Abstract
Transformation with an isopentenyl transferase (ipt) gene controlling cytokinin (CK) synthesis has been shown to enhance plant drought tolerance. The objective of this study was to identify differentially-expressed genes (DEGs) in creeping bentgrass (Agrostis stolonifera) overexpressing ipt compared to non-transgenic plants. The ipt transgene was controlled by a senescence-activated promoter (SAG12). Both a null transformed line (NT) and SAG12-ipt plants were exposed to drought stress in an environmentally-controlled growth chamber until the soil water content declined to approximately 5% and leaf relative water content declined to 47%, which were both significantly below the well-watered controls. RNA was extracted from leaf samples of both well-watered and drought-stressed plants. Eight sets of subtractive hybridizations were performed for detection of up-regulated and down-regulated genes due to the presence of the transgene and due to drought stress in both NT and transgenic plants. Sequencing analysis revealed the identity of 252 DEGs due to either the transgene and drought stress. Sequencing analysis of 170 DEGs identified genes encoding for proteins that were related to energy production, metabolism, stress defense, signaling, protein synthesis and transport, and membrane transport could play major roles in the improved drought tolerance by overexpressing ipt in creeping bentgrass.
Introduction
Drought stress affects various physiological and metabolic processes, including the induction of leaf senescence which is known to be regulated by plant hormones, such as CK [1]. Increasing CK synthesis through overexpressing a gene encoding isopentyl transferase controlling CK synthesis (ipt) has been shown to increase drought tolerance in creeping bentgrass [2–4] and other species [5–7]. SAG12-ipt creeping bentgrass exhibits a greater ability to maintain CK content and improvement in major physiological characteristics governing drought tolerance, as manifested by reduced electrolyte leakage, maintenance of higher relative water content, improvement in photosynthetic rate, and reduction in oxidative damage, which provided strong evidence of improved drought tolerance by increasing endogenous CK [2–4]. Rivero et al. [8] examined transcript changes in tobacco overexpressing ipt and found an increase in transcripts related to chlorophyll biosynthesis, photosynthetic reactions and other pathways. Their results focused on elevated CK levels in transgenic tobacco plants promoting transcription of photosynthesis-associated gene transcripts, ABA and brassinosteroid associated responses. Little discussion of antioxidant enzymes or other stress associated transcripts is available. In creeping bentgrass, enhanced cytokinin content via SAG-ipt expression promoted the expression of specific stress responsive proteins such as antioxidants and chaperones [3]. Thus, identifying gene expression changes associated with drought stress and enhanced cytokinin biosynthesis for stress responses, particularly in a monocot grass species is needed. Additionally, the physiology of turfgrass species is significantly different than other species due to the unique nature of their culture, which includes mowing. Mowing has been found to have a significant effect on hormone accumulations in creeping bentgrass [9]. Thus, how cytokinins affect gene expression in a mowed plant deserves investigation.
Proteomic evaluation of SAG12-ipt creeping bentgrass revealed major changes in the proteome during drought stress compared to NT plants. Protein abundance and activity assays revealed that enhanced CK may elicit improved physiological characteristics associated with a reduction in leaf senescence and drought damage via maintenance of greater antioxidant enzyme activities (e.g. superoxide dismutase, peroxidases, 2-Cys peroxiredoxin, and catalase). In addition to stress protective proteins, proteins associated with major metabolic pathways such as energy production, carbon and nitrogen metabolism, protein synthesis and destination were generally maintained to a greater extent SAG12-ipt plants compared to the null transformed line (NT) [4]. Metabolomic analysis revealed similar metabolic changes to those found in proteomic analysis, as major metabolites such as carbohydrate, amino acids, and organic acids involved carbohydrate metabolism and energy production pathways accumulated to a greater extent in SAG12-ipt plants compared to NT [10]. However, the underlying transcript changes associated with the aforementioned metabolic processes found through metabolic and proteomic profiling due to ipt overexpression that could contribute to the improved drought tolerance have yet to be documented. Identification of gene changes may help in elucidating molecular factors accounting for proteomic and metabolic changes contributing to enhanced drought tolerance by CK.
Subtractive suppressive hybridization (SSH) remains to be an effective way to identify specific differentially-expressed genes or transcript accumulation for comparisons between different genotypes or environmental conditions, despite recent advances and availability of next-generation gene sequencing technologies [11–13]. SSH evaluation followed by sequence analysis has identified genes that play a role in conferring drought tolerance in a number of crop species such as cotton (Gossypium arboretum) [14], soybean (Glycine max) [15], and corn (Zea mays) [16]. In grass species, SSH has been performed to identify heat-responsive genes in Fescue sp. (Festuca sp.) [17] and drought stress in perennial ryegrass (Lolium perenne) [18]. In drought tolerant compared to sensitive accessions of perennial ryegrass, major differences detected during drought stress were associated with plant antioxidant systems [18]. Numerous drought-responsive genes have been identified through SSH and the next-generation sequencing, as mentioned above, which provides insights into molecular mechanisms of drought responses; however, transcripts altered by increasing CK through genetic transformation that contribute to improved drought tolerance are not well understood. Therefore, the objectives of the study were to identify differentially-expressed genes in creeping bentgrass overexpressing SAG12-ipt compared to the NT under both well-watered and drought stressed conditions using SSH analysis and to determine major metabolic processes involved in CK-regulation of drought tolerance.
Materials and Methods
Plant material
A null transformed line of creeping bentgrass ‘Penncross’ (NT) and a transgenic line containing the ipt gene linked to the SAG12 promoter (SAG12-ipt plants) were exposed to different watering treatments in an environmental growth chamber. The ipt gene was cloned from agrobacterium (Agrobacterium tumefaciens) and SAG12-ipt plants were transformed using the Agrobacterium method as described previously [2–4, 19–20]. In order for ease of comparison to previous results, the same plant material used for proteomic and metabolomic evaluation [3–4] was used for SSH analysis. Briefly, plants were vegetatively propagated by separating tillers in a greenhouse. The plants were grown in PVC tubes (40 cm in height x 10.16 cm in diameter) all containing an equal volume of 1:1 fine sand:soil mix (fine-loamy, mixed mesic Typic Hapludult type soil). Plants were watered and fertilized to optimum levels for creeping bentgrass growth in a greenhouse until a full canopy covering the 10.16 cm diameter was achieved. Subsequently, the plants were moved to a controlled-environment growth chamber (Conviron, Winnipeg, Canada). All plants were fertilized once per week with Hoagland’s nutrient solution [21] and grown in growth chamber conditions consisting of 20/15°C (day/night) temperatures, 12 h photoperiod, 60% relative humidity, and 500 μmol m-2 s-1 photosynthetic photon flux density at canopy height. After a 10 d acclimation period, watering treatments were imposed on 3 March 2009.
Watering treatments
Both NT and SAG12-ipt plant types were subjected to either well-watered control or water stress conditions (total 80 plants). A subset of 20 plants of NT and SAG12-ipt were watered once daily to maintain soil volumetric water content (SWC) at approximately 25–30% (well-watered control). SWC was determined with the Time Domain Reflectometry (TDR) method [22] using a Trase TDR instrument (Soil Moisture Equipment Corp., Santa Barbara, CA). SWC was measured with one three-pronged waveguide probes (20 cm in length, spaced 2.54 cm apart) installed vertically in each pot, four probes in the control treatment and four probes in the water stress treatment (four replicates in each line). Pot capacity of the soil water was approximately 25%. Water stress was imposed to a subset of 20 plants of each plant type by completely withholding irrigation. The well-watered control and drought treatments each included four replicates for the NT and transgenic line.
cDNA library construction and SSH analysis
Leaves collected from NT and SAG12-ipt plants maintained under well-watered conditions and drought stress with 47% RWC were used to extract total RNA. For total RNA extraction, leaf tissue was ground to a fine powder with mortar and pestle while constantly in the presence of liquid nitrogen. Total RNA was then extracted from the ground leaf tissue with Trizol Reagent, following the manufacturer’s instructions (Invitrogen, Carlsbad, CA, USA). The poly adenylated mRNA fraction was isolated from total RNA using an Oligotex isolation midi kit (Qiagen, Valencia, CA, USA). Total RNA content and mRNA concentrations were evaluated using a spectrophotometric method in a NanoDrop instrument (ThermoScientific Co, USA). DNase-treated total RNA (Turbo DNA-free kit, Ambion Inc., Austin, TX) was used for the SMARTer PCR cDNA synthesis kit in order to generate 4 ug of mRNA from each sample. The mRNA was then utilized in the PCR-Select cDNA subtraction kit (Clontech, Takara BIO, Inc., Mountainview, CA) using the protocols provided by the manufacturer. The PCR-Select cDNA subtraction kit was used to generate both forward and reverse subtraction libraries indicating differential expression between the NT and SAG12-ipt plants under both well-watered and drought stressed conditions (47% RWC). A total of 16 cDNA libraries were created in order for performing 16 subtractive hybridizations to generate the subtracted libraries [4 treatments x 2 libraries (forward and reverse) x 2 repetitions]. The treatment expected to contain mRNA of the more drought tolerant or less stressed type (e.g. well-watered treatment or transgenic line) were utilized as testers in the hybridizations and the more sensitive were used as drivers (e.g. drought stressed plants or NT). For instance, in the subtraction comparing both NT and SAG12-ipt under drought stressed conditions, the forward subtraction identified mRNA transcripts from the drought-stressed SAG12-ipt plants was used as the tester and the mRNA from NT was used as the driver.
