Abstract
Objectives
Human complement C4 is sophisticatedly complex with multiple layers of diversity. This study aims to elucidate the CNVs of C4A and C4B in disease risk of SLE, and compare the basis of race-specific C4A-deficiency in East-Asians (EA) and Europeans.
Patients and Methods
Our EA study-population included 999 SLE patients and 1,347 healthy subjects. Variations in gene copy-numbers (GCNs) for total C4, C4A, C4B, long and short genes were determined and validated rigorously by independent genotyping technologies. Genomic regions with C4B96 were investigated to determine the basis of the most basic C4B protein that is concurrent with C4A-deficiency.
Results
In EA, strong protective effects of high GCNs for total C4 and C4A against SLE were notable; low and medium GCNs for total C4 and C4A, and the absence of short genes were risk factors of SLE. Homozygous C4A-deficiency was infrequent but had an odds-ratio (OR) of 12.4 (p=0.0015). Patients who experienced very-low serum complement were associated with low GCNs of total C4 (OR=3.27, p=7.0×10−7) and C4B (OR=2.55, p=2.5×10−5). Patients with low complement had high frequencies of anti-dsDNA (OR=4.96, p=9.7×10−17), hemolytic anemia (OR=3.89, p=3.6×10−10) and renal disease (OR=2.18, p=8.5×10−6). The monomodular-short haplotype with C4A-deficiency and in linkage-disequilibrium with HLA-DRB1*0301 prevalent in European was scarce in EA. Instead, most EA-subjects with C4A-deficiency shared a recombinant haplotype with bimodular-LS encoding C4B1 and C4B96, which was linked to HLA-DRB1*1501. DNA sequencing revealed the E920K polymorphism for C4B96.
Conclusion
C4 CNVs and C4A-deficiency are important in the risk and manifestations of East-Asian and European SLE.
INTRODUCTION
Systemic lupus erythematosus (SLE) is a female dominant autoimmune disease characterized by the generation of autoantibodies against nuclear antigens including double-stranded DNA (dsDNA) (1-3). The formation of immune complexes between autoantibodies and self-antigens activates complement, causing systemic tissue injuries and organ damages (4,5). Reduced serum levels or depressed hemolytic activities of complement are common clinical laboratory findings in patients with SLE and glomerulonephritis (4,6-9). Such phenomenon is generally interpreted as a result of consumption due to SLE disease activities. However, low serum complement can both be a cause and an effect of SLE. Human subjects with a homozygous genetic deficiency in anyone of the early components for the classical pathway of complement activation (Supporting Information: Figure S1), C1q, C1r, C1s and C4 were almost always inflicted with SLE irrespective of race, gender and HLA haplotypes (10-14). Under susceptible backgrounds, mice with genetic knockout of complement C4 or C1q manifested lupus-like phenotypes with high titers of antinuclear antibodies (ANA) and anti-dsDNA and high frequency of glomerulonephritis (15-18). Cumulated information suggests that complement deficiency contributes to pathogenesis of SLE through impaired handling and clearance of immune complexes, deficient scavenging of apoptotic cells, aberrant induction of peripheral tolerance because of inefficient elimination of self-reactive B cells to produce high affinity autoantibodies with class switching, and defective control of cytokine production including type I interferons (5,19,20).
While a homozygous genetic deficiency for complement C1 or C4 can be a causal factor of SLE, their prevalence is extremely rare as only 122 cases have ever been documented (4). Genome-wide association studies led to the identification of numerous single nucleotide polymorphisms (SNPs) associated with increased risk of SLE, but most of those SNPs are of low effect-size with odds ratios (OR) between 1.1 and 1.5 (1-3,21).
SLE affects all racial groups but patients of Asian, African and Hispanic ancestries/ethnicities often have severe disease with renal involvement (22,23). While SLE risk factors identified in Asians are also germane to Europeans, there are remarkable differences between races with respect to effect-sizes and frequencies of risk alleles (24). In a case-control study of C4 gene copy-number variations (CNVs; online Figure S1) in SLE of European ancestry, we found that low GCNs of total C4 and C4A were risk factors for, and high GCNs of total C4 and C4A were protective factors against, SLE disease susceptibility (25). A common major histocompatibility (MHC, or HLA in human) haplotype with a single short C4B gene (mono-S) and the absence of C4A is prevalent among European SLE patients. This mono-S haplotype is in strong linkage disequilibrium (LD) with the HLA class II gene DRB1 allele *0301 (or DR3) (26,27). Although the increased risk for C4A-deficiency in Asians had been observed in earlier studies (28-30), the mechanism of C4A-deficiency and diversity associated with C4-CNVs have not been investigated meticulously. Here, we report an in-depth study to elucidate patterns of CNVs for complement C4 and associated variants in large cohorts of East-Asian (Chinese) SLE and race-matched healthy controls. This study unravels the molecular basis of C4A-deficiency and fine details pertinent to C4-CNVs in East-Asian and European SLE disease susceptibility and pathogenesis.
STUDY POPULATIONS AND METHODS
Study populations
Our study populations included 999 Chinese SLE patients and 1,347 race-matched healthy controls (Table 1). All patients fulfilled at least four of the eleven revised 1982-ACR diagnostic criteria for human SLE (31,32), The patients’ female to male sex ratio was 11.6 to 1. Their mean age (±SD) at disease diagnosis was 30.61±11.45 years old. Besides the presence of ANA that was almost universal among SLE patients, the frequencies for SLE diagnostic disorders with details on hematologic and immunologic disorders are tabulated in panel B of Table 1. None of the race-match healthy controls reported to have an autoimmune disease. The IRB of the Nationwide Children's Hospital (USA), the ethics committees from the University of Hong Kong (Hong Kong) and the Chang Gung Memorial Hospital (Taiwan) approved the study. All blood donors provided written consent.
Table 1.
