Abstract
The phosphoinositide 3-kinase (PI3K), which phosphorylates phosphatidylinositol and produces PI3P, has been implicated in protein trafficking, intracellular survival, and virulence in the pathogenic yeast Candida glabrata. Here, we demonstrate PI3-kinase (CgVps34) to be essential for maintenance of cellular iron homeostasis. We examine how CgVps34 regulates the fundamental process of iron acquisition, and underscore its function in vesicular trafficking as a central determinant. RNA sequencing analysis revealed iron homeostasis genes to be differentially expressed upon CgVps34 disruption. Consistently, the Cgvps34Δ mutant displayed growth attenuation in low- and high-iron media, increased intracellular iron content, elevated mitochondrial aconitase activity, impaired biofilm formation, and extenuated mouse organ colonization potential. Furthermore, we demonstrate for the first time that C. glabrata cells respond to iron limitation by expressing the iron permease CgFtr1 primarily on the cell membrane, and to iron excess via internalization of the plasma membrane-localized CgFtr1 to the vacuole. Our data show that CgVps34 is essential for the latter process. We also report that macrophage-internalized C. glabrata cells express CgFtr1 on the cell membrane indicative of an iron-restricted macrophage internal milieu, and Cgvps34Δ cells display better survival in iron-enriched medium-cultured macrophages. Overall, our data reveal the centrality of PI3K signaling in iron metabolism and host colonization.
Keywords: biofilm, fungi, iron metabolism, microbial pathogenesis, phosphatidylinositide 3-kinase (PI 3-kinase), Ftr1-Fet3 complex, intracellular and mitochondrial iron levels, lipid kinase Vps34, pathogenic yeasts
Introduction
Invasive mycoses pose a serious therapeutic challenge with mortality rates of 30–80% with Candida species being the most prevalent infectious agents (1, 2). Among Candida species, Candida glabrata and Candida albicans account for about 20 and 60%, respectively, of Candida bloodstream infections (3–5). C. glabrata, a constituent of the normal human microflora, is emerging as a clinically important opportunistic fungal pathogen, as the mortality rate of up to 60% has been associated with C. glabrata infections (5–7).
Virulence of C. glabrata has primarily been attributed to its ability to form biofilms, adhere to mammalian cells, survive and proliferate in murine and human macrophages, possess resistance to antifungal drugs, regulate the expression of a family of cell surface adhesins and aspartyl proteases, and maintain iron homeostasis in varied environmental conditions (8–11). Recently, CgVps15 and CgVps34, regulatory and catalytic subunits, respectively, of the only known class III phosphoinositide 3-kinase (PI3K), that phosphorylates the third hydroxyl group of phosphatidylinositol to produce phosphatidylinositol 3-phosphate (PI3P),3 have been shown to regulate several aspects of C. glabrata pathogenesis including vesicular trafficking, autophagy, host cell adherence, intracellular survival, and virulence in systemic infections in mice (12).
The class III PI3-kinases are ubiquitously present in all eukaryotes, and are pivotal to cell division and survival, intracellular signaling, membrane dynamics and cell motility, adhesion, and differentiation (13, 14). Specifically, they have been implicated in the regulation of mitogenic signaling, endosome to Golgi trafficking, vacuolar protein sorting, polarized endocytic sorting, phagosome maturation, and autophagy (14, 15). These multiple functions are predominantly mediated through effector proteins containing the FYVE zinc finger and PX domains, which upon binding to phosphatidylinositol 3-phosphate lipids, are recruited to various cell membranes to regulate vesicular traffic (15).
Iron, one of the most abundant metal ions on earth, is very rarely freely available in the human host, and is usually found bound to high-affinity iron-binding host proteins such as lactoferrin, transferrin, and ferritin (16). Besides acting as a cofactor for several metalloproteins involved in fundamental cellular processes of oxygen transport, respiration, energy metabolism, and DNA synthesis and repair, iron modulates the transcriptional activation of virulence genes of pathogenic fungi (16, 17). To survive and proliferate in varied iron host niches including the gastrointestinal tract, blood, oral cavity, and genitourinary tract, C. glabrata has developed at least four distinct types of iron acquisition systems (10).
