Abstract
The insulinoma-associated 1 (insm1) gene is involved in the differentiation of several neuronal and endoderm derived cell types. insm1 is expressed in the retina and brain of several vertebrates including Xenopus laevis. We report the detailed expression pattern of insm1 in the X. laevis tadpole retina and brain. X. laevis insm1 is expressed in most of the ciliary marginal zone of the mature retina and the optic tectum, dorsal pallium, hypothalamus and preoptic areas of the developing tadpole brain. Overall, insm1 is expressed in regions of the tadpole brain and retina harboring populations of progenitor cells.
Keywords: Xenopus laevis, insm1, ciliary marginal zone, optic tectum, dorsal pallium, hypothalamus
INTRODUCTION
The insulinoma-associated 1(insm1 or IA-1) gene is a conserved gene encoding a DNA-binding zinc finger transcription factor. insm1 has been studied in several model organisms. Insm1 proteins include seven N-terminal SNAG transcriptional repressor motifs, five C-terminal Cys2-His2 zinc fingers and a putative nuclear localization signal (Parlier et al. 2008). In the mouse nervous system, insm1 is involved in the differentiation or transition of progenitors into neurons or neurogenic progenitors. In mouse, it is expressed in all proliferating neural cells, including retina, spinal cord, and fore- mid-, and hindbrain and thought to be expressed late in progenitor cell development, during the final cell division as late progenitors give rise to neurons (Duggan et al. 2008). Regulation of mouse insm1 by ascl1 is important in the differentiation of central serotonergic and noradrenergic monoaminergic neurons (Jacob et al. 2009). In insm1 null mice serotonergic precursors begin to differentiate but fail to produce serotonin due lack of tryptophan hydroxylase 2, which is coordinately regulated by insm1 and asc1. The olfactory epithelium of embryonic insm1 null mice exhibits a decrease in basal progenitors and an increase in apical progenitors as well as fewer terminally dividing progenitors, suggesting that insm1 is involved in the transition of progenitors to a basal position, which favors neurogenic and terminal division (Rosenbaum et al. 2011).
In the zebrafish retina, insm1 regulates cell cycle kinetics of the rod progenitor cells, is needed for the differentiation of rod and cone photoreceptors, and, as in mice, is negatively regulated by Notch-Delta signaling (Forbes-Osborne et al. 2013). Zebrafish insm1 in the retina is genetically upstream of neuroD as well as ath5/atoh7 and photoreceptor specification genes, crx and nr2e3. insm1 was also studied in zebrafish retinal regeneration where, in the event of retinal damage, insm1a is necessary for müller glia dedifferentiation by suppressing ascl1a and itself (Ramachandran et al. 2012). Furthermore, Insm1 defines the zone of Müller glia activity via suppression of hb-efga expression and stimulation of Müller glia progenitor cell cycle exit. In the mouse retina, insm1 expression was upregulated upon inhibition of notch signaling by application of the inhibitor DAPT (Nelson et al. 2007). An Insm1:LacZ transgene was expressed almost exclusively in non-proliferating cells in the retina. These cells were primarily found at the ventricular surface, although some were found in the ganglion cell layer. The authors concluded that cells expressing the transgene were primarily nascent photoreceptors.
insm1 is normally expressed during development and is regulated by a heterodimer of neuroD and E47, which binds to an E-box found in the promoter (Breslin et al. 2003). However, insm1 expression can be reactivated in cancer. insm1 is strongly expressed in tumors of neuroendocrine origin (Goto et al. 1992; Lan et al. 1993; Wang et al. 2009). This, combined with the absence of insm1 in adult tissues, has lead to the evaluation of the insm1 promoter in gene therapy against small-cell lung cancer, pediatric medulloblastoma, neuroblastoma, and retinoblastoma tumors (Pedersen et al. 2006; Wang et al. 2009; Christensen et al. 2010; Akerstrom et al. 2013).
Xenopus insm1 has been mainly studied in endoderm-derived cells. It is expressed in all endocrine cells and acts downstream of ngn3 and upstream of pax6 and neuroD in the transcriptional cascade for differentiation, suggesting that insm1 gene hierarchy found in the zebrafish retina is conserved and utilized in other species and organ systems (Gierl et al. 2006; Mellitzer et al. 2006; Pearl et al. 2009). insm1 has also been studied in the Xenopus nervous system where it functions downstream of Xash1 (ascl1) in the formation of the noradrenergic primary neurons in the developing heart field near the cement gland (Parlier et al. 2008). Although insm1 has been studied in the Xenopus nervous system, there are few reports on insm1 expression in the Xenopus brain and no reports on insm1 expression in the mature Xenopus retina.
