Abstract
Ad vectors are promising delivery vehicles for cancer therapeutic interventions. However, their application is limited by promiscuous tissue tropism and hepatotoxicity. This limitation can be avoided by altering the native tropism of Ads so that they can be redirected to the target cells through alternate cellular receptors. The CXCR4 chemokine receptor belongs to a large superfamily of G-protein-coupled receptors and is known to be upregulated in a wide variety of cancers, including breast cancer and melanoma. These receptors have been associated with cancer cell survival, progression, and metastasis. In the current study, an Ad to cancer cells overexpressing CXCR4 by using a bispecific adapter, sCAR-CXCL12, was retargeted. The sCAR-CXCL12 adapter contained the soluble ectodomain form of the native Ad5 receptor (sCAR), which was fused to a mature human chemokine ligand, CXCL12, through a short peptide linker. A dramatic increase in the infectivity of cancer cells using a targeted Ad vector compared with an untargeted vector was observed. Furthermore, sCAR-CXCL12 attenuated Ad infection of liver ex vivo and in vivo and enhanced Ad vector infection of xenograft tumors implanted in immunodeficient SCID-bg mice. Thus, the sCAR-CXCL12 adapter could be used to retarget Ad vectors to chemokine receptor-positive tumors.
Keywords: adapter, adenovirus serotype 5, cancer, hCAR, human coxsackievirus and adenovirus receptor, chemokine receptor, CXCL12, CXCR4, gene therapy, retargeting, viral vector
Introduction
Despite an overall decline in the death rate over the past two decades, cancer remains the second leading cause of death in the US.1 Therefore, there is a need to explore novel therapeutic approaches for treating cancer. Ad-based therapies have captured considerable interest in recent years in developing novel cancer treatment regimens to improve the survival rates in patients with cancer. Attributes such as large DNA incorporation capacity, high gene transfer efficiency, systemic stability, and low pathogenicity in human beings make the Ad a suitable vector for a variety of gene delivery and oncolytic virotherapy applications.2,3 Although attractive as gene delivery vehicles, the efficacy of Ad vectors is compromised because of their broad tissue tropism that leads to off-target uptake of Ads by normal cells.
Ad infection is initiated by the binding of its fiber knob domain to the hCAR in the host cell.4 This receptor is highly, but not exclusively, expressed on parenchyma cells in the liver. Thus, upon systemic administration, Ad is sequestered mainly in the liver, leading to hepatotoxicity.5 In contrast, cancer cells that often represent prime targets for cancer gene therapy are poorly transduced by the Ad vector due to the low and heterogeneous levels of the hCAR present on their cell surface.6 Despite many clinical trials that have indicated vector safety, Ad vectors have shown limited therapeutic activity, in part, because of poor infection efficiency of tumors after systemic delivery.7 Thus, alternative Ad vector approaches that rely on hCAR-independent infection pathways are necessary.
One of the strategies that can be employed to retarget Ad vectors to a nonnative viral receptor to achieve cellular specificity relies on transductional untargeting and retargeting, wherein the initial interaction between the trimeric Ad fiber and hCAR is blocked. This strategy allows for Ad-specific uptake by an indirect cross-linkage formed between an Ad particle and a receptor-specific target cell. Selective transduction of the target cells can be achieved by bispecific fusion proteins or adapter molecules. These have a dual binding specificity that allows them to interact with the knob component of the Ad fiber as well as with the cellular receptors expressed preferentially on the target cell. This selective transduction helps in ablating native tissue tropism of Ad while simultaneously redirecting it to the cell of interest.
The feasibility and efficacy of the adapter-based approach for transductional untargeting and retargeting Ads have already been demonstrated in several studies.8–10 Ad retargeting to cancer cells has been achieved using molecular adapters toward different cellular receptors, including the epidermal growth factor (EGF) receptor,11 the c-ErbB-2 oncoprotein,12 the urokinase-type plasminogen activator receptor,13 the fibroblast growth factor receptor,14 and carcinoembryonic antigen.15 A bispecific adapter that retargeted an Ad to the IL-2 receptor was used to overcome resistance to infection of T lymphocytes.16 In addition, a bispecific adapter enhanced transduction of dendritic cells through CD40.17
In the current study, we directed our efforts to retarget the Ad vector toward CXCR4 chemokine-receptor-expressing cancer cells using the CXCL12 ligand. CXCL12 (also known as SDF-1) is a CXC chemokine that is widely expressed in a variety of tissue types and functions as a potent chemoattractant for immature and mature hematopoietic cells.18 CXCR4 is a seven-membrane spanning G-protein-coupled receptor for CXCL12, whose role is implicated in a wide variety of solid19 and hematopoietic tumors,20 including breast cancer21 and melanoma.22 Altered expression of the CXCR4 receptor drives the signaling process of cancer cell migration and invasion23 and is associated with metastasis.24 CXCR7 is an alternate receptor for CXCL12, which is overexpressed in endothelial cells associated with tumors.25 Similar to CXCR4, CXCR7 plays a role as a G-protein-coupled receptor in regulating immunity, angiogenesis, stem cell trafficking, and mediating organ-specific metastases of cancer.26 However, CXCR7 may also function as a decoy receptor in which G-protein signaling is not activated. CXCR7 can signal through non-G-protein pathways.27 In addition, CXCR7 can heterodimerize with CXCR4 to mediate CXCL12 signaling.27
To retarget Ads, a novel bispecific adapter was generated, which embodies fusing a sCAR and human chemokine ligand CXCL12. We hypothesized that sCAR-CXCL12 would redirect Ad vectors to the chemokine receptors overexpressed on cancer cells. This study reports that sCAR-CXCL12 recombinant fusion protein efficiently retargets Ad5 vector to human cancer cells overexpressing CXCR4. In vivo, our data demonstrate tumor-selective targeting based on chemokine receptor binding coupled with hepatic untargeting of Ad using sCAR-CXCL12 adapter molecule. Overall, sCAR-CXCL12 could be used to improve the transductional ability of Ad vectors toward receptor-positive tumors, thereby limiting hepatic infection.
Materials and methods
Approval to perform the experimental protocols using human tissue samples in this study was obtained from the institutional review board at the University of Alabama at Birmingham. All patients provided written informed consent for the collection of samples and subsequent analysis. Fresh human liver samples were collected from the Department of Surgery (the University of Alabama at Birmingham) at the time of a medically indicated therapeutic or diagnostic procedure. Only tissue samples no longer required for patient management or evaluation and to be discarded were used. All animals used in this study received humane care based on guidelines set by the American Veterinary Medical Association. The experimental protocols involving live animals were reviewed and approved by our Institutional Animal Care and Use Committee.
Cell lines
The human cancer cell lines, MDA-MB-435S, ZR-75-1, BT-20, and MCF-7, and a human immortalized non- tumorigenic epithelial cell line, MCF-12A, were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). While the ZR-75-1, BT-20, and MCF-7 cell lines are of human breast cancer origin, the MDA-MB-435S cell line is likely derived from a melanoma.28 All the cancer cell lines were maintained in DMEM (Corning Life Sciences, Oneonta, NY, USA), containing 10% FBS (Gemini Bioproducts, Woodland, CA, USA) and 1% antibiotic–antimycotic solution (Corning Life Sciences). MCF-12A cells were cultured in DMEM/F12 (Corning Life Sciences) containing 5% donor horse serum (Atlanta Biologicals, Lawrenceville, GA, USA), 0.5 μg/mL hydrocortisone (Sigma-Aldrich, St. Louis, MO, USA), 0.01 mg/mL bovine insulin (Sigma-Aldrich), 100 ng/mL cholera toxin (Sigma-Aldrich), and 20 ng/mL human EGF (Thermo Fisher Scientific, Waltham, MA, USA). All the cell lines were maintained at 37°C in a humidified 5% CO2 atmosphere.
Ad vectors
Purified Ad serotype 5 (Ad5) and Ad serotype 3 (Ad3) fiber knob proteins were produced as described earlier29 and provided by Dr Anton V Borovjagin (the University of Alabama at Birmingham). An Ad vector (Ad5-CMV-GFP-luc), which was E1, E2, and E3 deleted and carried the GFP and firefly luc reporter under the control of the cytomegalovirus (CMV) promoter, and a wild-type Ad5 vector were constructed as described earlier.29 The propagation of Ad vectors was performed using the permissive 293 human embryonic kidney cell line and the A549 human lung adenocarcinoma epithelial cell line followed by purification according to an established protocol.30
Construction and purification of a recombinant bispecific adapter
We generated a novel recombinant adapter molecule that was composed of an ectodomain portion of the human Coxsackie and Ad receptor (amino acid sequence 1–236; GenBank accession BC010536, bp 76–783) followed by a 5-amino-acid peptide linker (GGPGS), a 6-His tag sequence (for detection and purification), fused to the mature human chemokine CXCL12/SDF-1a sequence (amino acid sequence 22–89; GenBank accession NM_199168.3, bp 156–359). Amino acids 22–89 represent the mature form of CXCL12. CXCL12a is processed from an initial translation product of 89 amino acids, in which the first 21 amino acids at the N-terminus serve as a signal peptide for secretion that is subsequently cleaved to produce the mature form.
