Abstract
Adaptor proteins participate in selective autophagy, which is critical for cellular detoxification and stress relief. However, new evidence supports an autophagy-independent key role of the adaptor p62 (encoded by the gene Sqstm1) in signaling functions central to tumor initiation in the epithelium, and suppression of tumor progression in the stroma.
Autophagy fulfills two major cellular functions -detoxification through waste removal and conferring resistance to nutrient stress. Autophagy is activated by metabolic emergencies such as nutrient starvation, which results in AMPK activation and mTORC1 inhibition (Galluzzi et al., 2014). The purpose of this “metabolic autophagy” is to provide intracellular energy sources and anabolic building blocks during nutrient shortage, as often happens during cancer progression. In that respect, autophagy might promote tumor growth when cancer cells have limited access to extracellular metabolites and energy sources. This mechanism involves the bulk incorporation of organelles into the autophagosomes without apparent selectivity (Kaur and Debnath, 2015). In contrast, during basal detoxification or waste removal in response to stress, autophagy is thought to require adaptors to target misfolded proteins and dysfunctional organelles to the autophagosomes, while sparing functional cellular constituents (Green and Levine, 2014). This is important because this type of selective autophagy, and its adaptors, maintains cellular well-being by preventing endoplasmic reticulum (ER) and oxidative stress. Therefore, selective autophagy, and theoretically its adaptors, function as tumor suppressors by preventing genotoxicity and the accumulation of oncogenic mutations. Consequently, conditions that impair selective autophagy are expected to promote mutagenesis and cancer initiation (Kimmelman, 2011; White, 2012).
Therefore, the signals converging onto the two types of autophagy must be finely balanced to prevent tumor initiation and restrain tumor progression once the cancer cell progenitors have emerged. The incomplete understanding of the intricacies of these pathways is likely the root of the conflicting interpretations of currently available data on the role of autophagy in cancer. Also, the widely accepted conceptual segregation of starvation-induced “bulk” unselective autophagy from nutrient-independent but stress-dependent and adaptor-driven selective autophagy, collides with the evidence that the autophagy adaptor p62 is degraded via selective and non-selective autophagy. This, together with the observation that p62 is often upregulated in cancer cells, suggest that a third role of autophagy is to make sure that p62 does not exceed undesired levels. This is important to keep in mind when considering autophagy as a cancer therapeutic target.
Here we discuss very recent data supporting the notion that p62, in addition to its role in selective autophagy, is a key pro-oncogenic regulator thanks to its function as a signaling hub (Figure 1A). Importantly, high levels of p62 protein in epithelial cells are necessary and sufficient for inducing oncogenic transformation, independent of its autophagy-related functions (Umemura, 2016). This model establishes that one of the critical roles of autophagy as a tumor suppressor is to prevent p62-driven tumor initiation and malignant transformation. However, in the non-transformed components of the tumor microenvironment, such as fibroblasts and macrophages, p62 functions as a non-cell autonomous tumor suppressor that attenuates fibrosis and inflammation (Valencia et al., 2014) (Zhong et al., 2016). Therefore, we propose that the homeostatic maintenance of p62 levels in both tumor and stroma by autophagy-dependent or independent mechanisms will decisively contribute to the final outcome of the tumorigenic process. This has important implications for the design of prospective therapeutic strategies for cancer targeting autophagy or p62-regulated signaling pathways.
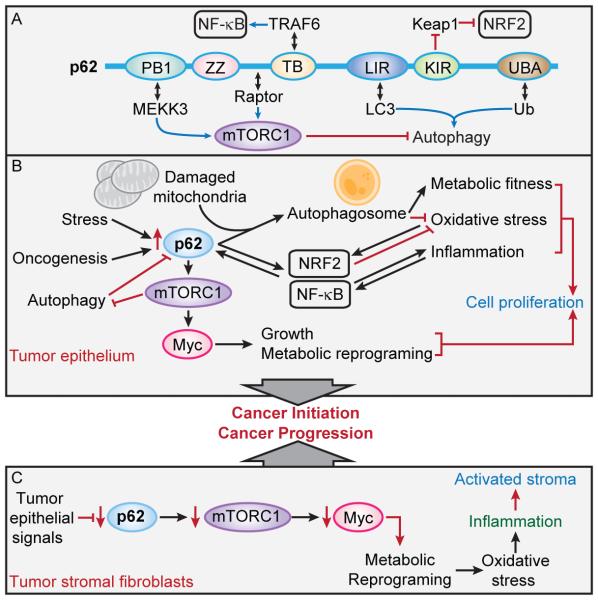
Figure 1. p62 signaling functions in the tumor epithelium and stroma.