Gene cloning, sequence analysis, and experimental design
Plants were arranged as appropriate for a split-plot design with water treatment as the main plots and plant materials as the sub-plots, with four replicates for each water treatment and plant material. Sequences obtained were evaluated and gene identity matches were selected for those that had a significantly low e value (e value < 10−5). A cloning procedure was performed with competent cells based on the manufacturer’s instructions (TOPO-TA cloning kit, Life Technologies, Carlsbad, CA). Colonies were plated in glycerol stock and kept at -80°C until sequencing. Colonies were selected and sent for sequencing utilizing M13 reverse primers (Genewiz, South Plainfield, NJ). Gene sequences were categorized based on their predicted function based on the system used previously in Bevan et al. [23] and [2–3]. Sequences were evaluated to remove redundant base pairs and vector sequences manually and through use of VecScreen BLAST database. Clean Blastnr and BLAST of EST databases were performed [24].
qPCR Analysis
qPCR confirmation of SSH analysis was performed on samples taken from WT and SAG12-ipt plants within a separate experiment. Genes were selected for qPCR out of the total sequenced forward and reverse libraries based on relevance to the proteomics, metabolomics, and SSH results. The experimental details and conditions are as follows: Leaf samples were ground in liquid nitrogen using pre-chilled mortar and pestle for four biological replicates. Total RNAs were extracted from all samples of leaf powder using TRIzol reagent and treated with DNase (TURBO DNA-free kit, Life Technologies, Carlsbad, CA). The quality and quantity of RNA were assessed with NanoDrop 1000 (Thermo Scientific, DE). 2 μg of RNA was used for cDNA synthesis using High-Capacity cDNA Reverse Transcription kit (Life Technologies, NY). PCR amplification was carried out using gene-specific primers and SYBR green master mix (Life Technologies, Carlsbad, CA), according to manufacturer’s instruction. Relative transcript abundance was calculated using Δ Δ Ct method with actin as an internal standard. Primers used are listed in Table 1.
Table 1. Primer sequences used in qPCR analysis of creeping bentgrass leaf tissues.
The primer design tool feature of BLAST was utilized based on sequences obtained from SSH analysis.
| Description | primer F | primer R |
|---|---|---|
| Actin | GATATGGAAAAGATCTGGCATCAC | TCATACTCGGCCTTGGAGATCCAC |
| LRR receptor-like kinase 1 | AGGGACCATACCTCCCATTC | CAGGTCTTGACACTGGCTCA |
| RuBisCo | GCTGCCGAATCTTCTACTGG | AGAGCACGTAGGGCTTTGAA |
| universal stress protein 5327 | GCAGGGACATGGGTGGGAGGA | GTACCCCGCTCTGATGCGCC |
| LRR receptor-like kinase 2 | GCCGTTTTGTGCGCTCCTGT | GGAGATCCAAAGCCGCCACCC |
| Oxygen-evolving enhancer protein 3–1, chloroplast precursor (OEE3) | AGGGCCTCCTACCTGCGCTAC | TGCCGGTGAGGTCCTTGAGGC |
Results
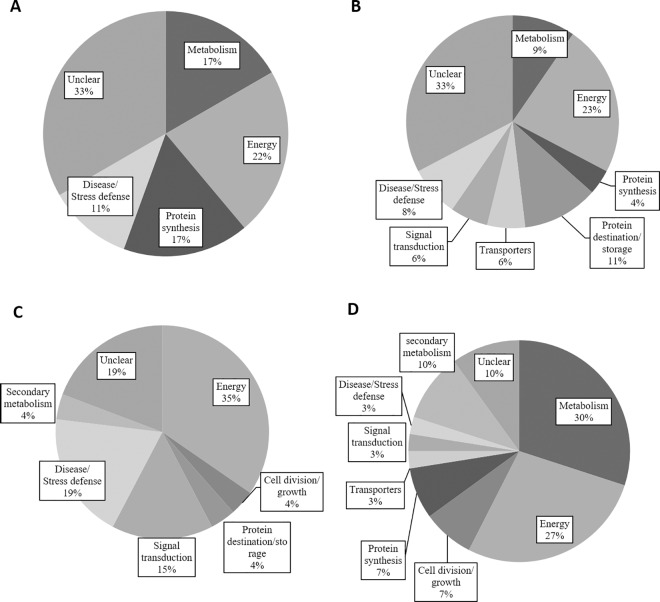
Comparing across hybridization libraries or genes that may have marginally differential gene expression can be difficult due to the potential for gene transcript loss during the experimental procedure and general limitations of sensitivity of the SSH approach. Therefore, specific changes will be discussed and interpreted only as they relate to the respective hybridization. The cDNA library construction and SSH subtraction successfully revealed 252 gene transcripts that were alternately regulated due to the ipt transgene or watering treatment. Of which, a total of 170 clones were selected for sequencing. After removal of redundant sequences and those that were unreadable and BLAST database searches, approximately 136 sequences were successfully identified in the GenBank database and grouped into functional categories (Fig 1). Differential gene regulation of 18 sequences occurred under well-watered conditions due to the ipt transgene, which were considered as ipt-responsive genes, including genes involved in the metabolism, energy, protein synthesis, stress defense, and unknown categories (Table 2). A total of 49 genes in the forward and reverse libraries comparing NT plants under the well-watered and drought stress condition were successfully sequenced (Table 3). For the SAG12-ipt libraries comparing well-watered to drought stress conditions, 26 genes exhibited differential expression (Table 4). A total of 75 genes were identified as drought-responsive genes in NT plants (49 genes) and SAG12-ipt plants (26 genes). The results of the libraries comparing NT and SAG12-ipt plants both under drought stress were a total of 38 genes being alternately regulated, including those that were up-regulating and down-regulated (Table 5).
Fig 1.
Percent total changes of gene transcripts (including forward and reverse libraries) in functional categories comparing A) NT to SAG12-ipt under well-watered conditions B) NT watered to NT drought stress C) SAG12-ipt well-watered to SAG12-ipt drought stress D) NT drought to SAG12-ipt drought stress conditions
Table 2. Genes categorized by protein function based on BLAST searches that were differentially expressed in SSH library A (NT compared to SAG12-ipt transgenic plants both growing under well-watered conditions) in the forward (up-regulated by ipt expression) and reverse (down-regulated by ipt expression) libraries.
Numbers in parentheses indicate number of transcript isoforms detected.
| Library | Description | Accession # | Best Match Accession # | E-value |
|---|---|---|---|---|
| Category 01 Metabolism | ||||
| Forward | β-glucosidase | JZ948409 | ACF22735.1 | 6.00E-40 |
| Category 02 Energy | ||||
| Forward | enolase (2-phosphoglycerate dehydratase) | JZ948410 | AAM69295.1 | 3.00E-60 |
| photosystem I P700 chlorophyll a apoprotein A2 | JZ948411 | ABG66207.1 | 1.00E-71 | |
| photosystem II precursor, chloroplast | JZ948412 | NP_001134061.1 | 9.00E-68 | |
| RuBisCO large subunit (3) | JZ948413 | ADU18941.1 | 4.00E-98 | |
| JZ948414 | ADU18941.1 | 5.00E-49 | ||
| JZ948415 | ADU18941.1 | 3.00E-76 | ||
| Reverse | Glyceraldehyde 3-phosphate dehydrogenase | JZ948416 | ACV86034.1 | 2.00E-133 |
| Category 05 Protein synthesis | ||||
| Forward | elongation factor 1-alpha-like protein | JZ948417 | ABB16977.1 | 2.00E-79 |
| nonribosomal peptide synthetase | JZ948418 | CBJ23772.1 | 1.00E-10 | |
| 40S ribosomal protein S21 | JZ948419 | NP_001105477.1 | 1.00E-47 | |
| Category 10 Signal transduction | ||||
| Forward | protein phosphatase 1 regulatory subunit 12b | JZ948420 | XP_001662854.1 | 4.00E-18 |
| Category 11 Disease/Stress defense | ||||
| Forward | catalase 2 | JZ948421 | A55092 | 1.00E-08 |
| catalase | JZ948422 | Q59296.1 | 3.00E-10 | |
| Category 20 Secondary metabolism | ||||
| oleosin 2 | JZ948423 | NP_198858.1 | 1.00E-76 | |
| S-adenosylmethionine synthase 4 (SAMS) | JZ948424 | Q4LB21.1 | 2.00E-60 | |
| Category 12 Unclear | ||||
| Forward | predicted protein | JZ948425 | BAJ87310.1 | 9.00E-104 |
| predicted protein | JZ948426 | BAJ96581.1 | 4.00E-117 | |
| predicted protein | JZ948427 | BAK02049.1 | 3.00E-36 | |
| SORBIDRAFT_01g032220 (2) | JZ948428 | XP_002467681.1 | 5.00E-58 | |
| JZ948434 | XP_002467681.1 | 3.00E-37 | ||
| Reverse | predicted protein | JZ948430 | BAJ93277.1 | 4.00E-16 |
| predicted: similar to CG18041 CG18041-PA | JZ948431 | XP_969418.1 | 4.00E-18 | |
Table 3. Genes categorized by protein function based on BLAST searches that were differentially expressed in SSH library B (NT under well-watered conditions compared to NT under drought stressed conditions) in the forward (up-regulated by drought) and reverse (down-regulated by drought) libraries.
