection the indicated concentrations of the furin inhibitor PCI, the serine protease inhibitor AEBSF and the SKI-1 inhibitor PF-429242 were added or the cells were mock treated. Cells were lysed 48 h post transfection and cell lysates were analyzed for LASV-GPC (panel A), EBOV-GP (panel B) or SFTSV-Gn/Gc (panel C) expression by Western blotting using a V5-specific antibody. Expression of β-actin was determined as loading control. Unprocessed glycoprotein precursor proteins are marked with filled triangles while asterisks indicate processing products. Similar results were obtained in at least three independent experiments.
The predicted signal peptide in Gc is essential for Gn/Gc processing
Evidence has been reported that the Gn/Gc proteins of certain bunyaviruses can be processed by signal peptidase during ER import [19–21] and in silico analysis (SignalP 4.0, [33]) also predicted the presence of a signal sequence at the N-terminus of Gc. Moreover, the sequences near the predicted cleavage site in the signal peptide of Gc proteins of other phleboviruses showed high similarity to the corresponding sequences of signal peptides deposited in the MEROPS database (Fig 6A). To investigate the role of the predicted Gc signal peptide in SFTSV-Gn/Gc processing, we deleted the predicted signal peptide or the sequence immediately preceding the predicted cleavage site. Notably, both deletions completely abrogated Gn/Gc processing (Fig 6B) and particle infectivity (Fig 6C) although they were compatible with robust expression and particle incorporation of the unprocessed Gn/Gc precursor (Fig 6C and 6D). Thus, processing of Gn/Gc into mature Gn and Gc proteins depends on the integrity of the predicted signal sequence in Gc and is required for viral infectivity.
Fig 6. Deletion of the Gc signal peptide abrogates Gn/Gc processing and Gn/Gc-driven entry.
(A) Predicted signal peptidase recognition and cleavage sites found in SFTSV-Gc variants (derived from Pubmed Acc. No. BAQ59274, AFK08659, JX462459, JX462462, KC392317). Depicted are the amino acid residues before (-4 to -1) and after (+1) the putative signal peptidase cleavage site. In comparison, the three most frequent amino acid residues for each position found in 103 phlebovirus Gn/Gc sequences and in the 1878 signal peptidase cleavage sites stored in the MEROPS database (http://merops.sanger.ac.uk/cgi-bin/pepsum?id=XS26-001;type=P) are shown. For the phlebovirus and MEROPS sequences the size of the letters corresponds to the frequency in percent of the respective amino acid found in this position. (B) 293T cells were transfected with empty plasmid (pCAGGS) or pCAGGS encoding SFTSV Gn/Gc wt or Gn/Gc with signal peptide deletions. Glycoprotein expression in cell lysates was analyzed by Western blotting using a V5-specific antibody (top) or a β-actin antibody (bottom). A single representative blot is shown from which irrelevant lanes were excised. Similar results were obtained in at least three independent experiments. (C) Rhabdoviral pseudotypes harboring the glycoproteins analyzed in panel A were used for transduction of 293T cells. The average of four independent experiments performed with triplicate samples is shown. Transduction measured with particles generated in Gn/Gc expressing cells was set as 1. Error bars indicate SEM. Asterisks denote a statistical significant decrease (p<0.05) of relative infectivity with respect to particles bearing Gn/Gc. (D) Western blot analysis of the pseudotypes analyzed in panel C employing V5-specific antibody (top) or a VSV M-specific antibody (bottom). Unprocessed glycoprotein precursor proteins are marked with filled triangles while asterisks indicate processing products.
Discussion
Bunyavirus entry into cells is the first essential step in viral replication and the choice of entry factors can constitute a pathogenic molecular signature [34,35]. The M segment of bunyaviruses encodes the proteins required for viral entry into target cells, Gn and Gc but many aspects regarding their biosynthesis and function are poorly understood. The present study focused on Gn and Gc of SFTSV, an emerging bunyavirus. It provides evidence that signal peptidase liberates Gc from the Gn/Gc precursor protein and it demonstrates that both Gn and Gc are required for infectious entry.