(A) Structural domain organization of p62, binding partners and signaling functions. p62 has a PB1 domain, a ZZ-type zinc finger domain, a TRAF6-binding (TB) domain, an LC3-interacting region (LIR), a Keap1-interacting region, and a ubiquitin-associated domain (UBA).
(B) Central role of p62 accumulation in the tumor epithelium during cancer initiation through detoxification and mTORC1 activation.
(C) Role of p62 downregulation in the tumor stroma through the inhibition of mTORC1 in metabolic reprogramming and inflammation
p62, an autophagy adaptor and signaling hub
Although p62 was the first identified autophagy adaptor, four other proteins have similar functions, including NBR1, TAX1BP1, NDP52, and OPTN. p62 was initially found as a signaling regulator residing in the late endosome-lysosome (Moscat and Diaz-Meco, 2009). Unlike other autophagy adaptors, with the exception of NBR1, p62 is also a central hub due to its ability to interact with key signaling proteins through well defined structural elements (Figure 1A)(Moscat and Diaz-Meco, 2009). Thus, p62 can promote the expression of inflammatory genes via NF-κB, which it activates through TRAF6 binding by its TRAF6-binding (TB) domain. p62 also activates the NRF2-dependent anti-oxidant response by sequestering Keap1 through its KIR domain (Figure 1A) (Moscat and Diaz-Meco, 2009). New data also describe that p62 activates mTORC1, which can upregulate c-Myc (Figure 1A) (Duran et al., 2011; Valencia et al., 2014). None of these functions depend on the ubiquitin-associated (UBA) or LC3-interacting region (LIR) domains of p62, which allow it to function as an autophagy adaptor (Figure 1A) (Moscat and Diaz-Meco, 2009). Autophagy, however, plays an important role in the control of p62 levels as it is constantly degraded via non-selective autophagy through its LIR domain that binds to LC3 on autophagosomes membranes (Moscat and Diaz-Meco, 2009) (Figure 1A). p62 expression is also subjected to positive transcriptional regulation since AP1, NF-κB and NRF2 can stimulate Sqstm1 gene transcription (Moscat and Diaz-Meco, 2009)(Figure 1B). Thus, oxidative stress and inflammation induce p62 through NRF2 and NF-κB, to promote selective autophagy and cell detoxification. However, just preventing cell death, although it helps tumorigenesis, is not sufficient to initiate cancer since the activation of growth promoting anabolic pathways are required to promote cancer cell growth. In this regard, more recent data demonstrate that p62, due to its location on lysosomes and its ability to bind Raptor and the Rag proteins, regulates mTORC1 activity (Duran et al., 2011). In the presence of growth factors, mTORC1 is activated by the GTPase Rheb (Jewell et al., 2013). Rheb is kept in the inactive GDP form by the tumor suppressive Tsc1/Tsc2 GTPase complex, which is inactivated by PI3K/AKT-mediated phosphorylation triggered by growth factors, and antagonized by the tumor suppressor PTEN (Jewell et al., 2013). Interestingly, p62 deficiency in several cell systems impairs the recruitment of mTORC1 to the lysosomes and its activation in response to amino acids (Duran et al., 2011), and Tsc1 ablation (Umemura, 2016).
Of special relevance for the p62-mTORC1 connection, p62 constitutively binds to MEKK3 through their respective PB1 domains (Linares et al., 2015). Upon amino acid stimulation, the p62-MEKK3 complex orchestrates a kinase cascade that includes the activation of MKK3/6 by MEKK3-mediated phosphorylation, and the subsequent activation of p38δ (Linares et al., 2015). This results in the direct phosphorylation of p62 at residues T269 and S272 by p38δ, which promotes the recruitment of TRAF6 to the mTORC1 complex resulting in the K63-type polyubiquitination of mTOR, which is important for its efficient activation (Linares et al., 2013)(Figure 1A). Since mTORC1 stimulates several anabolic pathways that promote cell growth and proliferation as well as c-Myc expression, these findings reveal that p62 not only controls cell survival of normal and cancer cells but also contributes to cell growth and, when upregulated, to cancer cell proliferation. A link between p62 accumulation and c-Myc expression is observed in prostate cancer stromal fibroblasts (Valencia et al., 2014), and hepatocellular carcinomas (HCC) (Umemura, 2016).
