Abstract
The maintenance of normal body weight is disrupted in patients with anorexia nervosa (AN) for prolonged periods of time. Prior to the onset of AN, premorbid body mass index (BMI) spans the entire range from underweight to obese. After recovery, patients have reduced rates of overweight and obesity. As such, loci involved in body weight regulation may also be relevant for AN and vice versa. Our primary analysis comprised a cross-trait analysis of the 1000 single nucleotide polymorphisms (SNPs) with the lowest p-values in a genome-wide association meta-analysis (GWAMA) of AN (GCAN) for evidence of association in the largest published GWAMA for BMI (GIANT). Subsequently we performed sex-stratified analyses for these 1000 SNPs. Functional ex vivo studies on four genes ensued. Lastly, a look-up of GWAMA-derived BMI related loci was performed in the AN GWAMA. We detected significant associations (p-values < 5×10−5, Bonferroni corrected p < 0.05) for 9 SNP alleles at 3 independent loci. Interestingly, all AN susceptibility alleles were consistently associated with increased BMI. None of the genes (chr. 10: CTBP2, chr. 19: CCNE1, chr. 2: CARF and NBEAL1; the latter is a region with high linkage disequilibrium) nearest to these SNPs has previously been associated with AN or obesity. Sex-stratified analyses revealed that the strongest BMI signal originated predominantly from females (chr. 10 rs1561589; poverall: 2.47 × 10−06/pfemales: 3.45 × 10−07/pmales: 0.043). Functional ex vivo studies in mice revealed reduced hypothalamic expression of Ctbp2 and Nbeal1 after fasting. Hypothalamic expression of Ctbp2 was increased in diet induced obese (DIO) mice as compared to age-matched lean controls. We observed no evidence for associations for the look-up of BMI related loci in the AN GWAMA. A cross-trait analysis of AN and BMI loci revealed variants at three chromosomal loci with potential joint impact. The chromosome 10 locus is particularly promising given that the association with obesity was primarily driven by females. In addition, the detected altered hypothalamic expression patterns of Ctbp2 and Nbeal1 as a result of fasting and DIO implicate these genes in weight regulation.
Keywords: obesity, loci, cross-disorder, shared, eating disorder, bulimia nervosa
Introduction
The joint analysis of GWAS data pertaining to different phenotypes/diseases with overlapping or co-morbid endophenotypes recently led to the discovery of novel genes that had escaped detection in single phenotype/disease analyses. For instance, by a cross-disorder analysis of five major psychiatric disorders common underlying biological mechanisms were revealed1,2. A number of genetic variants were associated with more than one psychiatric disorder, illustrating the usefulness of the approach. Other cross-disorder analyses have shown overlapping genetic risk factors for phenotypes that had not been expected to share risk factors (e.g., ulcerative colitis and bone density or white blood cell count3). Heritability of anorexia nervosa (AN) is moderately high4–8. However, the two published GWAMA9,10 were underpowered to detect signals of small effect sizes, which are characteristic of SNPs identified for other psychiatric disorders1,2. The largest GWAMA for AN was performed in 2,907 patients with AN and 14,860 controls by the Genetic Consortium for AN (GCAN) and the Wellcome Trust Case Control Consortium 3 (WTCCC3). Although a global meta-analysis comprised discovery and replication data sets on a total of 5,551 AN cases and 21,080 controls, genome-wide significance was not reached10. However, 76% of the variant effects were directionally consistent between discovery and replication groups. This observation was unlikely to be spurious (p = 4 × 10−6)10.
A substantial genetic contribution to the variance of body mass index (BMI) is implicated by twin, family, and adoption studies11,12. The largest currently published GWAMA pertaining to BMI variance revealed 97 genome-wide significant (p ≤ 5×10−08) gene loci13; we use the term ‘BMI SNPs’ for those SNPs associated with an increased BMI. As most of the respective genes are expressed in the brain, a largely central regulation of human body weight appears likely13,14. A region on chromosome 16p11.2 supports a possible genetic link between obesity and AN. Carriers of the respective deletion(s) are hyperphagic and obese, whereas the carriers of the duplication(s) are underweight and show restrictive/selective eating behavior15,16.
Sex-specific analyses have previously been conducted for BMI and related phenotypes. For instance, the weight increasing effect was more pronounced in female mice of the initial melano-cortin-4 receptor gene (Mc4r) knock-out strain17. In humans with MC4R mutations leading to reduced function, the weight increasing effect was also stronger in females18. Sex-stratified GWAMAs for waist-hip ratio variation and other anthropometric traits (height, weight, body mass index, waist circumference, and hip circumference) revealed a sexual dimorphism in the genetic effects for fat distribution and waist phenotypes19–22. For many of these, genome-wide significance was detected for females only19,20.
AN might be considered as an extreme weight condition23, potentially entailing that genetic factors involved in body weight regulation may overlap with those predisposing to AN as suggested by several groups8,10,23–30. Recent LD-score regression analyses revealed a negative genetic correlation between AN and obesity (and a similar genetic correlation with BMI) suggesting that the same genetic factors influence normal variation in BMI as well as dysregulated BMI in AN30. However, in the latest GWAMA for AN 89 SNPs with genome-wide significance for BMI variation and obesity31,32 and 15 SNPs related to extreme obesity31 were not associated with AN10.
There is no evidence for an aberrant body weight regulation prior to manifestation of AN; thus, recalled premorbid weight of AN patients seemingly covers the whole BMI range33–35. The BMI range of patients at medium term (five to ten years) follow-ups is shifted to the left (lower BMI); in recovered patients overweight occurs with a substantially lower probability than in the general population36,37.
Here we performed three cross-trait analyses involving AN risk and BMI variation in two GWAMAs. First, we performed a cross-trait analysis of the 1000 SNPs with the lowest p-values from the largest GWAMA for AN (GCAN10) for evidence of association in the largest published GWAMA for BMI variation (GIANT13). Second, we performed sensitivity analyses in sex-stratified data sets from the BMI GWAMA for the best cross-trait SNPs (Table 1) because of the profound female preponderance in AN38,39; furthermore, sex-stratified analyses have revealed BMI loci that had not been detected in sex-combined analyses13. Finally, we performed a look-up of GWAMA derived BMI, (childhood) obesity and waist-hip ratio (WHR) loci within the AN GWAMA.
Table 1.
Nine of the 1000 SNPs with the lowest p-values in a GWAS for AN risk (GCAN9) are associated with increased BMI (GIANT11, with Bonferroni-corrected P < 0.05 significance; sorted according to the nominal p-values for increased BMI in all GIANT participants)
| Chromosome/position SNP nearest gene(s) |
Location | Rank in AN GWAS | AN effect allele/frequency in AN cases | Odds Ratio (SE) | p-value for AN risk | Frequency of AN reference allele for BMI | β (se) for BMI for reference allele | Nominal p-value for increased BMI: all female/male | Bonferroni corrected p-valuea | Direction of effectb +/− |
|---|---|---|---|---|---|---|---|---|---|---|
| 10/126685663 rs1561589 CTBP2 |
Intron | 201 | A / 0.33 | 1.14 (0.04) | 7.74 × 10−05 | A / 0.34 | 0.0157 (0.0033) | 2.47 × 10−06 3.45 × 10−07 / 0.043 |
0.0025 | + |
| 10/126681170 rs12771627 CTBP2 |
Intron | 190 | G / 0.75 | 0.87 (0.03) | 7.28 × 10−05 | G / 0.74 | −0.0162 (0.0035) | 4.25 × 10−06 5.8 × 10−06 / 0.022 |
0.0043 | + |
| 10/126674064 rs11245456 CTBP2 |
Intron | 177 | C / 0.75 | 0.87 (0.03) | 6.79 × 10−05 | C / 0.75 | −0.0171 (0.0037) | 4.58 × 10−06 1.03 × 10−05 / 0.009 |
0.0046 | + |
| 19/34978662 rs17513613 CCNE1 |
Distant 5′ | 409 | T / 0.70 | 0.88 (0.03) | 0.0002 | T / 0.67 | −0.015 (0.0033) | 5.41 × 10−06 6.4 × 10−03 / 1.24 × 10−05 |
0.0054 | + |
| 2/203492447 rs17406900 CARF |
Intron | 709 | A / 0.48 | 0.90 (0.03) | 0.0003 | A / 0.49 | −0.0134 (0.0031) | 1.08 × 10−05 1.8 × 10−04 / 2.27 × 10−03 |
0.0108 | + |
| 2/203639257 rs7593917 NBEAL1 |
Intron | 444 | A / 0.46 | 0.89 (0.03) | 0.0002 | A / 0.46 | −0.0131 (0.0031) | 2.48 × 10−05 9.54 × 10−05 / 9.39 × 10−03 |
0.0248 | + |
| 2/203582157 rs11691351 NBEAL1 |
Distant 5′ | 412 | A / 0.46 | 0.89 (0.03) | 0.0002 | A / 0.46 | −0.0126 (0.0031) | 3.57 × 10−05 1.98 × 10−04 / 8.21 × 10−03 |
0.0357 | + |
| 19/34988693 rs8102137 CCNE1 |
Distant 5′ | 248 | T / 0.70 | 0.88 (0.03) | 9.45 × 10−05 | T / 0.67 | −0.0169 (0.0041) | 3.76 × 10−05 0.006 / 2.46 × 10−04 |
0.0376 | + |
| 2/203635796 rs7573079 NBEAL1 |
Intron | 401 | G / 0.46 | 0.89 (0.03) | 0.0002 | G / 0.46 | −0.0124 (0.0031) | 4.61 × 10−05 2.80 × 10−04 / 0.008 |
0.0461 | + |
primary analysis, sex-combined, correction for 1,000 tests
Direction of effect: + the effect/risk allele for increased BMI and AN risk are identical; − the effect/risk allele for increased BMI and AN risk are not identical.
Gene abbreviations (alphabetically): ALS2CR8 (amyotrophic lateral sclerosis 2 (juvenile) chromosome region, candidate 8), CARF (Calcium Responsive Transcription Factor); CCNE1 (cyclin E1), CTBP2 (C-terminal binding protein 2), NBEAL1 (neurobeachin-like 1)
Post hoc we also performed (1) a look-up of the best cross-trait SNPs (Table 1) in: (1) obese children and adolescents from the EGG Consortium40, and (2) the first GWAS for AN9 comprising 1,033 AN cases and 3,733 paediatric controls from the Price Foundation Collaborative Group and the Children’s Hospital of Pennsylvania. Finally, we performed functional studies of the four genes nearest to the best cross-trait findings.
Materials and Methods
Look-up of ‘AN SNPs’ (GCAN) in GIANT GWAMA for BMI including sex-specific analyses
Our primary analysis is based on the in silico look-up of the 1000 best hits according to p-value (SNPs in high linkage disequilibrium (LD) were not excluded) derived from the case-control AN GWAMA10 in the large scale GWAMA of up to 322,135 individuals from the population-based GIANT meta-analysis for BMI13. In light of the aforementioned results for obesity risk alleles in the AN GWAMA10, we did not pursue the directional hypothesis that AN susceptibility/risk alleles are protective of obesity (i.e. are expected to be BMI lowering); as a consequence, we report two-sided tests. Secondarily, we performed sex-stratified analyses for the best cross-trait SNPs in the BMI GWAMA13.
