Abstract
This paper describes the design and implementation of an application that parses and analyzes radiology report text to provide a radiologic differential diagnosis. The system was constructed using a combination of freely available web-based APIs and originally developed during the Society for Imaging Informatics in Medicine (SIIM) 2014 Hackathon. Continued development has refined and increased the accuracy of the algorithm. This project demonstrates the power and possibilities of combining existing technologies to solve unique problems as well as the stimulus of the hackathon setting to spur innovation.
Keywords: Computer-aided diagnosis (CAD), Decision support, Interpretation, Web technology
Background
The synthesis of patient history, physical examination findings, laboratory, and physiologic data to identify disease is a fundamental skill of a physician. Computer-aided diagnosis (CAD) can provide a second look at the patient data for unrealized associations and potential disease states. Commercial examples of clinical decision support systems exist such as Isabel (1) and IBM Watson Health (2).
Radiology CAD systems are often focused on computer-aided analysis of the imaging data, leaving the textual interpretation and reporting to the radiologist. A correlate system could accurately detect imaging findings described in a report, draw a correlation between those findings and disease, and integrate that information with the patient history to develop a differential diagnosis. This differential could then be provided to the radiologist for diagnostic assistance.
Several resources exist to aid in the development of such a tool. RadLex (3) is a lexicon of standardized radiologic terms with ontologic associations allowing for machine understanding of anatomy, physiology, and radiologic examinations. Similarly, Gamuts (4) is a collection of disease states linking symptoms, diseases, and causes. Together, these systems form the basis for basic conceptual mapping. With the addition of automated report retrieval and text annotation, a CAD system was developed to provide a supplementary differential diagnoses from the information gathered in radiology reports.
Methods
RadDDX (previously called “FHIRgamuts”) began as a hackathon project developed at the SIIM 2014 Hackathon. Several application programming interfaces (APIs) were available for use by hackathon participants including HL7 FHIR (5) and Gamuts. In addition, the National Center for Biomedical Ontology (NCBO) Bioportal (6) provides an API to annotate text with medical ontologies including RadLex. Using these resources, the described application was created over the three-day hackathon that could consume patient history and radiology report text and output a differential of possible diagnoses.
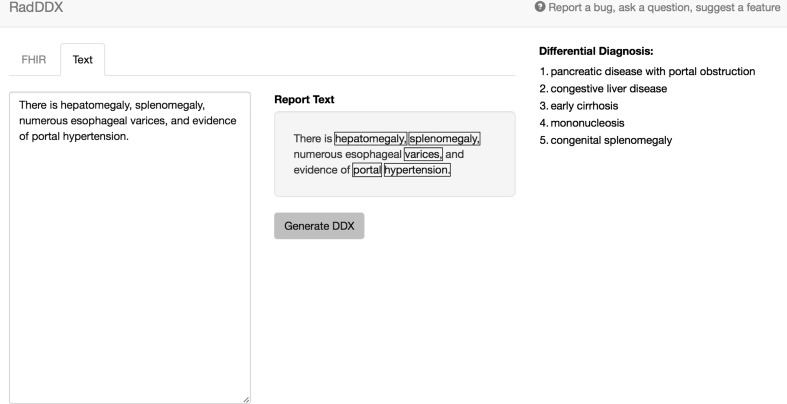
A web-based application was created using JavaScript and a NodeJS (7) framework to easily communicate with the open APIs from Bioportal, Gamuts, and HL7 FHIR. JavaScript is an interpreted programming language with a wide developer community support and extensive open-source libraries for web development. NodeJS allows both client and server coding in JavaScript with a scalable infrastructure designed for web-based application programming. The user interface was designed using Bootstrap (8), an open-source collection of HTML and cascading style sheet (CSS) tools for rapidly producing an organized, intuitive, and responsive interface (Fig. 1).
Fig. 1.
