Abstract
Obligate intracellular chlamydial bacteria of the Planctomycetes-Verrucomicrobia-Chlamydiae (PVC) superphylum are important pathogens of terrestrial and marine vertebrates, yet many features of their pathogenesis and host specificity are still unknown. This is particularly true for families such as the Waddliacea which, in addition to epithelia, cellular targets for nearly all Chlamydia, can infect and replicate in macrophages, an important arm of the innate immune system or in their free-living amoebal counterparts. An ideal pathogen model system should include both host and pathogen, which led us to develop the first larval zebrafish model for chlamydial infections with Waddlia chondrophila. By varying the means and sites of application, epithelial cells of the swim bladder, endothelial cells of the vasculature and phagocytosing cells of the innate immune system became preferred targets for infection in zebrafish larvae. Through the use of transgenic zebrafish, we could observe recruitment of neutrophils to the infection site and demonstrate for the first time that W. chondrophila is taken up and replicates in these phagocytic cells and not only in macrophages. Furthermore, we present evidence that myeloid differentiation factor 88 (MyD88) mediated signaling plays a role in the innate immune reaction to W. chondrophila, eventually by Toll-like receptor (TLRs) recognition. Infected larvae with depleted levels of MyD88 showed a higher infection load and a lower survival rate compared to control fish. This work presents a new and potentially powerful non-mammalian experimental model to study the pathology of chlamydial virulence in vivo and opens up new possibilities for investigation of other members of the PVC superphylum.
Keywords: zebrafish, PVC superphylum, Chlamydia, Waddlia, swim bladder infection, endothelial cells, neutrophils, MyD88
Introduction
The bacterial species Waddlia chondrophila is a purported abortifacient pathogen of cattle (Dilbeck-Robertson et al., 2003), first isolated from a cow abortion in the United States (Dilbeck et al., 1990) and subsequently from a similar case in Germany (Henning et al., 2002). The Waddliaceae is one of eight families described to date, within the phylum Chlamydiae (Collingro et al., 2011; Taylor-Brown et al., 2015), all of which are obligate intracellular pathogens able to infect a variety of hosts covering much of the animal kingdom. The best known family of this phylum is the Chlamydiaceae, classical pathogens of humans and animals, some of which are known for their high zoonotic potential and ability to cross species borders, such as Chlamydia psittaci, the agent of psittacosis in birds and humans and Chlamydia abortus, an agent of fetal death and abortion in ruminants and humans (Longbottom and Coulter, 2003). Waddlia chondrophila may similarly pose a zoonotic risk, based on evidence from serological tests and quantitative real-time PCR in cases of human miscarriage and respiratory disease (Baud et al., 2007, 2011, 2014; Haider et al., 2008; Goy et al., 2009). A marked difference to the Chlamydiaceae, however, is the additional ability of W. chondrophila to infect and replicate in phagocytic cells, including macrophages and free-living amoebae, at least in vitro (Goy et al., 2008; Lamoth and Greub, 2010). This considerably complicates experiments to uncover the infection mechanisms and routes W. chondrophila uses in vivo, adding to the intrinsic difficulty of tracing infectious processes in whole animals. This knowledge is, however, key to the design and application of effective treatment strategies for the Waddliaceae in particular and for the Chlamydiae as a whole.
In vitro data indicates that Waddlia may be able to infect quite different hosts, widening the choice available when considering model organisms. The first cultivation of W. chondrophila was achieved in bovine turbinate cells and mouse macrophages (Dilbeck et al., 1990; Kocan et al., 1990). Subsequent in vitro studies showed that W. chondrophila is potentially a highly versatile pathogen, able to infect and replicate in McCoy cells, buffalo green monkey cells, human fibroblasts (Henning et al., 2002), Vero cells, human pneumocytes and endometrial cells (Kebbi-Beghdadi et al., 2011b), as well as in human macrophages (Goy et al., 2008). During the infection of macrophages, W. chondrophila avoids degradation by successfully preventing the fusion of the endosome with a lysosome and relocating in a vacuole expressing endoplasmic reticulum (ER) markers (Croxatto and Greub, 2010). Freshwater amoebae of the genus Acanthamoeba are also susceptible to infection with W. chondrophila (Lamoth and Greub, 2010). Furthermore, W. chondrophila was recently found to be able to invade and proliferate in two fish cell lines derived from fathead minnow (Pimephales promelas; EPC-175) and rainbow trout (Oncorhynchus mykiss; RTG-2) (Kebbi-Beghdadi et al., 2011a). In addition to mammalian hosts, W. chondrophila has been isolated from aquatic environments as diverse as sediments from the eastern Mediterranean Sea (Polymenakou et al., 2005) and freshwater samples from well water sources in Spain (Codony et al., 2012). According to these findings, it has been speculated that freshwater protists and fish could potentially serve as an aquatic reservoir for W. chondrophila, that one possible transmission route is water-borne and by inference, that fish may not only be a valuable alternative model but also a natural host.
Indeed, chlamydial disease affects both marine as well as freshwater fishes causing the disease epitheliocystis (Hoffman et al., 1969; Draghi et al., 2004; Meijer et al., 2006; Stride et al., 2014) in which bacteria-filled intracellular inclusions are found infecting gill and skin epithelia. Several chlamydial agents of epitheliocystis have been described so far, some of which are closely related to the Waddliaceae such as Ca. Clavichlamydia salmonicola (Karlsen et al., 2008), Ca. Syngnamydia venezia (Fehr et al., 2013) and Ca. Syngnamydia salmonis (Nylund et al., 2015), whereas others are more distantly related such as members of the deep rooted Piscichlamydia clade, Ca. Piscichlamydia salmonis (Draghi et al., 2004; Schmidt-Posthaus et al., 2012), Ca. Parilichlamydiaceae (Stride et al., 2013a), Ca. Similichlamydiaceae (Stride et al., 2013b,c; Steigen et al., 2015; Seth-Smith et al., this issue), and Ca. Actinochlamydiaceae (Steigen et al., 2013). In a ground breaking study, Lagkouvardos and colleagues discovered up to 181 new putative families of the Chlamydiae from primarily marine and fresh water sources, of which the Waddliaceae formed a prominent clade (Lagkouvardos et al., 2014).
Expanding genomic information has greatly improved our understanding of potential mechanisms underlying host diversity and disease pathology and is an essential step for establishing a model pathogen-host system. The availability of the W. chondrophila genome provided precious information on putative virulence factors of these bacteria (Bertelli et al., 2010), however, there is a great need to develop animal models to test the ideas coming from these efforts (Bachmann et al., 2014). Ideally, such an animal model would lend itself to high throughput screening and share an immune system with close similarities to that of humans and other animal hosts. As a model organism in infection biology, the zebrafish (Danio rerio) has become increasingly popular. Many infection systems using larval and adult zebrafish have been successfully developed over the past decade (Kanther and Rawls, 2010; Meijer and Spaink, 2011; Fehr et al., 2015). Its small size, ease of breeding, high fertility and genetic tractability combined with transparent larval stages combine to make it an attractive model organism for science. The zebrafish immune system displays many similarities to that of mammals, with counterparts for most of the human immune cell types (Meeker and Trede, 2008) and have conserved mechanisms for the recognition of microbes like Toll-like receptors (TLRs) (Hall et al., 2009). The zebrafish innate immune system starts to develop as early as 24 h post fertilization (hpf) with a population of primitive macrophages which derive from cells located in a region of the yolk sac near the heart (Herbomel et al., 1999). At 2 days post fertilization (dpf), subpopulations of macrophages can already be observed throughout the organism along with neutrophils whose development initiates between 32 and 48 hpf. The development of the adaptive immune system lags behind, with the first lymphocytes observed from 4 dpf, although fully developed adaptive immunity takes another 4 weeks to mature (Meijer and Spaink, 2011). For this reason, it is possible to exclusively observe the reaction of the innate immune system within an infection during the first week of larval development. Previous studies have proposed that recognition by TLRs could mediate an efficient immune reaction against chlamydial infection leading to bacterial clearance (Naiki et al., 2005), while in other cases TLR dependent recruitment of innate immune cells had an adverse effect by enhancing the bacterial load during an infection (Rodriguez et al., 2005). The activation of TLRs initiates an inflammatory response through signaling cascades that lead to cytokine production which promote recruitment of leukocytes to the infection site and phagocytosis of invading pathogens (Newton and Dixit, 2012). A key factor for TLR signal transduction is the downstream adaptor molecule MyD88, which interacts with all known TLRs and members of the interleukin-1 receptor (IL-1R) family, resulting in the induction of nuclear factor-kappaB (NF-kB) and mitogen-activated protein kinase (MAPK) signaling (Medzhitov et al., 1998; Warner and Nunez, 2013).
Zebrafish larvae have been used to study infections of many different bacterial pathogens like Mycobacterium marinum, Salmonella typhimurium, Vibrio anguillarum, Listeria monocytogenes, Pseudomonas aeruginosa, Burkholderia cenocepacia, Staphylococcus aureus, Streptococcus iniae, Shigella flexneri, and Cronobacter turicensis (Herbomel et al., 1999; Davis et al., 2002; van der Sar et al., 2003; O'Toole et al., 2004; Brannon et al., 2009; Clatworthy et al., 2009; Levraud et al., 2009; Vergunst et al., 2010; Adams et al., 2011; Prajsnar et al., 2012; Harvie et al., 2013; Mostowy et al., 2013; Fehr et al., 2015). However, none of these pathogens are obligate intracellular bacteria. By developing an infection model with W. chondrophila, we demonstrate the first zebrafish infection model for an obligate intracellular pathogen and member of the Chlamydiae. We established alternate routes of infection and analyzed preferred target cells for Waddlia infection in vivo and have taken initial steps to elucidate molecular mechanisms regulating infection, including the impact of MyD88 signaling in knockdown larvae.
