Abstract
Mesenchymal stromal cells (MSCs) present in the bone marrow microenvironment secrete cytokines and angiogenic factors that support the maintenance and regenerative expansion of hematopoietic stem and progenitor cells (HSPCs). Here, we tested the hypothesis that extracellular vesicles (EVs) released by MSCs contribute to the paracrine crosstalk that shapes hematopoietic function. We systematically characterized EV release by murine stromal cells and demonstrate that MSC-derived EVs prompt a loss of HSPC quiescence with concomitant expansion of murine myeloid progenitors. Our studies reveal that HSPC expansion by MSC EVs is mediated via the MyD88 adapter protein and is partially blocked by treatment with a TLR4 inhibitor. Imaging of fluorescence protein-tagged MSC EVs corroborated their cellular co-localization with TLR4 and endosomal Rab5 compartments in HSPCs. The dissection of downstream responses to TLR4 activation reveals that the mechanism by which MSC EVs impact HSPCs involves canonical NF-κB signaling and downstream activation of Hif-1α and CCL2 target genes. Our aggregate data identify a previously unknown role for MSC-derived EVs in the regulation of hematopoiesis through innate immune mechanisms and illustrate the expansive cell-cell crosstalk in the bone marrow microenvironment.
Keywords: exosome (vesicle), extracellular vesicles, hematopoiesis, hematopoietic stem cells, mesenchymal stem cells (MSCs), microenvironment, Toll-like receptor (TLR)
Introduction
A small population of long-lived quiescent HSPCs3 residing in the bone marrow sustains lifelong hematopoietic function and provides regenerative capacity through cycles of self-renewal and differentiation (1). Cell fate commitment yields a cascade of successively more restricted progenitor populations that give rise to circulating blood and mature immune cells (2). HSPC activation relies on cell-autonomous programs as well as extrinsic cues from the surrounding bone marrow microenvironment where mesenchymal stromal cells, osteoprogenitors, and endothelial cells act through the paracrine action of secreted cytokines and angiogenic factors (3–6).
Recent studies indicate that HSPC self-renewal and emergence from quiescence are also regulated by type I and II interferons as well as Toll-like receptors (TLRs), signals associated with innate immunity (7–10), and known to contribute to regenerative HSPC responses (11–15). TLR signaling involves a number of transmembrane receptors and several critical adaptor molecules (16). Among them, the myeloid differentiation factor (MyD88) occupies a central role in transducing both surface and endosomal signals for most TLRs. Evidence supports TLR activation in the HSPC response to diverse stimuli, including radiation, bleeding, and infection (reviewed in Ref. 12). More recent reports, however, indicate that these mechanisms are also relevant for “tonic” homeostatic function and as well as developmental emergence of HSPCs (15, 17, 18). Indeed, many cytokines central to the inflammatory response are also critical to HSPC maintenance and differentiation (19–21).
Extracellular vesicles (EVs) are constitutively released from cells and are powerful paracrine regulators of bystander cell function (22–24). We recently described the role of exosomes (a subtype of EVs, 30–110 nm in diameter) released from leukemia cells and their ability to actively suppress HSPC function (25, 26). Mesenchymal stromal cells similarly release EVs that confer regenerative capacity in several (non-hematopoietic) tissues (reviewed in Ref. 27). Here, we test the hypothesis that EVs released from mesenchymal stromal cell (MSCs) present in the bone marrow can regulate HSPCs. Our in vitro and in vivo observations show, for the first time, that MSC-derived EVs expand myeloid-based (MPP2–4) progenitor cells. We confirm both TLR4 co-localization in endosomal compartments and HSPC activation by MSC EVs. Mechanistically, we show that these events coincide with activation of NF-κB and several characteristic downstream target genes.
Results
Bone Marrow- and Adipose-derived Stromal Cells Release a Heterogeneous Population of EVs
MSCs are critical to HSPC function, and several groups have reported the efficient release of EVs from bone marrow (BM)-derived MSCs (27, 28). Here, we set out to characterize three cellular sources of MSCs and their EVs. In addition to murine MSCs from a biorepository funded by the NHLBI, National Institutes of Health (validated by others (29, 30)), we analyzed BM-derived (primary mouse bone marrow-derived MSCs (PriMSCs)) as well as adipose-derived stromal cell (ADSC) MSCs from C57BL/6 animals, all extracted and propagated according to established protocols (31, 32). Using a broad panel of antibodies, we demonstrate the expression of characteristic MSC markers on cells from all sources (Fig. 1A) and confirm the predicted adipogenic and osteogenic differentiation induction via Oil Red O stain and Alizarin Red S, respectively, (Fig. 1B). Next, we generated EV preparations from all three sources according to protocols established in the lab (25, 26). We characterized these EVs by transmission electron microscopy for morphology and size, nanoparticle tracking analysis for size and concentration, and Western analysis for characteristic vesicle-associated proteins. Results reveal a mixed population of spherical and cup-shaped vesicles, ranging broadly from 100 to 400 nm, and expressing characteristic EV markers Rab5 and TSG101 (Fig. 1, C and D). Our aggregate data indicate that all three sources of stromal cells release a heterogeneous population of EVs.
FIGURE 1.
Mouse mesenchymal stromal cells are potent producers of extracellular vesicles. A, representative results showing expression of typical mesenchymal stromal cell surface markers on MSC (from Tulane University biorepository; TuMSC) (green), PriMSC (blue), and ADSC (red) assessed by flow cytometry. B, differentiation potential of each cell type was assessed by growing cells in osteogenic (O) or adipogenic (A) induction medium. Adipocytes and osteocytes were stained using Oil Red O and Alizarin Red S staining, respectively. C, conditioned medium from each cell type was assayed for EV size and concentration distribution using nanoparticle tracking analysis. Individual vesicles were imaged using transmission electron microscopy at 37,000× magnification (inset). Scale bar is 200 nm. D, EVs from each cell type were enriched using differential ultracentrifugation and assessed for typical exosome markers by immunoblotting.
In Vitro Exposure to MSC-derived EVs Activates Hematopoietic Progenitors via MyD88-dependent Signaling
To determine whether stromal derived EVs impact hematopoietic function, we exposed c-Kit-enriched murine HSPCs to concentrated EVs, prepared by serial ultracentrifugation, for 48 h, followed by immunophenotypic analysis. Results show a significant expansion of the HSPC KSL (c-Kit+, Sca-1+, lineage-negative) compartment following EV exposure, but not in those cultured in the presence of vesicle-depleted medium (termed: S100) that retains non-vesicle-associated factors, including cytokines, and angiogenic factors. To understand the downstream effects of MSC EV-mediated HSPC activation, we examined the fate of specific HSPC subsets following MSC EV exposure using a combination of antibodies that distinguish the earliest progenitors and that have validated functional attributes (2, 33). Principally, based on the absence of differentiated lineage markers and the presence of surface markers c-Kit, Sca-1, CD48, and CD150, we found prominent expansion of MPP2 and MPP3/4 progenitors (as defined by others (2, 33, 34)) but not the less mature, multipotent MPP1 progenitors (Fig. 2A). Differentiation and selective MPP2 and MPP3/4 expansion are anticipated to coincide with a loss of quiescence and increased entry into cell cycle. Indeed, when comparing EV- with S100-exposed HSPCs, we observe aggregate gains in G1/S/G2/M phase with a loss of quiescence (G0) following EV exposure of HSPCs (Fig. 2B). To determine the functional consequences of EV exposure, we evaluated progenitor clonogenicity in methylcellulose, scoring colony-forming progenitors (CFU-C) and subtypes. We found that the overall colony formation after exposure to EVs is equipotent in maintaining colony formation to that seen after S100 exposure of HSPCs. However, EV exposure results in the increased formation of CFU-M (monocyte colony-forming unit) and CFU-G (granulocyte colony-forming unit) when compared with the S100, suggesting an increase in the population of myeloid-committed progenitors (Fig. 2C).