We have a long-standing interest in the development of progenitor cells in the mature and regenerating X. laevis retina (Bailey et al. 2004; Kelly et al. 2007; Martinez-De Luna et al. 2011) and in gene expression in the developing brain (El-Hodiri et al. 2003; Kelly et al. 2007). Therefore, we sought to characterize insm1 expression in retinal progenitor cells. Here, we report that X. laevis insm1 is expressed in a subset of the progenitor cell population in the tadpole retina and brain.
RESULTS AND DISCUSSION
Expression in retinal progenitor cells
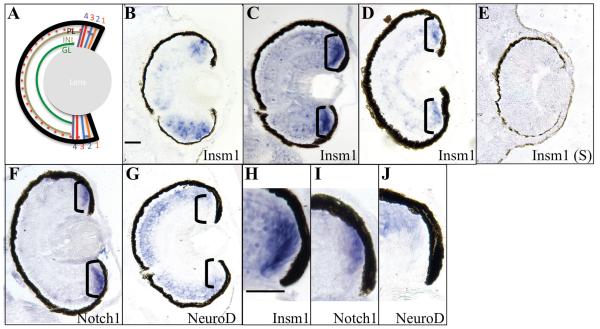
Xenopus laevis insm1 is expressed in the developing eye (Parlier et al. 2008). In order to more precisely characterize insm1 expression in the retina, we performed in situ hybridization using retinal sections from X. laevis embyros at stages 38 to 45 (Figure 1). insm1 is expressed in the ciliary marginal zone (CMZ) of the tadpole retina, a region of the peripheral retina containing retinal progenitor and stem cells (RPCs and RSCs, respectively). The CMZ can be divided into 4 zones, based on state of retinal neural development (Perron et al. 1998). The zones contain RPCs of increasing development in a distal – proximal arrangement (Figure 1A). At stage 38, insm1 is expressed in the retinal progenitor cells (RPCs) but absent from the retinal stem cells (RSCs) at the distal edge (zone 1) of the CMZ. insm1 expression is also seen in the undifferentiated neuroepithelium adjacent to the ventral CMZ (Figure 1A). Similar to stage 38, retinal insm1 expression in stage 41 and 45 embryos is restricted to the CMZ (Figure 1B-D). Further examination of stage 41 embryos demonstrates that insm1 expression begins distally in zone 2 of the CMZ similar to zone 2 marker, notch1 (compare Figure 1 C and H to F and I). insm1 expression expands proximally to zones 3 and 4 of the CMZ comparable to neuroD expression(compare Figure 1 C and H to G and J), which marks those zones (Perron et al., 1998). This pattern of expression in the CMZ is similar to that of zebrafish insm1a. insm1a expression is also observed outside the CMZ in the regenerating zebrafish retina (Morris et al. 2011), in the ONL and INL, in rod progenitors and glia-derived progenitors, respectively. The X. laevis expression pattern is markedly different from that of the mouse retina (Nelson et al. 2007) in that expression is not observed in the frog ONL. Retinal regeneration in Xenopus involves the recruitment of progenitor cells, primarily from dedifferentiated retinal pigmented epithelium (RPE) (Yoshii et al. 2007). It will be interesting to discover if insm1 is expressed in RPE-derived progenitor cells during X. laevis regeneration as it is in glia-derived progenitors of the regenerating zebrafish retina.
Figure 1. Insm1 is expressed in retinal progenitor cells (RPCs).
A. Schematic representation of the mature Xenopus tadpole retina. B – J. In situ hybridization performed using sectioned Xenopus laevis embryos. insm1 is expressed in retinal progenitor cells (RPCs) and undifferentiated neuroepithelium at stage 38. At stages 41 (C, H) and stage 45 (D), insm1 expression was detected in RPCs located in zones 2 and 3 of the ciliary marginal zone (CMZ) but not in the retinal stem cells of zone 1. Compare the expression of insm1 at stage 41 (C, H) to expression of notch1, a zone 2 marker, at stage 41 (F, I). Both are absent in the most distal zone 1 of the CMZ. Also compare the proximal expression of insm1 (C, H) to zone 3 and 4 marker, neuroD at stage 41 (G, J). The CMZ is indicated by black brackets. Scale bars (1 μ M) for (B-G) and (H-J) are indicated in (A) and (F), respectively. Abbreviations: GL – ganglion cell layer, INL – inner nuclear layer, PL – photoreceptor layer.