The high-affinity iron uptake system in C. glabrata is composed of an iron transporter (CgFtr1), a cell surface multicopper oxidase (CgFet3), a P-type Cu-ATPase (CgCcc2), and many members of the ferric reductase (CgFre) family, and is required for proliferation under in vitro iron-limited conditions as well as in murine disseminated candidiasis infection model (10). Conversely, the low-affinity iron transport system, typified by the nonspecific ion transporter, CgFet4, is dispensable for growth in the iron-poor environment (10). Although C. glabrata neither synthesizes nor secretes high-affinity iron-chelating compounds, siderophores, it does possess the sole siderophore transporter, CgSit1, which is essential for utilization of ferrichrome as an iron source under iron-deficient conditions, and for iron-dependent survival in macrophages (10, 18). Lastly, two components, identified so far, of the host-specific iron acquisition system are the putative hemolysin (CgMam3) and the cell wall structural protein (CgCcw14), which are required for survival in systemic infections in mice (10).
C. glabrata cells are known to respond to variations in the environmental iron content via differential expression of genes involved in DNA repair, autophagy, cellular respiration, and organonitrogen compound metabolism (19). In addition, two stress-responsive mitogen-activated protein kinases (MAPKs), CgHog1 and CgSlt2, are activated upon growth in the iron-rich medium (19). Environmental iron content also regulates expression of the major adhesin-encoding gene EPA1, as mirrored in elevated and diminished EPA1 transcript levels observed under high- and low-iron growth conditions, respectively (19).
Herein, using a combined approach of RNA sequencing based transcriptome profiling, phenotypic, microscopy, and biochemical analyses, we have elucidated a novel role for the C. glabrata phosphoinositide 3-kinase, CgVps34, in the maintenance of cellular iron homeostasis, mitochondrial functions, biofilm formation, and colonization of mouse organs in a disseminated candidiasis model. We demonstrate for the first time that expression and targeting of the CgFtr1 transporter to the cell membrane requires CgFet3 ferroxidase, and that CgVps34 is dispensable for iron limitation-responsive plasma membrane localization of CgFtr1 and CgFet3. Contrarily, CgVps34 is essential for retrograde trafficking of CgFtr1, upon exposure to iron-rich conditions, from the cell membrane. We report a pivotal requirement for CgVps34 for survival under iron-deficient and iron-sufficient growth conditions, and our data yield new insights into survival strategies of an emerging human fungal pathogen in environmental conditions of varied iron content.
Results
The Cgvps34Δ Mutant Displays Elevated Intracellular ATP Levels
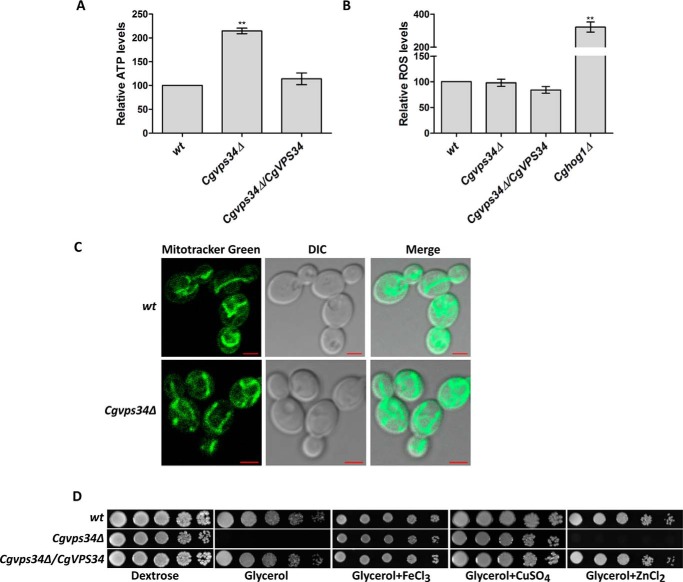
We have previously shown that deletion of C. glabrata phosphoinositide 3-kinase (CgVps34) results in cell death in human macrophages and highly attenuated virulence in mice (12). Lack of CgVps34 is also known to abrogate growth on non-fermentable carbon sources (12). As inefficient utilization of nonfermentable carbon sources and mitochondrial defects have previously been associated with hypovirulence of human fungal pathogens (20, 21), we sought to closely examine mitochondrial functions in the Cgvps34Δ mutant. For this, we first measured ATP and reactive oxygen species (ROS) levels in wild-type (wt) and mutant cells. The Cgvps34Δ mutant displayed 2.0-fold higher intracellular ATP levels in comparison to wt cells (Fig. 1A). However, intracellular ROS levels, as measured by the 2′,7′-dichlorodihydrofluorescein