FIGURE 2.
Bone marrow MSC-derived EVs induce HSPC activation and differentiation in a MyD88-dependent mechanism. A, representative flow cytometry dot plots demonstrating gating strategy used for immunophenotyping HSPCs. WT C57BL/6 bone marrow cells were harvested and enriched for HSPCs using a magnetic hematopoietic progenitor enrichment kit, and then cells were exposed to MSC-derived concentrated EVs (EV) or EV-depleted conditioned medium (S100) for 48 h. HSPCs were assessed by the presence of KSL (lineage−, C-Kit+, Sca-1+) and SLAM markers (CD48 and CD150). MPP3/4, KSL/CD48+/CD150−; MPP2, KSL/CD48+/CD150+; SLAM, KSL/CD48−/CD150−. B, the fate of the HSPCs after EV exposure was assayed by Ki67/Hoechst cell cycle analysis. Right panels show representative images of flow cytometry dot blots and histograms. C, to assay colony formation potential, HSPCs were plated in methylcellulose at 3,000 cells/ml, and total colony count and colony type scoring were done after 7 days of culture. Right-most panels depict representative images of colony types. D, to examine MyD88-dependent TLR4 involvement, MyD88−/− bone marrow cells were harvested and enriched for HSPCs using magnetic enrichment, and then exposed to EV or S100 conditions. All significance values were calculated using a two-tailed t test, and statistical significance was set at p ≤ 0.05. Error bars indicate means ± S.E. E, erythrocyte; GEMM, granulocyte, erythrocyte macrophage/monocyte, megakaryocyte; M, macrophage; GM, granulocyte, macrophage.
Because TLR signaling is a known contributor to HSPC activation and subsequent expansion (9, 10), we next turned our attention to MyD88, a TLR adaptor protein, to validate the observed phenotype and further delineate the mechanism by which this expansion occurs. We tested HSPCs from animals with genetic disruption of MyD88 by exposure to MSC-derived EVs and found that the absence of MyD88 reduces expansion of immunophenotypically defined HSPCs after EV exposure when compared with S100 (Fig. 2D).
EV-mediated Expansion Occurs via TLR 4 Engagement
Data to this point were consistent with the notion that MSC EV exposure might prompt the engagement of TLR4 as a specific candidate mechanism involved in HSPC expansion. When we used LPS, an extensively studied activator of TLR4, we readily replicated the observations made after EV exposure and demonstrated that treatment with TAK-242, a pharmacological inhibitor of TLR4, abrogates the effect of both EV exposure and LPS (Fig. 3A) (14, 20). Functionally, colony-forming analyses demonstrate that TLR4 inhibition is sufficient to reverse the bias in committed progenitor colony composition after EV exposure (Fig. 3B). We used confocal microscopy to examine EV interaction with cell surface TLR4 receptor and their ensuing intercellular fate. Using myristoylation to tag reporter proteins to the lipid bilayer membranes (35, 36), we marked MSCs to produce myristoylated/palmitoylated tomato protein (mTomato)-tagged EVs and utilized confocal immunofluorescent microscopy to determine co-localization partners of stably labeled EVs. Results demonstrate quantitative co-localization of EVs and TLR4. To further resolve the TLR4 activation by EVs in HSPCs, we additionally queried internalization of EVs, observed co-localization with Rab5-expressing early endosomal compartments (Fig. 3C), and showed that TLR4-EV co-localization occurs with fairly rapid kinetics over the course of 24 h (Fig. 3D). TLR4 activation is predicted to lead to potent induction of cytokines in HSPCs (37).
FIGURE 3.
HSPC differentiation is induced by TLR4-mediated signaling of MSC EVs. A, differentiation potentials of MSC EV-exposed HSPCs were analyzed by flow cytometry. The role of TRL4 receptor was tested by TLR4 inhibitor TAK-242 in HSPCs using LPS as a positive control and compared against EV-treated HSPCs, and differentiation potential was analyzed using flow cytometry. B, CFU assay to measure colony formation potential after TLR4 inhibition in EV-treated HSPCs. Cells were plated in methylcellulose at 3,000 cells/ml, and total colony count and colony type scoring were done after 7 days of culture. C, representative imaging of mTomato-tagged EVs (red) and their interaction with TLR4 receptor (green) in early and late endosomes. Rab5 and Rab7 (magenta) expressions as early and late endosome markers are used to depict endocytosis stages. Con, control. D, representative imaging of co-localized TLR4 and mTomato-tagged EVs. Association kinetics of TLR4 on HSPCs and mTomato-tagged MSC EVs are depicted as the Pearson's correlation coefficient plot, using co-localized voxels at 3-, 16-, and 24-h time points after EV exposure. Error bars indicate means ± S.E. SEM-Luc, spontaneously evolved microglia-luciferase; Exo, extracellular vesicles.
MSC EVs Lead to NF-κB Induction and Cytokine Dysregulation
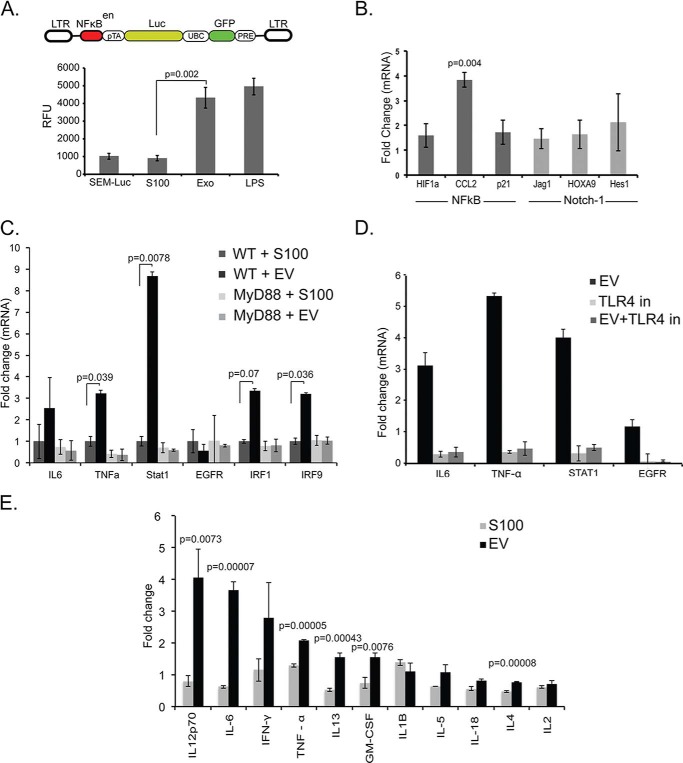
Given the involvement of TLR4 signaling, we proceeded to delineate HSPC-specific downstream signaling events with emphasis on NF-κB and Notch signaling, both with established relevance to HSPC function (9). NF-κB is a central transcription factor that can be activated in HSPCs by paracrine cues. To identify the putative functional axis of MSC EV-derived activation of NF-κB signaling, we used TLR4-responsive, stably transduced murine NF-κB reporter cells. These reporter cells were co-cultured in EV versus S100 conditions, and NF-κB expression was measured by luminescence emission 48 h later. Experiments show that NF-κB is functionally up-regulated following EV exposure (Fig. 4A). We also tested the well defined downstream NF-κB targets HIF1α and CCL2 and found increased expression after EV exposure when compared with S100. Because the TLR4-mediated NF-κB signaling cascade elicits downstream activation of Notch signaling in hematopoietic cells (15), we next tested Notch targets p21, Jag1, and the transcriptional repressor Hes1. Modest differences suggest that notch is unlikely to be a downstream target (Fig. 4B) (38, 39).
