Abstract
The bile acid-sensitive ion channel (BASIC) is a member of the degenerin/epithelial Na+ channel (Deg/ENaC) family of ion channels. It is mainly found in bile duct epithelial cells, the intestinal tract, and the cerebellum and is activated by alterations of its membrane environment. Bile acids, one class of putative physiological activators, exert their effect by changing membrane properties, leading to an opening of the channel. The physiological function of BASIC, however, is unknown. Deg/ENaC channels are characterized by a trimeric subunit composition. Each subunit is composed of two transmembrane segments, which are linked by a large extracellular domain. The termini of the channels protrude into the cytosol. Many Deg/ENaC channels contain regulatory domains and sequence motifs within their cytosolic domains. In this study, we show that BASIC contains an amphiphilic α-helical structure within its N-terminal domain. This α-helix binds to the cytosolic face of the plasma membrane and stabilizes a closed state. Truncation of this domain renders the channel hyperactive. Collectively, we identify a cytoplasmic domain, unique to BASIC, that controls channel activity via membrane interaction.
Keywords: bile acid, cholesterol, ion channel, patch clamp, X-ray scattering, Xenopus, BASIC, BLINaC, Deg/ENaC, amphiphilic helix
Introduction
In humans the degenerin/epithelial Na+ channel (Deg/ENaC)3 gene family comprises nine genes and in rodents eight genes, which give rise to different subunits of the acid-sensing ion channels (ASICs), to three to four subunits of the epithelial Na+ channel, and to the bile acid-sensitive ion channel (BASIC) (1–3). ASICs are mainly neuronal channels, which are activated by a sudden increase in the extracellular proton concentration (4). They are involved in the sensation of pain and mechanical stimuli, synaptic transmission, and various cognitive processes (5–12). ENaC is mainly involved in the absorption of Na+ in a variety of tissues. Best described is its role in the distal tubule and the collecting duct of the nephron, where it represents the rate-limiting step of Na+ absorption and thus plays a major role in Na+ homeostasis and blood volume control (13, 14). BASIC represents a third subgroup within the human and rodent Deg/ENaC family (3) and is sometimes referred to as ASIC5 (15), although it is not sensitive to protons (16, 17). Instead, BASIC is activated by millimolar concentrations of bile acids (18, 19).
Information on the structure and composition of Deg/ENaC channels is mainly based on the crystal structure of chicken ASIC1, which forms a homotrimeric complex, where each subunit is composed of two transmembrane domains, which are linked by a highly structured extracellular domain with various structural elements (20). The N- and C-terminal domains of each subunit are facing the cytosolic compartment (20–22).
BASIC was initially named brain-liver-intestine Na+ channel due to its expression pattern in these tissues (16). In addition, it is also found in testis, kidney, and lung (16). In the brain, it is predominantly expressed in a subset of cells within the cerebellum (15). In the liver, it is mainly found in cholangiocytes, epithelial cells that line the lumen of bile ducts (18). As its new name implies, BASIC is activated by bile acids and may therefore play a role in bile acid-dependent absorption and secretion in the bile ducts but also other epithelia along the intestinal tract that are in contact with bile acids (18). Because of their amphiphilic character, bile acids are membrane-active substances. Interestingly, other membrane-active substances, which are structurally unrelated to bile acids, also activate BASIC (23), suggesting that it is an ion channel that is sensitive to alterations of the biophysical properties of the plasma membrane (23). Furthermore, rat and human BASIC are characterized by a high affinity for extracellular Ca2+, which blocks the channels. Consequently, a reduction of the extracellular Ca2+ concentration leads to an activation of BASIC (17). The physiological function of BASIC has not been established yet; however, its expression in bile duct epithelial cells and its activation by bile acids point toward a role in epithelial transport in bile ducts and perhaps the intestine.
A large number of studies have shown that the cytosolic domains can either serve as targets for regulatory processes of Deg/ENaC channels or that they can modulate their biophysical properties (2). The epithelial Na+ channel (ENaC), for example, contains a conserved PY motif in the C-terminal domain, which serves as binding site for members of the Nedd4 family of ubiquitin ligases (24). Its surface expression is controlled via Nedd4-mediated ubiquitination of ENaC and thus the amount of Na+ absorption. Liddle's syndrome, an inherited form of hypertension, is caused by mutations within the Nedd4-binding site of ENaC, which alter ubiquitination and consequently surface expression (24–26). Several ASICs contain a PDZ motif at their C-terminal domain, which serve as interaction sites for a variety of regulatory proteins (2). ASIC4 was recently shown to traffic predominantly into early endosomes, a process that is mainly controlled by a sequence motif in the N-terminal domain of the channel (27). For ASIC2 and ASIC3, the selectivity between monovalent cations is influenced by the cytosolic N-terminal domain (28), and for ASIC1, the permeability for Ca2+ is affected by the analogous domain (29), showing that cytosolic domains can affect genuine biophysical pore properties.
Structural information on the cytosolic termini of cASIC1 and any other Deg/ENaC channel is lacking because these domains were not resolved by crystallization of cASIC1 (30). However, understanding the structure and the function of the cytosolic regions of BASIC could provide valuable information on the specific regulation of the channel.
In this study, we identified an amphiphilic α-helical domain in the cytosolic N-terminal sequence of BASIC, which inhibits the channel by binding to the plasma membrane and thus stabilizing the closed state of the channel. Taken together, our findings emphasize the role of the plasma membrane for the activity of the channel and provide a mechanistic insight into membrane sensitivity of BASIC.
Results
Removal of the Cytosolic N-terminal Region of rBASIC Increases Its Activity
Although the transmembrane and the extracellular domains are highly conserved between different members of the mammalian Deg/ENaC family, the cytosolic N-terminal domain shows a high degree of diversity (Fig. 1). Of the 65 cytosolic amino acid residues of rBASIC, only a few are conserved, including the HG motif, which is present in all mammalian Deg/ENaC channels (2). Therefore, we reasoned that the cytosolic N-terminal sequence of rBASIC may contain specific regulatory motifs.
FIGURE 1.
Alignment of the N-terminal cytosolic sequences of the members of the Deg/ENaC family from Rattus norvegicus. Residues conserved between all members are highlighted in red, and residues conserved between BASIC and at least five members are highlighted in green. The first transmembrane domain is indicated by a box (TMD1); only few amino acids close to TMD1 show high conservation. Accession numbers are as follows: NP_071563, BASIC; NP_077068, ASIC1a; XP_008764015, ASIC1b; NP_001029186, ASIC2a; NP_037024, ASIC2b; NP_775158, ASIC3; NP_071570, ASIC4; NP_113736, αENaC; NP_036780, βENaC; NP_058742, γENaC. Alignment was performed using Clustal W version 2.1.
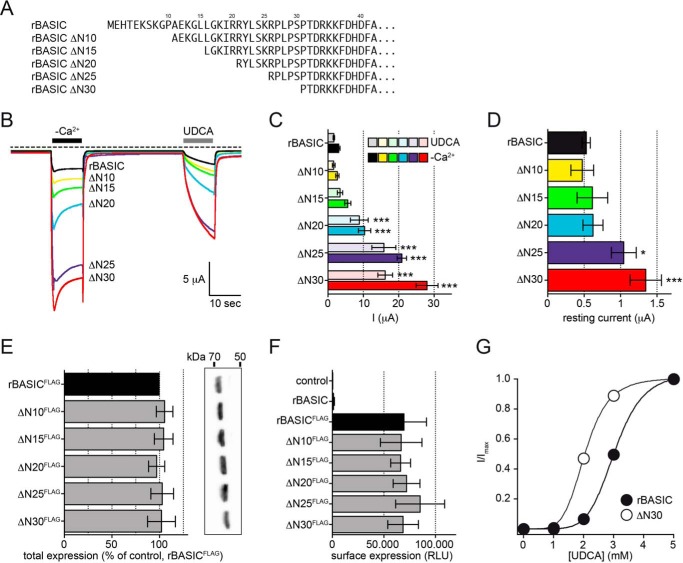
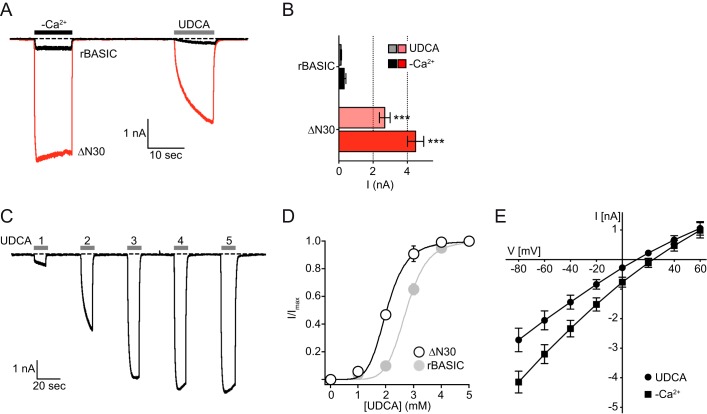
To identify such putative motifs, we generated a series of N-terminal truncations (ΔN10, -15, -20, -25, and 30, see Fig. 2A) of rBASIC, and we analyzed their effect by activating the channels successively by the removal of extracellular divalent cations (−Ca2+) and the application of 2 mm UDCA (Fig. 2), a potent activator of rat and human BASIC (19).
FIGURE 2.
Successive truncation of the N-terminal domain increases the activity of rBASIC. A, representation of the N-terminal sequences of WT rBASIC and the respective truncations. B, representative current traces illustrating the activation of WT rBASIC, rBASIC ΔN10, ΔN15, ΔN20, ΔN25, and ΔN30 by the removal of extracellular divalent cations (−Ca2+) and application of 2 mm UDCA. C, quantitative comparison of current amplitudes induced by the removal of divalent cations (−Ca2+, solid bars) or application of 2 mm UDCA (transparent bars) as shown in B). Error bars, S.E., n = 12, ***, p < 0.001 (ANOVA). D, quantitative comparison of the resting current amplitudes before divalent free activation as shown in B. Error bars, S.E., n = 12, *, p < 0.05; ***, p < 0.001 (ANOVA). E, expression levels of WT rBASICFLAG and the respective truncations as determined by Western blotting. Left, quantitative analysis of Western blotting results (n = 5); band intensities were determined using the software BioChem (Vilber Lourmat, Eberhardzell, Germany) and normalized against rBASICFLAG. Right, representative Western blot. No significant differences between WT rBASIC and the respective truncations were identified (ANOVA). F, surface expression levels of WT rBASICFLAG and the respective truncations as determined by oocyte luminescence assay. Uninjected oocytes and control oocytes expressing rBASIC without FLAG epitope served as controls. No significant differences between WT rBASIC and the truncations were observed (ANOVA). G, concentration-response curve for UDCA for WT rBASIC (closed circles) and rBASIC ΔN30 (open circles). Currents were normalized to the maximum current in the presence of 5 mm UDCA, which were 30.4 ± 5.9 μA for WT rBASIC and 57.8 ± 7.2 μA for rBASIC ΔN30. Error bars, S.E., curves were fitted to the Hill equation (n = 8).
