Abstract
Perturbations in skeletal development and bone degeneration may result in reduced bone mass and quality, leading to greater fracture risk. Bone loss is mitigated by bone protective therapies, but there is a clinical need for new bone-anabolic agents. Previous work has demonstrated that Ezh2 (enhancer of zeste homolog 2), a histone 3 lysine 27 (H3K27) methyltransferase, suppressed differentiation of osteogenic progenitors. Here, we investigated whether inhibition of Ezh2 can be leveraged for bone stimulatory applications. Pharmacologic inhibition and siRNA knockdown of Ezh2 enhanced osteogenic commitment of MC3T3 preosteoblasts. Next generation RNA sequencing of mRNAs and real time quantitative PCR profiling established that Ezh2 inactivation promotes expression of bone-related gene regulators and extracellular matrix proteins. Mechanistically, enhanced gene expression was linked to decreased H3K27 trimethylation (H3K27me3) near transcriptional start sites in genome-wide sequencing of chromatin immunoprecipitations assays. Administration of an Ezh2 inhibitor modestly increases bone density parameters of adult mice. Furthermore, Ezh2 inhibition also alleviated bone loss in an estrogen-deficient mammalian model for osteoporosis. Ezh2 inhibition enhanced expression of Wnt10b and Pth1r and increased the BMP-dependent phosphorylation of Smad1/5. Thus, these data suggest that inhibition of Ezh2 promotes paracrine signaling in osteoblasts and has bone-anabolic and osteoprotective potential in adults.
Keywords: epigenetics, histone, histone methylation, osteoblast, osteoporosis, methyltransferase, skeleton
Introduction
Decreased bone mineral density (BMD)3 and matrix material properties are associated with increased fracture risk and an imbalance in the biological activities of bone-forming osteoblasts and bone-resorbing osteoclasts (1–3). Loss of BMD observed in individuals with osteoporosis, a prevalent skeletal disease, can be mitigated by anti-resorptive agents including bisphosphonates, selective estrogen receptor modulators (raloxifene), or antibodies that inactivate the osteoclast-stimulatory specific ligand RANKL (Denosumab) (4). Therapeutics that stimulate bone formation include bone morphogenetic proteins (i.e. BMP2) and intermittent treatment with parathyroid hormone (PTH) or PTH-related protein (PTHLH), as well as antibody suppression of WNT inhibitors (e.g. SOST), which is both anabolic and anti-resorptive (5–7). Novel classes of bone-anabolic therapies used alone or in combination with current treatments can potentially increase BMD more effectively with fewer adverse effects than current clinically approved regimens. Therefore, we investigated new bone-anabolic mechanisms linked to regulation of osteoblast growth and differentiation.
Mesenchymal stromal cells (MSCs) reside in various locations in the body such as fat and bone marrow and can differentiate into a variety of skeletal tissues, including bone and cartilage (8). The commitment of MSCs into osteogenic differentiation is controlled by transcriptional and epigenetic events (9, 10). Several signaling pathways (e.g. Bmp, Pth, and Wnt pathways) result in the activation and expression of key osteogenic transcription factors (e.g. Runx2 and Sp7) that facilitate the commitment of MSC into the osteogenic lineage (11). Osteogenic differentiation is also modulated by epigenetic mechanisms such as microRNAs, DNA methylation, and post-translational modification of histones (12, 13). Some epigenetic events suppress whereas others enhance osteogenic differentiation of MSCs (12). Hence, it is important to characterize the epigenetic events that control osteoblast differentiation.
Reversible modifications of histones such as acetylation and methylation play a critical role in controlling gene transcription. Depending on the modification and site, these modifications permit or inhibit the transcriptional machinery in osteoblasts (12). For example, trimethylation of histone 3 lysine 4 (H3K4me3) is associated with transcriptionally active genes (14), whereas histone 3 lysine 27 trimethylation (H3K27me3) may epigenetically reduce chromatin accessibility and thus promote gene silencing (15). Because H3K27me3 suppresses gene expression, this mark has been extensively studied as a cancer therapeutic (16). More recently, altering the level of H3K27me3 has been explored in regenerative medicine (17).
Formation of H3K27me3 marks is mediated by Ezh2, the catalytic unit of polycomb-repressive complex 2 (PRC2) (18). The PRC2 complex may contain Ezh1 instead of Ezh2. However, Ezh1 possesses low methyltransferase activity and is believed to silence genes through alternative mechanisms (18, 19). The methyl-transferase activity of PRC2 is balanced by three major demethylases, Jhdm1d, Kdm6a, and Kdm6b, that catalyze the removal of methyl groups at H3K27 (20).
Recent studies have demonstrated that changes in H3K27me3 alter the phenotypic commitment of progenitor cells (21–25). For example, inhibition of Ezh2 and the resulting reduction of H3K27me3 promotes osteogenic differentiation and inhibits adipogenic differentiation of MSCs (21, 22). In this study, we assessed the role of Ezh2 and H3K27me3 levels in preosteoblasts in culture and in bone formation in vivo. We show that Ezh2 inhibition enhances osteogenic differentiation of preosteoblasts by reducing H3K27me3 near transcriptional start sites and enhances the expression of osteogenic genes. Administration of an Ezh2 inhibitor enhances bone formation and prevents bone loss associated with estrogen depletion in vivo.
Results
EZH2 Inhibition Enhances Osteogenic Differentiation of MC3T3 Cells
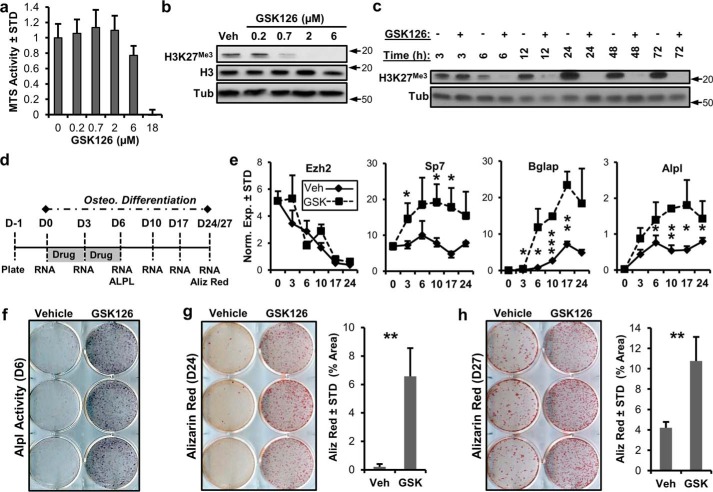
We utilized GSK126, a specific Ezh2 inhibitor, to assess whether enzymatic inhibition of this histone methyltransferase, and therefore suppression of H3K27 trimethylation (H3K27me3), promotes osteogenic differentiation of MC3T3 preosteoblasts. GSK126 exhibits concentration-dependent toxicity as measured by MTS assay (Fig. 1a). Subtoxic concentrations of this Ezh2 inhibitor decrease H3K27me3 levels in a concentration-dependent manner (Fig. 1b). The addition of GSK126 (2 μm) inhibits H3K27me3 6 h after drug administration, and this effect perseveres for at least 72 h (Fig. 1c). These results indicate that Ezh2 activity is rate-limiting for global H3K27me3 in MC3T3 cells, which parallels the established molecular function of this epigenetic regulator (18, 21).
FIGURE 1.
Ezh2 inhibition accelerates osteoblastic differentiation of MC3T3 cells. a, MTS toxicity assay of GSK126-treated MC3T3 cells for 3 days (n = 3) showing that 2 μm GSK126 does not affect total cellular metabolic activity. b and c, Western blotting showing dose-dependent (b) and time-dependent (c) modulation of H3K27me3 by GSK126 in MC3T3 cells. The results indicate rapid loss of H3K27me3 at a concentration of 2 μm GSK126 by 6 h in MC3T3 cells. The cells were treated with different concentrations of GSK126 1 day after plating and harvested 3 days later (b) or treated with 2 μm GSK126, with the protein lysates collected at the specified times (c). Arrows indicate molecular mass marker (kDa) location. d, diagram of the experimental protocol for treatment of MC3T3 cells with GSK126 (5 μm) shown in e–h. e–h, RT-qPCR of Ezh2 and osteogenic markers (n = 3) (e), alkaline phosphatase staining (f), and alizarin (Aliz) red staining (g and h) for MC3T3 cells treated with vehicle or GSK126. Alizarin red staining was quantified by ImageJ software. The experiments were repeated three times, and biological triplicates (means ± S.D.) are shown when applicable. We note that it is possible to detect appreciable residual H3K27me3 levels upon longer exposures in Western blots of cells treated with GSK126, indicating that inhibition is not absolute. Tub, tubulin; Veh, vehicle; Osteo., osteoblastic; STD, standard deviation; Norm. Exp., normalized expression.