The cDNA coding for the recombinant fusion protein was assembled in a baculovirus expression plasmid, and its integrity was confirmed by DNA sequencing. Linearized plasmid DNA was then used to establish stable insect Sf9 cell line producing sCAR-CXCL12 as a secreted protein. A positive clone, constitutively secreting the product, was expanded, and the fusion protein was purified by Ni column chromatography. Fractions were eluted from the column with 250 mM imidazole and dialyzed against buffer A (pH 8.0) containing 20 mM Tris–HCl, 300 mM NaCl, and 5% glycerol. Purified recombinant protein was subjected to a 12% sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) followed by Western blot analysis using anti-His tag (EMD Millipore, Billerica, MA, USA), anti-hCAR (R&D Systems, Minneapolis, MN, USA), and anti-CXCL12 (Novus Biologicals, Littleton, CO, USA) primary antibodies. This experiment was used to confirm the identity of the target protein regarding purity and size.
Flow cytometry analysis
The cell surface expression of chemokine receptors CXCR4 and CXCR7, as well as hCAR and integrins αvβ3 and αvβ5, were examined by a panel of human cancer and immortalized non-tumorigenic cell lines using flow cytometry. The cells were harvested from the cell culture flasks using 1× Versene (Thermo Fisher Scientific) and washed twice with PBS. Subsequently, cells were incubated in the dark with either PBS (unstained) or different primary antibodies, including 1) PE-labeled mouse antihuman CXCR4 monoclonal antibody (BD Biosciences, San Jose, CA, USA), 2) PE-labeled mouse antihuman CXCR7 monoclonal antibody (BD Biosciences), 3) PE-labeled mouse IgG2a κ isotype control (BD Biosciences), 4) PE-labeled mouse antihuman hCAR monoclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA), 5) PE-labeled mouse IgG1 κ isotype control (Abcam, Cambridge, MA, USA), 6) FITC-labeled mouse antihuman integrin αvβ3 monoclonal antibody (EMD Millipore), 7) FITC-labeled mouse antihuman integrin αvβ5 monoclonal antibody (EMD Millipore), and 8) FITC-labeled mouse IgG1 κ isotype control (BD Biosciences). The antibodies were diluted in PBS and incubated with the cells at 4°C for 30 min. Following incubation, the cells were washed twice with PBS, resuspended in 0.4 mL PBS, and analyzed by flow cytometry using a FACSCalibur instrument (BD Biosciences).
ELISA
The binding specificity of the sCAR-CXCL12 fusion protein to the Ad5 fiber knob protein was tested by an ELISA. Ad5 fiber knob protein was adsorbed on a Nunc Maxisorp 96-well plate followed by the overnight incubation at 4°C. Following extensive washes with Tris-buffered saline containing 0.05% Tween-20 (TBS-T) and blocking with 2% bovine serum albumin in TBS-T, the sCAR-CXCL12 fusion protein was added at varying concentrations ranging from 0 to 100 ng. This addition was followed by incubation with anti-His tag monoclonal antibody (Qiagen, Valencia, CA, USA) for 2 h at room temperature. After the addition of HRP-conjugated goat anti-mouse secondary antibody (Bio-Rad, Hercules, CA, USA) and incubation for 2 h at room temperature, colorimetric reaction was performed by TMB substrate (BD Biosciences) and absorbance was read at 450 nm on a micro-plate reader (Molecular Devices, Sunnyvale, CA, USA). A parallel experiment was run using fiber knob protein derived from Ad3 that served as a negative control.
Biological activity of the adapter protein
The biological activity to determine the optimum ratio of sCAR-CXCL12 adapter to Ad5-CMV-GFP-luc was determined by an infectivity assay. MDA-MB-435S cells (1 × 105 cells/well) growing in culture were plated in a 24-well tissue culture plate. The cells were allowed to attach by overnight incubation at 37°C. Following day, Ad5-CMV-GFP-luc (100 ifu/cell) was complexed with sCAR-CXCL12 at increasing concentrations (0, 1, 10, 100, and 1000 ng) for 20 min at room temperature. The sCAR-CXCL12–Ad conjugates (in a final volume of 250 μL with DMEM containing 2% FBS) were then added to monolayers of MDA-MB-435S cells and incubated for 4 h at 37°C. Afterward, the medium was replaced with 1 mL of DMEM containing 10% FBS. At 48 h post infection, quantitative analysis was performed by flow cytometry to determine the percentage of GFP reporter gene-expressing cells. To achieve maximum retargeting efficacy, the optimum time of Ad5 infection in the absence and presence of sCAR-CXCL12 adapter molecule was determined by infection of MDA-MB-435S cells. Briefly, 1 × 105 cells/well were infected in the presence and absence of retargeted Ad vector at various time points (0, 5, 15, 30, 60, 120, and 240 min) in a minimum volume of reduced serum culture medium (DMEM containing 2% FBS). Following incubation for the respective time periods at 37°C, medium containing virus or virus–adapter complexes was replaced with culture medium containing 10% FBS, and cells were allowed to incubate for 48 h at 37°C for maximal expression of the reporter gene. Infectivity was determined by fluorescence microscopy and flow cytometry.
Blocking assay
Before infection, MDA-MB-435S cells were pre-incubated with increasing concentrations of a mouse monoclonal CXCR4 blocking antibody (Abcam) for 30 min at 4°C. After washing with PBS, Ad5-CMV-GFP-luc (100 ifu/cell) pre-complexed with sCAR-CXCL12 (100 ng/108 ifu) was added to the cell monolayers and incubated for 1 h at 4°C. Following infection, cells were harvested, and DNA was extracted using QIAamp DNA Mini Kit (Qiagen) for the determination of viral E4 copy number using Ad5 E4-specific primers (upstream, 5′-d(GGGTCGCCACTTAATCTACCT)-3′; downstream, 5′-d(GCAAGGCGCTGTATCCAA)-3′) by real-time polymerase chain reaction (PCR).
Ad retargeting on cultured cell lines
To determine the retargeting efficacy of bispecific adapter, sCAR-CXCL12, to cells expressing the CXCR4 chemokine receptor, the cells (MDA-MB-435S, MCF-7, ZR-75-1, BT-20, and MCF-12A) were plated at a density of 1 × 105 cells/well in a 24-well tissue culture plate. Following overnight incubation, cell monolayers were infected with Ad5-CMV-GFP-luc at increasing MOI (0, 1, 10, 100, and 1000 ifu/cell) in the absence or presence of sCAR-CXCL12 (100 ng/well) in a minimal volume of medium containing 2% FBS. Before infection, Ad was allowed to form complexes with sCAR-CXCL12 protein for 20 min at room temperature. The medium was changed by aspiration after 4 h incubation at 37°C and replaced with fresh DMEM containing 10% serum. After 48 h incubation, the cells were examined by fluorescence microscopy and the percentage of GFP-positive cells was determined by flow cytometry.
Ad untargeting of the liver in a tissue slice model
The samples obtained were rinsed in ice-cold saline and stored in media on ice until brought to the laboratory for the experimental procedure. Thin liver slices were cut using the Krumdieck tissue slicing system (Alabama Research and Development, Munford, AL, USA) according to the manufacturer’s instructions as described earlier.31 Briefly, 8 mm coring tool was used to make 8 mm diameter core of human liver tissue. The tissue was then placed in a slicer filled with ice-cold culture media. Slices (250 μm thick) were cut with the reciprocating blade at 30 rpm. Cell number for tissue slices was estimated at 1×106 cells per slice based on a ten-cell thick slice (250 μm) and 8 mm slice diameter. For infectivity experiments, tissue slices were placed into six-well plates (one slice per well) containing 2 mL of complete culture media (William’s medium E with 1% antibiotics, 1% L-glutamine, and 10% FBS). Before infection of liver slices, Ad5-CMV-GFP-luc (~100 ifu/cell) was pre-complexed with sCAR-CXCL12 (100 ng/108 ifu) bispecific adapter. At 4 h post infection, virus–adapter complexes were removed, and slices were incubated in fresh culture medium for 48 h at 37°C. Infectivity was determined by using a standard luc assay. Alternatively, liver slices were infected with wild-type Ad5 virus (~100 ifu/cell) alone or conjugated with sCAR-CXCL12 (100 ng/108 ifu). Viral copy number was determined by measuring the E4 gene using real-time PCR from DNA samples extracted from the medium at day 2 and 4 post infection.
Systemic Ad administration to measure hepatic untargeting
Female SCID-bg mice at 4–6 weeks of age were obtained from Charles River Laboratories (Wilmington, MA, USA). Before i.v. administration, Ad5-CMV-GFP-luc (1 × 108 ifu/mouse) was pre-incubated for 1 h at room temperature in the absence or presence of sCAR-CXCL12 (5 μg) in a volume of 100 μL. Three days following virus administration, mice were anesthetized with ketamine/xylazine, injected intraperitoneally with D-luciferin (300 μL; 125 mg/kg body weight), and scanned using an IVIS 100 imaging system (PerkinElmer, Waltham, MA, USA). Bioluminescence images were analyzed using Living Image software version 3.0 (PerkinElmer). Regions of interest (ROI) were drawn over the tumor and liver area, and total photon counts were calculated. Following imaging, mice were sacrificed, and livers were extracted, snap frozen, and stored at −80°C. The frozen sections were processed later for DNA extraction and determination of viral E4 copy number by real-time PCR.
Ad retargeting following intratumoral (i.t.) injection into subcutaneous (s.c.) tumor xenografts
S.c. xenografts were established in 4–6-week-old female SCID-bg mice by injecting 4×106 MDA-MB-435S cells into the flanks. When tumors reached ~100 mm3 in size, Ad5-CMV-GFP-luc (1 × 108 ifu/mouse) pre-incubated in the absence or presence of sCAR-CXCL12 (5 μg) was injected intratumorally. At day 3, 6, and 9 post injection, animals were imaged noninvasively by an IVIS optical imaging system. Mice were sacrificed immediately after the final imaging, and tumors and livers were removed, snap frozen, and stored at -80°C. DNA was extracted from the frozen sections to determine viral E4 copy number by real-time PCR.