p62, epithelial cell stress and cancer
Accumulation of p62 upon impairment of autophagy may exert deleterious effects in normal epithelial cells. Genetic inactivation of autophagy by ablation of critical autophagy (atg) genes in liver parenchymal cells, including hepatocytes, results in a poorly understood chronic liver damage phenotype that is reversed upon global Sqstm1 gene ablation (Komatsu et al., 2007). Similarly, genetic inactivation of IKKα in pancreatic epithelial cells (PEC) results in chronic pancreatitis that is alleviated upon selective genetic inactivation of Sqstm1 in the same cells (Li et al., 2013). Those results are surprising given that p62 accumulation triggers the synthesis of a number of detoxifying enzymes that prevent oxidative stress, likely through the upregulation of NRF2, and one would expect that p62 inactivation under autophagy-deficient conditions should result in more damage. A confounding factor in the study examining autophagy deficiency in the liver is that autophagy is selectively inhibited in the liver parenchyma, whereas p62 is globally ablated in all cells (Komatsu et al., 2007). Therefore, in this case, it is very difficult to know if these effects result from p62 deficiency in the non-parenchyma. This potential shortcoming is rectified in a pancreatitis study in which IKKα and p62 are both knocked out in pancreatic epithelial cells (PEC), the same cell type in which p62 specifically accumulates upon IKKα ablation (Li et al., 2013). In that study, p62 removal does not cause any defects in autophagy but it does reduce ER stress induced by autophagy inhibition in IKKα-deficient PEC (Li et al., 2013). Although how p62 accumulation in autophagy deficient epithelia results in increased ER stress is not entirely clear yet, it is likely that unrestrained activation of mTORC1 driven by high p62 levels clogs the ER by excessive protein production, which will be an important contributor to the pancreatitis phenotype. Similar observations are made when p62 is specifically ablated in liver parenchymal cells (Umemura, 2016).
Interestingly, chronic p62 accumulation occurs in human pancreatitis (Li et al., 2013), and most liver degenerative diseases in which it is present within cytoplasmic inclusions known as Mallory-Denk Bodies (Umemura, 2016). High p62 levels promote the activation of NRF2 (Figure 1B), and increased NRF2 also results in the transcriptional activation of the Sqstm1 gene, further increasing p62 accumulation through a feed-forward loop that promotes cancer initiation (Umemura, 2016). This suggests that although a transient increase in NRF2 levels results in protective anti-oxidant responses (DeNicola et al., 2011), chronic NRF2 activation is a common occurrence in many epithelial cancers, including HCC (Umemura, 2016). The procarcinogenic p62-NRF2 autoregulatory loop has been recently elucidated by studying several models of HCC induction, either by Tsc1 ablation, which causes chronic mTORC1 activation, or non-alcoholic steatohepatitis (NASH), that is characterized by the accumulation of fat in the liver (like steatosis), along with inflammation and cell damage. Although Sqstm1 inactivation in liver parenchymal cells abrogates the expression of the NRF2-dependent anti-oxidant response, it also results in the disappearance of hepatocytes that accumulate reactive oxygen species (ROS), which presumably serve as HCC initiating cells (Umemura, 2016). The loss of these cells is most likely due to inactivation of the NRF2-mediated protective response as well as inhibition of mTORC1 activation and c-Myc expression (Umemura, 2016). Interestingly, although high-fat diet (HFD)-feeding of wildtype mice, which only develop simple steatosis, does not lead to substantial p62 accumulation, HFD-feeding of MUP-uPA mice, which develop NASH, does result in dramatic p62 accumulation and HCC (Nakagawa et al., 2014; Umemura, 2016). Altogether, these studies demonstrate that p62 accumulation in a chronically-damaged liver is one of the most important factors, together with compensatory proliferation, that leads to HCC development.