We estimated the percentage of AN GWAMA SNPs that met the same p-value threshold in the BMI GWAMA (Supplementary Figure S1). We estimated that the genetic overlap of BMI and AN can seemingly be demonstrated if the number of SNPs analyzed is larger than 500, so that the 1000 SNPs we had chosen is justified. We decided against a computational derivation of an “optimal” cut-off as this could inflate the type I error rate. We did not aim for a comprehensive assessment of the joint common SNP variation architecture of both traits.
Post hoc we performed analyses in the sub data sets of GIANT (a: full GWAS chip data on N~233,000; b: Metabochip on N~88,000) to analyze if the observed effects are confirmed for each sub data set.
Look-up of ‘BMI SNPs’ in GWAMA for AN susceptibility (GCAN)
We performed an in silico lookup of the 56 novel genome-wide significant ‘BMI SNPs’ detected by Locke et al.13 in the case-control AN GWAMA (GCAN10). Subsequently we also analyzed previously described SNPs for BMI, obesity, childhood obesity, and WHR (see supplementary Tables). A total of 2,916 quality controlled genotypes of controls were included in both GCAN and GIANT (n=1,437 NBS-WTCCC National Blood Service donors and n=1,479 British 1958 birth cohort-WTCCC). Balancing between consistency (i.e. running our analyses on the same data sets as those published) and the necessity of sample independence, we rendered a re-analysis excluding these overlapping samples unnecessary.
Subsequent look-ups in independent GWAS data sets
We performed a look-up of the best cross-trait SNPs in GWAMA data of the EGG Consortium40 consisting of 5,530 obese children and adolescents (BMI ≥ 95th percentile) and 8,318 controls (BMI < 50th percentile). Data on the childhood obesity trait has been contributed by the EGG Consortium and was downloaded from www.egg-consortium.org40. An additional look-up of the best cross-trait SNPs from GCAN and GIANT was performed in the first GWAS for AN9 consisting of 1,033 AN cases and 3,733 paediatric control subjects of European ancestry; five SNPs were available.
Written informed consent to take part in genetic association studies was given by all participants and in case of minors by their parents. Studies were approved by the respective institutional review boards or ethics committees and conducted in accordance with The Declaration of Helsinki9,10,13,40.
Statistical analyses
We performed two main analyses and one stratified analysis nested within the first main analysis focussing on European-descent individuals of two GWAMAs. The GWAMA for AN10 was performed as fixed-effect meta-analysis based on single-SNP case-control association analyses under an additive genetic model with control for population stratification at the discovery data set level. Similarly, the GWAMA for BMI variation13 also worked with a fixed-effect meta-analysis based on discovery data set results obtained under a linear regression model adjusted for age, age2, sex, and study-specific covariates including control for population stratification effects. For the first main analysis, we looked-up the 1000 SNPs of the GWAMA for AN10 with the lowest p-values (discovery p-values from 5.56×10−22 to 4.79×10−4, of note: the SNP with the lowest p-value in the initial discovery GWAS for AN was not confirmed by genotyping in the replication sample10) in the GWAMA for BMI13. We applied a conservative Bonferroni-correction to the uncorrected p-values of the GWAMA for BMI to address multiple testing (see Table 1 and Supplementary Tables), and accordingly regarded all associations as significant which met a nominal p-value ≤ 5×10−5 (Table 1). For the SNPs with significant associations in the GWAMA for BMI, we also report the results of sex-stratified sensitivity analyses (Table 1). For the second main analysis, we performed a look-up of the 97 BMI loci in the GWAMA for AN. The direction of effect was evaluated only for SNPs with a nominal p-value ≤ 0.05.
Post hoc we also analyzed genome-wide significant loci for BMI, obesity, childhood obesity not originally described in Locke et al.13 (reviewed in 41) and 68 genome-wide significant loci for waist-hip ratio (WHR) derived from a European GWAMA primary analysis (GIANT21) in the GWAMA for AN (GCAN10).
Animals and diet
Unless stated otherwise, male C57BL/6J mice were fed ad libitum with either a standard chow diet (Harlan Teklad LM-485; 5.6% kcal fat) or a high-fat diet (D12331; Research Diets, New Brunswick, NJ; 58% kcal fat). The mice had free access to water and were maintained under constant ambient conditions (22 ± 1°C, constant humidity, 12h/12h light/dark cycle). All animal studies were performed in Cincinnati, OH, USA and were approved by the Animal Ethics Committee of Cincinnati, OH, USA.
Gene expression analyses
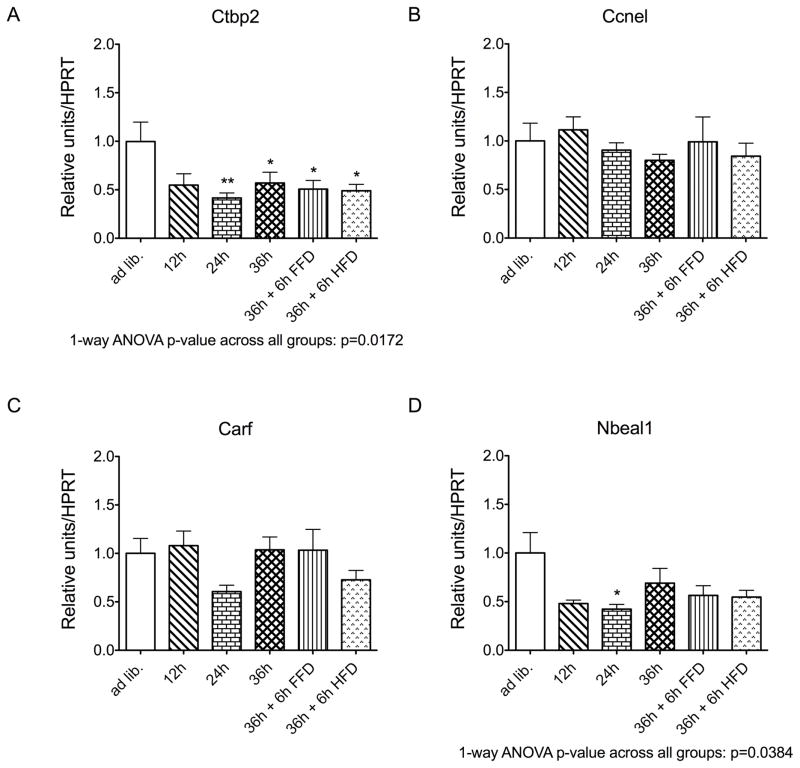
To assess effects on fasting and re-feeding, hypothalamic gene expression was profiled in male 27/28 week old C57BL/6J mice fed either ad libitum with a regular chow diet, or which had been fasted for 12h, 24h, or 36h, or which had been fasted for 36h and then re-fed for 6h using either a fat-free diet or a high-fat diet (N=6–8 mice per group). The use of existing ex vivo material is in agreement with the US and German guidelines of the Animal Welfare Committee to restrict animal experiments to an absolutely necessary minimum. Target genes were amplified using the ViiA 7 real-time PCR system (Life Technologies; Darmstadt, Germany); results were normalized to the housekeeping gene hypoxanthine guanine phosphoribo-syltransferase 1 (HPRT). The used primer sequences were CTBP2-F: 3′-TACCACACCATCACCCTCAC -5′; CTBP2-R: 3′-TGTGGCAGACTGTCGAATCT-5′; CCNEI-F:3′-AGCCTCGGAAAATCAGACCA-5′; CCNEI-R: 3′-CTTCGCACACctccattagc-5′, CARF-F: 3′-GTGGACGACAGATAGTGGGA-5′; CARF-R: 3′-GGAGAGGAGAGTCTTGGCTG-5′; NBEAL1-F: 3′-AGGAGAAGGAAATGGCTGATCA-5′, and NBEAL1-R: 3′-TCCACTGTGAGAGAAGCTGG-5′. Data represent means ± SEM. *P<0.05, **P<0.01, based on a one-way ANOVA with Dunnett’s Multiple Comparison post-hoc test.
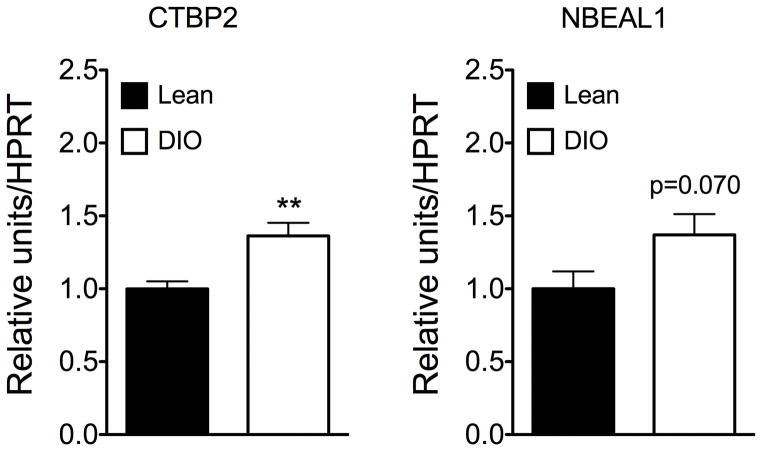
To additionally assess the effects of a high fat diet on hypothalamic expression of Nbeal1 and Ctbp2 was assessed in age matched male C57BL/6J mice fed either a regular chow diet (body weight 32.69g ± 0.45g) or a high-fat diet (body weight 54.72g ± 1.25g; N=7–8 mice per group). Data represent means ± SEM.
In silico analyses
Expression patterns and known variants in the coding regions (missense, nonsense and frameshift) were analyzed in silico (http://www.genecards.org/; http://exac.broadinstitute.org/about).
Results
Association of AN risk SNPs with increased BMI
We detected association (p-values < 5×10−5, Bonferroni corrected p < 0.05) at three independent chromosomal loci in the BMI GWAMA (chromosome 2: four SNPs in linkage disequilibrium [LD], r2 ≥ 0.819, D′ = 1; chromosome 10: three SNPs, r2 ≥ 0.363, D′ ≥ 0.728; and chromosome 19: two SNPs, r2 = 1, D′ = 1); the lowest p-value (rs1561589, 2.47×10−6, pcorrected = 0.0025) was observed at the chromosome 10 locus (Table 1). Within the GIANT13 data we post hoc also analyzed the data sets separately for (a) full GWAS chip data (HapMap imputed) on N~233,000 and (b) Metabochip on N~88,000 (Supplementary Table S1). Both independent data sets confirmed the association of the nine SNPs.
The nearest genes to these nine SNPs ordered from lowest to highest p-values are: (1) chromosome 10: CTBP2 (C-terminal binding protein 2 gene); (2) chromosome 19: CCNE1 (cyclin E1 gene); (3) chromosome 2: CARF (calcium responsive transcription factor gene) and (4) NBEAL1 (neurobeachin-like 1 gene). The third chromosomal locus included two genes, because the four SNPs are located in a region with high linkage disequilibrium (lowest LD for the four SNPs: r2 ≥ 0.819, D′ = 1). Interestingly, for all SNPs, the AN risk alleles were consistently associated with increased BMI (Table 1).