Statistical analysis
All experiments were performed at least three times. Data are presented as mean ± standard error of the mean of the data points. Statistical analysis was performed using Student’s t-test, using GraphPad Prism 5.0 software (GraphPad Software; La Jolla, CA, USA). Data were considered statistically significant when p<0.05.
Results
The human cancer cell line panel has high levels of CXCR4 chemokine receptor
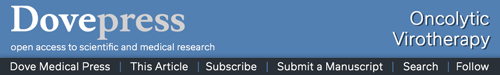
We analyzed the cell surface expression of the CXCR4 receptor in a panel of human cancer cell lines (MDA-MB-435S, ZR-75-1, BT-20, and MCF-7) by flow cytometry. Since hCAR represents the primary receptor for Ad5 vectors and αvβ3 and αvβ5 integrins are known to trigger virus internalization into the host cell, we also determined the surface expression of each molecule. A human epithelial cell line (MCF-12A) was used as a control. The results of the flow cytometry analysis are shown in Figure 1 and summarized in Table 1. These results demonstrated that the cancer cell lines tested expressed high to moderate levels of the CXCR4 receptor as shown in Figure 1A. However, CXCR7 expression in the cancer cell lines was found to be low (Figure 1B), with only the MCF-7 and ZR-75-1 cell lines exhibiting detectable levels. Likewise, hCAR expression in the cancer cell lines was found to be undetectable (Figure 1C). In addition, these cells showed low to moderate surface expression levels of αvβ3 and αvβ5 (Figure 1D). In contrast to these results, MCF-12A cells expressed low levels of the CXCR4, CXCR7, and integrins αvβ3 and αvβ5, while hCAR expression was high. Based on the low levels of CXCR7 expression detected in the cancer cell panel, we focused on targeting CXCR4 in cancer cells.
Figure 1.
Flow cytometry analysis of CXCR4, CXCR7, hCAR, and integrin αvβ3 and αvβ5 cell surface expression.
Notes: Evaluation of antigen expression using an anti-CXCR4 antibody (A), an anti-CXCR7 antibody (B), an anti-hCAR antibody (C), and anti-integrin αvβ3 and αvβ5 antibodies (D) in human cancer cell lines, MDA-MB-435S, ZR-75-1, BT-20, and MCF-7, and in the immortalized non-tumorigenic breast epithelial cell line MCF-12A.
Abbreviation: hCAR, human Coxsackie and adenovirus receptor.
Table 1.
Summary of surface antigen expression in cancer cell lines
| Antibody | Cell line
|
||||
|---|---|---|---|---|---|
| MDA-MB-435S | ZR-75-1 | BT-20 | MCF-7 | MCF-12A | |
| Unstained | 1.95 | 2.56 | 3.46 | 3.60 | 3.03 |
| Isotype (IgG2a κ) | 1.91 | 2.65 | 3.46 | 3.57 | 3.03 |
| Anti-CXCR4 | 6.41 | 6.48 | 11.27 | 11.17 | 3.78 |
| Unstained | 3.24 | 2.65 | 8.72 | 3.14 | 4.25 |
| Isotype (IgG2a κ) | 3.00 | 2.98 | 10.20 | 2.93 | 5.70 |
| Anti-CXCR7 | 3.03 | 4.19 | 10.68 | 4.99 | 4.86 |
| Unstained | 2.38 | 2.02 | 2.23 | 1.80 | 3.13 |
| Isotype (IgG1 κ) | 3.71 | 3.26 | 2.73 | 2.66 | 3.64 |
| Anti-hCAR | 3.98 | 2.52 | 2.63 | 2.81 | 9.80 |
| Unstained | 1.05 | 1.31 | 3.10 | 1.26 | 2.18 |
| Isotype (IgG1 κ) | 1.34 | 2.18 | 3.28 | 1.51 | 3.24 |
| Anti-anb3 | 4.38 | 2.42 | 3.56 | 2.09 | 5.48 |
| Anti-anb5 | 3.64 | 6.22 | 17.30 | 4.00 | 4.36 |
Notes: Each cancer cell line was analyzed for the cell surface expression of CXCR4, CXCR7, hCAR, integrin anb3, and integrin anb5. The data are represented as MFI of gated unstained cells and cells stained with antigen-specific antibodies or isotype control antibodies.
Abbreviations: hCAR, human Coxsackie and adenovirus receptor; MFI, mean fluorescence intensity.
Production and purification of a bispecific adapter protein sCAR-CXCL12
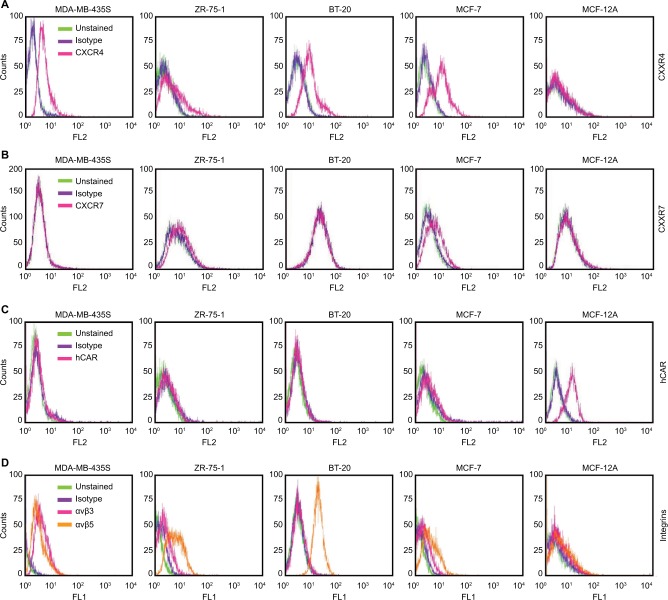
To target the cell surface expression of the CXCR4 receptor on the panel of cancer cells, we generated a recombinant bispecific adapter molecule, sCAR-CXCL12, by combining ectodomain of hCAR with CXCL12, the chemokine ligand of CXCR4 (Figure 2A). We assembled a recombinant cDNA coding for the recombinant fusion protein, sCAR-CXCL12, consisting of the 236-amino-acid C-terminal portion of human hCAR fused to a 72-amino-acid mature form of human CXCL12 ligand through a 5-amino-acid peptide linker and a 6-His detection/purification tag. We confirmed its integrity by DNA sequencing. Recombinant protein was expressed using a baculovirus vector system in Sf9 cells and purified from the supernatant by Ni column chromatography. Analysis of the purified protein by SDS-PAGE followed by Coomassie staining (Figure 2B) and Western blot (Figure 2C) showed a protein band of the expected size of ~35 kDa. A second higher band was also observed, which could be explained by glycosylation of the hCAR ectodomain. Glycosylation does occur in insect cells, which is similar but not identical to mammalian cells.32 Similar Western blot bands were obtained using an anti-CAR antibody and an anti-CXCL12 antibody (Figure 2D).
Figure 2.
Expression and purification of the bispecific adapter (sCAR-CXCL12).
Notes: The structure of the sCAR-CXCL12 recombinant molecule was composed of an ectodomain portion of the human Coxsackie and Ad receptor followed by a 5-amino-acid peptide linker, a 6-His tag, fused to the mature human chemokine CXCL12 amino acid sequence (A). SDS-PAGE analysis and Coomassie staining of the sCAR-CXCL12 recombinant adapter protein purified by Ni column chromatography (B) and Western blot analysis using an anti-His tag antibody (C). Comparison of hCAR, CXCL12, and His tag antigen detection by Western blot analysis in recombinant sCAR-CXCL12 protein (D). M, molecular weight marker; numbers indicate molecular weight in kDa.
Abbreviations: Ad, adenovirus; Ad5, Ad serotype 5; hCAR, human Coxsackie and adenovirus receptor; His, histidine; PAGE, polyacrylamide gel electrophoresis; sCAR, soluble ectodomain form of the native Ad5 receptor; SDS, sodium dodecyl sulfate.
sCAR-CXCL12 shows specificity toward Ad5 fiber knob domain
The specificity of interaction between the Ad5 vector and adapter is a critical factor that determines the efficacy and specificity of vector retargeting. To characterize the adapter protein and determine its binding specificity toward fiber knob domain derived from Ad5, we used an indirect ELISA. In the experiment shown in Figure 3, recombinant Ad5 or Ad3 knob was adsorbed to ELISA plates and the plates were subsequently incubated with dilutions of sCAR-CXCL12. The binding interaction was detected using an anti-His tag antibody. The results indicated a dose-dependent increase in the absorbance corresponding to the binding specificity of sCAR-CXCL12 adapter to the Ad5 knob protein. However, sCAR-CXCL12 failed to interact with the fiber knob domain from Ad3 that served as a negative control. These results suggest that sCAR-CXCL12 protein interacts with Ad5 fiber in a specific manner.
Figure 3.