The ultimate proof that p62 is an oncogenic protein is that its overexpression in vivo in the liver was sufficient to induce HCC without carcinogen administration or any other additional stimulus (Umemura, 2016). Notably, this correlated with increased mTORC1 and NRF2 activities. This effect of p62 is independent of autophagy because overexpression of a p62 variant lacking the UBA domain, and therefore stripped of its autophagy adaptor function, was also sufficient to drive HCC (Umemura, 2016). Therefore, therapies aimed at blocking p62 accumulation, or the activation of its downstream targets, would be a potentially new therapeutic approach for prevention of HCC in high-risk individuals. Targeting p62, or the ability of p62 to activate NRF2 and mTORC1, will have the advantage of not abolishing the basal activity of NRF2 or mTORC1, since both pathways are needed for suppression of liver toxicity and maintenance of hepatic integrity and function (Figure 1B) (Umemura et al., 2014).
p62 in the tumor microenvironment
In contrast to cancer cells, in which p62 expression is dramatically elevated, many tumors display reduced levels of p62 in their stroma, especially in cancer associated fibroblasts (CAF) (Valencia et al., 2014). This observation raises interesting questions regarding the function of p62 in the tumor microenvironment, which also plays a critical role in cancer progression. Investigating p62 function in prostate cancer, we find that p62-deficient stromal fibroblasts exhibit increased production of IL-6, which is critical for TGFβ synthesis that converts CAF into activated myofibroblasts that promote tumor progression (Valencia et al., 2014). Importantly, the downregulation of p62 in CAF is needed for acquiring their tumor promoting function (Valencia et al., 2014). Decreased mTORC1 activity due to the downregulation of p62 expression results in reduced c-Myc levels that leads to a failure in the synthesis of reduced glutathione due to defective metabolic reprogramming, and the concomitant increase in ROS levels under conditions of autophagy competence (Valencia et al., 2014). Interestingly, the increased oxidative stress in these stromal cells translated into higher levels of IL-6 that promoted TGFβ synthesis, being both required for CAF activation in p62-deficient prostate cancer stroma (Valencia et al., 2014) (Figure 1C). An important corollary of these studies is that blocking p62, or directly inhibiting mTORC1, in the stroma will favor tumor progression, which suggests that anti-cancer treatments aimed at reducing the pro-tumorigenic mTORC1 signaling in tumor cells will be counteracted by the undesired effects of mTORC1 inactivation in the stroma. This renders therapies that target mTORC1 in cancer inefficient or even counter-effective. Future studies in other cancer model systems, together with the analysis of more extensive patient’s samples will establish the generality of these observations.
The ability of p62 to act as a tumor suppressor in constituents of the tumor microenvironment might also apply to tumor associated macrophages (TAM), which serve as an important source of tumor promoting inflammatory signals (Grivennikov et al., 2010; Ruffell and Coussens, 2015). In this regard, selective autophagy dependent on p62 has been reported to be important for clearance of mitochondria that have been damaged due to exposure of macrophages to different stimuli capable of activating the NLRP3 inflammasome (Zhong et al., 2016). Such stimuli induce mitochondrial damage and the release of direct NLRP3-inflammasome activators such as mitochondrial DNA and ROS (Zhou et al., 2011). Damaged mitochondria undergo mitophagic clearance through p62 resulting in termination of inflammasome activation and reduced production of the tumor promoting cytokines IL-1β and IL-18 (Zhong et al., 2016). Many stimuli that lead to NLRP3 inflammasome activation, such as silica microcrystals and asbestos fibers are carcinogens even though they do not induce any oncogenic mutations. Altogether, these results can explain why genetic inactivation of p62 at an organismal level is associated with increased tumor progression (Valencia et al., 2014), whereas the selective inactivation of p62 in cancer epithelial cells restrains cancer initiation (Umemura, 2016).
Conclusions and outstanding questions
Although we now know a lot more about p62 and its role in cancer, many issues still need to be investigated. For example, why is p62 degraded during nutrient stress-induced autophagy if this process does not need adaptors? But even if it does, how can p62 distinguish between different cargos during “bulk” versus “selective autophagy”? And during selective autophagy, how is specificity of cargo recognition achieved? It is unlikely that just the PB1 and the UBA domains will be sufficient for cargo selection by p62. How about the other UBA-containing adaptors? Are they specific for different cargos or do they recognize the same cargos but the specificity is cell type-dependent? Do other adaptors have autophagy-independent signaling capabilities like in the case of p62? Our recent data demonstrating that NBR1 is a new scaffold for JNK activation in response to hyper-nutrition in macrophages suggest that this is the case (Hernandez et al., 2014). However, more data are required to address all these fundamental questions whose resolution will help us understand the function of these proteins in physiologically relevant models including cancer, and devise new therapeutics by targeting p62-regulated functions.