Expression in the developing tadpole brain
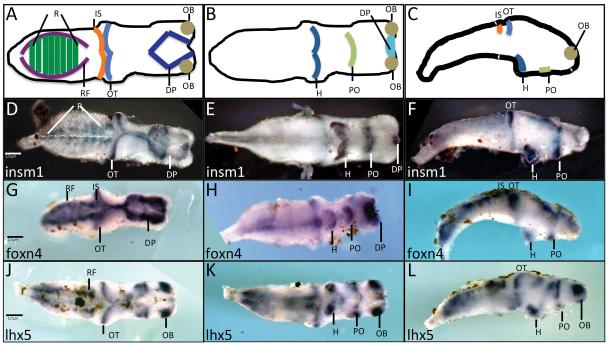
insm1 is also expressed in the developing tadpole brain (Parlier et al. 2008). To further characterize insm1 expression in Xenopus brain, we performed whole mount in situ hybridization on brains isolated from stage 45 tadpoles. insm1 is expressed in the optic tectum, dorsal pallium, rhombomeres, hypothalamus and preoptic area (Figure 2 D-F). This pattern is similar to that of foxn4 and lhx5, which are also expressed in similar progenitor cell-containing regions of the brain (Figure 2 G – L) (Moreno et al. 2004; Moreno et al. 2005; Kelly et al. 2007). These genes differ in expression pattern in the forebrain where insm1 and foxn4 are expressed in the dorsal pallium while lhx5 is expressed in the olfactory bulbs (compare Figure 2 D, G with J). The expression patterns of these genes also differ in the hindbrain: insm1 is expressed in the rhombomeres while foxn4 and lhx5 are expressed in the reticular formation (compare Figure 2 D with G, J). This is in general agreement with the expression of insm1 in late progenitor populations in the mouse brain (Duggan et al. 2008). It would be interesting to discover whether insm1 has a role in regulating proliferation in the brain, as has been demonstrated for the zebrafish retina (Ramachandran et al. 2012).
Figure 2. insm1 expression in the developing brain.
A – C. Diagrammatic representation of selected brain regions as viewed in dorsally (A), ventrally (B) or laterally (C). D – L. Whole mount in situ hybridizations of stage 45 dissected brains using probes for insm1 (D – F), foxn4 (G – I), or lhx5 (J – L). insm1 is expressed in the dorsal pallium, optic tectum and rhombomeres as viewed dorsally (D). insm1 expression was also detected ventrally (E) and laterally (F) in the hypothalamus, preoptic area and isthmus. Scale bars (1 μ M) are indicated in (D) for (D-F), (G) for (G-I) and (J) for (J-L). Abbreviations: DP – dorsal pallium, H – hypothalamus, IS – isthmus, OB – olfactory bulb, OT – optic tectum, PO – preoptic area, RF – reticular formation, R – rhombomeres.
MATERIALS AND METHODS
In situ hybridizations were performed using digoxigenin-labeled antisense riboprobes generated by in vitro transcription of linearized DNA templates. Probes for insm1 (Parlier et al. 2008), foxn4 (Kelly et al. 2007), lhx5 (Bachy et al. 2001), neurod (Lee et al. 1995), and notch1 (Coffman et al. 1990) were prepared as described. Embryos for in situ hybridization were generated and staged according to standard criteria (Nieuwkoop 1994; Sive 2000). Wholemount in situ hybridizations were performed using Xenopus laevis embryos were performed as described previously (Sive 2000). Section in situ hybridizations were performed using 8 μM sections of fixed, paraffin-embedded tadpoles (Viczian et al. 2003). Whole mount brain in situ hybridizations were performed on brains isolated from fixed stage 45 embryos (Colombo et al. 2004).