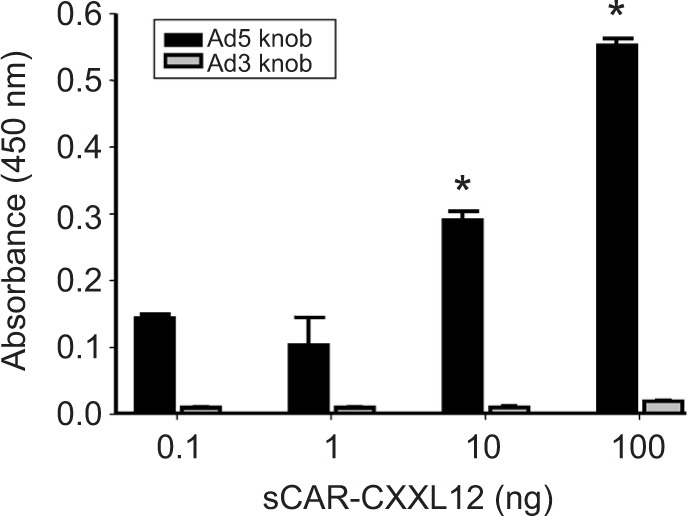
Binding specificity of the bispecific adapter (sCAR-CXCL12).
Notes: Purified sCAR-CXCL12 was tested in ELISA against recombinant Ad5 fiber knob (Ad5 knob) and recombinant Ad3 fiber knob (Ad3 knob). Each antigen was absorbed at 300 ng/well. The wells were incubated with dilutions of the adapter protein. The binding was detected by treating the wells with an anti-His tag antibody followed by an anti-rabbit IgG antibody conjugated with HRP and color reaction with TMB substrate. The color intensity was measured as optical density at 450 nm. Numbers indicate the mean ± SEM of absorbance from three replicate dishes. The concentration results were compared using a Student’s t-test; the differences were considered statistically significant (*) if p<0.05.
Abbreviation: Ad, adenovirus; Ad3, Ad serotype 3; Ad5, Ad serotype 5; ELISA, enzyme-linked immunosorbent assay; His, histidine; HRP, horseradish peroxidase; sCAR, soluble ectodomain form of the native Ad5 receptor; SEM, standard error of the mean; TMB, 3,3′,5,5′-tetramethylbenzidine.
Characterization of sCAR-CXCL12-mediated Ad infectivity
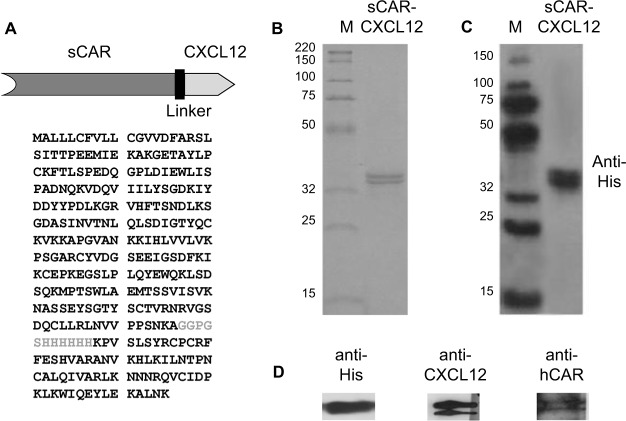
Determining an optimal adapter dose is important to achieve maximal transduction of CXCR4-expressing cancer cells. The optimal sCAR-CXCL12 to Ad ratio was determined by infecting MDA-MB-435S cells for 4 h with Ad5-CMV-GFP-luc (100 ifu/cell) in the presence of increasing concentrations of sCAR-CXCL12. The infected cells were observed for GFP expression following 48 h of infection. There was a remarkable increase in the number of GFP-infected cells from low to high amounts of the sCAR-CXCL12 adapter as evident by flow cytometry analysis (Figure 4A). The flow cytometry data showed that sCAR-CXCL12 to Ad ratio was optimal at 100 ng sCAR-CXCL12 per 1 × 108 ifu virus. At this ratio, sCAR-CXCL12 increased the infectivity of Ad5-CMV-GFP-luc virus in MDA-MB-435S cells at an multiplicity of infection (MOI) of 100 ifu/cell from 6.3% GFP-positive using an uncomplexed virus to ~80% GFP-positive using adapter-complexed virus. These results suggest that the genetically engineered adapter molecule is produced in the desired conformation with an intact CXCL12 ligand available to interact with the CXCR4 receptor. Using the sCAR-CXCL12 adapter led to an enhanced dose-dependent transduction of the CXCR4-positive cancer cells compared with the untargeted vector.
Figure 4.
Infection of a cancer cell line by an Ad pre-complexed with a bispecific adapter, sCAR-CXCL12, is concentration and time dependent.
Notes: After an initial 4 h infection with 100 ifu/cell Ad5-CMV-GFP-luc alone or pre-complexed with increasing concentrations of sCAR-CXCL12, the percent of MDA-MB-435S cells positive for GFP expression was quantified at 48 h by flow cytometry analysis (A). After infection with 100 ifu/cell Ad5-CMV-GFP-luc alone or pre-complexed with 100 ng/108 ifu of sCAR-CXCL12 for 5, 15, 30, 60, 120, and 240 min, the percent of MDA-MB-435S cells positive for GFP expression was quantified by flow cytometry analysis (B). Numbers indicate the mean ± SEM of percent GFP-positive cells from three replicate dishes.
Abbreviations: Ad, adenovirus; Ad5, Ad serotype 5; GFP, green fluorescent protein; ifu, infectious unit; luc, luciferase; sCAR, soluble ectodomain form of the native Ad5 receptor; SEM, standard error of the mean.
Another important parameter that was determined for the retargeting was the time of infection of cancer cells in the presence and absence of retargeted Ad5 vector. MDA-MB-435S cells growing in culture were seeded in a 24-well tissue culture plate before infection with Ad5-CMV-GFP-luc alone or complexed with 100 ng/well of sCAR-CXCL12 at increasing time points. A minimal increase in the number of reporter gene-expressing cells was observed at increasing time points of infection in the untargeted vector group (Figure 4B). Overall, infectivity was low in the untargeted vector group regardless of the time of Ad infection. In contrast to this result, an enhancement in Ad infectivity was observed with increasing time of infectivity in the presence of retargeted Ad vector (Figure 4B). While just 10% of the cells were transduced with the retargeted vector by incubation with the adapter-complexed virus for 1 h, the percentage of cells increased to ~60% when the cells were incubated with the retargeted virus for 4 h. These data suggest a correlation between time of incubation and infectivity using retargeted Ad5 vector. To confirm the involvement of the CXCR4 receptor in Ad retargeting through the sCAR-CXCL12 adapter, we pre-incubated the cells with an anti- CXCR4 antibody. This antibody allowed partial blocking in the binding of Ad5 vector to the CXCR4 overexpressing MDA-MB-435S cells as evident by the reduction in E4 copy number in Figure 5. The partial blocking effect evident in this experiment could be due to the involvement of alternate receptors, which might be playing a role in the mediating Ad transduction. However, a significant effect was observed by using 100 and 1000 ng of blocking antibody against CXCR4 receptor.
Figure 5.
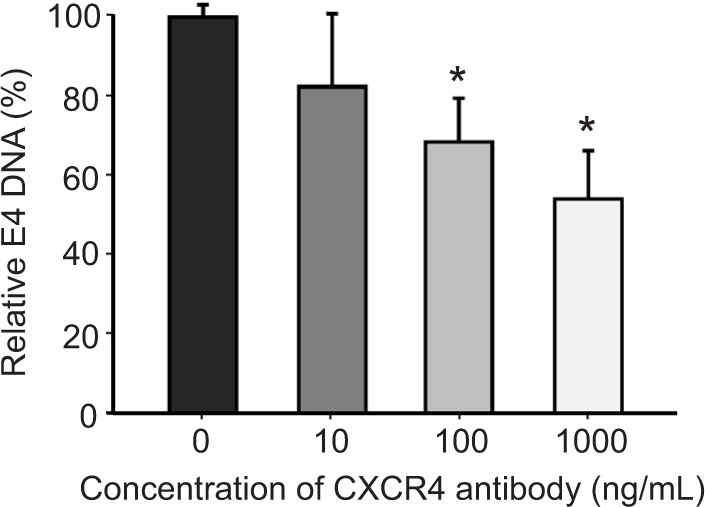
An anti-CXCR4 antibody blocks sCAR-CXCL12-mediated binding of Ad to CXCR4-positive cells.
Notes: Prior to the Ad infection, MDA-MB-435S cells were incubated with increasing concentrations of an anti-CXCR4 antibody for 30 min at 4°C. Subsequently, the cells were infected for 1 h at 4°C with 100 ifu/cell Ad5-CMV-GFP-luc pre-complexed with 100 ng/108 ifu of sCAR-CXCL12, and the amount of bound Ad5-CMV-GFP-luc DNA per dish was determined using a real-time PCR assay for the Ad5 E4 gene. Mean E4 copy number ± SEM from three replicate dishes is shown. The concentration results were compared using a Student’s t-test; the differences were considered statistically significant (*) if p<0.05.