Materials and methods
Zebrafish strains and husbandry
Zebrafish (D. rerio) strains used in this study were predominantly albino mutants (slc45a2b4/+) and transgenic fish of the Tg(lyz:DsRED2)nz50 line that produce red fluorescent protein in cells of the myelomonocytic lineage able to migrate to inflammatory sites and phagocytose bacteria (Hall et al., 2007), primarily neutrophils from 50 h post fertilization (hpf) (Clatworthy et al., 2009). In addition, the Tg(fli1a:eGFP) line which produces green fluorescent protein in endothelial cells, was used to visualize the vascular system (Lawson and Weinstein, 2002). Adult fish were kept at a 14/10 h light/dark cycle at a pH of 7.5 and 27°C. Eggs were obtained from natural spawning between adult fish which were set up pairwise in separate breeding tanks. Embryos were raised in petri dishes with E3 medium (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4) containing 0.3 μg/ml of methylene blue at 28°C. From 24 hpf, 0.003% 1-phenyl-2-thiourea (PTU) was added to prevent melanin synthesis. The albino mutants lack melanised melanophores, and for these PTU treatment was not necessary. Staging of embryos was performed according to Kimmel et al. (1995).
Research was conducted with ethics approval (no. 216/2012) from the animal research ethics committee of the Veterinary Office, Public Health Department, Canton of Zurich (Switzerland). Larvae were maintained up until 7 days post fertilization (dpf), at which time all were euthanized by applying an overdose of 4 g/l buffered tricaine (MS-222, Ethyl 3-aminobenzoate methanesulfonate, Sigma-Aldrich) in accordance with ethical procedures.
Bacterial cultures
Waddlia chondrophila strain WSU 86-1044 (ATCC VR-1470) were grown within Acanthamoeba castellanii strain ATCC 30010 in 25 cm2 cell culture flasks containing 10 ml of peptone yeast extract glucose (PYG) broth (Greub and Raoult, 2002). Cultures were harvested after 5 days and filtered through a 5 μm membrane to remove remaining amoebae. The flow-through was centrifuged at 7000 g for 15 min. The resulting pellet of bacterial elementary bodies (EBs) was then suspended in E3 medium for bath immersion or phosphate buffered saline (PBS) for microinjection experiments. The infectivity of W. chondrophila was tested in Epithelioma Papulosum Cyprini (EPC) cells, to investigate the infection process and morphology of the bacteria in these fish epithelial cells (Supplementary Material Figure S1). Subsequently, Inclusion Forming Units (IFU) of the cultures were determined by infecting monolayers of EPC cells in 24-well plates with a 10-fold dilution series of 1 μl of the bacterial suspension at 28°C. After 24 h cells were fixed with 4% paraformaldehyde and subsequently stained with a primary rabbit anti-Waddlia antibody and detected with a secondary goat anti-rabbit-IgG conjugated to a fluorescent AlexaFluor dye. After imaging with a fluorescent microscope, inclusions were counted in Imaris (Bitplane) to calculate a mean IFU value.
Bath immersion experiments
Zebrafish embryos between 24 hpf and 4 dpf were incubated in groups of 15 for each time point and each condition in 24-well plates with E3 medium containing 2 × 109 IFU/ml of W. chondrophila at 28°C. Embryos younger than 48 hpf were manually dechorionated prior to immersion. After 4 h of incubation embryos were washed twice in fresh E3 medium and transferred to 6-well plates containing 4 ml of E3 medium per well and further incubated at 28°C. Embryos were then observed under a binocular microscope for signs of disease and survival twice a day. At several time points embryos from each group were euthanized in E3 medium containing 4 mg/ml buffered tricaine (MS-222).
Microinjection experiments
For microinjections of W. chondrophila into zebrafish larvae bacteria were first harvested from a 5 days old amoebal co-culture as described above. The concentration of the W. chondrophila EBs was adjusted to 1000–2000 IFU/nl in PBS and 0.085% phenol red was added to visualize the injection procedure. Injections were done using pulled borosilicate glass microcapillary injection needles (Science Products, 1210332, 1 mm O.D. × 0.78 mm I.D.) and a PV830 Pneumatic PicoPump (World Precision Instruments). Prior to intravenious injections embryos of 2 dpf were manually dechorionated and anesthetised with 200 mg/l buffered tricaine (MS-222). Afterwards embryos were aligned on an agar plate and injected with 1 nl of the W. chondrophila suspension into the Duct of Cuvier, also known as common cardinal vein. Prior determination of the injected volume was performed by injection of a droplet into mineral oil and measurement of its approx. diameter over a scale bar. For swim bladder injections 4 dpf larvae were treated similarly but dechorionisation was not necessary because larvae were already hatched. 4 dpf larvae were then injected with a volume of 2 nl into the lumen of the swim bladder. After injections, infected larvae were allowed to recover in a petri dish with fresh E3 medium for 15 min. Subsequently, larvae were transferred in 6-well plates in groups of about 15 larvae in 4 ml E3 medium per well, incubated at 28°C and observed under a stereomicroscope twice a day. Samples for Immunofluorescence (IF), Transmission Electron Microscopy (TEM) and quantitative Polymerase Chain Reaction (qPCR) were taken at 0, 12, 24, 36, 48, 60 and 72 h post infection (hpi). Sampled larvae were euthanized with an overdose of 4 g/l buffered tricaine and transferred into different buffers and fixatives for subsequent analyses respectively.
Whole mount immunofluorescence (IF) and histological stainings
For IF stainings, whole zebrafish larvae were fixed in 4 % paraformaldehyde at 4°C overnight followed by 100 % methanol overnight at −20°C. Samples were rehydrated in 50% methanol for 5 min and subsequently in H2O for 1 h before blocking in PBDT (PBS containing 1% BSA, 1% DMSO, 0.5% Triton X-100, 2.5% goat serum) for 6–8 h at room temperature. Larvae were then incubated with primary antibody overnight at 4°C. Primary antibody was detected by incubation with a secondary goat anti-IgG antibody conjugated to a fluorescent AlexaFluor dye (Life Technologies) at 4°C overnight. Additionally 4′-6-diamin-2-phenylindole (DAPI) was added to visualize bacterial and host DNA. Stained larvae were prepared for microscopy on objective slides mounted in 1.5% agarose, 50% glycerol and screened under a fluorescence microscope. Positive samples were subsequently screened in more detail for W. chondrophila inclusions by Confocal Laser Scanning Microscopy (CLSM).
For histological examination, whole zebrafish larvae were fixed in 4% paraformaldehyde at 4°C and embedded in cubes of cooked egg white in order to position them correctly for histological sections. These cubes containing the larvae were dehydrated in an ascending alcohol series ending in xylene and afterwards embedded in paraffin. Paraffin blocks were cut in 2–3 μm thin sections, mounted on glass slides and stained using a routine protocol for haematoxylin and eosin (HE) staining.
Light-microscopy and image analysis
Overview images were taken with an upright light microscope (Olympus BX61) with both bright field and fluorescence modules. The fluorescence filter cube used was optimized for DAPI/FITC/TRIC. For higher resolution images, 3D-image stacks of whole mount samples were prepared using CLSM (Leica TCS SP5, Leica Microsystems). Various combinations of the fluorophors AlexaFluor dyes 594, 546, 488, GFP, dsRED, and DAPI were sequentially excited in descending series with the 594, 561, 488, and 405 nm laser lines, with emission signals collected within the respective range of wave lengths. Three-dimensional image stacks were collected sequentially (to prevent blue-green–red channel cross-talk) and according to Nyquist criteria and deconvolved using HuygensPro via the Huygens Remote Manager v2.1.2 (SVI, Netherlands). Images were further analyzed with Imaris 7.6.1 (Bitplane, Zurich, Switzerland) and aligned with Adobe Photoshop Elements 12. Fluorescent cells were quantified with the Imaris fluorescent spot counting tool.
Transmission electron microscopy
For electron microscopy, larvae were fixed in a mixed solution of 1% paraformaldehyde and 2.5% glutaraldehyde in 0.1 M sodium phosphate buffer, pH 7.5 at 4°C overnight. Samples were prepared by embedding into epoxy resin and for TEM microscopy according to standard procedures. Epoxy resin blocks were screened for larval location using semithin sections (1 μm) which were stained with toluidine blue (Sigma-Aldrich) to visualize tissue. Ultrathin sections (80 nm) were mounted on copper grids (Merck Eurolab AG, Dietlikon, Switzerland), contrasted with uranyl acetate dihydrate (Sigma-Aldrich) and lead citrate (Merck Eurolab AG) and investigated using a Philips CM10 transmission electron microscope. Images were processed with Imaris (Bitplane) and assembled for publication using Adobe Photoshop.
Quantitative PCR
A qPCR system was designed against the 16S rRNA gene from W. chondrophila (forward: 5′- AGTCCGGCTACACCAAGTATGC-3′, reverse: 5′-TGGCGAAGGCGGTTTTC-3′, probe: 5′-FAM-TTCGCTCCCCTAGCTTTCGGGCAT-TAMRA-3′), allowing quantification of the bacterial load of individual infected larvae. The TaqMan qPCR system was designed and validated against quantified serial dilutions of the target sequence cloned into the pCR2.1 plasmid and against total DNA extracts of non-infected larvae. Serial dilutions of pCR2.1 containing the target sequence were used as standards in each run. Total DNA of individual larvae was extracted with a MagNA Pure LC (Roche) robot and eluted in 100 μl elution buffer. Reactions were carried out on a StepOne Plus real-time PCR system (Applied Biosystems). TaqMan Fast Advanced reagents (Applied Biosystems) were used according to the manufacturer's instructions with 5 μl input DNA in a total reaction volume of 20 μl.
Morpholino knockdown
Knockdown of MyD88 expression was done by standard microinjection of 1 nl of a 5 mM solution of anti-Myd88 morpholino (5′-GTTAAACACTGACCCTGTGGATCAT-3′, Gene Tools; Bates et al., 2007; Cambier et al., 2014) into the early zygote immediately after fertilization using borosilicate glass microcapillary injection needles (Harvard Apparatus, 30-0019, 1 mm O.D. × 0.58 mm I.D.) and an Eppendorf FemtoJet. A group of control larvae was injected with 1 nl of a control morpholino (5′-CCTCTTACCTCAGTTACAATTTATA-3′, Gene Tools) with no known target in zebrafish at the same concentration in parallel. Levels of MyD88 protein in morphant and control larvae were analyzed by Western Blot at 3, 4, and 5 dpf with a mouse anti-myd88 antibody, detected with a goat anti-mouse-IgG antibody conjugated to horseradish peroxidase (HRP).
