Abstract
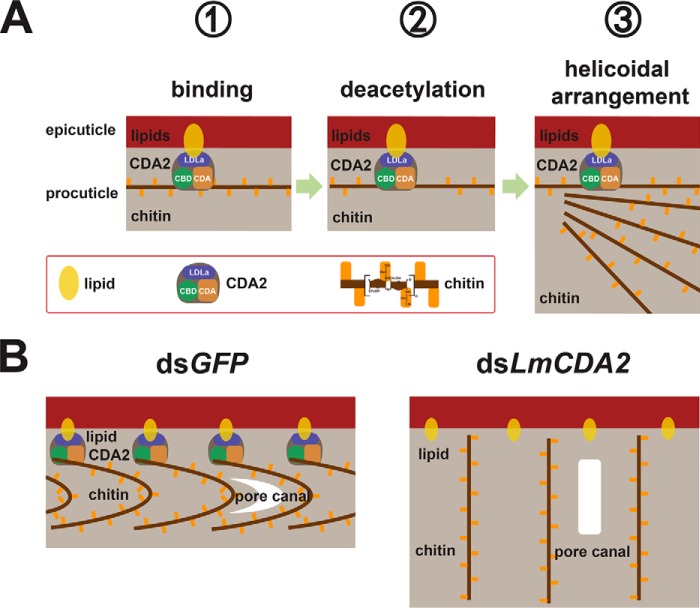
In the three-dimensional extracellular matrix of the insect cuticle, horizontally aligned microfibrils composed of the polysaccharide chitin and associated proteins are stacked either parallel to each other or helicoidally. The underlying molecular mechanisms that implement differential chitin organization are largely unknown. To learn more about cuticle organization, we sought to study the role of chitin deacetylases (CDA) in this process. In the body cuticle of nymphs of the migratory locust Locusta migratoria, helicoidal chitin organization is changed to an organization with unidirectional microfibril orientation when LmCDA2 expression is knocked down by RNA interference. In addition, the LmCDA2-deficient cuticle is less compact suggesting that LmCDA2 is needed for chitin packaging. Animals with reduced LmCDA2 activity die at molting, underlining that correct chitin organization is essential for survival. Interestingly, we find that LmCDA2 localizes only to the initially produced chitin microfibrils that constitute the apical site of the chitin stack. Based on our data, we hypothesize that LmCDA2-mediated chitin deacetylation at the beginning of chitin production is a decisive reaction that triggers helicoidal arrangement of subsequently assembled chitin-protein microfibrils.
Keywords: chitin, development, extracellular matrix, insect, lipid binding protein
Introduction
In the 60s and 70s of the last century, Neville (1) pioneered studies of chitin organization in the cuticle of insects. He elegantly described chitin organization levels by ultrastructural analyses using especially the migratory locust Locusta migratoria as a model insect. Chitin, the polymer of β-1,4-linked N-acetylglucosamine residues, interacts with proteins to form microfibrils that are aligned in parallel constituting horizontal sheets (laminae). These sheets are stacked either helicoidally or with unidirectional microfibril orientation along the vertical axis of the cuticle (2). In the locust tibia, helicoidal and non-helicoidal laminar organization alternate periodically reflecting different mechanisms of chitin production and orientation (2). This organization might have a yet unexplored impact on the mechanical properties of the cuticle. Cuticular pore canals connecting the cell surface with the cuticle surface follow chitin orientation; in helicoidally arranged laminae, they are crescentic (a term used by Neville), but in regions of non-helicoidal laminae they are straight (3, 4). Conceivably, chitin organization depends on cuticle proteins that interact with chitin. In a few recent studies, it was demonstrated that so-called CPR (cuticular protein with a Rebers and Riddiford consensus domain) proteins determine hardness in the elytra of the red flour beetle Tribolium castaneum (5, 6).
Neville added another level of complexity to the question of cuticle organization. It had been observed that every fifth or sixth sugar residue in chitin is deacetylated (7). He speculated that chitin and the partly deacetylated form of chitin, called chitosan, might be recognized by different sets of yet unidentified proteins. Chitin deacetylation is not a spontaneous event but requires the activity of chitin deacetylases (CDAs)4 that belong to the carbohydrate esterase family 4 (CE-4) (8).
Recent phylogenetic analyses of CDA sequences from four insect species showed that insect CDAs can be classified into five groups based on sequence similarity and domain diversity (9). Group I includes CDA1 and CDA2 that have a chitin-binding peritrophin-A domain (ChBD), a low-density lipoprotein receptor class A domain (LDLa), and a polysaccharide deacetylase-like catalytic domain. Group II is composed of CDA3 that has the same domain composition as members of group I CDAs. The overall sequence similarity is, however, as low as 38%. Groups III and IV include CDA4 and CDA5, respectively, which along with the deacetylase signature have a ChBD but no LDLa domain. They differ in the length of the linker sequence between these two domains. Group V harbors CDA6, CDA7, CDA8, and CDA9 that are only characterized by a deacetylase domain.
According to the localization of enzymes, insect CDAs can be separated into two subgroups comprising midgut-specific CDAs and non-gut (including epidermal) CDAs. Several midgut-specific chitin deacetylases supposedly modifying the structure of the chitinous barrier called the peritrophic matrix have been identified to date in various insects such as Trichoplusia ni (10), Helicoverpa armigera (11), Mamestra configurata (12, 13), Bombyx mori (14), and T. castaneum (15). However, the cellular function of these enzymes remains elusive.
More functional data are available on the non-gut CDA group proteins that are abundant in the cuticle or the tracheal system. In the fruit fly Drosophila melanogaster, the chitin deacetylases Serpentine (Serp, CDA1) and Vermiform (Verm, CDA2) contribute to the assembly of a luminal chitin rod that is involved in tracheal tube length regulation (16, 17). Indeed, it was shown that serp and verm mutant embryos displayed elongated and tortuous tracheal tubes (16); additionally, loss of Serp and Verm function causes disorganization of the larval cuticle (18). In the eastern spruce budworm Choristoneura fumiferana down-regulation of the CfCDA2 gene, which produces two transcripts CfCDA2a and CfCDA2b, causes molting failure (19). In the brown planthopper Nilaparvata lugens, down-regulation of NlCDA1 and NlCDA2 that are abundant in the integument is lethal (20). CDAs were intensively studied in the red flour beetle T. castaneum (15). TcCDA1, TcCDA2, and TcCDA5 are expressed in the carcass, and reduction of TcCDA1 and TcCDA2 expression impeded cuticle shedding. Despite these numerous reports, the impact of CDAs on cuticle architecture has not been investigated extensively.
To investigate CDA function in insect cuticle organization in detail, we used Neville's model insect L. migratoria, which is one of the most destructive agricultural pests worldwide (21). In this study, we focused on the L. migratoria Verm orthologue LmCDA2. By using the EST Database of Migratory Locust (22), transcriptomic data (23), and genomic data (24), we first identified the respective transcript and its expression profile at different developmental stages and in different tissues of the insect. For functional analyses, we examined the organization of the nymphal cuticle after injection of dsRNA to down-regulate the expression of LmCDA2. Our data indicate that LmCDA2 is needed for helicoidal chitin organization and procuticle compactness in the body cuticle. With this finding, we have identified LmCDA2 as a central factor in chitin organization, a fundamental biological process formulated by Neville 50 years ago (1, 7).