Clinical characteristics of East-Asian SLE patients
| Hong Kong | Taiwan | Combined | ||||
|---|---|---|---|---|---|---|
| Age of onset (yr) | 30.27±10.70 | 30.68±11.62 | 30.61±11.45 | |||
| Female/Male (N) | 172 / 8 | 748 / 71 | 920 / 79 | |||
| N (yes/no) | frequency | N (yes/no) | frequency | N (yes/no) | frequency | |
| Malar rash | 113 / 67 | 0.628 | 467 / 352 | 0.570 | 580 / 419 | 0.581 |
| Discoid rash | 39 / 141 | 0.217 | 158 / 661 | 0.193 | 197 / 802 | 0.197 |
| Photosensitivity | 62 / 118 | 0.344 | 189 / 630 | 0.231 | 251 / 748 | 0.251 |
| Oral ulcer | 23 / 157 | 0.128 | 224 / 595 | 0.274 | 247 / 752 | 0.247 |
| Arthritis | 147 / 33 | 0.817 | 520 / 299 | 0.635 | 667 / 332 | 0.668 |
| Serositis | 35 / 145 | 0.194 | 229 / 590 | 0.280 | 264 / 735 | 0.264 |
| Renal Disease | 52 / 128 | 0.289 | 464 / 355 | 0.567 | 516 / 483 | 0.517 |
| Neurologic disease | 11 / 169 | 0.061 | 137 / 682 | 0.167 | 148 / 851 | 0.148 |
| Immunologic disorder | 150 / 30 | 0.833 | 600 / 219 | 0.733 | 750 / 249 | 0.751 |
| Hematologic disease | 118 / 62 | 0.656 | 598 / 221 | 0.730 | 716 / 283 | 0.717 |
| leukopenia <3.5K | 471 /348 | 0.575 | ||||
| hemolytic anemia | 263 / 556 | 0.321 | ||||
| thrombocytopenia | 216 / 602 | 0.264 | ||||
| anti-dsDNA | 624 / 179 | 0.771 | ||||
| anti-RNP-1 | 295 / 374 | 0.441 | ||||
| anti-Sm | 262 / 408 | 0.391 | ||||
| anti-Ro (Ssa) | 362 / 189 | 0.657 | ||||
| anti-La (SSb) | 148 / 404 | 0.268 | ||||
| ACA-IgG | 188 / 461 | 0.290 | ||||
| ACA-IgM | 55 / 544 | 0.092 | ||||
| Low C3/C4 | 641 / 166 | 0.794 | ||||
| Pericarditis | 101 / 718 | 0.123 | ||||
| Pleuralitis | 159 / 660 | 0.194 | ||||
| Ascites | 43 / 776 | 0.053 | ||||
Healthy controls included 765 subjects (396 females and 369 males) from Taiwan, 371 subjects (204 females, 162 males, 5 unknown sex) from Hong Kong, and 211 subjects (122 females and 89 males) from central Ohio, USA.
Determination of C4 gene CNVs
Processing of blood samples for plasma, peripheral mononuclear cells (PBMC) and genomic DNA were as described previously (33). The copy-numbers of total C4, C4A and C4B genes, and long and short C4 genes were determined by TaqMan based, quantitative realtime PCR, and/or “hot-stop PCR” (33-35). Validation for CNV calls were achieved when GCNs of total C4=C4A+C4B. The RCCX haplotypes for the Ohio cohort were further determined by long-range mapping of PmeI and PacI-digested genomic DNA and resolved by pulsed-field gel electrophoresis (PFGE), and TaqI, PshAI/PvuII restriction fragment length polymorphisms (RFLPs).
Phenotyping of complement C4
Protein polymorphisms of C4A and C4B were determined by immunofixation of EDTA-plasma resolved by high-voltage agarose gel-electrophoresis (33,36,37). Ambiguous and intermediate allotypes migrating between C4B and C4A were further resolved by immunoblot analyses using monoclonal antibodies against Rodgers or Chido antigens (38,39). Interpretation of C4A and C4B allotypes were substantiated by their corresponding genotypes.
Autoantibody assays
Autoantibody titers were determined by ELISA. ANA was considered as positive when serum titers were >1:80 by Hep-2 cell assay. Anti-ENA (Ro/SSA, La/SSB, Sm, and RNP) and anti-cardiolipin were assessed by ELISA according to vendor's instructions (Pharmacia Diagnostics).
Statistics
Statistical analyses were performed using JMP Genomics version 6.0 (SAS Institute, Cary, NC) and Prism6 (GraphPad, San Diego, CA) software. Descriptive statistics were displayed as mean ± standard deviation (SD) for normally distributed data, and simple comparisons were made using Student's t-test for continuous data, or by χ2 analysis for categorical data. P-values were derived from Likelihood ratios. Odds ratios (OR) and 95% confidence levels were calculated by analysis of 2×2 tables through Fisher's exact test. For all analyses, p≤0.05 was considered significant.
RESULTS
CNVs of C4A, C4B, long genes, short genes and total C4
CNV of C4A
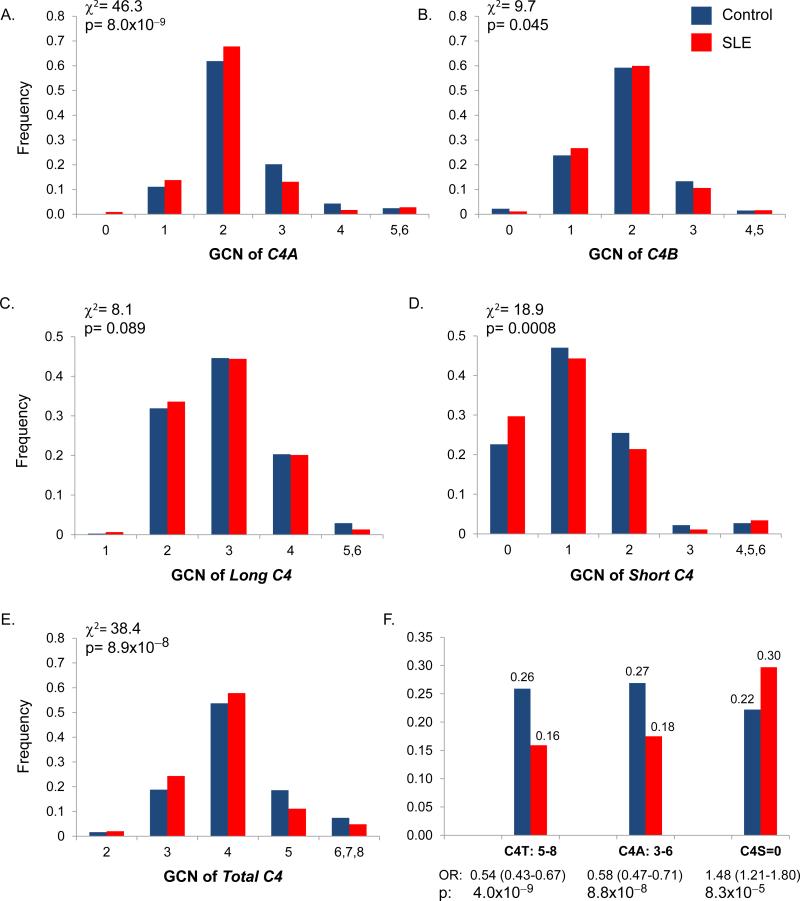
The number of C4A varied from 0 to 6 copies among different individuals (panel A, Fig. 1; Supporting Information: Table S1). There was a great difference in the distribution of GCN groups for C4A between SLE and controls (p=8.0×10−9). Homozygous deficiency of C4A (GCN=0) was found in nine patients and only one control, which translated into an OR of 12.4 (95% confidence interval, 1.57-97.9) and p=0.0015. The prevalence of heterozygous deficiency for C4A (GCN=1) was greater, 13.8% in SLE and 11.1% in controls, but its effect-size was modest: OR=1.28 (1.00-1.65), p=0.049. The most prevalent GCN group for C4A was two, which had a frequency of 67.8% in SLE and 61.9% in controls. Three and four copies of C4A had frequencies of 13.1% and 1.7%, respectively, in SLE; compared to 20.2% and 4.3%, respectively, in controls. For very high copy-number of C4A (GCN=5 and 6), the frequencies were low but similar between SLE and controls. The mean GCN (±SD) of C4A was 2.09±0.79 in SLE and 2.25±0.82 in controls (p=3.4×10−6, t-test).
Figure 1.