Numbers in parentheses indicate number of transcript isoforms detected.
| Library | Description | Accession # | Best Match Accession | E-value |
|---|---|---|---|---|
| Category 01 Metabolism | ||||
| Forward | malate dehydrogenase | JZ948432 | XP_001659012.1 | 2.00E-29 |
| glycogen synthase kinase-3 MsK-3 | JZ948433 | NP_001148880.1 | 6.00E-07 | |
| Reverse | nitrilase-associated protein, putative | JZ948434 | CAJ38376.1 | 4.00E-12 |
| thiamine biosythesis protein ThiC | JZ948435 | AAG49550.1 | 1.00E-54 | |
| Category 02 Energy | ||||
| Forward | RuBisCO large subunit (3) | JZ948436 | BAD20627.1 | 2.00E-44 |
| JZ948437 | BAD20627.1 | 8.00E-60 | ||
| JZ948438 | BAD20627.1 | 1.00E-53 | ||
| RuBisCO small subunit 1B | JZ948439 | NP_198659.1 | 0.00001 | |
| RuBisCO small chain c | JZ948440 | ABR26034.1 | 0.00013 | |
| Oxygen-evolving enhancer protein 3–1, chloroplast precursor | JZ948441 | BAC83128.1 | 1.00E-37 | |
| cytochrome c oxidase subunit III | JZ948442 | YP_003734710.1 | 8.00E-36 | |
| Photosystem II 10 kDa polypeptide, chloroplast | JZ948443 | NP_001134061.1 | 4.00E-60 | |
| thioredoxin-like 5, chloroplastic | JZ948444 | ABR26107.1 | 3.00E-30 | |
| ATP-citrate synthase, putative | JZ948445 | XP_002519229.1 | 5.00E-49 | |
| Reverse | RuBisCO large subunit (3) | JZ948446 | ADU18941.1 | 2.00E-76 |
| JZ948447 | ADU18941.1 | 2.00E-15 | ||
| JZ948448 | ADU18941.1 | 3.00E-08 | ||
| RuBisCO | JZ948448 | CBF07493.1 | 1.00E-26 | |
| glyceraldehyde-3-phosphate dehydrogenase 1 | JZ948450 | ACV86034.1 | 8.00E-107 | |
| oxygen-evolving enhancer protein 1, chloroplast | JZ948451 | ABQ52657.1 | 9.00E-20 | |
| Category 05: Protein synthesis | ||||
| Forward | elongation factor 1 alpha | JZ948417 | AEG78681.1 | 2.00E-112 |
| Category 06: Protein destination/storage | ||||
| Forward | E3 ubiquitin-protein ligase UBR5 | JZ948540 | EFN82877.1 | 2.00E-57 |
| thiol disulfide interchange protein txlA | JZ948453 | NP_001152226.1 | 2.00E-06 | |
| GTPase SAR1 | JZ948452 | ACD03831.1 | 3.00E-57 | |
| Reverse | ubiquitin-60S ribosomal protein L40-like | JZ948454 | XP_003465244.1 | 1.00E-12 |
| ubiquitin-conjugating enzyme E2-like protein | JZ948455 | ADB28900.1 | 2.00E-14 | |
| ATP-dependent Clp protease ATP-binding subunit clpA | JZ948456 | P31542.1 | 9.00E-34 | |
| Category 07 Transporters | ||||
| Forward | vacuolar H+-ATPase subunit B | JZ948457 | BAF38479.1 | 2.00E-61 |
| ABC transporter integral membrane protein | JZ948458 | ZP_07303354.1 | 0.00061 | |
| CDGSH iron sulfur domain 1 | JZ948459 | NP_001004811.1 | ||
| Category 10 Signal transduction | ||||
| Forward | uridine kinase | JZ948460 | ZP_00781761.1 | 0.00042 |
| serine/threonine kinase receptor precursor-like | JZ948461 | BAC57306.1 | 4.00E-39 | |
| Reverse | LRR receptor-like kinase (3) | JZ948462 | ACY30448.1 | 7.00E-70 |
| Category 11 Disease/Stress defense | ||||
| Forward | jasmonate-induced protein, putative | JZ948463 | ABA96835.1 | 3.00E-08 |
| universal stress protein 5327 | JZ948464 | ADB54812.1 | 4.00E-37 | |
| abscisic acid-responsive HVA22 family protein | JZ948465 | XP_002865508.1 | 3.00E-08 | |
| glyoxalase I | JZ948466 | AAW68026.1 | 1.00E-53 | |
| Category 20 Secondary metabolism | ||||
| Reverse | diphthine synthase, predicted | JZ948467 | XP_001604120.1 | 6.00E-75 |
| Category 12 Unclear | ||||
| Forward | predicted protein [Hordeum vulgare subsp. vulgare] | JZ948468 | BAK02049.1 | 1.00E-23 |
| predicted protein [Hordeum vulgare subsp. vulgare] | JZ948469 | BAJ85089.1 | 3.00E-42 | |
| predicted protein [Hordeum vulgare subsp. vulgare] | JZ948470 | BAJ96581.1 | 3.00E-21 | |
| SORBIDRAFT_10g001310 | JZ948471 | XP_002436374.1 | 0.00002 | |
| pBMB0558_00760 | JZ948472 | YP_004169259.1 | 5.00E-04 | |
| predicted protein [Hordeum vulgare subsp. vulgare] | JZ948473 | BAJ95019.1 | 1.00E-32 | |
| predicted protein [Hordeum vulgare subsp. vulgare] | JZ948474 | BAJ99280.1 | 5.00E-12 | |
| hypothetical protein MELLADRAFT_95019 | JZ948475 | EGF98952.1 | 1.00E-23 | |
| predicted protein [Hordeum vulgare subsp. vulgare] | JZ948476 | BAJ90401.1 | 3.00E-16 | |
| hypothetical protein OsJ_26652 | JZ948477 | EAZ42091.1 | 6.00E-17 | |
| Reverse | predicted protein | JZ948478 | XP_001770883.1 | 0.0004 |
| hypothetical protein OsJ_19720 | JZ948479 | EEE64863.1 | 1.00E-57 | |
| hypothetical protein Tc00.1047053508475.20 | JZ948480 | XP_804280.1 | 0.000031 | |
| hypothetical protein LOC100273563 | JZ948481 | NP_001141453.1 | 8.00E-12 | |
Table 4. Genes categorized by protein function based on BLAST searches that were differentially expressed in SSH library C (SAG12-ipt under well-watered conditions compared to SAG12-ipt under drought stressed conditions) in the forward (up-regulated by drought) and reverse (down-regulated by drought) libraries.