Results
Identification and Characterization of LmCDA2
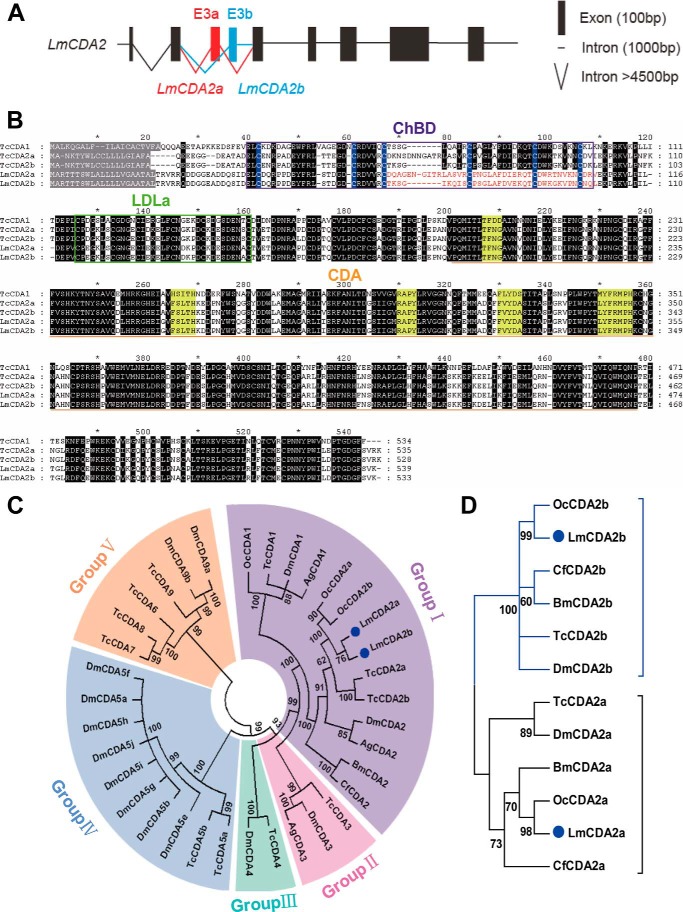
We identified LmCDA2 coding transcripts from the L. migratoria transcriptome database. Based on sequence analyses, we found two LmCDA2 alternative splicing variants. Comparing the cDNA sequences with the genomic sequence, nine exons spanning a 2.9-kb fragment were recognized. The alternative splicing variants were both composed of eight exons. The alternatively spliced exons were the third and fourth exons that we named exon 3a and 3b, respectively (Fig. 1A). The open reading frame of LmCDA2a and LmCDA2b was 1623 and 1605 bp encoding a protein of 541 and 535 amino acids, respectively. Both proteins, named LmCDA2a (KR537804) and LmCDA2b (KR537805), contain a signal peptide, a ChBD domain, an LDLa domain, and a chitin deacetylase (CDA) domain (Fig. 1B), which is similar to CDAs of T. castaneum (15). Phylogenetic analyses using full-length sequences revealed that LmCDA2a and LmCDA2b belong to insect group I CDAs (Fig. 1C). Additionally, to avoid masking effects of identical sequences in the alternative splicing CDA2 variants, we constructed a phylogenetic tree using only the sequences encoded by exon 3a/b, both of which code for a ChBD motif, and respective sequences in CDA2s of other insect species. This sequence comparison indicated that in all cases the CDA2a and CDA2b isoforms collectively grouped in two separate branches, suggesting early divergence of two different CDA2 variants in insects (Fig. 1D).
FIGURE 1.
Gene structure and the phylogenetic analyses. A, exon-intron organization of LmCDA2. Black boxes and lines represent common exons and introns. Red and blue solid boxes and lines indicate two alternative exons, 3a and 3b, and the respective introns. B, alignment of deduced amino acid sequences of TcCDA1, TcCDA2a, and TcCDA2b (T. castaneum) and LmCDA2a and LmCDA2b (L. migratoria). Predicted signal peptide residues are shaded in gray. The ChBD is boxed in purple. The blue shading highlights the conserved six cysteines in ChBD. Note that the spacing and amino acid composition between cysteines 3 and 4 (67–84 amino acids) and the sequence between cysteines 4 and 6 (84–106 amino acids) differ between LmCDA2a and LmCDA2b. These differences might have an influence on chitin-binding efficiency and by consequence enzyme processivity. The green box indicates the low-density LDLa domain. The CDA is underlined in orange. The regions shaded in yellow indicate five conserved motifs, which are essential for the catalytic activity. The black region shows conserved amino acids. The residues coded by alternative exons are highlighted in red. C, phylogenetic analysis of homologous CDAs from different insect species using the neighbor-joining method of the MEGA version 5 software. Protein sequences are encoded by whole full-length cDNA sequences. The five groups are represented in different colors. D, phylogenetic tree was constructed using the alternatively spliced exon 3 of insect CDA2s applying the neighbor-joining method of MEGA version 5. LmCDA2a and LmCDA2b are marked with blue dots. The brackets embrace the two types of alternatively spliced variants. GenBankTM accession numbers are listed in Table 2.
Tissue-specific and Developmental Expression Analysis of LmCDA2
To determine the tissue specificity of LmCDA2 expression, seven different tissues, including integument, foregut, midgut, hindgut, gastric caeca, Malpighian tubules, and fat body, were dissected from day 6 of the fifth-instar nymphs, and mRNA expression was examined by using RT-qPCR. The results of RT-qPCR showed that both LmCDA2a and LmCDA2b have the highest expression levels in the foregut, although their expression is low in the other tissues tested (Fig. 2, A and C). To explore the stage-dependent expression pattern of LmCDA2, mRNA expression levels in the integument at days 1–7 of fifth-instar nymphs were monitored by RT-qPCR. The expression of LmCDA2a and LmCDA2b was highest at the first 2 days and lower in the remaining days from day 3 to 7 (Fig. 2, B and D). In summary, the LmCDA2 transcripts were expressed in chitin-producing tissues, and the expression was dynamic underscoring the necessity for tight regulation of chitin modification during the development of the insect.
FIGURE 2.
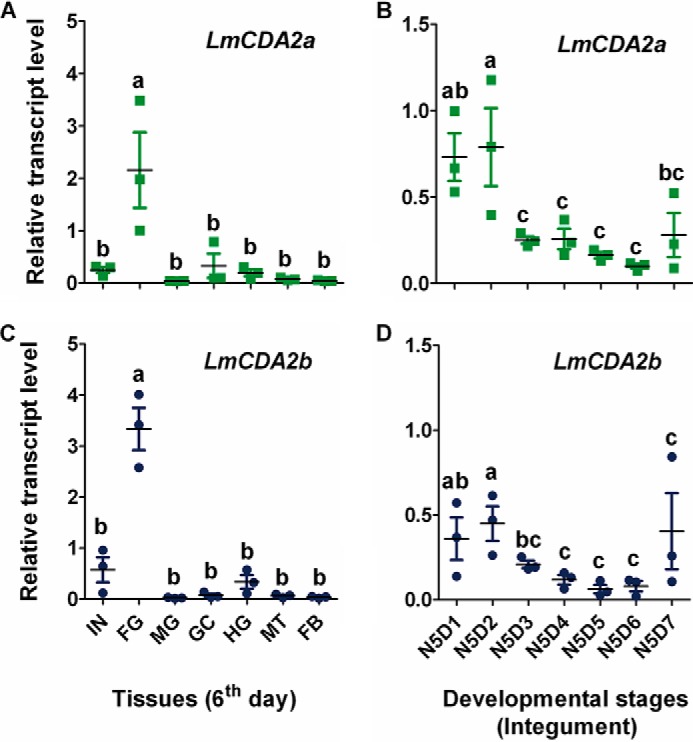
Expression patterns of LmCDA2a and LmCDA2b. A and C, expression of LmCDA2a and LmCDA2b in different tissues on day 6 of 5th-instar nymphs detected by RT-qPCR. β-Actin was used as reference control. IN, integument; FG, foregut; MG, midgut; GC, gastric caeca; HG, hindgut; MT, Malpighian tubules; FB, fat body. B and D, expression of LmCDA2a and LmCDA2b in integuments of fifth-instar nymphs at different days. N5D1 to N5D7, days 1–7 of fifth-instar nymphs. Data are reported as means ± S.E. of three independent biological replications. Statistical significance was calculated by the Tukey's test of one-way analyses of variance. Different letters (a–c) above the bars represent groups of significant difference (n = 3) (p < 0.05) among the different tissues and days of fifth-instar nymphs. a, b, and c mean that a is significantly different from b and c, respectively, and that b is significantly different from c.