diacetate fluorescence-based cellular ROS detection assay, were similar in both wt and mutant strains (Fig. 1B). As a positive control for ROS quantification, we used the Cghog1Δ mutant, which is deleted for the terminal mitogen-activated protein kinase CgHog1 of high osmolarity glycerol (HOG) pathway, and has previously been shown to possess high intracellular ROS levels (19) (Fig. 1B). Consistent with the earlier report (19), the Cghog1Δ mutant exhibited 3.2-fold higher ROS levels, thus, validating the assay (Fig. 1B). Next, we labeled log-phase wt and Cgvps34Δ cells with the MitoTracker Green stain, and visualized mitochondrial morphology using confocal microscopy. As shown in Fig. 1C, both wt and mutant cells exhibited normal reticular network of mitochondria. Furthermore, growth of the Cgvps34Δ mutant was also not perturbed under hypoxic conditions (data not shown). Together, these findings suggest that CgVPS34 disruption does not lead to generation of free radicals and gross structural defects in the mitochondria.
FIGURE 1.
The Cgvps34Δ mutant displays elevated ATP levels and tubular mitochondria. A, intracellular ATP levels of indicated log-phase C. glabrata cells were measured by a luciferin-luciferase luminescence assay. ATP concentrations in wild-type (wt) cells were set to 100, and data (mean ± S.E., n = 3) are expressed as the fold-change observed in Cgvps34Δ and complemented (Cgvps34Δ/CgVPS34) strains compared with the wt strain. Paired, two-tailed, Student's t test (**, p < 0.005). B, intracellular ROS levels, as detected by fluorimetry using the cell-permeant 2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA) probe (100 μm), in indicated log-phase C. glabrata cells. Data are expressed as ROS levels relative to ROS levels in wt cells (set to 100%), and represent mean ± S.E. from five independent experiments. Paired, two-tailed, Student's t test (**, p < 0.005). C, representative confocal microscopy images of MitoTracker Green (a membrane potential independent probe)-stained, log-phase wt and Cgvps34Δ cells showing extended tubular mitochondrial networks. DIC, differential interference contrast; bar, 2 μm. D, serial dilution spotting analysis. 3 μl of overnight YPD-medium grown, A600-normalized, 10-fold serially diluted cultures were spotted on the YPD medium containing either dextrose (2%) or glycerol (3%) as the sole carbon source. Ferric chloride, copper sulfate, and zinc chloride were added to the final concentration of 5 mm. Plate images were captured after growth at 30 °C for 3–5 days.
Growth defects on non-fermentable carbon sources in Saccharomyces cerevisiae have been associated with impaired functioning of the mitochondria and/or the vacuole (22–24). Furthermore, metal ion supplementation is known to result in the reversal of mitochondrial respiration defects (25, 26). Hence, we next examined if the incapacity of the Cgvps34Δ mutant to utilize non-fermentable carbon sources as the sole carbon source could be rescued by addition of cations, viz. Fe3+, Cu2+, and Zn2+ ions. As shown in Fig. 1D, Cgvps34Δ cells exhibited no growth in glycerol medium but robust growth in the glycerol medium supplemented with either Fe3+ or Cu2+. However, the mutant was deficient in utilization of glycerol as the sole carbon source in the zinc-containing medium (Fig. 1D) suggesting that cation-dependent rescue of growth in glycerol is specific to iron and copper. Altogether, these data are indicative of altered mitochondrial functions in the Cgvps34Δ mutant, and raise the possibility that respiratory fitness defects, upon CgVPS34 disruption, may arise from perturbed copper and iron homeostasis.
