Abstract
The chemical synthesis of new lipophilic polyphenols with improved properties presents technical difficulties. Here we describe the selection, isolation and identification of lipolytic bacteria from food-processing industrial wastes, and their use for tailoring a new set of compounds with great interest in the food industry. These bacteria were employed to produce lipolytic supernatants, which were applied without further purification as biocatalysts in the chemoselective and regioselective synthesis of lipophilic partially acetylated phenolic compounds derived from olive polyphenols. The chemoselectivity of polyphenols acylation/deacylation was analyzed, revealing the preference of the lipases for phenolic hydroxyl groups and phenolic esters. In addition, the alcoholysis of peracetylated 3,4-dihydroxyphenylglycol resulted in a series of lipophilic 2-alkoxy-2-(3,4-dihydroxyphenyl)ethyl acetate through an unexpected lipase-mediated etherification at the benzylic position. These new compounds are more lipophilic and retained their antioxidant properties. This approach can provide access to unprecedented derivatives of 3,4-dihydroxyphenylglycol with improved properties.
Introduction
The beneficial effects of the Mediterranean diet are partly due to its high content in antioxidant compounds [1]. In particular, the polyphenols present in the virgin olive oil display a strong antioxidant activity in vitro [2] and in vivo [3,4], which has impelled a growing interest in these compounds, especially those that can be obtained from by-products of the food industry [5]. Several epidemiological studies have shown the beneficial health effects arising from consumption of foods rich in antioxidants, preventing the damage caused by prolonged oxidative stress in certain biomolecules (nucleic acids, lipids, proteins), which is associated with an increased risk of chronic diseases [6]. The acylated polyphenols display improved properties as functional ingredients compared to the natural polyphenols [7] as they are lipophilic antioxidants with improved resistance against metabolic degradation, and they can also be incorporated into lipid food matrices such as fats and oils, processed foods, and margarines [8,9].
Protection and deprotection of functional groups are commonly employed in organic chemistry to carry out the synthesis of partially acylated derivatives [10]. However, despite of the many advances in organic synthesis during the last decades, the conventional chemical synthesis of mono acylated derivatives of polyphenols presents serious difficulties due to the high density of very similar functional groups, which requires extensive protection and deprotection sequences [10]. Thus, although some chemical approaches have been developed for accessing partially acylated polyphenols in a chemoselective way [11–13], most of such procedures are pure chemical synthesis, and involve the use of hazardous reagents or non-green conditions. As an alternative, the biotech industries have traditionally produced compounds of commercial interest by microbial fermentation, or by fermentation followed by subsequent chemical modification to improve specific properties, such as activity, solubility, absorption, pharmacokinetics or stability. In this context, enzymes and microorganisms have been efficiently used as biocatalysts in chemo-, regio- and stereoselective synthesis of bioactive compounds.
We reason that an enzyme-catalyzed approach could offer a viable alternative to traditional fermentation procedures and conventional chemical methods for the chemo- and regioselective synthesis of these new lipophilic compounds [14]. In this work, special attention has been focused on hydroxytyrosol (HT) and 3,4-dihydroxyphenylglycol (DHPG), which predominate in leaves and fruits of olive trees (Olea europea), either free or as acyl derivatives, and display antioxidant activity [15]. We have developed a novel enzymatic method to obtain targeted mono- or di-acylated derivatives of the polyphenols.
Materials and Methods
Site description and sample collection
The bacterial strains used in this study were isolated from locations in the provinces of Badajoz and Huelva (Spain) in 2010. Samples HR11 and HR12 contained semisolid fats from a meat curing factory (38.151216°N, -6.684258°E). Sample HR11 was obtained by collecting the dripping fat from the floor of the factory premises. Sample HR12 was obtained from a tank containing fat leftovers. Sample HR21 consisted of the fish dust that results after cutting fish into pieces before canning in a fish canning factory (37.20994°N, -7.26167°E). All samples were collected in 50 ml sterile plastic tubes and stored at 4°C until use.
Screening to detect lipolytic microorganisms (hydrolysis)
Fish sawdust samples (7.28 g) were suspended in 25 ml of sterile saline solution (NaCl 0.85% w/v). In the case of the sample from cured meat oil, 5 ml of each sample of fat were suspended in 20 ml of sterile saline solution (NaCl 0.85% w/v). Screening for lipolytic microorganisms was performed as previously described [16].
Transesterification assay of lipolytic activity
Transesterification activity of lipase was tested by a colorimetric method with minor modifications [16]. This method is based on the release of the yellow-colored compound p-nitrophenol (p-NP) after the transesterification of p-nitrophenyl palmitate (p-NPP; Sigma-Aldrich) with ethanol, and the subsequent detection by using UV-Vis spectrophotometry. Strains producing the maximum lipase activity were selected for further studies.
Optimization of bacterial growth conditions and lipase production
In order to optimize the production of the bacterial lipases, the strains were grown in two different media (PYB or LB) in the presence or in the absence of 2% tributyrin (PYBT or LBT media). PYB medium contains 1% (w/v) peptone, 0.5% (w/v) yeast extract, 0.1% (w/v) K2HPO4, 0.02% (w/v) MgSO4 ·7H2O. pH was adjusted to 7.5. LB medium (1% (w/v) tryptone, 0.5% (w/v) yeast extract, 0.5% (w/v) NaCl) was supplemented with 2% (w/v) glucose. Bacteria were grown in 500 ml Erlenmeyer flasks containing 100 ml of medium. Growth was monitored by measuring the absorbance at 600 nm (OD600) in a Beckman DU640 spectrophotometer. The lipase activity was assayed employing the p-NPP method described at 37°C.
Purification of the supernatants
The cell-free supernatant was obtained by centrifugation of bacterial cultures at 4,500 rpm for 5 min at 4°C. These supernatants were concentrated in dialysis bags (12 kDa, Sigma) against polyethylene glycol (8 kDa) at 4°C overnight, and then, they were dialyzed in 0.05 M potassium phosphate buffer (pH 7.6). Finally, they were freeze-dried. The dry supernatants were employed as enzymatic cocktails for the lipase assays.
Isolation of DNA and 16S rRNA gene sequence analysis
Bacterial DNAs were isolated and used for the amplification of the 16S rRNA by PCR using the universal primers 16F27 (5′-AGAGTTTGATCMTGGCTCAG-3′) and 16R1488 (5′-CGGTTACCTTGTTAGGACTTCACC-3′) as previously reported [16]. 16S rRNA sequences corresponding to positions 53 to 667 of the 16S rRNA gene from Escherichia coli were obtained and analyzed as previously described [16].
Nucleotide sequence accession numbers
The nucleotide sequences were deposited under accession numbers KP212109 to KP212128 in the GenBank database.
General methods for the chemoenzymatic syntheses of acylated polyphenols
NMR spectra were recorded at 25°C on a Bruker Avance 300 spectrometer, on a Bruker Avance III 500 MHz, and on a Bruker Avance III 700 MHz instruments equipped with a cryogenically cooled 5 mm TCI gradient probe. Chemical shifts are reported in ppm (δ) and spectra were referenced to the residual protonated solvent (3.31 and 49.0 ppm for CD3OD, 7.26 and 77.2 ppm for CDCl3, 2.05 and 29.8 for (CD3)2CO, for 1H and 13C NMR, respectively). Coupling constants (J) are expressed in Hertz (Hz). The assignments of 1H and 13C signals were confirmed by 1D and 2D NMR experiments (COSY, HSQC, HMBC). High resolution mass spectra were obtained by LSIM using a Hewlett Packard 5989 A spectrometer coupled to a Hewlett Packard 5990 II gas chromatographer and a Micromass AutoSpec-Q spectrometer with a resolution of 1,000 or 10,000 (10% valley definition); a cesium gun, 1-thioglicerol as matrix and NaI as additive were used. Column chromatography was performed using Merck silica gel 60 (230–400 mesh). TLCs were performed on silica-coated aluminum sheets from Merck (silica gel 60 F254) using mixtures of CH2Cl2−MeOH and EtOAc−hexane as eluants; spots were visualized by UV light and by staining with vanillin/H2SO4 in EtOH (1.5 g of vanillin in 100 mL of 95% EtOH/conc H2SO4 100:1).