Variations of complement C4 gene copy-numbers (GCNs) in SLE patients (red) and controls (blue) of East-Asian ancestry. A-E, Distributions of GCN groups for C4A (A), C4B (B), long genes (C), short genes (D), total C4 (E). F. A summary of C4 genetic factors associated with SLE in East-Asians. High GCNs of total C4 and C4A were strong protective factors and deficiency of short genes is a risk factor for SLE.
CNV of C4B
The GCN of C4B varies from 0 to 5 copies among different individuals (panel B, Fig. 1). There was only slight difference in distribution of GCN groups for C4B between SLE and controls (p=0.045). The mean GCN of C4B was 1.85±0.68 in SLE and 1.88±0.71 in controls.
CNV of long-C4 genes
One to six copies of long-C4 genes were present in a diploid genome among different individuals but the distribution patterns of long C4 were similar between SLE and controls (panel C, Fig. 1). It is notable that no subject had an absence and only nine had one copy of long gene in the entire study population. The mean GCN of long-C4 was 2.88±0.79 in SLE and 2.94±0.82 in controls.
CNV of short-C4 genes
Zero to six copies of short-C4 genes was present and the distribution was different between SLE and controls (p=0.0008, χ2 analysis; panel D, Fig. 1). The absence of short gene existed in 29.7% of patients and 22.6% of controls; OR=1.48 (1.22-1.80), p=8.3×10−5. The mean copy-number of short genes was 1.05±0.97 in SLE and 1.17±0.94 in controls (p=0.005, t-test).
CNV of total C4
Two to eight copies of total C4 in a diploid genome were detectable (panel E, Fig. 1; Supporting Information: Table S1). The most prevalent GCN group for total C4 was four. Similar to C4A, there was a consistent shift for increased frequencies of the low and median copy-number groups, and reduced frequencies of the high copy-number groups in SLE (p=3.7×10−7, χ2 analyses). The mean GCN of total C4 was 3.95±0.87 in SLE and 4.14±0.92 in controls (p=8.9×10−8, t-test).
Among the C4 genes, the proportions for C4A and C4B were 53.2%/46.8% in SLE and 54.8%/45.2% in controls (p=0.011). For long and short genes, the proportions were 75.0%/25.0% in SLE; and 73.0%/27.0% in controls (p=0.017). SLE patients had consistent reduction in mean GCNs of total-C4, C4A and short-C4 (Fig 1F, Supporting Information: Table S1). The reduction of GCNs for total C4 or C4A was attributable to a reduction of short genes. The protective effects for high copy-numbers of total-C4 and C4A against SLE were highly significant.
Complement C4 gene CNVs as risk factors for SLE diagnostic disorders
Intra-group analyses of C4 CNVs were performed to investigate their associations with diagnostic disorders of SLE. Three different associations emerged (Table 2).
Table 2.
CNVs of complement C4 as risk factors for SLE diagnostic disorders
| Yes | No | ||||
|---|---|---|---|---|---|
| Disorders | Group | N (freq.) / GCI | N (freq.) / GCI | OR (95% CI) | p |
| a. Potentially causal relationship | |||||
| Malar Rash | C4A = 0 | 9 (.016) | 0 (0) | na1 | 0.0016 |
| C4A ≥ 1 | 547 (.984) | 407 (1.0) | |||
| C4A GCI | 2.05±.82 | 2.16±.75 | 0.034 | ||
| Thrombocytopenia | C4B = 0 | 0 (0) | 11 (.019) | na2 | 0.0093 |
| C4B ≥ 1 | 205 (1.0) | 564 (.981) | |||
| C4B GCI | 1.83±.63 | 1.82±.69 | ns | ||
| b. Reduced GCN as a risk factor | |||||
| Low C3/C4 | C4-total = 2-3 | 183 (.306) | 18 (.118) | 3.27 (1.94-5.52) | 7.0×10−7 |
| C4-total ≥ 4 | 416 (.695) | 134 (.882) | |||
| C4-total GCI | 3.88±.89 | 4.16±.83 | 0.0003 | ||
| C4B = 0-1 | 194 (.324) | 24 (.158) | 2.55 (1.60-4.08) | 2.5×10−5 | |
| C4B = 2-5 | 405 (.676) | 128 (.842) | |||
| C4B GCI | 1.77±.67 | 2.03±.65 | 1.3×10−5 | ||
| C4-Long = 1-3 | 490 (.801) | 107 (.686) | 1.84 (1.24-2.72) | 0.0028 | |
| C4-Long = 4-6 | 122 (.199) | 49 .314) | |||
| C4-Long GCI | 2.87±.78 | 3.06±.83 | 0.0084 | ||
| Anticardiolipin-IgM | C4B = 0-1 | 23 (.426) | 145 (.282) | 1.89 (1.07-3.36) | 0.032 |
| C4B = 2-5 | 31 (.574) | 370 (.718) | |||
| C4B GCI | 1.63±.73 | 1.84±.68 | 0.029 | ||
| Discoid rash | C4-Short = 0-1 | 124 (.821) | 475 (.736) | 1.64 (1.05-2.58) | 0.026 |
| C4-Short = 2-6 | 27 (.179) | 170 (.264) | |||
| C4-Short GCI | 0.83±.87 | 1.08±1.03 | 0.0060 | ||
| Arthritis | C4-Short = 0 | 174 (.344) | 76 (.261) | 1.48 (1.07-2.03) | 0.016 |
| C4-Short ≥ 1 | 326 (.656) | 214 (.738) | |||
| C4-Short GCI | 0.97±.96 | 1.15±1.06 | 0.017 | ||
| c. Increased GCN as a risk factor | |||||
| Pericarditis | C4A = 3-6 | 28 (.292) | 106 (.154) | 2.26 (1.39-3.67) | 0.0017 |
| C4A = 0-2 | 68 (.708) | 581 (.846) | |||
| C4A GCI | 2.27±.79 | 2.08±.78 | 0.027 | ||
| Hemolytic anemia | C4A = 3-6 | 58 (.234) | 76 (.142) | 1.84 (1.26-2.70) | 0.0019 |
| C4A = 0-2 | 190 (.766) | 459 (.858) | |||
| C4A GCI | 2.22±.88 | 2.05±.74 | 0.0061 | ||
| Thrombocytopenia | C4-Short = 2-6 | 67 (.328) | 124 (.216) | 1.77 (1.24-2.52) | 0.0018 |
| C4-Short = 0-1 | 137 (.672) | 449 (.784) | |||
| C4-Short GCI | 1.27±1.17 | 0.94±.93 | 5.8×10−5 | ||
| C4A = 3-6 | 46 (.224) | 88 (.153) | 1.61 (1.08-2.40) | 0.022 | |
| C4A = 0-2 | 159 (.776) | 489 (.848) | |||
| C4A GCI | 2.26±.98 | 2.05±.70 | 0.0012 | ||
GCI, gene copy-index – the mean of gene copy-number in a population with standard deviation; OR, odds ratio p-values for continuous data were calculated by t-tests; p-values and odds ratios of categorical data were derived by χ2 analyses.
na1, odds ratio not applicable; all nine SLE patients with homozygous C4A deficiency had malar rash; na2, odds ratio not applicable; no subjects with thrombocytopenia had a homozygous C4B-deficiency; ns, not significant
a. Potentially causal relationship
Nine SLE patients had a homozygous C4A-deficiency and all of them had malar rash (p=0.0016). Also, eleven SLE patients had homozygous C4B-deficiency and none of them had thrombocytopenia (p=0.0093). Thus, the absence of C4A could be a causal factor for malar rash, and the presence of C4B might be required for thrombocytopenia.
b. Risks associated with low GCNs
The ever presence of very-low serum complement levels were recorded in 79.4% of SLE patients. Very-low serum complement was strongly correlated with low GCNs of total C4, C4B and long-C4. Low GCN of total C4 had the largest effect-size [OR=3.27 (1.94-5.52), p=7.0×10−7]. The OR for C4B was 2.55 (1.60-4.08), p=2.5×10−5, and for long-C4 was 1.84 (1.24-2.72), p=0.0028. Low GCNs of total C4 leading to very-low serum complement were attributable to low GCNs of C4B, which were likely long genes.