Removal of the first 10 or 15 amino acids did not significantly change the responses to divalent cation removal and UDCA compared with wild-type rBASIC (WT −Ca2+, 3.1 ± 0.4 μA; UDCA, 1.7 ± 0.2 μA; ΔN10 −Ca2+, 2.7 ± 0.5 μA; UDCA, 1.6 ± 0.4 μA; ΔN15 −Ca2+, 5.6 ± 0.9 μA; UDCA, 3.4 ± 0.8 μA, n = 12), and the removal of the first 20 amino acids, however, led to an approximate 3-fold increase of both responses (ΔN20 − Ca2+, 10.4 ± 1.7 μA; UDCA, 8.9 ± 2.5 μA). Removal of the first 25 and 30 amino acids even more dramatically increased the activity of the channel, for ΔN25 8–9-fold (−Ca2+, 20.9 ± 1.3 μA; UDCA, 15.8 ± 3.4 μA, n = 12) and for ΔN30 ∼10-fold compared with WT (−Ca2+, 28 ± 3.1 μA; UDCA, 16.2 ± 2.1 μA, n = 12) (Fig. 2, B and C).
rBASIC is characterized by a small resting activity when expressed in Xenopus oocytes (17). Similar to the currents induced by removal of divalent cations or by UDCA application, the resting activity was significantly increased when the first 25–30 amino acids were removed (Fig. 2D). Taken together, these data suggest that the amino acid sequence from position 15 to position 25 contains a crucial domain, which inhibits the channel if present. Its deletion consequently increases channel activity.
To address the mechanism behind this increased channel activity, we first tested whether the expression of the channels in Xenopus oocytes was altered by the truncations. Neither the total expression (Fig. 2E) nor the surface expression (Fig. 2F) was affected by the truncations implying that neither an increased forward trafficking nor a reduced turnover was responsible for the observed potentiation of channel activity. The truncation of the first 30 amino acids, however, increased the apparent affinity for UDCA compared with WT rBASIC (2.1 ± 0.05 mm and 3.0 ± 0.05 mm, n = 6, respectively, Fig. 2G). Based on these results we conclude that the sequence from position 15 to 25 contains an intrinsic inhibitory domain, which directly affects the activity of the channel.
The N-terminal domains of human BASIC and rat BASIC share ∼80% sequence identity. Therefore, we tested whether hBASIC activity could also be increased by the removal of the first 30 N-terminal amino acids. Indeed, ΔN30 hBASIC showed an approximate 4-fold potentiation upon application of 1.5 mm DCA or removal of extracellular divalent cations (Fig. 3).
FIGURE 3.
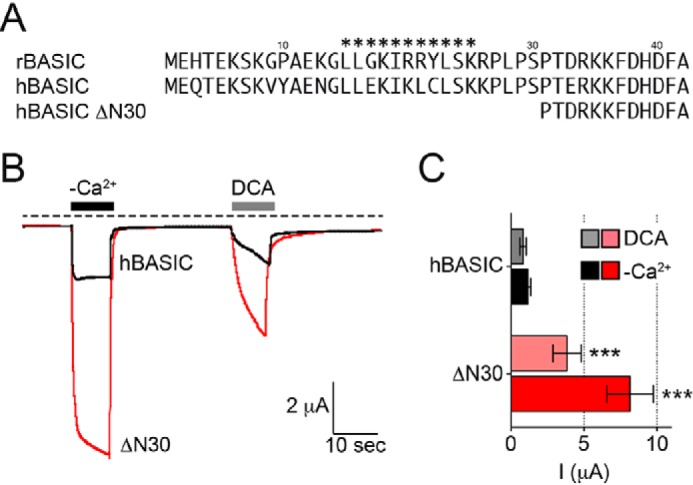
Removal of the N-terminal domain of hBASIC increases its activity. A, representation of the N-terminal sequences of rat and human BASIC and hBASIC with the ΔN30 truncation. B, representative current traces illustrating the activation of WT hBASIC and hBASIC ΔN30 by the removal of extracellular divalent cations (−Ca2+) or by the application of 1.5 mm DCA. C, quantitative comparison of current amplitudes induced by the removal of divalent cations (−Ca2+, solid bars) and 1.5 mm DCA (transparent bars) as shown in B. Error bars, S.E., n = 8, ***, p < 0.001 (ANOVA).
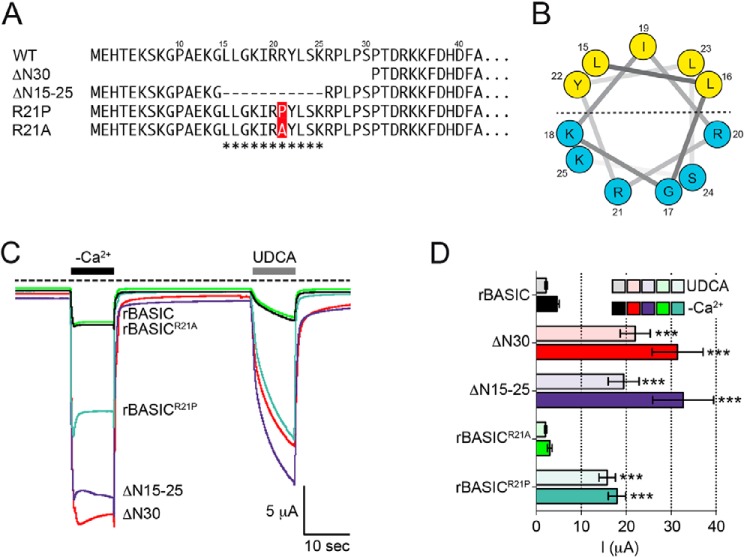
An Amphiphilic α-Helix in the N-terminal Region Forms an Inhibitory Domain
Next, we analyzed the N-terminal sequence of rBASIC for the presence of putative structural motifs using different structure prediction software (Protean DNASTAR; Predict Protein Open) (Fig. 4, A and B). Interestingly, we found that amino acids 15–25 can form a putative three-turn α-helix, which is characterized by an amphiphilic distribution of apolar and polar residues (Fig. 4B). To confirm the importance of the amino acid stretch from position 15 to 25, we removed these 11 residues (ΔN15–25), while leaving the rest of the sequence intact (Fig. 4A). Indeed, the deletion of these amino acids did mimic the effect of the full truncation of the first 30 amino acids (Fig. 4, C and D) confirming that within the N-terminal domain, amino acids 15–25 are the crucial sequence motif responsible for an inhibition of rBASIC. Next, we tested the α-helical structure of amino acids 15–25 by mutating the centrally located arginine residue at position 21 into a proline (Fig. 4A). The introduction of the proline residue should interfere with the helical structure by inducing a kink in the helix. Indeed, this mutation led to a strong potentiation of the divalent removal-induced current and the UDCA-induced current, similar to the truncation of the first 30 amino acids and the deletion of amino acids 15–25 (Fig. 4, C and D). In contrast, introduction of an alanine residue at position 21 did not affect the activity of the channel (Fig. 4, C and D). In summary, these data suggest that amino acids 15–25 form a three-turn amphiphilic α-helix, which serves as a cytosolic inhibitory domain for rBASIC.
FIGURE 4.
Amphiphilic α-helix in the N-terminal region forms an inhibitory domain. A, representation of the N-terminal sequences of rBASIC, rBASIC ΔN30, rBASIC ΔN15–25, rBASICR21P, and rBASICR21A; the putative α-helix is labeled with asterisks. B, helical wheel representation of the putative amphiphilic α-helix. The numbers represent the amino acid positions of WT rBASIC. Color code of amino acid residues: yellow, hydrophobic; blue, hydrophilic. C, representative current traces showing the activation of WT rBASIC, rBASIC ΔN30, rBASIC ΔN15–25, rBASICR21P, and rBASICR21A by the removal of extracellular divalent cations (−Ca2+) or the application of 2 mm UDCA. D, quantitative comparison of current amplitudes induced by the removal of divalent cations (−Ca2+, solid bars) or by the application of 2 mm UDCA (transparent bars) as shown in C. Error bars, S.E., n = 9, ***, p < 0.001 (ANOVA).