SFTSV is a member of the genus phlebovirus and is most closely related to the Bhanja serogroup viruses [36]. The M segment of SFTSV is predicted to encode the canonical Gn/Gc protein while an open reading frame for a NSm protein has not been identified. Our previous work indicated that Gc protein is efficiently expressed from an mRNA encoding Gn/Gc [15]. These findings were confirmed in the present study and raised the question how mature Gn and Gc are generated and whether production of the mature proteins is required for viral infectivity.
We first considered the possibility that two RRxR motifs located at positions 5 and 113, respectively, in SFTSV-Gc might constitute target sites for proprotein convertases, which are known to recognize the following consensus motif, (R/K)-2nX-R2 (with n 0–3 amino acids) [37–39]. Such considerations were supported by the well-established role of the proprotein convertase SKI-1 in the generation of Gn of CCHFV [22,23]. However, neither mutation of the RRxR motifs in the context of Gn/Gc nor inhibition of proprotein convertase activity reduced processing of Gn/Gc. Transduction efficiency was diminished upon mutation of the RRxR motifs but this effect was likely due to reduced virion incorporation of the Gc mutants. These results argue against a role of proprotein convertases in the generation of mature Gc. The impact of the methionine at position 1 of Gc on generation of Gc from Gn/Gc encoding mRNA was also examined. A role for the methionine in Gc expression would have been expected if Gn and Gc were separate translation products, with the first codon of Gc serving as a start codon. Such a scenario would apply, for instance, in case of an undetected IRES element in the Gn/Gc mRNA. However, mutation of the Gc methionine did not interfere with production of Gc from mRNA encoding Gn/Gc. These results indicate that proprotein convertase activity, RRxR motifs within Gc as well as the Gc methionine are dispensable for generation of mature Gc from its precursor, Gn/Gc.
We next asked whether signal peptidase might liberate Gc from Gn/Gc. This question was instigated by sequence analysis, which predicted the presence of a signal sequence at the N-terminus of Gc. Notably, the four amino acid residues preceding the predicted cleavage site in SFTSV-Gc (positions -4 to -1) as well as the amino acid residue at position +1 were found to be highly conserved among phleboviruses and residues -4 to -1 matched those most frequently found in signal peptides of cellular proteins. These findings are in line with previous reports documenting a role for signal peptidase in production of the Gc proteins of Uukuniemi virus, RVFV and Hantaan virus [19–21]. The deletion of the predicted Gc signal peptide in the context of SFTSV-Gn/Gc or the removal of the amino acids preceding the predicted cleavage site abrogated Gn/Gc processing, suggesting that signal peptidase might indeed generate the N-terminus of mature Gc. Although formal proof for this conclusion remains to be provided, our data are most compatible with the concept that SFTSV hijacks signal peptidase for liberation of Gc from its precursor, Gn/Gc.