To determine the effects of Ezh2 inhibition on osteogenic commitment of MC3T3 cells, 5 μm GSK126 was added to the cells for the first 6 days of osteogenic differentiation (Fig. 1d). This treatment regimen was selected because of the expression pattern of Ezh2 in differentiating MC3T3 cells (Fig. 1e). High expression of Ezh2 is observed in undifferentiated cells, whereas a significant decrease in expression occurs during osteogenic differentiation of MC3T3 cells. Ezh2 down-regulation during osteogenic differentiation could be due to transcriptional suppression by the bone master regulator Runx2 (26) and/or post-transcriptional inhibition by microRNA miR-101, which targets Ezh2 (27). Irrespective of exactly how cells regulate Ezh2, we tested whether inactivation of Ezh2 with GSK126 would have downstream functional consequences by biologically enhancing osteogenic differentiation of MC3T3 cells. The RT-qPCR results show that Ezh2 inhibition enhances the expression of several osteogenic markers including Sp7, Bglap, and Alpl (Fig. 1e). Similar expression of Ezh2 is observed in the vehicle and GSK126 treatment groups. Similar to Alpl mRNA expression, increased Alpl activity is observed in GSK126-treated MC3T3 cells (Fig. 1f). Alizarin red staining demonstrates that Ezh2 inhibition accelerates calcium deposition of MC3T3 cells. We note that alizarin red staining is very robust in GSK126-treated cells on day 24 of osteogenic differentiation, whereas few nodules are present in vehicle-treated cells (Fig. 1g). On day 27, more calcium deposition is observed in vehicle-treated cells, but this is significantly less when compared with GSK126-treated cultures (Fig. 1h). Similar to GSK126, another inhibitor of Ezh2, UNC1999, enhances osteogenic differentiation of MC3T3 cells (supplemental Fig. S1).
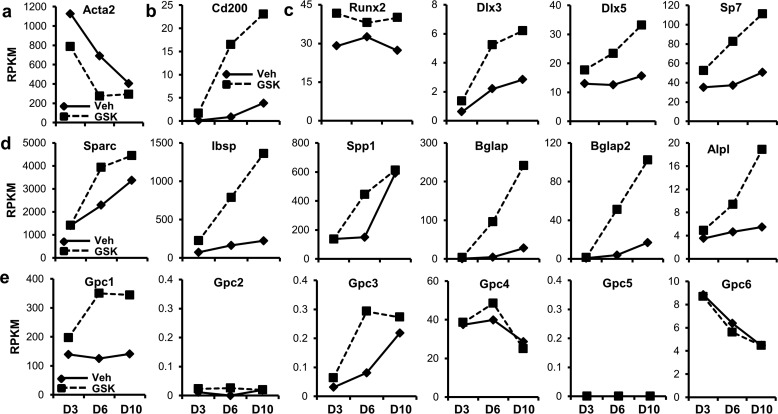
RT-qPCR analysis (Fig. 1e) suggests that Ezh2 inhibition promotes expression of osteogenic genes at early stages of MC3T3 differentiation. To assess mechanistic consequences at a broader scale, we assessed global gene expression by RNA-seq during osteogenic commitment of MC3T3 treated with 5 μm GSK126 and vehicle (Fig. 1d). Acta2, a mesenchymal progenitor marker, is down-regulated during the differentiation of MC3T3 cells and is further suppressed when Ezh2 function is inhibited (Fig. 2a). For comparison, the cluster of differentiation marker Cd200 is up-regulated during the differentiation time course and is further enhanced with the presence of Ezh2 inhibitor (Fig. 2b). MC3T3 differentiation results in enhanced expression of several osteogenic transcription factors including Dlx3, Dlx5, and Sp7 (Fig. 2c). The presence of GSK126 further increases expression of these transcription factors, as well as the bone master regulator Runx2. Similarly, extracellular matrix-related genes (e.g. Sparc, Ibsp, Spp1, Bglap, Bglap2, and Alpl) rise in expression over the differentiation time course, whereas their levels are dramatically increased in the presence of Ezh2 inhibitor (Fig. 2d). Glypicans (Gpc1–6), several of which are implicated in BMP signaling, are also modulated by Ezh2 inhibition. More specifically, the expression of Gpc1 and Gpc3 is enhanced with GSK126 (Fig. 2e).
FIGURE 2.
Ezh2 inhibition activates osteoblast-related gene expression programs. Early stages (D3, D6, and D10) of osteogenic differentiation of MC3T3 cells in the presence of vehicle (Veh) or GSK126 was assessed by RNA-seq (see treatment diagram in Fig. 1d). a and b, expression of the mesenchymal stem cell marker Acta2 is reduced (a), whereas the cluster of differentiation marker Cd200 is up-regulated (b) with Ezh2 inhibition. c–e, osteogenic transcription factors (c) and extracellular matrix-related genes (d), including glypicans (Gpc1 and Gpc3) (e), are up-regulated with Ezh2 inhibition. Three biological replicates were pooled to generate a single RNA-seq value (RPKM) for each condition time point. Gene expression for many bone-related markers trends upward during differentiation (solid lines) and GSK126 typically increases the slope of this trend (dotted lines).
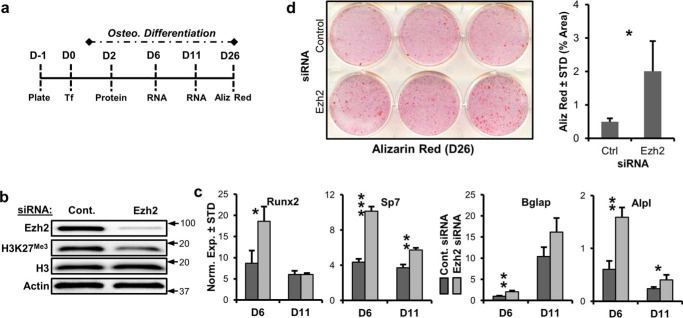
To control for potential nonspecific effects of GSK126 and UNC1999, we performed siRNA transfection targeting Ezh2 in MC3T3 cells using “smart pool siRNA” (GE Healthcare) (Fig. 3a). Two days after transfection, Ezh2 was depleted and coincided with reduced H3K27me3 (Fig. 3b). Similar to enzymatic inhibition via GSK126 and UNC1999, the knockdown of Ezh2 enhances the expression of several osteogenic markers including Runx2, Sp7, Alpl, and Bglap after 6 and/or 11 days of osteogenic differentiation (Fig. 3c). The transfection of Ezh2 siRNA also results in enhanced alizarin red staining of MC3T3 cultures (Fig. 3d). Collectively, our results indicate that Ezh2 inhibition, and thus reduction of H3K27me3, promotes osteogenic differentiation of MC3T3 preosteoblasts.
FIGURE 3.
siRNA depletion of Ezh2 promotes MC3T3 osteoblast differentiation. a, illustration of the experimental protocol for siRNA transfection of MC3T3 cells with control and Ezh2 siRNAs. b, Western blotting of Ezh2 protein and H3K27me3 relative to H3 and to β-actin 2 days after transfection. Arrows indicate molecular mass marker (kDa) location. c, RT-qPCR analysis of osteogenic genes for MC3T3 cells exposed to control or Ezh2 siRNA (n = 3). d, alizarin red staining for MC3T3 cells in the presence of control or Ezh2 siRNA (n = 3). The experiments were repeated three times, and biological triplicates (means ± S.D.) are shown when applicable. Aliz, alizarin; Cont. or Ctrl, control; Osteo., osteogenic; STD, standard deviation; Norm. Exp., normalized expression.
Ezh2 Inhibition Decreases Genome-wide Deposition of H3K27me3 Marks near TSSs
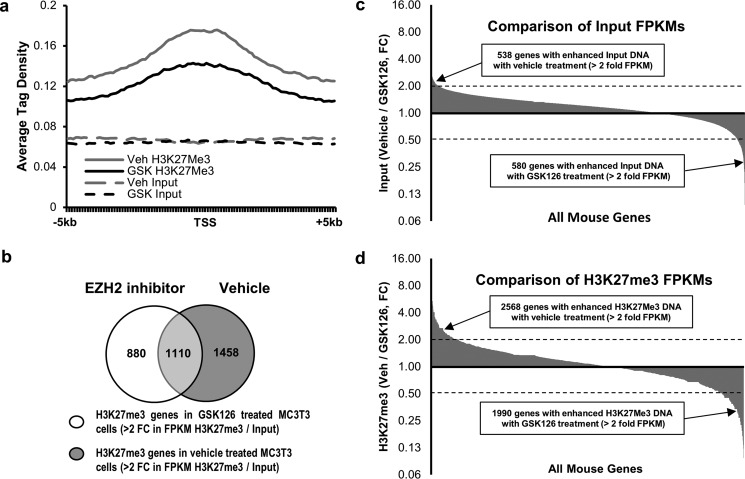
To assess the effect of Ezh2 inhibition on the epigenetic landscape in preosteoblasts, chromatin immunoprecipitation combined with next generation sequencing (ChIP-seq) analysis was performed utilizing a validated H3K27me3 antibody in MC3T3 cells treated for 24 h with vehicle or 5 μm GSK126. EZH2 inhibition using GSK126 rapidly reduces total H3K27me3 levels by severalfold within 6 h, and these reduced levels are sustained for at least 3 days (Fig. 1). Consequently, these data predict a genome-wide change in this histone modification. However, H3K27me3 peaks are typically found near transcriptional start sites (TSSs) throughout the genome, whereas H3K27me1 peaks, for example, are characteristic for distal transcriptional enhancers.