Sex-specific analyses for the best cross-trait SNPs (Table 1) revealed that the chromosome 10 association signal was primarily driven by females. Again, post hoc sex-specific analyses in the sub data sets of GIANT (a: full GWAS chip data on N~233,000; b: Metabochip on N~88,000; supplementary Table S1) confirmed the larger effect in females for the best locus.
Further look-ups
Because AN typically manifests during adolescence, we analyzed the identified SNPs in the EGG Consortium data set40, which includes only children and adolescents. The lookup of the nine cross-trait SNPs (Table 1) did not reveal significant findings at the five SNPs available (p-values from 0.0916 at rs11245456 to 0.6075 at rs1561589). However, the direction of effect was the same between AN risk and early onset extreme obesity in all five SNPs.
The look-up of the nine cross-trait SNPs in the first GWAS for AN9 comprising 1,033 AN cases and 3,733 paediatric controls (five SNPs were available, each locus was represented) showed nominally significant results for two SNPs at chromosome 2 (rs17406900, nominal p = 0.03; rs7573079, nominal p = 0.04), our second best locus. However, for these SNPs the direction of effect was opposite to the effect in GCAN.
Association of ‘BMI SNPs’ with AN
The look-up of the ‘BMI SNPs’ in the AN GWAMA did not reveal (Bonferroni-corrected for 97 SNPs) significant results (Supplementary Tables S2–S4). Similarly, post hoc lookups of additional genome-wide significant loci for BMI, obesity, childhood obesity41 (Supplementary Table S5) and WHR21 (Supplementary Table S6) in the GWAMA for AN (GCAN10) did not reveal statistically significant findings after correction for multiple testing.
In silico analyses
All four genes located at the three identified loci are widely expressed in brain tissues, including the hypothalamus (http://www.genecards.org/). A spectrum of different, potentially functionally relevant variants (missense, nonsense and frameshift) was detected for all four genes (Supplementary Table S7).
Mouse model
Gene expression profiling of Ctbp2, Ccne1, Carf, Nbeal1 and in male C57BL/6J mice revealed that hypothalamic expression of both Ctbp2 and Nbeal1 was decreased by fasting (one-way ANOVA p<0.05 for both targets; Figure 1). Notably, hypothalamic expression of Ctbp2 and Nbeal1 remained decreased after 36h fasting followed by 6h re-feeding with either a fat-free diet or a high-fat diet relative to control mice fed ad libitum (Figure 1). In line with the down-regulation of hypothalamic expression of Ctbp2 and Nbeal1 in response to nutrient availability, expression of Ctbp2 was increased in diet-induced obese compared to age-matched lean control mice (p < 0.01; Figure 2); for Nbeal1 we noted a trend for increased expression in obese compared to lean mice (p = 0.070; Figure 2).
Figure 1.
Hypothalamic expression of Ctbp2 (a), Ccne1 (b), Carf (c), Nbeal1 (d), or in response to fasting for 12, 24 or 36 h, and after re-feeding for 6 h with either a high-fat diet (HFD) or a fat-free diet (FFD, N = 6–8 mice per group) (c). *P<0.05, **P<0.01, based on a one-way ANOVA with Dunnett’s Multiple Comparison post-hoc test
Figure 2.
Hypothalamic expression of Ctbp2 and Nbeal1 in diet induced obesity (DIO) as compared to age-matched lean control mice.
Discussion
Among the 1000 SNPs with the lowest p-values in the GCAN GWAMA for AN10 we identified nine SNPs in three chromosomal regions with significant p-values in the currently largest GWAMA for BMI variation13 using a conservative Bonferroni correction.
The relevance of these three loci is uncertain, because none of the nine SNPs have previously been identified for either AN or BMI/obesity or other psychiatric disorders. Two NBEAL1 SNPs (intronic and 3′UTR, rs16839626, rs6733725; no detectable LD to the SNPs identified here; http://www.broadinstitute.org/mpg/snap/ldsearchpw.php) had been detected in a GWAS for obesity related traits in 815 Hispanic children from 263 families42. Nominal associations (not genome-wide significant) for energy storage and fat mass deposition (p = 2×10−7), fat mass change (4×10−7) and weight change (3×10−6) were shown42. Central (including hypothalamic) expression of all four genes was detected. We did not detect association of the previously published GWAMA SNPs for BMI, (childhood) obesity, or WHR with AN (Supplementary Tables).
The following results do not readily substantiate the relevance of our association findings: a) The analysis of the five out of nine available cross-trait SNPs in 5,530 obese children and adolescents (BMI ≥ 95th percentile) versus 8,318 controls (BMI < 50th percentile) from the EGG Consortium40 did not reveal significant findings. However, for all available SNPs the direction of effect was identical to that observed in the GIANT GWAMA. Because the EGG Consortium GWAMA is substantially smaller than the recent GIANT approach13, true signals may not have been detectable. b) The look-up of the same cross-trait SNPs in the first GWAS for AN9 did not support our findings. This might partly be explained by the lower sample size in the analysis of the Price Foundation Collaborative Group and Children’s Hospital of Pennsylvania samples9 (1,033 AN cases and 3,733 paediatric controls) compared to the latest GWAS10 (2,907 cases with AN and 14,860 controls). In conclusion, we cannot exclude that our detected associations for the nine SNPs represent false positive associations.
The following lines of evidence do however support that we have indeed detected SNPs associated with both AN and obesity: The identification of the three loci with nominal p-values in the range of 10−5 to 10−6 for association with BMI is quite unexpected. Accordingly, at least one and maximally all three loci are involved in body weight regulation; the same potentially holds true for AN. If this assumption is correct, future larger GWAMAs for both AN and BMI/obesity will pick up the respective loci. It is also of interest that all risk alleles were directionally consistent for AN risk and higher BMI. This is especially unexpected as (a) patients with AN do not have an elevated premorbid BMI33; (b) BMI-values of followed up patients only infrequently exceed the cutoff for overweight (BMI ≥ 25 kg/m2)36 and (c) LD-score regression analyses revealed a negative genetic correlation between AN and obesity30. It is unlikely that the overlap between the AN (controls) and GIANT GWAMAs explains our results.
Sex-specific analyses
We also performed look-ups in sex-stratified analyses for the best cross-trait SNPs in the BMI GWAMA, because (i) AN predominantly occurs in females38,39 and (ii) sex-specific analyses rendered BMI loci that had not been picked up by sex-combined analyses13. We found that the three AN risk SNPs at the chromosome 10 locus with the lowest p-values in the BMI GWAMA (sex-combined) mainly originated from the female participants (Table 1). This finding provides additional indirect evidence that particularly this locus is involved in both AN and body weight regulation in females mainly.
Animal model
The hypothalamic expression data obtained in male mice clearly substantiate that the detected associations at two loci may indeed represent true positive findings. The cDNA of the fasting/re-feeding experiment described in this manuscript is commonly used in the Müller/Tschöp lab to assess regulation of target genes. Whereas unfortunately there is no documentation on the total number of previously analyzed targets, it can be confirmed that only few of the previously analyzed genes have been found to be differentially regulated under the conditions reported here (e.g. 43). Expression of Ctbp2, whose locus represented our strongest association signal (Table 1), proved to be inversely regulated by fasting and diet induced obesity. Thus, hypothalamic gene expression was reduced for this gene and additionally in fasted (12, 24 or 36 h) mice; this down-regulation persisted 6 hours after renewed access to adlibitum feeding (re-feeding for 6 h with either a high-fat diet or a fat-free diet). Genes, whose expression is down-regulated in fasting, are usually anorexigenic (e.g., leptin44,45), while expression of orexigenic genes (e.g., ghrelin46) is increased in fasting. Hence it is likely that both Ctbp2 and Nbeal1 have an anorexigenic effect. In accordance with this assumption, both genes were up-regulated in diet induced obesity (DIO; Figure 2).
BDNF signalling
It is of interest to point out that the two genes CTBP2 and CARF are involved in BDNF signaling pathways47–63. The leptinergic-melanocortinergic-BDNF pathway includes genes with known genetic variation underlying both monogenic and polygenic obesity64. Multiple SNPs near BDNF are genome-wide significantly associated with obesity (e.g. 13,65). Evidence for an involvement of BDNF in AN stems from studies on (a) animal models, (b) genetics, and (c) serum or brain levels of BDNF. However, some of the data are equivocal. In more detail: (a) in animal models the central infusion of BDNF induces weight loss66,67. The suppressive effects of BDNF on feeding behavior and body weight are mediated by corticotropin-releasing factor (CRF) and hypothalamic neuronal histamine in mice68. BDNF signaling is altered by reduced BDNF expression in the hippocampus, in activity-based anorexia in mice69 and in immobilization stress induced anorexia in rats70. Deletion of the Bdnf gene in the PVH resulted in hyperphagia, reduced locomotor activity, impaired thermogenesis, and severe obesity. Additionally, in response to cold exposure BDNF expression in the PVH was increased71. (b) Association of variation in BDNF with AN was shown by some but not all studies72–84. For the widely studied BDNF Val66Met variant, a recent meta-analysis showed no association of the infrequent 66Met allele with AN82. (c) Decreased serum and brain levels of BDNF had unequivocally been reported in patients with AN66,85–100. This was recently confirmed in a meta-analysis97. While only one study has suggested an interaction between CTBP2 and BDNF47, the interaction of CARF and BDNF has been substantiated in numerous studies (see above). Thus, again as BDNF might be involved in both AN and obesity27,101,102 this gene is biologically highly plausible.
In the following we provide additional information on the genes located nearest to the three loci identified via the nine SNPs starting with the chromosome harboring the SNPs with the lowest p-values:
Chromosome 10
The three intronic SNPs in the CTBP2 gene (C-terminal binding protein 2) show the lowest p-values in our BMI GWAMA13 look-up (Table 1); as stated above the effect is almost only due to females. The two alternative CTBP2 transcripts lead to two distinct proteins, one of which is a transcriptional repressor, while the other is a major component of synaptic ribbons, a specialized form of synapses. A NAD+ binding domain is common to both isoforms. There is evidence that the gene/protein is involved in brown adipose tissue function and regulation103–109. Ctbp2 knock-out mice displayed abnormal phenotypes in the cardiovascular and central nervous systems, in addition to having effects on embryogenesis, growth/size/body, and mortality/aging (http://www.informatics.jax.org/allele/ MGI:2183646110). Recently, a miRNA that was up-regulated during the development of obesity in mice (miR-342-3p) was described to promote a suppressing effect on CtBP2111, again underscoring the relevance of the gene for weight regulation.
Chromosome 19
The cyclin E1 gene (CCNE1) identified via the two SNPs 5′ to this gene encodes a protein that belongs to the highly conserved cyclin family. Cyclins act as (i) regulators of specific kinases and (ii) contribute to the coordination of mitotic events. In many tumors overexpression of this gene has been observed112. It was recently shown that proliferation of 3T3-L1 preadipocytes promoted by recombinant myostatin increased expression of proliferation related genes (e.g. cyclin E1 by 20.5 %113).