FIGURE 4.
MSC-derived EVs activate NF-κB and Notch signaling to promote HSPC differentiation. A, diagrammatic representation of the lentiviral vector design used to generate the NF-κB enhancer-luciferase reporter cell line. Stably transduced and sorted SIM-A9 microglial cells were used for EV exposure followed by luciferase assay. The SIM-A9-NF-κB-Luc cells were exposed with MSC EVs and incubated for 48 h followed by luciferase assay. Non-treated SIM-A9-NF-κB-Luc and LPS-treated SIM-A9-NF-κB-Luc cells were used as negative and positive controls, respectively. Luciferase activity was measured as relative florescence units (RFU). B, quantitative mRNA expression of NF-κB and Notch-1 signaling and downstream targets involved in cell proliferation in MSC EV-exposed HSPCs. C, cytokines released by HSPCs after 48 h co-culture with MSC EVs when compared with S100 were measured using Luminex. D, quantitative mRNA expression of targets relevant for hematopoiesis and TLR4-mediated activation was analyzed in MSC EV-exposed WT and MyD88−/− HSPCs. E, TLR4 signaling was specifically inhibited in WT MSC EV-exposed HSPCs using TAK-242. Error bars indicate means ± S.E.
We next tested a candidate panel of canonical TLR4-responsive cytokines for both transcriptional activation and secretion, after either EV or S100 exposure. Transcriptional analysis of EV-exposed HSPCs reveals an up-regulation of several known TLR4-responsive cytokine genes (IL6, TNFα, Stat1, and EGFR) in WT but not MyD88−/− HSPCs (Fig. 4C). This EV-mediated increase is abrogated after inhibition of TLR4 by TAK-242 (Fig. 4D), further supporting EV involvement in HSPC expansion. Next, we examined the cytokine profile secreted by HSPCs after EV or S100 exposure. We observed increased secretion of proinflammatory cytokines such as IL-6, IFNγ, TNFα, and IL-1β, which have been shown to drive HSPC myeloid differentiation fate (Fig. 4E) (19, 21), consistent with transcriptional analyses.
MSC EVs Mediate HSPC Activation and Differentiation
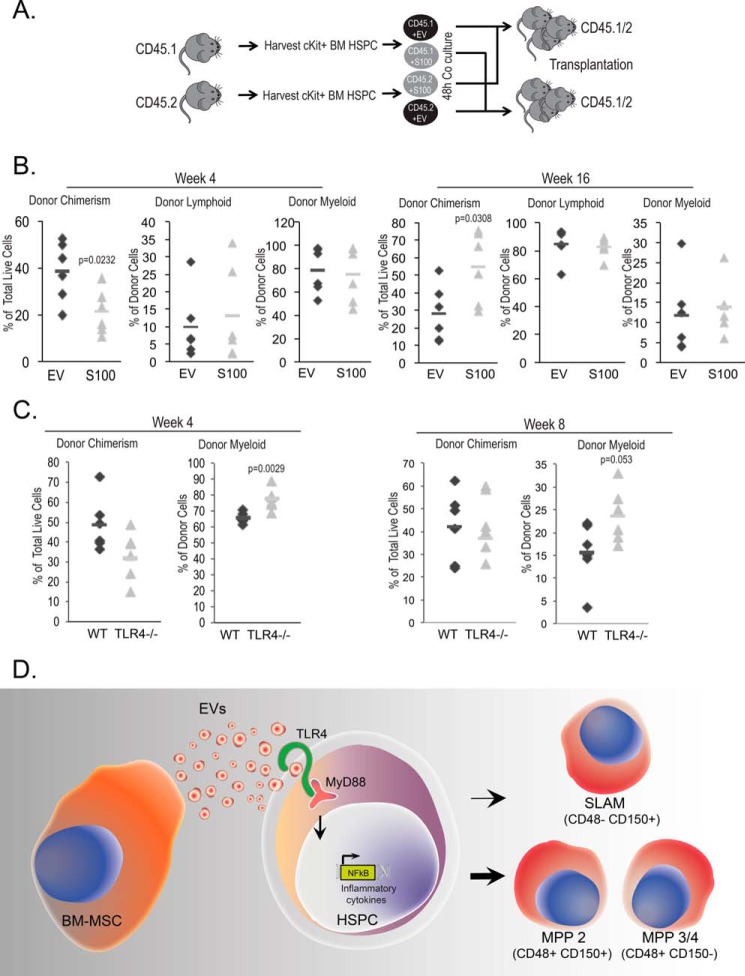
Finally, to determine the impact of the progenitor expansion on hematopoietic repopulation, we transplanted EV- and S100-exposed lineage-depleted HSPCs into myeloablated CD45-mismatched recipients (Fig. 5A). We used a crossover design to exclude strain-specific CD45.1 versus CD45.2 differences in repopulation capacity. We systematically tracked overall and leukocyte subset chimerism in recipient cohorts over time. Results demonstrate that the in vitro myeloid bias after EV exposure is rapidly lost and instead leads to a significant overall engraftment deficit at 16 weeks when compared with S100-exposed HSPCs (Fig. 5B). To specifically test the TLR4-responsive myelopoiesis response to MSC EV exposure, we next determined the competitive repopulation capacity of Tlr4−/− and WT grafts, respectively, using equal numbers of input donor HSPCs. In this model, significant deficits in the WT myeloid compartment became apparent early after transplantation when compared with Tlr4−/− contributions (Fig. 5C). We also undertook serial xenografting in this model and found consistent loss of chimerism in WT EV-exposed HSPCs (Fig. 5D). Unfortunately, statistical significance in the overall cohort was not achieved, a likely reflection of the recently described engraftment defect in CD45.1 recipients (40). Combined, the data corroborate recent reports of TLR4-mediated myelosuppression and long-term HSC exhaustion (41).
FIGURE 5.
MSC EV exposure induces myelosuppression and HSC exhaustion via TLR4. A, to examine HSPC repopulation potential, EV- or S100-exposed HSPCs were competitively transplanted into irradiation-conditioned CD45-mismatched recipients. B, donor chimerism and lymphoid versus myeloid contribution were examined at 4 weeks (short-term HSC) and 16 weeks (long-term HSC) after engraftment. C, competitively transplanted WT and Tlr4−/− donor chimerism and myeloid contribution were examined at 4 and 8 weeks after engraftment. D, model of BM-MSC-derived EVs effect on HSPCs. The HSPC transcriptome toward differentiation and proliferation undergoes activation after MSC EV exposure. The NF-κB signaling pathway up-regulation by MSC EVs results the entry into G1 cell cycle phase followed by MPP2 and MPP3/4 expansion.