The availability of SFTSV-Gn/Gc mutants allowed us to investigate whether Gn/Gc processing is required for infectivity. Unprocessed Gn/Gc was incorporated into particles but failed to facilitate transduction of target cells, demonstrating that unprocessed SFTSV-Gn/Gc is inactive. This finding is in line with the observation that expression of Gn or Gc alone in particle producing cells was not sufficient for particle infectivity, while particles generated in the presence of both proteins were infectious. This finding raised the question how Gn and Gc contributed to particle infectivity. Based on in silico analyses and the X-ray structure of the Rift valley fever virus Gc, bunyavirus Gc proteins were identified as class II membrane fusion proteins.[40]. Therefore, a requirement for SFTSV-Gc for particle infectivity was expected while the contribution of SFTSV-Gn was less clear. The observation that Gn expression was required for particle incorporation of Gc suggested that Gn might function as a chaperon, which facilitates appropriate trafficking of Gc. Indeed, immunofluorescence analysis revealed that Gc was mainly localized in the ER when expressed alone while coexpression of Gn resulted in accumulation of Gc in the Golgi apparatus, the site of bunyavirus assembly and budding. These observations are in keeping with previous studies, which ascribed a chaperon function to the Gn protein of Bunyamwera and Uukuniemi virus [41–43]. One can speculate that a hydrophobic domain in the SFTSV-Gn cytoplasmic tail is responsible for the Golgi localization of the protein, since such a domain is also present in the Gn proteins of other phleboviruses and is known to serve as a Golgi retention signal [44–46]. Moreover, it is conceivable that the long cytoplasmic tail of SFTSV-Gn masks a lysine at position -3 in the cytoplasmic tail of SFTSV-Gc, which is conserved between diverse bunyaviruses and is required for ER localization [46]. The chaperon activity of Gn might depend on Gn-Gc interactions formed with high efficiency only after proteolytic separation of these proteins from the Gn/Gc precursor during glycoprotein biogenesis. Such a scenario might explain why particles generated in Gn/Gc expressing cells incorporated more Gc and were more infectious than their counterparts produced in cells transfected with separate Gn and Gc expression plasmids, despite increased Gn expression and particle incorporation under the latter conditions. However, the role of Gn in SFTSV infection might not be limited to that of a chaperon, since a previous study reported that SFTSV-Gn binds to susceptible cells and interacts with the entry factor non-muscle myosin heavy chain IIA [17]. It is therefore conceivable that SFTSV-Gn facilitates trafficking and maybe folding of Gc and binds to entry factors while Gc drives fusion of viral and cellular membranes, a possibility supported by the recent findings that the Gc proteins of RVFV and SFTSV possess a class II membrane fusion protein architecture [47,48].
In sum, the present study highlights similarities between the biogenesis and biological functions of the Gn and Gc proteins of SFTSV and other bunyaviruses and reports an experimental system which allows the convenient analysis of these proteins.
Acknowledgments
We thank M. Winkler for STR DNA typing and F. van Kuppeveld for plasmids pE-GFP-ER and pE-GFP-Golgi.
Data Availability
All relevant data are within the paper.
Funding Statement
SP was supported by the Leibniz Association (Graduate School Emerging Infectious Diseases). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Mansfield KL, Banyard AC, McElhinney L, Johnson N, Horton DL, Hernandez-Triana LM, et al. (2015) Rift Valley fever virus: A review of diagnosis and vaccination, and implications for emergence in Europe. Vaccine 33: 5520–5531. 10.1016/j.vaccine.2015.08.020 [DOI] [PubMed] [Google Scholar]