The application interface. The input text is seen at the left, with the recognized terms highlighted in the center box, and differential diagnosis listed on the right
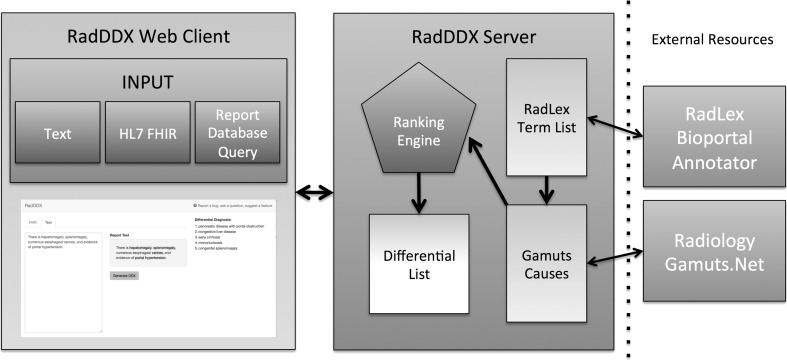
Radiology reports can be gathered automatically through HL7 FHIR requests from an electronic medical record database, by database queries, or entered directly as text into the web application. The application parses the supplied textual information and sends it to the NCBO Bioportal Annotator API to identify RadLex concepts. Gamuts is then queried with these concepts to identify any matching pathology/disease contained within the Gamuts database. Each term recognized by Gamuts is then queried for its details, which yields relationship information including synonyms, superclasses, subclasses, causes, and causal entities. This detailed information is aggregated for all of the matched terms in the input text. A ranking algorithm multiplies how many times each causal entity occurred by the inverse of the number of diseases the causal entity may cause. The causal entities are then ordered based upon rank into a differential diagnosis list and displayed for the user (Fig. 2).
Fig. 2.
The application structure. Input accepted as text, HL7 FHIR reports, or reports from a database query. Information returned from Bioportal produces a term list that is sent to Gamuts. The causal information returned from Gamuts is then entered into the ranking algorithm that produces the differential diagnosis list
Following HIPAA protocols for the protection of patient data, 50 CT abdomen and pelvis reports were obtained from our reporting database and anonymized. The RadLex concepts were annotated via a local instance of the NCBO Virtual Appliance to further ensure HIPAA compliance. The report text was used to test the accuracy of the differential diagnosis lists obtained when the “Clinical History” and “Findings” sections were submitted for analysis. The report “Impression” was used as the gold standard for comparison. To evaluate the quality of the differential diagnoses produced by the application a three-level grading scale was developed (Table 1). The results were compiled for each of the raw reports. Manual parsing of the input text was performed to remove titles (e.g., “Abdomen” or “Liver” in the text body), negations (e.g., “No biliary ductal dilation”), and normal (e.g., “The appendix is normal”). The parsed text was then input into RadDDX, and the results again compiled and compared to the report “Impression”.
Table 1.
Scoring system for evaluation of RadDDX differential results compared to the original report “Impression”
| Quality of DDX score | |
|---|---|
| High | Output includes exact match as seen in “Impression” |
| Medium | Logical differential output in similar category as “Impression” |
| Low | Output vastly different from expected |
Results
Beginning with simplified pathologic findings, the application responds appropriately with differential diagnosis lists produced by evaluation of the supplied text. For example, when given the input text, “right lower lobe consolidation,” the differential created is as follows (only top 5 listed for brevity):
Obstructive pneumonia
Bronchopneumonia
Infectious pneumonia
Localized pulmonary edema
Round pneumonia
With more complex text and increased number of matching terms, the application continues to provide an appropriately correlated differential diagnoses. For example, input text of:
“There is hepatomegaly, splenomegaly, numerous esophageal varices, and evidence of portal hypertension.”
Yields the differential diagnosis list (only top five listed for brevity):
Pancreatic disease with portal obstruction
Congestive liver disease
Early cirrhosis
Mononucelosis
Congenital splenomegaly
The complex text and concepts present in the CT abdomen and pelvis reports presented a greater challenge to our software. The results demonstrated a wider variety of differentials that were often poorly coordinated with the gold standard “Impression”. After manual parsing of the report text prior to RadDDX submission, the quality of the results improved (Table 2). An example of the differential diagnoses produced from raw report and parsed text compared to the gold standard is provided in Table 3.
Table 2.
Results of the raw report text versus the manually parsed text comparing the differential diagnosis supplied by RadDDX as compared to the original report ‘Impression’.
| Raw reports | Parsed reports | ||
|---|---|---|---|
| Scoring | High | 1 | 10 |
| Medium | 1 | 21 | |
| Low | 48 | 19 | |
Table 3.