Numbers in parentheses indicate number of transcript isoforms detected.
| Library | Description | Accession # | Best Match Accession | E value |
|---|---|---|---|---|
| Category 02 Energy | ||||
| Forward | Mg-protoporphyrin IX | JZ948482 | CAB58179.1 | 2.00E-60 |
| RuBisCo large subunit | JZ948483 | ADU18941.1 | 3.00E-76 | |
| Oxygen-evolving enhancer protein 3–1, chloroplast precursor (OEE3) | JZ948484 | BAC83128.1 | 4.00E-76 | |
| Thioredoxin-like 5, chloroplastic | JZ948485 | ABR26107.1 | 2E-14 | |
| Reverse | Fructose-bisphosphate aldolase, class I | JZ948486 | AT3G52930 | 1.00E-109 |
| Photosystem II 10 kDa polypeptide, chloroplast | JZ948487 | NP_001134061.1 | 2.00E-17 | |
| Cytochrome c oxidase subunit III | JZ948488 | ADO60570.1 | 2.00E-15 | |
| Category 03 Cell growth/division | ||||
| Reverse | tubulin α-3 chain | JZ948489 | NP_001167663.1 | 2.00E-58 |
| Category 06 Protein destination/storage | ||||
| Reverse | ubiquitin-conjugating enzyme | JZ948490 | ADX86831.1 | 7.00E-51 |
| Category 10 Signal transduction | ||||
| Forward | HVA22-like protein | JZ948491 | JK340582.1 | 7.00E-31 |
| F-box/LRR repeat protein | JZ948492 | ACY30448.1 | 5.00E-40 | |
| uridine kinase | JZ948493 | ZP_00781761.1 | 6.00E-30 | |
| Reverse | Os01g0629400 (phosphatase-like) | JZ948494 | NM_001050175.2 | 8.00E-60 |
| ctd-phosphatase-like protein | JZ948495 | ABR26130.1 | 8.00E-60 | |
| Category 11 Stress/disease defense | ||||
| Forward | glyoxalase I (3) | JZ948496 | AAW68026.1 | 4.00E-32 |
| jasmonate-induced protein, putative | JZ948497 | ABA96835.1 | 5.00E-30 | |
| Reverse | DELLA protein RGL1 | JZ948541 | EG429076.1 | 0.00E+00 |
| Category 20 Secondary metabolism | ||||
| Forward | spermidine synthase | JZ948498 | AEL33692.1 | 1.00E-109 |
| Category 12 Unclear | ||||
| Forward | hypothetical protein NCLIV_068840 | JZ948499 | CCA30004.1 | 5.00E-31 |
| SORBIDRAFT_06g021780 | JZ948500 | XP_002448130.1 | 0.011 | |
| OSJNBa0014K14.7 | JZ948501 | CAE02935.3 | 2.00E-91 | |
| Reverse | hypothetical protein | JZ948502 | CAM36311.1 | 2.00E-08 |
| predicted protein | JZ948503 | BAJ90401.1 | 5.00E-16 | |
Table 5. Genes categorized by protein function based on BLAST searches that were differentially expressed in SSH library D, which compared NT to SAG12-ipt under drought stressed conditions equal in cellular water deficit (47% RWC) in the forward (up-regulated by transgene during drought stress) and reverse (down-regulated by transgene during drought stress) libraries.
Numbers in parentheses indicate number of transcript isoforms detected.
| Library | Description | Accession # | Best Match # | E value |
|---|---|---|---|---|
| Category 01 Metabolism | ||||
| Forward | GDP-mannose 3,5-epimerase | JZ948450 | XM_003577361.1 | 3.00E-27 |
| mannose-1-phosphate guanylyltransferase 3 | JZ948505 | Q6Z9A3.1 | 3.00E-149 | |
| UDP-arabinopyranose mutase 1 | JZ948506 | Q6Z9A3.1 | 1.00E-15 | |
| glycosyl hydrolase family 19 protein | JZ948507 | AAQ84319.1 | 6.00E-44 | |
| UTP—glucose-1-phosphate uridylyltransferase | JZ948508 | Q43772.1 | 1.00E-15 | |
| UDP-glucose dehydrogenase | JZ948509 | AAX08057.1 | 7.00E-25 | |
| xylose isomerase-like | JZ948510 | XM_003562448.1 | 4.00E-23 | |
| xylose isomerase | JZ948511 | CAA64544.1 | 3.00E-25 | |
| Reverse | glycine decarboxylase P subunit | JZ948512 | AAB82711.1 | 8.00E-12 |
| acetyl-CoA carboxylase | JZ948513 | NP_001185143.1 | 4.00E-21 | |
| Category 02 Energy | ||||
| Forward | RuBisCo large subunit (3) | JZ948514 | AAQ08331.1 | 5.00E-61 |
| JZ948515 | AAQ08331.1 | 1.00E-26 | ||
| JZ948538 | AAQ08331.1 | 2.00E-06 | ||
| chloroplast-localized Ptr ToxA-binding protein1 | JZ948539 | AAR24582.1 | 6.00E-54 | |
| Reverse | RuBisCo large subunit (4) | JZ948516 | ACO35581.1 | 4.00E-46 |
| JZ948517 | ACO35581.1 | 5.00E-61 | ||
| JZ948518 | ACO35581.1 | 2.00E-17 | ||
| JZ948519 | ACO35581.1 | 3.00E-76 | ||
| thioredoxin-like 5, chloroplastic | JZ948484 | ABR26107.1 | 3.00E-30 | |
| ATP-citrate synthase, putative | JZ948445 | XP_002519229.1 | 5.00E-49 | |
| Category 04 Transcription | ||||
| Forward | partial 16S rRNA gene | JZ948520 | FN421445.1 | 2E-15 |
| histone H3 | JZ948521 | XP_001752178.1 | 1.00E-121 | |
| translational initiation factor eIF1 | JZ948522 | BAF63490.1 | 2.00E-11 | |
| Category 05 Protein synthesis | ||||
| Forward | Os07g0609766 | JZ948523 | NP_001175295.1 | 3.00E-05 |
| Category 07 Transporters | ||||
| Forward | aquaporin PIP1-2 | JZ948524 | NP_001078067.1 | 1.00E-32 |
| coatomer subunit beta'-2 | JZ948525 | NP_175645.1 | 1.00E-49 | |
| Reverse | plasma membrane H+-ATPase | JZ948526 | CAC50884.1 | 1.00E-28 |
| Category 10 Signal transduction | ||||
| Forward | receptor kinase ORK14 | JZ948527 | AAM09948.1 | 1.00E-5.8 |
| Category 11 Disease/Stress defense | ||||
| Forward | Chloroplast Ptr ToxA-binding protein | JZ948529 | AK332987.1 | 1.00E-45 |
| glyoxalase I | JZ948528 | BAB71741.1 | 9.00E-26 | |
| Category 20 Secondary metabolism | ||||
| Forward | cruciferin cru4 subunit | JZ948530 | X57848.1 | 2.00E-11 |
| isoflavone reductase | JZ948531 | ACH72670.1 | 4.00E-57 | |
| Category 12 Unclear | ||||
| Forward | Os11g0169100 | JZ948532 | NP_001065849.1 | 7.00E-50 |
| hypothetical protein | JZ948533 | ACG32534.1 | 5.00E-13 | |
| predicted protein | JZ948534 | BAK07943.1 | 1.00E-41 | |
| predicted protein | JZ948536 | BAK05471.1 | 5.00E-06 | |
| Reverse | hypothetical protein | JZ948537 | CAM36311.1 | 8.00E-07 |
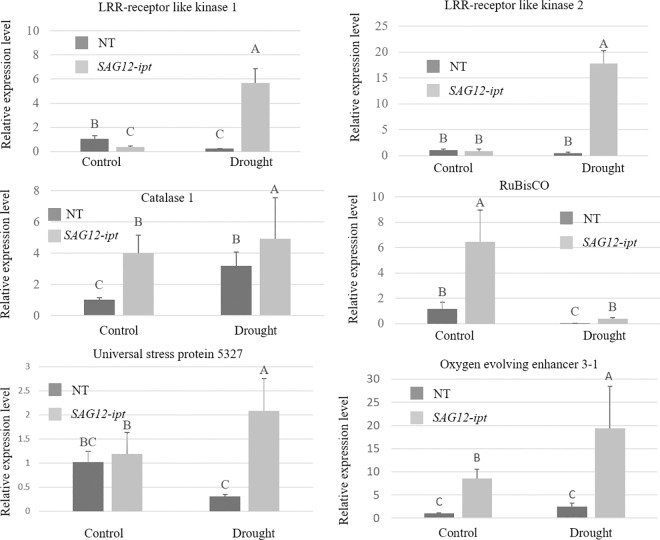
qPCR analysis of transcript levels of selected genes confirmed the validity of SSH screening for differentially-expressed genes between the NT and ipt-transgenic plants in response to drought stress, as results from qPCR were mostly consistent with the gene expression data through SSH analysis (Fig 2). The set of 6 genes with primers are listed in Table 5. Genes such as OEE, LRR receptor like kinase 1, and USP were increased by drought stress in SAG12-ipt plants. Elongation factor 1α and a RuBiSCo subunit were decreased by drought stress in both SAG12-ipt plants and NT due to drought stress.