LmCDA2 Is Required for Locust Molting and Development
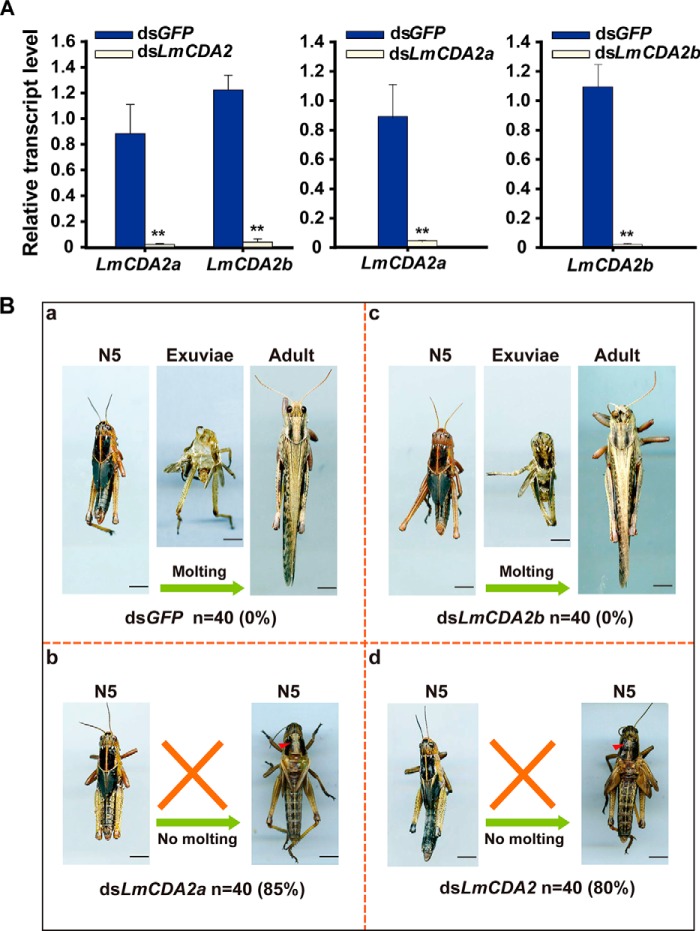
To investigate the function of LmCDA2, LmCDA2a, and LmCDA2b at molting, we sought to suppress their expression by injection of dsRNA against LmCDA2 into 2-day-old fifth-instar nymphs. The expression profiles of target genes were examined at 24 h after dsRNA injection by RT-qPCR. Double-stranded GFP (dsGFP) was used as control. The results showed that the expression of LmCDA2a and LmCDA2b were substantially down-regulated in the respective animals with a silencing efficiency higher than 95% compared with transcript levels in dsGFP-injected insects (Fig. 3A). The dsGFP-injected nymphs could molt to adults successfully (Fig. 3B, panel a), whereas the dsLmCDA2-injected nymphs failed to shed the old cuticle and were trapped within the exuviae until they died. Their mortality was 80% (Fig. 3B, panel d). This phenotype was provoked also by injecting dsRNA against the splicing variant LmCDA2a, with a mortality of 85% (Fig. 3B, panel b). However, injection of dsRNA against the splicing variant LmCDA2b had no effect on nymph development, nymph-adult molting, and viability (Fig. 3B, panel c). Injection of dsLmCDA2 into 2nd instar nymphs caused a similar phenotype (supplemental Fig. 5). These results indicate that LmCDA2a plays a vital role during L. migratoria development and molting.
FIGURE 3.
Effect of dsLmCDA2 on nymph-adult molts. A, determination of silencing efficiency at 24 h after injection of dsGFP, dsLmCDA2, dsLmCDA2a, and dsLmCDA2b by RT-qPCR assay. Expression of the reference gene β-actin was used to normalize transcript levels. Data were reported as the means ± S.E. of three independent biological replications. Statistical significance was analyzed with Student's t test. **, p < 0.01. B, phenotypes of L. migratoria after injection of dsLmCDA2, dsLmCDA2a, dsLmCDA2b, or dsGFP (control) into fifth-instar nymphs (n = 40). Locusts injected with dsGFP or dsLmCDA2b were able to molt to the adult stage successfully. By contrast, locusts injected with dsLmCDA2 or dsLmCDA2a were trapped within their exuviae with a slight split in the old dorsal cuticle of the thorax (red arrowheads) and finally died. Number of animals tested (n) and mortalities (%) are given below each experiment. N5, fifth instar nymphs. Scale bar, 5 mm.
LmCDA2 Localizes to the Apical Site of the Procuticle
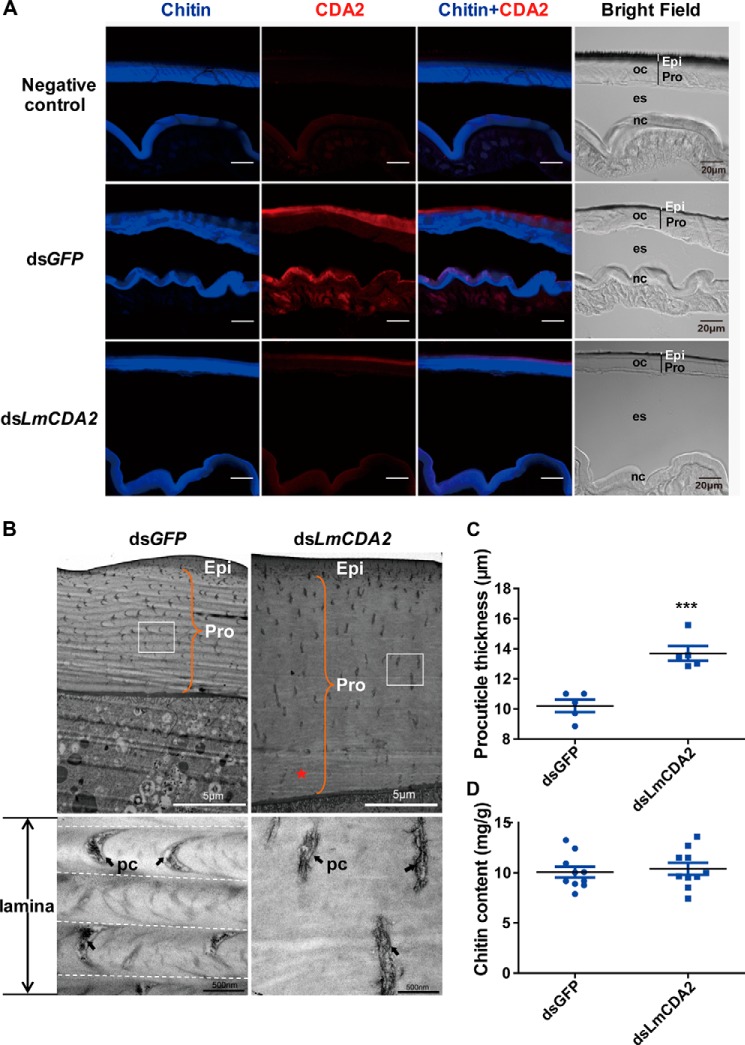
To scrutinize the biological role of LmCDA2, we sought to analyze its localization by immunofluorescence study of the integument. We first showed that the Drosophila CDA2 (Verm)-specific antibody recognizes LmCDA2 by mass spectrometry and Western blotting analyses (supplemental Figs. 1–4 and supplemental Table 1, see also “Experimental Procedures”). Then, we used this antibody for immunodetection experiments. A positive signal was found in both old and newly formed cuticles of the dsGFP-injected nymphs. This signal was visible at the apical site of the procuticle, but only a weak signal was detected in the dsLmCDA2-injected nymphs (Fig. 4A). Taken together, we conclude that dsLmCDA2 injection not only suppressed LmCDA2 transcript levels but consequently reduced LmCDA2 protein accumulation (Fig. 4A and supplemental Fig. 4). We also conclude that LmCDA2 localizes to the apical site of the procuticle (Fig. 4A).
FIGURE 4.