- 2.Bente DA, Forrester NL, Watts DM, McAuley AJ, Whitehouse CA, Bray M. (2013) Crimean-Congo hemorrhagic fever: history, epidemiology, pathogenesis, clinical syndrome and genetic diversity. Antiviral Res 100: 159–189. 10.1016/j.antiviral.2013.07.006 [DOI] [PubMed] [Google Scholar]
- 3.Xu B, Liu L, Huang X, Ma H, Zhang Y, Du Y, et al. (2011) Metagenomic analysis of fever, thrombocytopenia and leukopenia syndrome (FTLS) in Henan Province, China: discovery of a new bunyavirus. PLoS Pathog 7: e1002369 10.1371/journal.ppat.1002369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu XJ, Liang MF, Zhang SY, Liu Y, Li JD, Sun YL, et al. (2011) Fever with thrombocytopenia associated with a novel bunyavirus in China. N Engl J Med 364: 1523–1532. 10.1056/NEJMoa1010095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Q, He B, Huang SY, Wei F, Zhu XQ. (2014) Severe fever with thrombocytopenia syndrome, an emerging tick-borne zoonosis. Lancet Infect Dis 14: 763–772. 10.1016/S1473-3099(14)70718-2 [DOI] [PubMed] [Google Scholar]
- 6.Liu S, Chai C, Wang C, Amer S, Lv H, He H, et al. (2014) Systematic review of severe fever with thrombocytopenia syndrome: virology, epidemiology, and clinical characteristics. Rev Med Virol 24: 90–102. 10.1002/rmv.1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bao CJ, Guo XL, Qi X, Hu JL, Zhou MH, Varma JK, et al. (2011) A family cluster of infections by a newly recognized bunyavirus in eastern China, 2007: further evidence of person-to-person transmission. Clin Infect Dis 53: 1208–1214. 10.1093/cid/cir732 [DOI] [PubMed] [Google Scholar]
- 8.Liu Y, Li Q, Hu W, Wu J, Wang Y, Mei L, et al. (2012) Person-to-person transmission of severe fever with thrombocytopenia syndrome virus. Vector Borne Zoonotic Dis 12: 156–160. 10.1089/vbz.2011.0758 [DOI] [PubMed] [Google Scholar]
- 9.Tang X, Wu W, Wang H, Du Y, Liu L, Kang K, et al. (2013) Human-to-human transmission of severe fever with thrombocytopenia syndrome bunyavirus through contact with infectious blood. J Infect Dis 207: 736–739. 10.1093/infdis/jis748 [DOI] [PubMed] [Google Scholar]
- 10.Kim KH, Yi J, Kim G, Choi SJ, Jun KI, Kim NH, et al. (2013) Severe fever with thrombocytopenia syndrome, South Korea, 2012. Emerg Infect Dis 19: 1892–1894. 10.3201/eid1911.130792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takahashi T, Maeda K, Suzuki T, Ishido A, Shigeoka T, Tominaga T, et al. (2014) The first identification and retrospective study of Severe Fever with Thrombocytopenia Syndrome in Japan. J Infect Dis 209: 816–827. 10.1093/infdis/jit603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McMullan LK, Folk SM, Kelly AJ, MacNeil A, Goldsmith CS, Metcalfe MG, et al. (2012) A new phlebovirus associated with severe febrile illness in Missouri. N Engl J Med 367: 834–841. 10.1056/NEJMoa1203378 [DOI] [PubMed] [Google Scholar]
- 13.Elliott RM, Schmaljohn CS. (2013) Bunyaviruses In: Knipe DM, Howley PM, editors. Fields Virology. 5th edition ed. Lipincott Williams &Wilkins. [Google Scholar]
- 14.Elliott RM, Brennan B. (2014) Emerging phleboviruses. Curr Opin Virol 5: 50–57. 10.1016/j.coviro.2014.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hofmann H, Li X, Zhang X, Liu W, Kuhl A, Kaup F, et al. (2013) Severe fever with thrombocytopenia virus glycoproteins are targeted by neutralizing antibodies and can use DC-SIGN as a receptor for pH-dependent entry into human and animal cell lines. J Virol 87: 4384–4394. 10.1128/JVI.02628-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tani H, Shimojima M, Fukushi S, Yoshikawa T, Fukuma A, Taniguchi S, et al. (2016) Characterization of Glycoprotein-Mediated Entry of Severe Fever with Thrombocytopenia Syndrome Virus. J Virol 90: 5292–5301. 10.1128/JVI.00110-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun Y, Qi Y, Liu C, Gao W, Chen P, Fu L, et al. (2014) Nonmuscle myosin heavy chain IIA is a critical factor contributing to the efficiency of early infection of severe fever with thrombocytopenia syndrome virus. J Virol 88: 237–248. 10.1128/JVI.02141-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walter CT, Barr JN. (2011) Recent advances in the molecular and cellular biology of bunyaviruses. J Gen Virol 92: 2467–2484. 10.1099/vir.0.035105-0 [DOI] [PubMed] [Google Scholar]