Fig 2. qPCR of creeping bentgrass leaf tissue in null transformed plants (NT) and plants with an ipt gene for cytokinin biosynthesis (SAG12-ipt) of select genes for confirmation of SSH results.
Standard error bars indicate variation among replicates and different letters indicate statistical differences between treatments.
Discussion
The results of SSH analysis have revealed valuable information regarding alterations in gene expression due to the ipt gene or drought stress in creeping bentgrass in this study.A large number of DEGs was detected in plants in response to drought stress in this study and previous work has already extensively examined numerous drought-responsive genes [25] particularly in related grass species such as rice (Oryza sativa) [26] and wheat [27]. Therefore, the following discussion focuses on those genes that were uniquely up- or down-regulated in the ipt-transgenic creeping bentgrass under drought stress (or ipt-responsive genes) and that corresponding changes occurred at proteomic and metabolomics levels, as reported in our previous studies [4, 10]. The ipt-responsive genes were classified into the functional category of metabolism, energy production, stress defense, transporters, protein metabolism, signaling transduction, and secondary metabolite synthesis. The biological functions of those ipt-responsive genes related to drought tolerance are discussed below.
The functional categories having the greatest number of genes differentially expressed in the ipt-transgenic plants in the majority of the libraries were in the metabolism and energy production. Proteomic and metabolic analysis also identified changes in proteins and metabolites mainly involved in metabolism and energy production due to the ipt overexpression in creeping bentgrass [2–4]. The combined results of gene expression in this study with protein and metabolic data from our previous studies suggest that increased endogenous CK could mainly regulate metabolism and energy production, contributing to improved drought tolerance in the SAG12-ipt creeping bentgrass.
SSH results showed that almost twice the number of gene changes occurred due to drought stress in NT plants (52) compared to changes in SAG12-ipt plants (26) relative to their respective controls. In addition, NT plants generally had a greater number of gene changes in the metabolism and stress defense categories whereas SAG12-ipt exhibited less change in these categories to drought stress. Zhang et al. [17] found that the more heat-tolerant plants of tall fescue (Poa pratensis L.) had higher levels of transcripts related to energy production (particularly photosynthesis) protein synthesis, signaling and transcription factors compared to sensitive plants. These data suggested that NT plants that were more sensitive to drought stress exhibited greater transcript changes in metabolism and stress defense categories.
Gene transcripts that were differentially expressed under well-watered conditions due to the ipt transgene in the metabolism category had homology to a β-glucosidase gene, SAMS, and a protein phosphatase, which were in greater levels in the SAG12-ipt plants compared to NT. The glucosidase proteins are a family of enzymes involved in various cellular functions, including regulating cellular structure, defense, and hormone metabolism [28]. Higher protein abundance of SAMS and protein phosphatase were also detected in SAG12-ipt plants compared to NT plants. SAMS activity may be a source of methyl groups for compounds involved in osmotic adjustment under stress conditions [29]. The regulation of SAMS proteins may play a major role in controlling the accumulation of free amino acids or of those destined for proteogenesis, affecting plant growth regulated by CK.
Malate dehydrogenase is a key enzyme in the citric acid cycle, and its transcript level was increased by drought stress in NT plants in this study, but lower accumulation of this protein and malic acid content were detected in NT plants exposed to drought stress in our previous studies [4, 10]. The up-regulation of malate dehydrogenase transcript could function to compensate for the damaged proteins and enzymes induced by drought stress; however, this cannot be directly concluded from the results of this study. Similar results regarding malic acid content have been found in other plant species [30]. The gene for thiamine biosynthesis protein was found to be down-regulated by drought stress only in NT plants in this study. Thiamine is used in the biosynthesis of secondary metabolites such as gamma-aminobutyric acid (GABA). Previous metabolomics analysis found that the content of GABA and other secondary metabolites was lower NT plants compared to that in SAG12-ipt plants under drought stress [10], which could be related to the lower transcript level of thiamine biosynthesis in NT plants. Similarly, the differential regulation of other transcripts in the metabolism category such as glycogen synthase and malate dehydrogenase could be associated with the differential sugar accumulation and glycolytic activities detected between NT and SAG12-ipt plants [3–4]. Transcripts involved in respiration pathways, such as GAPDH were down-regulated in NT plants. A corresponding decrease in abundance of GAPDH proteins and intermediates in respiratory pathways also occurred in response to drought in NT plants [3]. These results suggested that improved drought tolerance in SAG12-ipt plants could be related to CK-regulation of respiratory metabolism involving changes of transcripts controlling synthesis of aforementioned metabolites in respiration pathways; however, more work would be needed to specifically determine ipt effects on the metabolites described here.
Differences in carbon requirements and allocation may be a major factor determining the differential drought response of NT and SAG12-ipt plants as they relate to carbon metabolism. Drought stress causing an up-regulation of transcripts related to carbon metabolism could be related to carbon availability and partitioning associated with drought damage [31]. Drought stress has also been shown to increase the expression of genes controlling the central processes of carbon metabolism. This is due to a need for carbon mobilization or due to the increased need for replacing damaged enzymes to allow for metabolic processes to remain functional. Increased CK content under drought stress could play a role in regulating carbon availability.
The increase in glycogen synthase kinase (GSK) transcripts in NT plants in response to drought could be associated with an increased need for sugar for stress defense. In rice plants, an increase in GSK was also exhibited due to drought stress [32]. It is not clear why GSK transcript expression was up-regulated while in NT plants a loss of photosynthetic proteins and reduction in photosystem health was observed in our previous studies [4, 10]. GSK may inactivate glycogen synthase or play a role in other metabolic functions such as the inactivation or activation of other transcription factors. Xylose isomerase like protein that exhibited greater transcript level in SAG12-ipt transgenic plants compared to NT under drought stress could be directly related to the expression of the ipt transgene. O-xylosyltransferases are involved in the regulation of CK hormone biosynthesis via reversible inactivation [33].
Transcripts coding for a Mg-protoporphyrin IX type domain were identified in the forward library of SAG12-ipt plants under drought. Based on sequence match results it is not known which enzyme the Mg-protoporphyrin IX domain may be a part of; however, in plants these IX type domains are most commonly a part of the chlorophyll biosynthetic enzyme, Mg-protoporphyrin IX monomethyl ester cyclase [34]. The gene expression and content of this protein is typically reduced by drought stress and maintenance of this enzyme has been associated with improved drought tolerance in transgenic rice [34]. The SAG12-ipt plants maintained greater chlorophyll content and photosynthetic activity than NT plants during drought stress [2]. Therefore, the increased expression of these transcripts in the ipt plants may be related to chlorophyll maintenance in plants with elevated CK content.
Chloroplast localized ToxA binding protein (Pr ToxA) was detected in the forward library of NT compared to SAG12-ipt under drought stressed conditions, meaning a greater number of Pr ToxA were in SAG12-ipt plants. Ptr ToxA function involving ToxA are also differentially responsive to cold in frost tolerant and sensitive Festuca grass plants and has been found to be required for PSII stability in chloroplasts [35–36]. How Pr ToxA may be related to drought tolerance is not well known. Perhaps, the consistent promotion of PSII health and maintenance of accumulation of photosynthetic compounds in SAG12-ipt plants coupled with the increased Pr Tox transcripts under drought stress could be associated with the CK gene regulatory network.
DELLA proteins play a role in various stress responses as they are components of regulatory pathways that are closely associated with plant hormones such as ethylene and gibberellins [37]. A transcript coding for a DELLA protein was reduced by drought stress in SAG12-ipt plants. DELLAs are repressors of GA signaling and degradation of DELLA proteins is known to alleviate growth repression [37]. Leaf biomass accumulation under drought stress was doubled in plants containing un-functional DELLA proteins caused by knock-out mutation [38]. Therefore, the reduction of leaf senescence due to CK and the maintenance of metabolic activities in SAG12-ipt plants could be related to this differential gene expression of DELLA proteins
Catalase (CAT) is an antioxidant enzyme that detoxifies hydrogen peroxide that can accumulate in plant cells and is an essential enzyme for stress tolerance. CAT expression was greater in SAG12-ipt plants compared to NT plants under well-watered condition. Tobacco plants deficient in the CAT enzyme were found to be greatly susceptible to abiotic stress [39]. Enhancement of the expression and activity of CAT promotes stress tolerance [40]. CAT expression in ipt transgenic tobacco was enhanced by drought stress to a greater extent than in non-transgenic plants [5]. Several grass species consistently show stable expression of CAT under drought stress [41–42, 3–4]. In addition, leaf senescence is correlated with a decline in CAT activity [43] and CKs have been shown to increase CAT activity [44]. In addition, CAT is also known to stimulate the activity of protoporphyrin cyclase involved in chlorophyll metabolism [45]. The positive roles of CAT for drought tolerance could be due to its antioxidant activity and involvement in chlorophyll metabolism. Therefore, the prevention of natural leaf senescence in the well-watered condition and the maintenance of protein content and activity of CAT under drought stress [3–4] may be a main factor promoting greater drought stress tolerance in SAG12-ipt transgenic creeping bentgrass with enhanced CK content. The stability of CAT during drought stress and its great capacity to reduce oxidative stress in grass species may make this a highly effective and valuable molecular target for enhancing or selecting drought tolerant grass germplasm in breeding programs.