Chromosome 2
The third chromosomal locus includes two genes, because the four SNPs are located in a region with high linkage disequilibrium (lowest LD for the four SNPs: r2 ≥ 0.819, D′ = 1). Three of the SNPs are located in an intron, one is 5′ to NBEAL1 (Table 1): (1) The calcium-response factor gene (CARF or as an alias name amyotrophic lateral sclerosis 2 (juvenile) chromosome region, candidate 8 gene: ALS2CR8) acts as a transcriptional activator that mediates the calcium- and neuron-selective induction of BDNF expression52. Lack of Carf (Als2cr8) in knock-out mice results in deficits associated with learning and memory56. Functionally relevant recessive mutations in the gene have been described in patients with amyotrophic lateral sclerosis 2 (ALS2114).
In sum, in a cross-trait analysis for genetic loci involved in AN risk and increased BMI three chromosomal loci with potential relevance for both traits were detected. Apart from the identification of these loci, their role in both AN and body weight regulation was particularly substantiated by ex vivo data of mouse models for fasting and DIO suggesting an anorexic role of CTBP2 and NBEAL1, by the sex specific results for CTBP2 and the finding that CTBP2 and CARF are involved in BDNF regulation. Further in depth molecular genetic and biological analyses are essential to understand the relevance of these loci and the genes they contain in the etiology of AN and in body weight regulation/obesity. The association of AN alleles with increased BMI might imply that a specific genetic variant (allele) can either increase or decrease BMI depending on presence or absence of additional factors with an influence of the body weight (e.g. occurrence of an eating disorder), or the variant predisposes to dysregulation and other genes or environmental factors determine its direction). If true, this general concept has implications for gene mapping approaches in genetic epidemiology calling for more hypothesis-driven stratified analyses. A spectrum of different variants (missense, nonsense and frameshift) has been described for the four genes (Supplementary Table 7), so that a mutation screen in these genes in study groups of patients with AN or extreme obesity is warranted.
Supplementary Material
Acknowledgments
The authors express their gratitude to all participants. We thank the following sources for funding or research: the German Ministry for Education and Research (National Genome Research Net-Plus 01GS0820 and 01KU0903; AS, MK and the CSCC were supported by 01EO1002, 01EO1502), the German Research Foundation (DFG; HI865/2-1, SFB940/1, SCHE1648/1-3, TS226/3-1), the European Community’s Seventh Framework Programme (FP7/2007–2013) under grant agreement n°245009 and n°262055, and the National Institutes of Health (NIH; R01DK075787), funding to MHT from the Alexander von Humboldt Foundation, the Helmholtz Alliance ICEMED – Imaging and Curing Environmental Metabolic Diseases, through the Initiative and Networking Fund of the Helmholtz Association, the Helmholtz cross-program topic “Metabolic Dysfunction”, the WTCCC3 WT088827/Z/09 entitled “A genome wide association study of anorexia nervosa”. The authors would like to thank the Exome Aggregation Consortium and the groups that provided exome variant data for comparison. A full list of contributing groups can be found at http://exac.broadinstitute.org/about. AH was supported by the ‘Landesprogramm für Geschlechtergerechte Hochschulen - Programmstrang Förderung von Denominationen in der Genderforschung’.
Genetic Consortium for Anorexia Nervosa (GCAN):
Vesna Boraska Perica, Christopher S Franklin, James A B Floyd, Laura M Thornton, Laura M Huckins, Lorraine Southam, N William Rayner, Ioanna Tachmazidou, Kelly L Klump, Janet Treasure, Cathryn M Lewis, Ulrike Schmidt, Federica Tozzi, Kirsty Kiezebrink, Johannes Hebebrand, Philip Gorwood, Roger A H Adan, Martien J H Kas, Angela Favaro, Paolo Santonastaso, Fernando Fernández-Aranda, Monica Gratacos, Filip Rybakowski, Monika Dmitrzak-Weglarz, Jaakko Kaprio, Anna Keski-Rahkonen, Anu Raevuori-Helkamaa, Eric F Van Furth, Margarita C T Slof-Op’t Landt, James I Hudson, Ted Reichborn-Kjennerud, Gun Peggy S Knudsen, Palmiero Monteleone, Allan S Kaplan, Andreas Karwautz, Hakon Hakonarson, Wade H Berrettini, Yiran Guo, Dong Li, Nicholas J Schork, Gen Komaki, Tetsuya Ando, Hidetoshi Inoko, Tõnu Esko, Krista Fischer, Katrin Männik, Andres Metspalu, Jessica H Baker, Roger D Cone, Jennifer Dackor, Janiece E DeSocio, Christopher E Hilliard, Julie K O’Toole, Jacques Pantel, Jin P Szatkiewicz, Chrysecolla Taico, Stephanie Zerwas, Sara E Trace, Oliver S P Davis, Sietske Helder, Katharina Bühren, Roland Burghardt, Martina de Zwaan, Karin Egberts, Stefan Ehrlich, Beate Herpertz-Dahlmann, Wolfgang Herzog, Hartmut Imgart, André Scherag, Susann Scherag, Stephan Zipfel, Claudette Boni, Nicolas Ramoz, Audrey Versini, Marek K Brandys, Unna N Danner, Carolien de Kove, Judith Hendriks, Bobby P C Koeleman, Roel A Ophoff, Eric Strengman, Annemarie A van Elburg, Alice Bruson, Maurizio Clementi, Daniela Degortes, Monica Forzan, Elena Tenconi, Elisa Docampo, Geòrgia Escaramí Susana Jiménez-Murcia, Jolanta Lissowska, Andrzej Rajewski, Neonila Szeszenia-Dabrowska, Agnieszka Slopien, Joanna Hauser, Leila Karhunen, Ingrid Meulenbelt, P Eline Slagboom, Alfonso Tortorella, Mario Maj, George Dedoussis, Dimitris Dikeos, Fragiskos Gonidakis, Konstantinos Tziouvas, Artemis Tsitsika, Hana Papezova, Lenka Slachtova, Debora Martaskova, James L Kennedy, Robert D Levitan, Zeynep Yilmaz, Julia Huemer, Doris Koubek, Elisabeth Merl, Gudrun Wagner, Paul Lichtenstein, Gerome Breen, Sarah Cohen-Woods, Anne Farmer, Peter McGuffin, Sven Cichon, Ina Giegling, Stefan Herms, Dan Rujescu, Stefan Schreiber, H-Erich Wichmann, Christian Dina, Rob Sladek, Giovanni Gambaro, Nicole Soranzo, Antonio Julia, Sara Marsal, Raquel Rabionet, Valerie Gaborieau, Danielle M Dick, Aarno Palotie, Samuli Ripatti, Elisabeth Widén, Ole A Andreassen, Thomas Espeseth, Astri Lundervold, Ivar Reinvang, Vidar M Steen, Stephanie Le Hellard, Morten Mattingsdal, Ioanna Ntalla, Vladimir Bencko, Lenka Foretova, Vladimir Janout, Marie Navratilova, Steven Gallinger, Dalila Pinto, Stephen W Scherer, Harald Aschauer, Laura Carlberg, Alexandra Schosser, Lars Alfredsson, Bo Ding, Lars Klareskog, Leonid Padyukov, Chris Finan, Gursharan Kalsi, Marion Roberts, Darren W Logan, Leena Peltonen, Graham R S Ritchie, Jeff C Barrett, Xavier Estivill, Anke Hinney, Patrick F Sullivan, David A Collier, Eleftheria Zeggini, and Cynthia M Bulik
Wellcome Trust Case Control Consortium 3 (WTCCC3):
Carl A Anderson, Jeffrey C Barrett, James A B Floyd, Christopher S Franklin, Ralph McGinnis, Nicole Soranzo, Eleftheria Zeggini, Jennifer Sambrook, Jonathan Stephens, Willem H Ouwehand, Wendy L McArdle, Susan M Ring, David P Strachan, Graeme Alexander, Cynthia M Bulik, David A Collier, Peter J Conlon, Anna Dominiczak, Audrey Duncanson, Adrian Hill, Cordelia Langford, Graham Lord, Alexander P Maxwell, Linda Morgan, Leena Peltonen, Richard N Sandford, Neil Sheerin, Frederik O Vannberg, Hannah Blackburn, Wei-Min Chen, Sarah Edkins, Mathew Gillman, Emma Gray, Sarah E Hunt, Suna Nengut-Gumuscu, Simon Potter, Stephen S Rich, Douglas Simpkin, and Pamela Whittaker
Genetic Investigation of ANthropometric Traits Consortium (GIANT):