Localization of LmCDA2 in the locust integument and effects of dsLmCDA2 injection on locust cuticular structure. A, immunofluorescence detection of LmCDA2 using a polyclonal antibody against CDA2 (Verm) of the fruit fly D. melanogaster on cross-sections of locust abdominal cuticle by confocal laser-scanning microscopy. A secondary Cy3-Affinipure donkey anti-rabbit antibody was applied to detect the anti-Verm antibody (red). Sections were counterstained with FB28 to visualize chitin in the procuticle (blue signal). In dsGFP-injected locusts, LmCDA2 protein (red) localizes apical to the chitin signal (blue). Some red signal is visible in the epidermal cells, probably representing CDA2-containing vesicles. In dsLmCDA2-injected animals, only traces of LmCDA2 protein can be detected. The preimmune rabbit serum was used as negative control. Please note that in our experience the folding of the skin is random and therefore different between the specimens. Epi, epicuticle; Pro, procuticle; oc, old cuticle; nc, new cuticle; es, exuvial space. Moreover, because of resolution limits, the procuticle and epicuticle of the new cuticle cannot be assigned. Scale bar, 20 μm. B, ultrastructure observation by TEM. The cuticle structure was observed after dsLmCDA2 injection using TEM. Boxes in the upper panels indicate the region magnified in the lower panels. The orange brace indicates procuticle thickness. The black arrows indicate the chitin lamina. The chitin laminar organization seen in dsGFP-treated animals (dotted lines delimit laminae) was absent after injection of dsLmCDA2. Some faint striation (*) especially at the basal site of the cuticle close to the epidermal cell can be discerned in these animals. pc, pore canal. Epi, epicuticle; Pro, procuticle. Scale bar, 5 μm and 500 nm. C, thickness of the locust cuticle. The new procuticle between the lower border of the epicuticle and the upper surface of the epidermal cells (orange brace in B) was measured using a ruler after dsGFP or dsLmCDA2 injection. Data were reported as means ± S.E. of five independent biological replications. Statistical significance was analyzed with Student's t test. ***, p < 0.001. D, chitin content analyses of abdominal cuticles of dsGFP- or dsLmCDA2-injected nymphs. Data are presented as means ± S.E. of 10 independent biological replicates. Amounts are given as milligram/g cuticle sample (mg/g). Statistical significance was analyzed with Student's t test. There is no significant difference between chitin amounts in dsGFP- or dsLmCDA2-injected animals.
LmCDA2 Is Needed for Chitin Organization during Cuticle Differentiation
To address why locusts failed to molt after dsLmCDA2 injection, we analyzed the ultrastructure of the nymphal new cuticle with reduced LmCDA2 expression by transmission electron microscopy (TEM). In the nymphs injected with dsGFP, as traced by the crescentic orientation of the pore canals, the procuticle of the body cuticle harbored helicoidally arranged chitin laminae that according to Bouligand (25) appear as alternating electron-dense and electron-lucid horizontal stripes (Fig. 4B). In the dsLmCDA2-injected nymphs, the procuticle had lost its laminar organization. Some faint striations close to the epidermal cells were occasionally visible, however. These probably reflected incomplete down-regulation of LmCDA2 expression. In addition, as judged by the straight pore canals, these laminae were not stacked helicoidally. Furthermore, the procuticle of the dsLmCDA2-injected insects was thicker than the procuticle of the dsGFP-injected insects (Fig. 4, B and C), although the epicuticle was normal.
Chitin Amount Is Unchanged in dsLmCDA2-treated Nymphs
To determine whether the thicker procuticle of dsLmCDA2-injected nymphs is due to increased chitin content, we compared the chitin amounts in dsGFP- and dsLmCDA2-injected insects. Clearly, there were no significant differences in chitin content between the two treatments (Fig. 4D). Thus, increased thickness of procuticle does not correlate with the chitin content.
LmCDA2 Is Needed for Tracheal Cuticle Formation and Feeding
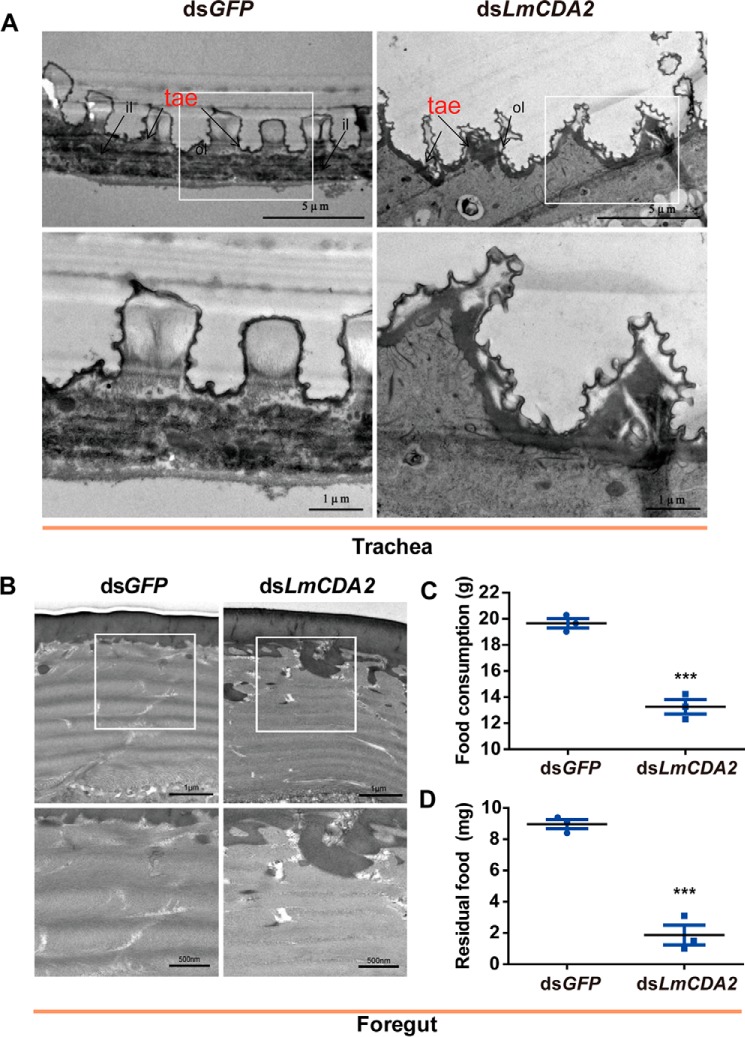
To further evaluate the role of LmCDA2 in cuticle formation, we analyzed the tracheal and the foregut cuticle that are produced by epithelial cells. In dsLmCDA2-injected insects, the taenidiae, which constitute the spiral cuticle of the tracheal system running perpendicular to the length of tracheal tubes, lost their regular spacing and shape. Moreover, their cuticular layers are, in contrast to the plain layers in untreated animals, uneven (Fig. 5A). In particular, the regular shape of the chitinous procuticle appeared to be deformed. By contrast, we were unable to discern any defect in the foregut cuticle in dsLmCDA2-injected insects compared with control insects (Fig. 5B). However, in our feeding experiments, we demonstrated that dsLmCDA2-injected insects consumed less food than control insects and that their foregut contained less food than their control counterparts (Fig. 5C). Thus, LmCDA2 plays an important role in feeding, probably due to foregut malfunction, but it apparently is not needed for foregut procuticle organization.
FIGURE 5.
Morphological observations of trachea and foregut after dsLmCDA2 injection by TEM. A, transmission electron micrographs of trachea after dsGFP and dsLmCDA2 injection. White open boxes indicate the region magnified in the lower panel. ol, outer layer; il, inner layer; tae, taenidiae. Scale bar, 5 and 1 μm. B, transmission electron micrographs of the foregut after dsLmCDA2 injection. White open boxes indicate the region magnified in the lower panel. Scale bar, 1 μm and 500 nm. C, food consumption; D, residual food in the foregut. Food content in the foregut was measured using an electronic balance at 5 days after injection of dsGFP or dsLmCDA2. Data are presented as mean ± S.E. of three independent biological replicates. Statistical significance was analyzed with Student's t test. ***, p < 0.001.