Lipase-mediated acetylations of polyphenols 1, 4 and 8
The O-acetylations of the phenolic compounds were performed using the four lipolytic bacterial extracts, in a substrate-lipase extract 1:1 ratio in weight (40 mg). Isopropenyl acetate was used as solvent and acylating agent, 40 equiv (0.52–0.63 mL). The mixture was stirred at 40°C for 24 h in darkness. In the case of glycol derivative 4, DMF was used as co-solvent because of solubility problems, isopropenyl acetate-DMF 1:1 in volume (1.0 mL).
Lipase-mediated deacylation of compounds 2, 5 and 9
For the deacylation reactions of peracetylated polyphenols, to a solution of 2, 5 and 9 (50 mg) in a primary linear aliphatic alcohol (MeOH, EtOH, propan-1-ol or butan-1-ol) was added silica gel and the lipase extract (from Bacillus sp. HR21-6 or Terribacillus sp. 2B122), in a substrate-alcohol-lipase extract-silica gel 1:50:2:2 ratio in weight. The mixture was heated at 60°C for 24–168 h until total conversion (in the case of Terribacillus sp. 2B122 lipase a second addition of lipase extract was needed when the reaction rate is markedly reduced). Finally, the solvent was evaporated and the residue was purified by column chromatography to give 3, 6a-d and 10, respectively, using EtOAc-hexane or CH2Cl2-MeOH gradients as eluants (see S1 Supplementary Experimental Procedures).
Hydroxytyrosyl acetate (3) from compound 2
Eluted with EtOAc-hexane 1:1 gave 3 (90% with Bacillus sp. HR21-6 or Terribacillus sp. 2B122) as a colorless syrup. Spectroscopic data for 3 are in agreement with those reported in literature [17].
Deacetylation reactions of compounds 6a-d
To a solution of 6 (40 mg) in dry MeOH (1 mL) were added Cs2CO3 (2 equiv.) and sodium ascorbate (1 equiv.). The mixture was stirred in the darkness and in the presence of Ar at room temperature for 2–3 h. Then, the product was purified without evaporation of the solvent by column chromatography to give 7a-d, using CH2Cl2−MeOH, CH2Cl2−EtOH, and EtOAc−hexane gradients as eluants (see S1 Supplementary Experimental Procedures).
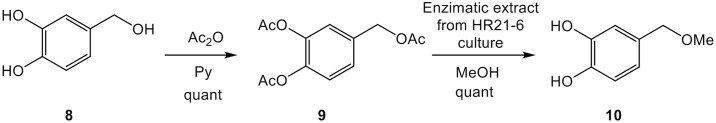
4-(Methoxymethyl)benzene-1,2-diol (10) from compound 9
Eluted with EtOAc-hexane 1:1 gave 10 (quant. with Bacillus sp. HR21-6) as a colorless syrup. Spectroscopic data for 10 are in agreement with those reported in literature [18]. Supplementary experimental data procedures include additional information for the compounds 6a-d and 7a-d.
DPPH radical scavenging activity
The antiradical activity of 6a-d and 7a-d has been evaluated by the DPPH method [19,20].
Results
Lipase-mediated O-deacylation of peracetylated HT
In order to obtain lipophilic phenolic esters it is essential to achieve the esterification of primary alcoholic groups without affecting the catechol moiety, which is known to be essential for the antioxidant effects. This process requires a chemoselective procedure.
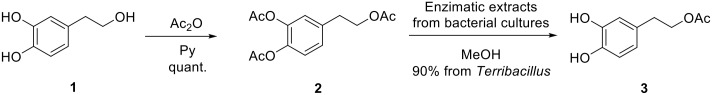
In a previous work we isolated bacteria capable of performing transesterification reactions in a regioselective fashion [16]. We reasoned that those bacterial isolates could also be used for the regioselective acylation/deacylation of polyphenols. For an initial analysis, HT (1) was firstly peracetylated with acetic anhydride in pyridine to obtain 2, as previously described [21]. The deacetylation of the peracetylated HT was efficiently achieved using the supernatants of a bacterial culture obtained from our best performing strain (Terribacillus sp. 2B122) and an aliphatic alcohol as solvent (MeOH). The transesterification reaction was completely chemoselective in the phenolic positions, and gave a monoacetylated derivative in the aliphatic position (compound 3, [22] as depicted in Fig 1, a natural compound present in extra virgin olive oil [23]).
Fig 1. Regioselective deacetylation of peracetylated HT (2) catalyzed by the supernatants from bacterial cultures.
Screening for additional microorganisms showing lipolytic activity
In the previous reaction, the deacetylation was completely selective towards the phenolic esters, but we did not observe any selectivity towards the aliphatic ester moieties with this or other strains previously isolated. Therefore, we designed a new screening aiming at finding microorganisms capable of transesterification the aliphatic ester selectively. We reasoned that food industries involved in the processing of foods with high fat content would provide the best environment. Since a vegetable oil-rich location was screened previously, a fish and a meat industries located near Isla Cristina (Huelva, Spain) and Higuera la Real (Badajoz, Spain), respectively, were selected for sampling this time. After a first round of selection over 4,000 colonies (fungi and bacteria) were isolated from fish sawdust and from cured meat fat. Out of those, 459 were considered as positive (S1 Table). Interestingly, no lipolytic bacterial isolates were selected from the sample HR12 obtained from a meat curing factory, where the number of microorganisms obtained was 378. The lipolytic bacteria were grown in a non-selective medium (i.e. in the absence of tributyrin), and the lipolytic activity was then confirmed on LB supplemented with 0.5% tributyrin. These samples were transferred to the secondary screening.
Screening for microorganisms capable of performing transesterification reactions among the selected lipolytic strains
The second step of the screening was performed using freeze-dried supernatants of the 66 bacterial isolates selected after the first step of the screening. The supernatants were assayed for transesterification activity by quantification of the yellow-colored p-nitrophenol (p-NP) that is released after the transesterification of p-nitrophenyl palmitate (p-NPP) with ethanol to give acetyl-palmitate ester and p-NP. We performed control assays as previously described [16]. The maximum value obtained in these control reactions was 0.119 and it was set as the threshold. Out of the 66 lipolytic bacterial strains in this second screening, 20 supernatants produced absorbance values higher than the cut-off of 0.119 (S2 Table). Most of the supernatants from the bacterial isolates displayed values between the cut-off and 0.300 and only very few were capable of displaying higher activities (4 isolates).
Phylogenetic classification of the selected lipolytic bacteria
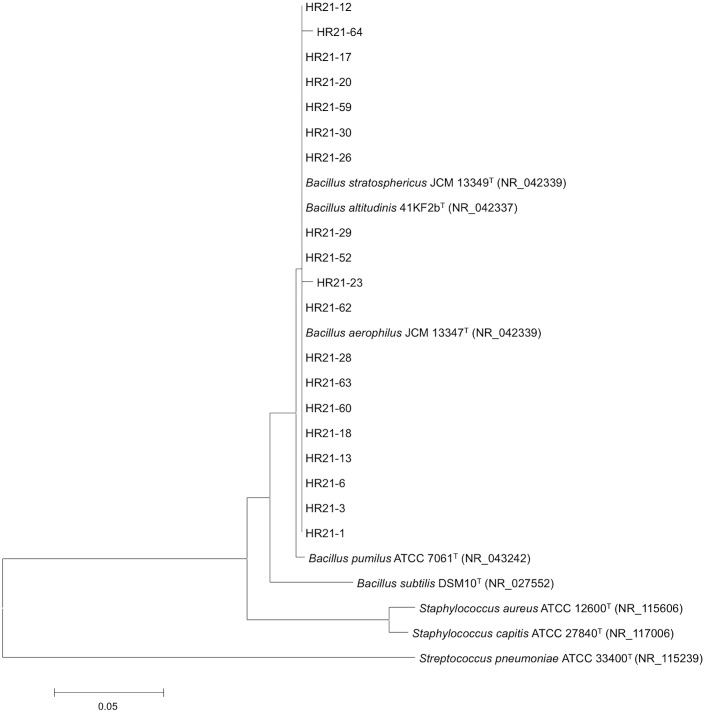
Phylogenetic studies were conducted using partial 16S rRNA sequences with the aim of identifying the bacterial isolates. Significantly, all strains isolated belong to the Gram-positive bacteria and were closely related to members of the genus Bacillus exhibiting similarity values ranging from 98% to 100%. The phylogenetic reconstruction carried out with different methods was consistent and suggested that all the strains (HR21-1, HR21-3, HR21-6, HR21-12, HR21-13, HR21-17, HR21-18, HR21-20, HR21-23, HR21-26, HR21-28, HR21-29, HR21-30, HR21-52, HR21-59, HR21-60, HR21-62, HR21-63, HR11-64 and HR11-65) clustered together, exhibiting a high similarity (98% to 100%) to the 16S rRNA sequences of Bacillus aerophilus, Bacillus stratosphericus and Bacillus altitudinis (Fig 2).