Example of output produced from testing of a CT Chest, Abdomen, and Pelvis report.
| Raw report DDX | Parsed report DDX | Gold standard “Impression” | |
|---|---|---|---|
| DDX | 1. Contraction of choledochal sphincter | 1. Viral pneumonia | In the lower chest, there is a moderate sized pericardial effusion with enhancement of the visceral and parietal pericardium. Bilateral pleural effusions. Mild bibasilar atelectasis. Liver enhances heterogeneously with a nutmeg appearance, which may be secondary to increased right-sided heart pressures from the pericardial effusion. Mild peri-portal edema |
| 2. Air bubble | 2. Amebic pericarditis | ||
| 3. Choledocholithiasis | 3. Bacterial pericarditis | ||
| 4. Intussusception | 4. Infectious pericarditis | ||
| 5. Chiliaditi syndrome | 5. Neoplasm of the mediastinum | ||
| 6. Dysplastic nodule | |||
| 7. Compression atelectasis | |||
| 8. Contraction atelectasis | |||
| 9. Peripheral mucus plug | |||
| 10. Postoperative adhesive atelectasis | |||
| Grade | Low | High |
Discussion
RadDDX was successfully developed in the short timeframe of the SIIM Hackathon, demonstrating that unique and usable tools can be rapidly produced from existing open-source APIs. As RadLex and Gamuts continue to be developed, refined, and updated, the developed CAD system will immediately incorporate those changes, possibly becoming more accurate.
This project also shows the potential of hackathons as a tool for solving problems and using existing technologies in new or unusual ways. While this application was born without a particular problem in mind, it is not practical without applications. Potential future use cases include the following: (1) Use of the tool in real time to aid/support radiologists as they make findings and lead them to possible diagnoses, (2) Data mine old radiology reports to develop a medical “Problem List” for each patient, (3) Data mine clinical data from other sources to help summarize clinical information for radiologist use.
The use of freely available APIs allows for rapid development of the application; however, the system is also limited by/dependent upon the information contained within these resources. In addition, the application does not currently differentiate between positive or negative findings in reports so erroneous terms may be included in the differential. For example, “No evidence of appendicitis,” will still generate the term appendicitis and its causal relationships will be contained and processed by the ranking algorithm. The same scenario exists for “Normal” findings and section or organ headings used in structured reporting such as “Spleen:” or “Liver:.” This situation could be improved by development of a text parser that could eliminate negative statements, normal findings, and titles/headings from the analyses as demonstrated by our manual parsing analysis. Reports can be entered directly into the interface, but is tedious for large volume analysis. Future goals include integration with dictation software to provide differential lists on-the-fly and further refinement of the language processing engine and differential ranking algorithm to increase diagnosis accuracy.
Conclusion
Automated analysis of medical records and radiology reporting is in its infancy but the opportunity exists for computer-aided diagnosis to play a large role in assisting diagnostic interpretation. Ontologic resources such as RadLex and Gamuts are instrumental in developing language-processing applications that can accurately parse report text and identify relevant concepts and terms. Open source APIs like HL7 FHIR lower the barrier to data acquisition, analysis, and evaluation of the electronic health record. The inaugural SIIM Hackathon provided fertile ground to bring together these disparate technologies, proving that medical hackathons are ripe for creating sophisticated clinical tools.
References
- 1.Isabel Healthcare. Isabel. Accessible at: http://www.isabelhealthcare.com/. (Accessed May 11, 2015).
- 2.International Business Machines. IBM Watson Health. Accessible at: http://www.ibm.com/smarterplanet/us/en/ibmwatson/health/. (Accessed May 11, 2015).
- 3.Radiological Society of North America. RadLex. Accessible at: www.radlex.org. (Accessed on September 12, 2014).
- 4.Khan, Charles. Gamuts. Accessible at: www.gamuts.net. (Accessed on September 12, 2014).
- 5.Health Level Seven International. HL7 FHIR. Available at: http://www.hl7.org/implement/standards/fhir/. (Accessed on September 12, 2014).
- 6.Whetzel PL, Noy NF, Shah NH, et al. BioPortal: enhanced functionality via new Web services from the National Center for Biomedical Ontology to access and use ontologies in software applications. Nucleic Acids Res. 2011 Jul;39(Web Server issue):W541-5. Epub 2011 Jun 14. [DOI] [PMC free article] [PubMed]
- 7.Joyent Inc. NodeJS. Available from: http://www.nodejs.org. (Accessed on September 12, 2014).
- 8.Otto M, Thornton J. Bootstrap. Available from: http://www.getbootstrap.com. (Accessed on September 12, 2014).