Discussion
Extrinsic cues shape the hematopoietic response to stress and injury (8), and inflammatory signaling via interferon and tumor necrosis factor is a recently described physiological regulator of HSPCs during development (9, 10). Indeed, TLR engagement is a powerful stimulus for the release of inflammatory cytokines by murine HSPCs, leading to their expansion and ultimate exhaustion (15, 20). Here, we confirm a role for extracellular vesicles in signaling via TLRs, and present the first functional and immunophenotypic evidence for their impact on progenitor differentiation.
Hematopoietic stem cells give rise to mature, immunophenotypically defined, lineage-restricted progenitors (2, 20). Postnatally, the bone marrow and MSCs within provide critical stimuli for HSPC maintenance (5, 28). The release of EVs from MSCs is a constitutive cellular function and confers regenerative potential following experimental cardiac or kidney injury (27, 42, 43). Here, we carefully characterize murine bone marrow-derived stromal cells using extensive immunophenotyping as well as osteogenic and adipogenic differentiation assays and the vesicle populations released by MSCs. These studies provide a diverse picture of EVs released from MSCs, beyond an exosome-sized population (44–46). As they are a presumably more physiological representation of the BM microenvironment, we deliberately and abstained from up-front isolation of more rare MSC subpopulations or their EVs, although their contributions are likely to add additional nuance to the regulatory cascade described here (47). Although we noted differences in incorporation of Rab5, TSG101, and GAPDH in EVs between different MSC sources, we have no evidence to directly link this to the observed TLR4-mediated activation of HSPCs.
MSC exosomes regulate the phenotypic behavior of THP-1 cells via MyD88 restricted mechanisms (45, 48), and we therefore systematically dissected key TLR signaling events. Our observations were consistent with prior reports of HSPC activation after in vitro exposure to LPS and a recent report of G-CSF action, in both cases via TLR4 (15, 49). Indeed, LPS endotoxemia and G-CSF action enhance both short-term hematopoietic progenitor colony formation and transient expansion of murine HSPC progenitors with downstream exhaustion of the compartment and differentiation consistent with our findings herein (18, 20, 49). We show that EV disruption by incubation with TLR4 inhibitor TAK-242 and mouse MSC EV exposure of HSPCs from mice with genetic inactivation of the adaptor protein MyD88 both interfere with multipotent progenitor (MPP) expansion and differential colony formation, again consistent with the involvement of TLR4. Further downstream of MyD88, we tested the NF-κB-driven HSPC cytokine response and saw a significant up-regulation of targets IL-6, TNFα, G-CSF, and to a lesser extent, IL-1α and IL-10 but not IL-2, IL-12, or GM-CSF.
EVs can act through their cargo or via cell surface interaction on cells. We performed co-localization studies to further dissect the EV action via TLR4, as well as the specific association with internal endosomal compartments marked by Rab5 and Rab7, respectively, in murine HSPCs (50–52). The observed co-localizing events appear at the cell surface during the early time points consistent with studies by others (48), followed by successive internalization of TLR4 alongside EVs at the later time points up to 24 h. Rapid kinetics, TLR4 restriction, and initial surface engagement are most consistent with EV membrane interaction in accounting for downstream signaling events, rather than EV RNA content.
Recent studies reveal cellular regulatory networks unique to specific immunophenotypically defined MPP subsets (2). Here, we relied on those MPP definitions to classify the significant expansion of immunophenotypically defined progenitors after 48 h of liquid culture with EVs from marrow-derived murine MSC (31). The observed EV-mediated culture expansion of HSC SLAM cells was surprisingly similar in magnitude to the EV-free fraction of the cell culture supernatant (S100) and medium controls, both replete with cytokines known to support HSPCs. Remarkably, we observed significant MPP2 and MPP3/4 gains that are functionally complemented by differential in vitro colony formation and coincide with decreased quiescence (G0). This is consistent with our overall observation of skewed differentiation and underscores the importance of TLR4 regulation of myeloid progenitor commitment.
EVs can evoke regeneration after experimental kidney and cardiac injury, or in response to stress (42, 43, 53). In HSPCs, canonical inflammatory signals via IFNs, TNFα, or TLR not only shape the adult regenerative response (15), but appear to be part of hematopoietic physiology and fetal development (9, 10). The aggregate data reinforce an emerging picture of a highly context-dependent HSPC response whereby activation of different components of the innate immune response leads to unique, and not always deleterious, functional outcomes (41). Indeed, our observations are more consistent with a role for TLR activation and downstream NF-κB signaling in a physiological response mediated by the microenvironment (8, 13, 54). Altogether, these studies provide a new level of understanding for the complexity of compartmental hematopoietic regulation involving the paracrine trafficking of EVs to HSPCs with myeloid-biased progenitor expansion activation via TLR4-MyD88-NF-κB (Fig. 5E).
Experimental Procedures
Cell Culture
PriMSCs were grown in MSC medium: IMDM, 15% FBS, 1% penicillin/streptomycin. Mouse MSCs were obtained from the Tulane Center for Gene Therapy and maintained as reported previously (29, 30). Primary mouse ADSCs were isolated from the omentum of C57BL/6 mice and cultured in ADSC medium: IMDM, 20% FBS, 1% penicillin/streptomycin, 10−5 m hydrocortisone, 10−4 m β-mercaptoethanol. MSC in vitro differentiation was induced using the following medium conditions: osteogenesis (20 mm β-glycerol phosphate, 50 μm thyroxine, 1 nm dexamethasone, 0.5 μm ascorbate-2-phosphate); adipogenesis (5 mm insulin, 50 μm indomethacin, 0.5 μm 3-isobutyl-1-methylxanthine). Vesicle-free (VF) FBS was produced by centrifugation of FBS (Gemini Bio-Products) at 100,000 × g for 6 h.
Extracellular Vesicle Isolation and Labeling
MSCs were grown to confluency in four 15-cm cell culture plates (Corning) in VF MSC medium for 72 h. The medium from cultured plates was collected for serial centrifugation at 2,000 × g for 20 min and 10,000 × g for 20 min, followed by supernatant centrifugation at 100,000 × g for 2 h. EV pellets were resuspended in 15% VF MSC medium for 4 h with vigorous shaking at 4 °C. Supernatant collected from EV preparations after the 100,000 × g spin is defined as S100 control. We measured endotoxin levels and found that levels were below the allowable limits in commercially available cell culture medium (Pierce LAL Chromogenic Endotoxin Quantitation Kit; Thermo Scientific, catalog number 88282).
Transmission Electron Microscopy
EV preparations (10 μl) were deposited onto UV-activated carbon Formvar 400-mesh copper grids (Ted Pella 01822-F) for 3 min, rinsed, and stained in filtered 1.33% uranyl acetate, and then air-dried. Samples were imaged at 100 kV on a Philips CM120 transmission electron microscope operated by the Oregon Health & Science University Electron Microscope Core facility. Images were collected as 1,024 × 1,024-pixel 14-bit grayscale. Gatan Digital Micrograph 3 (DM3) files on a Gatan 794CCD multiscan camera were converted into 8-bit grayscale TIF images using the program Digital Micrograph 3.4.