Results
W. chondrophila is capable of infecting zebrafish larvae via bath-immersion
In order to first establish whether W. chondrophila can infect zebrafish through an oral or dermal route, embryos and larvae of different stages were incubated in a suspension of W. chondrophila infectious particles or EBs, in a bath-immersion experiment. While younger embryos and larvae of up to 72 hpf (hours post fertilization) stages could not be infected with this method, 4 dpf (days post fertilization) larvae appeared to swallow (Figure 1A) the bacteria and interestingly, succumbed to an infection of their swim bladder which we detected by IF staining (Figure 1B).
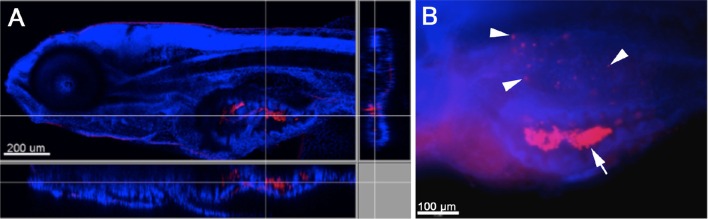
Figure 1.
Infection of zebrafish larvae with W. chondrophila via bath-immersion at 4 dpf followed by IF staining with an anti-Waddlia antibody (red) and DAPI (blue). Section view of a CLSM acquired 3D stack of a larva at 4 hpi. The section view exhibits a large amount of swallowed W. chondrophila inside the lumen of the intestine (A). Fluorescence light-microscope appearance of the trunk region of a larva at 48 hpi shows in addition to the accumulation of W. chondrophila in the gut lumen (arrow), an infection of the swim bladder with bacterial inclusions (arrow heads) visible inside the epithelium (B).
Swim bladder infection can be provoked via microinjection
The swim bladder provides the only possibility to examine infections of air-exposed epithelium in fish and, as such, is a putative model for equivalent epithelia in humans, in particular the lungs. In this context, the spontaneous bath-immersion induced infection of the swim bladder and especially the attachment to the cilia of the epithelium was an exciting finding, although infection via bath immersion carried inherent experimental variability due to varying local concentrations of W. chondrophila and different gulping rates of individual larvae. Therefore, we tried direct microinjection of 103 W. chondrophila EBs into the swim bladder lumen of 4 dpf larvae (Figure 2A). The resulting infection was comparable to that seen using bath-immersion but under more controlled conditions, confirming that directed injection can be used as a more reproducible method of establishing infection. In addition to live W. chondrophila, two groups of control embryos were injected with either heat-inactivated W. chondrophila or PBS in parallel. Samples were taken at 4, 24, 48, 72, and 96 hpi and screened for the presence of W. chondrophila inclusions by confocal laser scanning microscopy CLSM (Laser Scanning Microscopy) and TEM (Transmission Electron Microscopy). W. chondrophila inclusions were detected in the epithelium of the swim bladder as well as in adjacent tissues at 24–48 hpi (Figure 2B). In haematoxylin and eosin (HE) stained samples, the swim bladder walls of infected larvae were markedly thickened when compared with control animals (Figures 2C,D). The epithelial cells lining the cavity were piled up and were no longer a single cell row, with the cells themselves exhibiting a cuboidal instead of a flattened morphology (Figure 2E). The epithelium was surrounded by a moderate proliferation of fibroblasts resulting in a fibrosis, as has been described in mice for chlamydial lung infections (Jupelli et al., 2013). Neutrophils were also found to have migrated into the epithelial cell layers (Figure 2F). No increase in mortality was observed up until the end of the observation period of 3 days post infection (dpi) and infected larvae showed no altered behavior compared to control larvae.
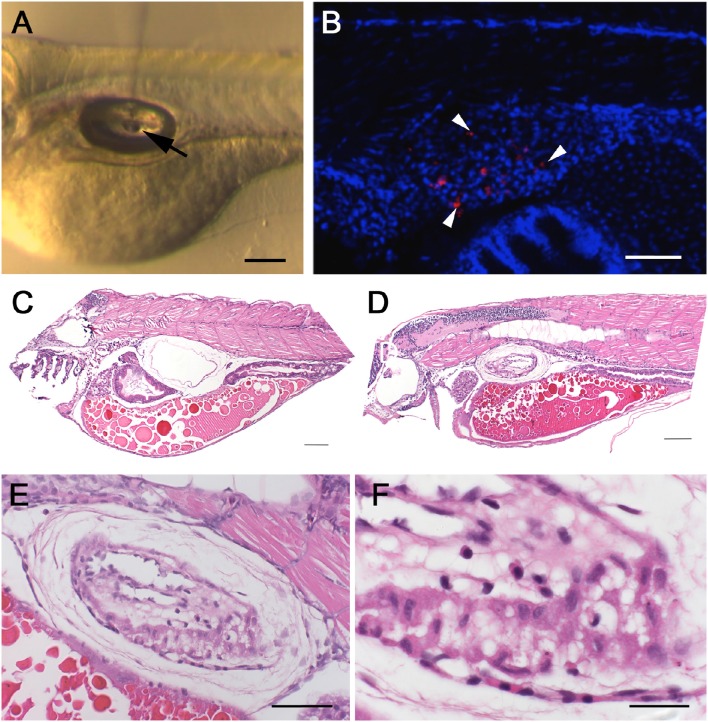
Figure 2.
Swim bladder infection in 4–5 dpf old larvae. Microinjection of W. chondrophila EBs directly into the lumen of the swim bladder of 4 dpf larvae. A drop of approx. 1 nl of the bacterial suspension can be seen hanging on the tip of the injection needle (arrow) inside the air filled lumen of the swim bladder (A). CLSM acquired 3D stack of the trunk region of a larva at 36 hpi, after IF staining with an anti-Waddlia antibody (red) and DAPI (blue). The swim bladder in the center of the image exhibits several bacterial inclusions (arrow heads) inside the epithelium (B). Histology on HE-stained sections of the swim bladder of a PBS injected control larva (C) with normal appearance and of an infected larva (D) showing clear pathological changes, including thickening of the epithelium (E) and infiltration of innate immune cells like macrophages and neutrophils (F). Scale bars (A–D) 100 μm, (E) 50 μm, and (F) 20 μm.
Intravenous microinjection of W. chondrophila causes a systemic infection
Chlamydial infection of vascular tissue has been an area of intense interest in the past, and not without controversy in respect to atherosclerosis (recently reviewed by Campbell and Rosenfeld (2014). It is also an aspect which is difficult to follow in vivo in other animal models but which is readily accessible in younger zebrafish larval stages by intravenous injection. We injected 48 hpf embryos intravenously via the Duct of Cuvier, also known as common cardinal vein with 103–104 W. chondrophila EBs. In comparison to the non-lethal swim bladder infection, a systemic infection caused a dosage dependent mortality rate of injected larvae (Figures 3A,B). The injection resulted in a rapid systemic infection with mortalities up to 100% within the first 24 h for the highest tested dosage (104) and a LD50 within 48 hpi of approximately 5 × 103. A dosage of 2 × 103 W. chondrophila EBs was chosen for the following experiments to produce moderate mortalities of between 20 and 30% at 72 hpi. With that dosage larvae showed an increasingly impaired blood circulation between 24 and 48 hpi, resulting in the formation of a pericardial oedema (Figure 4A). Histologically, no tissue alterations could be seen.
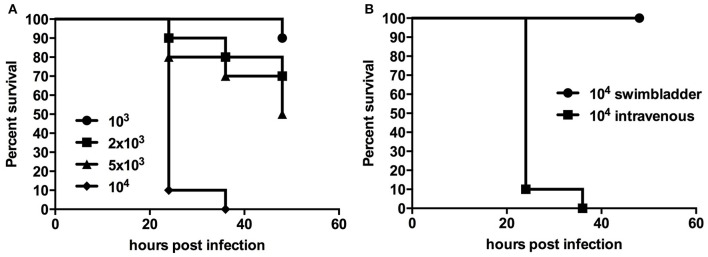
Figure 3.
Survival rates of larvae with a systemic infection after injection of different dosages of W. chondrophila show diminished survival with increasing dosage (A), while injection into the swim bladder has no impact on larval survival (B).
Figure 4.