Abbreviations: Ad, adenovirus; Ad5, Ad serotype 5; GFP, green fluorescent protein; ifu, infectious unit; luc, luciferase; PCR, polymerase chain reaction; sCAR, soluble ectodomain form of the native Ad5 receptor; SEM, standard error of the mean.
sCAR-CXCL12 decreases Ad infectivity and replication in an ex vivo liver slice model
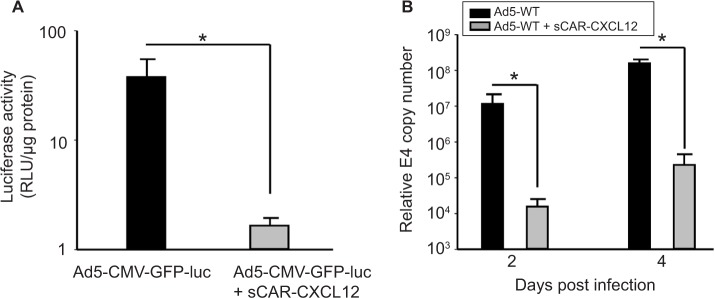
Precision cut tissue slice technique allows stringent evaluation of the specificity and efficacy of a viral vector system using human tissue. Therefore, we determined the effect of the sCAR-CXCL12 adapter on the Ad infectivity and replication using human liver slices. Our data indicated a considerable decrease in luc activity in the liver slices following ex vivo infection with Ad5 virus pre-conjugated with sCAR-CXCL12. Using sCAR-CXCL12 resulted in an approximately tenfold reduction in the luc activity following infection with Ad5-CMV-GFP-luc compared with the untargeted Ad vector (Figure 6A). In addition, we examined the ability of the Ad pre-complexed with the sCAR-CXCL12 adapter to attenuate wild-type Ad initial infection in an ex vivo system using human liver slices. These data showed a reduction in wild-type Ad5 replication at both day 2 and 4 post infection using the sCAR-CXCL12 adapter (Figure 6B). Infection of the liver slices with the adapter-complexed vector led to an ~100-fold reduction in the viral E4 copy number when compared with the Ad wild-type virus as determined by the real-time PCR assay. Thus, wild-type Ad5 virus pre-incubated with the sCAR-CXCL12 adapter demonstrated a “liver-off ” profile that suggested an inefficient initial viral infection in the human liver tissue resulting in decreased replication. The native Ad receptor has been shown to be highly expressed in liver, at cell–cell contacts between hepatocytes, which form the bile canaliculi, as well as in epithelial cholangiocytes that line the bile ducts.33 These results are consistent with the sCAR-CXCL12 adapter ability in vivo to block interaction with the hCAR receptor in liver.
Figure 6.
Ablation of native liver tropism of Ad using a bispecific adapter (sCAR-CXCL12).
Notes: Luc assay at 48 h after infection of primary human liver slices with Ad5-CMV-GFP-luc in the absence or presence of 100 ng/108 ifu sCAR-CXCL12. (A) Mean RLU/μg protein ± SEM of three replicate dishes. Real-time PCR assay for the Ad5 E4 gene in extracts from human primary liver slices with Ad5-WT in the absence or presence of 100 ng/108 ifu sCAR-CXCL12 at 2 and 4 days after infection. (B) Mean E4 copy number ± SEM in extracts from three replicate dishes. The treatment results were compared using a Student’s t-test; the differences were considered statistically significant (*) if p<0.05.
Abbreviations: Ad, adenovirus; Ad5, Ad serotype 5; GFP, green fluorescent protein; ifu, infectious unit; luc, luciferase; PCR, polymerase chain reaction; sCAR, soluble ectodomain form of the native Ad5 receptor; SEM, standard error of the mean; WT, wild type; RLU, relative light unit.
sCAR-CXCL12 enhances Ad infectivity in cancer cell lines
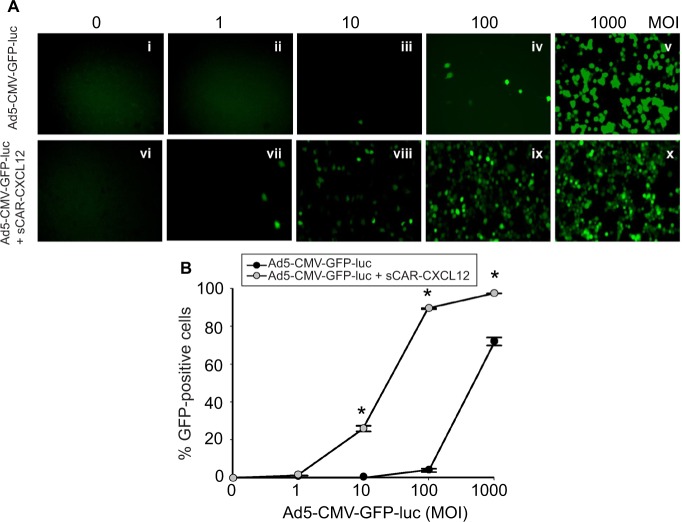
The retargeting efficacy of bispecific adapter molecule sCAR-CXCL12 was determined in human cancer cell lines expressing varying levels of CXCR4 and hCAR receptors on their cell surface. Cells were infected with increasing titers of Ad5-CMV-GFP-luc alone or pre-incubated with an optimum concentration of sCAR-CXCL12. At 48 h post infection, GFP reporter gene expression was determined in the infected cells by fluorescence microscopy and by flow cytometry. By flow cytometry analysis, there was a dramatic increase in Ad infectivity in MDA-MB-435S cells using a CXCR4-retargeted vector compared with an untargeted vector (Figure 7A). When quantified by flow cytometry, Ad5-CMV-GFP-luc virus showed a significant increase in the infection of MDA-MB-435S cells from 5% using untar-geted Ad vector to ~89% using Ad vector complexed with sCAR-CXCL12 at an MOI of 100 ifu/cell (Figure 7B). The infection was further increased to 97% by using 1000 ifu/cell of Ad vector complexed with sCAR-CXCL12. Another interesting observation in this experiment was the reduction in the viral dose that resulted in optimum transduction of CXCR4-positive cancer cells. The MOI required to achieve ~50% of transduction of MDA-MB-435S cells was 500 ifu/cell in the case of the untargeted vector. However, a similar level of transduction was observed by using 50 ifu/cell of retargeted viral vector.
Figure 7.
Ad targeted to CXCR4 by sCAR-CXCL12 enhances the efficacy of gene transfer to MDA-MB-435S cells.
Notes: MDA-MB-435S cells were plated at 1×105 cells/well in a 24-well plate. The indicated amounts of Ad5-CMV-GFP-luc pre-complexed in the absence or presence of sCAR-CXCL12 (100 ng/well) were incubated for 4 h at 37°C followed by a medium change. (A) At 48 h post infection, GFP expression was examined by fluorescence microscopy (40 × magnification) in cells infected with increasing titers of Ad: (i and vi); 0 ifu/cell (ii and vii) 1 ifu/cell; (iii and viii) 10 ifu/cell; (iv and ix) 100 ifu/cell; (v and x) 1000 ifu/cell. (B) The percentage of GFP-positive cells was determined by flow cytometry. Numbers indicate the mean ± SEM of percent GFP-positive cells from three replicate dishes. The treatment results were compared using a Student’s t-test; the differences were considered statistically significant (*) if p<0.05.
Abbreviations: Ad, adenovirus; Ad5, Ad serotype 5; GFP, green fluorescent protein; ifu, infectious unit; luc, luciferase; MOI, multiplicity of infection; sCAR, soluble ectodomain form of the native Ad5 receptor; SEM, standard error of the mean.
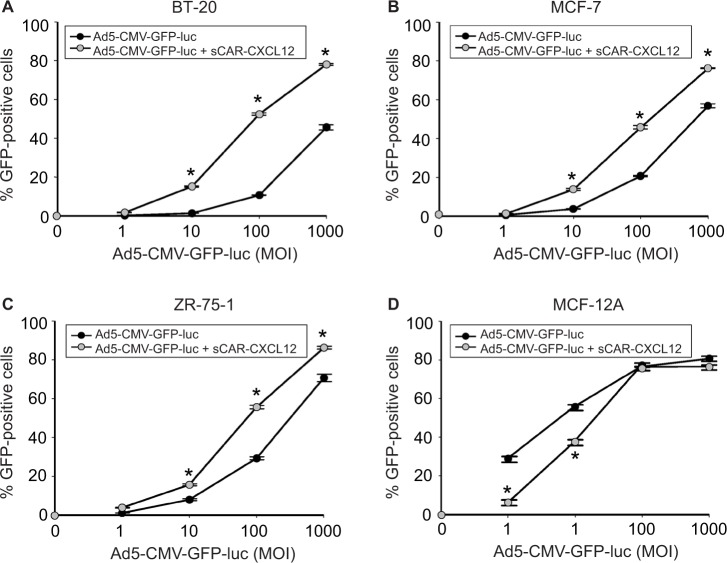
Three additional cancer cell lines were tested: BT-20 (Figure 8A), MCF-7 (Figure 8B), and ZR-75-1 (Figure 8C). All the cancer cell lines that were tested using sCAR-CXCL12-retargeted Ad vector showed increased infection compared with untargeted Ad vector. In MCF-7 cell line, the percentage of GFP-positive cells also increased from 20% (using untargeted Ad5 vector) to ~50% (using sCAR-CXCL12-retargeted Ad vector) at 100 ifu/cell. A similar increase in infectivity was observed in ZR-75-1 and BT-20 cell lines. In contrast to this, a decrease in Ad infectivity was observed in MCF-12A cell line (Figure 8D) in the presence of sCAR-CXCL12 adapter when compared with vector alone. The difference in Ad infectivity seen in these cell lines can be attributed to the differential cell surface expression of the chemokine receptor and native Ad receptor on these cells. In addition, inefficient transduction of CXCR4-negative MCF-12A cells by the Ad vector in the presence of adapter suggests cancer specificity for binding of sCAR-CXCL12 with CXCR4-positive cancer cells. Overall, the CXCR4-retargeted Ad vector not only resulted in a significant enhancement of the reporter gene expression in the target cells but also helped in reducing viral vector doses required to achieve optimal transduction compared with the untargeted vector.