de Bakker P, Bültmann U, Geleijnse M, Harst Pv, Koppelman G, Rosmalen JG, van Rossum L, Smidt H, Swertz MA, Stolk RP, Alizadeh B, de Boer R, Boezen HM, Bruinenberg M, Franke L, van der Harst P, Hillege H, van der Klauw M, Navis G, Ormel J, Postma D, Rosmalen J, Slaets J, Snieder H, Stolk R, Wolffenbuttel B, Wijmenga C, Berg J, Blackwood D, Campbell H, Cavanagh J, Connell J, Connor M, Cunningham-Burley S, Deary I, Dominiczak A, Ellis P, FitzPatrick B, Ford I, Gertz R, Grau A, Haddow G, Jackson C, Kerr S, Lindsay R, McGilchrist M, McIntyre D, Morris A, Morton R, Muir W, Murray G, Palmer C, Pell J, Philp A, Porteous D, Porteous M, Procter R, Ralston S, Reid D, Sinnott R, Smith B, Clair DS, Sullivan F, Sweetland M, Ure J, Watt G, Wolf R, Wright A, Berndt SI, Gustafsson S, Mägi R, Ganna A, Wheeler E, Feitosa MF, Justice AE, Monda KL, Croteau-Chonka DC, Day FR, Esko T, Fall T, Ferreira T, Gentilini D, Jackson AU, Luan J, Randall JC, Vedantam S, Willer CJ, Winkler TW, Wood AR, Workalemahu T, Hu YJ, Lee SH, Liang L, Lin DY, Min JL, Neale BM, Thorleifsson G, Yang J, Albrecht E, Amin N, Bragg-Gresham JL, Cadby G, den Heijer M, Eklund N, Fischer K, Goel A, Hottenga JJ, Huffman JE, Jarick I, Johansson A, Johnson T, Kanoni S, Kleber ME, König IR, Kristiansson K, Kutalik Z, Lamina C, Lecoeur C, Li G, Mangino M, McArdle WL, Medina-Gomez C, Müller-Nurasyid M, Ngwa JS, Nolte IM, Paternoster L, Pechlivanis S, Perola M, Peters MJ, Preuss M, Rose LM, Shi J, Shungin D, Smith AV, Strawbridge RJ, Surakka I, Teumer A, Trip MD, Tyrer J, Van Vliet-Ostaptchouk JV, Vandenput L, Waite LL, Zhao JH, Absher D, Asselbergs FW, Atalay M, Attwood AP, Balmforth AJ, Basart H, Beilby J, Bonnycastle LL, Brambilla P, Bruinenberg M, Campbell H, Chasman DI, Chines PS, Collins FS, Connell JM, Cookson W, de Faire U, de Vegt F, Dei M, Dimitriou M, Edkins S, Estrada K, Evans DM, Farrall M, Ferrario MM, Ferrières J, Franke L, Frau F, Gejman PV, Grallert H, Grönberg H, Gudnason V, Hall AS, Hall P, Hartikainen AL, Hayward C, Heard-Costa NL, Heath AC, Hebebrand J, Homuth G, Hu FB, Hunt SE, Hyppönen E, Iribarren C, Jacobs KB, Jansson JO, Jula A, Kähönen M, Kathiresan S, Kee F, Khaw KT, Kivimaki M, Koenig W, Kraja AT, Kumari M, Kuulasmaa K, Kuusisto J, Laitinen JH, Lakka TA, Langenberg C, Launer LJ, Lind L, Lindström J, Liu J, Liuzzi A, Lokki ML, Lorentzon M, Madden PA, Magnusson PK, Manunta P, Marek D, März W, Mateo Leach I, McKnight B, Medland SE, Mihailov E, Milani L, Montgomery GW, Mooser V, Mühleisen TW, Munroe PB, Musk AW, Narisu N, Navis G, Nicholson G, Nohr EA, Ong KK, Oostra BA, Palmer CN, Palotie A, Peden JF, Pedersen N, Peters A, Polasek O, Pouta A, Pramstaller PP, Prokopenko I, Pütter C, Radhakrishnan A, Raitakari O, Rendon A, Rivadeneira F, Rudan I, Saaristo TE, Sambrook JG, Sanders AR, Sanna S, Saramies J, Schipf S, Schreiber S, Schunkert H, Shin SY, Signorini S, Sinisalo J, Skrobek B, Soranzo N, Stancakova A, Stark K, Stephens JC, Stirrups K, Stolk RP, Stumvoll M, Swift AJ, Theodoraki EV, Thorand B, Tregouet DA, Tremoli E, Van der Klauw MM, van Meurs JB, Vermeulen SH, Viikari J, Virtamo J, Vitart V, Waeber G, Wang Z, Widen E, Wild SH, Willemsen G, Winkelmann BR, Witteman JC, Wolffenbuttel BH, Wong A, Wright AF, Zillikens M, Amouyel P, Boehm BO, Boerwinkle E, Boomsma DI, Caulfield MJ, Chanock SJ, Cupples L, Cusi D, Dedoussis GV, Erdmann J, Eriksson JG, Franks PW, Froguel P, Gieger C, Gyllensten U, Hamsten A, Harris TB, Hengstenberg C, Hicks AA, Hingorani A, Hinney A, Hofman A, Hovingh KG, Hveem K, Illig T, Jarvelin MR, Jöckel KH, Keinanen-Kiukaanniemi SM, Kiemeney LA, Kuh D, Laakso M, Lehtimäki T, Levinson DF, Martin NG, Metspalu A, Morris AD, Nieminen MS, Njølstad I, Ohlsson C, Oldehinkel AJ, Ouwehand WH, Palmer LJ, Penninx B, Power C, Province MA, Psaty BM, Qi L, Rauramaa R, Ridker PM, Ripatti S, Salomaa V, Samani NJ, Snieder H, Sørensen TI, Spector TD, Stefansson K, Tönjes A, Tuomilehto J, Uitterlinden AG, Uusitupa M, van der Harst P, Vollenweider P, Wallaschofski H, Wareham NJ, Watkins H, Wichmann H-, Wilson JF, Abecasis GR, Assimes TL, Barroso I, Boehnke M, Borecki IB, Deloukas P, Fox CS, Frayling T, Groop LC, Haritunian T, Heid IM, Hunter D, Kaplan RC, Karpe F, Moffatt M, Mohlke KL, O’Connell JR, Pawitan Y, Schadt EE, Schlessinger D, Steinthorsdottir V, Strachan DP, Thorsteinsdottir U, van Duijn CM, Visscher PM, Di Blasio AM, Hirschhorn JN, Lindgren CM, Morris AP, Meyre D, Scherag A, McCarthy MI, Speliotes EK, North KE, Loos RJ, Ingelsson E.
Early Growth Genetics Consortium (EGG):
Adair LS, Ang W, Atalay M, van Beijsterveldt T, Bergen N, Benke K, Berry DJ, Boomsma DI, Bradfield JP, Charoen P, Coin L, Cooper C, Cousminer DL, Das S, Davis OS, Dedoussis GV, Elliott P, Estivill X, Evans DM, Feenstra B, Flexeder C, Frayling T, Freathy RM, Gaillard R, Geller F, Gillman M, Grant SF, Groen-Blokhuis M, Goh LK, Guxens M, Hakonarson H, Hattersley AT, Haworth CM, Hadley D, Hedebrand J, Heinrich J, Hinney A, Hirschhorn JN, Hocher B, Holloway JW, Holst C, Hottenga JJ, Horikoshi M, Huikari V, Hypponen E, Iñiguez C, Jaddoe VW, Jarvelin MR, Kaakinen M, Kilpeläinen TO, Kirin M, Kowgier M, Lakka HM, Lakka TA, Lange LA, Lawlor DA, Lehtimäki T, Lewin A, Lindgren C, Lindi V, Maggi R, Marsh J, McCarthy MI, Melbye M, Middeldorp C, Millwood I, Mohlke KL, Mook-Kanamori DO, Murray JC, Nivard M, Nohr EA, Ntalla I, Oken E, Ong KK, O’Reilly PF, Palmer LJ, Panoutsopoulou K, Pararajasingham J, Pearson ER, Pennell CE, Power C, Price TS, Prokopenko I, Raitakari OT, Rodriguez A, Salem RM, Saw SM, Scherag A, Sebert S, Siitonen N, Simell O, Sørensen TI, Sovio U, Pourcain BS, Strachan DP, Sunyer J, Taal HR, Teo YY, Thiering E, Tiesler C, Timpson NJ, Uitterlinden AG, Valcárcel B, Warrington NM, White S, Widén E, Willemsen G, Wilson JF, Yaghootkar H, Zeggini E, Elks CE, Perry JR, Sulem P, Chasman DI, Franceschini N, He C, Lunetta KL, Visser JA, Byrne EM, Cousminer DL, Gudbjartsson DF, Esko T, Feenstra B, Hottenga JJ, Koller DL, Kutalik Z, Lin P, Mangino M, Marongiu M, McArdle PF, Smith AV, Stolk L, van Wingerden SH, Zhao JH, Albrecht E, Corre T, Ingelsson E, Hayward C, Magnusson PK, Smith EN, Ulivi S, Warrington M, Zgaga L, Alavere H, Amin N, Aspelund T, Bandinelli S, Barroso I, Berenson GS, Bergmann S, Blackburn H, Boerwinkle E, Buring JE, Busonero F, Campbell H, Chanock SJ, Chen W, Cornelis MC, Couper D, Coviello AD, d’Adamo P, de Faire U, de Geus EJ, Deloukas P, Döring A, Davey Smith G, Easton DF, Eiriksdottir G, Emilsson V, Eriksson J, Ferrucci L, Folsom AR, Foroud T, Garcia M, Gasparini P, Geller F, Gieger C, Gudnason V, Hall P, Hankinson SE, Ferreli L, Heath AC, Hernandez DG, Hofman A, Hu FB, Illig T, Järvelin MR, Johnson AD, Karasik D, Khaw KT, Kiel DP, Kilpeläinen TO, Kolcic I, Kraft P, Launer LJ, Laven JS, Li S, Liu J, Levy D, Martin NG, McArdle WL, Melbye M, Mooser V, Murray JC, Murray SS, Nalls MA, Navarro P, Nelis M, Ness AR, Northstone K, Oostra BA, Peacock M, Palmer LJ, Palotie A, Paré G, Parker AN, Pedersen NL, Peltonen L, Pennell CE, Pharoah P, Polasek O, Plump AS, Pouta A, Porcu E, Rafnar T, Rice JP, Ring SM, Rivadeneira F, Rudan I, Sala C, Salomaa V, Sanna S, Schlessinger D, Schork NJ, Scuteri A, Segrè AV, Shuldiner AR, Soranzo N, Sovio U, Srinivasan SR, Strachan DP, Tammesoo ML, Tikkanen E, Toniolo D, Tsui K, Tryggvadottir L, Tyrer J, Uda M, van Dam RM, van Meurs JB, Vollenweider P, Waeber G, Wareham NJ, Waterworth DM, Weedon MN, Wichmann HE, Willemsen G, Wilson JF, Wright AF, Young L, Zhai G, Zhuang WV, Bierut LJ, Boyd HA, Crisponi L, Demerath EW, van Duijn CM, Econs MJ, Harris TB, Hunter DJ, Loos RJ, Metspalu A, Montgomery GW, Ridker PM, Spector TD, Streeten EA, Stefansson K, Thorsteinsdottir U, Uitterlinden AG, Widen E, Murabito JM, Ong KK, Murray A.
Children’s Hospital of Philadelphia/Price Foundation:
We gratefully thank all the patients and their families who were enrolled in this study, as well as all the control subjects who donated blood samples to Children’s Hospital of Philadelphia (CHOP) for genetic research purposes. We thank the Price Foundation for their support of the Collaborative Group effort that was responsible for recruitment of patients, collection of clinical information and provision of the DNA samples used in this study. The authors also thank the Klarman Family Foundation for supporting the study. We thank the technical staff at the Center for Applied Genomics at CHOP for producing the genotypes used for analyses and the nursing, medical assistant and medical staff for their invaluable help with sample recruitments. CTB and NJS are funded in part by the Scripps Translational Sciences Institute Clinical Translational Science Award [Grant Number U54 RR0252204-01]. All genome-wide genotyping was funded by an Institute Development Award to the Center for Applied Genomics from the CHOP. 2011 – 2014 Davis Foundation Postdoctoral Fellowship Program in Eating Disorders Research Award, Yiran Guo; 2012 – 2015 Davis Foundation Postdoctoral Fellowship Program in Eating Disorders Research Award, Dong Li.
The Price Foundation Collaborative Group:
Harry Brandt, Steve Crawford, Scott Crow, Manfred M. Fichter, Katherine A. Halmi, Craig Johnson, Allan S. Kaplan, Maria La Via, James Mitchell, Michael Strober, Alessandro Rotondo, Janet Treasure, D. Blake Woodside, Cynthia M. Bulik, Pamela Keel, Kelly L. Klump, Lisa Lilenfeld, Laura M. Thornton, Kathy Plotnicov, Andrew W. Bergen, Wade Berrettini, Walter Kaye, and Pierre Magistretti.
Footnotes
Conflict of Interest
Dr. Bulik is a grant recipient from Shire Pharmaceuticals. None of the other authors declared conflicts of interest.