Average tag density from 5 kb upstream to 5 kb downstream of the TSSs for high confidence methylation peaks (false discovery rate ≤ 1e-10) based on ChIP-seq analysis upon treatment with either vehicle or GSK126 are plotted (Fig. 4a). Ezh2 inhibition reduces the average tag density in the H3K27me3 plot near TSSs when compared with vehicle (compare Veh H3K27me3 with GSK H3K27me3). The average tag densities for the input DNA are similar between the two treatment groups (compare Veh Input with GSK Input). A comparison of genes showing a greater than 2-fold increase in fragments/kilobase pair/million mapped reads (FPKM) values between input DNA and the corresponding DNA after H3K27me3 ChIP indicates that there are fewer genes showing H3K27me3 in MC3T3 cells after GSK126 treatment (Fig. 4b). Comparison of FPKM values for the input DNA from MC3T3 cells treated with vehicle or GSK126 demonstrates that less than 1% of all genes show greater than 2-fold difference in FPKMs (Fig. 4c). The small changes in input DNA that are observed may be accounted for by differences in DNA accessibility resulting from changes in chromatin structure following GSK126 treatment. Comparison of FPKM values from vehicle- and GSK126-treated MC3T3 cells after H3K27me3 ChIP shows an increased number of genes with >2-fold difference between the two groups, indicating that Ezh2 inhibition changes the status of H3K27me3 marks near TSSs (Fig. 4d). These data support the concept that enzymatic inhibition of Ezh2 decreases the deposition of H3K27me3 marks across the genome and in particular near TSSs.
FIGURE 4.
Ezh2 inhibition reduces H3K27me3 marks near transcriptional start sites in MC3T3 cells. a, a plot of high confidence methylation peaks (FDR ≤ 1e-10) based on ChIP-seq analysis. Graph shows the average tag density from 5 kb upstream to 5 kb downstream of the TSS upon treatment with either vehicle or GSK126. b, a comparison of genes showing a greater than 2-fold increase in FPKM values between input DNA (before H3K27me3 ChIP) and the corresponding DNA after H3K27me3 ChIP. The data indicate that there are fewer genes showing H3K27me3 in MC3T3 cells after GSK126 treatment. c, comparison of FPKM values for the input DNA from MC3T3 cells treated with vehicle or GSK126. Less than 1% of all genes show greater than 2-fold difference in FPKMs between the two treatment groups. d, comparison of FPKM values from vehicle- and GSK126-treated MC3T3 cells after H3K27me3 ChIP. These data show an increased number of genes with greater than 2-fold difference between treatment groups indicating that Ezh2 inhibition changes the status of H3K27me3 marks near TSSs. After quality control, sequencing was performed on a single sample for pulldowns and inputs for each treatment condition. Veh, vehicle.
Correlation between H3K27me3 and Gene Expression
To correlate H3K27 trimethylation patterns with changes in gene expression, we compared ChIP-seq for H3K27me3 to gene expression by RNA-seq in MC3T3 cells treated with vehicle or 5 μm GSK126. Genes that show H3K27me3 marks in vehicle- and GSK126-treated cells (Fig. 4b) were compared with genes that are up-regulated at least 1.4-fold in GSK126-treated cells on the indicated days of differentiation (Fig. 5a). These experiments show that Ezh2 inhibition results in H3K27 demethylation and subsequent up-regulation of a number of genes with H3K27me3 marks for key osteogenic transcription factors, growth factors and genes that modulate BMP signaling (Figs. 5, b–d, and 6). Thus, the osteogenic effect of enzymatic inhibition of Ezh2 using GSK126 is mechanistically linked to selective changes in the deposition of H3K27me3 marks near TSSs of genes that encode components of principal gene regulatory signaling pathways.
FIGURE 5.
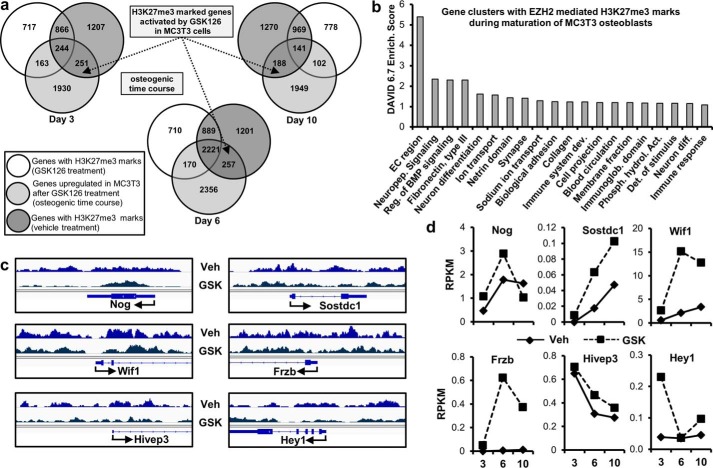
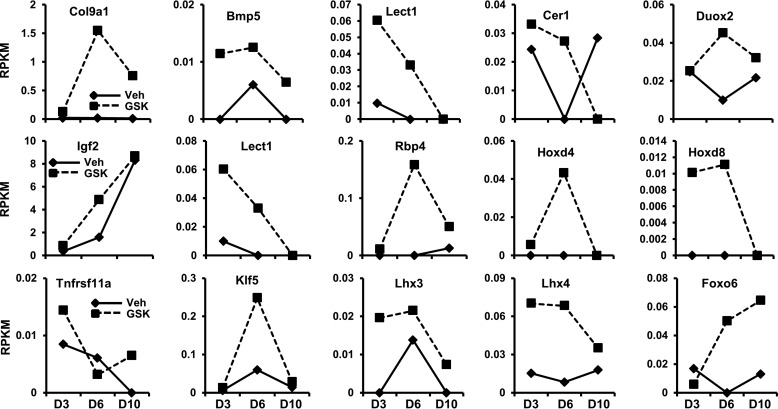
Reduction in H3K27me3 enhances gene expression in MC3T3 cells. a, genes with H3K27me3 marks by ChIP-seq (Fig. 4) showing a greater than 1.4-fold increase in expression by RNA-seq (see Fig. 2) after GSK126 treatment during osteogenic differentiation in MC3T3 cells. b, gene ontology analysis using DAVID 6.7 (52) of all genes enhanced by GSK126 and showing a reduction in H3K27me3 marks (n = 696; see dotted arrows in a). c and d, examples of genes showing decreased H3K27me3 marks near the TSS (c) and enhanced expression (d) with GSK126 in MC3T3 cells. See Figs. 2 and 4 for data acquisition. Veh, vehicle.
FIGURE 6.
Osteogenic genes with a reduction in H3K27me3 and exhibiting enhanced expression with Ezh2 inhibition. We examined RNA-seq data (see Fig. 2) for genes exhibiting up-regulation in gene expression after GSK126 treatment. The genes presented here show decreased H3K27me3 levels (based on ChIP-seq data from Fig. 4) and focuses primarily on genes that modulate osteogenesis through transcriptional regulation and cell signaling mechanisms.