Discussion
The insect cuticle is a complex apical extracellular matrix formed by the underlying epidermal cells (37). The main component of the cuticle is the polysaccharide chitin that adopts an ordered organization to contribute to the physical properties of the cuticle. Despite some considerable amount of data, the molecular mechanisms of chitin organization are not well understood. Here, we show that the chitin deacetylase LmCDA2 in L. migratoria is indispensable for chitin organization in the body cuticle and the tracheal system but not in the foregut.
LmCDA2 Function Is Essential in the Migratory Locust
Injection of dsRNA against LmCDA2 into L. migratoria nymphs arrests fifth-instar eclosure. The respective insects are unable to molt and eventually die. Thus, LmCDA2 is responsible for the nymph-adult molts. Similarly, in D. melanogaster, mutations in the verm gene coding for a CDA2-type enzyme are embryonic lethal. D. melanogaster mutant embryos for both verm and serp (coding for CDA1) display a stronger phenotype than either single mutant embryo (16). This indicates a partially redundant function of CDA1- and CDA2-type chitin deacetylases in D. melanogaster. To what extent LmCDA1 and LmCDA2 have redundant functions remains to be elucidated.
Redundancy among chitin deacetylases is apparently common in insects. Our data indicate that the isoform LmCDA2a is essential, whereas activity of the isoform LmCDA2b seems to be dispensable for survival, although the patterns of LmCD2a and LmCD2b RNA accumulation are similar. Therefore, one may speculate that LmCDA2a (or any other chitin deacetylase) is able to replace LmCDA2b, although LmCDA2b is unable to rescue LmCDA2a suppression. A different complex situation involving chitin deacetylase isoforms is encountered in the red flour beetle T. castaneum. Despite similar expression patterns of the alternative spliced transcripts of TcCDA2, TcCDA2a, and TcCDA2b, the respective enzymes seem to have distinct functions. TcCDA2a is needed for the establishment of the soft femoral-tibial joint cuticle, and TcCDA2b is involved in the formation of the hard elytra (15). Overall, these functional differences are surprising, because the respective isoforms differ only in their chitin-binding domain. The chitin-binding efficiencies of some chitinases may actually depend on differences in their ChBD sequences (26). In turn, this might have an impact on enzyme processivity. Biochemical analyses of the CDA2 ChBDs shall shed light on this problem.
There are more examples of non-redundant relationships between chitin deacetylases. Non-redundancy is, for instance, observed in the honeybee Apis mellifera where the genes encoding CDA1 and CDA2 are expressed at different stages of development (27). In L. migratoria, some cuticle types such as the foregut and the hindgut are not fully disrupted upon dsLmCDA2 injection. If chitin organization in these tissues requires deacetylase function, this finding suggests that different non-redundant deacetylases are active in them. Taken together, to assess the full impact of chitin deacetylases in L. migratoria, identification and characterization of all LmCDAs are needed.
LmCDA2 Is Necessary to Establish Chitin Organization
Consistent with the data in D. melanogaster and T. castaneum, lethality of LmCDA2 down-regulation is correlated with tracheal and cuticular defects. Indeed, in D. melanogaster, the tracheal system is dysfunctional, and the helicoidal chitin organization in the body cuticle is lost when verm and serp are mutated (16–18). The newly adopted organization of chitin in these animals was not studied in detail.
In this work to interpret chitin organization in dsLmCDA2-treated locusts, we followed the arguments of Neville et al. (3, 4), who showed that helicoidal chitin arrangement is accompanied by crescent pore canals, although straight pore canals are found in cuticle types with unidirectional chitin microfibril orientation. Following this line of argument, we show that helicoidal chitin arrangement is lost when LmCDA2 expression is knocked down by RNAi in L. migratoria. This phenotype is also observed in the verm and serp mutant D. melanogaster larval cuticle (18). Instead, chitin microfibrils along the vertical axis of the cuticle, as judged by the straight pore canals, are oriented in parallel. This change in chitin architecture is accompanied by a loss of procuticle compactness.
These findings allow two fundamental conclusions. First, helicoidal packaging of chitin laminae is tighter than non-helicoidal packaging. Second, helicoidal arrangement of chitin laminae requires chitin deacetylation by LmCDA2. The aberrant chitin architecture may explain the lethal phenotype of LmCDA2 knocked down insects. Forces needed for eclosion may require mechanical properties of the cuticle conferred by helicoidally arranged chitin laminae rather than by unidirectional chitin laminae. By consequence, nymphs with reduced LmCDA2 function are unable to shed their old cuticle and die.
The situation is different in the foregut. In this tissue LmCDA2 is not needed for chitin organization. Nevertheless, nymphs with reduced LmCDA2 function consume less food than control nymphs. We reckon that LmCDA2 function in the foregut is required for correct mechanical properties of the cuticle without having a visible impact on chitin organization.
Model for Chitin Organization
What is the function of LmCDA2 during chitin lamina orientation? LmCDA2 localizes to the apical site of the procuticle. This observation argues that LmCDA2 is present at the initially produced chitin fibers that end up at the distal site of the procuticle. One possible interpretation is that deacetylation of chitin at the beginning of procuticle formation is necessary and sufficient for helicoidal chitin organization. Hence, LmCDA2 may modify the first chitin fibers produced but not be associated with further chitin fiber modifications. In our model (Fig. 6, A and B), we speculate that the first laminae may serve as a template for a helicoidal arrangement of the entire procuticle. Presumably, in this scenario and according to the hypothesis of Neville (7), proteins (yet unidentified) that specifically recognize and bind partially deacetylated chitin fix a microfibril conformation that dictates a helicoidal laminar architecture. Localization of LmCDA2 at the site between the pro- and epicuticle may rely on both the chitin-binding and the LDLa domains. The chitin-binding domain ensures association with the procuticle, while at the same time the LDLa domain anchors the proteins to the epicuticle, which contains lipids. Of course, this model has to be tested in appropriate genetic experiments in D. melanogaster and L. migratoria.
FIGURE 6.
Model illustrating the biological role of LmCDA2 in L. migratoria. A, helicoidal organization of chitin microfibrils in the locust cuticle relies on a sequence of molecular interactions starting with the localization of LmCDA2 between the epicuticle and the procuticle that contain lipids and chitin, respectively. Association with lipids and chitin is mediated by the LDLa (blue box) and the chitin-binding domains (green box), respectively. At the beginning, LmCDA2 deacetylases newly produced chitin through its CDA domain (brown box). How many chitin fibers along the vertical axis of the growing procuticle are modified is unclear. B, this partially deacetylated chitin serves as a template for the subsequent helicoidal arrangement of chitin laminae. The pore canals follow this organization and appear as crescentic structures. When LmCDA2 protein is absent, chitin laminae became disturbed and subsequently affect the structure of pore canals.
In summary, our data answer a fundamental question in insect biology, the organization of the cuticle formulated over 50 years ago by Neville (1). The alternating production of helicoidally and non-helicoidally stacked chitin laminae in the locust tibia as observed by Neville (1) possibly depends on the circadian expression and deposition of LmCDA2 into the growing cuticle. Understanding the molecular mechanisms of cuticle construction in general and chitin organization in particular will potentially have a double impact on molecular agricultural sciences seeking to optimize insect pest control and on material sciences that are inspired by biological materials such as the insect cuticle.
Experimental Procedures
Insects
The egg masses of L. migratoria were incubated at 28 ± 1 °C, 50% relative humidity, and with a 14-h light/10-h dark photoperiod in the laboratory. After 10 days, the nymphs were reared on fresh wheat sprouts under the same conditions (28). The wheat bran was supplemented after the nymphs reached third instar. Fifth-instar nymphs were used in this study.
Validation of cDNAs Putatively Encoding Chitin Deacetylases
Putative chitin deacetylase2 cDNA sequences were retrieved from the L. migratoria transcriptome by using bioinformatics methods (23). Total RNA was extracted from the integument of fifth-instar nymphs by using RNAisoTM Plus (TaKaRa, Japan). The first-strand cDNA was synthesized from 1 μg of total RNA by using Moloney murine leukemia virus reverse transcriptase (TaKaRa, Japan) and oligo(dT)18 primer (Thermo Fisher Scientific). PCR was performed with the cDNA and a pair of gene-specific primers (Table 1). After specific fragments were purified with the gel purification kit (Omega), each purified product was ligated into the pEASY-T3 vector (TransGen, China). The recombinant plasmid was used to transform Escherichia coli, and positive colonies were identified by bacterial colony PCR. The cDNAs were sequenced in both directions by the Thermo Fisher Scientific.