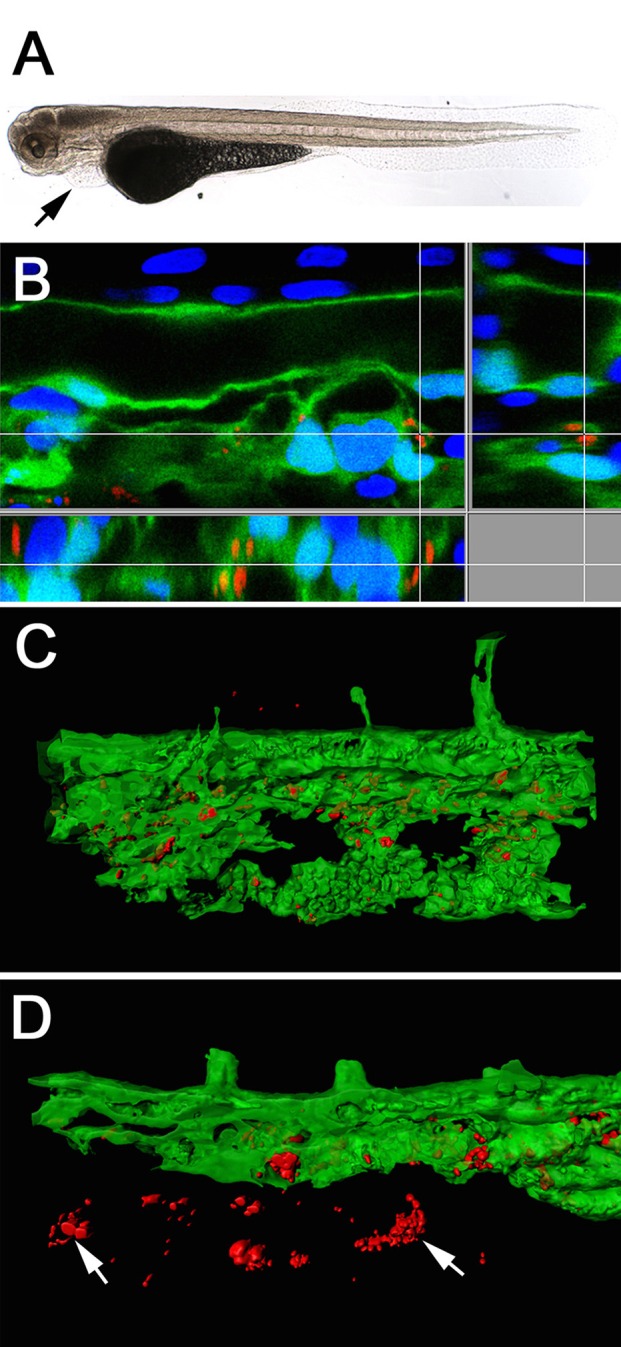
Light-microscope appearance of a transgenic Tg(fli1a:eGFP) larva at 48 hpi after intravenous injection of live bacteria (A), showing an impaired blood flow ending in an almost completely stopped circulation, accompanied by pericardial oedema formation (arrow). Section view of a CLSM acquired 3D stack (B) after IF staining with an anti-Waddlia antibody, showing the infection of GFP expressing endothelial cells (green) with W. chondrophila (red). Host cell nuclei were stained with DAPI (blue). Surface rendering of the tail artery and caudal vein shows the distribution of the inclusions inside the vasculature at 24 hpi (C) and 36 hpi (D), showing bacterial spread across endothelial bounds (arrows).
To follow the distribution of W. chondrophila in the vascular system we used the transgenic Tg(fli1a:eGFP) line, which expresses green fluorescent protein in all endothelial cells. Tracking the infection with IF using an anti-Waddlia antibody, the maximal number of W. chondrophila inclusions were found at 36–48 hpi, distributed throughout the whole embryo, and with inclusions located both within the vasculature and beyond the bounds of the endothelium (Figures 4B–D). Cell types that were identified to be susceptible to W. chondrophila invasion during a systemic infection were predominantly endothelial cells and phagocytosing innate immune cells. Especially in regions where the blood is flowing more slowly, such as the fine capillary network of the tail muscles and veins, infection of the endothelium was more common.
Morphology of W. chondrophila inside zebrafish host cells
To investigate the morphological features of W. chondrophila infection in zebrafish larvae, we performed detailed TEM and CLSM analyses of infected larvae after IF staining with an anti-Waddlia antibody and an anti-OxPhosIV antibody to stain mitochondria. W. chondrophila could be found to infect different cells of the zebrafish, predominantly epithelial cells of the swim bladder, phagocytes of the innate immune system and endothelial cells (Figure 5). The chlamydial inclusions inside these cells exhibited features typical for a Waddlia infection, including the transformation from the round shaped smaller (up to 0.5 μm in diameter) metabolically almost inactive but infectious EBs with highly condensed DNA, to the larger (about 0.9 μm in diameter) replicating reticulate bodies (RBs) with finely distributed chromatin inside a bacteria containing vacuole. Further, host cell mitochondria were readily recruited and closely associated with the inclusion, a characteristic for Waddlia infection in vitro (Croxatto and Greub, 2010) and now shown in vivo. While epithelial and endothelial cells usually contained a single perinuclear inclusion, individual phagocytes could harbor several inclusions of dividing bacteria. In TEM images of larvae that were injected into the swim bladder, some bacteria were in close relation to microvilli on the epithelial cell surface while others were found within the cytoplasm of these cells. The morphological structure of these bacteria on the surface resembled as well the typical morphology known for infectious particles of W. chondrophila (Rusconi et al., 2013). The perinuclear bacteria containing vacuoles inside epithelial cells were closely associated with host cell mitochondria and ER and are typical for actively replicating W. chondrophila observed in vitro (Rusconi et al., 2013).
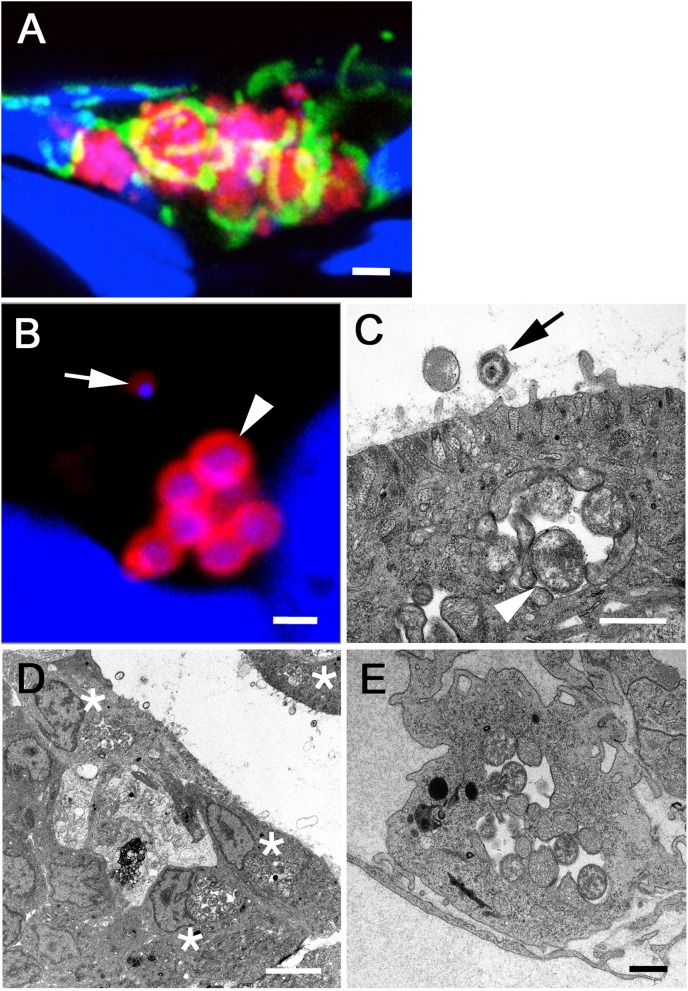
Figure 5.
CLSM acquired 3D image of W. chondrophila inclusions inside the swim bladder epithelium (A) after IF staining of Waddlia (red), mitochondria (green) and DAPI staining of host cell and bacterial DNA (blue). The image shows the formation of several bacteria containing vacuoles inside epithelial cells, accompanied by the recruitment and close association with host cell mitochondria. Close-ups of single inclusions with CLSM (B) and TEM (C) reveal typical features of the chlamydial life cycle, such as transformation from the smaller infectious EB with condensed DNA (arrow) to the larger and metabolically active RB form with finely distributed chromatin (arrow head), dividing inside the vacuole. TEM images show also the morphology of infected epithelial cells of the swim bladder (D) and endothelial cells of blood vessels (E), usually containing a single perinuclear inclusion (asterisks), strongly associated with host cell mitochondria. Scale bars (A) 2 μm, (B,C,E) 1 μm and (D) 5 μm.
Innate immune reaction to W. chondrophila includes a strong recruitment of neutrophils
The zebrafish innate immune system reacts to W. chondrophila with a strong recruitment of inflammatory cells to the infection site, as we already observed in our histology analysis (Figure 2).
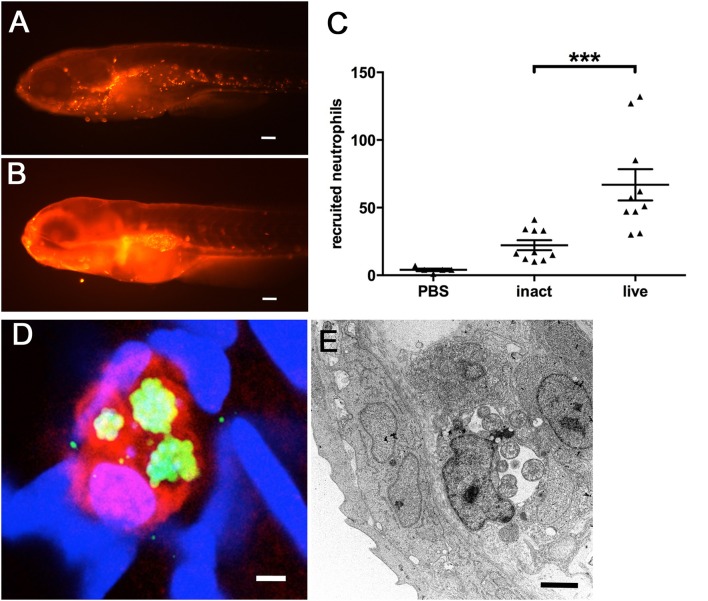
This recruitment of inflammatory cells could be confirmed by using larvae of the transgenic Tg(lyzC:dsRednz50) line, whose neutrophils express red fluorescent protein, easily assessable by fluorescence microscopy. Since the zebrafish swim bladder is an enclosed compartment, usually devoid of immune cells, we used our swim bladder infection assay in order to quantify the induced recruitment of neutrophils following microinjection. In these larvae, large numbers of neutrophils could be detected clustered inside the swim bladder (Figure 6B), compared to PBS injected control larvae (Figure 6A). We quantified the recruitment by counting the fluorescent neutrophils using the image analysis package of Imaris (Video S1). The results show further a significantly increased response of neutrophil invasion to live W. chondrophila compared to heat-inactivated bacteria (Figure 6C). Phagocytosed W. chondrophila show the ability to survive and replicate inside neutrophils (Figures 6D,E).