Ezh2 Inhibition Stimulates Paracrine Signaling in Osteoblast
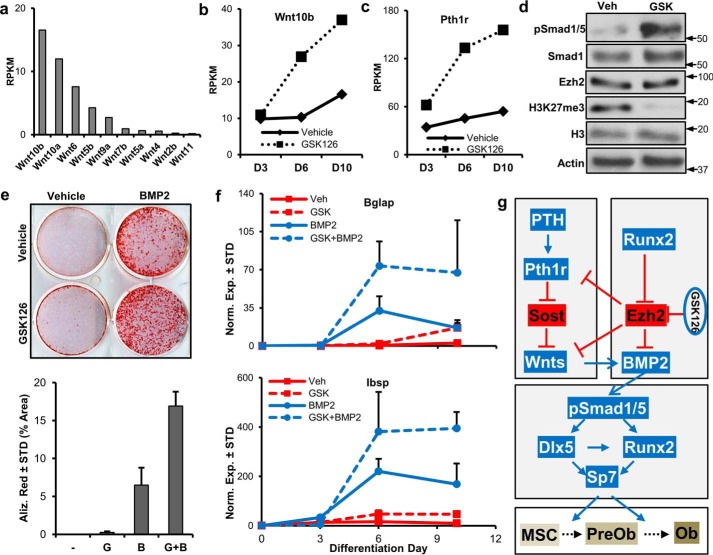
Our initial RNA-seq analysis suggested that Ezh2 inhibition modulates components of the WNT and BMP signaling pathways (Fig. 5). We therefore performed additional analyses and experiments to address whether Ezh2 inhibition affects paracrine signaling in osteoblasts. Several Wnt ligands (e.g. Wnt10b, Wnt10a, and Wnt6) are robustly expressed in differentiating MC3T3 cells (Fig. 7a). Interestingly, the pro-osteogenic Wnt10b is greatly up-regulated by Ezh2 inhibition (Fig. 7b). Similarly, the PTH receptor (Pthr1h) is also enhanced by GSK126 in MC3T3 (Fig. 7c). Western blot analysis demonstrates that Ezh2 inhibition enhances Smad1/5 phosphorylation, a well established biomarker for the activation of BMP2 signaling, in MC3T3 cells (Fig. 7d). As a result of these findings, we assessed combination treatment of GSK126 and BMP2. As anticipated, GSK126 and BMP2 result in a faster acquisition of alizarin red-positive colonies in MC3T3 cells (Fig. 7e). Interestingly, the addition of GSK126 to BMP2-treated cells further enhances the mineral deposition. Similarly, combination of GSK126 and BMP2 enhances the expression of Bglap and Ibsp, two key osteogenic markers (Fig. 7f). Based on the combined results from our study, including RNA-seq and ChIP-seq data, we propose a mechanistic working model for Ezh2 as an epigenetic suppressor of paracrine signaling in osteoblasts (Fig. 7g). The exciting ramification of our study is that inhibitors of Ezh2, which include well tolerated and orally available drugs, may be effective by supporting the endogenous local activation of natural bone stimulatory ligands at physiological doses in bone.
FIGURE 7.
Potential mechanisms by which Ezh2 inhibition promotes osteogenic differentiation. a, expression pattern of the highest expressing Wnts in differentiating (d7) MC3T3 cells. b and c, Wnt10b (b) and Pth1r (c) are highly up-regulated with Ezh2 inhibition in differentiating MC3T3 cells. Western blotting analysis demonstrating enhanced Smad1/5 phosphorylation after Ezh2 inhibition in MC3T3 cells (5 μm GSK126, 24 h) (d). Arrows indicate molecular mass marker (kDa) location. e and f, MC3T3 cells were treated and differentiated as described under “Experimental Procedures” and Fig. 1d with GSK126 (5 μm), BMP2 (50 ng/ml), and the combination of GSK126 and BMP2. Alizarin red staining (D20) and stain quantification by ImageJ software. g, GSK126; B, BMP2. f, RT-qPCR analysis of Bglap and Ibsp at indicated days of differentiation (n = 3). g, proposed model by which Ezh2 inhibition activates osteogenic cascades in preosteoblasts. Pro-osteogenic factors are in blue, whereas anti-osteogenic factors are in red. MSC, mesenchymal stem cell; PreOb, preosteoblast; Ob, osteoblast. Western blotting, alizarin red staining, and RT-qPCR analysis experiments were repeated three times. See Fig. 2 for RNA-seq information. Veh, vehicle.
Ezh2 Inhibition Is Bone-anabolic and Osteoprotective in Vivo
RNA-seq data obtained during osteogenic differentiation of MSCs (21) or osteoblasts (Fig. 2) consistently indicate that pharmacological inhibition of Ezh2 is pro-osteogenic and enhances expression of skeletal ECM proteins. We therefore assessed the biological effects of decreasing Ezh2 activity on bone homeostasis in adult mice. Because our in vivo studies encompass multiple comparisons, we performed statistical analyses using the Wilcoxon test or Wilcoxon rank sums test (supplemental Tables S2 and S3). Only the most relevant comparisons are presented in the bar graphs (Figs. 8 and 9).
FIGURE 8.
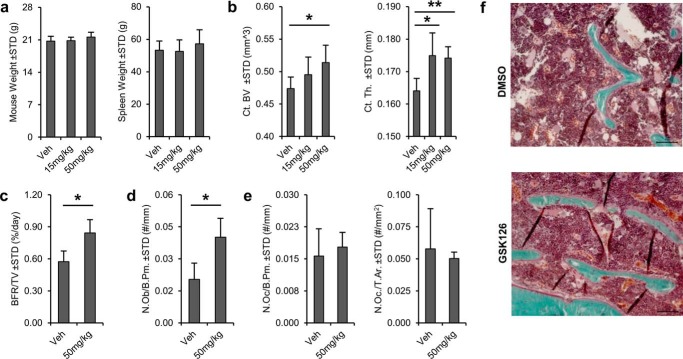
Bone-anabolic effects of Ezh2 inhibition in mice. Systemic administration of Ezh2 inhibitor GSK126 promotes bone formation and maintains cortical structure in vivo. C57Bl/6 mice were treated with vehicle (Veh), 15 mg/kg GSK126, or 50 mg/kg GSK126 for 5 weeks. a, similarities in body or spleen weight at sacrifice indicate absence of major adverse reactions. b, Ezh2 inhibition has bone-anabolic effects based on μCT analysis of the femoral diaphysis. Cortical bone volume (Ct.BV) and cortical thickness (Ct.Th.) are significantly enhanced following treatment with 50 mg/kg GSK126 compared with the vehicle (n = 6 each). c, dynamic histomorphometric analysis performed in the distal femoral metaphysis reveals enhancement of bone formation rate per tissue volume (BFR/TV) in both GSK126-treated groups (vehicle n = 3 and 50 mg/kg n = 5). d, similarly, static histomorphometry analysis of the same region shows increased number of osteoblasts per bone perimeter (N.Ob./B.Pm). e, number of osteoclasts per bone perimeter (N.Oc/B.Pm.) and osteoclasts number per tissue area (N.Oc/T.Ar.) are unaffected by GSK126 treatment. f, an increase in osteoblasts and osteocytes is evidenced by Goldner's trichrome staining of the distal femoral diaphysis.
FIGURE 9.
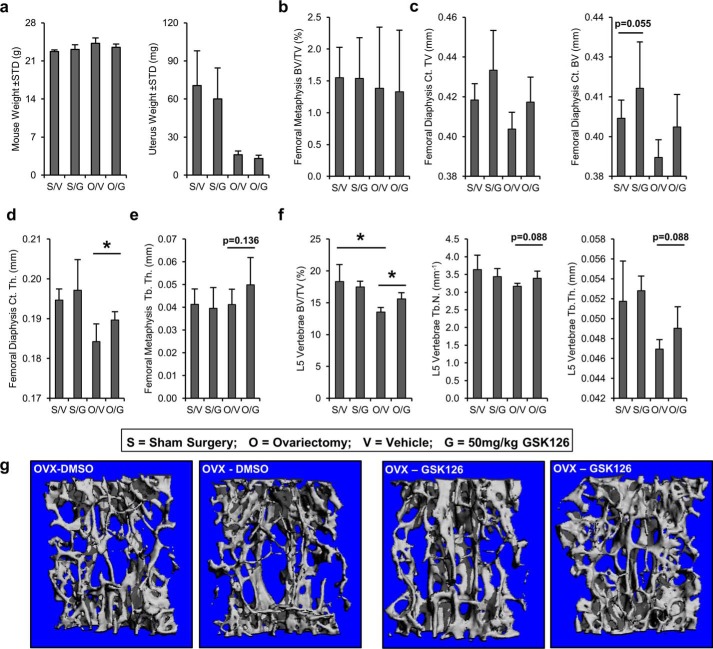
Osteoprotective effects of Ezh2 inhibition skeletally mature mice. The effect of GSK126 was also analyzed using μCT analysis in a mouse ovariectomy model of hormone deficiency osteoporosis. Daily injections were performed for 6 weeks on each treatment group (n = 4–6/group). S, sham surgery; O, ovariectomy surgery; V, vehicle treatment (DMSO); G, 50 mg/kg GSK126 treatment. a, at sacrifice, body weights were similar between surgical and treatment groups, and uterus weights are reduced in the ovariectomized groups. b and c, analysis of bone volume in the femoral metaphysis (b) and femoral diaphysis (c). The data reveal a trend in cortical volume (Ct. TV) and cortical bone volume (Ct. BV) between vehicle- and GSK126-treated animals in both sham and OVX groups (compare S/V with S/G and O/V with O/G). d, assessment of femoral diaphysis shows increased cortical thickness (Ct. Th.) in OVX mice treated with GSK126 (compare O/V with O/G). e, analysis of femoral metaphysis trabecular thickness (Tb. Th.) shows a trend between vehicle- and GSK126-treated OVX animals (compare O/V with O/G). f, evaluation of L5 vertebrae. A reduction in bone volume fraction (BV/TV) in the spine is observed between sham and OVX animals (compare S/V with O/V), whereas GSK126 partially restores the loss of bone as a result of OVX surgery (compare O/V and O/G). g, examples of L5 spine μCT reconstructions OVX animals treated with vehicle (DMSO) or Ezh2 inhibitor (GSK126).