ACKNOWLEDGEMENTS
We thank Eric J. Bellefroid for the Xenopus insm1 probe. We also thank Lisa E. Kelly for technical assistance and Jessica L. Buescher for critical reading of this manuscript. This work was funded, in part, by NIH grant EY015480 to HME.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES CITED
- Akerstrom V, Chen C, Lan MS, Breslin MB. Adenoviral insulinoma-associated protein 1 promoter-driven suicide gene therapy with enhanced selectivity for treatment of neuroendocrine cancers. Ochsner J. 2013;13(1):91–99. [PMC free article] [PubMed] [Google Scholar]
- Bachy I, Vernier P, Retaux S. The LIM-homeodomain gene family in the developing Xenopus brain: conservation and divergences with the mouse related to the evolution of the forebrain. J Neurosci. 2001;21(19):7620–7629. doi: 10.1523/JNEUROSCI.21-19-07620.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TJ, El-Hodiri H, Zhang L, Shah R, Mathers PH, Jamrich M. Regulation of vertebrate eye development by Rx genes. Int J Dev Biol. 2004;48(8-9):761–770. doi: 10.1387/ijdb.041878tb. [DOI] [PubMed] [Google Scholar]
- Breslin MB, Zhu M, Lan MS. NeuroD1/E47 regulates the E-box element of a novel zinc finger transcription factor, IA-1, in developing nervous system. J Biol Chem. 2003;278(40):38991–38997. doi: 10.1074/jbc.M306795200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen CL, Gjetting T, Poulsen TT, Cramer F, Roth JA, Poulsen HS. Targeted cytosine deaminase-uracil phosphoribosyl transferase suicide gene therapy induces small cell lung cancer-specific cytotoxicity and tumor growth delay. Clin Cancer Res. 2010;16(8):2308–2319. doi: 10.1158/1078-0432.CCR-09-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman C, Harris W, Kintner C. Xotch, the Xenopus homolog of Drosophila notch. Science. 1990;249(4975):1438–1441. doi: 10.1126/science.2402639. [DOI] [PubMed] [Google Scholar]
- Colombo E, Galli R, Cossu G, Gecz J, Broccoli V. Mouse orthologue of ARX, a gene mutated in several X-linked forms of mental retardation and epilepsy, is a marker of adult neural stem cells and forebrain GABAergic neurons. Dev Dyn. 2004;231(3):631–639. doi: 10.1002/dvdy.20164. [DOI] [PubMed] [Google Scholar]
- Duggan A, Madathany T, de Castro SC, Gerrelli D, Guddati K, Garcia-Anoveros J. Transient expression of the conserved zinc finger gene INSM1 in progenitors and nascent neurons throughout embryonic and adult neurogenesis. J Comp Neurol. 2008;507(4):1497–1520. doi: 10.1002/cne.21629. [DOI] [PubMed] [Google Scholar]
- El-Hodiri HM, Qi XL, Seufert DW. The Xenopus arx gene is expressed in the developing rostral forebrain. Dev Genes Evol. 2003;212(12):608–612. doi: 10.1007/s00427-002-0282-8. [DOI] [PubMed] [Google Scholar]
- Forbes-Osborne MA, Wilson SG, Morris AC. Insulinoma-associated 1a (Insm1a) is required for photoreceptor differentiation in the zebrafish retina. Dev Biol. 2013;380(2):157–171. doi: 10.1016/j.ydbio.2013.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierl MS, Karoulias N, Wende H, Strehle M, Birchmeier C. The zinc-finger factor Insm1 (IA-1) is essential for the development of pancreatic beta cells and intestinal endocrine cells. Genes Dev. 2006;20(17):2465–2478. doi: 10.1101/gad.381806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto Y, De Silva MG, Toscani A, Prabhakar BS, Notkins AL, Lan MS. A novel human insulinoma-associated cDNA, IA-1, encodes a protein with "zinc-finger" DNA-binding motifs. J Biol Chem. 1992;267(21):15252–15257. [PubMed] [Google Scholar]
- Jacob J, Storm R, Castro DS, Milton C, Pla P, Guillemot F, Birchmeier C, Briscoe J. Insm1 (IA-1) is an essential component of the regulatory network that specifies monoaminergic neuronal phenotypes in the vertebrate hindbrain. Development. 2009;136(14):2477–2485. doi: 10.1242/dev.034546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly LE, Nekkalapudi S, El-Hodiri HM. Expression of the forkhead transcription factor FoxN4 in progenitor cells in the developing Xenopus laevis retina and brain. Gene Expr Patterns. 2007;7(3):233–238. doi: 10.1016/j.modgep.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan MS, Russell EK, Lu J, Johnson BE, Notkins AL. IA-1, a new marker for neuroendocrine differentiation in human lung cancer cell lines. Cancer Res. 1993;53(18):4169–4171. [PubMed] [Google Scholar]
- Lee JE, Hollenberg SM, Snider L, Turner DL, Lipnick N, Weintraub H. Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix-loop-helix protein. Science. 1995;268(5212):836–844. doi: 10.1126/science.7754368. [DOI] [PubMed] [Google Scholar]