Glyoxalase is another stress protective enzyme that accumulates in response to drought conditions in order to break down a toxic by-product of glycolysis, methyl-glyoxyl. In the drought tolerant resurrection grass (Sporobolus stapfianus), glyoxylase transcript expression was up-regulated in response to drought [46]. The accumulation of glycolytic proteins [3–4] and transcripts involved in respiration detected in SAG12-ipt plants such as GAPDH compared to NT may be related to an increased flux through glycolysis and increased need for the glyoxalase enzyme. In addition, metabolite analysis revealed the accumulation of intermediates in the glycolysis pathway to a greater extent in SAG12-ipt plants than in NT plants during drought stress [3–4]. A stimulation of glycolysis is thought to be a beneficial early drought stress tolerance mechanism, as also evident in another resurrection plant Craterostigma plantagineum [47], for readily available free sugars for osmotic adjustment and energy production. This increased flux through glycolysis may play a role in CK regulation of drought tolerance.
The transcript levels of several transporters were greater in the SAG12-ipt line compared to NT plants under drought stress. An aquaporin Pip1-2, water transporting channel, was up-regulated by drought stress in SAG12-ipt plants. A greater expression of aquaporin transcripts occurred in drought-tolerant lines compared to a drought sensitive cultivar of chickpea (Cicer arietinum) [48]. The up-regulation of aquaporins during drought stress has been reported in Arabidopsis [49] and rice [50]. In addition to water movement, aquaporins also play an important role in mobilizing other metabolites throughout the plant, including those of small molecular weight such as glycerol and urea and gases such as NH3 and CO2 [51]. Metabolites that were important in the drought response in SAG12-ipt plants included several small molecular weight compounds including glycerol. The up-regulation of these transports may play an important role in allowing water and metabolite movement during stress periods. The maintenance of greater RWC in SAG12-ipt plants than the NT under drought stress could be related to this type of differential aquaporin or transporter expression.
Another type of transporter, an ABC transporter was found to be up-regulated in NT plants. ABC transporter transcripts were greater in tall fescue plants sensitive to drought stress [14]. Other transcripts that were increased by drought stress in NT plants but not in SAG12-ipt plants include one coding for a CDGSH iron sulfur domain 1 protein and a vacuolar ATPase. CDGSH iron sulfur domains are typically located in mitochondrial membranes, serving as transport channels for electron gradient regulation and iron transport [52]. Vacuolar ATPases are primarily involved in transporting ions across the plasma membranes such as Ca2+ ions [53]. The requirement of up-regulation of these transcripts could be related to the need for maintenance of osmotic balance or osmotic adjustment in plants exposed to drought stress.
Transcripts coding for proteins related to protein transport or protein degradation were the main types of transcripts differentially regulated in the protein destination/storage category and more changes in these transcripts were detected in NT plants than in SAG12-ipt in response to drought. For example, an ATP-dependent Clp protease ATP-binding subunit clpA and several transcripts associated with ubiquitin were found to be either up- or down-regulated in NT plants. The ATP-dependent Clp protease ATP-binding subunit clpA homolog may interact with a clpP-like protease involved in degradation of denatured proteins in the chloroplast [54]. Ubiquitin pathways mark proteins for degradation by the proteasome or can function to regulate membrane bound proteasome associated signaling transduction caused by CK during cell division processes [55]. Stability of ubiquitin transcripts have been associated with drought tolerance in a dessication tolerant grass species (Sporobolus stapfianus). Transcripts coding for an ubiquitin-conjugating protein was found to be down-regulated due to drought stress in barley (Hordeum vulgare) [30]. Tian et al. [56] found the ubiquitin proteasome pathway to be up-regulated under heat stress conditions in a grass species with superior heat tolerance. Alternate mechanisms could be employed in regard to protein degradation and stress tolerance since increased accumulation of proteases is important for stress tolerance to facilitate utilization of nutrients in protein turnover whereas a reduction of protease activity could allow for maintenance of protein health and functionality under stress. Transcripts such as GTPase SAR1 are necessary for the initiation of the process of transporting proteins from the exit sites of the Golgi apparatus [57]. This could reflect the requirement of newly synthesized proteins to replace those damaged by drought stress.
Leu-rich repeat (LRR) receptor kinases were decreased in response to drought stress in NT plants but exhibited enhanced expression in SAG12-ipt. LRR receptor kinases are a large group of transmembrane receptor proteins that have diverse functions in developmental and defense related processes. LRRs are typically localized to plasma membranes and a limited number of them can be up-regulated by increased ABA content [58]. LRR receptor kinases function is also closely tied to regulation by peptide hormones (e.g. phytosulfokine) that are less well understood [59]. Differential expression of LRR in NT and SAG12-ipt plants suggested the involvement of this gene in the signal transduction process for the differential drought responses of the two plant types.
A universal stress protein (USP) was detected in NT plants due to drought stress in SSH analysis, but more USP transcripts were detected in SAG12-ipt plants. An accumulation of the USP 1 transcripts was also detected due to drought stress in rice [60] and is responsive to other abiotic stress such as cold [35]. Different studies have found contrasting results regarding the expression or accumulation of USP proteins as they relate to drought sensitivity or tolerance [35, 60–61]. Species and stress duration and differences in USP isoforms may be the cause of whether an up-regulation or a down-regulation of gene transcripts and differences in USP protein accumulation are correlated with stress tolerance. Similar to our results, various USP genes were more highly expressed in the salt sensitive variety compared to the tolerant type [61]. USP transcripts and protein generally has been associated with stress incidence and accumulate in the cytoplasm when cells are undergoing stress damage [62–63]. Therefore, the NT plants may be up-regulating the USP protein due to cellular damage. Genetic manipulation of USP proteins may be a viable method to improve plant stress tolerance [62]. How CK may regulate USP proteins is not conclusive from our studies. However, since the response occurred in the more drought-sensitive NT plants, further investigation into the responses of USPs in turfgrass species may be warranted and enhancement of the response of USP under drought stress in grass species may help promote drought tolerance.
Transcripts coding for an isoflavone reductase-like protein 5 were found to be greater in SAG12-ipt plants compared NT under drought stress. Isoflavone reductase proteins are known to decrease due to drought stress [64]. Maintenance of greater levels of isoflavone reductase enzymes is associated with drought tolerance [65]. Our results are consistent with those found in loblolly pine (Pinus taeda), where isoflavone transcripts were up-regulated in response to mild drought stress but not in the severe stress state [66]. Thus, this suggests less stress damage in SAG12-ipt plants at the same level of cellular water deficit than in NT plants, which is consistent with our physiological results [3–4]. Transcripts encoding iosoflavone reductase were also found to exhibit differential regulation in SSH libraries constructed comparing differential tall fescue cultivars in response to heat stress [14]. Transcripts coding for a putative dipthine synthase enzyme were down-regulated by drought stress only in NT plants. However, not much information is available regarding dipthine synthase and drought tolerance in plants.
In conclusion, genes of particular interest in how CK may enhance the drought tolerance response were primarily coding for proteins associated with major metabolic functions such as those regulating energy production, metabolism, and stress defense. Further studies evaluating the downstream effects of differentially expressed genes discussed here that were associated with CK maintenance under drought stress in creeping bentgrass may be beneficial for further understanding of how these genes may relate to CK regulation under drought stress in creeping bentgrass.
Acknowledgments
The authors wish to thank the OJ Noer Foundation and Rutgers Center of Turfgrass Science for funding support for this research. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations
- NT
null transformant plant line
- WT
wild-type
- SAG12
senescence activated promoter 12
- ipt
isopentenyl transferase gene
- RWC
relative water content
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- RuBisCO
ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit
- CAT
catalase
- OEE
oxygen evolving enhancer
- LRR
Leucine-rich-repeat
- CK
cytokinin
- SSH
suppression subtractive hybridization
Data Availability
All relevant data are within the manuscript and supporting information files.