- 19.Andersson AM, Melin L, Persson R, Raschperger E, Wikstrom L, Pettersson RF. (1997) Processing and membrane topology of the spike proteins G1 and G2 of Uukuniemi virus. J Virol 71: 218–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerrard SR, Nichol ST. (2007) Synthesis, proteolytic processing and complex formation of N-terminally nested precursor proteins of the Rift Valley fever virus glycoproteins. Virology 357: 124–133. 10.1016/j.virol.2006.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lober C, Anheier B, Lindow S, Klenk HD, Feldmann H. (2001) The Hantaan virus glycoprotein precursor is cleaved at the conserved pentapeptide WAASA. Virology 289: 224–229. 10.1006/viro.2001.1171 [DOI] [PubMed] [Google Scholar]
- 22.Sanchez AJ, Vincent MJ, Erickson BR, Nichol ST. (2006) Crimean-congo hemorrhagic fever virus glycoprotein precursor is cleaved by Furin-like and SKI-1 proteases to generate a novel 38-kilodalton glycoprotein. J Virol 80: 514–525. 10.1128/JVI.80.1.514-525.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vincent MJ, Sanchez AJ, Erickson BR, Basak A, Chretien M, Seidah NG, et al. (2003) Crimean-Congo hemorrhagic fever virus glycoprotein proteolytic processing by subtilase SKI-1. J Virol 77: 8640–8649. 10.1128/JVI.77.16.8640-8649.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dirks WG, Drexler HG. (2013) STR DNA typing of human cell lines: detection of intra- and interspecies cross-contamination. Methods Mol Biol 946: 27–38. 10.1007/978-1-62703-128-8_3 [DOI] [PubMed] [Google Scholar]
- 25.Simmons G, Reeves JD, Grogan CC, Vandenberghe LH, Baribaud F, Whitbeck JC, et al. (2003) DC-SIGN and DC-SIGNR bind ebola glycoproteins and enhance infection of macrophages and endothelial cells. Virology 305: 115–123. [DOI] [PubMed] [Google Scholar]
- 26.Gierer S, Muller MA, Heurich A, Ritz D, Springstein BL, Karsten CB, et al. (2015) Inhibition of proprotein convertases abrogates processing of the middle eastern respiratory syndrome coronavirus spike protein in infected cells but does not reduce viral infectivity. J Infect Dis 211: 889–897. 10.1093/infdis/jiu407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Jong AS, de MF, Van Dommelen MM, Lanke K, Melchers WJ, Willems PH, et al. (2008) Functional analysis of picornavirus 2B proteins: effects on calcium homeostasis and intracellular protein trafficking. J Virol 82: 3782–3790. 10.1128/JVI.02076-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Jong AS, Visch HJ, de MF, Van Dommelen MM, Swarts HG, Luyten T, et al. (2006) The coxsackievirus 2B protein increases efflux of ions from the endoplasmic reticulum and Golgi, thereby inhibiting protein trafficking through the Golgi. J Biol Chem 281: 14144–14150. 10.1074/jbc.M511766200 [DOI] [PubMed] [Google Scholar]
- 29.Mueller SM, Jung R, Weiler S, Lang SM. (2004) Vpx proteins of SIVmac239 and HIV-2ROD interact with the cytoskeletal protein alpha-actinin 1. J Gen Virol 85: 3291–3303. 10.1099/vir.0.80198-0 [DOI] [PubMed] [Google Scholar]
- 30.Welch BL. (1947) The generalisation of student's problems when several different population variances are involved. Biometrika 34: 28–35. [DOI] [PubMed] [Google Scholar]