Low GCN of C4B was also associated with the presence of anticardiolipin-IgM [C4B=0 or 1, OR=1.89 (1.07-3.36), p=0.032]. The absence or low copy-numbers of short-C4 were associated with discoid rash and arthritis with moderate effect-sizes [discoid rash: OR=1.64 (1.05-2.58), p=0.026; arthritis: OR=1.48 (1.07-2.03), p=0.016].
c. Risks associated with high GCNs
High GCN of C4A (GCN=3-6) appeared to be a risk factor for pericarditis [OR=2.26 (1.39-3.67), p=0.0017], hemolytic anemia [OR=1.84 (1.26-2.700, p=0.0019] and thrombocytopenia [OR=1.61 (2.08-2.40), p=0.022]. High GCN of short-C4 genes (≥2) appeared as a risk factor of thrombocytopenia [OR=1.77 (1.24-2.52), p=0.0018].
Associations of low serum C3/C4 levels with SLE diagnostic disorders
Besides low GCNs of total C4, C4B or long genes, low serum C3 and C4 levels could also reflect systemic complement consumption or SLE disease states (Table 3). Patients with very-low C3/C4 had an earlier age of disease-onset (mean age ± SD: 30.0±11.0 with low C3/C4 versus 33.3±13.8 without low C3/C4; p=0.0011). Very strong associations with large effect-sizes were found between very-low C3/C4 and (a) immunologic disorders such as the presence of anti-dsDNA [OR: 4.96 (3.41-7.22), p=9.7×10−17], anti-Sm [OR: 2.37 (1.54-3.64), p=3.6×10−5], and IgG-anticardiolipin [OR: 2.33 (1.41-3.87), p=0.0004]; (b) hematologic disorders such as hemolytic anemia [OR: 3.89 (2.42-6.27), p=3.6×10−10] and leukopenia [OR:1.92 (1.36-2.71), p=0.0002]; (c) renal disease [OR: 2.32 (1.63-3.31), p=2.2×10−6]; and (d) serositis including ascites [OR: 3.62 (1.10-11.8), p=0.011] and pericarditis [OR: 2.55 (1.29-5.01), p=0.0027]. Very-low C3/C4 also significantly associated with increased risks of serositis, thrombocytopenia, anti-RNP1, and anti-La/SSb, although their effect-sizes were modest.
Table 3.
Associations of low serum complement protein levels (C4/C3)* with clinical and immunologic features of EA-SLE.
| Yes (N=641) frequency | No (N=166) frequency | OR (95% CI) | p | |
|---|---|---|---|---|
| Malar rash | 0.577 | 0.530 | ns | |
| Discoid rash | 0.184 | 0.217 | ns | |
| Photosensitivity | 0.225 | 0.259 | ns | |
| Oral ulcers | 0.270 | 0.295 | ns | |
| Arthritis | 0.619 | 0.693 | 0.08 | |
| Neurologic disorder | 0.181 | 0.120 | 0.055 | |
| Immunologic disorders** | ||||
| Anti-dsDNA | 0.846 | 0.524 | 4.96 (3.41-7.22) | 9.7×10–17 |
| Anti-Sm | 0.429 | 0.241 | 2.37 (1.54-3.64) | 3.6×10–5 |
| Anti-RNP-1 | 0.474 | 0.314 | 1.97 (1.32-2.94) | 0.0007 |
| Anti-Ro/SSa | 0.676 | 0.600 | ns | |
| Anti-La/SSb | 0.843 | 0.748 | 1.80 (1.10-2.98) | 0.016 |
| Aca-IgG | 0.320 | 0.168 | 2.33 (1.41-3.87) | 0.0004 |
| Aca-IgM | 0.097 | 0.068 | ns | |
| Hematologic disorders | ||||
| Hemolytic anemia | 0.373 | 0.133 | 3.89 (2.42-6.27) | 3.6×10–10 |
| Leukopenia ≤3.5k | 0.607 | 0.446 | 1.92 (1.36-2.71) | 0.0002 |
| Thrombocytopenia | 0.281 | 0.206 | 1.50 (0.99-2.27) | 0.048 |
| Renal disease | 0.608 | 0.416 | 2.18 (1.54-3.09) | 8.5×10–6 |
| Serositis | ||||
| Ascites | 0.062 | 0.018 | 3.62 (1.10-11.8) | 0.011 |
| Pericarditis | 0.140 | 0.060 | 2.55 (1.29-5.01) | 0.0027 |
| Age of onset (yrs±SD) | 29.98±10.95 | 33.29±13.75 | 0.0011 |
Low C3 was defined as a documentation of serum C3 concentration <700 mg/L; low C4 was defined as a documentation of serum C4 concentration <100 mg/L.
As proposed by SLICC in 2012, low C4/C3 is one of the diagnostic factors for an immunologic disorder in SLE.