Figure 6.
Neutrophil recruitment and uptake of W. chondrophila monitored in transgenic Tg(lyzC:dsRednz50) larvae at 8 hpi. Whilst the swim bladder of a healthy larva is usually nearly devoid of innate immue cells (A), the injection of W. chondrophila into the swim bladder activates strong neutrophil recruitment (B). Quantification of recruited neutrophils (C) shows a significantly increased reaction to live W. chondrophila (live) compared to heat-inactivated (inact) bacteria or with sterile PBS injected control larvae, ***p < 0.001. After the uptake by a neutrophil W. chondrophila can successfully avoid its degradation and instead start replicating inside the phagosome to form an inclusion shown by 3D-CLSM (D) and TEM (E). (D) shows a 3D acquired z-stack image of a dsRed expressing neutrophil (red) of the transgenic Tg(lyzC:dsRednz50) line, harboring three W. chondrophila inclusions, visualized by antibody staining (green). DNA was stained with DAPI (blue). The TEM image in (E) shows a zebrafish phagocyte containing two inclusions of replicating W. chondrophila RBs. Scale bars (A,B) 100 μm, (D) 1 μm and (E) 2 μm.
Systemically infected larvae can be rescued by antibiotic treatment with tetracycline
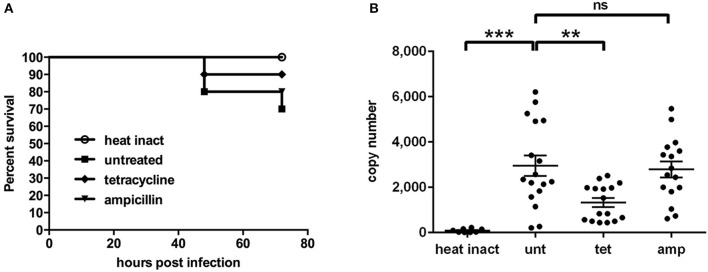
In vitro studies have previously shown that W. chondrophila is susceptible to antibiotics of the tetracycline group, but is resistant to β-lactam antibiotics (Goy and Greub, 2009). Therefore we compared the effect of tetracycline and ampicillin on W. chondrophila infections in vivo by using our systemic infection assay, for which we compared the survival rates of differently treated larvae. In order to additionally quantify and compare the bacterial burden of individual larvae during the infection, we furthermore developed a specific quantitative PCR targeting the two-copy W. chondrophila 16S rRNA gene.
While treatment with tetracycline significantly (p < 0.01) reduced W. chondrophila replication in vivo and increased the survival rate (Figure 7), treatment with ampicillin had a small but non-significant impact on the bacterial load compared to untreated larvae. Formation and distribution of W. chondrophila inclusions in untreated and ampicillin treated larvae were similar. In tetracycline treated larvae inclusion formation was strongly attenuated. Furthermore heat-inactivated bacteria were quickly cleared from the system.
Figure 7.
Survival rates (A) and bacterial load (B) are shown for larvae after intravenous injection of live W. chondrophila and subsequent treatment with 30 μg/ml tetracycline (tet) or 30 μg/ml ampicillin (amp) or left untreated (unt). Another group was injected with heat inactivated W. chondrophila (heat inact) for comparison. For survival rates (A) 10 larvae of each condition were observed in three independent experiments, respectively. The bacterial load (B) was determined at 36 hpi by qPCR of total DNA extracts from homogenates of individual larvae, targeting the W. chondrophila 16S rRNA sequence. Statistical analysis was done by one-way ANOVA with Bonferroni's posttest. ***p < 0.001, **p < 0.01, ns = not significant. Mean values ± SEM are shown by horizontal bars.
Effect of MyD88 on infection with W. chondrophila
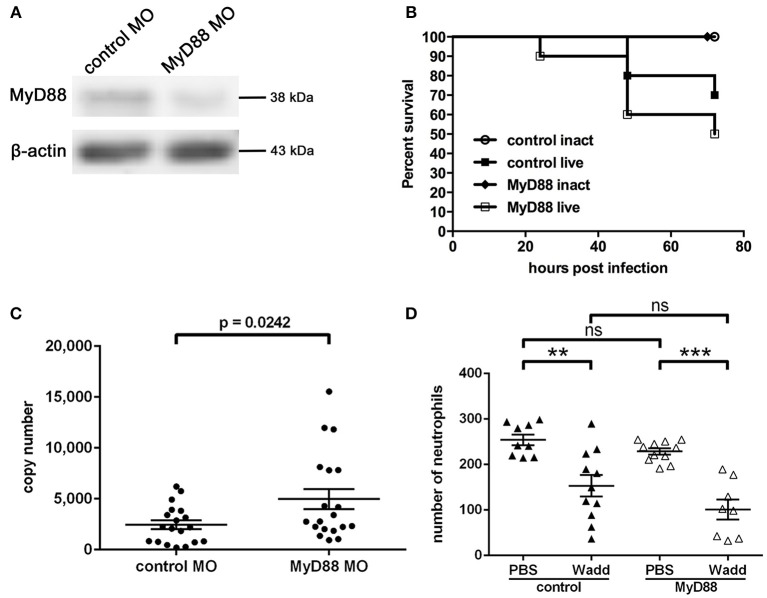
The signaling molecule MyD88 has been shown to play a central role for pathogen recognition, immune reaction and survival of infected individuals during Chlamydia infection in several in vivo studies. Now we wished to address how MyD88 mediated signaling affects the infection with W. chondrophila in our zebrafish model. To investigate whether MyD88 signaling has a function during W. chondrophila infection in zebrafish, we knocked down MyD88 expression by injection of a specific morpholino at the 1-cell stage. The resulting knockdown lasted for up to 4 dpf as determined by Western Blot (Figure 8A). Survival rates and bacterial loads were compared between control and MyD88 knockdown larvae during systemic infection. Our results show, that MyD88 morphant larvae exhibited a 20% lower survival rate compared to control larvae with a similar initial injection dose of 2 × 103 W. chondrophila EBs (Figure 8B). Furthermore, the bacterial load of MyD88 morphant larvae at 36 hpi was higher compared to control larvae (Figure 8C).
Figure 8.
Morpholino knockdown of MyD88, verified by Western Blot analysis (A) of zebrafish MyD88 at 4 dpf, showing a depleted level of the protein in morphant fish. β-actin served as loading control. Survival rates of control and MyD88 depleted larvae at 36 hpi (B) show differences after intravenous injection of W. chondrophila. Morphant larvae show a slightly reduced survival rate compared to control larvae. Survival rates of 10 larvae for each condition were observed in three independent experiments, respectively. The bacterial load, determined by qPCR, (C) of MyD88 depleted larvae at 36 hpi is significantly (p = 0.0242) higher compared to control larvae. Statistical analysis was done by Student's unpaired t-test. Mean values ± SEM are shown by horizontal bars. Total numbers of neutrophils of whole individual control and MyD88 depleted larvae (D) were determined in the transgenic Tg(lyzC:dsRed)nz50 line at 36 hpi after intravenous injection of PBS or live W. chondrophila by counting dsRed expressing neutrophils after IF staining with an anti-dsRED antibody and subsequent CLSM analysis. The acquired 3D stacks of whole larvae were analyzed with the fluorescent spot counting tool in Imaris (Bitplane). The results show a significant depletion of neutrophils after injection of W. chondrophila compared to PBS injection in both normal and MyD88 depleted larvae with no significant differences between the two groups. Statistical analysis was done by one-way ANOVA with Bonferroni's posttest. ***p < 0.001, **p < 0.01, ns = not significant. Mean values ± SEM are shown by horizontal bars.
Additionally, to compare the total numbers of neutrophils during a systemic infection between morphant and control fish we injected transgenic Tg(lyzC:dsRednz50) larvae either with sterile PBS, heat-inactivated W. chondrophila or live W. chondrophila (103 EBs) at 2 dpf. Subsequently, total numbers of neutrophils within individual larvae were counted with the cell counting tool in Imaris. At 36 hpi, these numbers were found to be significantly depleted upon the systemic infection with W. chondrophila compared to PBS injected larvae, although no differences between MyD88 morphant and control fish were evident (Figure 8D).
Discussion
We present the first zebrafish infection model for an obligate intracellular pathogen and member of the PVC superphylum, W. chondrophila, which produces a non-lethal swim bladder infection and a lethal systemic infection in zebrafish larvae. Primary target cells are epithelial cells of the swim bladder, endothelial cells of the vascular system and innate immune phagocytes. Moreover, in this study, we show for the first time that W. chondrophila can successfully survive and grow within zebrafish neutrophils. By using zebrafish larvae between 2 and 5 dpf our model provides the opportunity to study specifically the reaction of the innate immune system to a W. chondrophila infection.
The zebrafish swim bladder is an air-exposed epithelium, regarded as a homologous organ to the mammalian lung, having similar developmental and molecular ontogeny (Winata et al., 2009; Flores et al., 2010). Oral uptake of W. chondrophila by 4 dpf larvae results in a swim bladder infection, which can be reproducibly replicated by direct microinjection of W. chondrophila into the swim bladder. Since W. chondrophila has been linked to cases of pneumonia in humans (Haider et al., 2008), as have related organisms such as Chlamydia pneumoniae and C. psittaci, our model could provide a new approach to investigate aspects of human respiratory disease caused by these pathogens in vivo. Even more we could demonstrate that W. chondrophila enters the swim bladder epithelial cells and goes into the life cycle typical for these organisms forming metabolic active RB as described in mice with C. pneumoniae infections. A fibrosis surrounding the swim bladder was found to develop within few days after inoculation (Jupelli et al., 2013). Similarly to experimental infections of rodent lungs with other members of the order chlamydiales (Roger et al., 2010), the injection of live W. chondrophila into the swim bladder causes a strong recruitment of neutrophils to the infection site, compared to lack of neutrophil recruitment when using heat-inactivated bacteria. While zebrafish macrophages are known to react to invading bacteria at all locations, neutrophils are able to phagocytise surface associated bacteria in a vacuum-cleaner-like manner (Colucci-Guyon et al., 2011). Thus from our findings, it is possible that live W. chondrophila EBs adhere efficiently to the epithelium of the swim bladder, while heat-inactivated bacteria are unable to do so. The adhesion of live bacteria could lead to an increased neutrophil recruitment to the infection site.