TABLE 1.
Primers for PCR amplification and dsRNA synthesis
| Application of primers | Gene | Primer sequence (5′-3′) | Product |
|---|---|---|---|
| bp | |||
| cDNA cloning | LmCDA2a | F, GGTGAAAGTGAAGCGGTCCA | 1806 |
| R, TCCTGGTGTATTTTGGGACTGG | |||
| LmCDA2b | F, GGTGAAAGTGAAGCGGTCCA | 1788 | |
| R, TCCTGGTGTATTTTGGGACTGG | |||
| mRNA expression | β-actin | F, CGAAGCACAGTCAAAGAGAGGTA | 156 |
| R, GCTTCAGTCAAGAGAACAGGATG | |||
| LmCDA2a | F, GGATTAGCTTTTGATATTGAACGAC | 148 | |
| R, CATTGCCACAGGACAGTTTGC | |||
| LmCDA2b | F, CGACGTTGAAAAGCAAACTTGT | 108 | |
| R, TTGCAGAAGAGTTCCTTCTCAATG | |||
| dsRNA | GFP | F, taatacgactcactatagggGTGGAGAGGGTGAAGG | 571 |
| synthesis | R, taatacgactcactatagggGGGCAGATTGTGTGGAC | ||
| LmCDA2 | F, taatacgactcactatagggGGTGCCACAGATGATCACAC | 496 | |
| R, taatacgactcactatagggGTTATGGGCATTACCGTTGC | |||
| LmCDA2a | F, taatacgactcactatagggTGCGACCAAGCGGGAGA | 136 | |
| R, taatacgactcactatagggGTGGCTTCTCTACCCTATCACAGTT | |||
| LmCDA2b | F, taatacgactcactatagggGGTGCACAAAGTCTGGTATAAAGC | 108 | |
| R, taatacgactcactatagggGCTGGTTACAGTTCGGCACTTTAC |
Bioinformatic Analysis
Conceptual translation of cDNA sequences was carried out with translation tools at ExPASy. Alignments of deduced amino acid sequences of LmCDA2 were conducted with GeneDoc software. Signal peptide was predicted using SignalP version 4.1 software. Analyses of conserved regions, including the chitin-binding/peritrophin-A domain (ChBD), a low-density lipoprotein receptor class A domain (LDLa), and polysaccharide deacetylase-like catalytic domain (CDA), were conducted with BlastP tools at the NCBI website. Multiple alignments of sequences were conducted by using ClustalX version 1.81 software. MEGA version 5.0 was utilized to construct the phylogenetic tree with the neighbor-joining method (29). Bootstrap analysis was performed by 1000 replications at the cutoff of 50% similarity. The name of genes and the accession numbers are listed in Table 2 .
TABLE 2.
Name of gene and accession number for phylogenetic analysis from NCBI
| Homologous gene | Insects | Accession number |
|---|---|---|
| DmCDA1 | D. melanogaster | NP730444 |
| DmCDA2 | D. melanogaster | NP730442 |
| DmCDA2a | D. melanogaster | NP730441 |
| DmCDA2b | D. melanogaster | NM168810 |
| DmCDA3 | D. melanogaster | NP609806 |
| DmCDA4 | D. melanogaster | NP728468 |
| DmCDA5a | D. melanogaster | AAF51567 |
| DmCDA5b | D. melanogaster | AAF51568 |
| DmCDA5e | D. melanogaster | ABI31281 |
| DmCDA5f | D. melanogaster | AFH03482 |
| DmCDA5g | D. melanogaster | AFH03483 |
| DmCDA5h | D. melanogaster | AFH03484 |
| DmCDA5i | D. melanogaster | AFH03485 |
| DmCDA5j | D. melanogaster | ABV53594 |
| DmCDA9a | D. melanogaster | NP611192 |
| DmCDA9b | D. melanogaster | NP001286519 |
| TcCDA1 | T. castaneum | NP001095946 |
| TcCDA2a | T. castaneum | NP001096047 |
| TcCDA2b | T. castaneum | NP001116303 |
| TcCDA3 | T. castaneum | NP001103905 |
| TcCDA4 | T. castaneum | NP001103903 |
| TcCDA5a | T. castaneum | NP001103739 |
| TcCDA5b | T. castaneum | NP001107799 |
| TcCDA6 | T. castaneum | NP001103905 |
| TcCDA7 | T. castaneum | ABW74150 |
| TcCDA8 | T. castaneum | NP001103906 |
| TcCDA9 | T. castaneum | NP001103904 |
| AgCDA1 | Anopheles gambiae | XP320597 |
| AgCDA2 | A. gambiae | XP320596 |
| AgCDA3 | A. gambiae | EJH28929 |
| OcCDA1 | Oxya chinensis | KP271171 |
| OcCDA2a | O. chinensis | KR537801 |
| OcCDA2b | O. chinensis | KR537802 |
| CfCDA2 | C. fumiferana | KC295591 |
| CfCDA2a | C. fumiferana | KC295540 |
| CfCDA2b | C. fumiferana | KC295591 |
| BmCDA2 | Bombyx mori | NP001103796 |
| BmCDA2a | B. mori | NP001103795 |
| BmCDA2b | B. mori | NP001103796 |
| LmCDA2a | L. migratoria | KR537804 |
| LmCDA2b | L. migratoria | KR537805 |
The gene structure was determined by searching the L. migratoria genome with LmCDA2 cDNA sequences as queries by using the NCBI Blast tool. Exons and introns were determined by comparing the genomic DNA and cDNA sequences, and the GT-AG rule was applied to determine the exon-intron boundaries. The exon-intron organization was graphed using the Adobe Illustrator CS3 software (Adobe).
Tissue-specific and Developmental Expression Analysis of LmCDAs
Total RNA was extracted using RNAisoTM Plus (TaKaRa, Japan) from each of seven different tissues, including the integument, foregut, midgut, gastric caeca, hindgut, Malpighian tubules, and fat body, dissected from fifth-instar nymphs. A total of 45 nymphs were collected and grouped into three biological replications. The quality and quantity of total RNA were evaluated on 1% agarose gel and NanoDrop 2000 (Thermo). One μg of total RNA was used to synthesize first-strand cDNA by using Moloney murine leukemia virus reverse transcriptase (TaKaRa, Japan). Each cDNA sample was diluted 20-fold for RT-qPCR analysis, and SYBR Green reagents were used according to the manufacturer (TOYOBO, Japan) with primers listed in Table 1. The reaction consisted of 10 μl of SYBR Green qPCR Master Mix (TOYOBO, Japan), 6.4 μl of deionized water, 2 μl of diluted template, and 0.8 μl of 0.4 μm forward and reverse primers, followed by the following conditions: denaturation at 95 °C for 1 min followed by 40 cycles at 95 °C for 15 s, 60 °C for 31 s with an ABI 7300 real time PCR machine (Applied Biosystems) using FastStart Universal SYBR Green Master. A melting curve was determined for each sample to detect the gene-specific peak and checked for the absence of primer-dimers. Relative transcript levels of target genes were calculated with the 2−ΔΔCt method (30), and the target gene expression level was normalized to the expression of the internal marker gene β-actin (KC118986) (31) that is stably expressed at all stages and in all tissues. Three independent biological replicates were performed.