ns, not significant
A specific Asian haplotype with C4B96 and C4A-deficiency
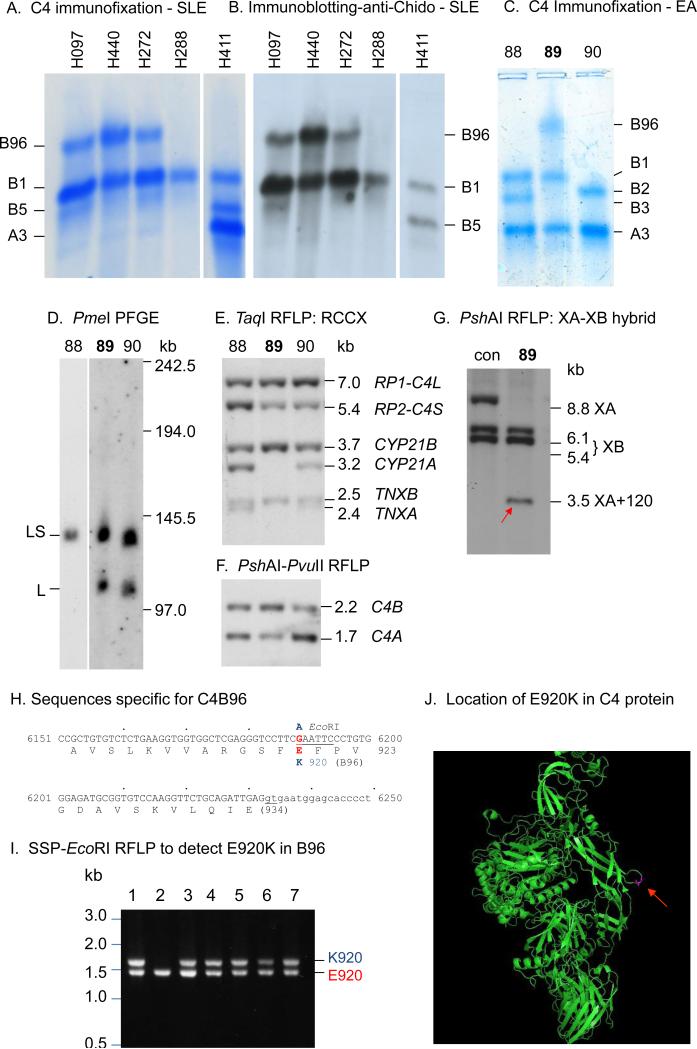
Immunochemical analyses of EDTA-plasma from four SLE patients with homozygous C4A deficiency and one patient with both C4A and C4B present were shown in panels A and B of Figure 2. Immunofixation experiment revealed that three out of four C4A-deficient patients possessed the slowest-migrating C4 allotype, C4B96, in addition to C4B1. The fourth C4A-deficient patient had C4B1 only (panel A). Immunoblot experiment revealed that C4B96 and C4B1 (and C4A1) but not C4A3 were associated with Chido blood-group antigens (panel B) (38,39). C4A-deficient patients had two to four copies of C4 genes in a diploid genome and they all coded for C4B protein. Those C4 genes either existed in (a) monomodular RCCX haplotype with a single long gene coding for C4B1, or (b) bimodular RCCX coding for C4B1-B96 or C4B1-B1. The limited accessibility of patient samples restricted further characterization of homozygous C4A deficiency.
Figure 2.
C4A-deficiency in East-Asians. A. Immunofixation of C4 allotypes showing the homozygous deficiency of C4A protein in four SLE patients (H097, H440, H288 and H288) and one control (H411). B. Immunoblot analysis of C4 plasma protein from the same subjects shown in panel A using monoclonal antibodies against Chido antigens that is generally associated with C4B protein. CH, Characterization of C4B96 protein by immunofixation (C), RCCX haplotypes by long range mapping of PmeI-digested genomic DNA (D), TaqI RFLP of RP-C4-CYP21-TNX modules (E), PshAI-PvuII RFLP for relative dosage of C4B and C4A genes (F) and rearrangement of RCCX in Subject-89 (G). A red-arrow indicates the presence of a TNXA-XB recombinant characterized by a 3.5 kb PshAI restriction fragment after Southern blot analysis using a TNX 3’ probe. H. DNA and amino acid sequences specific for C4B96 highlighting the E920K polymorphism with EcoRI RFLP. I. EcoRI RFLP of genomic PCR fragments from six subjects with C4B96 (lanes 1 and 3-7) and one subject without C4B96 (lane 2). J. The location of E920K in 3-dimensional structure of native C4 protein (indicated by a red arrow) (44). X-ray crystal structure of C4 was downloaded from RCSB PDB (http://www.rcsb.org/pdb/home/home.do; entry “4FXK”, and visualized by PyMOL).
Seven subjects in our American EA study-cohort contained C4B96 and Subject-89 was chosen for detailed analysis because of its relative simplicity (panels C-G, Fig. 2). PmeI-PFGE and TaqI-RFLP revealed that Subject-89 had heterozygous RCCX haplotypes with bimodular-LS and monomodular-L (panels D and E), and a total of two C4B and one C4A (panel F). The LS haplotype coding for C4B1-B96 was a recombinant between tenascin TNXB and TNXA, as characterized by the absence of pseudogene steroid 21-hydroxylase CYP21A, and the presence of an 120-bp insertion in TNXA, or XA+120, as documented previously (Supporting Information: Fig. S2) (40-42). Examinations of other Asian subjects with C4B96 revealed that they all shared the same bimodular-LS haplotype encoding C4B1-B96, with two CYP21B (no CYP21A) and the presence of an XA+120 recombinant. The coding sequences of C4 genes in Subject-89 were amplified, sequenced to completion and compared with known C4 sequences (Supporting Information: Figs. S2-S4; Table S2) (39,43). We identified a novel, non-synonymous G→A nucleotide change at exon 21 from the short gene coding for C4B96, which attributed to the E920K polymorphism. The negatively-charged glutamic acid-920 was changed to the positively-charged lysine-920 in C4B96. This basic residue is located at the MG7 domain of the C4 protein structure (panel J, Fig. 2) (44). The DNA sequence for K920 ablated a restriction site for EcoRI (GAATTC→AAATTC; panel H, Fig. 2). Thus, 1.7 kb PCR fragments spanning from intron 20 to exon 26 of C4B were generated from six subjects with C4B96 plus one control without this allotype. The PCR products were digested with EcoRI resolved by electrophoresis. All subjects with B96 displayed the 1.7 kb fragment in addition to the 1.5 kb from C4B1 gene (lanes 1 and 3-7, panel I). C4 protein allotyping and/or EcoRI RFLP of PCR-amplified DNA revealed that 75% of EA-SLE with homozygous C4A-deficiency contained C4B96. HLA-DRB1 genotyping revealed that all subjects with C4B96 had DRB1*1501 (DR2).
DISCUSSIONS
The phenomenon of common CNVs is gaining appreciation but their impacts on rheumatic diseases among different racial groups await accurate and in-depth investigations. Here we report the continuous variation in GCN of human complement C4 and polymorphisms for C4A, C4B, long genes and short genes in association with SLE disease susceptibility and clinical manifestations, and compare them to those of European Americans.
In East-Asians, homozygous deficiency of C4A was present only in ~1% of SLE patients but it had a very large effect-size on disease susceptibility (OR=12.4). Heterozygous C4A-deficiency and low GCNs for total C4 were more prevalent but their effects on SLE disease risk were modest (OR=1.28 and 1.45, respectively). On the other end, the protective effects for high copy-numbers C4A or total C4 against SLE were conspicuous and highly significant. Over ¼ of healthy controls had high copy-numbers of total C4 or C4A, and their frequencies were reduced in SLE (p=8.8×10−8 and 4.0×10−9). Overall, there were reductions in SLE on the mean copy-numbers of total C4 by 0.19, C4A by 0.15 and short genes by 0.12.
Compared with subjects of European ancestry, East-Asians have significantly higher GCNs of total C4 and its associated variants (Table 4, Supporting Information: Fig. S5). In European-SLE, homozygous and heterozygous deficiencies of C4A, low GCNs of total C4 and long genes were common and medium-to-high effect-size risk factors of SLE (25). Notably, 6.5% of had a homozygous C4A deficiency (OR=8.57), 27.6% had a heterozygous C4A deficiency (OR=1.97), 42.2% had low GCNs of total C4 (OR=1.77). The variability of total C4 or C4A in Europeans was driven by changes in copy-number of long genes instead of short genes as in East-Asians. Remarkably, 13.9% of European SLE patients had zero or one copy of long genes, which were infrequent in Asians (p=2.9×10−19, Supporting Information: Fig. S5, panel H).