N-terminal Domain Stabilizes the Closed State of rBASIC
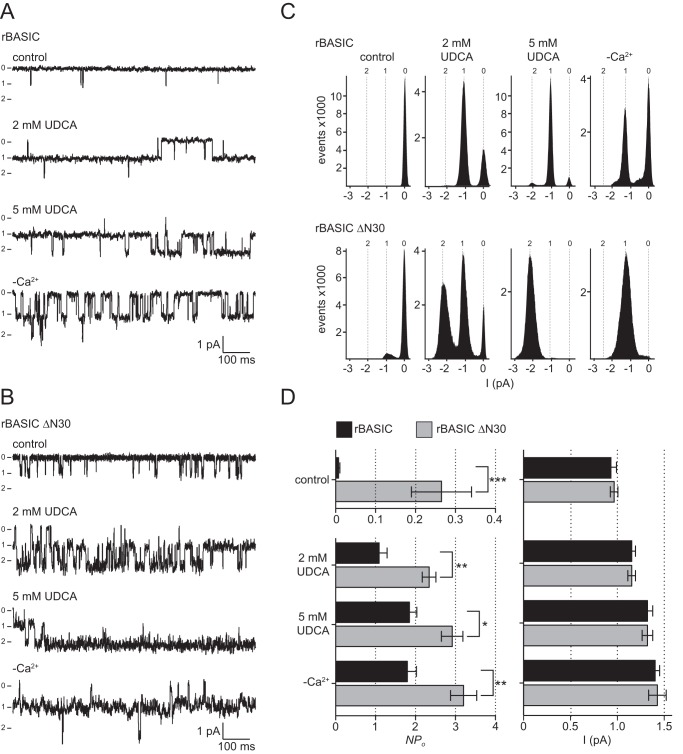
To address the functional difference between WT rBASIC and ΔN30 rBASIC, we performed single channel patch clamp recordings in the outside-out configuration from Xenopus oocytes. We investigated the effect of application of 2 mm UDCA and 5 mm UDCA and extracellular divalent cation removal on WT rBASIC and ΔN30 rBASIC. At a holding potential of −60 mV, single channel transitions of WT rBASIC and ΔN30 rBASIC were detected (Fig. 5, A–C). WT rBASIC showed a very low activity (open probability, NPo) of 0.007 ± 0.002, similar to the previously described single channel characteristics of hBASIC (NPo = 0.011) (31); in contrast, the hyperactive ΔN30 rBASIC showed a dramatically increased activity (NPo) of 0.23 ± 0.07 compared with WT rBASIC (n = 8, p < 0.001) (Fig. 5D). The current amplitudes at a holding potential of −60 mV did not differ significantly between WT rBASIC and ΔN30 rBASIC (WT rBASIC, I−60 mV = 0.93 ± 0.06 pA; ΔN30 rBASIC I−60 mV = 0.97 ± 0.04 pA) and were similar to hBASIC as described previously (I−70 mV = 0.95 pA) (Fig. 5D) (31).
FIGURE 5.
Truncation of the N-terminal domain of rBASIC increases the open probability of the channel. A and B, representative segments of single channel current traces from outside-out patches from oocytes expressing rBASIC WT (A) or rBASIC ΔN30 (B). The traces were recorded at a holding potential of −60 mV in the absence of an activating stimulus (control), the presence of 2 mm UDCA or 5 mm UDCA, or in the absence of extracellular divalent cations (−Ca2+) (0 = closed state and 1, 2 = open states). Currents were recorded for 30 s after each solution exchange. C, single channel binned amplitude histograms; data points were obtained from 10-s segments, including the current traces shown in A and B. Histograms were used to determine the channel activity (NPo). D, left panel, summary of calculated NPo values for rBASIC WT and rBASIC ΔN30 under control condition and after activation with 2 or 5 mm UDCA or by removal of extracellular divalent cations (−Ca2+) obtained from eight recordings similar to the recordings shown in A. Right panel, summary of single channel current amplitudes of rBASIC WT and rBASIC ΔN30 under control conditions and after activation with 2 or 5 mm UDCA or by removal of extracellular divalent cations (−Ca2+) from similar recordings as shown in A or B. n = 8; *, p < 0.05; **, p < 0.01; *, p < 0.001* (ANOVA).
Upon application of 2 or 5 mm UDCA, the activity (NPo) of WT rBASIC and ΔN30 rBASIC increased dramatically; however, ΔN30 rBASIC showed a significantly higher activity than WT rBASIC (WT rBASIC, NPo = 1.1 ± 0.2 and 1.8 ± 0.2, respectively; ΔN30 rBASIC, NPo = 2.3 ± 0.2 and 2.9 ± 0.3, respectively) (Fig. 5). The single channel amplitude of WT rBASIC and ΔN30 rBASIC slightly increased upon activation with 2 and 5 mm UDCA (WT rBASIC, 1.1 ± 0.03 and 1.3 ± 0.09 pA, respectively; ΔN30 rBASIC, 1.2 ± 0.04 and 1.3 ± 0.05 pA, respectively); however, no significant difference in single channel amplitude between WT rBASIC and ΔN30 rBASIC was observed. Similar to UDCA, removal of extracellular divalent cations strongly increased the activity of both WT rBASIC and ΔN30 rBASIC, and similar to the control condition or the activation by UDCA, activity of ΔN30 rBASIC was significantly more strongly increased compared with WT rBASIC (NPo = 2.5 ± 0.5 and 1.7 ± 0.4, respectively). The single channel amplitude was increased to 1.4 pA for both channels by removal of divalent cations (WT rBASIC, 1.4 ± 0.05 pA; ΔN30 rBASIC, 1.4 ± 0.08 pA), but no significant difference between WT rBASIC and ΔN30 rBASIC was observed (Fig. 5). Interestingly, the current recordings of ΔN30 rBASIC had a higher background noise (Fig. 5, A and B), possibly indicating that fast single channel transitions occur, which cannot be resolved using this experimental setup.
Taken together, the single channel data suggest that removal of the N-terminal domain increases the activity of rBASIC by increasing the open probability of the channel but not by affecting the single channel conductance. We conclude that the N-terminal α-helix stabilizes the closed state of rBASIC.
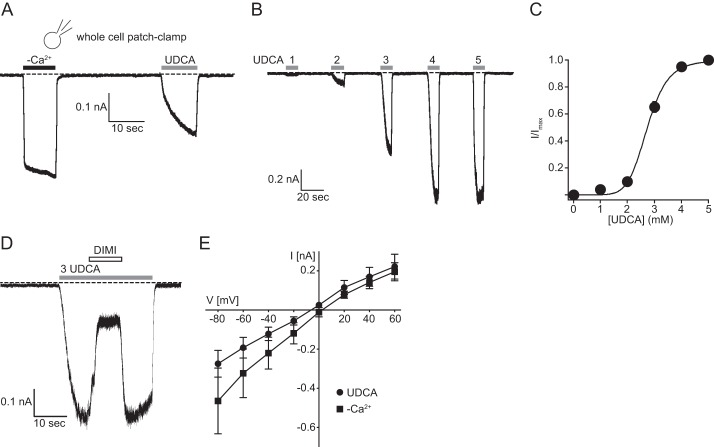
Patch Clamp Recordings from HEK293 Cells Confirm Biophysical Characteristics of rBASIC Obtained from Xenopus laevis Oocytes
Most previous measurements of WT BASIC were generated using the X. laevis oocyte expression system (16–19, 31, 32). Patch clamp recordings of BASIC expressed in mammalian cells have only been reported for BASIC containing a gain-of-function mutation at the crucial degenerin position (16). We expressed WT rBASIC transiently in HEK293 cells and recorded currents induced by both: application of bile acids and divalent cation removal (Fig. 6A). As a first step, we analyzed the biophysical characteristics of rBASIC expressed in HEK293 cells and compared it with the previously described properties of rBASIC expressed in Xenopus oocytes. In contrast to Xenopus oocytes, rBASIC did not show a significant activity at rest when expressed in HEK293 cells. rBASIC was activated by UDCA, however, and the apparent affinity of rBASIC was 2.7 ± 0.07 mm, which is very similar to the apparent affinity observed for rBASIC when expressed in Xenopus oocytes (2.5 mm ± 0.04) (Fig. 6, B and C). Furthermore, the UDCA-induced current could be inhibited by 10 μm diminazene (Fig. 6D), a potent inhibitor for rBASIC (32). WT rBASIC expressed in Xenopus oocytes is characterized by unselectivity for monovalent cations, a rather uncommon feature of Deg/ENaC channels (17). The reversal potential of the UDCA-activated rBASIC in HEK293 cells was −3.9 ± 2.7 mV, and the reversal potential of rBASIC activated by divalent cation removal was slightly higher (5.4 ± 1.6 mV; p < 0.05, n = 7) (Fig. 6E), indicating that rBASIC expressed in HEK293 cells is also characterized by unselectivity for monovalent cations. Taken together, these data suggest that rBASIC has very similar functional characteristics when expressed in Xenopus oocytes or in HEK293 cells.
FIGURE 6.
Whole cell patch clamp recordings from HEK293 cells expressing rBASIC. A, representative current trace illustrating the activation of rBASIC expressed in HEK293 cells by the removal of extracellular divalent cations (−Ca2+) or by the application of 2 mm UDCA. B, representative current trace illustrating the concentration-dependent activation of rBASIC by UDCA. C, concentration-response curve for UDCA. Currents were normalized to the maximum current in the presence of 5 mm UDCA, which was 1.27 ± 0.25 nA. Error bars, S.E., curves were fitted to the Hill equation (n = 8). D, representative current trace illustrating the diminazene-dependent inhibition (10 μm diminazene) of rBASIC activated by 3 mm UDCA. E, mean current-voltage relationships of rBASIC in the absence of extracellular divalent cations (squares) or in the presence of UDCA (circles). The holding potential was increased stepwise from −80 to +60 mV in 20-mV steps. Error bars, S.E.; n = 8.
Furthermore, we analyzed the role of the amphiphilic α-helix in the N-terminal domain of rBASIC. Similar to the results obtained in Xenopus oocytes, removal of the first 30 amino acids dramatically increased the responses to divalent cation removal and UDCA application. Although the increase in amplitude of WT rBASIC currents was 0.31 ± 0.11 nA to divalent cation removal and 0.13 ± 0.03 nA to application of 2 mm UDCA, the increases in current amplitude of ΔN30 rBASIC currents were significantly higher, 4.48 ± 0.53 and 2.68 ± 0.40 nA (p < 0.005, n = 8), respectively (Fig. 7, A and B). Furthermore, the apparent affinity of ΔN30 rBASIC was 2.1 ± 0.04 mm and thus significantly higher than the apparent affinity of WT rBASIC to UDCA, partly explaining the stronger response to UDCA (Fig. 7, C and D). Interestingly, the reversal potential of ΔN30 rBASIC activated by divalent cation removal and the UDCA-activated state was shifted to more positive values (divalent free, 10.6 ± 2.7 mV; UDCA, 24.6 ± 5.2 mV) (Fig. 7E), indicative of an increase in selectivity for Na+ over K+.
FIGURE 7.