Western Blotting
Cell and EV samples were washed with PBS and lysed using radioimmunoprecipitation assay buffer with protease and phosphatase inhibitor (Thermo Scientific), followed by ultra-sonication (Cole-Parmer CPX-130). Lysates were prepared in LDS sample buffer (Life Technologies) with 8% β-mercaptoethanol and boiled 5 min for denaturation. Denatured protein lysates were loaded on 4–15% Tris-HCl PAGE gel (Bio-Rad) for gel electrophoresis separation (50 min at 200 V) and transfer (overnight at 25 V). Transferred membranes were blocked with 5% nonfat dry milk (Carnation, Nestlé) in TBST and blotted with anti-GAPDH (1:5000, Novus Biologicals), anti-TSG101 (1:500, Santa Cruz Biotechnology) and anti-Rab5 (1:500, Cell Signaling Technology), followed by anti-rabbit IgG HRP conjugate (Promega) and anti-mouse IgG HRP conjugate (Promega). Blots were imaged in G-Box using GeneSys after a 5-min exposure to the SuperSignal West Pico Chemiluminescent HRP substrate (Thermo Scientific).
Nanoparticle Tracking Analysis (NTA)
EV samples were resuspended, and serial dilutions were prepared in nanofiltered (Whatman Anotop 25, 0.02-μm) molecular-grade water (Thermo Scientific) using low-adhesion 1.7-ml tubes (GeneMate). Diluted samples were loaded into the NanoSight LM10 chamber (Malvern), the laser was engaged, and microparticles were visualized. Sixty-second videos were acquired with a Hamamatsu C11440 ORCA-Flash 2.8 camera and analyzed by NanoSight NTA 2.3 software. Individual samples were diluted to 1 × 108−1 × 109 particles/ml.
RNA Analysis and Quantitative RT-PCR
RNA was extracted using miRNeasy or RNeasy (Qiagen) and quantified using a NanoDrop 2000c (Thermo) and Agilent Bioanalyzer (Agilent). cDNA was synthesized using a SuperScript III First Strand Synthesis Kit (Invitrogen) with oligo(dT) priming, followed by PCR. SYBR Green PCR (Applied Biosystems) was used for quantitative RT-PCR analysis. The ΔΔCT method was used for quantification. Primer sequences were: Hif1-a F, GCGTGTGAGGAAACTTCTGG; Hif1-a R, CACAAGGCCATTTCTGTGTGT; Tnf-a F, GAACTGGCAGAAGAGGCACT; Tnf-a R, GGTCTGGGCCATAGAACTGA; Il6 F, TTCCATCCAGTTGCCTTCTT; Il6 R, CAGAATTGCCATTGCACAAC; Ccl2 F, CCCAATGAGTAGGCTGGAGA; Ccl2 R, CCTTAGGGCAGATGCAGTTT; Egfr F, GAAGCCACATCTCCAAAAGC; Egfr R, AGGAGGTACTGGGAGCCAAT; p21 F, GTATGTCCCCTGAGCAGAGC; p21 R, AAAAACCCAAACTGCACCTG; Stat1 F, GAGGTGAACCTGACTTCCA; Stat1 R, TCTGGTGCTTCCTTTGGTCT; Irf1 F, CCTGGGTCAGGACTTGGATA; Irf1 R, TTCGGCTATCTTCCCTTCCT; Irf9 F, TCCTGGAGCATCAACTTCCT; Irf9 R, ATTCAGCAGGTGCTGGGTAG; Hoxa9 F, TCTCCGGGATGCATAGATTC; Hoxa9 R, TAGCAACCCTCTGCACACAC; Hes1 F, AAGTCCCTAGCCCACCTCTC; Hes1 R, AGGCGCAATCCAATATGAAC; Jag1 F, CAGTGCCTCTGTGAGACCAA; and Jag1 R, GTTATGGCAGGGGTCAGAGA.
ELISA
Major proinflammatory cytokines and chemokines contained in the EVs and S100 medium were analyzed by ELISA. The Mouse Inflammatory Cytokines Multi-Analyte ELISArray Kit (Qiagen) is used to analyze the panel of 12 pro-inflammatory cytokines using by the ELISA protocol. The cytokines and chemokines represented by this array are IL-1A, IL-1B, IL-2, IL-4, IL-6, IL-10, IL-12, IL-17A, IFNγ, TNFα, G-CSF, and GM-CSF. The ELISA was conducted as per the manufacturer's protocol.
Analysis of MSC Secreted Cytokine Content
Cell culture supernatants were analyzed for cytokines using a mouse Th1/Th2 Luminex panel (eBioscience, San Diego, CA), which included IL-12, GM-CSF, IFNγ, IL-1β, IL-13, IL-18, IL-2, IL-4, IL-5, IL-6, and TNFα. This analysis was performed by the Endocrine Technologies Support Core (ETSC) at the Oregon National Primate Research Center (ONPRC) using the manufacturer's instructions. Briefly, 25 μl of each cell culture supernatant was incubated overnight with antibody-coated, fluorescent-dyed capture microspheres specific for each, followed by detection antibodies and streptavidin-phycoerythrin. Washed microspheres with bound analytes were resuspended in reading buffer and analyzed on a Milliplex Analyzer (EMD Millipore, Billerica, MA) bead sorter with xPONENT software version 3.1 (Luminex, Austin, TX). Data were calculated using Milliplex Analyst software version 5.1 (EMD Millipore). Intra-assay CVs were as follows: IL-1β, 14.2%; IL-2, 10.4%; IL-4, 6.9%; IL-5, 18.1%; IL-6, 5.7%; IL-13, 7.2%; IFNγ, 25.4%; IL-12, 14.0%; GM-CSF, 24.5%; TNFα, 3.8%; IL-18, 6.4%. Because all samples were analyzed in a single assay, no inter-assay variation was calculated.
Generation of NF-κB Luc GFP Reporter Cells
Human embryonic kidney (HEK-293T) cells were seeded at a density of 1.5 × 107 cells per 15-cm tissue culture dish (Corning), precoated with 0.01% poly-l-lysine (Sigma), and transfected by using the transducing plasmid pHAGE NF-κB-TA-luc-UBC-GFP-W and helper constructs pLP1, pLP2, and pLP/VSVG (Gibco-Invitrogen) (55, 56). Calcium phosphate transfection was performed in the presence of DMEM (Gibco), 10% FBS (Gibco), 1% penicillin/streptomycin (Pen/Strep, Gibco). Vector supernatant was harvested 36, 48, and 72 h later, filtered through a 0.45-μm filter, and pooled. For transduction, SIM-A9 microglia cells (57) were washed and resuspended in medium with 8 μg/ml protamine sulfate (MP Biomedicals). Transductions took place overnight at 37 °C followed by wash and maintenance in culture. After 48 h, GFP positive cells were sorted by Influx Cell Sorter (BD Biosciences). Sorted cells were washed twice, propagated, plated in a 10-cm plate, and used for luciferase assay after EV exposure.
Immunofluorescence Microscopy
mTomato-tagged EVs were prepared as described above. Primary murine MSCs were generated from mT/mG mice that constitutively express membrane-targeted mTomato protein, which allows visualization of membranous vesicles (35). For imaging, lineage-negative cells were exposed to mTomato-labeled EVs overnight, washed twice in PBS, and labeled with anti-TLR4 followed by secondary staining by anti-mouse IgG Alexa Fluor 488 (Life Technologies) and PacBlue-anti-Sca-1 (Clone D7, BioLegend). Slides were mounted with Fluoromount G (Southern Biotech). Microscopy was performed on an Olympus IX71 with a 60 × 1.4 NA oil lens; Z-stacks were acquired every 0.2 μm with a Z-stack center bright-field reference, and images were processed with DeltaVision softWoRx Explorer.