References
- 1.Cross-Disorder Group of the Psychiatric Genomics Consortium. Smoller JW, Craddock N, Kendler K, Lee PH, Neale BM, et al. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381:1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Serretti A, Fabbri C. Shared genetics among major psychiatric disorders. Lancet. 2013;381:1339–1341. doi: 10.1016/S0140-6736(13)60223-8. [DOI] [PubMed] [Google Scholar]
- 3.Anderson CA, Boucher G, Lees CW, Franke A, D’Amato M, Taylor KD, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011;43:246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gorwood P, Kipman A, Foulon C. The human genetics of anorexia nervosa. Eur J Pharmacol. 2003;480:163–170. doi: 10.1016/j.ejphar.2003.08.103. [DOI] [PubMed] [Google Scholar]
- 5.Helder SG, Collier DA. The genetics of eating disorders. Curr Top Behav Neurosci. 2011;6:157–175. doi: 10.1007/7854_2010_79. [DOI] [PubMed] [Google Scholar]
- 6.Thornton LM, Mazzeo SE, Bulik CM. The heritability of eating disorders: methods and current findings. Curr Top Behav Neurosci. 2011;6:141–156. doi: 10.1007/7854_2010_91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clarke TK, Weiss AR, Berrettini WH. The genetics of anorexia nervosa. Clin Pharmacol Ther. 2012;91:181–188. doi: 10.1038/clpt.2011.253. [DOI] [PubMed] [Google Scholar]
- 8.Treasure J, Zipfel S, Micali N, Wade T, Stice E, Claudino A, et al. Anorexia nervosa. Nature Reviews Disease Primers. 2015 doi: 10.1038/nrdp.2015.74. [DOI] [PubMed] [Google Scholar]
- 9.Wang K, Zhang H, Bloss CS, Duvvuri V, Kaye W, Schork NJ, et al. A genome-wide association study on common SNPs and rare CNVs in anorexia nervosa. Mol Psychiatry. 2011;16:949–959. doi: 10.1038/mp.2010.107. [DOI] [PubMed] [Google Scholar]
- 10.Boraska V, Franklin CS, Floyd JA, Thornton LM, Huckins LM, Southam L, et al. A genome-wide association study of anorexia nervosa. Mol Psychiatry. 2014;19:1085–1094. doi: 10.1038/mp.2013.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maes HH, Neale MC, Eaves LJ. Genetic and environmental factors in relative body weight and human adiposity. Behav Genet. 1997;27:325–351. doi: 10.1023/a:1025635913927. [DOI] [PubMed] [Google Scholar]
- 12.Hinney A, Vogel CI, Hebebrand J. From monogenic to polygenic obesity: recent advances. Eur Child Adolesc Psychiatry. 2010;19:297–310. doi: 10.1007/s00787-010-0096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Willer CJ, Speliotes EK, Loos RJ, Li S, Lindgren CM, Heid IM, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walters RG, Jacquemont S, Valsesia A, de Smith AJ, Martinet D, Andersson J, et al. A new highly penetrant form of obesity due to deletions on chromosome 16p11.2. Nature. 2010;463:671–675. doi: 10.1038/nature08727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacquemont S, Reymond A, Zufferey F, Harewood L, Walters RG, Kutalik Z, et al. Mirror extreme BMI phenotypes associated with gene dosage at the chromosome 16p11.2 locus. Nature. 2011;478:97–102. doi: 10.1038/nature10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huszar D, Lynch CA, Fairchild-Huntress V, Dunmore JH, Fang Q, Berkemeier LR, et al. Targeted disruption of the melanocortin-4 receptor results in obesity in mice. Cell. 1997;88:131–141. doi: 10.1016/s0092-8674(00)81865-6. [DOI] [PubMed] [Google Scholar]
- 18.Dempfle A, Hinney A, Heinzel-Gutenbrunner M, Raab M, Geller F, Gudermann T, et al. Large quantitative effect of melanocortin-4 receptor gene mutations on body mass index. J Med Genet. 2004;41:795–800. doi: 10.1136/jmg.2004.018614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heid IM, Jackson AU, Randall JC, Winkler TW, Qi L, Steinthorsdottir V, et al. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat Genet. 2010;42:949–960. doi: 10.1038/ng.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Randall JC, Winkler TW, Kutalik Z, Berndt SI, Jackson AU, Monda KL, et al. Anorexia nervosa viewed as an extreme weight condition: genetic implications. Hum Genet. 1995;95:1–11. doi: 10.1007/BF00225065. [DOI] [PubMed] [Google Scholar]
- 21.Shungin D, Winkler TW, Croteau-Chonka DC, Ferreira T, Locke AE, et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518:187–196. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winkler TW, Justice AE, Graff M, Barata L, Feitosa MF, Chu S, et al. The influence of age and sex on genetic associations with adult body size and shape: a large-scale genome-wide interaction study. PLoS Genet. 2015;11:e1005378. doi: 10.1371/journal.pgen.1005378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hebebrand J, Remschmidt H. Anorexia nervosa viewed as an extreme weight condition: genetic implications. Hum Genet. 1995;95:1–11. doi: 10.1007/BF00225065. [DOI] [PubMed] [Google Scholar]
- 24.Hinney A, Friedel S, Remschmidt H, Hebebrand J. Genetic risk factors in eating disorders. Am J Pharmacogenomics. 2004;4:209–223. doi: 10.2165/00129785-200404040-00001. [DOI] [PubMed] [Google Scholar]
- 25.Pinheiro AP, Sullivan PF, Bacaltchuck J, Prado-Lima PA, Bulik CM. Genetics in eating disorders: extending the boundaries of research. Rev Bras Psiquiatr. 2006;28:218–225. doi: 10.1590/s1516-44462006000300015. [DOI] [PubMed] [Google Scholar]
- 26.Sulek S, Lacinová Z, Dolinková M, Haluzik M. Genetic polymorphisms as a risk factor for anorexia nervosa. Prague Med Rep. 2007;108:215–225. [PubMed] [Google Scholar]
- 27.Day J, Ternouth A, Collier DA. Eating disorders and obesity: two sides of the same coin? Epidemiol Psichiatr Soc. 2009;18:96–100. [PubMed] [Google Scholar]
- 28.Scherag A, Dina C, Hinney A, Vatin V, Scherag S, Vogel CI, et al. Two new Loci for body-weight regulation identified in a joint analysis of genome-wide association studies for early-onset extreme obesity in French and German study groups. PLoS Genet. 2010;6:e1000916. doi: 10.1371/journal.pgen.1000916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gervasini G, Gamero-Villarroel C. Discussing the putative role of obesity-associated genes in the etiopathogenesis of eating disorders. Pharmacogenomics. 2015;16:1287–1305. doi: 10.2217/pgs.15.77. [DOI] [PubMed] [Google Scholar]
- 30.Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, et al. ReproGen Consortium, Psychiatric Genomics Consortium. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fall T, Ingelsson E. Genome-wide association studies of obesity and metabolic syndrome. Mol Cell Endocrinol. 2014;382:740–757. doi: 10.1016/j.mce.2012.08.018. [DOI] [PubMed] [Google Scholar]
- 32.Guo Y, Lanktree MB, Taylor KC, Hakonarson H, Lange LA, Keating BJ, et al. Gene-centric meta-analyses of 108 912 individuals confirm known body mass index loci and reveal three novel signals. Hum Mol Genet. 2013;22:184–201. doi: 10.1093/hmg/dds396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coners H, Remschmidt H, Hebebrand J. The relationship between premorbid body weight, weight loss, and weight at referral in adolescent patients with anorexia nervosa. Int J Eat Disord. 1999;26:171–8. doi: 10.1002/(sici)1098-108x(199909)26:2<171::aid-eat6>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 34.Föcker M, Bühren K, Timmesfeld N, Dempfle A, Knoll S, Schwarte R, et al. The relationship between premorbid body weight and weight at referral, at discharge and at 1-year follow-up in anorexia nervosa. Eur Child Adolesc Psychiatry. 2015;24:537–544. doi: 10.1007/s00787-014-0605-0. [DOI] [PubMed] [Google Scholar]
- 35.Hebebrand J. Identification of determinants of referral and follow-up body mass index of adolescent patients with anorexia nervosa: evidence for the role of premorbid body weight. Eur Child Adolesc Psychiatry. 2015;24:471–475. doi: 10.1007/s00787-015-0711-7. [DOI] [PubMed] [Google Scholar]
- 36.Hebebrand J, Himmelmann GW, Herzog W, Herpertz-Dahlmann BM, Steinhausen HC, Amstein M, et al. Prediction of low body weight at long-term follow-up in acute anorexia nervosa by low body weight at referral. Am J Psychiatry. 1997;154:566–569. doi: 10.1176/ajp.154.4.566. [DOI] [PubMed] [Google Scholar]
- 37.Keski-Rahkonen A, Raevuori A, Bulik CM, Hoek HW, Rissanen A, Kaprio J. Factors associated with recovery from anorexia nervosa: a population-based study. Int J Eat Disord. 2014;47:117–123. doi: 10.1002/eat.22168. [DOI] [PubMed] [Google Scholar]
- 38.Steinhausen HC, Jensen CM. Time trends in lifetime incidence rates of first-time diagnosed anorexia nervosa and bulimia nervosa across 16 years in a danish nationwide psychiatric registry study. Int J Eat Disord. 2015;48:845–850. doi: 10.1002/eat.22402. [DOI] [PubMed] [Google Scholar]
- 39.Knoll S, Föcker M, Hebebrand J. Clinical problems encountered in the treatment of adolescents with anorexia nervosa. Z Kinder Jugendpsychiatr Psychother. 2013;41:433–446. doi: 10.1024/1422-4917/a000259. [DOI] [PubMed] [Google Scholar]
- 40.Bradfield JP, Taal HR, Timpson NJ, Scherag A, Lecoeur C, Warrington NM, et al. A genome-wide association meta-analysis identifies new childhood obesity loci. Nat Genet. 2012;44:526–531. doi: 10.1038/ng.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yazdi FT, Clee SM, Meyre D. Obesity genetics in mouse and human: back and forth, and back again. PeerJ. 2015;3:e856. doi: 10.7717/peerj.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Comuzzie AG, Cole SA, Laston SL, Voruganti VS, Haack K, Gibbs RA, et al. Novel genetic loci identified for the pathophysiology of childhood obesity in the Hispanic population. PLoS One. 2012;7:e51954. doi: 10.1371/journal.pone.0051954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Müller TD, Müller A, Yi CX, Habegger KM, Meyer CW, Gaylinn BD, et al. The orphan receptor Gpr83 regulates systemic energy metabolism via ghrelin-dependent and ghrelin-independent mechanisms. Nat Commun. 2013;4:1968. doi: 10.1038/ncomms2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Müller TD, Föcker M, Holtkamp K, Herpertz-Dahlmann B, Hebebrand J. Leptin-mediated neuroendocrine alterations in anorexia nervosa: somatic and behavioral implications. Child Adolesc Psychiatr Clin N Am. 2009;18:117–129. doi: 10.1016/j.chc.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 45.Hebebrand J, Müller TD, Holtkamp K, Herpertz-Dahlmann B. The role of leptin in anorexia nervosa: clinical implications. Mol Psychiatry. 2007;12:23–35. doi: 10.1038/sj.mp.4001909. [DOI] [PubMed] [Google Scholar]