Fig 2. Evolutionary relationships of the selected strains.
The phylogenetic tree shows the position of the isolates displaying lipolytic activity with respect to other type strains of genus Bacillus and an external bacterial group. The distances were calculated using Maximum Composite Likelihood. 16S rRNA gene sequences from the isolates correspond to 614 bps.
O-Deacylation of peracetylated HT by Bacillus lipases
Since most of the isolates belonged to the Bacillus genus and were closely related, we only tested two random isolates for deacylation of HT. However, the results were similar to those obtained previously in Fig 1. Since the phylogenetic reconstruction using partial 16S rRNA sequences did not clearly differentiate between several species belonging to the genus Bacillus, we selected one of those two isolates for further studies and sequenced the complete 16S RNA. After further analysis, HR21-6 appeared to cluster together with B. pumilus and not with B. stratosphericus, B. aerophilus or B. altitudinis (S1 Fig).
Optimization of the bacterial growth conditions for the production of lipases
The study was continued with four isolates belonging to different genera for tests in acylation and deacylation reactions of natural polyphenols. Three of the strains were previously isolated by our research group (the Gram negative Pseudomonas sp. 2B120 and Enterobacter sp. 1B89, and the Gram positive Terribacillus sp. 2B122) [16] and the fourth strain, Bacillus HR21-6, was isolated during this work (as described above).
An important factor in this study was to define the optimal conditions for the production of the lipase-rich supernatants. Two parameters were selected for designing the optimization variables: medium and incubation time. At this stage it was not known whether the lipases responsible for the transesterification activity observed were induced by lipidic substances or constitutively expressed. Therefore, we also added the lipidic substrate tributyrin (previously used for the screening) to the media. The effect of the different culture media in lipase activity at various time intervals is shown in Fig 3. In general, the activity of the supernatants of Bacillus sp. HR21-6 was higher when the microorganism was grown in PYB than in LB medium. The maximum activity was obtained at 48 h in PYB medium, which corresponded with the late stationary phase. In LB medium, the maximum activity was reached after 24 h of growth (Fig 3A). In both cases, addition of tributyrin produced a decrease in the lipase activity.
Fig 3. Optimization of growth conditions of the selected strains for the detection of lipase activity.
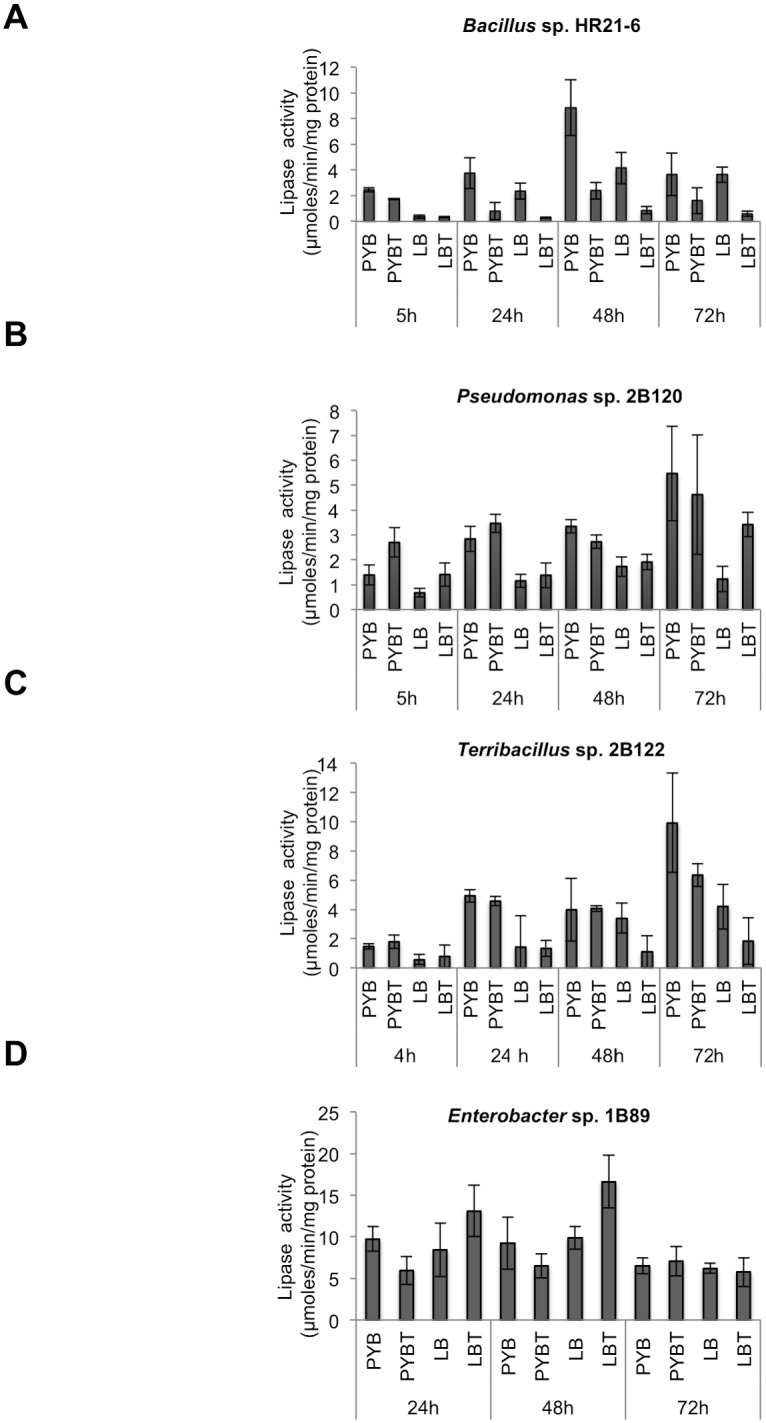
To determine de lipase activity the bacterial strains, Bacillus sp. HR21-6 (A), Pseudomonas sp. 2B120 (B), Terribacillus sp. 2B122 (C) and Enterobacter sp. 1B89 (D) were grown in PYB or LB media with (PYBT or LBT) or without (PYB or LB) tributyrin for the indicated times. Lipase activity in the supernatants was quantified by using the p-NPP method. Data shown are the average of at least 3 independent experiments and the standard deviation of the mean.
For Pseudomonas sp. 2B120 the lipase production started at late stationary phase of the bacterial grown. The maximum lipase activity was obtained in PYB medium after 72 h of growth (Fig 3B). The addition of tributyrin did not increase the lipase activity of the supernatants. For Terribacillus sp. 2B122 the maximum lipase activity was also detected in PYB medium at 72 h (Fig 3C). Surprisingly the results of the Terribacillus sp. 2B122 lipase activity in LB medium from 48 h to 72 h showed a significantly high error, which may be due to the cell lysis experienced by this strain in LB and LBT at around 24 h of cultivation. Optimal conditions for the lipase activity of Enterobacter sp. 1B89 were determined to be maximal in LBT medium at 48 h cultivation. In this case, addition of tributyrin to the culture medium allowed an increase in the lipase activity (Fig 3D).