Flow Cytometry Analysis
All data were analyzed on FACSCANTOTM or LSR IITM cytometers (BD Biosciences). In vivo chimerism was studied using CD45.1 (BioLegend; A20) or CD45.2 (BioLegend; 104). Lymphoid/myeloid analysis was performed with CD3 (BD Pharmingen; 17A2), CD45R/B220 (BD Pharmingen; RA3-6B2), or CD11b/Mac-1 (BioLegend; M1/70). HSPC immunophenotyping was performed with c-Kit (BioLegend; 2B8), Sca-1 (eBioscience; D7), CD150 (BioLegend; TC15-12F12.2), or CD48 (BioLegend; HM48-1). Cell cycle analysis was performed with Ki67 (BD Pharmingen; 550609) or Hoechst 33342 Solution (BD Pharmingen; 561908). MSC immunophenotyping was performed with CD44 (BioLegend; IM7), CD45 (BD Pharmingen; 30-F11), CD51 (BioLegend; RMV-7), CD105 (BioLegend; MJ7/18), CD29 (BD Pharmingen; KMI6), or nestin (eBioscience; Rat-401). All antibodies are anti-mouse, and all collected data were analyzed with FlowJo (Tree Star).
In Vitro Co-culture and CFU Assay
Whole bone marrow cells were collected by flushing femurs and tibias from 6–10-week-old C57BL/6 mice, with Iscove's modified Dulbecco's medium. Samples were depleted of red cells by hemolysis and c-Kit-enriched using an EasySep Mouse Hematopoietic Progenitor Cell Enrichment Kit according to the manufacturer's instructions (StemCell Technologies Inc., Vancouver, British Columbia, Canada). c-Kit-enriched primary mouse HSPCs were co-cultured with EVs or S100 in IMDM supplemented with 15% VF FBS, 1% penicillin/streptomycin, 50 ng/ml mouse stem cell factor, and 50 ng/ml mIL3 for 48 h. For experiments involving the TLR-4 inhibitor (58), mouse HSPCs were precultured with 1 μm TAK-242 (CLI-095, InvivoGen) for 30 min at 37 °C, and lipopolysaccharide (Sigma-Aldrich) was added at 10 ng/ml as a positive control. After a 48-h co-culture, HSPCs were plated at 3E+03 cells/ml in Mouse Methylcellulose Complete Medium (R&D Systems) in triplicates for CFU-C assays (37 °C, 5% CO2, ≥95% humidity, × 7 days).
Animal Husbandry
C57BL/6 mice were group-housed and allowed ad libitum access to standard chow pellets (Purina Laboratory Rodent Diet 5001; Ralston Purina Co., St Louis, MO). Following EV exposure, HSPCs were washed twice and resuspended in 200 μl of Hanks' balanced salt solution, and 1.0E+04 cells were injected intravenously into myeloablated (1,000 centigrays by Shepherd137 cesium irradiator) recipients (CD45.1; B6.SJL-Ptprca Pepcb/BoyJ). Following transplantation, retro-orbital eye bleeds were performed at intervals, and white blood cells were analyzed for CD45.1/-.2 chimerism by flow-cytometry (28). All animal studies were approved by the Oregon Health & Science University (OHSU) Institutional Animal Care and Use Committee. Myd88 (stock number 009088; B6.129P2 (SJL)-Myd88tm1.1Defr/J)-deficient animals and Tlr4 (stock number 007227; B6.B10ScN-Tlr4lps-del/JthJ)-deficient animals were obtained from The Jackson Laboratory.
Statistical Analysis
Continuous variables are summarized as means ± S.E. Analysis of variance and subsequent two-tailed Student's t tests were employed for comparison between different conditions. Statistical significance was set at p < 0.05.
Author Contributions
N. A. G. designed and performed experiments, interpreted data, and prepared the manuscript; S. C. V. designed and performed experiments and interpreted data; Y. m. Y. performed experiments and interpreted data; D. L. M. contributed to experimental design interpreted data and edited the manuscript; O. T. assisted with experiments and edited the manuscript; and P. K. designed experiments, interpreted data, and edited the manuscript.
Acknowledgments
Some of the materials employed in this work were provided by the Tulane Center for Gene Therapy through the National Center for Research Resources (NCRR) Grant P40RR017447. We gratefully acknowledge the assistance of Dr. Stephanie Krasnow, Dr. Xinxia Zhu, Pete Levasseur, Dr. Claudia Lopez, John Butler, Dr. Shelton Viola, Ben Doron, and Merna Labib with select experiments. We also thank Dr. David W. Erikson at The Endocrine Technologies Support Core (ETSC) at the Oregon National Primate Research Center (ONPRC) for help with the Luminex experiment. The ETSC at the ONPRC is supported by National Institutes of Health Grant P51 OD011092 awarded to ONPRC.
Preliminary accounts of this work were presented at the annual meeting of the International Society for Extracellular Vesicles, Bethesda, MD on April 23–26, 2015, in Washington, D. C. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- HSPC
- hematopoietic stem and progenitor cell
- MSC
- mesenchymal stromal cell
- EV
- extracellular vesicle
- BM
- bone marrow
- ADSC
- adipose-derived stromal cell
- PriMSC
- primary mouse bone marrow-derived MSC
- HSC
- hematopoietic stem cell
- TLR
- Toll-like receptor
- MPP
- mitochondrial processing peptidase
- VF
- vesicle-free
- IMDM
- Iscove's modified Dulbecco's medium
- NTA
- nanoparticle tracking analysis
- SLAM
- signaling lymphocyte activation molecule.