Funding Statement
The authors wish to thank the OJ Noer Foundation and Rutgers Center of Turfgrass Science for funding support for this research. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Novakova M, Dobrev P, Motyka V, Gaudinova A, Malbeck J, Pospisiolova J, et al. Cytokinin function in drought stress response and subsequent recovery Biotechnology and Sustainable Agriculture 2006 and Beyond. Springer, Netherlands: 2007; p. 171–174. [Google Scholar]
- 2.Merewitz E, Gianfagna T, Huang B. Effects of SAG12-ipt and HSP18.2-ipt expression on cytokinin production, root growth and leaf senescence in creeping bentgrass exposed to drought stress. J. Amer. Soc. Hort. Sci. 2010; 135: 230–239. [Google Scholar]
- 3.Merewitz E, Gianfagna T, Huang B. Photosynthesis, water use, and root viability under water stress as affected by expression of SAG12-ipt controlling cytokinin synthesis in Agrostis stolonifera. J. Exp. Bot. 2011a; 62: 383–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Merewitz E, Gianfagna T, Huang B. Protein accumulation in leaves and roots associated with improved drought tolerance in creeping bentgrass expressing an ipt gene for cytokinin synthesis. J. Expt. Bot. 2011b; 8: 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rivero RM, Kojima M, Gepstein A, Sakakibara H, Mittler R, Gepstein S, et al. Delayed leaf senescence induces extreme drought tolerance in a flowering plant. P.N.A.S. 2007; 104: 19631–19636. 10.1073/pnas.0709453104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark DG, Dervinis C, Barrett JE. Klee H, Jones M. Drought-induced leaf senescence and horticultural performance of transgenic PSAG12-IPT petunias. J. Amer. Soc. Hort. Sci. 2004; 129: 193–199. [Google Scholar]
- 7.Zhang P, Gruissem W . Leaf senescence-inducible expression of isopentenyl transferase in cassava rendering it resistant to drought stress In: Tielkes E, Hülsebusch C, Häuser I, Deininger A, Becker K, eds. The global food and product chain: dynamics, innovations, conflicts, strategies. Stuttgart, Germany: MDD Media Digitaldruck Copy Shop Büromaschinen GmbH; 2005. pp: 277. [Google Scholar]
- 8.Rivero RM, Gimeno J, Deynze AV, Walia H, Blumwald E. Enhanced cytokinin synthesis in tobacco plants expressing PSARK: IPT prevents the degradation of photosynthetic protein complexes during drought. Plant Cell Physiol. 2010; 51: 1929–1941. 10.1093/pcp/pcq143 [DOI] [PubMed] [Google Scholar]
- 9.Krishnan S, Ma Y, Merewitz E. Leaf trimming and high temperature regulation of phytohormones and polyamines in creeping bentgrass leaves. J. Amer. Soc. Hort. Sci. 2016; 141: 66–75. [Google Scholar]
- 10.Merewitz E, Gianfagna T, Huang B. Elevated cytokinin content in creeping bentgrass may promote drought tolerance by regulation of the metabolite profile. J. Exp. Bot 2012; 63: 1315–1328. 10.1093/jxb/err372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desai S, Hill J, Trelogan S, Diatchenko L, Siebert P. Identification of differentially expressed genes by suppression subtractive hybridization In: Hunt S Livesey R, editors. Functional genomics, a practical approach. Oxford University Press; 2000. pp. 81–112. [Google Scholar]
- 12.Diatchenko L, Lau YF, Campbell AP, Chenchik A, Moqadam F, Huang B, et al. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. P.N.A.S. 1996; 93: 6025–6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miao H, Ye Z, Hu G, Qin Y. Comparative transcript profiling of gene expression between self-incompatible and self-compatible mandarins by suppression subtractive hybridization and cDNA microarray. Mol. Breeding. 2015; 35: 47–52. [Google Scholar]
- 14.Zhang L, Li FG, Liu CL, Zhang CJ, Zhang XY. Construction and analysis of cotton (Gossypium arboreum L.) drought-related cDNA library. BMC Research Notes. 2009; 2: 120–128. 10.1186/1756-0500-2-120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clement M, Lambert A, Herouart D, Boncompagni E. Identification of new up-regulated genes under drought stress in soybean nodules. Gene. 2008; 426: 15–22. 10.1016/j.gene.2008.08.016 [DOI] [PubMed] [Google Scholar]
- 16.Li HY, Huang S, Shi Y, Song Y, Zhao J, Wang F, et al. Isolating soil drought-induced genes from maize seedling leaves through suppression subtractive hybridization. Agricultural Sciences in China. 2007; 6: 647–651. [Google Scholar]
- 17.Zhang Y, Rouf Mian MA, Chekhovskiy K, So S, Kupfer D, Lai H, et al. Differential gene expression in Festuca under heat stress conditions. J. Expt. Bot. 2005; 56: 897–907. [DOI] [PubMed] [Google Scholar]
- 18.Liu S, Jiang Y. Identification of differentially expressed genes under drought stress in perennial ryegrass. Physiol. Plant. 2010; 139: 375–387. 10.1111/j.1399-3054.2010.01374.x [DOI] [PubMed] [Google Scholar]
- 19.Xing J, Xu Y, Tian J, Gianfagna T, Huang B. Transformation of a perennial grass species with ipt gene controlling cytokinin synthesis associated with suppression of shade or heat-induced leaf senescence. J. Amer. Soc. Hort. Sci. 2010; 134:602–609. [Google Scholar]
- 20.Xu Y, Tian J, Gianfagna T, Huang B. Effects of SAG12-ipt expression on cytokinin production, growth and senescence of creeping bentgrass (A. stolonifera) under heat stress. Plant Growth Regulat. 2009; 57: 281–291. [Google Scholar]
- 21.Hoagland DR, Arnon DI. The water-culture method for growing plants without soil. California Agricultural Experiment Station 1950; 347: 2nd edit pp. 32. [Google Scholar]
- 22.Topp GC. Electromagnetic determination of soil water content: Measurements in coaxial transmission lines. Water Resour. Res. 1980; 16: 574–582. [Google Scholar]
- 23.Bevan M, Bancroft I, Bent E, Love K, Goodman H, Dean C, et al. Analysis of 1.9|[thinsp]|Mb of contiguous sequence from chromosome 4 of Arabidopsis thaliana. Nature 1998; 391: 485–488. 10.1038/35140 [DOI] [PubMed] [Google Scholar]
- 24.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990; 215: 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 25.Bray EA. Genes commonly regulated by water-deficit stress in Arabidopsis thaliana. J. Expt. Bot. 2004; 55: 2331–2341. [DOI] [PubMed] [Google Scholar]
- 26.Huang L, Zhang F, Zhang F, Wang W, Zhou Y1, Fu B, et al. Comparative transcriptome sequencing of tolerant rice introgression line and its parents in response to drought stress. BMC Genomics. 2014; 15: 1026–1032. 10.1186/1471-2164-15-1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aprile A, Mastrangelo AM, De Leonardis AM, Galiba G, Roncaglia E, Ferrari F, et al. Transcriptional profiling in response to terminal drought stress reveals differential responses along the wheat genome. BMC Genomics. 2009; 10: 279–297. 10.1186/1471-2164-10-279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu Z, Escamilla-Treviño L, Zeng L, Lalgondar M, Bevan D, Winkel B, et al. Functional genomic analysis of Arabidopsis thaliana glycoside hydrolase family. Plant Mol. Biol. 2004; 55: 343–367. 10.1007/s11103-004-0790-1 [DOI] [PubMed] [Google Scholar]
- 29.Bohnert HJ, Sheveleva E. Plant stress adaptations -making metabolism move. Current Opinion in Plant Biology. 1998; 1: 267–274. [DOI] [PubMed] [Google Scholar]
- 30.Talame V, Ozturk NZ, Bohnert HJ, Tuberosa R. Barley transcript profiles under dehydration shock and drought stress treatments: a comparative analysis. J. Expt. Bot. 2007; 58: 229–240. [DOI] [PubMed] [Google Scholar]
- 31.Hummel I, Pantin F, Sulpice R, Piques M, Rolland G, Dauzat M, et al. Arabidopsis plants acclimate to water deficit at low cost through changes of carbon usage: an integrated perspective using growth, metabolite, enzyme and gene expression analysis. Plant Physiol. 2010; 154: 357–372. 10.1104/pp.110.157008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei M, Xiong J, Li Y, Fu B. Study on glycogen synthase kinase gene expression variation under drought stress in rice by real-time pcr. Chinese Journal of Rice Science. 2006; 20: 567–571. [Google Scholar]
- 33.Mok DWS, Mok MC. Cytokinin metabolism and action. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001; 52: 89–118. 10.1146/annurev.arplant.52.1.89 [DOI] [PubMed] [Google Scholar]