Figure 8.
Determination of Ad targeting to CXCR4 by sCAR-CXCL12 in tumorigenic and immortalized non-tumorigenic cells.
Notes: Tumorigenic BT-20 (A), MCF-7 (B), and ZR-75-1 (C) cancer cells or non-tumorigenic MCF-12A epithelial cells (D) were plated at 1 × 105 cells/well in a 24-well plate. Indicated amounts of Ad5-CMV-GFP-luc pre-complexed in the absence or presence of sCAR-CXCL12 (100 ng/well) were incubated for 4 h at 37°C followed by a medium change. At 48 h post infection, the percentage of GFP-positive cells was determined by flow cytometry. Numbers indicate the mean ± SEM of percent GFP-positive cells from three replicate dishes. The treatment results were compared using a Student’s t-test; the differences were considered statistically significant (*) if p<0.05.
Abbreviations: Ad, adenovirus; Ad5, Ad serotype 5; GFP, green fluorescent protein; luc, luciferase; MOI, multiplicity of infection; sCAR, soluble ectodomain form of the native Ad5 receptor; SEM, standard error of the mean.
sCAR-CXCL12 untargets Ad from the liver infection in an in vivo model
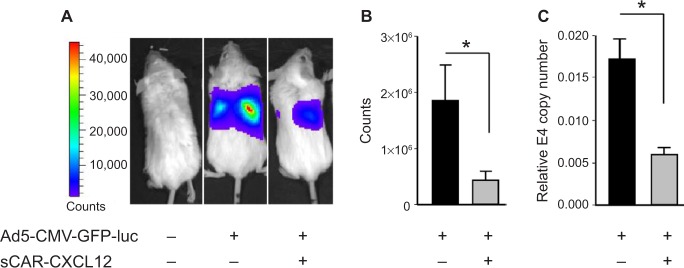
Liver sequestration following systemic administration of Ad is a major limitation that needs to be addressed to allow successful application of Ad vectors for cancer gene therapy. To examine the ability of sCAR-CXCL12 to prevent liver infection, Ad5-CMV-GFP-luc was administered with or without sCAR-CXCL12 into SCID-bg mice by tail vein injection. Bioluminescent images were obtained at 3 days post injection by IVIS imaging. Immediately after imaging, animals were sacrificed and livers were extracted and snap frozen in −80°C for further analysis by real-time PCR. The results obtained showed maximum signal in the liver in the group of animals injected with Ad5-CMV-GFP-luc alone. In contrast, animals that received Ad5-CMV-GFP-luc conjugated to sCAR-CXCL12 showed comparatively lower signal indicative of the untargeting effect of the adapter from liver tissue (Figure 9A). The charge-coupled device (CCD) images were quantified to demonstrate the difference in the liver uptake of the Ad5 vector in the presence and absence of sCAR-CXCL12. More than 50% reduction in the bioluminescence signal was observed in the targeted vector group compared with the untargeted counterparts (Figure 9B). Furthermore, real-time PCR data showed that in vivo Ad E4 copy number was reduced by approximately threefold in the liver following systemic administration of Ad5-CMV-GFP-luc + sCAR-CXCL12 vector compared with Ad5-CMV-GFP-luc (Figure 9C). These in vivo results were in agreement with the data obtained from the liver slice experiment.
Figure 9.
Liver untargeting of Ad infection using a bispecific adapter, sCAR-CXCL12, in SCID-bg mice in vivo.
Notes: Ad5-CMV-GFP-luc (1 × 108 ifu/mouse) pre-incubated in the absence or presence of sCAR-CXCL12 (5 μg) was injected intravenously into SCID-bg mice. Ad-directed luc expression was monitored in living animals by bioluminescence imaging at 3 days after injection. (A) Examples of an untreated mouse, a mouse injected with Ad5-CMV-GFP-luc, and a mouse injected with Ad5-CMV-GFP-luc + sCAR-CXCL12. Luc expression was quantified from ROI of the liver regions. (B) Mean count ± SEM from ten animals. After bioluminescent imaging, the mice were sacrificed, liver extracts were prepared, and Ad was quantified by real-time PCR. (C) Mean E4 copy number ± SEM in livers from ten animals. The treatment results were compared using a Student’s t-test; the differences were considered statistically significant (*) if p<0.05. Abbreviations: Ad, adenovirus; Ad5, Ad serotype 5; GFP, green fluorescent protein; ifu, infectious unit; luc, luciferase; PCR, polymerase chain reaction; ROI, regions of interest; sCAR, soluble ectodomain form of the native Ad5 receptor; SCID-bg, severe combined immunodeficiency-beige; SEM, standard error of the mean.
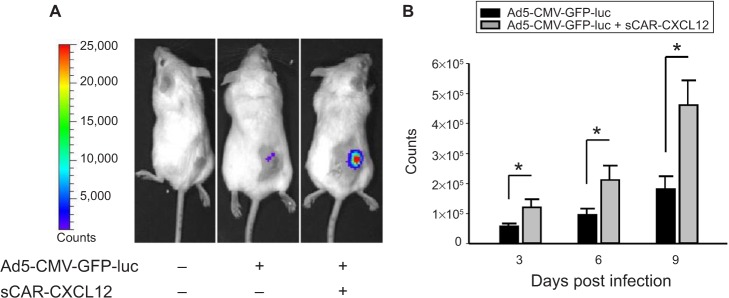
sCAR-CXCL12 retargets Ad to CXCR4-positive s.c. tumor xenografts
Following a demonstration of hepatic untargeting by the adapter, we determined the retargeting ability of sCAR-CXCL12 using an in vivo model of cancer. In this experiment, tumors were implanted in SCID-bg mice by s.c. injection of MDA-MB-435S cells. After the tumors had reached ~100 mm3 in size, Ad5-CMV-GFP-luc was injected either alone or pre-complexed with sCAR-CXCL12 into the centers of the tumor. IVIS imaging was performed at days 3, 6, and 9 post injection to monitor the bioluminescence. A strong signal was observed in the tumors in animals that received retargeted vector compared with the untargeted vector. This result can be attributed to the uptake of Ad complexed with the CXCL12 adapter by the CXCR4-positive tumor cells. An enhanced bioluminescence signal was observed in the MDA-MB-435S tumor grafts that were injected with retargeted Ad vector from day 3 to day 9. In contrast, animals injected with the untargeted vector displayed minimal bioluminescence in the tumors as evident by the IVIS images (Figure 10A). At each time point, quantitative analysis of the CCD images demonstrated a significant increase in the bioluminescence signal in the tumors injected with sCAR-CXCL12-targeted Ad vector compared with the tumors injected with untargeted Ad vector (Figure 10B). Furthermore, real-time PCR analysis of Ad E4 copy number showed a similar increase in the tumor uptake on day 9 in the group of mice that received an i.t. injection of sCAR-CXCL12-retargeted Ad vector compared with the group of mice that received an i.t. injection of untargeted Ad vector (data not shown).
Figure 10.
Enhancement of Ad infection in xenograft tumors in vivo using a bispecific adapter (sCAR-CXCL12).
Notes: S.c. tumors were injected with Ad5-CMV-GFP-luc (1 × 108 ifu/mouse) pre-incubated in the absence or presence of sCAR-CXCL12 (5 μg). Ad-directed luc expression was monitored in living animals by bioluminescence imaging at 3, 6, and 9 days after injection. (A) Examples of an untreated mouse, a mouse injected with Ad5-CMV-GFP-luc, and a mouse injected with Ad5-CMV-GFP-luc + sCAR-CXCL12. Luc expression was quantified from ROI of the tumor regions. (B) Mean count ± SEM from ten animals. The treatment results were compared using a Student’s t-test; the differences were considered statistically significant (*) if p<0.05.
Abbreviations: Ad, adenovirus; Ad5, Ad serotype 5; GFP, green fluorescent protein; ifu, infectious unit; luc, luciferase; ROI, regions of interest; sCAR, soluble ectodomain form of the native Ad5 receptor; s.c., subcutaneous; SEM, standard error of the mean.
Discussion
Ad gene therapy offers tremendous potential, but targeting the vector and delivering the payload to the diseased cell or tissue have been a major challenge. The widespread distribution of its natural receptor hCAR not only poses a hurdle in targeting specific cell types but also prevents exploiting these vectors to their full potential for cancer gene therapy applications. This distribution pattern presents the need to modify native tropism of Ads and simultaneously introduce a novel tropism. The current study tests the validity of an approach involving targeted Ad vectors with altered tropism and cell-specific gene delivery ability using a nonnative Ad receptor. Since CXCR4 is a relevant tumor marker that plays a crucial role in cancer cell growth, proliferation, migration, invasion, and metastasis,23 targeting this chemokine receptor using tropism-modified Ad seems to be a promising strategy for cancer gene therapy interventions. In this study, CXCR4 was utilized as a candidate receptor to demonstrate enhanced transductional targeting toward human cancer cells using sCAR-CXCL12 adapter molecule. The bispecific adapter, sCAR-CXCL12, confers a novel binding specificity to an Ad vector that redirects it to a nonnative cellular receptor (CXCR4) present on target cancer cells. This redirected specificity is particularly relevant for Ad-refractory tumor cells that are deficient in hCAR. In addition, the adapter protein allows hepatic untargeting of Ad, thus minimizing nonspecific uptake by the liver. Although CXCR7 was not examined due to its low expression in the cell lines used in this study, future studies will need to address the use of sCAR-CXCL12 to target this receptor.