Table 4.
CNVs of complement C4 as a risk or protective factor in human SLE: a comparison between East-Asian and European-American.¶
| East-Asian | European-American* | |
|---|---|---|
| Total C4 | ||
| SLE - GCI: | 3.95 ± 0.87 | 3.56 ± 0.78 |
| Controls - GCI: | 4.14 ± 0.92 | 3.83 ± 0.69 |
| difference | −0.19 | −0.27 |
| p | 3.7×10–7 | 3.6×10–6 |
| C4T=2+3, OR: | 1.45 (1.20-1.77) | 1.77 (1.28-2.45) |
| C4T=5-8, OR: | 0.55 (0.45-0.68) | 0.53 (0.30-0.94) |
| C4A | ||
| SLE - GCI: | 2.09 ± 0.79 | 1.80 ± 0.90 |
| Controls - GCI: | 2.25 ± 0.82 | 2.09 ± 0.75 |
| difference | −0.16 | −0.30 |
| p | 3.4×10–6 | 3.4×10–6 |
| C4A=0, OR: | 12.4 (1.57-97.9) | 8.57 (2.81-26.1) |
| C4A=1, OR: | 1.28 (1.00-1.65) | 1.97 (1.36-2.85) |
| C4A≥3, OR: | 0.58 (0.47-0.71) | 0.55 (0.37-0.83) |
| C4B | ||
| SLE - GCI: | 1.85 ± 0.68 | 1.78 ± 0.59 |
| Controls - GCI: | 1.88 ± 0.71 | 1.73 ± 0.63 |
| difference | −0.03 | −0.04 |
| p | ns | ns |
| Long C4 | ||
| SLE - GCI: | 2.88 ± 0.79 | 2.63 ± 1.16 |
| Controls - GCI: | 2.94 ± 0.82 | 2.95 ± 0.98 |
| difference | −0.06 | −0.31 |
| p | ns | 0.0002 |
| C4L≤2, OR: | na | 1.66 (1.26-2.18) |
| Short C4 | ||
| SLE - GCI: | 1.05 ± 0.97 | 0.98 ± 0.85 |
| Controls - GCI: | 1.17 ± 0.94 | 0.89 ± 0.79 |
| difference | −0.12 | 0.09 |
| p | 0.005 | ns |
| C4S=0, OR: | 1.48 (1.22-1.80) | na |
See also Supporting Information Figure S5.
European-American data derived from White Ohio healthy subjects (N=500) and 232 White Ohio SLE patients (25).
na, not applicable; ns, not significant. GCI: gene copy-index - the mean of gene copy-number (± standard deviation).
The main cause for C4A-deficiency among Europeans is the presence of mono-S coding for C4B1 and the absence of a long C4A gene in haplotypes with HLA-DRB1*0301. This mono-S haplotype has a frequency of 0.113 in healthy subjects and 0.169 in SLE (25). Strikingly, in our study cohort >2000 Asian subjects, none had a homozygous mono-S (which is a homozygous deficiency of long genes). A new and predominant mechanism for C4A-deficiency in Asians unraveled in this study is a bimodular-LS haplotype coding for C4B1-B96 with markers characteristic of an ancient recombination and is linked to DRB1*1501 (or DR2).
Among SLE patients who ever experienced very-low serum levels of C4 and C3, low GCNs of total C4, C4B and long genes were major risk factors. Low GCN of total C4 would result in lower rate of biosynthesis and therefore a lower reservoir of C4 protein. During an active disease, high reactivity and high turnover of activated C4B protein would lead to fast depletion and therefore very low levels complement. Very-low serum complement levels were also strongly correlated with the presence of dsDNA autoantibodies, IgG-anticadiolipin, hemolytic anemia, renal disease and younger age of disease onset. The association between low C3/C4 and the presence of anti-dsDNA was remarkably strong, with OR=4.96 and p=9.7×10−17. Renal disease occurred in greater than half of East-Asian SLE patients. Among SLE patients with low GCN of total C4 (GCN=2 or 3), low C3/C4 protein levels had greater effects on the occurrence of renal disease [OR=3.95 (1.37-11.4), p=0.0067] than those with medium and high GCNs of total C4 [OR=1.45 (0.98-2.16), p=0.064]. Associations of low complement with renal disease and/or hematologic disease of SLE were also observed earlier in US patients (9,45). Other serologic factors that correlate with renal disease of SLE are anti-dsDNA and anti-C1q. In a recent multicenter study, it was found that simultaneous positivity of anti-C1q, anti-dsDNA and low complement associated with renal SLE with a combined odds ratio of 14.9 (5.8-38.4) (46). Measuring serum levels of C3 and C4 and anti-C1q, cell-bound levels of processed complement (e.g., erythrocyte-C4d) (47), and CNV genotyping of C4A and C4B are desirable laboratory tests to facilitate precision management of SLE and minimize renal disease.
The co-existence of C4A-deficiency with HLA risk alleles DRB1*0301 in Europeans and DRB1*1501 in East-Asians in SLE and other autoimmune diseases continue to be a fascinating topic on whether C4-CNVs or C4A-deficiency and DRB1 genetic variants are independent, additive or confounding risk factors (4,48-50). The pathologic effects for genetic and/or acquired deficiencies of C4, C1q, C1r and C1s on SLE have been overwhelming (4). Thus, it would be appropriate to investigate the feasibility of complement-guided therapeutics for SLE.
Supplementary Material
ACKNOWLEDGMENTS
We are indebted to volunteer blood donors and SLE patients for participation in this study. We are grateful to Jeannie Shaw (Columbus OH) for helps in recruiting healthy subjects and the Hong Kong Red Cross for sharing expired blood samples. This work was supported in part by the National Institute of Arthritis and Musculoskeletal and Skin Disease of the National Institutes of Health grant 1R01 AR054459 (CYY), Institutional grant from the Research Institute of the Nationwide Children's Hospital, Columbus Ohio (CYY), Ministry of Science and Technology grant 101-2314-B-182A-062-MY3 (JYC), and Chang Gung Memorial Hospital Grant CMRPG3E0531 (JYC).
Footnotes
AUTHOR CONTRIBUTIONS
JYC, YLW, EKC, YY, CSL and CYY designed project. JYC, CSL, YLL, CHS, YJJW, YY and MYM performed diagnosis and clinical studies. JYC, YLW, MYM, YY, YJJW, CMW, EKC, KEL, BZ, KJ, CHS, YLL, CSL and CYY recruited study subjects. YLW, YJJW, KEL, CMW, EKC, YY, BZ, HW, DY, AA, HNN, JYC and CYY performed experiments and data analyses. CYY, JYC, YLW, EKC and YY drafted the manuscript. All authors read and approved the manuscript.
REFERENCES
- 1.Schur PH, Hahn BH. Pisetsky DS, editor. Epidemiology and pathogenesis of systemic lupus erythematosus. 2015 www.uptodate.com/contents/epidemiology-and-pathogenesis-of-systemic-lupus-erythematosus.