- 34.Phung TH, Jung H, Park JH, Kim JG, Back K, Jung S. Porphyrin biosynthesis control under water stress: sustained porphyrin status correlates with drought tolerance in transgenic rice. Plant Physiol. 2011; 157: 1746–1764. 10.1104/pp.111.188276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kosmala A, Bocian A, Rapacz M, Jurczyk B, Zwierzykowski Z. Identification of leaf proteins differentially accumulated during cold acclimation between Festuca pratensis plants with distinct levels of frost tolerance. J. Expt. Bot. 2009; 60: 3595–3609. [DOI] [PubMed] [Google Scholar]
- 36.Manning VA, Hardison LK, Ciuffetti LM. Ptr ToxA interacts with a chloroplast-localized protein. Mol Plant Microbe Interact. 2007; 20: 168–177. 10.1094/MPMI-20-2-0168 [DOI] [PubMed] [Google Scholar]
- 37.Neumann PM. Coping mechanisms for crop plants in drought-prone environments. Ann. Bot. 2008; 101: 901–907. 10.1093/aob/mcn018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Achard P, Cheng H, De Grauwe L, Decat J, Schoutteten H, Moritz T, et al. Integration of plant responses to environmentally activated phytohormonal signals. Science. 2006; 311: 91–94. 10.1126/science.1118642 [DOI] [PubMed] [Google Scholar]
- 39.Willekens H, Chamnongpol S, Davey M, Schraudner M, Langebartels C, Van Montagu M, et al. Catalase is a sink for H2O2 and is indispensable for stress defense in C3 plants. EMBO J. 1997; 16: 4806–4816. 10.1093/emboj/16.16.4806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.França MB, Panek AD, Eleutherio ECA. The role of cytoplasmic catalase in dehydration tolerance of Saccharomyces cerevisiae. Cell Stress Chaperones. 2005; 10: 167–170. 10.1379/CSC-103R.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bian S, Jiang Y. Reactive oxygen species, antioxidant enzyme activities and gene expression patterns in leaves and roots of kentucky bluegrass in response to drought stress and recovery. Scientia Horticulturae. 2009; 120: 264–270. [Google Scholar]
- 42.Jiang Y, Watkins E, Liu S, Yu X, Luo N. Antioxidative responses and candidate gene expression in prairie junegrass under drought stress. J Amer. Soc Hort. Sci. 2010; 135: 303–309. [Google Scholar]
- 43.Dhindsa RS, Plumb-Dhindsa P, Thorpe T. Leaf senescence: correlated with increased levels of membrane permeability and lipid peroxidation, and decreased levels of superoxide dismutase and catalase. J. Exp. Bot. 1981; 32: 93–101. [Google Scholar]
- 44.Zavaleta-Mancera HA, López-Delgado H, Loza-Tavera H, Mora-Herrera M, Trevilla-García C, Vargas-Suárez M, et al. Cytokinin promotes catalase and ascorbate peroxidase activities and preserves the chloroplast integrity during dark-senescence. J Plant Physiol. 2007; 164: 1572–1582. 10.1016/j.jplph.2007.02.003 [DOI] [PubMed] [Google Scholar]
- 45.Bollivar DW, Beale SI. The chlorophyll biosynthetic enzyme mg-protoporphyrin IX monomethyl ester (oxidative) cyclase (characterization and partial purification from Chlamydomonas reinhardtii and Synechocystis sp. PCC 6803). Plant Physiol. 1996; 112: 105–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blomstedt CK, Gianello RD, Hamill JD, Neale AD, Gaff DF. Drought-stimulated genes correlated with desiccation tolerance of the resurrection grass Sporobolus stapfianus. Plant Growth Regulation. 1998; 24: 153–161. [Google Scholar]
- 47.Velasco R, Salamini F, Bartels D. Dehydration and ABA increase mRNA levels and enzyme activity of cytosolic GAPDH in the resurrection plant Craterostigma plantagineum. Plant Mol. Biology. 1994; 26: 541–546. [DOI] [PubMed] [Google Scholar]
- 48.Jain D, Chattopadhyay D. Analysis of gene expression in response to water deficit of chickpea (Cicer arietinum L.) varieties differing in drought tolerance. BMC Plant Biology. 2010; 10: 24–38. 10.1186/1471-2229-10-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, et al. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. The Plant J. 2002; 31: 279–292. [DOI] [PubMed] [Google Scholar]
- 50.Gorlanta M, Babu PR, Lachagari R, Feltus FA, Paterson AH, Reddy AR. Functional genomics of drought stress response in rice: Transcript mapping of annotated unigenes of an indica rice (Oryza sativa L. cv. Nagina 22). Current Sci. 2005; 89: 496–514. [Google Scholar]
- 51.Maurel C. Plant aquaporins: Novel functions and regulation properties. FEBS Letters. 2007; 581: 2227–2236. 10.1016/j.febslet.2007.03.021 [DOI] [PubMed] [Google Scholar]
- 52.Lin J, Zhang L, Lai S, Ye K. Structure and molecular evolution of CDGSH iron-sulfur domains. PLoS ONE. 2011; 6: e24790 10.1371/journal.pone.0024790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sze H. H+-translocating ATPases of the plasma membrane and tonoplast of plant cells. Physiologia Plantarum. 1984; 61: 683–691. [Google Scholar]
- 54.Sjögren LLE, Tara M. Stanne TM, Zheng B, Sutinen S, Clarke AK. Structural and functional insights into the chloroplast atp-dependent clp protease in Arabidopsis. Plant Cell. 2006; 18: 2635–2649. 10.1105/tpc.106.044594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim YS, Park CM. Membrane regulation of cytokinin-mediated cell division in Arabidopsis. Plant Signal. Behav. 2007; 2: 15–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tian J, Belanger FC, Huang B. Identification of heat stress-responsive genes in heat-adapted thermal Agrostis scabra by suppression subtractive hybridization. J. Plant Phys. 2009; 166: 588–601. [DOI] [PubMed] [Google Scholar]
- 57.Aridor M, Fish KN, Bannykh S, Weissman J, Roberts TH, Lippincott-Schwartz J, et al. The Sar1 GTPase coordinates biosynthetic cargo selection with endoplasmic reticulum export site assembly. J Cell Biol. 2001; 152: 213–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Osakabe Y, Maruyama K, Seki M, Satou M, Shinozaki K, Yamaguchi-Shinozaki K. Leucine-rich repeat receptor-like kinase 1 is a key membrane-bound regulator of abscisic acid early signaling in Arabidopsis. Plant Cell. 2005; 17: 1105–1119. 10.1105/tpc.104.027474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Matsubayashi Y, Ogawa M, Morita A, Sakagami Y. An LRR receptor kinase involved in perception of a peptide plant hormone, phytosulfokine. Science. 2002; 296: 1470–1472. 10.1126/science.1069607 [DOI] [PubMed] [Google Scholar]
- 60.Hazen SP, Pathan MS, Sanchez A, Baxter I, Dunn M, Estes B, et al. Expression profiling of rice segregating for drought tolerance QTLs using a rice genome array. Funct. Integr. Genomics. 2005; 5: 104–116. 10.1007/s10142-004-0126-x [DOI] [PubMed] [Google Scholar]
- 61.Li WT, Wei YM, Wang JR, Liu CJ, Lan XJ, Jiang QT, et al. Identification, localization, and characterization of putative USP genes in barley. Theor. Appl. Genet. 2010; 121: 907–917. 10.1007/s00122-010-1359-9 [DOI] [PubMed] [Google Scholar]
- 62.Isokpehi RD, Simmons SS, Cohly HHP, Ekunwe SIN, Begonia GB, Ayensu WK. Identification of drought-responsive universal stress proteins in Viridiplantae. Bioinform Biol. Insights. 2011; 5: 41–58. 10.4137/BBI.S6061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Harb A, Krishnan A, Ambavaram MMR, Pereira A. Molecular and physiological analysis of drought stress in Arabidopsis reveals early responses leading to acclimation in plant growth. Plant Physiol. 2010; 154: 1254–1271. 10.1104/pp.110.161752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Salekdeh GH, Siopongco J, Wade LJ, Ghareyazie B, Bennett J. Proteomic analysis of rice leaves during drought stress and recovery. Proteomics. 2002; 2: 1131–1145. [DOI] [PubMed] [Google Scholar]
- 65.Watkinson JI, Sioson AA, Vasquez-Robinet C, Shukla M, Kumar D, Ellis M, et al. Photosynthetic acclimation is reflected in specific patterns of gene expression in drought-stressed loblolly pine. Plant Physiol. 2003; 133: 1702–1716. 10.1104/pp.103.026914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Heath LS, Ramakrishnan N, Sederoff RR, Whetten RW, Chevone BI, Struble CA, et al. Studying the functional genomics of stress responses in loblolly pine with the Expresso microarray experiment management system. Comput. Funct. Genomics. 2002; 3: 226–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the manuscript and supporting information files.