- 31.Lenz O, ter MJ, Klenk HD, Seidah NG, Garten W. (2001) The Lassa virus glycoprotein precursor GP-C is proteolytically processed by subtilase SKI-1/S1P. Proc Natl Acad Sci U S A 98: 12701–12705. 10.1073/pnas.221447598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Volchkov VE, Feldmann H, Volchkova VA, Klenk HD. (1998) Processing of the Ebola virus glycoprotein by the proprotein convertase furin. Proc Natl Acad Sci U S A 95: 5762–5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petersen TN, Brunak S, von HG, Nielsen H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8: 785–786. 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- 34.Gavrilovskaya IN, Shepley M, Shaw R, Ginsberg MH, Mackow ER. (1998) beta3 Integrins mediate the cellular entry of hantaviruses that cause respiratory failure. Proc Natl Acad Sci U S A 95: 7074–7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gavrilovskaya IN, Brown EJ, Ginsberg MH, Mackow ER. (1999) Cellular entry of hantaviruses which cause hemorrhagic fever with renal syndrome is mediated by beta3 integrins. J Virol 73: 3951–3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsuno K, Weisend C, Travassos da Rosa AP, Anzick SL, Dahlstrom E, Porcella SF, et al. (2013) Characterization of the Bhanja serogroup viruses (Bunyaviridae): a novel species of the genus Phlebovirus and its relationship with other emerging tick-borne phleboviruses. J Virol 87: 3719–3728. 10.1128/JVI.02845-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seidah NG, Sadr MS, Chretien M, Mbikay M. (2013) The multifaceted proprotein convertases: their unique, redundant, complementary, and opposite functions. J Biol Chem 288: 21473–21481. 10.1074/jbc.R113.481549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seidah NG, Prat A. (2012) The biology and therapeutic targeting of the proprotein convertases. Nat Rev Drug Discov 11: 367–383. [DOI] [PubMed] [Google Scholar]
- 39.Rawlings ND, Barrett AJ, Finn R. (2016) Twenty years of the MEROPS database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res 44: D343–D350. 10.1093/nar/gkv1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spiegel M, Plegge T, Pöhlmann S. (2016) The Role of Phlebovirus Glycoproteins in Viral Entry, Assembly and Release. Viruses 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Melin L, Persson R, Andersson A, Bergstrom A, Ronnholm R, Pettersson RF. (1995) The membrane glycoprotein G1 of Uukuniemi virus contains a signal for localization to the Golgi complex. Virus Res 36: 49–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ronnholm R. (1992) Localization to the Golgi complex of Uukuniemi virus glycoproteins G1 and G2 expressed from cloned cDNAs. J Virol 66: 4525–4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi X, Lappin DF, Elliott RM. (2004) Mapping the Golgi targeting and retention signal of Bunyamwera virus glycoproteins. J Virol 78: 10793–10802. 10.1128/JVI.78.19.10793-10802.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gerrard SR, Nichol ST. (2002) Characterization of the Golgi retention motif of Rift Valley fever virus G(N) glycoprotein. Journal of virology 76: 12200–12210. 10.1128/JVI.76.23.12200-12210.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matsuoka Y, Chen SY, Compans RW. (1994) A signal for Golgi retention in the bunyavirus G1 glycoprotein. The Journal of biological chemistry 269: 22565–22573. [PubMed] [Google Scholar]
- 46.Overby AK, Popov VL, Pettersson RF, Neve EP. (2007) The cytoplasmic tails of Uukuniemi Virus (Bunyaviridae) G(N) and G(C) glycoproteins are important for intracellular targeting and the budding of virus-like particles. Journal of virology 81: 11381–11391. 10.1128/JVI.00767-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dessau M, Modis Y. (2013) Crystal structure of glycoprotein C from Rift Valley fever virus. Proc Natl Acad Sci U S A 110: 1696–1701. 10.1073/pnas.1217780110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Halldorsson S, Behrens AJ, Harlos K, Huiskonen JT, Elliott RM, Crispin M, et al. (2016) Structure of a phleboviral envelope glycoprotein reveals a consolidated model of membrane fusion. Proc Natl Acad Sci U S A. 113:7154–9. 10.1073/pnas.1603827113 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.