Removal of the N-terminal domain of rBASIC increases its activity in HEK293 cells. A, representative current traces illustrating the activation of WT rBASIC and rBASIC ΔN30 expressed in HEK293 cells by the removal of extracellular divalent cations (−Ca2+) or by the application of 2 mm UDCA. B, quantitative comparison of current amplitudes induced by the removal of divalent cations (−Ca2+, solid bars) or by the application of 2 mm UDCA (transparent bars) as shown in A. Error bars, S.E., n = 8, ***, p < 0.001 (ANOVA). C, representative current trace illustrating the concentration-dependent activation of rBASIC ΔN30 by UDCA. D, concentration-response curve of rBASIC ΔN30 for UDCA. Currents were normalized to the maximum current in the presence of 5 mm UDCA, which was 5.72 ± 0.63 nA. Error bars, S.E., curves were fitted to the Hill equation (n = 8). For comparison, the concentration-response curve of WT rBASIC for UDCA is shown in light gray. E, mean current-voltage relationships of rBASIC ΔN30 in the absence of extracellular divalent cations (squares) and the presence of UDCA (circles). The holding potential was increased stepwise from −80 to +60 mV in 20-mV steps. Error bars, S.E.; n = 8.
Amphiphilic α-Helix Mediates Membrane Association of the N-terminal Region of rBASIC
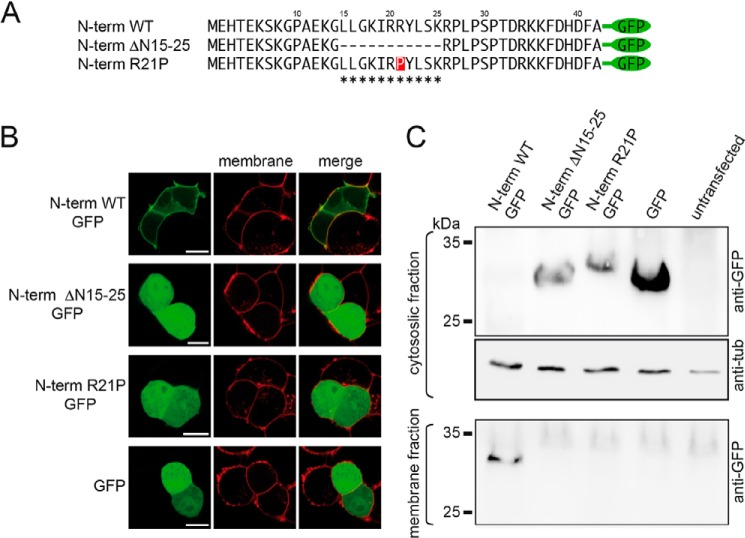
Based on the amphiphilic composition of the putative α-helix, we hypothesize that it may be tethered to the plasma membrane in a longitudinal manner with the polar residues facing the polar headgroups and the apolar residues facing the apolar tail groups of the lipid bilayer. To verify this hypothesis, we fused the first 42 N-terminal amino acids of the rBASIC N terminus of GFP (N-term WT GFP) (Fig. 8A) and expressed this fusion protein in HEK293 cells. Strikingly, the fusion protein localized almost completely to the plasma membrane (Fig. 8B), whereas GFP when expressed alone showed a typical diffuse cytosolic pattern, suggesting that the 40 N-terminal amino acids of rBASIC tether the soluble GFP to the plasma membrane, thus supporting the hypothesis that it binds to the plasma membrane. Removal of the α-helical domain from the N-terminal sequence (N-term ΔN15–25 GFP) (Fig. 8A) completely abolished the membrane localization of GFP (Fig. 8B). Furthermore, the introduction of a proline into the center of the putative α-helix (N-term R21P GFP) (Fig. 8A) also fully abolished the membrane localization (Fig. 8B). Taken together, these data imply that the cytosolic N-terminal domain of rBASIC binds the plasma membrane, and this binding is mediated by the putative amphiphilic α-helix at position 15–25.
FIGURE 8.
Amphiphilic α-helix mediates membrane association of the N-terminal region of BASIC. A, representation of the N-terminal sequences of rBASIC, rBASIC ΔN15–25, and rBASICR21P; the putative α-helix is labeled with asterisks. B, representative images of GFP fused to the N-terminal sequences of rBASIC or GFP alone in HEK293 cells (green, left panel). The plasma membrane was stained using CellMaskTM deep red plasma stain (red, middle panel). Right panel, merged images. Scale bars, 5 μm. C, representative Western blot showing the exclusive presence of the full N-terminal domain in the insoluble membrane fraction (lower panel), whereas the N-terminal domain lacking amino acids 15–25 and the N-terminal domain containing the R21P mutation are exclusively present in the cytosolic soluble fraction (upper panel). Loading control, tubulin (middle panel).
Using a biochemical approach, we confirmed membrane binding of the N-terminal domain. We expressed the GFP fusion proteins and GFP alone in HEK293 cells and separated membrane and cytosolic fractions. Although N-term WT GFP was almost exclusively found in the membrane fraction, N-term ΔN15–25 GFP and N-term R21P GFP were only found in the cytosolic fraction like GFP alone (Fig. 8C). These results perfectly match the results from the fluorescence localization study.
To verify the position of the N-terminal domain within the bilayer, we conducted X-ray diffraction of oriented and stacked artificial membranes in complex with a peptide representing the first 35 N-terminal amino acids of rBASIC. Analogous to protein crystallography, X-ray diffraction of oriented membranes permits structural refinement of the membrane on the angstrom level. In a further analogy to protein crystals, our membranes are measured in a dehydrated state, at T = 28 °C and 50% relative humidity, to further enhance structural features. Two membrane complexes were prepared as follows: a sample composed purely of 1,2-dimyristoyl-sn-glycero-3-phosphatidylcholine (DMPC), a 14-carbon saturated phospholipid, and DMPC membranes in complex with 0.6 mol % peptide. Out-of-plane X-ray diffraction was taken as shown in Fig. 9B. A series of evenly spaced Bragg peaks was observed from both membranes, indicating well ordered membrane stacks. The lamellar spacings for the membranes complexes without and with the peptide were determined to be 53.6 and 55 Å, respectively, in agreement with previous reports (33–36).
FIGURE 9.
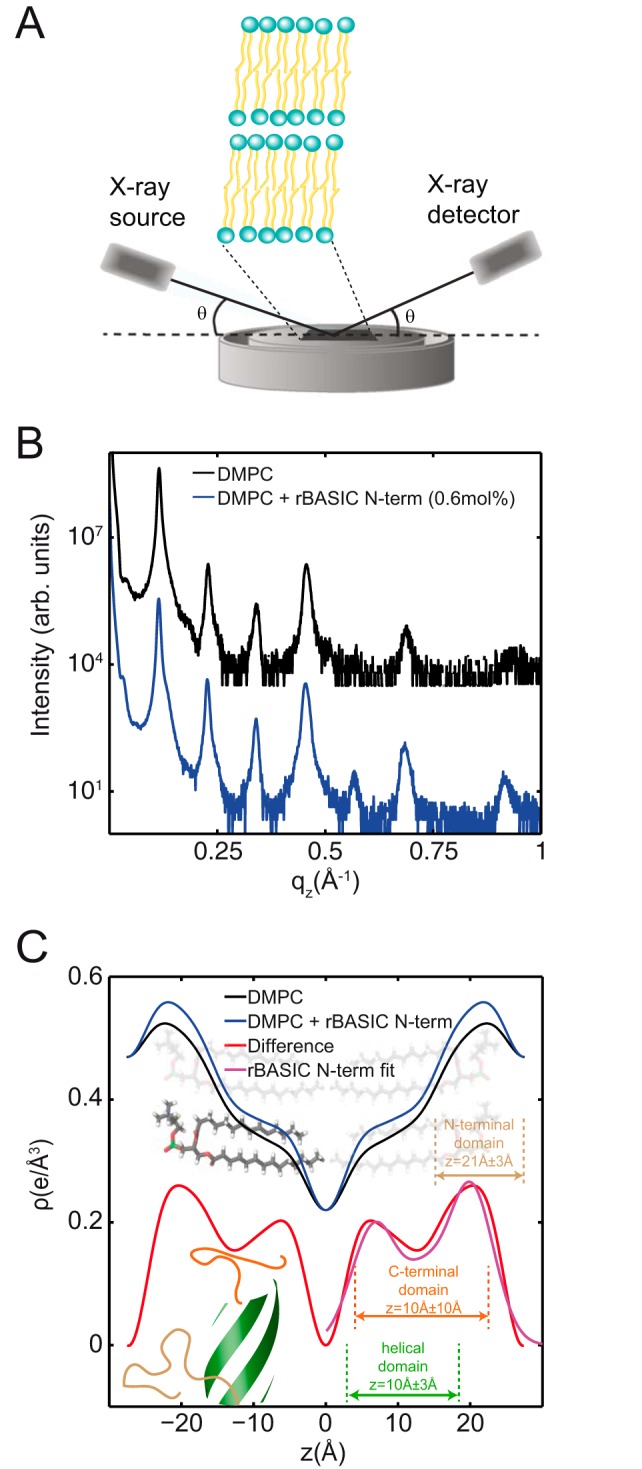
X-ray diffraction of synthetic membranes containing the N-terminal domain locate the peptide fragment within the membrane in a helical configuration. A, schematic of the experimental setup. The atomistic structure of oriented lipid bilayers was studied using X-ray diffraction in supported, highly aligned lipid membranes. B, out-of-plane diffraction measured from membrane complexes with and without the peptide fragment, rBASIC N-term. C, electron density profiles for each membrane complex, calculated by Fourier transformation of the Bragg peaks. The schematic lipids highlight the meaning of the features in the density profile: the peak at z ∼21 Å represents the electron-rich headgroup, although the bilayer center is between the tails of two lipid leaflets. The density of the peptide was calculated from ρrBASIC N-term(z) = ρrBASIC N-term + membrane(z), − ρmembrane(z) and is shown in the red curve. The electron density for a model of the peptide configuration and position is shown by the magenta curve.
The Bragg peaks were integrated and Fourier-transformed resulting in electron density profiles perpendicular to the bilayers. The density profiles, shown in Fig. 9C, represent the density of electrons at position z along the membrane normal. The membrane density, ρmembrane(z) in black in Fig. 9C, is highest at the position of the phosphate-rich lipid headgroups, at z values of z ∼21 Å, and decreases to the bilayer center at z = 0 Å. Changes to the electron density for a membrane in complex with the peptide, ρmembrane + rBASIC N-term(z), represent additional electrons added by the peptide. In this model, the peptide density, ρrBASIC N-term(z), is calculated as ρpeptide(z) = ρmembrane + rBASIC N-term (z) − ρmembrane(z) (red curve in Fig. 9C). Note that the red curve in Fig. 9C is vertically scaled for clarity. The data indicate the peptide fully embeds within the headgroups of the lipid bilayer, along 3 Å < z < 25 Å.