- 2.Tsokos GC. Systemic lupus erythematosus. N Engl J Med. 2011;365:2110–21. doi: 10.1056/NEJMra1100359. [DOI] [PubMed] [Google Scholar]
- 3.Tsao BP, Deng Y. Constitutive genes and lupus. In: Lahita RG, Tsokos G, Buyon J, Koike T, editors. Systemic Lupus Erythematosus. Fifth ed. Academic Press at Elsevier; Amsterdam: 2011. pp. 47–61. [Google Scholar]
- 4.Atkinson JP, Yu CY. The complement system in systemic lupus erythematosus. In: Tsokos GC, editor. Systemic Lupus Erythematosus. 1st ed. Elsevier/Academic Publisher; 2015. in press. [Google Scholar]
- 5.Sturfelt G, Truedsson L. Complement in the immunopathogenesis of rheumatic disease. Nat Rev Rheumatol. 2012;8:458–68. doi: 10.1038/nrrheum.2012.75. [DOI] [PubMed] [Google Scholar]
- 6.Elliot JA, Mathieson DR. Complement in disseminated (systemic) lupus erythematosus. A M A Arch Dermatol Syphilol. 1953;68:119–28. doi: 10.1001/archderm.1953.01540080003001. [DOI] [PubMed] [Google Scholar]
- 7.Lange K, Wasserman E, Slobody LB. The significance of serum complement levels for the diagnosis and prognosis of acute and subacute glomerulonephritis and lupus erythematosus disseminatus. Ann Intern Med. 1960;53:636–46. doi: 10.7326/0003-4819-53-4-636. [DOI] [PubMed] [Google Scholar]
- 8.Petri M, Orbai AM, Alarcon GS, Gordon C, Merrill JT, Fortin PR, et al. Derivation and validation of the Systemic Lupus International Collaborating Clinics classification criteria for systemic lupus erythematosus. Arthritis Rheum. 2012;64:2677–86. doi: 10.1002/art.34473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Birmingham DJ, Irshaid F, Nagaraja HN, Zou X, Tsao BP, Wu H, et al. The complex nature of serum C3 and C4 as biomarkers of lupus renal flare. Lupus. 2010;19:1272–80. doi: 10.1177/0961203310371154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pickering MC, Botto M, Taylor PR, Lachmann PJ, Walport MJ. Systemic lupus erythematosus, complement deficiency and apoptosis. Adv Immunol. 2001;76:227–324. doi: 10.1016/s0065-2776(01)76021-x. [DOI] [PubMed] [Google Scholar]
- 11.Lipsker D, Hauptmann G. Cutaneous manifestations of complement deficiencies. Lupus. 2010;19:1096–106. doi: 10.1177/0961203310373370. [DOI] [PubMed] [Google Scholar]
- 12.Yang Y, Lhotta K, Chung EK, Eder P, Neumair F, Yu CY. Complete complement components C4A and C4B deficiencies in human kidney diseases and systemic lupus erythematosus. J Immunol. 2004;173:2803–14. doi: 10.4049/jimmunol.173.4.2803. [DOI] [PubMed] [Google Scholar]
- 13.Wu YL, Hauptmann G, Viguier M, Yu CY. Molecular basis of complete complement C4 deficiency in two North-African families with systemic lupus erythematosus. Genes Immun. 2009;10:433–45. doi: 10.1038/gene.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu YL, Brookshire BP, Verani RR, Arnett FC, Yu CY. Clinical presentations and molecular basis of complement C1r deficiency in a male African-American patient with systemic lupus erythematosus. Lupus. 2011;20:1126–34. doi: 10.1177/0961203311404914. [DOI] [PubMed] [Google Scholar]
- 15.Chatterjee P, Agyemang AF, Alimzhanov MB, Degn S, Tsiftsoglou SA, Alicot E, et al. Complement C4 maintains peripheral B-cell tolerance in a myeloid cell dependent manner. Eur J Immunol. 2013;43:2441–50. doi: 10.1002/eji.201343412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paul E, Pozdnyakova OO, Mitchell E, Carroll MC. Anti-DNA autoreactivity in C4-deficient mice. Eur J Immunol. 2002;32:2672–9. doi: 10.1002/1521-4141(200209)32:9<2672::AID-IMMU2672>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 17.Prodeus AP, Goerg S, Shen LM, Pozdnyakova OO, Chu L, Alicot EM, et al. A critical role for complement in maintenance of self-tolerance. Immunity. 1998;9:721–31. doi: 10.1016/s1074-7613(00)80669-x. [DOI] [PubMed] [Google Scholar]
- 18.Botto M, Dell'Agnola C, Bygrave AE, Thompson EM, Cook HT, Petry F, et al. Homozygous C1q deficiency causes glomerulonephritis associated with multiple apoptotic bodies. Nat Genet. 1998;19:56–9. doi: 10.1038/ng0598-56. [DOI] [PubMed] [Google Scholar]
- 19.Navratil JS, Watkins SC, Wisnieski JJ, Ahearn JM. The Globular Heads of C1q Specifically Recognize Surface Blebs of Apoptotic Vascular Endothelial Cells. The Journal of Immunology. 2001;166:3231–9. doi: 10.4049/jimmunol.166.5.3231. [DOI] [PubMed] [Google Scholar]
- 20.Lood C, Gullstrand B, Truedsson L, Olin AI, Alm GV, Ronnblom L, et al. C1q inhibits immune complex-induced interferon-alpha production in plasmacytoid dendritic cells: a novel link between C1q deficiency and systemic lupus erythematosus pathogenesis. Arthritis Rheum. 2009;60:3081–90. doi: 10.1002/art.24852. [DOI] [PubMed] [Google Scholar]
- 21.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–53. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mok CC. Epidemiology and survival of systemic lupus erythematosus in Hong Kong Chinese. Lupus. 2011;20:767–71. doi: 10.1177/0961203310388447. [DOI] [PubMed] [Google Scholar]
- 23.Lau CS, Yin G, Mok MY. Ethnic and geographical differences in systemic lupus erythematosus: an overview. Lupus. 2006;15:715–9. doi: 10.1177/0961203306072311. [DOI] [PubMed] [Google Scholar]
- 24.Wang C, Ahlford A, Jarvinen TM, Nordmark G, Eloranta ML, Gunnarsson I, et al. Genes identified in Asian SLE GWASs are also associated with SLE in Caucasian populations. Eur J Hum Genet. 2013;21:994–9. doi: 10.1038/ejhg.2012.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Y, Chung EK, Wu YL, Savelli SL, Nagaraja HN, Zhou B, et al. Gene copy number variation and associated polymorphisms of complement component C4 in human systemic erythematosus (SLE): low copy number is a risk factor for and high copy number is a protective factor against European American SLE disease susceptibility. Am J Hum Genet. 2007;80:1037–54. doi: 10.1086/518257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horton R, Gibson R, Coggill P, Miretti M, Allcock RJ, Almeida J, et al. Variation analysis and gene annotation of eight MHC haplotypes: the MHC Haplotype Project. Immunogenetics. 2008;60:1–18. doi: 10.1007/s00251-007-0262-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu CY, Whitacre CC. Sex, MHC and complement C4 in autoimmune diseases. Trends Immunol. 2004;25:694–9. doi: 10.1016/j.it.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 28.Lv Y, He S, Zhang Z, Li Y, Hu D, Zhu K, et al. Confirmation of C4 gene copy number variation and the association with systemic lupus erythematosus in Chinese Han population. Rheumatol Int. 2012;32:3047–53. doi: 10.1007/s00296-011-2023-7. [DOI] [PubMed] [Google Scholar]