Many studies have reported retargeting of Ad vectors to heterologous cellular receptors to achieve efficient transduction of the Ad-resistant tumor cells. In this context, Dmitriev et al12 demonstrated a ninefold increase in gene transfer by targeting Ad vector to the EGF receptor. Consistent with these reports, we observed a substantial increase in the infectivity of CXCR4-positive cancer cells by using the sCAR-CXCL12 adapter with Ad5 vector in a dose- and time-dependent manner. Importantly, the retargeted Ad also showed decreased infectivity of immortalized non-tumorigenic epithelial cells (MCF-12A), which could be attributed to the negligible expression of the CXCR4, CXCR7, and integrin receptors on the cell surface (Figure 1). Besides the enhanced transduction, there is an added advantage of using retargeted vectors: a lower dose of viral vector is required to gain optimal levels of transduction. This result was evident in our in vitro studies in which pre-complexing Ad5 vector with the targeting adapter resulted in a remarkable decrease in the vector dose to effect similar levels of transduction observed with the untargeted control vector.
Considering a narrow therapeutic window between efficacy and potential toxicity, a clear understanding of the viral dose is necessary that would result in a therapeutic response and avoid severe side effects. In this context, it is logically anticipated that vectors with increased transduction ability may trigger weak immune response resultant of the low doses of the viral vector used. Therefore, the reduction of viral vector dose, as observed in our retargeting experiments, may have future implications on minimizing vector-related toxicities and undesirable effects. The transductional targeting gain observed in vitro was further extended to a murine model of cancer. In our xenograft studies, we noted that physically targeted Ad5 vector conjugated to CXCL12 fusion protein retained its specificity in vivo and successfully retargeted to CXCR4-positive tumors implanted in SCID-bg mice. This specificity was evident from the noninvasive imaging studies performed to track Ad localization in the tumor grafts. However, the fold increase in the tumor transduction using targeted Ad was modest, which could be due to the presence of local barriers affecting viral transduction.
The underlying mechanism by which adapter molecules help in rerouting the Ad toward receptor overexpressed on cancer cells is not well understood. Ad infection begins with the binding of the fiber knob domain to the hCAR receptor expressed on the host cell surface followed by internalization by receptor-mediated endocytosis.34 Since the steps involving initial binding of Ad to the hCAR and its internalization into the host cell are uncoupled, the first step in the Ad infection cycle can be easily modulated without affecting the following steps to achieve cell-specific targeting. We speculate that the bispecific adapters modify the initial interaction step in the Ad infection pathway so that the Ad vectors can be redirected toward the target cell of interest through alternate pathways.
Another interesting aspect of the current study is adapter-mediated hepatic untargeting. Sequestration of Ad in the liver following i.v. administration is one of the major concerns in the field of Ad gene therapy. Most of the Ad injected by the i.v. route is taken up by the Kupffer cells.35–37 This uptake not only leads to adverse liver toxicity but also reduces the viral uptake by the relevant target tissue resulting in a suboptimal therapeutic response. Thus, hepatic untargeting provides a key to minimizing nonspecific transduction of the liver. We used precision-cut liver slices as a model system, which has been used to validate the potency of targeted Ad vectors to attenuate liver infection in preclinical studies.38,39 Our ex vivo data indicate a significant decrease in the viral infection and replication in the human liver slices while using the retargeted vector. This result is consistent with the previous report by Kirby et al,40 in which the reduction of viral replication was observed in human liver slices using a conditionally replicating Ad5/3 COX-2 virus. Furthermore, we observed a significant decrease in liver transduction following systemic i.v. administration of tropism-modified Ad5 vector in vivo. The untargeted vector, on the other side, still retained the natural tropism and accumulated in the liver as confirmed by the bioluminescence imaging and real-time PCR analysis. These studies, in which replication of a viral vector can be examined in the context of a major target organ, enhance our understanding and play a critical role to assess the therapeutic index at the preclinical level. It will be important in future studies to study the biodistribution and toxicity profiles of the sCAR-CXCL12-retargeted Ad to determine whether the hepatic untargeting is sufficient to reduce vector-mediated hepatotoxicity.
The process by which adapter molecules disrupt Ad binding to the hCAR-expressing cells, particularly liver, is not clear. Ad biodistribution, in vivo, is an intricate process and is driven by multiple interactions. Recent evidence has suggested that, besides hCAR, several blood coagulation factors, which interact with the Ad capsid components, also play a role in the hepatic uptake resulting in liver infection.41,42 In particular, coagulation factor X and complement component C4-binding protein can bind to the Ad fiber knob domain, and the complex formed between the two is redirected to the hepatic cells using alternate hepatocellular receptors.42 This interaction ultimately results in Ad infection of hepatic cells. We hypothesize that sCAR-CXCL12 adapter blocks those potential binding sites to prevent the interaction of Ad with coagulation factor proteins. However, future studies are warranted to explore the mechanistic pathway involved in adapter-mediated hepatic untargeting. Regardless of the mechanism, our data clearly underscore the importance of adapter-based strategies in minimizing hepatic transduction by Ads.
Conclusion
We report exploiting the CXCL12 chemokine for retargeting an Ad vector toward cancer cells overexpressing the cognate chemokine receptor. The results illustrated in this study also provide a strong rationale to apply this approach to developing Ad-based therapies targeting metastatic disease. In addition, transductional targeting described in this study can be combined with additional transcriptional or translational targeting methods to aid Ad vector specificity. Combining these approaches not only would offer extensive flexibility of developing novel recombinant adapters using different sets of Ad untargeting and retargeting moieties but will also result in generating Ad vectors with an improved safety profile favorable for use in the clinical settings. This result would have a field-wide impact that would encourage the use of Ad vectors in targeting different pathological conditions including cancer.
Acknowledgments
The authors would like to thank the following people for their contributions to this work: Bing Cheng, Larry Smart, and Tracee Terry for their expert technical assistance. This work was supported, in part, by the Louisiana Gene Therapy Research Consortium, and a grant from the NIH-NCI (5R01-CA154697). These funders had no role in the study design, data collection, analysis, decision to publish, or preparation of the manuscript.
Abbreviations
- Ad
Adenovirus
- Ad5
Ad serotype 5
- DMEM
Dulbecco’s Modified Eagle’s Medium
- ELISA
enzyme-linked immunosorbent assay
- FBS
fetal bovine serum
- FITC
fluorescein isothiocyanate
- F12
Ham’s F12 Medium
- GFP
green fluorescent protein
- hCAR
human Coxsackie and adenovirus receptor
- His
histidine
- HRP
horseradish peroxidase
- ifu
infectious unit
- i.v.
intravenous
- IVIS
in vivo imaging system
- i.t.
intratumoral
- luc
luciferase
- MOI
multiplicity of infection
- PBS
phosphate-buffered saline
- PE
phycoerythrin
- ROI
regions of interest
- sCAR
soluble ectodomain form of the native Ad5 receptor
- SCID-bg
severe combined immunodeficiency-beige
- s.c.