Functional Analysis of LmCDA2 by RNA Interference (RNAi)
RNAi was performed to reveal the biological role of LmCDA2 during development and the molting process of L. migratoria. The forward and reverse primers harboring T7 RNA polymerase promoter sequences for synthesizing dsRNA of LmCDA2 (dsLmCDA2) and GFP (dsGFP, control) genes were designed using the E-RNAi web service (Table 1). The region (588–1083 bp) common for both variants was used for dsLmCDA2 synthesis, the splicing region “186–325 bp” for dsLmCDA2a and “190–316 bp” for dsLmCDA2b synthesis. PCR was performed to prepare the cDNA templates for synthesis of the dsLmCDA2 and dsGFP constructs, and single bands of expected PCR product size were verified on a 1% agarose gel. Concentrations of PCR products were determined by NanoDrop 2000 (Thermo Scientific). Two micrograms of PCR product was used as template to synthesize dsLmCDA2 and dsGFP using T7 RiboMAXTM Express RNAi System (Promega) (32). The synthesized dsLmCDA2 and dsGFP were dissolved in the appropriate volumes of deionized water, and the concentration was determined and adjusted to 2.5 μg/μl. Integrity of dsLmCDA2 and dsGFP were confirmed using 1% agarose gel analysis.
The 2nd-day-old fifth-instar nymphs were randomly divided into four groups (each with 40 nymphs) for injection of each dsRNA sample. Aliquots of 2.5 μl (6.25 μg) of dsLmCDA2, dsLmCDA2a, dsLmCDA2b, or dsGFP were injected into the hemocoel between the second and third abdominal segments by using a microinjector (Ningbo, China). To determine silencing efficiency, relative transcript levels of LmCDA2, LmCDA2a, and LmCDA2b were measured at 24 h after the injection of dsRNA by RT-qPCR as described above. Three biological and two technical replicates were applied for dsGFP, dsLmCDA2, dsLmCDA2a, and dsLmCDA2b injections. The remaining nymphs were maintained under the same conditions as described above. The visible phenotype changes were recorded every day until the nymphs started to molt to adults.
Immunohistochemistry
To analyze LmCDA2 protein localization, immune staining was performed as described (33). In brief, paraffin sections (5 μm) of the third abdominal segments from nymphs 9 days after injection of dsGFP or dsLmCDA2 in 2-day-old 5th-instar nymphs were prepared. The LmCDA2 protein was detected in paraffin section with the Drosophila Verm rabbit antiserum (1:300) (16) as a primary antibody at 4 °C overnight followed by washing with PBS three times for 5 min each. Then tissues were incubated with Cy3-Affinipure donkey anti-rabbit (Jackson ImmunoResearch) as secondary antibody for 1 h at room temperature. After washing the tissues three times with PBS, specimens were incubated with Fluorescent Brightener 28 (FB28) (Sigma) (1 mg/ml) for 5 s to detect chitin (34). The preimmune rabbit serum was used as negative control. We verified that the D. melanogaster Verm antibody cross-reacts with LmCDA2 in Western blotting and mass spectrometry experiments as shown in supplemental Figs. 1–4. Images were captured on an LSM 880 confocal laser-scanning microscope (Zeiss, Germany) equipped with a UV (405 nm) and a visible (561 nm) laser to excite FB28 and Cy3-Affinipure donkey anti-rabbit (Jackson ImmunoResearch), respectively.
TEM Examination after RNAi
TEM analysis was performed according to the described method (35). In brief, the cuticular structure of fifth-instar nymphs was examined at day 9 after dsRNA injection. The integument of the third abdominal segment and the foregut were dissected from each of three locusts and fixed with 3% glutaraldehyde in 0.2 m phosphate buffer (pH 7.2) for 48 h at 4 °C. The samples were then rinsed three times with the phosphate buffer followed by post-fixation in 1% osmium tetroxide for 3 h at 4 °C. The integument samples were washed twice, each for 10 min, and put into a series of ascending concentrations of acetone (50, 70, 80, 90, and 100%) for dehydration. They were embedded in Epon 812 at room temperature for 2 h. The samples were trimmed to prepare ultrathin sections. Sections were collected on copper grids, and images were captured with a JEM-1200EX transmission electron microscope (JEOL, Japan).
Chitin Content Analysis
The first, second, and third abdominal segments were dissected from the fifth-instar nymphs at day 9 for chitin content analysis. After the segments were dried at 90 °C until cuticle weight was stable, chitin preparation and chitin content measurements were performed based on the method described (36). Ten biological replicates (each with three technical replicates) were used for each of the dsLmCDA2- and dsGFP (control)-injected groups.
Assessment of Food Consumption
To determine the food consumption of the fifth-instar nymphs after treating with dsRNA, fresh wheat was provided to insects, and the consumed food of nymphs was measured after 5 days. In addition, the content of the foregut of these nymphs was determined. Twelve 2-day-old fifth-instar nymphs were treated with dsGFP (control) or dsLmCDA2, respectively. Three biological replicates were used for each group.
Statistical Analysis
The one-way analysis of variance test of the SPSS software (SPSS Inc., Chicago) was applied to analyze differences between tissues and developmental stages. Statistical significance was determined using the Tukey test. The t test was carried out for silencing efficiency, cuticle thickness, and food uptake after RNAi. Asterisks indicate significant differences (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
Author Contributions
J. Z. and B. M. conceived and coordinated the study and wrote the paper. R. Y. designed, performed, and analyzed the experiments shown in Figs. 1–4. W. L. designed, performed, and analyzed the experiments shown in Figs. 4 and 5. D. L., X. Z., G. D., M. Z., E. M., S. L., and K. Y. Z. provided technical assistance and contributed to the preparation of the figures. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Prof. Le Kang at the Institute of Zoology, Chinese Academy of Sciences, for sharing the resource of the locust EST and genome databases.
This work was supported by National Natural Science Foundation of China Grants 31272380 and 31672364, National Basic Research Program of China Grant 2012CB114102, Natural Science Foundation of Shanxi Province, China, Grant 2015011070, and Shanxi Scholarship Council of China Grant 2015-007. The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Table S1 and Figs. S1–S5.
- CDA
- chitin deacetylase
- ChBD
- chitin-binding peritrophin-A domain
- LDLa
- lipoprotein receptor class A
- Serp
- Serpentine
- Verm
- Vermiform
- TEM
- transmission electron microscope
- qPCR
- quantitative PCR
- dsRNA
- double-stranded RNA.