References
- 1. Purton L. E., and Scadden D. T. (2008) The hematopoietic stem cell niche. in StemBook, Harvard Stem Cell Institute, Cambridge, MA, 10.3824/stembook.1.28.1 [DOI] [PubMed] [Google Scholar]
- 2. Cabezas-Wallscheid N., Klimmeck D., Hansson J., Lipka D. B., Reyes A., Wang Q., Weichenhan D., Lier A., von Paleske L., Renders S., Wünsche P., Zeisberger P., Brocks D., Gu L., Herrmann C., et al. (2014) Identification of regulatory networks in HSCs and their immediate progeny via integrated proteome, transcriptome, and DNA methylome analysis. Cell Stem Cell 15, 507–522 [DOI] [PubMed] [Google Scholar]
- 3. Cosgun K. N., Rahmig S., Mende N., Reinke S., Hauber I., Schäfer C., Petzold A., Weisbach H., Heidkamp G., Purbojo A., Cesnjevar R., Platz A., Bornhäuser M., Schmitz M., Dudziak D., Hauber J., Kirberg J., and Waskow C. (2014) Kit regulates HSC engraftment across the human-mouse species barrier. Cell Stem Cell 15, 227–238 [DOI] [PubMed] [Google Scholar]
- 4. Greenbaum A., Hsu Y. M., Day R. B., Schuettpelz L. G., Christopher M. J., Borgerding J. N., Nagasawa T., and Link D. C. (2013) CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance. Nature 495, 227–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Morrison S. J., and Scadden D. T. (2014) The bone marrow niche for haematopoietic stem cells. Nature 505, 327–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhou B. O., Ding L., and Morrison S. J. (2015) Hematopoietic stem and progenitor cells regulate the regeneration of their niche by secreting Angiopoietin-1. Elife 4, e05521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. King K. Y., and Goodell M. A. (2011) Inflammatory modulation of HSCs: viewing the HSC as a foundation for the immune response. Nat. Rev. Immunol. 11, 685–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Baldridge M. T., King K. Y., and Goodell M. A. (2011) Inflammatory signals regulate hematopoietic stem cells. Trends Immunol. 32, 57–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Espín-Palazón R., Stachura D. L., Campbell C. A., García-Moreno D., Del Cid N., Kim A. D., Candel S., Meseguer J., Mulero V., and Traver D. (2014) Proinflammatory signaling regulates hematopoietic stem cell emergence. Cell 159, 1070–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sawamiphak S., Kontarakis Z., and Stainier D. Y. (2014) Interferon γ signaling positively regulates hematopoietic stem cell emergence. Dev. Cell 31, 640–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Boettcher S., Gerosa R. C., Radpour R., Bauer J., Ampenberger F., Heikenwalder M., Kopf M., and Manz M. G. (2014) Endothelial cells translate pathogen signals into G-CSF-driven emergency granulopoiesis. Blood 124, 1393–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Boiko J. R., and Borghesi L. (2012) Hematopoiesis sculpted by pathogens: Toll-like receptors and inflammatory mediators directly activate stem cells. Cytokine 57, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. He Q., Zhang C., Wang L., Zhang P., Ma D., Lv J., and Liu F. (2015) Inflammatory signaling regulates hematopoietic stem and progenitor cell emergence in vertebrates. Blood 125, 1098–1106 [DOI] [PubMed] [Google Scholar]
- 14. Nagai Y., Garrett K. P., Ohta S., Bahrun U., Kouro T., Akira S., Takatsu K., and Kincade P. W. (2006) Toll-like receptors on hematopoietic progenitor cells stimulate innate immune system replenishment. Immunity 24, 801–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schuettpelz L. G., Borgerding J. N., Christopher M. J., Gopalan P. K., Romine M. P., Herman A. C., Woloszynek J. R., Greenbaum A. M., and Link D. C. (2014) G-CSF regulates hematopoietic stem cell activity, in part, through activation of Toll-like receptor signaling. Leukemia 28, 1851–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Takeuchi O., and Akira S. (2010) Pattern recognition receptors and inflammation. Cell 140, 805–820 [DOI] [PubMed] [Google Scholar]
- 17. Fiedler K., Kokai E., Bresch S., and Brunner C. (2013) MyD88 is involved in myeloid as well as lymphoid hematopoiesis independent of the presence of a pathogen. Am. J. Blood Res. 3, 124–140 [PMC free article] [PubMed] [Google Scholar]
- 18. Grassinger J., Williams B., Olsen G. H., Haylock D. N., and Nilsson S. K. (2012) Granulocyte colony stimulating factor expands hematopoietic stem cells within the central but not endosteal bone marrow region. Cytokine 58, 218–225 [DOI] [PubMed] [Google Scholar]
- 19. Mirantes C., Passegué E., and Pietras E. M. (2014) Pro-inflammatory cytokines: emerging players regulating HSC function in normal and diseased hematopoiesis. Exp. Cell Res. 329, 248–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhao J. L., Ma C., O'Connell R. M., Mehta A., DiLoreto R., Heath J. R., and Baltimore D. (2014) Conversion of danger signals into cytokine signals by hematopoietic stem and progenitor cells for regulation of stress-induced hematopoiesis. Cell Stem Cell 14, 445–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pietras E. M., Mirantes-Barbeito C., Fong S., Loeffler D., Kovtonyuk L. V., Zhang S., Lakshminarasimhan R., Chin C. P., Techner J. M., Will B., Nerlov C., Steidl U., Manz M. G., Schroeder T., and Passegué E. (2016) Chronic interleukin-1 exposure drives haematopoietic stem cells towards precocious myeloid differentiation at the expense of self-renewal. Nat. Cell Biol. 18, 607–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Colombo M., Raposo G., and Théry C. (2014) Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 30, 255–289 [DOI] [PubMed] [Google Scholar]
- 23. Costa-Silva B., Aiello N. M., Ocean A. J., Singh S., Zhang H., Thakur B. K., Becker A., Hoshino A., Mark M. T., Molina H., Xiang J., Zhang T., Theilen T. M., García-Santos G., Williams C., et al. (2015) Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 17, 816–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Peinado H., Alečković M., Lavotshkin S., Matei I., Costa-Silva B., Moreno-Bueno G., Hergueta-Redondo M., Williams C., García-Santos G., Ghajar C., Nitadori-Hoshino A., Hoffman C., Badal K., Garcia B. A., Callahan M. K., et al. (2012) Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 18, 883–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huan J., Hornick N. I., Goloviznina N. A., Kamimae-Lanning A. N., David L. L., Wilmarth P. A., Mori T., Chevillet J. R., Narla A., Roberts C. T. Jr., Loriaux M. M., Chang B. H., and Kurre P. (2015) Coordinate regulation of residual bone marrow function by paracrine trafficking of AML exosomes. Leukemia 29, 2285–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huan J., Hornick N. I., Shurtleff M. J., Skinner A. M., Goloviznina N. A., Roberts C. T. Jr, and Kurre P. (2013) RNA trafficking by acute myelogenous leukemia exosomes. Cancer Res. 73, 918–929 [DOI] [PubMed] [Google Scholar]
- 27. Lai R. C., Yeo R. W., and Lim S. K. (2015) Mesenchymal stem cell exosomes. Semin. Cell Dev. Biol. 40, 82–88 [DOI] [PubMed] [Google Scholar]
- 28. Frenette P. S., Pinho S., Lucas D., and Scheiermann C. (2013) Mesenchymal stem cell: keystone of the hematopoietic stem cell niche and a stepping-stone for regenerative medicine. Annu. Rev. Immunol. 31, 285–316 [DOI] [PubMed] [Google Scholar]