Our first study examined whether the Ezh2 inhibitor GSK126 is bone-anabolic after skeletal patterning. We investigated biological effects in mice at 2 months of age (i.e. prior to skeletal maturation) (Fig. 8). Daily intraperitoneal administration of 15 and 50 mg/kg GSK126 for 5 weeks does not result in gross adverse reactions as suggested by similarities in body and spleen weight among the treatment groups (Fig. 8a). Analysis by μCT shows a significant increase in cortical bone volume and thickness of the femoral diaphysis, while also revealing a trend toward increased cancellous bone thickness in the distal femoral metaphysis with GSK126 treatment (Fig. 8b and supplemental Table S2). Corroborating these results, histomorphological analysis of the distal femoral metaphysis reveals a significant increase in bone formation rate per tissue volume (Fig. 8c), number of osteoblasts per bone perimeter (Fig. 8d), and mineral apposition rate in the 50 mg/kg GSK126 treatment group (supplemental Table S2). Osteoclast number per bone perimeter and tissue area is not significantly different between the groups, indicating that GSK126 stimulates bone formation without affecting bone resorption (Fig. 8e and supplemental Table S2). We conclude that pharmacological inactivation of Ezh2 has bone-anabolic effects in adult mice.
Based on the bone-anabolic effects in normal adult mice, a second study assessed whether GSK126 can mitigate bone loss in female mice with a fully mature skeleton at peak bone mass (Fig. 9 and supplemental Table S3). We used an ovariectomy (OVX) model for post-menopausal osteoporosis in female mice starting at 3 months of age and administered 50 mg/kg of GSK126 daily for 6 weeks. Body weight or spleen weight are similar between groups, whereas uterus weight is reduced in ovariectomized mice as expected (Fig. 9a and data not shown). As observed in the first study, femoral metaphyseal bone volume is not affected by Ezh2 inhibition in sham and OVX mice treated with GSK126 (Fig. 9b). However, there is a trend for increased cortical thickness and bone volume in the diaphysis of sham and OVX mice following GSK126 administration (Fig. 9c). In OVX mice, cortical thickness of the femoral diaphysis (Fig. 9d) and trabecular thickness of the femoral metaphysis (Fig. 9e) are increased upon administration of GSK126 compared with mice treated with vehicle. L5 vertebral bone volume, trabecular number, and trabecular thickness are reduced in OVX mice compared with sham animals (Fig. 9f). These parameters are at least partially restored in the presence of GSK126 in OVX mice (Fig. 9, f and g). Mitigation of the bone phenotype in OVX mice upon treatment by GSK126 suggests that inhibition of Ezh2 in adult females has osteoprotective properties.
Intraperitoneal administration of the Ezh2 inhibitor GSK126 is not overtly toxic in mice, because we did not experience lethality in any of our cohorts. In addition, we have examined multiple soft tissues (e.g. heart, liver, kidney, and spleen) from mice treated with intraperitoneal doses of for up to 6 weeks. These studies did not reveal any obvious adverse effects at the level of gross anatomy, body weight, and spleen weight. The latter results indicate that GSK126 is well tolerated as previously suggested (28).
Discussion
The present study assessed the role of Ezh2 on osteoblast differentiation in vitro and whether inhibition of this epigenetic enzyme alters bone parameters in vivo. Knockdown and inhibition of Ezh2 enhances osteogenic differentiation of MC3T3 preosteoblasts. RNA-seq and ChIP-Seq analyses suggest that Ezh2 inhibition enhances expression of osteogenic genes by reducing H3K27me3 near TTSs. It remains to be established whether Ezh2 binds near TSSs in immature osteoblasts. We consider it likely that Ezh2 remains bound to the promoters of a number of genes that suppress cell growth or support osteogenic lineage-progression, consistent with data on Ezh2 binding in non-osseous cell types (18).
The loss of H3K27me3 upon Ezh2 inhibition is expected to perturb the dynamic balance between H3K27 methylation and demethylation. The very rapid loss of H3K27me3 we observed in cultured osteoblasts indicates that the corresponding demethylases (e.g. Kdm6a/Utx, Kdm6b/Jmjd3, Kdm7a/Jhdm1d) are highly active. Selective localization of Ezh2 and H3K27 demethylases could further modify local methylation kinetics at gene promoters. It is conceivable that the equilibrium between H3K27 methylation and demethylation changes during osteoblast differentiation, consistent with our observation that mRNA levels for Ezh2 and the H3K27 demethylase Jhdm1d are modulated during early stages of differentiation in mesenchymal stromal cells (21).
Administration of an Ezh2 inhibitor increases bone density both in wild type adult mice and an estrogen-deficient mammalian model for osteoporosis, although these quantitative effects are relatively modest. Our study does not formally demonstrate a direct reduction of H3K27me3 in vivo at specific gene promoters in bone, but it has been established in other studies (28). Hence, the potential bone-anabolic effects of GSK126 in vivo may be limited by incomplete demethylation of H3K27me3, partial inhibition of Ezh2, and compensatory mechanisms by other enzymes (e.g. Ezh1).
Nevertheless, molecular studies demonstrate that Ezh2 inhibition promotes paracrine signaling by enhancing expression and phosphorylation of key osteogenic signaling pathways. Thus, inhibition of Ezh2 has bone-anabolic and osteoprotective potential (presumably by reducing H3K27me3), leading to enhanced expression and activation of pro-osteogenic pathways.
Our results are consistent with studies demonstrating that Ezh2 plays a critical role in maintaining proliferation and multilineage differentiation potential of mesenchymal and other progenitor cells (18, 21, 22, 29–34). Additionally, phosphorylation of Ezh2 promotes osteogenic differentiation of progenitor cells (23). These data collectively indicate that H3K27me3, which is balanced by Ezh2 and the corresponding demethylases (e.g. Jhdm1d and Kdm6a), controls osteogenic lineage commitment. Interestingly, Ezh2 expression is interlocked with the bone-related master regulator Runx2 (26) and long non-coding RNA LncRNA-ANCR (25). Additional regulation of Ezh2 may be attributed to miR-101, which was shown to target Ezh2 in other biological systems (27, 35). Together, these regulatory feedback mechanisms may contribute to the observed osteogenic effects of Ezh2 inhibition.
Current treatment options for osteoporosis, which affects 200 million people worldwide and is responsible for more than 8.9 million fractures annually, rely on drugs with therapeutic limitations, including anti-resorptive bisphosphonates (linked to pathologic femur fractures and osteonecrosis of the jaw) or the PTH-related bone-anabolic drug teriparatide. Use of the latter is restricted to 18–24 months because of safety concerns with onset of osteosarcoma, even though this risk is very slight (36). Use of the bone-anabolic agent BMP2 is limited to spine fusion and fracture healing, but its potency provokes heterotopic ossification (37). The critical finding of our studies is that although the loss of Ezh2 function creates abnormalities in skeletal patterning and bone formation in young animals (21), Ezh2 inhibition in older and skeletally mature animals has both bone-anabolic and osteoprotective biological effects.
Mechanistically, our results show that epigenetic modifications altered by Ezh2 inhibition promote osteogenic differentiation by stimulating pathways related to WNT, PTH, and BMP2. The latter mechanisms may proceed via paracrine physiological effects that are more controlled than treatment with exogenous ligands that are administered at supraphysiological levels. The more balanced endogenous activation of these pathways by Ezh2 inhibition may perhaps assuage some of the clinical concerns related to the therapeutic use of the corresponding ligands. Interestingly, our studies show that the Ezh2 inhibitor GSK126 enhances BMP2-induced osteogenic differentiation. This finding suggests that GSK126 may perhaps have utility as an adjuvant therapy in current clinical applications for BMP2.