To determine the structure of the peptide in the bilayer based on ρrBASIC N-term(z), a model for the peptide electron density was derived. This procedure has previously been used to determine the position of the amyloid-β peptide in various lipid membranes (36, 37). In this model, residues 1–14 and 26–35 were modeled as amorphous, and their density was described by individual Gaussian profiles. Residues 15–25 were modeled by a three-turn helix, which was positioned with the long axis of the helix in the plane of the membrane. The resultant fit is shown by the magenta curve in Fig. 9C. In this model, the more hydrophobic C-terminal domain (residues 26–35) embeds within the membrane tails at 0 Å < z < 20 Å from the center. The helix then embeds in a similar location, 7 Å < z < 13 Å, with the hydrophilic residues facing the headgroups. The amorphous N-terminal domain was found in the headgroup region, 18 Å < z < 24 Å. As highlighted in Fig. 9C, the model electron density well describes the experimental ρrBASIC N-term(z), presenting strong evidence the peptide that embeds within the membrane takes the proposed helical configuration in the synthetic membrane system.
rBASIC Activity Is Dependent on the Cholesterol Content of the Plasma Membrane
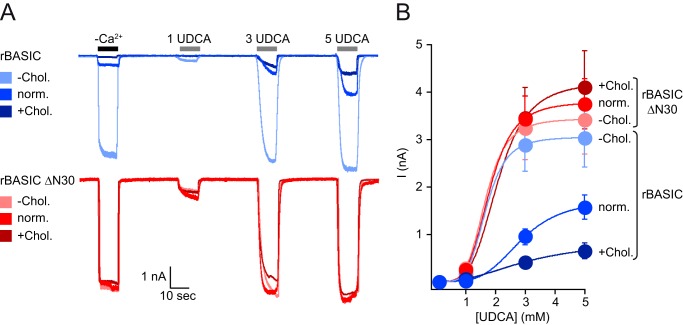
As we have shown in our previous study, rBASIC is sensitive for its membrane environment (23), and bile acids most likely activate rBASIC by altering membrane properties. Other membrane-active substances, such as N-lauroylsarcosine or SDS, are also potent activators of rBASIC (23). Because cholesterol is a major component of the plasma membrane, we reasoned that altering the cholesterol content of the membrane may affect the activity of the channel. Manipulation of the cholesterol content in Xenopus oocytes did not affect rBASIC activity (data not shown). Therefore, we studied the effect of cholesterol on rBASIC activity in HEK293 cells. WT rBASIC expressing HEK293 cell membranes were either depleted of cholesterol by incubation with 10 mm MβCD or enriched by incubation with 10 mm water-soluble cholesterol. rBASIC activity was then studied using an application protocol, which included divalent free activation and successive activation by increasing concentrations of UDCA (1, 3, and 5 mm). Interestingly, cholesterol depletion strongly potentiated WT rBASIC activity. The divalent free-induced current amplitude increased from 0.45 ± 0.07 to 2.97 ± 0.57 nA (p < 0.05; n = 10), and the UDCA (3 mm)-induced current amplitude increased from 1.56 ± 0.26 to 3.03 ± 0.62 nA (p < 0.05; n = 8). Conversely, cholesterol enrichment strongly decreased both the divalent free current amplitude to 0.20 ± 0.42 nA (p < 0.05; n = 8) and the UDCA-induced current amplitude to 0.65 ± 0.12 nA (p < 0.05; n = 8) (Fig. 10A).
FIGURE 10.
rBASIC activity is dependent on the cholesterol content of the plasma membrane. A, representative current traces showing the activation of WT rBASIC (upper panel, blue traces) and rBASIC ΔN30 (lower panel, red traces) expressed in HEK293 cells by the removal of extracellular divalent cations (−Ca2+) and the application of increasing concentrations UDCA. Before current recordings, HEK293 cells were either incubated for 1 h in 10 mm MβCD to remove cholesterol from the plasma membrane (−Chol.) or incubated for 1 h in 10 mm water-soluble cholesterol to increase the cholesterol content of the plasma membrane (+Chol.). Untreated HEK293 cells served as control (norm.). B, concentration-response curves for UDCA for WT rBASIC and rBASIC ΔN30 recorded from cells treated with 10 mm MβCD or 10 mm water-soluble cholesterol or untreated as shown in A. Color coding matches traces shown in A. Error bars, S.E., curves were fitted to the Hill equation (n = 8).
The apparent affinity of WT rBASIC for UDCA in the presence of increased membrane cholesterol content was significantly decreased compared with the apparent UDCA affinity of WT rBASIC in the presence of normal cholesterol (norm chol., 2.7 ± 0.1 mm; +chol., 3.1 ± 0.1 mm; p < 0.05, n = 10), although the apparent UDCA affinity was increased in the absence of membrane cholesterol (−chol., 1.7 ± 0.05 mm; p < 0.005, n = 10) (Fig. 10B).
To address whether the amphiphilic α-helix also mediates the cholesterol sensitivity of rBASIC, we repeated the cholesterol depletion and enrichment experiments with ΔN30 rBASIC. Strikingly, activity of ΔN30 rBASIC was not significantly different between normal, low, or high cholesterol conditions. Neither the current amplitudes induced by divalent cation removal nor the current amplitudes induced by 3 mm UDCA changed significantly in the presence of different membrane cholesterol concentrations (I−Ca2+, norm chol., 3.20 ± 0.44 nA; low chol., 3.09 ± 0.63 nA; high chol., 2.90 ± 0.28 nA; n.s., n = 10; I3 mM UDCA, norm chol., 3.42 ± 0.73 nA; low chol., 3.75 ± 0.53; high chol., 4.10 ± 0.76 nA; n.s., n = 10) (Fig. 10A).
The apparent affinity for UDCA was also not affected (norm chol., 1.8 ± 0.08 mm; +chol., 2.1 ± 0.1 mm; −chol., 1.7 ± 0.07 mm; n.s., n = 10) (Fig. 10B). This finding clearly suggests that the cholesterol sensitivity of rBASIC is mediated by the N-terminal domain of rBASIC, most likely the amphiphilic α-helix formed by residues 15–25.
In summary, cholesterol negatively regulates rBASIC activity, and cholesterol requires the presence of the amphiphilic α-helix to exert its inhibitory effect. Furthermore, these data support our previous finding that rBASIC is sensitive for alterations of its membrane environment (23).
Discussion
Our study on the role of the cytosolic N-terminal domain of BASIC has two major findings. First, we show that this domain inhibits the activity of rat and human BASIC. Second, we show that the structural basis of this inhibitory domain is a three-turn α-helix, which shows an amphiphilic amino acid composition. We propose that this α-helix is longitudinally embedded into the plasma membrane with the polar residues facing the charged headgroups and the apolar residues facing the tail groups of the lipids in the plasma membrane. In addition, the α-helical domain is important for the newly identified sensitivity of BASIC for the cholesterol content of the plasma membrane.
The evidence for the proposed model is severalfold. Removal of the first 30 amino acids renders the channel hyperactive, without affecting the expression of the channel (Figs. 2, 3, 6, and 7). Within the first 30 amino acid residues, we identified a putative α-helix-forming sequence motif ranging from amino acids 15 to 25. Removal of these amino acids had the same effect as removal of the first 30 amino acids (Fig. 4), suggesting that these 11 amino acids are responsible for the observed hyperactivity. We verified the α-helical nature of this sequence motif by introducing a helix-breaking proline mutation (R21P) and observed a similar effect as for the ΔN30 truncation or the ΔN15–25 deletion, whereas an alanine mutation was without any effect (R21A) (Fig. 4). This finding supports the assumption that residues 15–25 form an α-helix. However, other structural conformations or sequence motifs, which may also be disrupted by the proline mutation, may also explain the inhibitory effect.
To analyze the molecular basis for the hyperactivity of the ΔN30 rBASIC, we compared the single channel properties of WT rBASIC and ΔN30 rBASIC. The single channel open probability was increased by the removal of the first 30 amino acids, whereas the single channel amplitude was not significantly affected (Fig. 5), indicating that the closed state of rBASIC is stabilized by the N-terminal domain.
Based on the amphiphilic amino acid composition of the putative α-helix, we hypothesized that it may direct and tether the N-terminal domain to the plasma membrane. We provide evidence that the N-terminal domain indeed localizes to the plasma membrane in HEK293 cells and that the α-helix is crucial for this localization (Fig. 8). Furthermore, we show that in artificial membranes the N-terminal domain embeds into the membrane in a manner that perfectly matches our model (Fig. 9), where the amphiphilic helix is oriented parallel to the membrane lipid bilayer.
In addition to the functional and structural information, we provide evidence that rBASIC is negatively controlled by membrane cholesterol. Upon removal of the N-terminal domain, this inhibitory effect of cholesterol is abolished (Fig. 10). We conclude that the N-terminal domain mediates the cholesterol sensitivity of rBASIC.
BASIC and Membrane Sensitivity
rBASIC is activated by bile acids. This activation is mediated by bile acid-induced membrane alterations. The membrane sensitivity of BASIC is not specific to bile acids but rather is a general feature because other membrane-active substances also activate rBASIC in a similar fashion (23). In our previous study, we speculate that the physiological function of BASIC may be associated with its membrane sensitivity, and we showed that the structural basis of membrane sensitivity are various regions of BASIC, including the TMDs and the ECD. Here, we present data that show that the cytosolic N-terminal domain is involved in membrane binding and membrane-based stabilization of the channel in the closed state. This is perfectly in line with our previous conclusion of the membrane sensitivity of BASIC (23). Furthermore, it supports membrane sensitivity as a genuine functional feature of BASIC.
In a recent study, a putative binding site for bile acids in the pore region of ENaC was identified (38). Whether this binding site is also present in BASIC remains unknown. However, this binding site is localized in the transmembrane region of the channel and thus can only be reached via the lipid bilayer. An alteration of the membrane is therefore occurring before the possible binding may occur and may actually be a prerequisite for binding.
What Is the Mechanistic Basis for Closed State Stabilization?