Intravenous injection of W. chondrophila into zebrafish larvae leads to a strongly impaired blood circulation and subsequent oedema formation, as is typical for severe systemic inflammatory states such as sepsis, with the ensuing increased mortality correlating with increased dosage. This reaction was only provoked by injection of live infectious EBs and not by the use of heat-inactivated W. chondrophila. The infection triggers a strong innate immune response, evidenced by the initial recruitment and subsequent depletion of phagocytes. In addition to macrophages, we show for the first time that epithelial cells and neutrophils are among the preferred target cells for infection by W. chondrophila in vivo. The involvement of neutrophils in the host response to W. chondrophila was unexpected and is a new finding of this study which will stimulate a re-evaluation of the relative importance of macrophages and neutrophils in the initial clearance of chlamydial infections. Infection of endothelial cells has already been shown in vitro for Chlamydia pneumoniae (Godzik et al., 1995; Gaydos et al., 1996), suggesting at the time that these chlamydial pathogens may play a possible role in the development of vascular disease. Our findings with intravenously injected W. chondrophila now provide in vivo evidence to support this idea with another chlamydial pathogen and by using the zebrafish, a readily tractable model for future investigations.
W. chondrophila was first proposed as an abortifacient agent in ruminants (Dilbeck et al., 1990; Henning et al., 2002; Dilbeck-Robertson et al., 2003) and more recently in humans (Baud et al., 2007, 2011, 2014; Haider et al., 2008; Goy et al., 2009; Goy and Greub, 2009). The pathological mechanisms for this are unknown, although other chlamydial abortifacient agents are thought to act via a permanent infection of the intestine and from there via sepsis infecting the placenta and fetus. Aborted placentas of ewes after infections with C. abortus show, in addition to large areas of necrotic trophoblasts, a severe necrotizing vasculitis (Buxton et al., 2002). That W. chondrophila is able to establish an infection within vessel endothelium is a new finding and offers a potential mechanism to promote invasion of the placental tissues from the blood circulation.
Treatment of the infected larvae with tetracycline significantly reduces the bacterial load and increases the larval survival rate, while treatment with ampicillin is ineffective. Nevertheless, even without antibiotic treatment, the bacterial load starts to decrease after 72 hpi in surviving larvae. The infection is greatly reduced by 4 dpi although isolated inclusions or infection loci can remain. This experimental zebrafish model may thus be used to test new antibiotics in vivo during the initial infection window and may be used to investigate the effect of anti-virulence compounds such as Type III secretion system (T3SS) inhibitors, recently shown to be effective in vitro (Bertelli et al., 2010).
Our results show that although phagocytes are susceptible to infection with W. chondrophila, the innate immune system on its own is able to mount an effective initial counter strike against W. chondrophila infection in surviving larvae. Furthermore, we found that MyD88 mediated signaling contributes to an increased survival and a reduced bacterial load in control larvae compared to MyD88 morphants. These findings indicate a possible role of MyD88 dependent activation and recruitment of innate immune cells, including neutrophils, for an efficient reaction to W. chondrophila infection. On the other hand, we could not observe differences in phagocyte depletion between control and morphant larvae, which indicates that recruitment and phagocytosis of W. chondrophila could basically also be mediated by a MyD88 independent manner. MyD88 mediated signaling has also been found to contribute to the generation of an effective early immune response and increased host survival in a mouse model for Chlamydia pneumoniae infection (Naiki et al., 2005). Furthermore, it has been shown that Parachlamydia acanthamoebae is recognized and internalized by macrophages in a MyD88 independent manner (Roger et al., 2010), while on the other hand in another mouse model it was shown that neutrophil recruitment to Chlamydia pneumoniae infection is strongly depending on MyD88 signaling, although in this case, recruitment of neutrophils initially increased the bacterial load (Rodriguez et al., 2005).
The potential role of MyD88 signaling at later time points of an infection needs to be further investigated. Possible subsequent recognition of the W. chondrophila bacteria containing vacuole inside infected phagocytes by endosomal TLRs or cytoplasmic nucleotide binding oligomerization domain (NOD)-like receptors acting in association with MyD88 mediated signaling could lead to a more efficient response of the innate immune system to W. chondrophila infection. An essential role for the recognition and defensive reaction induced by NOD-like receptors has already been shown for Chlamydia pneumoniae and Chlamydia trachomatis (Buchholz and Stephens, 2008; Shimada et al., 2009). Whether W. chondrophila is also recognized by intracellular pattern recognition receptors is another key aspect for future studies.
Taken together, our results complement those from mouse models, but offers new insights into pathogenesis and immune response during Chlamydia infections. In particular, the use of different injection sites permits a staged analysis of separate events in an infection and the opportunity to understand the molecular mechanisms guiding these processes. The genetic tractability of the model is a particularly opportune addition to the tools available to the Chlamydia field, being potentially high throughput for screening of novel anti-chlamydial agents and especially useful given the recent progress in development of transgenic Chlamydia (Gérard et al., 2013; Song et al., 2013). The zebrafish model presented here offers chlamydial researchers in particular, and the PVC field in general, a powerful new experimental tool.
Author contributions
AF, AL, AV, HS, LV, MR, PC, and SN conceived and designed the experiments. AF, AL, HS, LN, LV, MR, and SN performed the experiments. AF, AL, AV, GG, HS, LN, LV, MR, PC, and SN analyzed the data. AF, GG, HS, LV, MR, and PC wrote the manuscript.
Funding
This study was supported by the EU through Marie Curie IEF grant number 332058, the SBF through COST Action 867: Fish Welfare in European Aquaculture and by the Swiss National Science Foundation (SNF), grant number 310030_138533/1. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Kara Dannenhauer for fish maintenance and the Center of Microscopy and Image Analysis of the University of Zurich for providing microscopic equipment and support. The laboratory work was partly performed using the logistics of the Center for Clinical Studies at the Vetsuisse Faculty of the University of Zurich.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01829/full#supplementary-material
References
- Adams K. N., Takaki K., Connolly L. E., Wiedenhoft H., Winglee K., Humbert O., et al. (2011). Drug tolerance in replicating mycobacteria mediated by a macrophage-induced efflux mechanism. Cell 145, 39–53. 10.1016/j.cell.2011.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann N. L., Polkinghorne A., Timms P. (2014). Chlamydia genomics: providing novel insights into chlamydial biology. Trends Microbiol. 22, 464–472. 10.1016/j.tim.2014.04.013 [DOI] [PubMed] [Google Scholar]
- Bates J. M., Akerlund J., Mittge E., Guillemin K. (2007). Intestinal alkaline phosphatase detoxifies lipopolysaccharide and prevents inflammation in zebrafish in response to the gut microbiota. Cell Host Microbe 2, 371–382. 10.1016/j.chom.2007.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud D., Goy G., Osterheld M. C., Borel N., Vial Y., Pospischil A., et al. (2011). Waddlia chondrophila: from bovine abortion to human miscarriage. Clin. Infect. Dis. 52, 1469–1471. 10.1093/cid/cir205 [DOI] [PubMed] [Google Scholar]
- Baud D., Goy G., Osterheld M. C., Croxatto A., Borel N., Vial Y., et al. (2014). Role of Waddlia chondrophila placental infection in miscarriage. Emerging Infect. Dis. 20, 460–464. 10.3201/eid2003.131019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud D., Thomas V., Arafa A., Regan L., Greub G. (2007). Waddlia chondrophila, a potential agent of human fetal death. Emerging Infect. Dis. 13, 1239–1243. 10.3201/eid1308.070315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertelli C., Collyn F., Croxatto A., Rückert C., Polkinghorne A., Kebbi-Beghdadi C., et al. (2010). The Waddlia genome: a window into chlamydial biology. PLoS ONE 5:e10890. 10.1371/journal.pone.0010890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannon M. K., Davis J. M., Mathias J. R., Hall C. J., Emerson J. C., Crosier P. S., et al. (2009). Pseudomonas aeruginosa Type III secretion system interacts with phagocytes to modulate systemic infection of zebrafish embryos. Cell. Microbiol. 11, 755–768. 10.1111/j.1462-5822.2009.01288.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz K. R., Stephens R. S. (2008). The cytosolic pattern recognition receptor NOD1 induces inflammatory interleukin-8 during Chlamydia trachomatis infection. Infect. Immun. 76, 3150–3155. 10.1128/IAI.00104-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxton D., Anderson I. E., Longbottom D., Livingstone M., Wattegedera S., Entrican G. (2002). Ovine Chlamydial abortion: characterization of the inflammatory immune response in placental tissues. J. Comp. Pathol. 127, 133–141. 10.1053/jcpa.2002.0573 [DOI] [PubMed] [Google Scholar]