ACKNOWLEDGEMENTS
Our research is funded by NIH grants R01DK108743 (J.M.), R01CA172025 (J.M.), R01CA163798 (M.K.), R01CA118165 (M.K.), P01DK098108 (M.K.), R01CA192642 (M.T.D.-M.) and 5P30CA030199 (M.T.D-M. and J.M), and by DoD grants W81XWH-13-1-0354 (J.M.) and W81XWH-13-1-0353 (M.T.D-.M.) MK holds the Ben and Wanda Hildyard Chair for Mitochondrial and Metabolic Diseases. We thank Diantha LaVine for the artwork.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- DeNicola GM, Karreth FA, Humpton TJ, Gopinathan A, Wei C, Frese K, Mangal D, Yu KH, Yeo CJ, Calhoun ES, et al. Nature. 2011;475:106–109. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran A, Amanchy R, Linares JF, Joshi J, Abu-Baker S, Porollo A, Hansen M, Moscat J, Diaz-Meco MT. Mol Cell. 2011;44:134–146. doi: 10.1016/j.molcel.2011.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Pietrocola F, Levine B, Kroemer G. Cell. 2014;159:1263–1276. doi: 10.1016/j.cell.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green DR, Levine B. Cell. 2014;157:65–75. doi: 10.1016/j.cell.2014.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov SI, Greten FR, Karin M. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez ED, Lee SJ, Kim JY, Duran A, Linares JF, Yajima T, Muller TD, Tschop MH, Smith SR, Diaz-Meco MT, et al. Cell Metab. 2014 doi: 10.1016/j.cmet.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewell JL, Russell RC, Guan KL. Nat Rev Mol Cell Biol. 2013;14:133–139. doi: 10.1038/nrm3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur J, Debnath J. Nat Rev Mol Cell Biol. 2015;16:461–472. doi: 10.1038/nrm4024. [DOI] [PubMed] [Google Scholar]
- Kimmelman AC. Genes & development. 2011;25:1999–2010. doi: 10.1101/gad.17558811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata J, Ezaki J, Murata S, et al. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- Li N, Wu X, Holzer RG, Lee JH, Todoric J, Park EJ, Ogata H, Gukovskaya AS, Gukovsky I, Pizzo DP, et al. J Clin Invest. 2013;123:2231–2243. doi: 10.1172/JCI64498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linares JF, Duran A, Reina-Campos M, Aza-Blanc P, Campos A, Moscat J, Diaz-Meco MT. Cell reports. 2015;12:1339–1352. doi: 10.1016/j.celrep.2015.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linares JF, Duran A, Yajima T, Pasparakis M, Moscat J, Diaz-Meco MT. Mol Cell. 2013;51:283–296. doi: 10.1016/j.molcel.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscat J, Diaz-Meco MT. Cell. 2009;137:1001–1004. doi: 10.1016/j.cell.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa H, Umemura A, Taniguchi K, Font-Burgada J, Dhar D, Ogata H, Zhong Z, Valasek MA, Seki E, Hidalgo J, et al. Cancer Cell. 2014;26:331–343. doi: 10.1016/j.ccr.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffell B, Coussens LM. Cancer Cell. 2015;27:462–472. doi: 10.1016/j.ccell.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umemura A, He F, Taniguchi K, Nakagawa H, Yamachika S, Font-Burgada J, Zhong Z, Subramaniam S, Raghunandan S, Duran A, Linares JF, Reina-Campos M, Umemura S, Valasek MA, Seki E, Yamaguchi K, Koike K, Itoh Y, Diaz-Meco MT, Moscat J, Karin M. Cancer Cell. 2016 doi: 10.1016/j.ccell.2016.04.006. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umemura A, Park EJ, Taniguchi K, Lee JH, Shalapour S, Valasek MA, Aghajan M, Nakagawa H, Seki E, Hall MN, et al. Cell Metab. 2014;20:133–144. doi: 10.1016/j.cmet.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia T, Kim JY, Abu-Baker S, Moscat-Pardos J, Ahn CS, Reina-Campos M, Duran A, Castilla EA, Metallo CM, Diaz-Meco MT, et al. Cancer Cell. 2014;26:121–135. doi: 10.1016/j.ccr.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E. Nature reviews Cancer. 2012;12:401–410. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z, Umemura A, Sanchez-Lopez E, Liang S, Shalapour S, Wong J, He F, Boassa D, Perkins G, Ali SR, et al. Cell. 2016;164:896–910. doi: 10.1016/j.cell.2015.12.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Yazdi AS, Menu P, Tschopp J. Nature. 2011;469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]