The hyperactivity of ΔN30 BASIC is due to an increased open probability and not increased single channel amplitude. Based on our results, we speculate that the amphiphilic α-helices of the three subunits of a functional channel, embed longitudinally within the bilayer, forming an additional membrane anchor. For an opening of the channel, the transmembrane helices, which form the pore, have to move (20, 39). This movement could be hindered allosterically by the membrane-anchored N-terminal domain. In this model, removal of the N-terminal domain would allow an easier movement of the transmembrane domains and thus an easier opening of the pore.
An alternative mechanism could involve proteins, which may bind to the N-terminal domain and link the channel to solid structures within the cytosol, for example, the cytoskeleton. Additional experiments are required to unequivocally solve the mechanistic basis for the inhibitory effect of the N-terminal domain.
What Is the Possible Role of This Newly Identified Regulatory Mechanism?
A large variety of ion channels are regulated and modulated by their cytosolic domains. The inhibitory domain of BASIC, which stabilizes the closed state, may serve as a target for a large variety of regulatory processes.
Phosphorylation or any other kind of post-translational modification within the α-helix or its vicinity may interfere with its ability to bind to the membrane, which may eventually lead to an increased activity of the channel. The ENaC is also a target for a variety of post-translational modifications, including proteolytic cleavage in its ECD, which increases its activity (40). Similar to this activating mechanism, it is possible that intracellular proteases may cleave off the N-terminal part of BASIC upon maturation or application of a certain stimulus and thus lead to a more active channel compared with resting conditions. We screened the N-terminal sequence for conserved cleavage sites but did not identify any. The absence of such a motif does not speak in favor of such a mechanism but cannot rule out such a possibility.
The N-terminal domain of BASIC, including the α-helix, may serve as interaction site for regulatory proteins. Such interaction sites are not without precedent within the Deg/ENaC family (2) and, for example, are important for the proper function of ENaC (26). The hyperactivity of the channel, which is induced by the removal of the N-terminal domain, may therefore also be caused by abolishing interaction with inhibitory proteins. However, the fact that we observe these results in different cellular systems may speak in favor of the previously described membrane binding of the domain rather than a possible function as an interaction site for regulatory proteins.
Electrophysiological Properties of BASIC in Different Cellular Systems
Most information on the electrophysiological properties of BASIC originate from the X. laevis oocyte expression system (16–18, 32). In COS cells, BASIC only produced detectable currents when gain-of-function mutations in the degenerin site were introduced (16). Here, we present for the first time functional data for BASIC expressed in mammalian cells. These patch clamp data reveal the inhibition by extracellular divalent cations, sensitivity to bile acids, and inhibition by diminazene; hence, BASIC shows similar characteristics when expressed in different cellular systems. These finding are important for two different reasons. First, the bile acid sensitivity of BASIC, which in fact represents membrane sensitivity, is not an expression system-specific phenomenon but is a general feature of the channel. This supports the possibility that membrane sensitivity may represent a genuine functional feature of BASIC. Second, it will enable experiments that were not possible in oocytes, for example screening for interaction partners and regulatory proteins.
Besides the common electrophysiological and pharmacological similarities of BASIC expressed in Xenopus oocytes and mammalian cells, we observed a clear difference. BASIC expressed in HEK293 cells does not show a resting activity as in oocytes. We conclude that the plasma membrane of Xenopus oocytes and HEK293 cells has slightly different physical properties and hence affects the activity of the channel slightly differently. It is possible that the plasma membrane of HEK293 cells is more rigid compared with the Xenopus oocyte membrane, thus stabilizing the closed state of the channel more efficiently. For the future, it would be interesting to address the question of how different membranes may affect the channel differently. Because BASIC is expressed in a variety of tissues like the bile duct or neurons in the cerebellum, where the membrane composition is likely to be different, the properties of the channel could be tissues or cell type-specific.
Modulation of BASIC by Cholesterol
We made use of the ability to measure functional BASIC expressed in HEK293 cells to analyze the effect of cholesterol on BASIC activity. Interestingly, the channel is negatively modulated by cholesterol. This modulation may have two different implications. First, cholesterol as part of the plasma membrane affects general physical membrane properties and may thus influence the activity of BASIC. Second, lipid bilayer microdomains, like lipid rafts, are characterized by increased cholesterol content, and BASIC may be regulated in its activity by shifting between lipid raft regions and non-lipid raft regions. It would be interesting to test BASIC for a possible lipid raft localization and lipid raft-dependent activity.
In summary, our results highlight the importance of the cytosolic N terminus of BASIC and suggest the presence of a membrane-binding amphiphilic α-helical structure. These findings may eventually contribute to understanding the physiological function of the channel.
Experimental Procedures
Molecular Biology
Cloning of rBASIC (GenBankTM accession no. NM_022227) and hBASIC (GenBankTM accession no. NM_017419) was described previously (17, 31). For expression analysis of rBASIC, a FLAG epitope (DYKDDDDK) was introduced in the extracellular domain (ECD) between amino acids Asp156 and Phe157 by overlap PCR (17). Truncations were produced by PCR; fragments were cloned into the oocyte expression vector pRSSP6009 (29), and single amino acid substitutions were generated by site-directed mutagenesis with KAPA HiFi polymerase (Peqlab, Erlangen, Germany) using standard protocols. cDNA inserts were completely sequenced. The SP6 mMessage mMachine kit (Ambion, Austin, TX) was used to amplify capped cRNA from linearized plasmids. For the generation of GFP fusion proteins, the N-terminal sequences of rBASIC were inserted into pEGFP-C1 (Clontech, Saint-Germain-en-Laye, France).
Two Electrode Voltage Clamp in X. laevis Oocytes
Surgical removal of oocytes was performed as described previously (17). Anesthetized frogs were killed after the final oocyte collection by decapitation. Animal care and experiments followed approved institutional guidelines at RWTH Aachen University. cRNAs (8 ng) were injected into stage V or VI oocytes of X. laevis. Oocytes were kept in low-Na+ OR-2 medium (in mm, 5 NaCl, 77.5 N-methyl-d-glucamine, 2.5 KCl, 1 Na2HPO4, HEPES, 1.0 MgCl2, 1 CaCl2, and 0.5 mg/ml polyvinylpyrrolidone) at 19 °C. Whole cell currents were recorded 24–48 h post-injection at room temperature (20–25 °C) with a Turbo Tec 03× amplifier (npi electronic, Tamm, Germany). Oocytes were superfused using an automated, fast, pump-driven solution exchange system (npi electronic) (41, 42). Data acquisition and solution exchange were controlled using Cellworks 5.1.1 (npi electronic). Data were filtered at 20 Hz and acquired at 1 kHz. Holding potential was −70 mV, if not stated otherwise. Standard bath solution for two electrode voltage clamp measurements contained (in mm) 140 NaCl, 1.8 CaCl2, 1 MgCl2, and 10 HEPES. Low Ca2+ (10 nm) bath solution (denoted as −Ca2+ in figures) contained (in mm) 140 NaCl, 10 HEPES, 2 EDTA, and 0.56 of CaCl2 and was supplemented with 100 μm FFA to block the conductance induced in Xenopus oocytes by divalent free extracellular solutions.
Single Channel Patch Clamp Recordings
Single channel recordings from oocytes were performed as described previously (43). Briefly, 24–48 h prior to experiments, oocytes were injected with WT rBASIC or ΔN30 rBASIC coding cRNA (0.008 ng). The oocyte vitelline membrane was manually removed before it was placed in the recording chamber containing standard bath solution containing (in mm) 140 NaCl, 1.8 CaCl2, 1 MgCl2, and 10 HEPES. Patch pipettes with a resistance of 7–9 MΩ made from thick-walled borosilicate glass capillaries (Harvard Apparatus, Holliston, MA) with dental wax-coated tips were used. Pipettes were filled with pipette solution containing (in mm) 90 K+ gluconate, 5 NaCl, 2 MgCl2, and 2 EGTA, pH 7.3. Seals were formed, and the outside-out configuration was established. Outside-out patches were clamped at −60 mV, and solution was exchanged manually using a gravity-driven perfusion system. Data were filtered at 5 kHz and digitized at 20 kHz with an Axopatch 200B amplifier (Axon Instruments, Molecular Devices, Sunnyvale, CA). All single channel experiments were carried out at room temperature.
Determination of Total and Surface Expression in X. laevis Oocytes
For total expression, microsomal membranes were prepared from oocytes 2 days after injection, as described previously (44). Proteins from microsomal membranes of 2–5 oocytes were separated by SDS-PAGE and transferred to a PVDF membrane (PolyScreen; PerkinElmer Life Sciences, Rodgau, Germany). The membrane was then probed using a mouse anti-FLAG antibody (Sigma, Munich, Germany, catalog no. F3165, LOT 21K9230) in combination with a peroxidase-coupled goat anti-mouse antibody (Sigma, catalog no. A4416, LOT SLBL3207).
The surface expression of rBASICFLAG and the rBASICFLAG truncations was determined as described previously (45). Mouse anti-FLAG antibody and peroxidase-coupled anti-mouse antibody (Sigma) were employed to label the surface-expressed FLAG-tagged rBASIC constructs. Oocytes were injected with 8 ng of cRNA. Relative light units were calculated as a measure of surface-expressed channels. The results are from at least three independent experiments with oocytes from different frogs; at least eight oocytes were analyzed for each experiment and each condition.
Cell Culture, Confocal Microscopy, Cholesterol Manipulation, and Membrane Isolation
HEK293 cells were cultured on poly-l-lysine-coated 35-mm Petri dishes in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 5% l-glutamine, and 1% pyruvate (Biochrom, Berlin, Germany) and kept in humidified atmosphere with 5% CO2 at 37 °C. Cell culture material, including media and supplements, was purchased from Biochrom. HEK293 cells were transfected using the calcium-phosphate method. 2 μg of DNA of each construct were used for the transfection of a 35-mm Petri dish of HEK293 cells.
For live cell imaging, HEK293 cells were grown on poly-l-lysine-coated coverslips, which were placed in a custom made microscope chamber. Confocal fluorescence live cell imaging was performed at room temperature using the LSM 700 confocal microscope (Carl Zeiss, Jena, Germany) with a ×63 oil immersion objective. Images were acquired using the ZEN software (Carl Zeiss). Plasma membrane staining was achieved by a 5-min incubation prior to microscope analysis with CellMaskTM Deep Red plasma stain (Life Technologies, Inc., Paisley, UK), following the manufacturer's recommendations.