- 29.Kim JH, Jung SH, Bae JS, Lee HS, Yim SH, Park SY, et al. Deletion variants of RABGAP1L, 10q21.3, and C4 are associated with the risk of systemic lupus erythematosus in Korean women. Arthritis Rheum. 2013;65:1055–63. doi: 10.1002/art.37854. [DOI] [PubMed] [Google Scholar]
- 30.Dunckley H, Gatenby PA, Hawkins B, Naito S, Serjeantson SW. Deficiency of C4A is a genetic determinant of systemic lupus erythematosus in three ethnic groups. J Immunogenet. 1987;14:209–18. doi: 10.1111/j.1744-313x.1987.tb00383.x. [DOI] [PubMed] [Google Scholar]
- 31.Tan EM, Cohen AS, Fries JF, Masi AT, McShane DJ, Rothfield NF, et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1982;25:1271–7. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 32.Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40:1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- 33.Chung EK, Wu YL, Yang Y, Zhou B, Yu CY. Human complement components C4A and C4B genetic diversities: complex genotypes and phenotypes. In: Coligan JE, Bierer BE, Margulis DH, Shevach EM, Strober W, editors. Current Protocols in Immunology. John Wiley & Sons, Inc.; Edison, NJ: 2005. pp. 13.8.1–13.8.36. [DOI] [PubMed] [Google Scholar]
- 34.Chung EK, Yang Y, Rupert KL, Jones KN, Rennebohm RM, Blanchong CA, et al. Determining the one, two, three or four long and short loci of human complement C4 in a major histocompatibility complex haplotype encoding for C4A or C4B proteins. Am J Hum Genet. 2002;71:810–22. doi: 10.1086/342778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu YL, Savelli SL, Yang Y, Zhou B, Rovin BH, Birmingham DJ, et al. Sensitive and specific real-time PCR Assays to accurately determine copy-number variations (CNVs) of human complement C4A, C4B, C4-Long, C4-Short and RCCX modules: Elucidation of C4 CNVs in 50 consanguineous subjects with defined HLA genotypes. J Immunol. 2007;179:3012–25. doi: 10.4049/jimmunol.179.5.3012. [DOI] [PubMed] [Google Scholar]
- 36.Mauff G, Brenden M, Braun-Stilwell M, Doxiadis G, Giles CM, Hauptmann G, et al. C4 reference typing report. Complement Inflamm. 1990;7:193–212. doi: 10.1159/000463148. [DOI] [PubMed] [Google Scholar]
- 37.Sim E, Cross S. Phenotyping of human complement component C4, a class III HLA antigen. Biochem J. 1986;239:763–7. doi: 10.1042/bj2390763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu CY, Campbell RD, Porter RR. A structural model for the location of the Rodgers and the Chido antigenic determinants and their correlation with the human complement C4A/C4B isotypes. Immunogenetics. 1988;27:399–405. doi: 10.1007/BF00364425. [DOI] [PubMed] [Google Scholar]
- 39.Yu CY, Belt KT, Giles CM, Campbell RD, Porter RR. Structural basis of the polymorphism of human complement component C4A and C4B: gene size, reactivity and antigenicity. EMBO J. 1986;5:2873–81. doi: 10.1002/j.1460-2075.1986.tb04582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rupert KL, Rennebohm RM, Yu CY. An unequal crossover between the RCCX modules of the human MHC leading to the presence of a CYP21B gene and a tenascin TNXB/TNXA-RP2 recombinant between C4A and C4B genes in a patient with juvenile rheumatoid arthritis. Exp Clin Immunogenet. 1999;16:81–97. doi: 10.1159/000019099. [DOI] [PubMed] [Google Scholar]
- 41.Yang Z, Mendoza AR, Welch TR, Zipf WB, Yu CY. Modular variations of HLA class III genes for serine/threonine kinase RP, complement C4, steroid 21-hydroxylase CYP21 and tenascin TNX (RCCX): a mechanism for gene deletions and disease associations. J Biol Chem. 1999;274:12147–56. doi: 10.1074/jbc.274.17.12147. [DOI] [PubMed] [Google Scholar]
- 42.Saxena K, Kitzmiller KJ, Wu YL, Zhou B, Esack N, Hiremath L, et al. Great genotypic and phenotypic diversities associated with copy-number variations of complement C4 and RP-C4-CYP21-TNX (RCCX) modules: A comparison of Asian-Indian and European American populations. Mol Immunol. 2009;46:1289–303. doi: 10.1016/j.molimm.2008.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu CY. The complete exon-intron structure of a human complement component C4A gene: DNA sequences, polymorphism, and linkage to the 21-hydroxylase gene. J Immunol. 1991;146:1057–66. [PubMed] [Google Scholar]
- 44.Kidmose RT, Laursen NS, Dobo J, Kjaer TR, Sirotkina S, Yatime L, et al. Structural basis for activation of the complement system by component C4 cleavage. Proc Natl Acad Sci U S A. 2012;109:15425–30. doi: 10.1073/pnas.1208031109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ho A, Barr SG, Magder LS, Petri M. A decrease in complement is associated with increased renal and hemologic activity in patients with systemic lupus erythematosus. Arthritis Rheum. 2001;44:2350–7. doi: 10.1002/1529-0131(200110)44:10<2350::aid-art398>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 46.Orbai AM, Truedsson L, Sturfelt G, Nived O, Fang H, Alarcon GS, et al. Anti-C1q antibodies in systemic lupus erythematosus. Lupus. 2015;24:42–9. doi: 10.1177/0961203314547791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manzi S, Navratil JS, Ruffing MJ, Liu CC, Danchenko N, Nilson SE, et al. Measurement of erythrocyte C4d and complement receptor 1 in systemic lupus erythematosus. Arthritis Rheum. 2004;50:3596–604. doi: 10.1002/art.20561. [DOI] [PubMed] [Google Scholar]
- 48.Fernando MM, Stevens CR, Sabeti PC, Walsh EC, McWhinnie AJ, Shah A, et al. Identification of two independent risk factors for lupus within the MHC in United Kingdom families. PLoS Genet. 2007;3:e192. doi: 10.1371/journal.pgen.0030192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boteva L, Morris DL, Cortes-Hernandez J, Martin J, Vyse TJ, Fernando MM. Genetically determined partial complement C4 deficiency states are not independent risk factors for SLE in UK and Spanish populations. Am J Hum Genet. 2012;90:445–56. doi: 10.1016/j.ajhg.2012.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fielder AHL, Walport MJ, Batchelor JR, Rynes RI, Black CM, Dodi IA, et al. Family study of the major histocompatibility complex in patients with systemic lupus erythematosus: importance of null alleles of C4A and C4B in determining disease susceptibility. Br Med J. 1983;286:425–8. doi: 10.1136/bmj.286.6363.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.