RNA Sequencing Analysis Reveals Deregulation of Iron Metabolism Genes in the Cgvps34Δ Mutant
To investigate the molecular basis underlying perturbed mitochondrial respiration in the Cgvps34Δ mutant, we performed global transcriptome profiling of YPD-grown logarithmic (log)-phase wt and Cgvps34Δ cells using RNA Sequencing (RNA-seq) analysis. CgVPS34 disruption led to differential regulation of 160 genes (≥1.5-fold change and a false discovery rate adjusted p value of ≤0.05). Of these genes, 96 were up-regulated and 64 were down-regulated in the Cgvps34Δ mutant (supplemental Table S1). Gene Ontology (GO)-Slim Mapper analysis, using the Candida Genome Database, revealed genes involved in biological processes of “transport” and “response to stress” to be differentially expressed in the Cgvps34Δ mutant (supplemental Table S1). Next, we employed the DAVID (Database for Annotation, Visualization and Integrated Discovery) and the FungiFun2 tool, to identify significantly enriched GO terms, which were classified according to biological process annotations, in differentially expressed genes (supplemental Table S1). GO terms, cofactor metabolic process, aerobic respiration, and iron-sulfur cluster assembly were enriched in the up-regulated gene list in the DAVID analysis (supplemental Table S1). GO categories, iron ion transmembrane transport, and cellular response to zinc ion starvation were found to be significantly enriched in the down-regulated gene list in the FungiFun2 analysis (supplemental Table S1). Pathway analysis using the KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway database showed that the citrate cycle, and glycine, serine, and threonine metabolism were the only pathways that were significantly enriched among up-regulated and down-regulated genes, respectively (supplemental Table S1). These transcriptome data are in agreement with previous observations that CgVPS34 disruption may alter certain aspects of mitochondrial physiology.
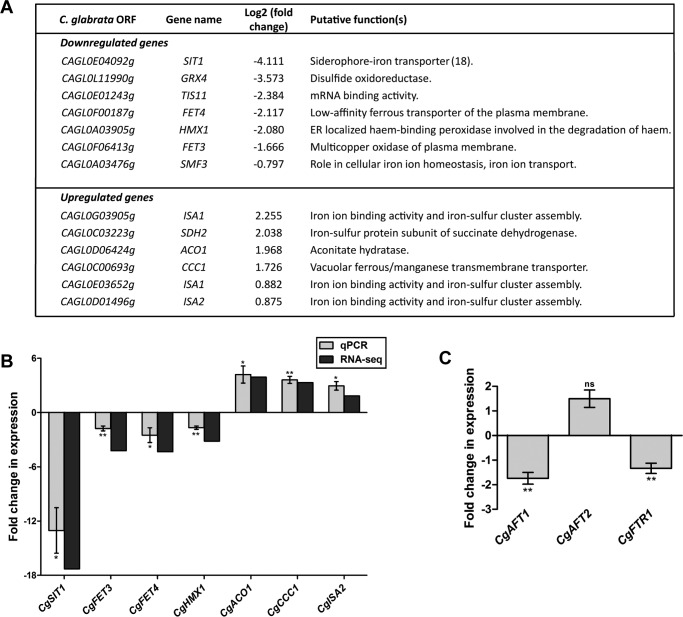
Iron, an essential cofactor of many mitochondrial, nuclear, and cytosolic enzymes, aids several cellular processes (16). Based upon the environmental iron source and the content, three types of iron uptake systems in C. glabrata, the high-affinity system consisting of CgFtr1, CgFet3, and CgCcc2, the low-affinity system composed of CgFet4, and the siderophore uptake system comprised of CgSit1 transporter, are required for iron acquisition (10). As shown in Fig. 2A, 13 iron homeostasis genes including genes encoding proteins involved in high-affinity iron uptake (CgFet3, a multicopper oxidase), low-affinity ion transport (CgFet4, a low-affinity ion transporter), and siderophore-iron acquisition (CgSit1, a siderophore-iron transporter) were differentially regulated in the Cgvps34Δ mutant. Of note, the high-affinity iron transport system in C. glabrata is activated and repressed under iron-depleted and iron-repleted conditions, respectively (10, 19). Overall, our RNA-Seq analysis revealed that genes involved in iron transport were down-regulated, whereas genes implicated in iron utilization/iron-sulfur (Fe-S) cluster binding were up-regulated in the Cgvps34Δ mutant (Fig. 2A).
FIGURE 2.