- Martinez-De Luna RI, Kelly LE, El-Hodiri HM. The Retinal Homeobox (Rx) gene is necessary for retinal regeneration. Dev Biol. 2011;353(1):10–18. doi: 10.1016/j.ydbio.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellitzer G, Bonne S, Luco RF, Van De Casteele M, Lenne-Samuel N, Collombat P, Mansouri A, Lee J, Lan M, Pipeleers D, Nielsen FC, Ferrer J, Gradwohl G, Heimberg H. IA1 is NGN3-dependent and essential for differentiation of the endocrine pancreas. EMBO J. 2006;25(6):1344–1352. doi: 10.1038/sj.emboj.7601011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno N, Bachy I, Retaux S, Gonzalez A. LIM-homeodomain genes as developmental and adult genetic markers of Xenopus forebrain functional subdivisions. J Comp Neurol. 2004;472(1):52–72. doi: 10.1002/cne.20046. [DOI] [PubMed] [Google Scholar]
- Moreno N, Bachy I, Retaux S, Gonzalez A. LIM-homeodomain genes as territory markers in the brainstem of adult and developing Xenopus laevis. J Comp Neurol. 2005;485(3):240–254. doi: 10.1002/cne.20498. [DOI] [PubMed] [Google Scholar]
- Morris AC, Forbes-Osborne MA, Pillai LS, Fadool JM. Microarray analysis of XOPS-mCFP zebrafish retina identifies genes associated with rod photoreceptor degeneration and regeneration. Invest Ophthalmol Vis Sci. 2011;52(5):2255–2266. doi: 10.1167/iovs.10-6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BR, Hartman BH, Georgi SA, Lan MS, Reh TA. Transient inactivation of Notch signaling synchronizes differentiation of neural progenitor cells. Dev Biol. 2007;304(2):479–498. doi: 10.1016/j.ydbio.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal Table of Xenopus Laevis (Daudin): A Systematical & Chronological Survey of the Development from the Fertilized Egg till the End of Metamorphosis. Garland Science; 1994. [Google Scholar]
- Parlier D, Ariza A, Christulia F, Genco F, Vanhomwegen J, Kricha S, Souopgui J, Bellefroid EJ. Xenopus zinc finger transcription factor IA1 (Insm1) expression marks anteroventral noradrenergic neuron progenitors in Xenopus embryos. Dev Dyn. 2008;237(8):2147–2157. doi: 10.1002/dvdy.21621. [DOI] [PubMed] [Google Scholar]
- Pearl EJ, Bilogan CK, Mukhi S, Brown DD, Horb ME. Xenopus pancreas development. Dev Dyn. 2009;238(6):1271–1286. doi: 10.1002/dvdy.21935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen N, Pedersen MW, Lan MS, Breslin MB, Poulsen HS. The insulinoma-associated 1: a novel promoter for targeted cancer gene therapy for small-cell lung cancer. Cancer Gene Ther. 2006;13(4):375–384. doi: 10.1038/sj.cgt.7700887. [DOI] [PubMed] [Google Scholar]
- Perron M, Kanekar S, Vetter ML, Harris WA. The genetic sequence of retinal development in the ciliary margin of the Xenopus eye. Dev Biol. 1998;199(2):185–200. doi: 10.1006/dbio.1998.8939. [DOI] [PubMed] [Google Scholar]
- Ramachandran R, Zhao XF, Goldman D. Insm1a-mediated gene repression is essential for the formation and differentiation of Muller glia-derived progenitors in the injured retina. Nat Cell Biol. 2012;14(10):1013–1023. doi: 10.1038/ncb2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum JN, Duggan A, Garcia-Anoveros J. Insm1 promotes the transition of olfactory progenitors from apical and proliferative to basal, terminally dividing and neuronogenic. Neural Dev. 2011;6 doi: 10.1186/1749-8104-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sive HLG, Harland RM. Early Development of Xenopus laevis: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. R.M. [Google Scholar]
- Viczian AS, Vignali R, Zuber ME, Barsacchi G, Harris WA. XOtx5b and XOtx2 regulate photoreceptor and bipolar fates in the Xenopus retina. Development. 2003;130(7):1281–1294. doi: 10.1242/dev.00343. [DOI] [PubMed] [Google Scholar]
- Wang HW, Breslin MB, Chen C, Akerstrom V, Zhong Q, Lan MS. INSM1 promoter-driven adenoviral herpes simplex virus thymidine kinase cancer gene therapy for the treatment of primitive neuroectodermal tumors. Hum Gene Ther. 2009;20(11):1308–1318. doi: 10.1089/hum.2008.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii C, Ueda Y, Okamoto M, Araki M. Neural retinal regeneration in the anuran amphibian Xenopus laevis post-metamorphosis: transdifferentiation of retinal pigmented epithelium regenerates the neural retina. Dev Biol. 2007;303(1):45–56. doi: 10.1016/j.ydbio.2006.11.024. [DOI] [PubMed] [Google Scholar]