Consistent with our studies, Jing et al. (38) demonstrated that Ezh2 is up-regulated in osteoporotic MSCs and treatment of mice with 3-deazaneplanocin A increased bone formation in osteoporotic mice. Because 3-deazaneplanocin A is an inhibitor of S-adenosyl homocysteine hydrolase (39) that globally inhibits several methylation sites on histones (40, 41) and does not specifically inhibit Ezh2 methyltransferase activity (16, 41, 42), it remains to be established whether their work is directly related to effects on Ezh2.
In conclusion, a principal finding of our study is that specific enzymatic inhibition of Ezh2 has bone-anabolic and bone-protective effects in vivo. Mechanistically, our data suggest that Ezh2 inhibition promotes paracrine signaling in osteoblasts by up-regulating genes (e.g. Wnt10b and Pth1r) and enhancing phosphorylation of key osteogenic intermediates (Smad1/5), as suggested by other studies (30, 38). These encouraging findings may lead to new therapeutic bone regenerative strategies to treat osteoporosis.
Experimental Procedures
MC3T3 Cell Culture
MC3T3 sc4 murine calvarial osteoblasts (43) were purchased from American Type Culture Collection and maintained in α-minimal essential medium without ascorbic acid (Invitrogen) containing 10% FBS (Gibco), 100 units/ml penicillin, and 100 μg/ml streptomycin.
MTS Activity Assay
MC3T3 cells were plated in 96-well plates in maintenance medium (5,000 cells/well). The following day, vehicle (DMSO) or Ezh2 inhibitor (GSK126 and UNC1999) in fresh maintenance medium was added to the cells. Three days later, MTS activity was assayed according to the manufacturer's protocol (Promega). Absorbance was measured at 490 nm using a SpectraMAX Plus spectrophotometer (Molecular Devices).
mRNA RT-qPCR
RNA was isolated using the miRNeasy kit (Qiagen). Isolated RNA was reverse transcribed into cDNA using the SuperScript III first strand synthesis system (Invitrogen). Gene expression was quantified as described (21). Transcript levels were quantified using the 2ΔΔCt method and normalized to the housekeeping gene Gapdh (set at 100). Gene specific primers are shown (supplemental Table S1).
Western Blotting
MC3T3 cells were plated in 6-well plates at a density of 10,000 cells/cm2 in maintenance medium and treated with vehicle or Ezh2 inhibitor (e.g. GSK126). Cell lysis and Western blotting was performed as described (21). Proteins were visualized using an ECL Prime detection kit. The primary antibodies used were: actin (1:10,000; sc-1616; Santa Cruz), H3 (1:10,000; 05-928; Millipore), H3K27me3 (1:5,000; 17–622; Millipore), Ezh2 (1:10,000; 5246; Cell Signaling), tubulin (1:10,000; E7; University of Iowa Hybridoma Bank), p-Smad1/5 (Ser463/5) (1:2,000; 700047; Thermo Scientific), and Smad1 (1:1,000; ab63356; Abcam).
Osteogenic Differentiation
MC3T3 cells were seeded in 6-well plates in maintenance medium (10,000 cells/cm2). The following day, maintenance medium was replaced with osteogenic medium (α-minimal essential medium supplemented with 50 μg/ml ascorbic acid (Sigma) and 4 mm β-glycerol phosphate (Sigma)) containing vehicle or Ezh2 inhibitor (GSK126 or UNC1999). Three days later, vehicle or Ezh2 inhibitors were added again with osteogenic medium. When relevant, BMP2 (50 ng/ml; R&D Systems) was added and removed on the same days as GSK126. On day 6, Ezh2 inhibitor and vehicle were removed, and fresh osteogenic medium was added with medium changes scheduled every 3 days until RNA harvest at the indicated times. On day 6, a subset of the cell cultures were fixed in 10% neutral buffered formalin and stained with 5-bromo-4-chloro-3-indolyl-phosphate/nitro blue tetrazolium to monitor the enzymatic activity of alkaline phosphatase (Promega). Between days 21 and 28 of osteogenic differentiation, the cells were also fixed in 10% neutral buffered formalin and stained with 2% alizarin red to visualize calcium deposition. Absorption of alizarin red stain was quantified with ImageJ software (44).
Ezh2 Knockdown and Osteogenic Differentiation
MC3T3 cells were seeded in 6- or 12-well plates in maintenance medium (10,000 cells/cm2). The following day, siRNA transfections with control (D-001810-10-20; GE Healthcare) and mouse Ezh2 (L-040882-00; GE Healthcare) ON-TARGETplus siRNA SMARTpools were performed using RNAiMAX as instructed by the manufacturer (Invitrogen). The next day, MC3T3 osteogenic medium was added, and the cells were cultured until harvest.
High Throughput RNA Sequencing and Bioinformatic Analysis
RNA-seq of mRNAs was performed using RNA isolated at days 3, 6, and 10 from MC3T3 treated with vehicle or 5 μm GSK126. To improve sample representation, we pooled three distinct RNA samples (biological triplicates) for each treatment group at each time point. We note that pooling reduces biological variation to yield a single “averaged” sample (n = 1) that does not permit visualization of statistical variation (e.g. error bars) in our figures. High throughput read mapping and bioinformatic analyses for RNA-seq were performed as previously reported (21, 45). Gene expression is expressed in reads/kilobase pair/million mapped reads. RNA-seq data were deposited in the Gene Expression Omnibus of the National Institute for Biotechnology Information (GSE83506).
ChIP-seq and Bioinformatics Analysis
MC3T3 cells (10,000 cells/cm2) were plated in 10 cm plates in maintenance medium. Two days later, 5 μm GSK126 or vehicle was added to the cells in osteogenic medium. The cells were harvested 24 h later by trypsin and analyzed using a ChIP assay as described previously (46) using H3K27me3 (17-622, lot 2213948; Millipore) and control IgG (PP64B, lot 2056666A; Millipore) antibodies. Sequencing libraries were prepared and massively parallel high throughput sequencing was performed on a Illumina HiSeq2000 system. The alignment, quality assessment, peak calling, and visualization was performed with the HiChIP analysis pipeline (47). Briefly, 50 base pair reads were aligned to the mm10 reference genome using the Burrows-Wheeler Aligner, and Picard was used to mark duplicates. Read pairs without a unique alignment were filtered out using SAMTools (48) and a custom script that only retains pairs with one or both ends uniquely mapped. Enriched regions were identified using SICER (49). Peaks were identified from vehicle- and GSK126-treated cells using the SICER package (49) at 1% FDR. A subset of high confident peaks with FDR < 1e-10 was extracted from each library and merged into a single list of peaks if peaks from the two libraries are within 100 bp from each other. The average tag density (normalized to 1 million mapped reads) from upstream 5 kb to downstream 5 kb around the middle of all the merged peaks was estimated using the ngsplot package (50). ChIP-seq data were deposited along with RNA-seq data (see above) with accession number GSE83506.
Animal Welfare
All animal studies were conducted according to guidelines provided by the National Institutes of Health and the Institute of Laboratory Animal Resources, National Research Council. The Mayo Clinic Institutional Animal Care and Use Committee approved all animal studies. The animals were housed in an accredited facility under a 12-h light/dark cycle and provided water and food (PicoLab Rodent Diet 20, LabDiet) ad libitum.
In Vivo Ezh2 Inhibition Studies
Female C57Bl/6 mice were purchased from Harlan Laboratories. Sample sizes used in this study were determined based on previous studies with bone-anabolic drugs (51). For efficacy studies, 6-week-old mice received daily i.p. injections of vehicle (DMSO) or 50 mg/kg GSK126 in 20% Captisol adjusted to pH 4–4.5 with 1 n acetic acid (28) for 5 weeks. The dosage, delivery schedule, and administration route were selected based on previous studies demonstrating the anti-cancer effects of GSK126 in mice (28). The animals were weighed daily. To label mineralizing bone surfaces, the mice received subcutaneous injections of calcein (10 mg/kg) 5 days and 24 h before euthanasia. The effects of GSK126 administration on the skeleton were evaluated in an estrogen-deficient OVX model. At ∼12 weeks of age, female C57BL/6 mice underwent either sham or OVX surgeries. The following day, the animals received daily i.p. injections of vehicle (DMSO) or GSK126 (50 mg/kg body weight) for 6 weeks, as described above. Mice with body weights greater than 1 S.D. from the mean from each group were excluded from further analysis. For both studies, mice were randomly allocated to each group. Investigators performing tissue analysis (μCT and histomorphology) were blinded from the group assignments.