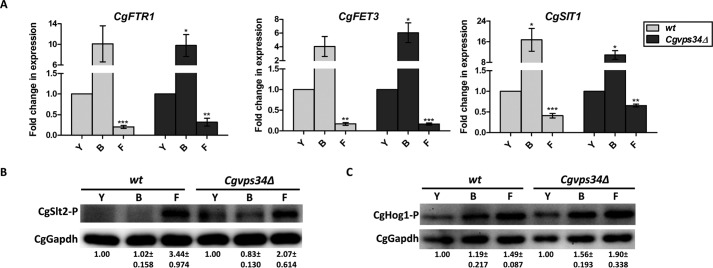
Expression of genes involved in iron homeostasis is deregulated in the Cgvps34Δ mutant. A, list of iron homeostasis genes that are differentially expressed upon CgVPS34 disruption. B, validation of RNA-seq data by qPCR. Wild-type and Cgvps34Δ cells were grown in the YPD medium for 4 h, RNA was harvested, and transcript levels of seven selected genes (4 down-regulated and 3 up-regulated in the RNA-seq experiment) were measured by qPCR. The qPCR data are shown as expression fold-changes after normalization against the CgACT1 mRNA control, and represent the mean ± S.E. (n = 3–4). *, p < 0.05; **, p < 0.005, paired two-tailed Student's t test. C, qPCR analysis of CgAFT1, CgAFT2, and CgFTR1 gene expression in log-phase, YPD medium-grown Cgvps34Δ cells. Data (mean ± S.E., n = 3–4) were normalized to an internal CgACT1 mRNA control, and represent fold-change in expression in the Cgvps34Δ mutant compared with the wt strain. Paired, two-tailed, Student's t test was used (**, p < 0.005; ns, not significant).
Next, we verified the gene expression data obtained from the RNA-Seq experiment by performing quantitative real time-PCR (qPCR) analysis, and observed good correlation in expression levels between RNA-Seq and qPCR data for three up-regulated (CgACO1, CgCCC1, and CgISA2) and four down-regulated (CgSIT1, CgFET3, CgFET4, and CgHMX1) tested genes (Fig. 2B). In addition, we also checked the transcript levels of CgFTR1 (codes for a high-affinity iron permease), CgAFT1 (encodes a major iron-responsive transcriptional activator), and CgAFT2 (codes for a putative iron-responsive transcriptional factor) genes, in the Cgvps34Δ mutant. In accordance with reduced expression of iron uptake genes, transcription of CgFTR1 and CgAFT1 genes was also down-regulated in the Cgvps34Δ mutant (Fig. 2C). It is worth noting that CgFtr1 is an essential component of the high-affinity iron uptake system, and CgAft1 is the principal regulator of high-affinity iron uptake gene expression (10). However, CgAFT2 expression was found to be similar between wt and Cgvps34Δ cells (Fig. 2C), which is consistent with earlier findings reporting no effect of environmental iron content on the transcription of the CgAFT2 gene (10). Collectively, these data indicate that CgVPS34 deletion leads to transcriptional induction and repression of the Fe-S cluster insertion and high-affinity iron uptake system, respectively, in C. glabrata.