- Cambier C. J., Takaki K. K., Larson R. P., Hernandez R. E., Tobin D. M., Urdahl K. B., et al. (2014). Mycobacteria manipulate macrophage recruitment through coordinated use of membrane lipids. Nature 505, 218–222. 10.1038/nature12799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell L. A., Rosenfeld M. E. (2014). Persistent C. pneumoniae infection in atherosclerotic lesions: rethinking the clinical trials. Front. Cell Infect. Microbiol. 4:34. 10.3389/fcimb.2014.00034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clatworthy A. E., Lee J. S., Leibman M., Kostun Z., Davidson A. J., Hung D. T. (2009). Pseudomonas aeruginosa infection of zebrafish involves both host and pathogen determinants. Infect. Immun. 77, 1293–1303. 10.1128/IAI.01181-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codony F., Fittipaldi M., López E., Morató J., Agustí G. (2012). Well water as a possible source of Waddlia chondrophila infections. Microb. Environ. 27, 529–532. 10.1264/jsme2.ME12048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collingro A., Tischler P., Weinmaier T., Penz T., Heinz E., Brunham R. C., et al. (2011). Unity in variety–the pan-genome of the Chlamydiae. Mol. Biol. Evol. 28, 3253–3270. 10.1093/molbev/msr161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colucci-Guyon E., Tinevez J. Y., Renshaw S. A., Herbomel P. (2011). Strategies of professional phagocytes in vivo: unlike macrophages, neutrophils engulf only surface-associated microbes. J. Cell Sci. 124(Pt 18), 3053–3059. 10.1242/jcs.082792 [DOI] [PubMed] [Google Scholar]
- Croxatto A., Greub G. (2010). Early intracellular trafficking of Waddlia chondrophila in human macrophages. Microbiology 156(Pt 2), 340–355. 10.1099/mic.0.034546-0 [DOI] [PubMed] [Google Scholar]
- Davis J. M., Clay H., Lewis J. L., Ghori N., Herbomel P., Ramakrishnan L. (2002). Real-time visualization of Mycobacterium-macrophage interactions leading to initiation of granuloma formation in zebrafish embryos. Immunity 17, 693–702. 10.1016/S1074-7613(02)00475-2 [DOI] [PubMed] [Google Scholar]
- Dilbeck P. M., Evermann J. F., Crawford T. B., Ward A. C. S., Leathers C. W., Holland C. J., et al. (1990). Isolation of a previously undescribed Rickettsia from an aborted bovine fetus. J. Clin. Microbiol. 28, 814–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilbeck-Robertson P., McAllister M. M., Bradway D., Evermann J. F. (2003). Results of a new serologic test suggest an association of Waddlia Chondrophila with Bovine Abortion. J. Veterin. Diagnost. Investig. 15, 568–569. 10.1177/104063870301500609 [DOI] [PubMed] [Google Scholar]
- Draghi A., II., Popov V. L., Kahl M. M., Stanton J. B., Brown C. C., Tsongalis G. J., et al. (2004). Characterization of “Candidatus piscichlamydia salmonis” (order Chlamydiales), a chlamydia-like bacterium associated with epitheliocystis in farmed Atlantic salmon (Salmo salar). J. Clin. Microbiol. 42, 5286–5297. 10.1128/JCM.42.11.5286-5297.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A., Eshwar A. K., Neuhauss S. C. F., Ruetten M., Lehner A., Vaughan L. (2015). Evaluation of zebrafish as a model to study the pathogenesis of the opportunistic pathogen Cronobacter turicensis. Emerg. Microb Infect 4:e30 10.1038/emi.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A., Walther E., Schmidt-Posthaus H., Nufer L., Wilson A., Svercel M., et al. (2013). Candidatus Syngnamydia venezia, a novel member of the phylum Chlamydiae from the broad nosed pipefish, Syngnathus typhle. PLoS ONE 8:e70853. 10.1371/journal.pone.0070853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores M. V., Crawford K. C., Pullin L. M., Hall C. J., Crosier K. E., Crosier P. S. (2010). Dual oxidase in the intestinal epithelium of zebrafish larvae has anti-bacterial properties. Biochem. Biophys. Res. Commun. 400, 164–168. 10.1016/j.bbrc.2010.08.037 [DOI] [PubMed] [Google Scholar]
- Gaydos C. A., Summersgill J. T., Sahney N. N., Ramirez J. A., Quinn T. C. (1996). Replication of Chlamydia pneumoniae in vitro in human macrophages, endothelial cells, and aortic artery smooth muscle cells. Infect. Immun. 64, 1614–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gérard H. C., Mishra M. K., Mao G., Wang S., Hali M., Whittum-Hudson J. A., et al. (2013). Dendrimer-enabled DNA delivery and transformation of Chlamydia pneumoniae. Nanomedicine 9, 996–1008. 10.1016/j.nano.2013.04.004 [DOI] [PubMed] [Google Scholar]
- Godzik K. L., O'Brien E. R., Wang S., Kuo C. (1995). In vitro susceptibility of human vascular wall cells to infection with Chlamydia pneumoniae. J. Clin. Microbiol. 33, 2411–2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goy G., Croxatto A., Greub G. (2008). Waddlia chondrophila enters and multiplies within human macrophages. Microbes Infect. 10, 556–562. 10.1016/j.micinf.2008.02.003 [DOI] [PubMed] [Google Scholar]
- Goy G., Croxatto A., Posfay-Barbe K. M., Gervaix A., Greub G. (2009). Development of a real-time PCR for the specific detection of Waddlia chondrophila in clinical samples. Eur. J. Clin. Microbiol. Infect. Dis. 28, 1483–1486. 10.1007/s10096-009-0804-7 [DOI] [PubMed] [Google Scholar]
- Goy G., Greub G. (2009). Antibiotic susceptibility of Waddlia chondrophila in Acanthamoeba castellanii amoebae. Antimicrob. Agents Chemother. 53, 2663–2666. 10.1128/AAC.00046-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greub G., Raoult D. (2002). Crescent bodies of Parachlamydia acanthamoeba and its life cycle within Acanthamoeba polyphaga: an electron micrograph study. Appl. Environ. Microbiol. 68, 3076–3084. 10.1128/aem.68.6.3076-3084.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haider S., Collingro A., Walochnik J., Wagner M., Horn M. (2008). Chlamydia-like bacteria in respiratory samples of community-acquired pneumonia patients. FEMS Microbiol. Lett. 281, 198–202. 10.1111/j.1574-6968.2008.01099.x [DOI] [PubMed] [Google Scholar]
- Hall C., Flores M. V., Chien A., Davidson A., Crosier K., Crosier P. (2009). Transgenic zebrafish reporter lines reveal conserved Toll-like receptor signaling potential in embryonic myeloid leukocytes and adult immune cell lineages. J. Leukoc. Biol. 85, 751–765. 10.1189/jlb.0708405 [DOI] [PubMed] [Google Scholar]
- Hall C., Flores M. V., Storm T., Crosier K., Crosier P. (2007). The zebrafish lysozyme C promoter drives myeloid-specific expression in transgenic fish. BMC Dev. Biol. 7:42. 10.1186/1471-213X-7-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvie E. A., Green J. M., Neely M. N., Huttenlocher A. (2013). Innate immune response to Streptococcus iniae infection in zebrafish larvae. Infect. Immun. 81, 110–121. 10.1128/IAI.00642-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henning K., Schares G., Granzow H., Polster U., Hartmann M., Hotzel H., et al. (2002). Neospora caninum and Waddlia chondrophila strain 2032/99 in a septic stillborn calf. Vet. Microbiol. 85, 285–292. 10.1016/S0378-1135(01)00510-7 [DOI] [PubMed] [Google Scholar]
- Herbomel P., Thisse B., Thisse C. (1999). Ontogeny and behaviour of early macrophages in the zebrafish embryo. Development 126, 3735–3745. [DOI] [PubMed] [Google Scholar]
- Hoffman G. L., Dunbar C. E., Wolf K., Zwillenberg L. O. (1969). Epitheliocystis, a new infectious disease of the bluegill (Lepomis macrochirus). Antonie Van Leeuwenhoek 35, 146–158. [DOI] [PubMed] [Google Scholar]
- Jupelli M., Shimada K., Chiba N., Slepenkin A., Alsabeh R., Jones H. D., et al. (2013). Chlamydia pneumoniae infection in mice induces chronic lung inflammation, iBALT formation, and fibrosis. PLoS ONE 8:e77447. 10.1371/journal.pone.0077447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanther M., Rawls J. F. (2010). Host-microbe interactions in the developing zebrafish. Curr. Opin. Immunol. 22, 10–19. 10.1016/j.coi.2010.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsen M., Nylund A., Watanabe K., Helvik J. V., Nylund S., Plarre H. (2008). Characterization of 'Candidatus Clavochlamydia salmonicola': an intracellular bacterium infecting salmonid fish. Environ. Microbiol. 10, 208–218. 10.1111/j.1462-2920.2007.01445.x [DOI] [PubMed] [Google Scholar]
- Kebbi-Beghdadi C., Batista C., Greub G. (2011a). Permissivity of fish cell lines to three Chlamydia-related bacteria: Waddlia chondrophila, Estrella lausannensis and Parachlamydia acanthamoebae. FEMS Immunol. Med. Microbiol. 63, 339–345. 10.1111/j.1574-695X.2011.00856.x [DOI] [PubMed] [Google Scholar]
- Kebbi-Beghdadi C., Cisse O., Greub G. (2011b). Permissivity of Vero cells, human pneumocytes and human endometrial cells to Waddlia chondrophila. Microbes Infect. 13, 566–574. 10.1016/j.micinf.2011.01.020 [DOI] [PubMed] [Google Scholar]
- Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B., Schilling T. F. (1995). Stages of embryonic development of the zebrafish. Develop. Dyn. 203, 253–310. [DOI] [PubMed] [Google Scholar]
- Kocan K. M., Crawford T. B., Dilbeck P. M., Evermann J. F., McGuire T. C. (1990). Development of a Rickettsia isolated from an aborted bovine fetus. J. Bacteriol. 172, 5949–5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagkouvardos I., Weinmaier T., Lauro F. M., Cavicchioli R., Rattei T., Horn M. (2014). Integrating metagenomic and amplicon databases to resolve the phylogenetic and ecological diversity of the Chlamydiae. ISME J. 8, 115–125. 10.1038/ismej.2013.142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamoth F., Greub G. (2010). Amoebal pathogens as emerging causal agents of pneumonia. FEMS Microbiol. Rev. 34, 260–280. 10.1111/j.1574-6976.2009.00207.x [DOI] [PubMed] [Google Scholar]