Bone Histomorphometry
The right distal femur was processed for static and dynamic histomorphometry as previously described (51). Thin (5 μm) sections were stained with Von Kossa/McNeal's tetrachrome to highlight osteoblast surfaces, Goldner's trichrome to examine mineralizing surface and bone surfaces, and tartrate-resistant alkaline phosphatase to detect osteoclasts. Unstained sections were used for assessment of dynamic histomorphometry and bone area. Beginning 450 μm proximal to the growth plate, mineralizing surface (MS/BS, %/day), mineral apposition rate (MAR, μm/day), bone formation rate (BFR/BV, %/day), osteoblast surface/bone surface (Ob.S/BS, %), osteoblast number/bone perimeter (N.Ob/B.Pm, #/mm), osteoclast surface/bone surface (Oc.S/BS, %), osteoclast number/bone perimeter (N.Oc/B.Pm, #/mm), osteoclast number/tissue area (N.Oc/T.Ar), trabecular separation (Tb. Separation), and trabecular number (Tb. N) were quantified using the OsteoMeasure image analysis system (OsteoMetrics Inc.).
Microcomputed Analysis of in Vivo Ezh2 Inhibition Studies
Quantitative analyses of the femoral metaphysis and fifth lumbar vertebra (L5) were performed using a VivaCT 40 scanner (SCANCO Medical AG) with the following parameters: E = 55 kVp, I = 145 μA, and integration time = 300 ms. A voxel size of 10.5 μm using a threshold of 220 units was applied to all scans at high resolution. Two-dimensional data from scanned slices were used for a three-dimensional interpolation and calculation of morphometric parameters that define cortical and trabecular bone mass and micro-architecture.
Statistics
When applicable, the data are shown as mean ± S.D. For in vitro studies, statistical analysis was performed with unpaired Student's t test. Significance is noted in the figures, when applicable (*, p < 0.05; **, p < 0.01; and ***, p < 0.001). For in vivo studies, statistical analysis was performed using Wilcoxon test or Wilcoxon rank sums test for multiple comparisons with the statistical software JMP Pro.
Author Contributions
A. D., E. T. C., S. M. R., G. S. S., M. A. M., J. J. W., and A. J. v. W. designed the study. A. D., E. T. C., S. M. R., C. R. P., M. G., T. M. O., R. T., M. S., J. R. H., and M. E. M.-L. performed the biological experiments. J. M. E. and H. Y. performed bioinformatic analysis. G. S. S., M. A. M., and A. J. v. W. collectively developed key epigenetic concepts for gene regulation during osteoblast differentiation that provided the foundation for the current study. A. D. and A. J. v. W. wrote the paper with comments from all authors.
Supplementary Material
Acknowledgments
We thank Oksana Pichurin, Bashar Hasan, David Razidlo, and Bridget Stensgard for technical support and the members of our laboratory, as well as our colleagues Anne Gingery, Alexey Leontovich, Sanjeev Kakar, Jay Smith, Aaron Krych, John Sperling, Wenchun Qu, Jane Lian, and Janet Stein for sharing reagents and/or stimulating discussions. We also acknowledge the support of Asha Nair and Jared Evans (Bioinformatics Core, Medical Genome Facility, Biomaterials Characterization and Quantitative Histomorphometry Core Facility and the Arlene Kogod Center on Aging at Mayo Clinic).
This work was supported, in whole or in part, by National Institutes of Health Grants F32 AR066508 (to A. D.), R01 AR049069 (to A. J. v. W.), R01 AR068103 (to J. J. W.), and R01 DE020194 (to J. J. W.), Fondo de Financiamiento de Centros de Investigación en Áreas Prioritarias 15090007 (to M. A. M.), and generous philanthropic support from William H. and Karen J. Eby and the charitable foundation in their names. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Tables S1–S3 and Fig. S1.
- BMD
- bone mineral density
- RNA-seq
- RNA sequencing
- ChIP-seq
- sequencing of chromatin immunoprecipitations
- PTH
- parathyroid hormone
- MSC
- mesenchymal stromal cell
- PRC
- polycomb-repressive complex
- RT-qPCR
- quantitative real time reverse transcriptase PCR
- TSS
- transcriptional start site
- FPKM
- fragments/kilobase pair/million mapped reads
- OVX
- ovariectomy
- Dn
- day n
- μCT
- microcomputed topography
- FDR
- false discovery rate.
References
- 1. (1993) Consensus development conference: diagnosis, prophylaxis, and treatment of osteoporosis. Am. J. Med. 94, 646–650 [DOI] [PubMed] [Google Scholar]
- 2. Burge R., Dawson-Hughes B., Solomon D. H., Wong J. B., King A., and Tosteson A. (2007) Incidence and economic burden of osteoporosis-related fractures in the United States, 2005–2025. J. Bone Miner. Res. 22, 465–475 [DOI] [PubMed] [Google Scholar]
- 3. Dennison E., and Cooper C. (2000) Epidemiology of osteoporotic fractures. Horm. Res. 54, 58–63 [DOI] [PubMed] [Google Scholar]
- 4. MacLean C., Newberry S., Maglione M., McMahon M., Ranganath V., Suttorp M., Mojica W., Timmer M., Alexander A., McNamara M., Desai S. B., Zhou A., Chen S., Carter J., Tringale C., et al. (2008) Systematic review: comparative effectiveness of treatments to prevent fractures in men and women with low bone density or osteoporosis. Ann. Intern. Med. 148, 197–213 [DOI] [PubMed] [Google Scholar]
- 5. Rosen V. (2009) BMP2 signaling in bone development and repair. Cytokine Growth Factor Rev. 20, 475–480 [DOI] [PubMed] [Google Scholar]
- 6. Bellido T., Saini V., and Pajevic P. D. (2013) Effects of PTH on osteocyte function. Bone 54, 250–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Canalis E. (2013) Wnt signalling in osteoporosis: mechanisms and novel therapeutic approaches. Nat. Rev. Endocrinol. 9, 575–583 [DOI] [PubMed] [Google Scholar]
- 8. Jiang Y., Jahagirdar B. N., Reinhardt R. L., Schwartz R. E., Keene C. D., Ortiz-Gonzalez X. R., Reyes M., Lenvik T., Lund T., Blackstad M., Du J., Aldrich S., Lisberg A., Low W. C., Largaespada D. A., et al. (2002) Pluripotency of mesenchymal stem cells derived from adult marrow. Nature 418, 41–49 [DOI] [PubMed] [Google Scholar]
- 9. Pike J. W. (2011) Genome-scale techniques highlight the epigenome and redefine fundamental principles of gene regulation. J. Bone Miner. Res. 26, 1155–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lian J. B., Stein G. S., van Wijnen A. J., Stein J. L., Hassan M. Q., Gaur T., and Zhang Y. (2012) MicroRNA control of bone formation and homeostasis. Nat. Rev. Endocrinol 8, 212–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. de Gorter D. J. J., and ten Dijke P. (2013) Signal transduction cascades controlling osteoblast differentiation. In Primer on the Metabolic Bone Diseases and Disorders of Mineral Metabolism, pp. 15–24, John Wiley & Sons, Inc. [Google Scholar]
- 12. Gordon J. A., Stein J. L., Westendorf J. J., and van Wijnen A. J. (2015) Chromatin modifiers and histone modifications in bone formation, regeneration, and therapeutic intervention for bone-related disease. Bone 81, 739–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. van Wijnen A. J., van de Peppel J., van Leeuwen J. P., Lian J. B., Stein G. S., Westendorf J. J., Oursler M. J., Im H. J., Taipaleenmäki H., Hesse E., Riester S., and Kakar S. (2013) MicroRNA functions in osteogenesis and dysfunctions in osteoporosis. Curr. Osteoporos Rep. 11, 72–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee J. S., Smith E., and Shilatifard A. (2010) The language of histone crosstalk. Cell 142, 682–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Margueron R., and Reinberg D. (2011) The polycomb complex PRC2 and its mark in life. Nature 469, 343–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim K. H., and Roberts C. W. (2016) Targeting EZH2 in cancer. Nat. Med. 22, 128–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chou R. H., Chiu L., Yu Y. L., and Shyu W. C. (2015) The potential roles of EZH2 in regenerative medicine. Cell Transplant 24, 313–317 [DOI] [PubMed] [Google Scholar]
- 18. Margueron R., Li G., Sarma K., Blais A., Zavadil J., Woodcock C. L., Dynlacht B. D., and Reinberg D. (2008) Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol. Cell 32, 503–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Marchesi I., Giordano A., and Bagella L. (2014) Roles of enhancer of zeste homolog 2: from skeletal muscle differentiation to rhabdomyosarcoma carcinogenesis. Cell Cycle 13, 516–527 [DOI] [PubMed] [Google Scholar]