References
- 1. Neville A. C. (1965) Chitin lamellogenesis in locust cuticle. Q. J. Microsc. Sci. 106, 269–286 [PubMed] [Google Scholar]
- 2. Neville A. C. (1965) Circadian organization of chitin in some insect skeletons. Q. J. Microsc. Sci. 106, 315 [Google Scholar]
- 3. Neville A. C., and Luke B. M. (1969) A two-system model for chitin-protein complexes in insect cuticles. Tissue Cell 1, 689–707 [DOI] [PubMed] [Google Scholar]
- 4. Neville A. C., Thomas M. G., and Zelazny B. (1969) Pore canal shape related to molecular architecture of arthropod cuticle. Tissue Cell 1, 183–200 [DOI] [PubMed] [Google Scholar]
- 5. Noh M. Y., Kramer K. J., Muthukrishnan S., Kanost M. R., Beeman R. W., and Arakane Y. (2014) Two major cuticular proteins are required for assembly of horizontal laminae and vertical pore canals in rigid cuticle of Tribolium castaneum. Insect Biochem. Mol. Biol. 53, 22–29 [DOI] [PubMed] [Google Scholar]
- 6. Noh M. Y., Muthukrishnan S., Kramer K. J., and Arakane Y. (2015) Tribolium castaneum RR-1 cuticular protein TcCPR4 is required for formation of pore canals in rigid cuticle. PLoS Genet. 11, e1004963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Neville A. C. (1975) Biology of the Arthropod Cuticle, Zoophysiology and Ecology, pp. 71–73, Springer-Verlag, Berlin [Google Scholar]
- 8. Tsigos I., Martinou A., Kafetzopoulos D., and Bouriotis V. (2000) Chitin deacetylases: new, versatile tools in biotechnology. Trends Biotechnol. 18, 305–312 [DOI] [PubMed] [Google Scholar]
- 9. Dixit R., Arakane Y., Specht C. A., Richard C., Kramer K. J., Beeman R. W., and Muthukrishnan S. (2008) Domain organization and phylogenetic analysis of proteins from the chitin deacetylase gene family of Tribolium castaneum and three other species of insects. Insect. Biochem. Mol. Biol. 38, 440–451 [DOI] [PubMed] [Google Scholar]
- 10. Guo W., Li G., Pang Y., and Wang P. (2005) A novel chitin-binding protein identified from the peritrophic membrane of the cabbage looper, Trichoplusia ni. Insect Biochem. Mol. Biol. 35, 1224–1234 [DOI] [PubMed] [Google Scholar]
- 11. Campbell P. M., Cao A. T., Hines E. R., East P. D., and Gordon K. H. (2008) Proteomic analysis of the peritrophic matrix from the gut of the caterpillar, Helicoverpa armigera. Insect Biochem. Mol. Biol. 38, 950–958 [DOI] [PubMed] [Google Scholar]
- 12. Toprak U., Baldwin D., Erlandson M., Gillott C., Hou X., Coutu C., and Hegedus D. D. (2008) A chitin deacetylase and putative insect intestinal lipases are components of the Mamestra configurata (Lepidoptera: Noctuidae) peritrophic matrix. Insect Mol. Biol. 17, 573–585 [DOI] [PubMed] [Google Scholar]
- 13. Toprak U., Hegedus D. D., Baldwin D., Coutu C., and Erlandson M. (2014) Spatial and temporal synthesis of Mamestra configurata peritrophic matrix through a larval stadium. Insect Biochem. Mol. Biol. 54, 89–97 [DOI] [PubMed] [Google Scholar]
- 14. Zhong X. W., Wang X. H., Tan X., Xia Q. Y., Xiang Z. H., and Zhao P. (2014) Identification and molecular characterization of a chitin deacetylase from Bombyx mori peritrophic membrane. Int. J. Mol. Sci. 15, 1946–1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Arakane Y., Dixit R., Begum K., Park Y., Specht C. A., Merzendorfer H., Kramer K. J., Muthukrishnan S., and Beeman R. W. (2009) Analysis of functions of the chitin deacetylase gene family in Tribolium castaneum. Insect Biochem. Mol. Biol. 39, 355–365 [DOI] [PubMed] [Google Scholar]
- 16. Luschnig S., Bätz T., Armbruster K., and Krasnow M. A. (2006) Serpentine and vermiform encode matrix proteins with chitin binding and deacetylation domains that limit tracheal tube length in Drosophila. Curr. Biol. 16, 186–194 [DOI] [PubMed] [Google Scholar]
- 17. Wang S., Jayaram S. A., Hemphälä J., Senti K. A., Tsarouhas V., Jin H., and Samakovlis C. (2006) Septate-junction-dependent luminal deposition of chitin deacetylases restricts tube elongation in the Drosophila trachea. Curr. Biol. 16, 180–185 [DOI] [PubMed] [Google Scholar]
- 18. Gangishetti U., Veerkamp J., Bezdan D., Schwarz H., Lohmann I., and Moussian B. (2012) The transcription factor Grainy head and the steroid hormone ecdysone cooperate during differentiation of the skin of Drosophila melanogaster. Insect Mol. Biol. 21, 283–295 [DOI] [PubMed] [Google Scholar]
- 19. Quan G., Ladd T., Duan J., Wen F., Doucet D., Cusson M., and Krell P. J. (2013) Characterization of a spruce budworm chitin deacetylase gene: stage- and tissue-specific expression, and inhibition using RNA interference. Insect Biochem. Mol. Biol. 43, 683–691 [DOI] [PubMed] [Google Scholar]
- 20. Xi Y., Pan P. L., Ye Y. X., Yu B., and Zhang C. X. (2014) Chitin deacetylase family genes in the brown planthopper, Nilaparvata lugens (Hemiptera: Delphacidae). Insect Mol. Biol. 23, 695–705 [DOI] [PubMed] [Google Scholar]
- 21. Chen Y. L. (2000) Main achievement of research and management of migratory locust in China. Entomological. Knowledge 37, 50–55 [Google Scholar]
- 22. Kang L., Chen X., Zhou Y., Liu B., Zheng W., Li R., Wang J., and Yu J. (2004) The analysis of large-scale gene expression correlated to the phase changes of the migratory locust. Proc. Natl. Acad. Sci. U.S.A. 101, 17611–17615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ma Z., Yu J., and Kang L. (2006) LocustDB: a relational database for the transcriptome and biology of the migratory locust (Locusta migratoria). BMC Genomics 7, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang X., Fang X., Yang P., Jiang X., Jiang F., Zhao D., Li B., Cui F., Wei J., Ma C., Wang Y., He J., Luo Y., Wang Z., Guo X., et al. (2014) The locust genome provides insight into swarm formation and long-distance flight. Nat. Commun. 5, 2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bouligand Y. (1965) On a twisted fibrillar arrangement common to several biologic structures. C R Acad. Sci. Hebd. Seances Acad. Sci. D 261, 4864–4867 [PubMed] [Google Scholar]
- 26. Zhu Q., Arakane Y., Beeman R. W., Kramer K. J., and Muthukrishnan S. (2008) Characterization of recombinant chitinase-like proteins of Drosophila melanogaster and Tribolium castaneum. Insect Biochem. Mol. Biol. 38, 467–477 [DOI] [PubMed] [Google Scholar]
- 27. Wang Y. W., Odemer R., Rosenkranz P., and Moussian B. (2014) Putative orthologues of genetically identified Drosophila melanogaster chitin producing and organising genes in Apis mellifera. Apidologie 45, 733–747 [Google Scholar]
- 28. Guo Y., Zhang J., Yang M., Yan L., Zhu K. Y., Guo Y., and Ma E. (2012) Comparative analysis of cytochrome P450-like genes from Locusta migratoria manilensis: expression profiling and response to insecticide exposure. Insect Sci. 19, 75–85 [Google Scholar]
- 29. Tamura K., Peterson D., Peterson N., Stecher G., Nei M., and Kumar S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Livak K. J., and Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)) method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 31. Liu X., Li F., Li D., Ma E., Zhang W., Zhu K. Y., and Zhang J. (2013) Molecular and functional analysis of UDP-N-acetylglucosamine pyrophosphorylases from the migratory locust, Locusta migratoria. PLoS ONE 8, e71970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang J., Liu X., Zhang J., Li D., Sun Y., Guo Y., Ma E., and Zhu K. Y. (2010) Silencing of two alternative splicing-derived mRNA variants of chitin synthase 1 gene by RNAi is lethal to the oriental migratory locust, Locusta migratoria manilensis (Meyen). Insect Biochem. Mol. Biol. 40, 824–833 [DOI] [PubMed] [Google Scholar]
- 33. Yang M. L., Zhang J. Z., Zhu K. Y., Xuan T., Liu X. J., Guo Y. P., and Ma E. B. (2009) Mechanisms of organophosphate resistance in a field population of oriental migratory locust, Locusta migratoria manilensis (Meyen). Arch. Insect Biochem. Physiol. 71, 3–15 [DOI] [PubMed] [Google Scholar]
- 34. Toprak U., Baldwin D., Erlandson M., Gillott C., and Hegedus D. D. (2010) Insect intestinal mucins and serine proteases associated with the peritrophic matrix from feeding, starved and molting Mamestra configurata larvae. Insect Mol. Biol. 19, 163–175 [DOI] [PubMed] [Google Scholar]
- 35. Liu W., Xie Y., Xue J., Gao Y., Zhang Y., Zhang X., and Tan J. (2009) Histopathological changes of Ceroplastes japonicus infected by Lecanicillium lecanii. J. Invertebr. Pathol. 101, 96–105 [DOI] [PubMed] [Google Scholar]
- 36. Zhang J., and Zhu K. Y. (2006) Characterization of a chitin synthase cDNA and its increased mRNA level associated with decreased chitin synthesis in Anopheles quadrimaculatus exposed to diflubenzuron. Insect Biochem. Mol. Biol. 36, 712–725 [DOI] [PubMed] [Google Scholar]
- 37. Moussian B. (2010) Recent advances in understanding mechanisms of insect cuticle differentiation. Insect Biochem. Mol. Biol. 40, 363–375 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.