- 29. Platt M. O., Roman A. J., Wells A., Lauffenburger D. A., and Griffith L. G. (2009) Sustained epidermal growth factor receptor levels and activation by tethered ligand binding enhances osteogenic differentiation of multi-potent marrow stromal cells. J. Cell. Physiol. 221, 306–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zielske S. P., Livant D. L., and Lawrence T. S. (2009) Radiation increases invasion of gene-modified mesenchymal stem cells into tumors. Int. J. Radiat. Oncol. Biol. Phys. 75, 843–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Peister A., Mellad J. A., Larson B. L., Hall B. M., Gibson L. F., and Prockop D. J. (2004) Adult stem cells from bone marrow (MSCs) isolated from different strains of inbred mice vary in surface epitopes, rates of proliferation, and differentiation potential. Blood 103, 1662–1668 [DOI] [PubMed] [Google Scholar]
- 32. Soleimani M., and Nadri S. (2009) A protocol for isolation and culture of mesenchymal stem cells from mouse bone marrow. Nat. Protoc 4, 102–106 [DOI] [PubMed] [Google Scholar]
- 33. Pietras E. M., Reynaud D., Kang Y. A., Carlin D., Calero-Nieto F. J., Leavitt A. D., Stuart J. M., Göttgens B., and Passegué E. (2015) Functionally distinct subsets of lineage-biased multipotent progenitors control blood production in normal and regenerative conditions. Cell Stem Cell 17, 35–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Oguro H., Ding L., and Morrison S. J. (2013) SLAM family markers resolve functionally distinct subpopulations of hematopoietic stem cells and multipotent progenitors. Cell Stem Cell 13, 102–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Muzumdar M. D., Tasic B., Miyamichi K., Li L., and Luo L. (2007) A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605 [DOI] [PubMed] [Google Scholar]
- 36. Lai C. P., Kim E. Y., Badr C. E., Weissleder R., Mempel T. R., Tannous B. A., and Breakefield X. O. (2015) Visualization and tracking of tumour extracellular vesicle delivery and RNA translation using multiplexed reporters. Nat. Commun. 6, 7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Burberry A., Zeng M. Y., Ding L., Wicks I., Inohara N., Morrison S. J., and Núñez G. (2014) Infection mobilizes hematopoietic stem cells through cooperative NOD-like receptor and Toll-like receptor signaling. Cell Host Microbe 15, 779–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shi C., Jia T., Mendez-Ferrer S., Hohl T. M., Serbina N. V., Lipuma L., Leiner I., Li M. O., Frenette P. S., and Pamer E. G. (2011) Bone marrow mesenchymal stem and progenitor cells induce monocyte emigration in response to circulating Toll-like receptor ligands. Immunity 34, 590–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Buza-Vidas N., Duarte S., Luc S., Bouriez-Jones T., Woll P. S., and Jacobsen S. E. (2011) GATA3 is redundant for maintenance and self-renewal of hematopoietic stem cells. Blood 118, 1291–1293 [DOI] [PubMed] [Google Scholar]
- 40. Mercier F. E., Sykes D. B., and Scadden D. T. (2016) Single targeted exon mutation creates a true congenic mouse for competitive hematopoietic stem cell transplantation: the C57BL/6-CD45.1STEM mouse. Stem Cell Reports 6, 985–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang H., Rodriguez S., Wang L., Wang S., Serezani H., Kapur R., Cardoso A. A., and Carlesso N. (2016) Sepsis induces hematopoietic stem cell exhaustion and myelosuppression through distinct contributions of TRIF and MYD88. Stem Cell Reports 6, 940–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cantaluppi V., Gatti S., Medica D., Figliolini F., Bruno S., Deregibus M. C., Sordi A., Biancone L., Tetta C., and Camussi G. (2012) Microvesicles derived from endothelial progenitor cells protect the kidney from ischemia-reperfusion injury by microRNA-dependent reprogramming of resident renal cells. Kidney Int. 82, 412–427 [DOI] [PubMed] [Google Scholar]
- 43. Vicencio J. M., Yellon D. M., Sivaraman V., Das D., Boi-Doku C., Arjun S., Zheng Y., Riquelme J. A., Kearney J., Sharma V., Multhoff G., Hall A. R., and Davidson S. M. (2015) Plasma exosomes protect the myocardium from ischemia-reperfusion injury. J. Am. Coll. Cardiol. 65, 1525–1536 [DOI] [PubMed] [Google Scholar]
- 44. Anderson P., Carrillo-Gálvez A. B., García-Pérez A., Cobo M., and Martín F. (2013) CD105 (endoglin)-negative murine mesenchymal stromal cells define a new multipotent subpopulation with distinct differentiation and immunomodulatory capacities. PLoS ONE 8, e76979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang B., Yin Y., Lai R. C., Tan S. S., Choo A. B., and Lim S. K. (2014) Mesenchymal stem cells secrete immunologically active exosomes. Stem Cells Dev. 23, 1233–1244 [DOI] [PubMed] [Google Scholar]
- 46. Zhu Y. G., Feng X. M., Abbott J., Fang X. H., Hao Q., Monsel A., Qu J. M., Matthay M. A., and Lee J. W. (2014) Human mesenchymal stem cell microvesicles for treatment of Escherichia coli endotoxin-induced acute lung injury in mice. Stem Cells 32, 116–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pinho S., Lacombe J., Hanoun M., Mizoguchi T., Bruns I., Kunisaki Y., and Frenette P. S. (2013) PDGFRα and CD51 mark human nestin+ sphere-forming mesenchymal stem cells capable of hematopoietic progenitor cell expansion. J. Exp. Med. 210, 1351–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bretz N. P., Ridinger J., Rupp A. K., Rimbach K., Keller S., Rupp C., Marmé F., Umansky L., Umansky V., Eigenbrod T., Sammar M., and Altevogt P. (2013) Body fluid exosomes promote secretion of inflammatory cytokines in monocytic cells via Toll-like receptor signaling. J. Biol. Chem. 288, 36691–36702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kaulen D. R., Sanin A. V., Khorobrykh V. V., Malikov V. E., and Dranovskaia E. A. (1983) [Stimulating action of Brucella melitensis lipopolysaccharide on hemopoiesis in mice]. Zh. Mikrobiol. Epidemiol. Immunobiol. 84–87 [PubMed] [Google Scholar]
- 50. Nielsen E., Severin F., Backer J. M., Hyman A. A., and Zerial M. (1999) Rab5 regulates motility of early endosomes on microtubules. Nat. Cell Biol. 1, 376–382 [DOI] [PubMed] [Google Scholar]
- 51. Vanlandingham P. A., and Ceresa B. P. (2009) Rab7 regulates late endocytic trafficking downstream of multivesicular body biogenesis and cargo sequestration. J. Biol. Chem. 284, 12110–12124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Simon N. C., and Barbieri J. T. (2014) Exoenzyme S ADP-ribosylates Rab5 effector sites to uncouple intracellular trafficking. Infect. Immun. 82, 21–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sahoo S., and Losordo D. W. (2014) Exosomes and cardiac repair after myocardial infarction. Circ. Res. 114, 333–344 [DOI] [PubMed] [Google Scholar]
- 54. Borggrefe T., and Oswald F. (2009) The Notch signaling pathway: transcriptional regulation at Notch target genes. Cell. Mol. Life Sci. 66, 1631–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wilson A. A., Kwok L. W., Porter E. L., Payne J. G., McElroy G. S., Ohle S. J., Greenhill S. R., Blahna M. T., Yamamoto K., Jean J. C., Mizgerd J. P., and Kotton D. N. (2013) Lentiviral delivery of RNAi for in vivo lineage-specific modulation of gene expression in mouse lung macrophages. Mol. Ther. 21, 825–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ciuculescu M. F., Brendel C., Harris C. E., and Williams D. A. (2014) Retroviral transduction of murine and human hematopoietic progenitors and stem cells. Methods Mol. Biol. 1185, 287–309 [DOI] [PubMed] [Google Scholar]
- 57. Nagamoto-Combs K., Kulas J., and Combs C. K. (2014) A novel cell line from spontaneously immortalized murine microglia. J. Neurosci. Methods 233, 187–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Matsunaga N., Tsuchimori N., Matsumoto T., and Ii M. (2011) TAK-242 (resatorvid), a small-molecule inhibitor of Toll-like receptor (TLR) 4 signaling, binds selectively to TLR4 and interferes with interactions between TLR4 and its adaptor molecules. Mol. Pharmacol. 79, 34–41 [DOI] [PubMed] [Google Scholar]