- 20. Ge K. (2012) Epigenetic regulation of adipogenesis by histone methylation. Biochim. Biophys. Acta 7, 727–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dudakovic A., Camilleri E. T., Xu F., Riester S. M., McGee-Lawrence M. E., Bradley E. W., Paradise C. R., Lewallen E. A., Thaler R., Deyle D. R., Larson A. N., Lewallen D. G., Dietz A. B., Stein G. S., Montecino M. A., et al. (2015) Epigenetic control of skeletal development by the histone methyltransferase Ezh2. J. Biol. Chem. 290, 27604–27617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hemming S., Cakouros D., Isenmann S., Cooper L., Menicanin D., Zannettino A., and Gronthos S. (2014) EZH2 and KDM6A act as an epigenetic switch to regulate mesenchymal stem cell lineage specification. Stem Cells 32, 802–815 [DOI] [PubMed] [Google Scholar]
- 23. Wei Y., Chen Y. H., Li L. Y., Lang J., Yeh S. P., Shi B., Yang C. C., Yang J. Y., Lin C. Y., Lai C. C., and Hung M. C. (2011) CDK1-dependent phosphorylation of EZH2 suppresses methylation of H3K27 and promotes osteogenic differentiation of human mesenchymal stem cells. Nat. Cell Biol. 13, 87–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hui T., Zhao Y., Wang C., Gao B., Zhang P., Wang J., Zhou X., and Ye L. (2014) EZH2, a potential regulator of dental pulp inflammation and regeneration. J. Endod. 40, 1132–1138 [DOI] [PubMed] [Google Scholar]
- 25. Zhu L., and Xu P. C. (2013) Downregulated LncRNA-ANCR promotes osteoblast differentiation by targeting EZH2 and regulating Runx2 expression. Biochem. Biophys. Res. Commun. 432, 612–617 [DOI] [PubMed] [Google Scholar]
- 26. Wu H., Whitfield T. W., Gordon J. A., Dobson J. R., Tai P. W., van Wijnen A. J., Stein J. L., Stein G. S., and Lian J. B. (2014) Genomic occupancy of Runx2 with global expression profiling identifies a novel dimension to control of osteoblastogenesis. Genome Biol. 15, R52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Varambally S., Cao Q., Mani R. S., Shankar S., Wang X., Ateeq B., Laxman B., Cao X., Jing X., Ramnarayanan K., Brenner J. C., Yu J., Kim J. H., Han B., Tan P., Kumar-Sinha C., Lonigro R. J., Palanisamy N., Maher C. A., and Chinnaiyan A. M. (2008) Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science 322, 1695–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McCabe M. T., Ott H. M., Ganji G., Korenchuk S., Thompson C., Van Aller G. S., Liu Y., Graves A. P., Della Pietra A. 3rd, Diaz E., LaFrance L. V., Mellinger M., Duquenne C., Tian X., Kruger R. G., et al. (2012) EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 492, 108–112 [DOI] [PubMed] [Google Scholar]
- 29. Caretti G., Di Padova M., Micales B., Lyons G. E., and Sartorelli V. (2004) The polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 18, 2627–2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang L., Jin Q., Lee J. E., Su I. H., and Ge K. (2010) Histone H3K27 methyltransferase Ezh2 represses Wnt genes to facilitate adipogenesis. Proc. Natl. Acad. Sci. U.S.A. 107, 7317–7322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bernstein B. E., Mikkelsen T. S., Xie X., Kamal M., Huebert D. J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Jaenisch R., Wagschal A., Feil R., Schreiber S. L., and Lander E. S. (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326 [DOI] [PubMed] [Google Scholar]
- 32. Pereira J. D., Sansom S. N., Smith J., Dobenecker M. W., Tarakhovsky A., and Livesey F. J. (2010) Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc. Natl. Acad. Sci. U.S.A. 107, 15957–15962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schellenberg A., Lin Q., Schüler H., Koch C. M., Joussen S., Denecke B., Walenda G., Pallua N., Suschek C. V., Zenke M., and Wagner W. (2011) Replicative senescence of mesenchymal stem cells causes DNA-methylation changes which correlate with repressive histone marks. Aging 3, 873–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cakouros D., Isenmann S., Cooper L., Zannettino A., Anderson P., Glackin C., and Gronthos S. (2012) Twist-1 induces Ezh2 recruitment regulating histone methylation along the Ink4A/Arf locus in mesenchymal stem cells. Mol. Cell. Biol. 32, 1433–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Huang D., Wang X., Zhuang C., Shi W., Liu M., Tu Q., Zhang D., and Hu L. (2016) Reciprocal negative feedback loop between EZH2 and miR-101–1 contributes to miR-101 deregulation in hepatocellular carcinoma. Oncol. Rep. 35, 1083–1090 [DOI] [PubMed] [Google Scholar]
- 36. Black D. M., and Rosen C. J. (2016) Clinical practice: postmenopausal osteoporosis. N. Engl. J. Med. 374, 254–262 [DOI] [PubMed] [Google Scholar]
- 37. Sánchez-Duffhues G., Hiepen C., Knaus P., and Ten Dijke P. (2015) Bone morphogenetic protein signaling in bone homeostasis. Bone 80, 43–59 [DOI] [PubMed] [Google Scholar]
- 38. Jing H., Liao L., An Y., Su X., Liu S., Shuai Y., Zhang X., and Jin Y. (2016) Suppression of EZH2 prevents the shift of osteoporotic MSC fate to adipocyte and enhances bone formation during osteoporosis. Mol. Ther. 24, 217–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Glazer R. I., Hartman K. D., Knode M. C., Richard M. M., Chiang P. K., Tseng C. K., and Marquez V. E. (1986) 3-Deazaneplanocin: a new and potent inhibitor of S-adenosylhomocysteine hydrolase and its effects on human promyelocytic leukemia cell line HL-60. Biochem. Biophys. Res. Commun. 135, 688–694 [DOI] [PubMed] [Google Scholar]
- 40. Tan J., Yang X., Zhuang L., Jiang X., Chen W., Lee P. L., Karuturi R. K., Tan P. B., Liu E. T., and Yu Q. (2007) Pharmacologic disruption of polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 21, 1050–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Miranda T. B., Cortez C. C., Yoo C. B., Liang G., Abe M., Kelly T. K., Marquez V. E., and Jones P. A. (2009) DZNep is a global histone methylation inhibitor that reactivates developmental genes not silenced by DNA methylation. Mol. Cancer Ther. 8, 1579–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Arrowsmith C. H., Audia J. E., Austin C., Baell J., Bennett J., Blagg J., Bountra C., Brennan P. E., Brown P. J., Bunnage M. E., Buser-Doepner C., Campbell R. M., Carter A. J., Cohen P., Copeland R. A., et al. (2015) The promise and peril of chemical probes. Nat. Chem. Biol 11, 536–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang D., Christensen K., Chawla K., Xiao G., Krebsbach P. H., and Franceschi R. T. (1999) Isolation and characterization of MC3T3-E1 preosteoblast subclones with distinct in vitro and in vivo differentiation/mineralization potential. J. Bone Miner. Res. 14, 893–903 [DOI] [PubMed] [Google Scholar]
- 44. Schneider C. A., Rasband W. S., and Eliceiri K. W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dudakovic A., Camilleri E., Riester S. M., Lewallen E. A., Kvasha S., Chen X., Radel D. J., Anderson J. M., Nair A. A., Evans J. M., Krych A. J., Smith J., Deyle D. R., Stein J. L., Stein G. S., et al. (2014) High-resolution molecular validation of self-renewal and spontaneous differentiation in clinical-grade adipose-tissue derived human mesenchymal stem cells. J. Cell. Biochem. 115, 1816–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dudakovic A., Evans J. M., Li Y., Middha S., McGee-Lawrence M. E., van Wijnen A. J., and Westendorf J. J. (2013) Histone deacetylase inhibition promotes osteoblast maturation by altering the histone H4 epigenome and reduces Akt phosphorylation. J. Biol. Chem. 288, 28783–28791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yan H., Evans J., Kalmbach M., Moore R., Middha S., Luban S., Wang L., Bhagwate A., Li Y., Sun Z., Chen X., and Kocher J. P. (2014) HiChIP: a high-throughput pipeline for integrative analysis of ChIP-Seq data. BMC Bioinformatics 15, 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., and 1000 Genome Project Data Processing Subgroup (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zang C., Schones D. E., Zeng C., Cui K., Zhao K., and Peng W. (2009) A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics 25, 1952–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shen L., Shao N., Liu X., and Nestler E. (2014) ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics 15, 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. McGee-Lawrence M. E., McCleary-Wheeler A. L., Secreto F. J., Razidlo D. F., Zhang M., Stensgard B. A., Li X., Stein G. S., Lian J. B., and Westendorf J. J. (2011) Suberoylanilide hydroxamic acid (SAHA; vorinostat) causes bone loss by inhibiting immature osteoblasts. Bone 48, 1117–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Huang da W., Sherman B. T., and and Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.