Lipase-mediated O-deacylation of polyphenols
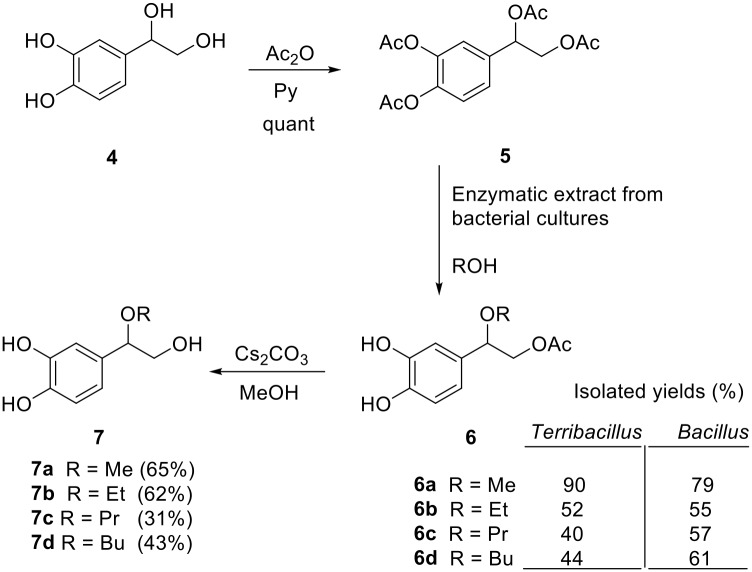
During this work we also aimed at studying another interesting polyphenol, 3,4-dihydroxyphenylglycol (DHPG), which is also present in olives and shows excellent antioxidant characteristics. In a first step, the peracetylated DHPG (compound 5) was prepared from DHPG 4 [24]. The phenolic hydroxyl groups were regioselectively deacetylated by a transacetylation process with a primary linear aliphatic alcohol (methanol, ethanol, propan-1-ol or butan-1-ol) in a reaction mixture containing the bacterial supernatants and silica gel, using a 1:50:2:2 substrate−alcohol−lipase extract−silica gel ratio. The acetoxy group at the primary position remained unchanged, whereas the acetoxy group at benzylic position was, unexpectedly, substituted by the corresponding alkoxy group (Figs 4 and 5A). The progress of the reactions was monitored by TLC, and it resulted in the synthesis of monoacetylated etherified derivatives 6a-d (see S1 Supplementary Experimental Procedures) with a yield ranging from 40 to 90% depending on the bacterial isolate (Fig 5B). This procedure constitutes the first synthesis of 3,4-dihydroxyphenylglycol ethers 6. The enzymes did not only catalyze the alcoholysis of the acetoxy groups on the aromatic ring, but also the substitution of the acetoxy group at benzylic position by an alkoxy group (–OR). The corresponding deacetylated derivatives 7a-d were prepared by a basic methanolysis in the presence of Cs2CO3 as a weak base (with a yield of 31–65%, see S1 Supplementary Experimental Procedures). Due to the easy degradation of the catechol group at the high pH required for the deacetylation reaction, sodium ascorbate (1 equiv) was added to the reaction mixture to prevent extensive degradation.
Fig 4. Regioselective deacetylation of peracetylated DHPG (5) with bacterial supernatants.
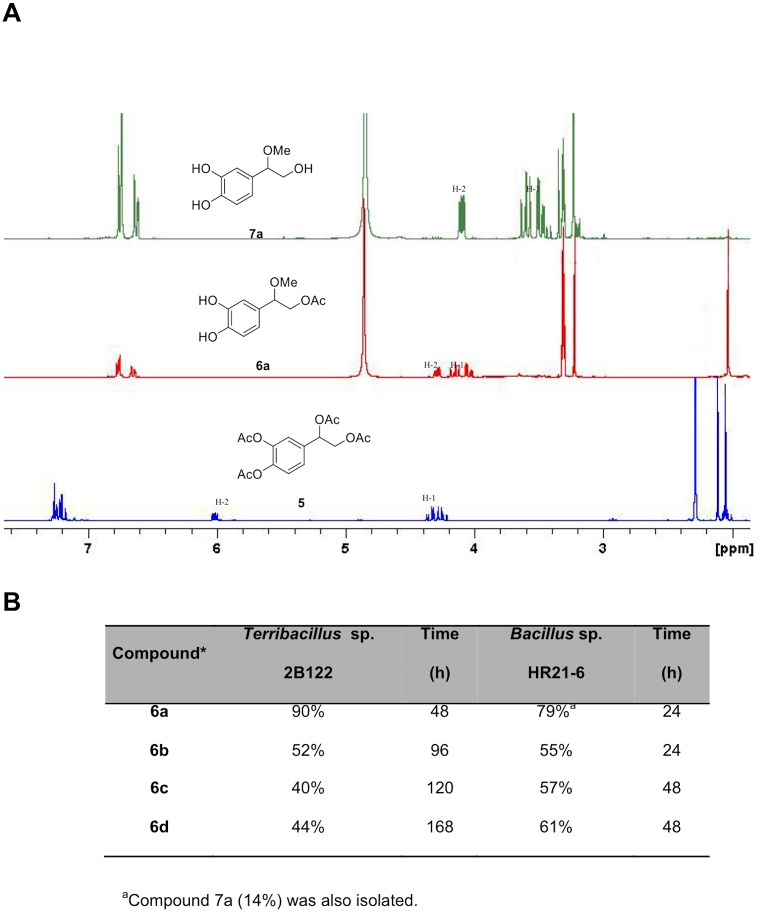
Fig 5. Lipase catalized deacetylation of peracetylated DHPG (5).
A) The figure depicts the 1H-NMR (300 MHz) spectra of compounds 5 (CDCl3), 6a (CD3OD), and 7a (CD3OD). B) Regioselective deacylation of compound 5 using Terribacillus sp. 2B122 and Bacillus sp. HR21-6 lipases to give compounds 6a-d at the indicated reaction times.
The previous reaction on DHPG constituted an outstanding case of an enzymatic conversion of an ester into an ether. In order to further check this reaction, we assayed peracetylated protocatechuic alcohol 9, which is a compound very similar to peracetylated DHPG 5, harbouring a benzylic alcohol but lacking the acetoxymethyl group at the end of the aliphatic side chain. Protocatechuic alcohol 8 is a very potent antioxidant molecule, also found in virgin olive oil [25]. As described above, we first obtained the peracetylated derivative of protocatechuic alcohol (compound 9 [26]), and then the deacetylation reaction proceeded in the presence of the bacterial supernatants and MeOH as solvent (Fig 6). Similarly to the reaction described for peracetylated DHPG 5 in Fig 4, the acetoxy group at the benzylic position was substituted by a methoxy group, giving ether 10.
Fig 6. Regioselective deacetylation of peracetylated protocatechuic alcohol (9) catalyzed by the bacterial supernatant of Bacillus sp. HR21-6.
Analysis of lipase-mediated O-acylation of polyphenols
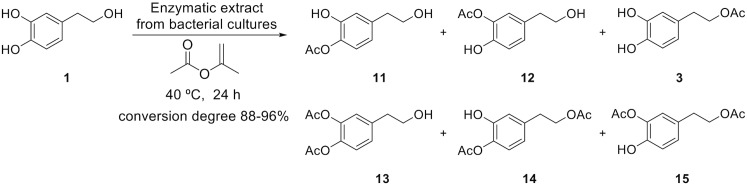
All the previous reactions were based on the deacetylation of a peracetylated derivative of the corresponding polyphenol. In order to check the selectivity of the enzymes in the bacterial supernatants during esterification reactions, we also tested the direct O-acylation of polyphenols 1, 4 and 8. In this case, a commercial lipase obtained from Candida antarctica (Novozyme 435) used previously in this type of reactions [27] was also studied in parallel with our substrates for comparative purposes. The O-acylations of the phenolic compounds 1, 4 and 8 were carried out with the four lipolytic bacterial extracts and isopropenyl acetate, as solvent and acylating agent, by heating at 40°C for 24 h. The enzymes in the bacterial supernatants were adsorbed on celita for 1 and 8.
The conversion degree for the acetylation of HT (1) by the lipase extracts was in the range from 96% for the Bacillus sp HR21-6 extract to 88% for the Enterobacter sp. 1B89 extract (Fig 7 and Table 1). The OH-4 of the catechol moiety is the more reactive hydroxyl group, and the formation of the monoacetylated derivative at that position (compound 11) ranged from 44% for Pseudomonas sp. 2B120 extract to 34% for Enterobacter sp. 1B89 extract. This compound and its regioisomer in position 3 (compound 12) were formed in a ratio 2:1 after 24 h of reaction (Fig 7 and Table 1). The three diacetylated derivatives 13−15 were formed in low proportion (2–10%), and the triacetylated derivative could not be detected (S2 Fig). In contrast, the acetylation was completely regioselective towards the lateral chain with C. antarctica lipase, yielding only derivative 3 (quant.). Most of the substrate was converted into the acetylated version after 24 h of reaction (88–96% for the bacterial supernatants and 100% for the Candida lipase).