For membrane cholesterol manipulation, HEK293 cells expressing WT rBASIC and ΔN30 rBASIC were treated for 60 min at 37 °C with either 10 mm MβCD or 10 mm water-soluble cholesterol (Sigma), dissolved in DMEM. HEK293 cells were then analyzed using the whole cell patch clamp technique. For separation of membrane and cytosolic fractions from GFP-transfected HEK293 cells, cells were lysed in PBS-T (in mm: 137 NaCl, 2.7 KCl, 10 Na2HPO4, 1.8 NaH2PO4, 1 CaCl2, 0.5 MgCl2, 1% (v/v) Triton X-100) at 4 °C for 1 h. Lysates were centrifuged for 5 min at 14,000 × g. Proteins from supernatant and pellet were separated by SDS-PAGE and transferred to a PVDF membrane (PolyScreen). The membrane was then probed using a mouse anti-GFP (Sigma, catalog no. G1546, lot 073M4809V) antibody or a mouse anti-tubulin antibody (Sigma, catalog no. T7451, lot 118K4821) in combination with a peroxidase-coupled goat anti-mouse antibody (Sigma, catalog no. A4416, lot SLBL3207).
Whole Cell Patch Clamp Recordings
Whole cell patch clamp measurements on cultured HEK293 cells used patch pipettes of 4–6 MΩ resistance filled with (in mm) 10 NaCl, 121 KCl, 2 MgCl2, 5 EGTA, 2 Na2-ATP, 10 HEPES, pH 7.25. The bath solution contained (in mm) 128 NaCl, 5.4 KCl, 1 MgCl2, 2 CaCl2, 5.55 glucose, 10 HEPES, pH 7.4, with NaOH. Osmolarity was adjusted with sucrose to 280 mosmol/liter for the pipette solution and to 290 mosmol/liter for the extracellular solution. Data were acquired using an Axon Digidata 1440A and an Axon Axopatch 200B and recorded and analyzed using the pClamp software (Axon Instruments, Molecular Devices, Sunnyvale, CA).
Data Analysis
Data were collected and pooled from at least two preparations of oocytes isolated on different days from different animals or were collected and pooled from different passages of HEK293 cells, if not stated otherwise.
Single channel analyses of 10-s recording segments were conducted as described previously (43). Data were analyzed with IgorPro 6.02 (WaveMetrics, Lake Oswego, OR) and are presented as means ± S.E. Statistical significance was calculated using one-way ANOVA followed by a Dunnett's test (PRISM GraphPad, La Jolla, CA).
Preparation of Synthetic Membrane Samples
Highly oriented multilamellar membranes were prepared on polished 2 × 2-cm2 silicon wafers. The wafers were first pre-treated by sonication in dichloromethane at 310 K for 25 min to remove all organic contamination and to create a hydrophobic substrate. After removal from the dichloromethane post-sonication, each wafer was thoroughly rinsed three times by alternating with ∼50 ml of ultra-pure water (18 MΩ cm) and methanol. DMPC was obtained from Avanti Polar Lipids (Alabaster, AL) and dissolved in a 1:1 mixture of chloroform and trifluoroethanol. A peptide fragment, 35 amino acids long and representing the N-terminal region of the protein, was synthesized by Proteogenix (Schiltigheim, France) and also dissolved in a mixture of 1:1 chloroform and trifluoroethanol. The DMPC/peptide solutions were then mixed in the appropriate ratios to produce membranes composed of pure DMPC and DMPC + 0.6 mol % peptide.
A tilting incubator was heated to 313 K, and the lipid solutions were placed inside to equilibrate. 200 μl of lipid solution was deposited on each wafer, and the solvent was then allowed to slowly evaporate for ∼10 min while being gently rocked, such that the lipid solution spread evenly on the wafers. After drying, the membrane samples were placed in vacuum at 313 K for 12 h to remove all traces of solvent. Samples were then placed in a sealed container containing an open vial of pure water and incubated at 303 K for 24 h. This procedure results in highly oriented multilamellar membrane stacks and a uniform coverage of the silicon substrates (34, 36, 46, 47). About 3000 highly oriented stacked membranes with a total thickness of ∼10 μm are produced using this protocol. The high sample quality and high degree of order is a prerequisite to determine the position of the peptide in the membrane stack with angstrom resolution.
X-ray Scattering Experiments
Out-of-plane X-ray scattering data were obtained using the Biological Large Angle Diffraction Experiment (BLADE) in the Laboratory for Membrane and Protein Dynamics at McMaster University. BLADE uses a 9-kilowatt (45 kV, 200 mA) CuKα Rigaku Smartlab rotating anode at a wavelength of 1.5418 Å. Both source and detector are mounted on movable arms, such that the membranes stay horizontal during measurements. Focusing multilayer optics provide a high intensity parallel beam with monochromatic X-ray intensities up to 1010 counts/(s mm2). This beam geometry provides optimal illumination of the membrane samples to maximize the scattered signal. By using highly oriented stacks, up to 8 out-of-plane Bragg peaks were observed, permitting a detailed structural refinement. A sketch of the scattering geometry is depicted in Fig. 9A.
Out-of-Plane Structure and Electron Densities
The out-of-plane structure of the membranes was determined using the out-of-plane X-ray diffraction in Fig. 9B. The membrane electron density, ρ(z), is approximated by a one-dimensional Fourier analysis (35, 48) as shown in Equation 1,
| (Eq. 1) |
where N is the highest order of the Bragg peaks observed in the experiment. The integrated peak intensities, In, are multiplied by qn to generate the form factors, F(qn). The bilayer form factor, which is in general a complex quantity, is real-valued when the structure is centro-symmetric. The phase problem of crystallography therefore simplifies to the sign problem F(qz) = ± |F(qz)|, and the phases, νn, can only take the values ±1. To determine the phases for this study, a fitting procedure was used, as described previously (33–35).
The calculated electron densities, ρ(z), which are initially on an arbitrary scale, were vertically shifted to fulfill the condition ρ(0) = 0.22 e−/Å3 (the electron density of a CH3 group) in the center of the bilayer. The curves were then scaled until the total number of electrons, e−, across a membrane leaflet as shown in Equation 2,
| (Eq. 2) |
agreed with the total number of electrons calculated based on the sample composition, with the addition of seven water molecules, in agreement with Refs. 33, 36.
The spacing between two neighboring membranes in the stack, dz, was determined from the distance between the Bragg reflections (dz = 2π/Δqz) along the out-of-plane axis, qz.
Author Contributions
A. S., D. L., R. J. A., P. L., A. O.-B., M. W., and D. W. performed the experiments. A. S., D. L., R. J. A., M. C. R., S. G., and D. W. analyzed the data and prepared the figures. A. S., D. L., S. G., and D. W. designed the study and interpreted the data. D. W., M. C. R., and R. J. A. wrote the paper. All authors approved the final version of the manuscript.
Acknowledgments
We thank Dr. Verena Schwartz and Dr. Anna Reska for helpful discussions and valuable comments on the manuscript.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG) (to D. W.). Part of this work was presented in abstract form in Abstracts of the 95th Meeting of the German Physiological Society, March 3–5, 2016, Lübeck, Germany (Wiemuth D., Lenzig P., and Gründer S. (2016) Acta Physiol. 216, OS10-8 (Suppl. 707)). The authors declare that they have no conflicts of interest with the contents of this article.
- Deg/ENaC
- degenerin/epithelial Na+ channel
- ASIC
- acid-sensing ion channel
- BASIC
- bile acid-sensitive ion channel
- ECD
- extracellular domain
- TMD
- transmembrane domain
- ANOVA
- analysis of variance
- DMPC
- 1,2-dimyristoyl-sn-glycero-3-phosphatidylcholine
- r
- rat
- h
- human
- MβCD
- methyl-β-cyclodextrin
- norm chol.
- normal cholesterol
- chol.
- cholesterol
- MΩ
- megohm
- r
- rat
- h
- human
- UDCA
- tauroursodeoxycholic acid
- DCA
- taurodeoxycholic acid
- n.s.
- not significant.