- 46.Müller TD, Nogueiras R, Andermann ML, Andrews ZB, Anker SD, Argente J, et al. Ghrelin. Mol Metab. 2015;4:437–460. doi: 10.1016/j.molmet.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tong M, Brugeaud A, Edge AS. Regenerated synapses between postnatal hair cells and auditory neurons. J Assoc Res Otolaryngol. 2013;14:321–329. doi: 10.1007/s10162-013-0374-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shieh PB, Hu SC, Bobb K, Timmusk T, Ghosh A. Identification of a signaling pathway involved in calcium regulation of BDNF expression. Neuron. 1998;20:727–740. doi: 10.1016/s0896-6273(00)81011-9. [DOI] [PubMed] [Google Scholar]
- 49.Shieh PB, Ghosh A. Molecular mechanisms underlying activity-dependent regulation of BDNF expression. J Neurobiol. 1999;41:127–134. [PubMed] [Google Scholar]
- 50.Finkbeiner S. Calcium regulation of the brain-derived neurotrophic factor gene. Cell Mol Life Sci. 2000;57:394–401. doi: 10.1007/PL00000701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xia Z, Storm DR. CaRF: a neuronal transcription factor that CaREs. Neuron. 2002;33:315–316. doi: 10.1016/s0896-6273(02)00578-0. [DOI] [PubMed] [Google Scholar]
- 52.Tao X, West AE, Chen WG, Corfas G, Greenberg ME. A calcium-responsive transcription factor, CaRF, that regulates neuronal activity-dependent expression of BDNF. Neuron. 2002;33:383–395. doi: 10.1016/s0896-6273(01)00561-x. [DOI] [PubMed] [Google Scholar]
- 53.Williams EJ, Walsh FS, Doherty P. The FGF receptor uses the endocannabinoid signaling system to couple to an axonal growth response. J Cell Biol. 2003;160:481–486. doi: 10.1083/jcb.200210164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Merighi A, Bardoni R, Salio C, Lossi L, Ferrini F, Prandini M, et al. Presynaptic functional trkB receptors mediate the release of excitatory neurotransmitters from primary afferent terminals in lamina II (substantia gelatinosa) of postnatal rat spinal cord. Dev Neurobiol. 2008;68:457–475. doi: 10.1002/dneu.20605. [DOI] [PubMed] [Google Scholar]
- 55.Singer W, Panford-Walsh R, Watermann D, Hendrich O, Zimmermann U, Köpschall I, et al. Salicylate alters the expression of calcium response transcription factor 1 in the cochlea: implications for brain-derived neurotrophic factor transcriptional regulation. Mol Pharmacol. 2008;73:1085–1091. doi: 10.1124/mol.107.041814. [DOI] [PubMed] [Google Scholar]
- 56.McDowell KA, Hutchinson AN, Wong-Goodrich SJ, Presby MM, Su D, Rodriguiz RM, et al. Reduced cortical BDNF expression and aberrant memory in Carf knock-out mice. J Neurosci. 2010;30:7453–7465. doi: 10.1523/JNEUROSCI.3997-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pfenning AR, Kim TK, Spotts JM, Hemberg M, Su D, West AE. Genome-wide identification of calcium-response factor (CaRF) binding sites predicts a role in regulation of neuronal signaling pathways. PLoS One. 2010;5:e10870. doi: 10.1371/journal.pone.0010870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alboni S, Benatti C, Capone G, Corsini D, Caggia F, Tascedda F, et al. Time-dependent effects of escitalopram on brain derived neurotrophic factor (BDNF) and neuroplasticity related targets in the central nervous system of rats. Eur J Pharmacol. 2010;643:180–187. doi: 10.1016/j.ejphar.2010.06.028. [DOI] [PubMed] [Google Scholar]
- 59.West AE. Biological functions and transcriptional targets of CaRF in neurons. Cell Calcium. 2011;49:290–295. doi: 10.1016/j.ceca.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lyons MR, Schwarz CM, West AE. Members of the myocyte enhancer factor 2 transcription factor family differentially regulate Bdnf transcription in response to neuronal depolarization. J Neurosci. 2012;32:12780–12785. doi: 10.1523/JNEUROSCI.0534-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Calabrese F, Guidotti G, Middelman A, Racagni G, Homberg J, Riva MA. Lack of serotonin transporter alters BDNF expression in the rat brain during early postnatal development. Mol Neurobiol. 2013;48:244–256. doi: 10.1007/s12035-013-8449-z. [DOI] [PubMed] [Google Scholar]
- 62.Calabrese F, Guidotti G, Racagni G, Riva MA. Reduced neuroplasticity in aged rats: a role for the neurotrophin brain-derived neurotrophic factor. Neurobiol Aging. 2013;34:2768–2776. doi: 10.1016/j.neurobiolaging.2013.06.014. [DOI] [PubMed] [Google Scholar]
- 63.Ji JF, Ji SJ, Sun R, Li K, Zhang Y, Zhang LY, et al. Forced running exercise attenuates hippocampal neurogenesis impairment and the neurocognitive deficits induced by whole-brain irradiation via the BDNF-mediated pathway. Biochem Biophys Res Commun. 2014;443:646–651. doi: 10.1016/j.bbrc.2013.12.031. [DOI] [PubMed] [Google Scholar]
- 64.Hinney A, Volckmar AL, Antel J. Genes and the hypothalamic control of metabolism in humans. Best Pract Res Clin Endocrinol Metab. 2014;28:635–647. doi: 10.1016/j.beem.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 65.Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pelleymounter MA, Cullen MJ, Wellman CL. Characteristics of BDNF-induced weight loss. Exp Neurol. 1995;131:229–238. doi: 10.1016/0014-4886(95)90045-4. [DOI] [PubMed] [Google Scholar]
- 67.Bariohay B, Lebrun B, Moyse E, Jean A. Brain-derived neurotrophic factor plays a role as an anorexigenic factor in the dorsal vagal complex. Endocrinology. 2005;146:5612–5620. doi: 10.1210/en.2005-0419. [DOI] [PubMed] [Google Scholar]
- 68.Gotoh K, Masaki T, Chiba S, Ando H, Fujiwara K, Shimasaki T, et al. Brain-derived neurotrophic factor, corticotropin-releasing factor, and hypothalamic neuronal histamine interact to regulate feeding behavior. J Neurochem. 2013;125:588–598. doi: 10.1111/jnc.12213. [DOI] [PubMed] [Google Scholar]
- 69.Gelegen C, van den Heuvel J, Collier DA, Campbell IC, Oppelaar H, Hessel E, et al. Dopaminergic and brain-derived neurotrophic factor signalling in inbred mice exposed to a restricted feeding schedule. Genes Brain Behav. 2008;7:552–559. doi: 10.1111/j.1601-183X.2008.00394.x. [DOI] [PubMed] [Google Scholar]
- 70.Charrier C, Chigr F, Tardivel C, Mahaut S, Jean A, Najimi M, et al. BDNF regulation in the rat dorsal vagal complex during stress-induced anorexia. Brain Res. 2006;1107:52–57. doi: 10.1016/j.brainres.2006.05.099. [DOI] [PubMed] [Google Scholar]
- 71.An JJ, Liao GY, Kinney CE, Sahibzada N, Xu B. Discrete BDNF Neurons in the Paraventricular Hypothalamus Control Feeding and Energy Expenditure. Cell Metab. 2015;22:175–188. doi: 10.1016/j.cmet.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ribasés M, Gratacòs M, Armengol L, de Cid R, Badía A, Jiménez L, et al. Met66 in the brain-derived neurotrophic factor (BDNF) precursor is associated with anorexia nervosa restrictive type. Mol Psychiatry. 2003;8:745–751. doi: 10.1038/sj.mp.4001281. [DOI] [PubMed] [Google Scholar]
- 73.Ribasés M, Gratacòs M, Fernández-Aranda F, Bellodi L, Boni C, Anderluh M, et al. Association of BDNF with anorexia, bulimia and age of onset of weight loss in six European populations. Hum Mol Genet. 2004;13:1205–1212. doi: 10.1093/hmg/ddh137. [DOI] [PubMed] [Google Scholar]
- 74.Ribasés M, Gratacòs M, Fernández-Aranda F, Bellodi L, Boni C, Anderluh M, et al. Association of BDNF with restricting anorexia nervosa and minimum body mass index: a family-based association study of eight European populations. Eur J Hum Genet. 2005;13:428–434. doi: 10.1038/sj.ejhg.5201351. [DOI] [PubMed] [Google Scholar]
- 75.de Krom M, Bakker SC, Hendriks J, van Elburg A, Hoogendoorn M, Verduijn W, et al. Polymorphisms in the brain-derived neurotrophic factor gene are not associated with either anorexia nervosa or schizophrenia in Dutch patients. Psychiatr Genet. 2005;15:81. doi: 10.1097/00041444-200506000-00003. [DOI] [PubMed] [Google Scholar]
- 76.Friedel S, Horro FF, Wermter AK, Geller F, Dempfle A, Reichwald K, et al. Mutation screen of the brain derived neurotrophic factor gene (BDNF): identification of several genetic variants and association studies in patients with obesity, eating disorders, and attention-deficit/hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet. 2005;132B:96–99. doi: 10.1002/ajmg.b.30090. [DOI] [PubMed] [Google Scholar]
- 77.Dardennes RM, Zizzari P, Tolle V, Foulon C, Kipman A, Romo L, et al. Family trios analysis of common polymorphisms in the obestatin/ghrelin, BDNF and AGRP genes in patients with Anorexia nervosa: association with subtype, body-mass index, severity and age of onset. Psychoneuroendocrinology. 2007;32:106–113. doi: 10.1016/j.psyneuen.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 78.Rybakowski F, Dmitrzak-Weglarz M, Szczepankiewicz A, Skibinska M, Slopien A, Rajewski A, Hauser J. Brain derived neurotrophic factor gene Val66Met and −270C/T polymorphisms and personality traits predisposing to anorexia nervosa. Neuro Endocrinol Lett. 2007;28:153–158. [PubMed] [Google Scholar]
- 79.Dmitrzak-Weglarz M, Skibinska M, Slopien A, Szczepankiewicz A, Rybakowski F, Kramer L, et al. BDNF Met66 allele is associated with anorexia nervosa in the Polish population. Psychiatr Genet. 2007;17:245–246. doi: 10.1097/YPG.0b013e3280991229. [DOI] [PubMed] [Google Scholar]
- 80.Slof-Op ‘t Landt MC, Meulenbelt I, Bartels M, Suchiman E, Middeldorp CM, Houwing-Duistermaat JJ, et al. Association study in eating disorders: TPH2 associates with anorexia nervosa and self-induced vomiting. Genes Brain Behav. 2011;10:236–243. doi: 10.1111/j.1601-183X.2010.00660.x. [DOI] [PubMed] [Google Scholar]