Fig 7. O-Acetylation of HT (1) with isopropenyl acetate catalyzed by lipases from the supernatant of bacterial cultures.
Table 1. Regioselective acetylation of compounds 1, 4 and 8 with bacterial supernatants or commercial lipase from C. antarctica (Novozyme 435).
| Compound | Terribacillus sp. 2B122 | Enterobacter sp. 1B89 | Pseudomonas sp. 2B120 | Bacillus sp. HR21-6 | Commercial lipase (C. antarctica) |
|---|---|---|---|---|---|
| 1 | 8% | 12% | 6% | 4% | - |
| 11 | 36% | 34% | 44% | 38% | - |
| 12 | 20% | 16% | 23% | 21% | - |
| 3 | 19% | 21% | 11% | 18% | 100% |
| 13 | 2% | 2% | 3% | 3% | - |
| 14 | 10% | 9% | 5% | 9% | - |
| 15 | 5% | 6% | 8% | 7% | - |
| 4 | 39% | 30% | 32% | 34% | 47% |
| 16 | 17% | 16% | 14% | 12% | 10% |
| 17 | 16% | 16% | 13% | 11% | 10% |
| 18 | 15% | 18% | 22% | 23% | 20% |
| 19 | 7% | 11% | 10% | 11% | 7% |
| 20 | 7% | 9% | 9% | 9% | 6% |
| 8 | 22% | 12% | 10% | 10% | 5% |
| 21 | 22% | 24% | 35% | 27% | - |
| 22 | 19% | 20% | 29% | 23% | - |
| 23 | 20% | 24% | 13% | 21% | 87% |
| 24 | 3% | 1% | 2% | 1% | - |
| 25 | 7% | 10% | 6% | 9% | 9% |
| 26 | 7% | 9% | 5% | 9% | - |
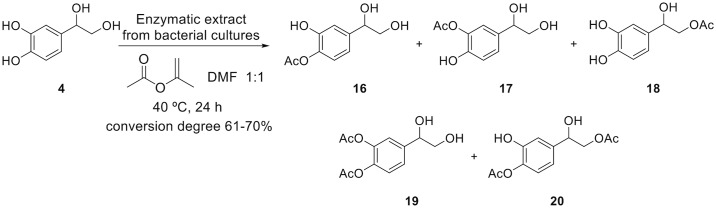
When the acetylation reaction was carried out on DHGP (compound 4), both monoacetylated and diacetylated derivatives were also obtained (Fig 8 and S2D and S2E Fig). A low conversion degree for the acetylation of 4 is found for the lipase extracts, in a range of from 61% to 70% (Table 1). The monoacetylated derivatives in the catechol moiety 16, 17 and in the primary aliphatic alcohol 18 were formed in similar proportions (23–11%). The ratios between the monoacylation in the aromatic ring and the aliphatic chain were 1:1:1 when the supernatants from Terribacillus sp. 2B122 or Enterobacter sp. 1B89 were used, and 1:1:2 when the supernatants from Pseudomonas sp. 2B120 or Bacillus sp. HR21-6 were employed (Table 1). Surprisingly, no acylation of the benzylic hydroxyl group was observed in any case (Fig 8 and S2D and S2E Fig). The acylation in the presence of the C. antarctica lipase showed a low regioselectivity, and the monoacylation in the primary hydroxyl group only took place with a 20% yield. Remarkably the conversion of the original substrate achieved with the lipase from C. antarctica was lower (53%) than those with the bacterial lipases extracts (61–70%).
Fig 8. O-Acetylation of DHPG (4) with isopropenyl acetate catalyzed by lipases from the supernatant of bacterial cultures.
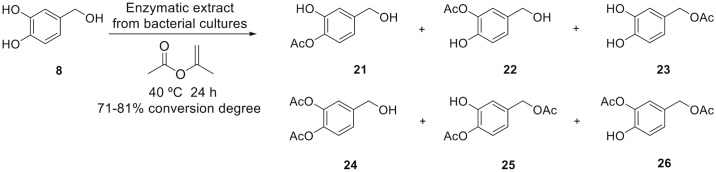
Since it was very interesting that the benzylic alcohol in DHPG was not acetylated, we also tested the protocatechuic alcohol 8 for regioselective acetylation of the benzylic alcohol. In this case, a mixture of all the possible mono- and di-acetylated derivatives, including the benzylic position, was observed regardless of the bacterial isolate (Fig 9 and S2F–S2H Fig). The OH-4 of the catechol moiety was the most reactive hydroxyl group of 8 (Fig 9), and the monoacetylation at that position ranged from 35% to 22%. The ratio of the three possible monoacetylated compounds 21–23 was different depending on the isolate: 1:1:1 in the case of Terribacillus sp. 2B122, 1:2:2 in the case of Enterobacter sp. 1B89, 1:3.5:3 in the case of Pseudomonas sp. 2B120, and approx. 1:3:2 in the case of Bacillus sp. HR21-6. The degree of conversion of the protochatechuic alcohol 8 (78–90%) was lower than the HT 1 (88–96%) but higher than the DHGP 4 (61–70%). The three diacetylated derivatives 24–26 were formed in low proportion (1–10%) and triacetylated derivative 9 was not detected as happened with HT and DHPG. The acylation with C. antarctica lipase is highly regioselective towards the lateral chain to form 23, although less efficient than the acylation of 1.
Fig 9. O-Acetylation of protocatechuic alcohol (8) with isopropenyl acetate catalyzed by lipases from the supernatant of bacterial cultures.
DPPH radical scavenging activity assay
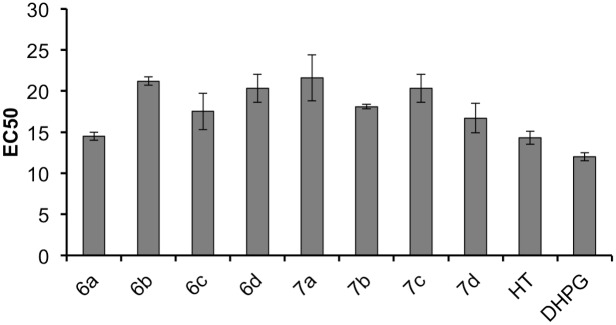
DHPG displays a strong antioxidant activity. Acetylation of the non-catecholic alcohols increases its lypophilic character, which expands the possibilities of formulations for this compound in the cosmetics, food and pharmacology industries. Therefore, the antioxidant capabilities of phenolic compounds 6a-d and 7a-d was evaluated using the DPPH method [19,20]. The EC50 value was calculated through the lineal representation of each compound in 5 different concentrations between 5 and 25 μM, and it represents the concentration of the compound necessary to reduce the amount of the DPPH radical to 50%, i.e. lower concentrations of the compound under test indicate a higher antioxidant capacity. For comparative purposes, HT 1 and DHPG 4, two antioxidants with remarkable activity were included. As shown in Fig 10, all the compounds have a similar antiradical activity (14–21 μM). In addition, the compounds 6a and 7d showed the best activity with values comparable to that of HT (14.5 ± 0.5 and 16.7 ± 1.8 μM, respectively).
Fig 10. DPPH radical scavenging activity of lipophilic derivatives of DHPG (compounds 6a-d and 7a-d) compared to HT and DHPG.
Discussion
Previous reports have shown that the lipophilic derivatives and analogs of one of the olive phenols, HT, with esters or ethers functionalities display a better hydrophilic/lipophilic balance [28], an increased bioavailability compared with their unprotected analogs [29] and stability to oxygen compared with the underivatized HT, which make these molecules more attractive for the food industry [30]. These findings prompted us to develop three innovative aspects. Firstly, we introduce a novel concept of biocatalyst for the widespread use in industry, employing extracts of lipolytic bacterial strains. These extracts were only partially purified and consequently very cheap to obtain, but displaying performance levels similar or even higher than commercially available purified lipases. Additionally, the use of these lipolytic extracts allowed us to obtain unprecedented analogs in one single step.