- Lawson N. D., Weinstein B. M. (2002). In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev. Biol. 248, 307–318. 10.1006/dbio.2002.0711 [DOI] [PubMed] [Google Scholar]
- Levraud J. P., Disson O., Kissa K., Bonne I., Cossart P., Herbomel P., et al. (2009). Real-time observation of listeria monocytogenes-phagocyte interactions in living zebrafish larvae. Infect. Immun. 77, 3651–3660. 10.1128/IAI.00408-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longbottom D., Coulter L. J. (2003). Animal Chlamydioses and Zoonotic implications. J. Comp. Pathol. 128, 217–244. 10.1053/jcpa.2002.0629 [DOI] [PubMed] [Google Scholar]
- Medzhitov R., Preston-Hurlburt P., Kopp E., Stadlen A., Chen C., Ghosh S., et al. (1998). MyD88 is an adaptor protein in the hToll/IL-1 receptor family signaling pathways. Mol. Cell 2, 253–258. [DOI] [PubMed] [Google Scholar]
- Meeker N. D., Trede N. S. (2008). Immunology and zebrafish: spawning new models of human disease. Dev. Comp. Immunol. 32, 745–757. 10.1016/j.dci.2007.11.011 [DOI] [PubMed] [Google Scholar]
- Meijer A. H., Spaink H. P. (2011). Host-pathogen interactions made transparent with the zebrafish model. Curr. Drug Targets 12, 1000–1017. 10.2174/138945011795677809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer A., Roholl P. J., Ossewaarde J. M., Jones B., Nowak B. F. (2006). Molecular evidence for association of chlamydiales bacteria with epitheliocystis in leafy seadragon (Phycodurus eques), silver perch (Bidyanus bidyanus), and barramundi (Lates calcarifer). Appl. Environ. Microbiol. 72, 284–290. 10.1128/AEM.72.1.284-290.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostowy S., Boucontet L., Mazon Moya M. J., Sirianni A., Boudinot P., Hollinshead M., et al. (2013). The zebrafish as a new model for the in vivo study of Shigella flexneri interaction with phagocytes and bacterial autophagy. PLoS Pathog. 9:e1003588. 10.1371/journal.ppat.1003588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naiki Y., Michelsen K. S., Schröder N. W., Alsabeh R., Slepenkin A., Zhang W., et al. (2005). MyD88 is pivotal for the early inflammatory response and subsequent bacterial clearance and survival in a mouse model of Chlamydia pneumoniae pneumonia. J. Biol. Chem. 280, 29242–29249. 10.1074/jbc.M503225200 [DOI] [PubMed] [Google Scholar]
- Newton K., Dixit V. M. (2012). Signaling in innate immunity and inflammation. Cold Spring Harb. Perspect. Biol. 4:a006049. 10.1101/cshperspect.a006049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylund S., Steigen A., Karlsbakk E., Plarre H., Andersen L., Karlsen M., et al. (2015). Characterization of ‘Candidatus Syngnamydia salmonis’ (Chlamydiales, Simkaniaceae), a bacterium associated with epitheliocystis in Atlantic salmon (Salmo salar L.). Arch. Microbiol. 197, 17–25. 10.1007/s00203-014-1038-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole R., Von Hofsten J., Rosqvist R., Olsson P. E., Wolf-Watz H. (2004). Visualisation of zebrafish infection by GFP-labelled Vibrio anguillarum. Microb. Pathog. 37, 41–46. 10.1016/j.micpath.2004.03.001 [DOI] [PubMed] [Google Scholar]
- Polymenakou P. N., Bertilsson S., Tselepides A., Stephanou E. G. (2005). Bacterial community composition in different sediments from the Eastern Mediterranean Sea: a comparison of four 16S ribosomal DNA clone libraries. Microb. Ecol. 50, 447–462. 10.1007/s00248-005-0005-6 [DOI] [PubMed] [Google Scholar]
- Prajsnar T. K., Hamilton R., Garcia-Lara J., McVicker G., Williams A., Boots M., et al. (2012). A privileged intraphagocyte niche is responsible for disseminated infection of Staphylococcus aureus in a zebrafish model. Cell. Microbiol. 14, 1600–1619. 10.1111/j.1462-5822.2012.01826.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez N., Fend F., Jennen L., Schiemann M., Wantia N., da Costa C. U. P., et al. (2005). Polymorphonuclear neutrophils improve replication of Chlamydia pneumoniae In vivo upon MyD88-Dependent Attraction. J. Immunol. 174, 4836–4844. 10.4049/jimmunol.174.8.4836 [DOI] [PubMed] [Google Scholar]
- Roger T., Casson N., Croxatto A., Entenza J. M., Pusztaszeri M., Akira S., et al. (2010). Role of MyD88 and Toll-like receptors 2 and 4 in the sensing of Parachlamydia acanthamoebae. Infect. Immun. 78, 5195–5201. 10.1128/IAI.00786-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusconi B., Lienard J., Aeby S., Croxatto A., Bertelli C., Greub G. (2013). Crescent and star shapes of members of the Chlamydiales order: impact of fixative methods. Antonie Van Leeuwenhoek 104, 521–532. 10.1007/s10482-013-9999-9 [DOI] [PubMed] [Google Scholar]
- Schmidt-Posthaus H., Polkinghorne A., Nufer L., Schifferli A., Zimmermann D. R., Segner H., et al. (2012). A natural freshwater origin for two chlamydial species, Candidatus Piscichlamydia salmonis and Candidatus Clavochlamydia salmonicola, causing mixed infections in wild brown trout (Salmo trutta). Environ. Microbiol. 14, 2048–2057. 10.1111/j.1462-2920.2011.02670.x [DOI] [PubMed] [Google Scholar]
- Shimada K., Chen S., Dempsey P. W., Sorrentino R., Alsabeh R., Slepenkin A. V., et al. (2009). The NOD/RIP2 pathway is essential for host defenses against Chlamydophila pneumoniae lung infection. PLoS Pathog. 5:e1000379. 10.1371/journal.ppat.1000379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L., Carlson J. H., Whitmire W. M., Kari L., Virtaneva K., Sturdevant D. E., et al. (2013). Chlamydia trachomatis plasmid-encoded Pgp4 is a transcriptional regulator of virulence-associated genes. Infect. Immun. 81, 636–644. 10.1128/IAI.01305-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steigen A., Karlsbakk E., Plarre H., Watanabe K., Øvergård A. C., Brevik Ø., et al. (2015). A new intracellular bacterium, Candidatus Similichlamydia labri sp. nov. (Chlamydiaceae) producing epitheliocysts in ballan wrasse, Labrus bergylta (Pisces, Labridae). Arch. Microbiol. 197, 311–318. 10.1007/s00203-014-1061-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steigen A., Nylund A., Karlsbakk E., Akoll P., Fiksdal I. U., Nyland S., et al. (2013). ‘Cand. Actinochlamydia clariae’ gen. nov., sp. nov., a Unique Intracellular Bacterium Causing Epitheliocystis in Catfish (Clarias gariepinus) in Uganda. PLoS ONE 8:e66840. 10.1371/journal.pone.0066840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stride M. C., Polkinghorne A., Miller T. L., Groff J. M., LaPatra S. E., Nowak B. F. (2013a). Molecular characterization of “Candidatus Parilichlamydia carangidicola”, a Novel Chlamydia-Like Epitheliocystis Agent in Yellowtail Kingfish, Seriola lalandi (Valenciennes), and the Proposal of a New Family, “Candidatus Parilichlamydiaceae” fam. nov. (Order Chlamydiales). App. Environ. Microbiol. 79, 1590–1597. 10.1128/AEM.02899-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stride M. C., Polkinghorne A., Miller T. L., Nowak B. F. (2013b). Molecular characterization of “Candidatus Similichlamydia latridicola” gen. nov., sp. nov. (Chlamydiales: “Candidatus Parilichlamydiaceae”), a novel Chlamydia-like epitheliocystis agent in the striped trumpeter, Latris lineata (Forster). App. Environ. Microbiol. 79, 4914–4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stride M. C., Polkinghorne A., Powell M. D., Nowak B. F. (2013c). “Candidatus similichlamydia laticola,” a novel Chlamydia-like agent of epitheliocystis in seven consecutive cohorts of farmed Australian barramundi, Lates calcarifer (Bloch). PLoS ONE 8:e82889 10.1371/journal.pone.0082889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stride M. C., Polkinghorne A., Nowak B. F. (2014). Chlamydial infections of fish: diverse pathogens and emerging causes of disease in aquaculture species. Vet. Microbiol. 170, 19–27. 10.1016/j.vetmic.2014.01.022 [DOI] [PubMed] [Google Scholar]
- Taylor-Brown A., Vaughan L., Greub G., Timms P., Polkinghorne A. (2015). Twenty years of research into Chlamydia-like organisms: a revolution in our understanding of the biology and pathogenicity of members of the phylum Chlamydiae. Pathog. Dis. 73, 1–15. 10.1093/femspd/ftu009 [DOI] [PubMed] [Google Scholar]
- van der Sar A. M., Musters R. J. P., van Eeden F. J. M., Appelmelk B. J., Vandenbroucke-Grauls C. M. J. E., Bitter W. (2003). Zebrafish embryos as a model host for the real time analysis of Salmonella typhimurium infections. Cell. Microbiol. 5, 601–611. 10.1046/j.1462-5822.2003.00303.x [DOI] [PubMed] [Google Scholar]
- Vergunst A. C., Meijer A. H., Renshaw S. A., O'Callaghan D. (2010). Burkholderia cenocepacia creates an intramacrophage replication niche in zebrafish embryos, followed by bacterial dissemination and establishment of systemic infection. Infect. Immun. 78, 1495–1508. 10.1128/IAI.00743-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner N., Nunez G. (2013). MyD88: a critical adaptor protein in innate immunity signal transduction. J. Immunol. 190, 3–4. 10.4049/jimmunol.1203103 [DOI] [PubMed] [Google Scholar]
- Winata C. L., Korzh S., Kondrychyn I., Zheng W., Korzh V., Gong Z. (2009). Development of zebrafish swimbladder: the requirement of Hedgehog signaling in specification and organization of the three tissue layers. Dev. Biol. 331, 222–236. 10.1016/j.ydbio.2009.04.035 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.