Iron Homeostasis Is Perturbed in the Cgvps34Δ Mutant
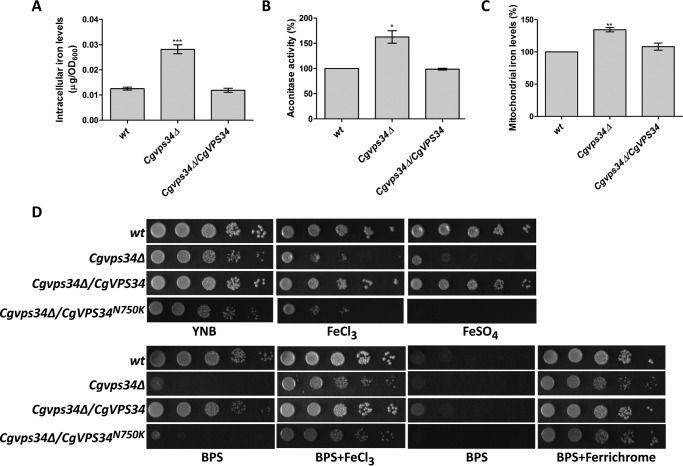
To examine the relationship between differential gene expression of the iron homeostatic system and respiration defects of the Cgvps34Δ mutant, we first measured intracellular iron levels using inductively coupled plasma atomic emission spectrometry (ICP-AES) in log-phase wt and Cgvps34Δ cells. The Cgvps34Δ mutant contained ∼3.0-fold higher intracellular iron levels compared with wt cells, which were restored back to normal levels in the complemented-mutant strain (Fig. 3A). Efficient Fe-S cluster assembly (maturation of Fe-S clusters) is dependent upon the abundance of intracellular iron, and alterations in intracellular iron levels significantly affect activity of Fe-S cluster-containing enzymes (27, 28). Hence, we next measured the activity of the mitochondrial aconitase, which has a 4Fe-4S cluster at its active site, in the Cgvps34Δ mutant, and found it to be 1.6-fold higher than the wt strain (Fig. 3B) indicating that the high intracellular iron content in the Cgvps34Δ mutant may modify mitochondrial functions. Consistent with the elevated mitochondrial aconitase activity, the mitochondrial iron content was also found to be 40% higher in the Cgvps34Δ mutant in comparison to wt cells (Fig. 3C). Importantly, all iron homeostasis-related defects were rescued upon ectopic expression of CgVPS34 in the Cgvps34Δ mutant (Fig. 3, A–C). It is also worth noting that elevated mitochondrial iron content and aconitase activity are consistent with higher transcript levels of genes coding for Fe-S cluster proteins observed in the global transcriptional profiling analysis (Fig. 2A). Together, these results are indicative of a significantly perturbed iron metabolism upon CgVPS34 disruption, and raise the possibility that the higher mitochondrial iron content may lead to elevated Fe-S cluster generation, thereby, acting as a signal for transcriptional down-regulation of the iron uptake machinery in the Cgvps34Δ mutant.
FIGURE 3.
CgVPS34 disruption results in perturbed iron homeostasis. A, intracellular iron levels of the indicated, YPD medium-grown, log-phase C. glabrata cells, as determined by ICP-AES. Data (mean ± S.E., n = 6) are presented as iron (μg) present in cells equivalent to one A600. Unpaired, two-tailed, Student's t test was used (***, p < 0.0001). B, mitochondrial aconitase activity, as measured by the reduced nicotinamide adenine dinucleotide-coupled assay, in crude mitochondrial extracts of the indicated YPD medium-grown, log-phase C. glabrata cells. Data represent mean ± S.E. (n = 3). *, p < 0.05; paired two-tailed Student's t test was used. C, mitochondrial iron levels, as determined by the BPS-iron complex absorbance, in purified mitochondrial preparations of the indicated YPD medium-grown, log-phase C. glabrata cells. Data represent mean ± S.E. (n = 4). **, p < 0.005; paired two-tailed Student's t test was used. D, serial dilution spotting growth analysis of the indicated C. glabrata strains in YNB medium lacking or containing ferric chloride (3.5 mm), ferrous sulfate (30 mm), BPS (35 μm), BPS (35 μm) plus ferric chloride (50 μm), BPS (100 μm), and BPS (100 μm) plus ferrichrome (10 μm). Plate images were captured after growth at 30 °C for 2–5 days.
Due to elevated intracellular and mitochondrial iron content, we hypothesized that the growth of the Cgvps34Δ mutant will be impaired in the high-iron environment. To test this, we checked the susceptibility of the Cgvps34Δ mutant to surplus iron caused by addition of FeSO4 and FeCl3 to the medium (Fig. 3D). As a control, we also monitored growth of the Cgvps34Δ mutant on regular YNB and iron-limited medium (Fig. 3D). As expected, the Cgvps34Δ mutant was sensitive to iron-repleted conditions (Fig. 3D). However, to our surprise, iron limitation caused by extracellular iron chelators bathophenanthrolinedisulfonic acid (BPS) and ferrozine also led to severe growth attenuation of the Cgvps34Δ mutant (Fig. 3D and data not shown). Importantly, retarded growth in iron-restricted medium was rescued by supplementing medium with extra iron either in the form of ferric chloride or the siderophore ferrichrome (Fig. 3D), which suggests that iron scarcity is the cause for diminished growth of the Cgvps34Δ mutant, and Cgvps34Δ cells are able to scavenge iron from both types of iron sources. Of note, the growth defect in both high- and low-iron medium was complemented when CgVPS34 was expressed ectopically in the Cgvps34Δ mutant (Fig. 3D). These findings indicate that CgVPS34 disruption renders cells susceptible to both iron-limited and iron-surplus environmental conditions pointing to a requirement for CgVps34 for growth under both iron-poor and iron-rich conditions.