Secondly, in this study, in addition to HT, we have also employed DHPG, a compound also present in olives [23]. Indeed, our data confirmed that DHPG is more efficient as antioxidant than HT. DHPG is a simple phenol structurally similar to HT, but with an additional hydroxyl group at the benzylic position. The reason to use this poorly studied polyphenol relies on its antioxidant efficiency in water, which is 2–3 times higher than that of ascorbic acid or HT, whereas in lipid medium it is comparable to that of vitamin E despite its high polarity [31]. Interestingly, this compound could be obtained from olive-mill wastes (alperujo) using a simple method [32]. The wastes of the olive oil industry are considered a cheap and easily available source of bioactive compounds, whose activity is reflected in the olive oil itself. DHPG displays excellent possibilities to be modified, containing different free OH groups on its structure that can be acylated, obtaining a battery of novel bioactive lipophilic derivatives with improved activity and bioavailability. Despite of its advantages, to the best of our knowledge this compound has never been reported as an ingredient for the formulation of functional foods, and no semisynthetic derivatives of this compound have been reported in the literature. During this work, the deacetylation of fully acetylated DHPG 5 in different aliphatic alcohols gave not only deprotection of the phenolic hydroxyl groups, but also an unexpected substitution of the acetoxy group at the benzylic position by an alkoxy group, which allowed access in one single synthetic step to a new family of lipophilic polyphenols 6a-6d, bearing both ester and ether functionalities, that can be tailored to get the best pharmacological profile.
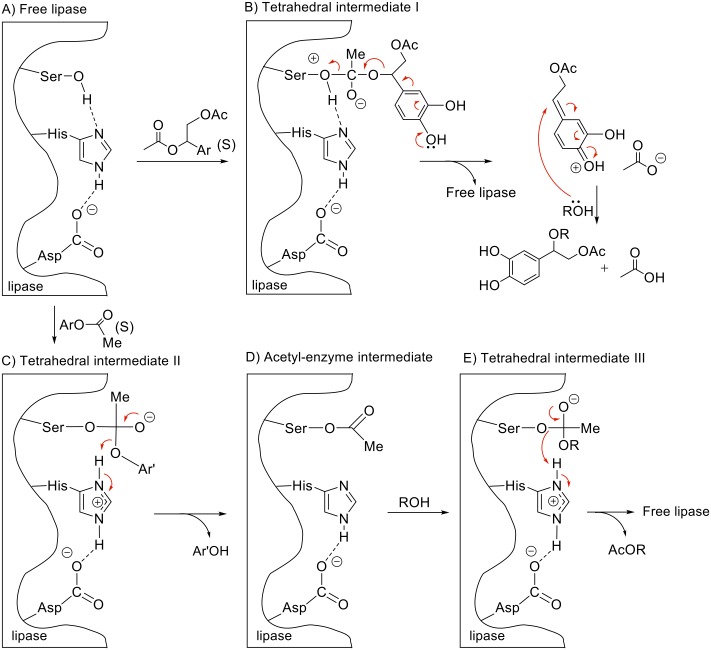
A generally accepted mechanism for lipase-catalyzed transesterification reactions involves Ser, His and Asp residues as the catalytic triad, and the intermediates II and III shown in Fig 11 [33]. For the alcoholysis of the acetate group at the benzylic position, the tetrahedral intermediate I might be the key step. The electron donating character of the p-hydroxyl group helps to eliminate the acetate group simultaneously to the release of the free enzyme. Finally, the stabilized oxocarbenium cation reacts at the benzylic position with ROH, what might explain the outcome of the reaction.
Fig 11. Proposed mechanism for the lipase catalysed formation of 3,4-dihydroxyphenylglycol ethers 6.
An increase in the reaction rate and the complete disappearance of the starting material was observed when silica gel was added to the reaction mixture. On the contrary, using only MeOH and lipase we observed a slower reaction rate and a low final conversion. These results are in agreement with the significant improvement of the conversion yield when silica gel-adsorbed substrates were used in lipase-catalyzed esterification reactions [34]. The critical role of silica gel in these kinds of reactions was associated with its behavior as polar substrate reservoir and with the protection of the enzyme avoiding its blockage [35]
Furthermore, ethers 7a-7d bearing a free hydroxyl group at the end of the side chain were easily accessed by chemical hydrolysis of the acetate of 6a-6d, expanding even more the possibilities for these compounds. Some of these compounds have already been detected before. In particular, compound 7a was detected in olive (Olea europaea) tree leaves [36] and compound 10 in the fermentation broth of the marine fungus Y26-02 [18].
And lastly, and also to the best of our knowledge it is the first time that a lipase mediated reaction has catalyzed not only the deacylation of phenolic esters, but also the deacetoxylation of an ester group at benzylic position. At the same time, the aliphatic ester at the end of the side chain of peracetylated DHPG 5 remained untouched. On the contrary, peracetylated hydroxytyrosol 2 could not be transformed into ether because it lacks the acetoxy group at the right position. However, a series of HT alkyl ethers, with an alkoxy group at the end of the side chain, have been previously prepared in three steps starting from HT [37]. These methyl, ethyl, propyl and butyl ethers reduced ROS generation, malondialdehyde formation, and glutathione depletion in HepG2 human hepatocarcinoma cells treated [38]. Similarly, HT alkyl ethers (C2-C12) inhibited lipid peroxidation and reduced glutathione depletion in rat brain slices, after hypoxia and re-oxygenation [39]. Esters and ethers derived from HT exert higher anti-proliferative and pro-apoptotic activity than HT itself. The highest cytotoxic activity was found by the ethers with the longest alkyl chain [40]. Similarly, these lipophilic derivatives inhibit both monocyte and macrophage pro-tumorigenic inflammatory activities and platelet function in a more effective way than HT [41], which reflects the broad applicability and importance of these type of molecules.
In conclusion, this study opens new avenues for the food industries to obtain unprecedented derivatives of antioxidants with expanded physico-chemical properties and utilities. Families of DHPG derivatives could be more suited for these purposes than the extensively studied HT. Consequently, biological and pharmacological studies of both series of DHPG derivatives will be undertaken, in order to elucidate the more effective compounds in cancer and chronic degenerative disease prevention, and as anti-inflammatory compounds.
Supporting Information
The 16S rRNA sequence of Streptococcus pneumoniae was used as an external group. 16S rRNA gene sequences from the isolates correspond to 1380 bps. The phylogenetic reconstruction carried out with different methods was consistent, and consequently only the tree obtained with Neighbor-Joining, for the evolutionary history, and Maximum Composite Likelihood, for evolutionary distances is shown.
(TIF)
Partial 1H-NMR spectra for the lipase-mediated acetylation of HT (A) to give mono- and di-acetylated derivatives of HT (C), compared to the peracetylated HT (B). Partial 1H-NMR spectra for the lipase-mediated acetylation of DHPG (D) to give mono- and di-acetylated derivatives of DHPG (E). Partial 1H-NMR spectra for the lipase-mediated acetylation of protocatechuic alcohol (F) to give mono- and di-acetylated derivatives of protocatechuic alcohol (H), compared to the peracetylated protocatechuic alcohol (G).
(TIF)
2-(3,4-Dihydroxyphenyl)-2-metoxyethyl acetate (6a) from compound 5; 2-(3,4-Dihydroxyphenyl)-2-ethoxyethyl acetate (6b); 2-(3,4-Dihydroxyphenyl)-2-propoxyethyl acetate (6c); 2-Butoxy-2-(3,4-dihydroxyphenyl)ethyl acetate (6d); 2-(3,4-Dihydroxyphenyl)-2-methoxyethanol (7a); 2-(3,4-Dihydroxyphenyl)-2-ethoxyethanol (7b); 2-(3,4-Dihydroxyphenyl)-2-propoxyethanol (7c); 2-Butoxy-2-(3,4-dihydroxyphenyl)ethanol (7d); Spectroscopic data for crude reaction depicted in Fig 7; Spectroscopic data for crude reaction depicted in Fig 8; Spectroscopic data for crude reaction depicted in Fig 9.
(PDF)
(PDF)
In red, the isolates selected with absorbance values higher than the highest hydrolysis control.