References
- 1. Kellenberger S., and Schild L. (2002) Epithelial sodium channel/degenerin family of ion channels: a variety of functions for a shared structure. Physiol. Rev. 82, 735–767 [DOI] [PubMed] [Google Scholar]
- 2. Kellenberger S., and Schild L. (2015) International Union of Basic and Clinical Pharmacology. XCI. structure, function, and pharmacology of acid-sensing ion channels and the epithelial Na+ channel. Pharmacol. Rev. 67, 1–35 [DOI] [PubMed] [Google Scholar]
- 3. Wiemuth D., Assmann M., and Gründer S. (2014) The bile acid-sensitive ion channel (BASIC), the ignored cousin of ASICs and ENaC. Channels 8, 29–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Waldmann R., Champigny G., Bassilana F., Heurteaux C., and Lazdunski M. (1997) A proton-gated cation channel involved in acid-sensing. Nature 386, 173–177 [DOI] [PubMed] [Google Scholar]
- 5. Bohlen C. J., Chesler A. T., Sharif-Naeini R., Medzihradszky K. F., Zhou S., King D., Sánchez E. E., Burlingame A. L., Basbaum A. I., and Julius D. (2011) A heteromeric Texas coral snake toxin targets acid-sensing ion channels to produce pain. Nature 479, 410–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen C. C., and Wong C. W. (2013) Neurosensory mechanotransduction through acid-sensing ion channels. J. Cell. Mol. Med. 17, 337–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Coryell M. W., Wunsch A. M., Haenfler J. M., Allen J. E., McBride J. L., Davidson B. L., and Wemmie J. A. (2008) Restoring acid-sensing ion channel-1a in the amygdala of knock-out mice rescues fear memory but not unconditioned fear responses. J. Neurosci. 28, 13738–13741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Deval E., Noël J., Lay N., Alloui A., Diochot S., Friend V., Jodar M., Lazdunski M., and Lingueglia E. (2008) ASIC3, a sensor of acidic and primary inflammatory pain. EMBO J. 27, 3047–3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Diochot S., Baron A., Salinas M., Douguet D., Scarzello S., Dabert-Gay A. S., Debayle D., Friend V., Alloui A., Lazdunski M., and Lingueglia E. (2012) Black mamba venom peptides target acid-sensing ion channels to abolish pain. Nature 490, 552–555 [DOI] [PubMed] [Google Scholar]
- 10. Du J., Reznikov L. R., Price M. P., Zha X. M., Lu Y., Moninger T. O., Wemmie J. A., and Welsh M. J. (2014) Protons are a neurotransmitter that regulates synaptic plasticity in the lateral amygdala. Proc. Natl. Acad. Sci. U.S.A. 111, 8961–8966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jalalvand E., Robertson B., Wallén P., and Grillner S. (2016) Ciliated neurons lining the central canal sense both fluid movement and pH through ASIC3. Nat. Commun. 7, 10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lin S. H., Cheng Y. R., Banks R. W., Min M. Y., Bewick G. S., and Chen C. C. (2016) Evidence for the involvement of ASIC3 in sensory mechanotransduction in proprioceptors. Nat. Commun. 7, 11460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Palmer L. G., Patel A., and Frindt G. (2012) Regulation and dysregulation of epithelial Na+ channels. Clin. Exp. Nephrol. 16, 35–43 [DOI] [PubMed] [Google Scholar]
- 14. Rossier B. C. (2014) Epithelial sodium channel (ENaC) and the control of blood pressure. Curr. Opin. Pharmacol. 15, 33–46 [DOI] [PubMed] [Google Scholar]
- 15. Boiko N., Kucher V., Wang B., and Stockand J. D. (2014) Restrictive expression of acid-sensing ion channel 5 (asic5) in unipolar brush cells of the vestibulocerebellum. PLoS ONE 9, e91326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sakai H., Lingueglia E., Champigny G., Mattei M. G., and Lazdunski M. (1999) Cloning and functional expression of a novel degenerin-like Na+ channel gene in mammals. J. Physiol. 519, 323–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wiemuth D., and Gründer S. (2010) A single amino acid tunes Ca2+ inhibition of brain liver intestine Na+ channel (BLINaC). J. Biol. Chem. 285, 30404–30410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wiemuth D., Sahin H., Falkenburger B. H., Lefèvre C. M., Wasmuth H. E., and Gründer S. (2012) BASIC–a bile acid-sensitive ion channel highly expressed in bile ducts. FASEB J. 26, 4122–4130 [DOI] [PubMed] [Google Scholar]
- 19. Wiemuth D., Sahin H., Lefèvre C. M., Wasmuth H. E., and Gründer S. (2013) Strong activation of bile acid-sensitive ion channel (BASIC) by ursodeoxycholic acid. Channels 7, 38–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jasti J., Furukawa H., Gonzales E. B., and Gouaux E. (2007) Structure of acid-sensing ion channel 1 at 1.9 A resolution and low pH. Nature 449, 316–323 [DOI] [PubMed] [Google Scholar]
- 21. Snyder P. M., McDonald F. J., Stokes J. B., and Welsh M. J. (1994) Membrane topology of the amiloride-sensitive epithelial sodium channel. J. Biol. Chem. 269, 24379–24383 [PubMed] [Google Scholar]
- 22. Saugstad J. A., Roberts J. A., Dong J., Zeitouni S., and Evans R. J. (2004) Analysis of the membrane topology of the acid-sensing ion channel 2a. J. Biol. Chem. 279, 55514–55519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schmidt A., Lenzig P., Oslender-Bujotzek A., Kusch J., Lucas S. D., Gründer S., and Wiemuth D. (2014) The bile acid-sensitive ion channel (BASIC) is activated by alterations of its membrane environment. PLoS ONE 9, e111549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Staub O., Dho S., Henry P., Correa J., Ishikawa T., McGlade J., and Rotin D. (1996) WW domains of Nedd4 bind to the proline-rich PY motifs in the epithelial Na+ channel deleted in Liddle's syndrome. EMBO J. 15, 2371–2380 [PMC free article] [PubMed] [Google Scholar]
- 25. Shimkets R. A., Warnock D. G., Bositis C. M., Nelson-Williams C., Hansson J. H., Schambelan M., Gill J. R. Jr, Ulick S., Milora R. V., and Findling J. W. (1994) Liddle's syndrome: heritable human hypertension caused by mutations in the β subunit of the epithelial sodium channel. Cell 79, 407–414 [DOI] [PubMed] [Google Scholar]
- 26. Staub O., Gautschi I., Ishikawa T., Breitschopf K., Ciechanover A., Schild L., and Rotin D. (1997) Regulation of stability and function of the epithelial Na+ channel (ENaC) by ubiquitination. EMBO J. 16, 6325–6336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schwartz V., Friedrich K., Polleichtner G., and Gründer S. (2015) Acid-sensing ion channel (ASIC) 4 predominantly localizes to an early endosome-related organelle upon heterologous expression. Sci. Rep. 5, 18242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Coscoy S., de Weille J. R., Lingueglia E., and Lazdunski M. (1999) The pre-transmembrane 1 domain of acid-sensing ion channels participates in the ion pore. J. Biol. Chem. 274, 10129–10132 [DOI] [PubMed] [Google Scholar]
- 29. Bässler E. L., Ngo-Anh T. J., Geisler H. S., Ruppersberg J. P., and Gründer S. (2001) Molecular and functional characterization of acid-sensing ion channel (ASIC) 1b. J. Biol. Chem. 276, 33782–33787 [DOI] [PubMed] [Google Scholar]
- 30. Gonzales E. B., Kawate T., and Gouaux E. (2009) Pore architecture and ion sites in acid-sensing ion channels and P2X receptors. Nature 460, 599–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lefèvre C. M., Diakov A., Haerteis S., Korbmacher C., Gründer S., and Wiemuth D. (2014) Pharmacological and electrophysiological characterization of the human bile acid-sensitive ion channel (hBASIC). Pflugers Arch. 466, 253–263 [DOI] [PubMed] [Google Scholar]
- 32. Wiemuth D., and Gründer S. (2011) The pharmacological profile of brain liver intestine Na+ channel: inhibition by diarylamidines and activation by fenamates. Mol. Pharmacol. 80, 911–919 [DOI] [PubMed] [Google Scholar]
- 33. Alsop R. J., Armstrong C. L., Maqbool A., Toppozini L., Dies H., and Rheinstädter M. C. (2015) Cholesterol expels ibuprofen from the hydrophobic membrane core and stabilizes lamellar phases in lipid membranes containing ibuprofen. Soft Matter 11, 4756–4767 [DOI] [PubMed] [Google Scholar]
- 34. Alsop R. J., Maria Schober R., and Rheinstädter M. C. (2016) Swelling of phospholipid membranes by divalent metal ions depends on the location of the ions in the bilayers. Soft Matter 12, 6737–6748 [DOI] [PubMed] [Google Scholar]
- 35. Barrett M. A., Zheng S., Roshankar G., Alsop R. J., Belanger R. K., Huynh C., Kučerka N., and Rheinstädter M. C. (2012) Interaction of aspirin (acetylsalicylic acid) with lipid membranes. PLoS ONE 7, e34357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dies H., Toppozini L., and Rheinstädter M. C. (2014) The interaction between amyloid-β peptides and anionic lipid membranes containing cholesterol and melatonin. PLoS ONE 9, e99124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Barrett M. A., Alsop R. J., Hauss T., and Rheinstädter M. C. (2015) The position of Aβ22–40 and Aβ1–42 in anionic lipid membranes containing cholesterol. Membranes 5, 824–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ilyaskin A. V., Diakov A., Korbmacher C., and Haerteis S. (2016) Activation of the human epithelial sodium channel (ENaC) by bile acids involves the degenerin site. J. Biol. Chem. 291, 19835–19847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Baconguis I., Bohlen C. J., Goehring A., Julius D., and Gouaux E. (2014) X-ray structure of acid-sensing ion channel 1-snake toxin complex reveals open state of a Na+-selective channel. Cell 156, 717–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hughey R. P., Mueller G. M., Bruns J. B., Kinlough C. L., Poland P. A., Harkleroad K. L., Carattino M. D., and Kleyman T. R. (2003) Maturation of the epithelial Na+ channel involves proteolytic processing of the α- and γ-subunits. J. Biol. Chem. 278, 37073–37082 [DOI] [PubMed] [Google Scholar]
- 41. Chen X., Paukert M., Kadurin I., Pusch M., and Gründer S. (2006) Strong modulation by RFamide neuropeptides of the ASIC1b/3 heteromer in competition with extracellular calcium. Neuropharmacology 50, 964–974 [DOI] [PubMed] [Google Scholar]
- 42. Madeja M., Musshoff U., and Speckmann E. J. (1995) Improvement and testing of a concentration clamp system for oocytes of Xenopus laevis. J. Neurosci. Methods 63, 211–213 [DOI] [PubMed] [Google Scholar]
- 43. Wiemuth D., Lefèvre C. M., Heidtmann H., and Gründer S. (2014) Bile acids increase the activity of the epithelial Na+ channel. Pflugers Arch. 466, 1725–1733 [DOI] [PubMed] [Google Scholar]
- 44. Gründer S., Firsov D., Chang S. S., Jaeger N. F., Gautschi I., Schild L., Lifton R. P., and Rossier B. C. (1997) A mutation causing pseudohypoaldosteronism type 1 identifies a conserved glycine that is involved in the gating of the epithelial sodium channel. EMBO J. 16, 899–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen X., and Gründer S. (2007) Permeating protons contribute to tachyphylaxis of the acid-sensing ion channel (ASIC) 1a. J. Physiol. 579, 657–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Alsop R. J., Khondker A., Hub J. S., and Rheinstädter M. C. (2016) The lipid bilayer provides a site for cortisone crystallization at high cortisone concentrations. Sci. Rep. 6, 22425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tang J., Alsop R. J., Backholm M., Dies H., Shi A. C., and Rheinstädter M. C. (2016) Amyloid-β(25–35) peptides aggregate into cross-β sheets in unsaturated anionic lipid membranes at high peptide concentrations. Soft Matter 12, 3165–3176 [DOI] [PubMed] [Google Scholar]
- 48. Adachi T. (2000) A new method for determining the phase in the X-ray diffraction structure analysis of phosphatidylcholine/alcohol. Chem. Phys. Lipids 107, 93–97 [DOI] [PubMed] [Google Scholar]