subcutaneous
- SDF-1α
stromal derived factor-1α
- TMB
3;3′;5;5′-tetramethylbenzidine
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Kasala D, Choi JW, Kim SW, Yun CO. Utilizing adenovirus vectors for gene delivery in cancer. Expert Opin Drug Deliv. 2014;11(3):379–392. doi: 10.1517/17425247.2014.874414. [DOI] [PubMed] [Google Scholar]
- 3.Wold WS, Toth K. Adenovirus vectors for gene therapy, vaccination and cancer gene therapy. Curr Gene Ther. 2013;13(6):421–433. doi: 10.2174/1566523213666131125095046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Freimuth P, Philipson L, Carson SD. The coxsackievirus and adenovirus receptor. Curr Top Microbiol Immunol. 2008;323:67–87. doi: 10.1007/978-3-540-75546-3_4. [DOI] [PubMed] [Google Scholar]
- 5.Imperiale MJ. Keeping adenovirus away from the liver. Cell Host Microbe. 2008;3(3):119–120. doi: 10.1016/j.chom.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 6.Reeh M, Bockhorn M, Görgens D, et al. Presence of the coxsackievirus and adenovirus receptor (CAR) in human neoplasms: a multitumour array analysis. Br J Cancer. 2013;109(7):1848–1858. doi: 10.1038/bjc.2013.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uusi-Kerttula H, Hulin-Curtis S, Davies J, Parker AL. Oncolytic adeno-virus: strategies and insights for vector design and immuno-oncolytic applications. Viruses. 2015;7(11):6009–6042. doi: 10.3390/v7112923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nettelbeck DM, Rivera AA, Kupsch J, et al. Retargeting of adenoviral infection to melanoma: combining genetic ablation of native tropism with a recombinant bispecific single-chain diabody (scDb) adapter that binds to fiber knob and HMWMAA. Int J Cancer. 2004;108(1):136–145. doi: 10.1002/ijc.11563. [DOI] [PubMed] [Google Scholar]
- 9.Dreier B, Mikheeva G, Belousova N, et al. Her2-specific multivalent adapters confer designed tropism to adenovirus for gene targeting. J Mol Biol. 2011;405(2):410–426. doi: 10.1016/j.jmb.2010.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Beusechem VW, Mastenbroek DC, Van den Doel PB, et al. Conditionally replicative adenovirus expressing a targeting adapter molecule exhibits enhanced oncolytic potency on CAR-deficient tumors. Gene Ther. 2003;10(23):1982–1991. doi: 10.1038/sj.gt.3302103. [DOI] [PubMed] [Google Scholar]
- 11.Kashentseva EA, Seki T, Curiel DT, Dmitriev IP. Adenovirus targeting to c-erbB-2 oncoprotein by single-chain antibody fused to trimeric form of adenovirus receptor ectodomain. Cancer Res. 2002;62(2):609–616. [PubMed] [Google Scholar]
- 12.Dmitriev I, Kashentseva E, Rogers BE, Krasnykh V, Curiel DT. Ectodo-main of coxsackievirus and adenovirus receptor genetically fused to epidermal growth factor mediates adenovirus targeting to epidermal growth factor receptor-positive cells. J Virol. 2000;74(15):6875–6884. doi: 10.1128/jvi.74.15.6875-6884.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harvey TJ, Burdon D, Steele L, et al. Retargeted adenoviral cancer gene therapy for tumour cells overexpressing epidermal growth factor receptor or urokinase-type plasminogen activator receptor. Gene Ther. 2010;17(8):1000–1010. doi: 10.1038/gt.2010.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang W, Zhu NL, Chua J, et al. Retargeting of adenoviral vector using basic fibroblast growth factor ligand for malignant glioma gene therapy. J Neurosurg. 2005;103(6):1058–1066. doi: 10.3171/jns.2005.103.6.1058. [DOI] [PubMed] [Google Scholar]
- 15.Li HJ, Everts M, Pereboeva L, et al. Adenovirus tumor targeting and hepatic untargeting by a coxsackie/adenovirus receptor ectodomain anti-carcinoembryonic antigen bispecific adapter. Cancer Res. 2007;67(11):5354–5361. doi: 10.1158/0008-5472.CAN-06-4679. [DOI] [PubMed] [Google Scholar]
- 16.Beatty MS, Timares L, Curiel DT. Augmented adenovirus transduction of murine T lymphocytes utilizing a bi-specific protein targeting murine interleukin 2 receptor. Cancer Gene Ther. 2013;20(8):445–452. doi: 10.1038/cgt.2013.39. [DOI] [PubMed] [Google Scholar]
- 17.Pereboev AV, Nagle JM, Shakhmatov MA, et al. Enhanced gene transfer to mouse dendritic cells using adenoviral vectors coated with a novel adapter molecule. Mol Ther. 2004;9(5):712–720. doi: 10.1016/j.ymthe.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 18.Nagasawa T. CXC chemokine ligand 12 (CXCL12) and its receptor CXCR4. J Mol Med (Berl) 2014;92(5):433–439. doi: 10.1007/s00109-014-1123-8. [DOI] [PubMed] [Google Scholar]
- 19.Zhao H, Guo L, Zhao H, Zhao J, Weng H, Zhao B. CXCR4 over-expression and survival in cancer: a system review and meta-analysis. Oncotarget. 2015;6(7):5022–5040. doi: 10.18632/oncotarget.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peled A, Tavor S. Role of CXCR4 in the pathogenesis of acute myeloid leukemia. Theranostics. 2013;3(1):34–39. doi: 10.7150/thno.5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Z, Ni C, Chen W, et al. Expression of CXCR4 and breast cancer prognosis: a systematic review and meta-analysis. BMC Cancer. 2014;14:49. doi: 10.1186/1471-2407-14-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitchell B, Mahalingam M. The CXCR4/CXCL12 axis in cutaneous malignancies with an emphasis on melanoma. Histol Histopathol. 2014;29(12):1539–1546. doi: 10.14670/HH-29.1539. [DOI] [PubMed] [Google Scholar]
- 23.Guo F, Wang Y, Liu J, Mok SC, Xue F, Zhang W. CXCL12/CXCR4: a symbiotic bridge linking cancer cells and their stromal neighbors in oncogenic communication networks. Oncogene. 2016;35(7):816–826. doi: 10.1038/onc.2015.139. [DOI] [PubMed] [Google Scholar]
- 24.Hung CS, Su HY, Liang HH, et al. High-level expression of CXCR4 in breast cancer is associated with early distant and bone metastases. Tumour Biol. 2014;35(2):1581–1588. doi: 10.1007/s13277-013-1218-9. [DOI] [PubMed] [Google Scholar]
- 25.Stacer AC, Fenner J, Cavnar SP, et al. Endothelial CXCR7 regulates breast cancer metastasis. Oncogene. 2016;35(13):1716–1724. doi: 10.1038/onc.2015.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sánchez-Martín L, Sánchez-Mateos P, Cabañas C. CXCR7 impact on CXCL12 biology and disease. Trends Mol Med. 2013;19(1):12–22. doi: 10.1016/j.molmed.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 27.Puchert M, Engele J. The peculiarities of the SDF-1/CXCL12 system: in some cells, CXCR4 and CXCR7 sing solos, in others, they sing duets. Cell Tissue Res. 2014;355(2):239–253. doi: 10.1007/s00441-013-1747-y. [DOI] [PubMed] [Google Scholar]
- 28.Rae JM, Ramus SJ, Waltham M, et al. Common origins of MDA-MB-435 cells from various sources with those shown to have melanoma properties. Clin Exp Metastasis. 2004;21(6):543–552. doi: 10.1007/s10585-004-3759-1. [DOI] [PubMed] [Google Scholar]
- 29.Takayama K, Reynolds PN, Short JJ, et al. A mosaic adenovirus possessing serotype Ad5 and serotype Ad3 knobs exhibits expanded tropism. Virology. 2003;309(2):282–293. doi: 10.1016/s0042-6822(03)00067-9. [DOI] [PubMed] [Google Scholar]
- 30.He TC, Zhou S, da Costa LT, Yu J, Kinzler KW, Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci U S A. 1998;95(5):2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olinga P, Meijer DK, Slooff MJ, Groothuis GM. Liver slices in in vitro pharmacotoxicology with special reference to the use of human liver tissue. Toxicol In Vitro. 1997;12(1):77–100. doi: 10.1016/s0887-2333(97)00097-0. [DOI] [PubMed] [Google Scholar]
- 32.Altmann F, Staudacher E, Wilson IB, Marz IB. Insect cells as hosts for the expression of recombinant glycoproteins. Glycoconj J. 1999;16(2):109–123. doi: 10.1023/a:1026488408951. [DOI] [PubMed] [Google Scholar]
- 33.Raschperger E, Thyberg J, Pettersson S, Philipson L, Fuxe J, Pettersson RF. The coxsackie-and adenovirus receptor (CAR) is an in vivo marker for epithelial tight junctions, with a potential role in regulating permeability and tissue homeostasis. Exp Cell Res. 2006;312(9):1566–1580. doi: 10.1016/j.yexcr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 34.Roelvink PW, Mi Lee G, Einfeld DA, Kovesdi I, Wickham TJ. Identification of a conserved receptor-binding site on the fiber proteins of CAR-recognizing adenoviridae. Science. 1999;286(5444):1568–1571. doi: 10.1126/science.286.5444.1568. [DOI] [PubMed] [Google Scholar]
- 35.Smith JS, Xu Z, Tian J, Stevenson SC, Byrnes AP. Interaction of systemically delivered adenovirus vectors with Kupffer cells in mouse liver. Hum Gene Ther. 2008;19(5):547–554. doi: 10.1089/hum.2008.004. [DOI] [PubMed] [Google Scholar]
- 36.Xu Z, Tian J, Smith JS, Byrnes AP. Clearance of adenovirus by Kupffer cells is mediated by scavenger receptors, natural antibodies, and complement. J Virol. 2008;82(23):11705–11713. doi: 10.1128/JVI.01320-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prill JM, Espenlaub S, Samen U, et al. Modifications of adenovirus hexon allow for either hepatocyte detargeting or targeting with potential evasion from Kupffer cells. Mol Ther. 2011;19(1):83–92. doi: 10.1038/mt.2010.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rots MG, Elferink MG, Gommans WM, et al. An ex vivo human model system to evaluate specificity of replicating and non-replicating gene therapy agents. J Gene Med. 2006;8(1):35–41. doi: 10.1002/jgm.815. [DOI] [PubMed] [Google Scholar]
- 39.Stoff-Khalili MA, Rivera AA, Le LP, et al. Employment of liver tissue slice analysis to assay hepatotoxicity linked to replicative and nonreplicative adenoviral agents. Cancer Gene Ther. 2006;13(6):606–618. doi: 10.1038/sj.cgt.7700934. [DOI] [PubMed] [Google Scholar]
- 40.Kirby TO, Rivera A, Rein D, et al. A novel ex vivo model system for evaluation of conditionally replicative adenoviruses therapeutic efficacy and toxicity. Clin Cancer Res. 2004;10(24):8697–8703. doi: 10.1158/1078-0432.CCR-04-1166. [DOI] [PubMed] [Google Scholar]
- 41.Bradshaw AC, Parker AL, Duffy MR, et al. Requirements for receptor engagement during infection by adenovirus complexed with blood coagulation factor X. PLoS Pathog. 2010;6(10):e1001142. doi: 10.1371/journal.ppat.1001142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu Z, Qiu Q, Tian J, et al. Coagulation factor X shields adenovirus type 5 from attack by natural antibodies and complement. Nat Med. 2013;19(4):452–457. doi: 10.1038/nm.3107. [DOI] [PubMed] [Google Scholar]