(PDF)
Acknowledgments
We would like to thank the Servicio de Resonancia Magnética Nuclear, CITIUS (University of Seville) for the performance of NMR experiments.
Data Availability
The nucleotide sequences reported in this work have been deposited under accession numbers KP212109 to KP212128 in the GenBank database.
Funding Statement
We thank the Junta de Andalucía (P08-NMR-3515, P11-CVI-7427 MO, FQM134 and BIO-213) and the European Regional Development Fund (FEDER) for financial support. AGB thanks the Spanish Ministerio de Economía y Competitividad for the award of a grant.
References
- 1.Pauwels EKJ. The protective effect of the Mediterranean diet: focus on cancer and cardiovascular risk. Med Princ Pract. 2011;20: 103–111. 10.1159/000321197 [DOI] [PubMed] [Google Scholar]
- 2.Deiana M, Corona G, Incani A, Loru D, Rosa A, Atzeri A, et al. Protective effect of simple phenols from extravirgin olive oil against lipid peroxidation in intestinal Caco-2 cells. Food Chem Toxicol. 2010;48: 3008–3016. 10.1016/j.fct.2010.07.041 [DOI] [PubMed] [Google Scholar]
- 3.Covas M-I, Nyyssönen K, Poulsen HE, Kaikkonen J, Zunft H-JF, Kiesewetter H, et al. The effect of polyphenols in olive oil on heart disease risk factors: a randomized trial. Ann Intern Med. 2006;145: 333–341. Available: http://www.ncbi.nlm.nih.gov/pubmed/16954359 [DOI] [PubMed] [Google Scholar]
- 4.Kalaiselvan I, Samuthirapandi M, Govindaraju A, Sheeja Malar D, Kasi PD. Olive oil and its phenolic compounds (hydroxytyrosol and tyrosol) ameliorated TCDD-induced heptotoxicity in rats via inhibition of oxidative stress and apoptosis. Pharm Biol. 2016;54: 338–346. 10.3109/13880209.2015.1042980 [DOI] [PubMed] [Google Scholar]
- 5.Teixeira A, Baenas N, Dominguez-Perles R, Barros A, Rosa E, Moreno DA, et al. Natural bioactive compounds from winery by-products as health promoters: a review. Int J Mol Sci. 2014;15: 15638–15678. 10.3390/ijms150915638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cilla A, De Palma G, Lagarda MJ, Barberá R, Farré R, Clemente G, et al. Impact of fruit beverage consumption on the antioxidant status in healthy women. Ann Nutr Metab. 2009;54: 35–42. 10.1159/000205318 [DOI] [PubMed] [Google Scholar]
- 7.Bohn T. Dietary factors affecting polyphenol bioavailability. Nutr Rev. 2014;72: 429–452. 10.1111/nure.12114 [DOI] [PubMed] [Google Scholar]
- 8.Medina I, Lois S, Alcántara D, Lucas R, Morales JC. Effect of lipophilization of hydroxytyrosol on its antioxidant activity in fish oils and fish oil-in-water emulsions. J Agric Food Chem. 2009;57: 9773–9779. 10.1021/jf9023867 [DOI] [PubMed] [Google Scholar]
- 9.Zhong Y, Shahidi F. Lipophilised epigallocatechin gallate (EGCG) derivatives and their antioxidant potential in food and biological systems. Food Chem. 2012;131: 22–30. 10.1016/j.foodchem.2011.07.089 [DOI] [Google Scholar]
- 10.Wuts PGM, Greene TW. Greene’s Protective Groups in Organic Synthesis. John Wiley & Sons; 2006. Available: https://books.google.com/books?id=KPyGzoxfAt0C&pgis=1 [Google Scholar]
- 11.Bernini R, Mincione E, Barontini M, Crisante F. Convenient synthesis of hydroxytyrosol and its lipophilic derivatives from tyrosol or homovanillyl alcohol. J Agric Food Chem. 2008;56: 8897–904. 10.1021/jf801558z [DOI] [PubMed] [Google Scholar]
- 12.Bernini R, Crisante F, Barontini M, Tofani D, Balducci V, Gambacorta A. Synthesis and structure/antioxidant activity relationship of novel catecholic antioxidant structural analogues to hydroxytyrosol and its lipophilic esters. J Agric Food Chem. 2012;60: 7408–7406. 10.1021/jf301131a [DOI] [PubMed] [Google Scholar]
- 13.Sánchez-Fidalgo S, Villegas I, Aparicio-Soto M, Cárdeno A, Rosillo MÁ, González-Benjumea A, et al. Effects of dietary virgin olive oil polyphenols: hydroxytyrosyl acetate and 3, 4-dihydroxyphenylglycol on DSS-induced acute colitis in mice. J Nutr Biochem. 2015;26: 513–520. 10.1016/j.jnutbio.2014.12.001 [DOI] [PubMed] [Google Scholar]
- 14.Gupta A, Khare SK. Enzymes from solvent-tolerant microbes: Useful biocatalysts for non-aqueous enzymology. Crit Rev Biotechnol. 2009;29: 44–54. 10.1080/07388550802688797 [DOI] [PubMed] [Google Scholar]
- 15.Fernandez-Bolanos J, Lopez O, Fernandez-Bolanos J, Rodriguez-Gutierrez G. Hydroxytyrosol and Derivatives: Isolation, Synthesis, and Biological Properties. Curr Org Chem. 2008;12: 442–463. 10.2174/138527208784083888 [DOI] [Google Scholar]
- 16.Escobar-Niño A, Luna C, Luna D, Marcos AT, Cánovas D, Mellado E. Selection and characterization of biofuel-producing environmental bacteria isolated from vegetable oil-rich wastes. PLoS One. 2014;9: e104063 10.1371/journal.pone.0104063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christophoridou S, Dais P, Tseng L-H, Spraul M. Separation and identification of phenolic compounds in olive oil by coupling high-performance liquid chromatography with postcolumn solid-phase extraction to nuclear magnetic resonance spectroscopy (LC-SPE-NMR). J Agric Food Chem. 2005;53: 4667–4679. 10.1021/jf040466r [DOI] [PubMed] [Google Scholar]
- 18.Wu H-H, Tian L, Chen G, Xu N, Wang Y-N, Sun S, et al. Six compounds from marine fungus Y26-02. J Asian Nat Prod Res. 2009;11: 748–751. 10.1080/10286020903025783 [DOI] [PubMed] [Google Scholar]
- 19.Bernini R, Crisante F, Fabrizi G, Gentili P. Convenient Synthesis of 1-Aryl-dihydroxyisochromans Exhibiting Antioxidant Activity. Curr Org Chem. 2012;16: 1051–1057. 10.2174/138527212800194700 [DOI] [Google Scholar]
- 20.Barontini M, Bernini R, Carastro I, Gentili P, Romani A, Nunomura A, et al. Synthesis and DPPH radical scavenging activity of novel compounds obtained from tyrosol and cinnamic acid derivatives. New J Chem. The Royal Society of Chemistry; 2014;38: 809–816. 10.1039/c3nj01180a [DOI] [Google Scholar]
- 21.Capasso R, Sannino F, De Martino A, Manna C. Production of Triacetylhydroxytyrosol from Olive Mill Waste Waters for Use as Stabilized Bioantioxidant. J Agric Food Chem. 2006;54: 9063–9070. 10.1021/jf061290r [DOI] [PubMed] [Google Scholar]
- 22.Sánchez-Fidalgo S, Villegas I, Aparicio-Soto M, Cárdeno A, Rosillo MÁ, González-Benjumea A, et al. Effects of dietary virgin olive oil polyphenols: hydroxytyrosyl acetate and 3, 4-dihydroxyphenylglycol on DSS-induced acute colitis in mice. J Nutr Biochem. 2015;26: 513–520. 10.1016/j.jnutbio.2014.12.001 [DOI] [PubMed] [Google Scholar]
- 23.Brenes M, García A, García P, Rios JJ, Garrido A. Phenolic Compounds in Spanish Olive Oils. J Agric Food Chem. American Chemical Society; 1999;47: 3535–3540. 10.1021/jf990009o [DOI] [PubMed] [Google Scholar]