- 81.Ando T, Ishikawa T, Hotta M, Naruo T, Okabe K, Nakahara T, et al. No association of brain-derived neurotrophic factor Val66Met polymorphism with anorexia nervosa in Japanese. Am J Med Genet B Neuropsychiatr Genet. 2012;159B:48–52. doi: 10.1002/ajmg.b.32000. [DOI] [PubMed] [Google Scholar]
- 82.Brandys MK, Kas MJ, van Elburg AA, Ophoff R, Slof-Op’t Landt MC, et al. The Val66Met polymorphism of the BDNF gene in anorexia nervosa: new data and a meta-analysis. World J Biol Psychiatry. 2013;14:441–451. doi: 10.3109/15622975.2011.605470. [DOI] [PubMed] [Google Scholar]
- 83.Pjetri E, Dempster E, Collier DA, Treasure J, Kas MJ, Mill J, et al. Quantitative promoter DNA methylation analysis of four candidate genes in anorexia nervosa: a pilot study. J Psychiatr Res. 2013;47:280–282. doi: 10.1016/j.jpsychires.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 84.Gamero-Villarroel C, Gordillo I, Carrillo JA, García-Herráiz A, Flores I, Jiménez M, et al. BDNF genetic variability modulates psychopathological symptoms in patients with eating disorders. Eur Child Adolesc Psychiatry. 2014;23:669–679. doi: 10.1007/s00787-013-0495-6. [DOI] [PubMed] [Google Scholar]
- 85.Nakazato M, Hashimoto K, Shimizu E, Kumakiri C, Koizumi H, Okamura N, et al. Decreased levels of serum brain-derived neurotrophic factor in female patients with eating disorders. Biol Psychiatry. 2003;54:485–490. doi: 10.1016/s0006-3223(02)01746-8. [DOI] [PubMed] [Google Scholar]
- 86.Nakazato M, Hashimoto K, Yoshimura K, Hashimoto T, Shimizu E, Iyo M. No change between the serum brain-derived neurotrophic factor in female patients with anorexia nervosa before and after partial weight recovery. Prog Neuropsychopharmacol Biol Psychiatry. 2006;30:1117–1121. doi: 10.1016/j.pnpbp.2006.04.017. [DOI] [PubMed] [Google Scholar]
- 87.Nakazato M, Tchanturia K, Schmidt U, Campbell IC, Treasure J, Collier DA, et al. Brain-derived neurotrophic factor (BDNF) and set-shifting in currently ill and recovered anorexia nervosa (AN) patients. Psychol Med. 2009;39:1029–1035. doi: 10.1017/S0033291708004108. [DOI] [PubMed] [Google Scholar]
- 88.Nakazato M, Hashimoto K, Shimizu E, Niitsu T, Iyo M. Possible involvement of brain-derived neurotrophic factor in eating disorders. IUBMB Life. 2012;64:355–361. doi: 10.1002/iub.1012. [DOI] [PubMed] [Google Scholar]
- 89.Monteleone P, Tortorella A, Martiadis V, Serritella C, Fuschino A, Maj M. Opposite changes in the serum brain-derived neurotrophic factor in anorexia nervosa and obesity. Psychosom Med. 2004;66:744–748. doi: 10.1097/01.psy.0000138119.12956.99. [DOI] [PubMed] [Google Scholar]
- 90.Monteleone P, Fabrazzo M, Martiadis V, Serritella C, Pannuto M, Maj M. Circulating brain-derived neurotrophic factor is decreased in women with anorexia and bulimia nervosa but not in women with binge-eating disorder: relationships to co-morbid depression, psychopathology and hormonal variables. Psychol Med. 2005;35:897–905. doi: 10.1017/s0033291704003368. [DOI] [PubMed] [Google Scholar]
- 91.Monteleone P, Maj M. Dysfunctions of leptin, ghrelin, BDNF and endocannabinoids in eating disorders: beyond the homeostatic control of food intake. Psychoneuroendocrinology. 2013;38:312–330. doi: 10.1016/j.psyneuen.2012.10.021. [DOI] [PubMed] [Google Scholar]
- 92.Mercader JM, Fernández-Aranda F, Gratacòs M, Ribasés M, Badía A, Villarejo C, et al. Blood levels of brain-derived neurotrophic factor correlate with several psychopathological symptoms in anorexia nervosa patients. Neuropsychobiology. 2007;56:185–190. doi: 10.1159/000120623. [DOI] [PubMed] [Google Scholar]
- 93.Mercader JM, Ribasés M, Gratacòs M, González JR, Bayés M, de Cid R, et al. Altered brain-derived neurotrophic factor blood levels and gene variability are associated with anorexia and bulimia. Genes Brain Behav. 2007;6:706–716. doi: 10.1111/j.1601-183X.2007.00301.x. [DOI] [PubMed] [Google Scholar]
- 94.Mercader JM, Fernández-Aranda F, Gratacòs M, Aguera Z, Forcano L, Ribasés M, et al. Correlation of BDNF blood levels with interoceptive awareness and maturity fears in anorexia and bulimia nervosa patients. J Neural Transm. 2010;117:505–512. doi: 10.1007/s00702-010-0377-8. [DOI] [PubMed] [Google Scholar]
- 95.Ehrlich S, Salbach-Andrae H, Eckart S, Merle JV, Burghardt R, Pfeiffer E, et al. Serum brain-derived neurotrophic factor and peripheral indicators of the serotonin system in underweight and weight-recovered adolescent girls and women with anorexia nervosa. J Psychiatry Neurosci. 2009;34:323–329. [PMC free article] [PubMed] [Google Scholar]
- 96.Saito S, Watanabe K, Hashimoto E, Saito T. Low serum BDNF and food intake regulation: a possible new explanation of the pathophysiology of eating disorders. Prog Neuropsychopharmacol Biol Psychiatry. 2009;33:312–316. doi: 10.1016/j.pnpbp.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 97.Brandys MK, Kas MJ, van Elburg AA, Campbell IC, Adan RA. A meta-analysis of circulating BDNF concentrations in anorexia nervosa. World J Biol Psychiatry. 2011;12:444–454. doi: 10.3109/15622975.2011.562244. [DOI] [PubMed] [Google Scholar]
- 98.Dmitrzak-Weglarz M, Skibinska M, Slopien A, Tyszkiewicz M, Pawlak J, Maciukiewicz M, et al. Serum neurotrophin concentrations in polish adolescent girls with anorexia nervosa. Neuropsychobiology. 2013;67:25–32. doi: 10.1159/000343500. [DOI] [PubMed] [Google Scholar]
- 99.Zwipp J, Hass J, Schober I, Geisler D, Ritschel F, Seidel M, et al. Serum brain-derived neurotrophic factor and cognitive functioning in underweight, weight-recovered and partially weight-recovered females with anorexia nervosa. Prog Neuropsychopharmacol Biol Psychiatry. 2014;54:163–169. doi: 10.1016/j.pnpbp.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 100.Eddy KT, Lawson EA, Meade C, Meenaghan E, Horton SE, Misra M, et al. Appetite regulatory hormones in women with anorexia nervosa: binge-eating/purging versus restricting type. J Clin Psychiatry. 2015;76:19–24. doi: 10.4088/JCP.13m08753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rask-Andersen M, Olszewski PK, Levine AS, Schiöth HB. Molecular mechanisms underlying anorexia nervosa: focus on human gene association studies and systems controlling food intake. Brain Res Rev. 2010;62:147–164. doi: 10.1016/j.brainresrev.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 102.Rosas-Vargas H, Martínez-Ezquerro JD, Bienvenu T. Brain-derived neurotrophic factor, food intake regulation, and obesity. Arch Med Res. 2011;42:482–494. doi: 10.1016/j.arcmed.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 103.Banke E, Rödström K, Ekelund M, Dalla-Riva J, Lagerstedt JO, Nilsson S, et al. Superantigen activates the gp130 receptor on adipocytes resulting in altered adipocyte metabolism. Metabolism. 2014;63:831–840. doi: 10.1016/j.metabol.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 104.Chornokur G, Amankwah EK, Davis SN, Phelan CM, Park JY, Pow-Sang J, et al. Variation in HNF1B and Obesity May Influence Prostate Cancer Risk in African American Men: A Pilot Study. Prostate Cancer. 2013;2013:384594. doi: 10.1155/2013/384594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Guirguis E, Hockman S, Chung YW, Ahmad F, Gavrilova O, Raghavachari N, et al. A role for phosphodiesterase 3B in acquisition of brown fat characteristics by white adipose tissue in male mice. Endocrinology. 2013;154:3152–3167. doi: 10.1210/en.2012-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Vernochet C, Peres SB, Davis KE, McDonald ME, Qiang L, Wang H, et al. C/EBPalpha and the corepressors CtBP1 and CtBP2 regulate repression of select visceral white adipose genes during induction of the brown phenotype in white adipocytes by peroxisome proliferator-activated receptor gamma agonists. Mol Cell Biol. 2009;29:4714–4728. doi: 10.1128/MCB.01899-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Vernochet C, Davis KE, Scherer PE, Farmer SR. Mechanisms regulating repression of haptoglobin production by peroxisome proliferator-activated receptor-gamma ligands in adipocytes. Endocrinology. 2010;151:586–594. doi: 10.1210/en.2009-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Farmer SR. Molecular determinants of brown adipocyte formation and function. Genes Dev. 2008;22:1269–1275. doi: 10.1101/gad.1681308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kajimura S, Seale P, Tomaru T, Erdjument-Bromage H, Cooper MP, Ruas JL, et al. Regulation of the brown and white fat gene programs through a PRDM16/CtBP transcriptional complex. Genes Dev. 2008;22:1397–1409. doi: 10.1101/gad.1666108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hildebrand JD, Soriano P. Overlapping and unique roles for C-terminal binding protein 1 (CtBP1) and CtBP2 during mouse development. Mol Cell Biol. 2002;22:5296–5307. doi: 10.1128/MCB.22.15.5296-5307.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang L, Xu L, Xu M, Liu G, Xing J, Sun C, Ding H. Obesity-Associated MiR-342-3p Promotes Adipogenesis of Mesenchymal Stem Cells by Suppressing CtBP2 and Releasing C/EBPα from CtBP2 Binding. Cell Physiol Biochem. 2015;35:2285–2298. doi: 10.1159/000374032. [DOI] [PubMed] [Google Scholar]
- 112.Li Y, Yang XH, Fang SJ, Qin CF, Sun RL, Liu ZY, et al. HOXA7 stimulates human hepatocellular carcinoma proliferation through cyclin E1/CDK2. Oncol Rep. 2015;33:990–996. doi: 10.3892/or.2014.3668. [DOI] [PubMed] [Google Scholar]
- 113.Zhu HJ, Pan H, Zhang XZ, Li NS, Wang LJ, Yang HB, et al. The effect of myostatin on the proliferation and lipid accumulation in 3T3-L1 preadipocytes. J Mol Endocrinol. 2015;54:217–226. doi: 10.1530/JME-15-0038. [DOI] [PubMed] [Google Scholar]
- 114.Hadano S, Hand CK, Osuga H, Yanagisawa Y, Otomo A, Devon RS, et al. A gene encoding a putative GTPase regulator is mutated in familial amyotrophic lateral sclerosis 2. Nat Genet. 2001;29:166–173. doi: 10.1038/ng1001-166. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.