- 24.Aparicio-Soto M, Sánchez-Fidalgo S, González-Benjumea A, Maya I, Fernández-Bolaños JG, Alarcón-de-la-Lastra C. Naturally occurring hydroxytyrosol derivatives: hydroxytyrosyl acetate and 3,4-dihydroxyphenylglycol modulate inflammatory response in murine peritoneal macrophages. Potential utility as new dietary supplements. J Agric Food Chem. 2015;63: 836–846. 10.1021/jf503357s [DOI] [PubMed] [Google Scholar]
- 25.Saitta M, Salvo F, Di Bella G, Dugo G, La Torre GL. Minor compounds in the phenolic fraction of virgin olive oils. Food Chem. 2009;112: 525–532. 10.1016/j.foodchem.2008.06.001 [DOI] [Google Scholar]
- 26.Du X.-p., Zhao B.-b., Zheng Z.-h., Xu Q.-y., Su W.-j. Study on the Isolation Identification and Antitumor Activity of a Phenol Derivative from Mangrove Fungus BYY-1. J Jimei Univ Sci. 2011;16: 424–428. [Google Scholar]
- 27.Aladedunye F, Niehaus K, Bednarz H, Thiyam-Hollander U, Fehling E, Matthäus B. Enzymatic lipophilization of phenolic extract from rowanberry (Sorbus aucuparia) and evaluation of antioxidative activity in edible oil. LWT—Food Sci Technol. 2015;60: 56–62. 10.1016/j.lwt.2014.08.008 [DOI] [Google Scholar]
- 28.Lorentz C, Dulac A, Pencreac’h G, Ergan F, Richomme P, Soultani-Vigneron S. Lipase-catalyzed synthesis of two new antioxidants: 4-O- and 3-O-palmitoyl chlorogenic acids. Biotechnol Lett. 2010;32: 1955–1960. 10.1007/s10529-010-0386-6 [DOI] [PubMed] [Google Scholar]
- 29.Fragopoulou E, Nomikos T, Karantonis HC, Apostolakis C, Pliakis E, Samiotaki M, et al. Biological activity of acetylated phenolic compounds. J Agric Food Chem. 2007;55: 80–89. 10.1021/jf0627221 [DOI] [PubMed] [Google Scholar]
- 30.Pereira-Caro G, Sarriá B, Madrona A, Espartero JL, Escuderos ME, Bravo L, et al. Digestive stability of hydroxytyrosol, hydroxytyrosyl acetate and alkyl hydroxytyrosyl ethers. Int J Food Sci Nutr. 2012;63: 703–707. 10.3109/09637486.2011.652943 [DOI] [PubMed] [Google Scholar]
- 31.Rodríguez G, Lama A, Jaramillo S, Fuentes-Alventosa JM, Guillén R, Jiménez-Araujo A, et al. 3,4-Dihydroxyphenylglycol (DHPG): an important phenolic compound present in natural table olives. J Agric Food Chem. 2009;57: 6298–6304. 10.1021/jf803512r [DOI] [PubMed] [Google Scholar]
- 32.Rodríguez G, Lama A, Trujillo M, Espartero JL, Fernández-Bolaños J. Isolation of a powerful antioxidant from Olea europaea fruit-mill waste: 3,4-Dihydroxyphenylglycol. LWT—Food Sci Technol. 2009;42: 483–490. 10.1016/j.lwt.2008.08.015 [DOI] [Google Scholar]
- 33.Kobayashi S. Lipase-catalyzed polyester synthesis—a green polymer chemistry. Proc Jpn Acad Ser B Phys Biol Sci. The Japan Academy; 2010;86: 338–365. 10.2183/pjab.86.338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bellot JC, Choisnard L, Castillo E, Marty A. Combining solvent engineering and thermodynamic modeling to enhance selectivity during monoglyceride synthesis by lipase-catalyzed esterification. Enzyme Microb Technol. 2001;28: 362–369. 10.1016/s0141-0229(00)00326-4 [DOI] [PubMed] [Google Scholar]
- 35.Castillo E, Dossat V, Marty A, Condoret JS, Combes D. The role of silica gel in lipase-catalyzed esterification reactions of high-polar substrates. J Am Oil Chem Soc. Springer-Verlag; 1997;74: 77–85. [Google Scholar]
- 36.Di Donna L, Mazzotti F, Naccarato A, Salerno R, Tagarelli A, Taverna D, et al. Secondary metabolites of Olea europaea leaves as markers for the discrimination of cultivars and cultivation zones by multivariate analysis. Food Chem. 2010;121: 492–496. 10.1016/j.foodchem.2009.12.070 [DOI] [Google Scholar]
- 37.Madrona A, Pereira-Caro G, Mateos R, Rodríguez G, Trujillo M, Fernández-Bolaños J, et al. Synthesis of hydroxytyrosyl alkyl ethers from olive oil waste waters. Molecules. 2009;14: 1762–1772. 10.3390/molecules14051762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pereira-Caro G, Sarriá B, Madrona A, Espartero JL, Goya L, Bravo L, et al. Alkyl hydroxytyrosyl ethers show protective effects against oxidative stress in HepG2 cells. J Agric Food Chem. 2011;59: 5964–5976. 10.1021/jf2002415 [DOI] [PubMed] [Google Scholar]
- 39.Guerrero A, De la Cruz JP, Muñoz-Marín J, López-Villodres JA, Madrona A, Espartero JL, et al. Neuroprotective effect of alkyl hydroxytyrosyl ethers in rat brain slices subjected to a hypoxia-reoxygenation model. Food Chem. 2012;134: 2176–2183. 10.1016/j.foodchem.2012.04.022 [DOI] [PubMed] [Google Scholar]
- 40.Calderón-Montaño JM, Madrona A, Burgos-Morón E, Orta ML, Mateos S, Espartero JL, et al. Selective Cytotoxic Activity of New Lipophilic Hydroxytyrosol Alkyl Ether Derivatives. J Agric Food Chem. 2013;61: 5046–5053. 10.1021/jf400796p [DOI] [PubMed] [Google Scholar]
- 41.Bernini R, Gilardini Montani MS, Merendino N, Romani A, Velotti F. Hydroxytyrosol-Derived Compounds: A Basis for the Creation of New Pharmacological Agents for Cancer Prevention and Therapy. J Med Chem. 2015;58: 9089–9107. 10.1021/acs.jmedchem.5b00669 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The 16S rRNA sequence of Streptococcus pneumoniae was used as an external group. 16S rRNA gene sequences from the isolates correspond to 1380 bps. The phylogenetic reconstruction carried out with different methods was consistent, and consequently only the tree obtained with Neighbor-Joining, for the evolutionary history, and Maximum Composite Likelihood, for evolutionary distances is shown.
(TIF)
Partial 1H-NMR spectra for the lipase-mediated acetylation of HT (A) to give mono- and di-acetylated derivatives of HT (C), compared to the peracetylated HT (B). Partial 1H-NMR spectra for the lipase-mediated acetylation of DHPG (D) to give mono- and di-acetylated derivatives of DHPG (E). Partial 1H-NMR spectra for the lipase-mediated acetylation of protocatechuic alcohol (F) to give mono- and di-acetylated derivatives of protocatechuic alcohol (H), compared to the peracetylated protocatechuic alcohol (G).
(TIF)
2-(3,4-Dihydroxyphenyl)-2-metoxyethyl acetate (6a) from compound 5; 2-(3,4-Dihydroxyphenyl)-2-ethoxyethyl acetate (6b); 2-(3,4-Dihydroxyphenyl)-2-propoxyethyl acetate (6c); 2-Butoxy-2-(3,4-dihydroxyphenyl)ethyl acetate (6d); 2-(3,4-Dihydroxyphenyl)-2-methoxyethanol (7a); 2-(3,4-Dihydroxyphenyl)-2-ethoxyethanol (7b); 2-(3,4-Dihydroxyphenyl)-2-propoxyethanol (7c); 2-Butoxy-2-(3,4-dihydroxyphenyl)ethanol (7d); Spectroscopic data for crude reaction depicted in Fig 7; Spectroscopic data for crude reaction depicted in Fig 8; Spectroscopic data for crude reaction depicted in Fig 9.
(PDF)
(PDF)
In red, the isolates selected with absorbance values higher than the highest hydrolysis control.
(PDF)
Data Availability Statement
The nucleotide sequences reported in this work have been deposited under accession numbers KP212109 to KP212128 in the GenBank database.