CgVps34 and CgVps15 are postulated to be catalytic and regulatory subunits of the PI3-kinase in C. glabrata (12). To determine whether lipid kinase activity of CgVps34 is required for its role in iron homeostasis, we checked growth of the Cgvps34Δ mutant expressing catalytically dead form of CgVps34 in the iron-limited and iron-excess medium. As shown in Fig. 3D, CgVps34 protein carrying a lysine residue in place of a highly conserved asparagine at position 750 (CgVps34N750K) in its catalytic domain could not rescue attenuated growth of the Cgvps34Δ mutant in either medium suggesting that the capacity of CgVps34 to phosphorylate PI is pivotal to the maintenance of cellular iron homeostasis in C. glabrata.
Next, to examine whether the mutant lacking the regulatory domain of PI3K, Cgvps15Δ, also displays iron homeostasis-related defects, we checked growth of the Cgvps15Δ mutant under iron-repleted and iron-depleted conditions, and quantified intracellular iron levels. Similar to the Cgvps34Δ mutant, the Cgvps15Δ mutant exhibited increased susceptibility to iron depletion and iron excess stress (Fig. 4A), and also possessed 1.5-fold higher intracellular iron content (Fig. 4B). Furthermore, reduced growth in the iron-limited medium was rescued with extra iron supplementation suggesting that growth attenuation is due to inadequate iron in the medium (Fig. 4A). Altogether, these data indicate that PI3K signaling is required for maintenance of iron homeostasis in C. glabrata, and that lack of either subunit of the kinase impedes the ability of C. glabrata cells to survive iron-poor and iron-rich environmental conditions.
FIGURE 4.
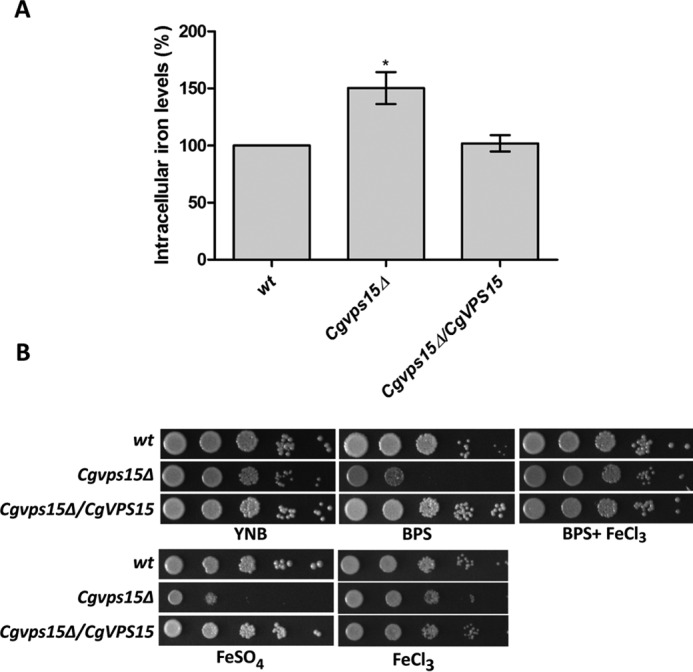
The Cgvps15Δ mutant displays elevated intracellular iron levels. A, intracellular iron levels in the Cgvps15Δ mutant as measured by BPS-iron absorbance-based assay. Data (mean ± S.E., n = 3) are expressed as intracellular iron levels relative to wt cells (considered as 100%). *, p < 0.05; two-tailed paired Student's t test. B, serial dilution spotting growth analysis of the indicated C. glabrata strains in YNB and YNB medium containing 35 μm BPS, 35 μm BPS plus 50 μm ferric chloride, 30 mm ferrous sulfate or 3.5 mm ferric chloride. Plate images were captured after 2–5 days of incubation at 30 °C.
Intracellular Levels of Zinc, Calcium, and Manganese Are Not Altered in the Cgvps34Δ Mutant
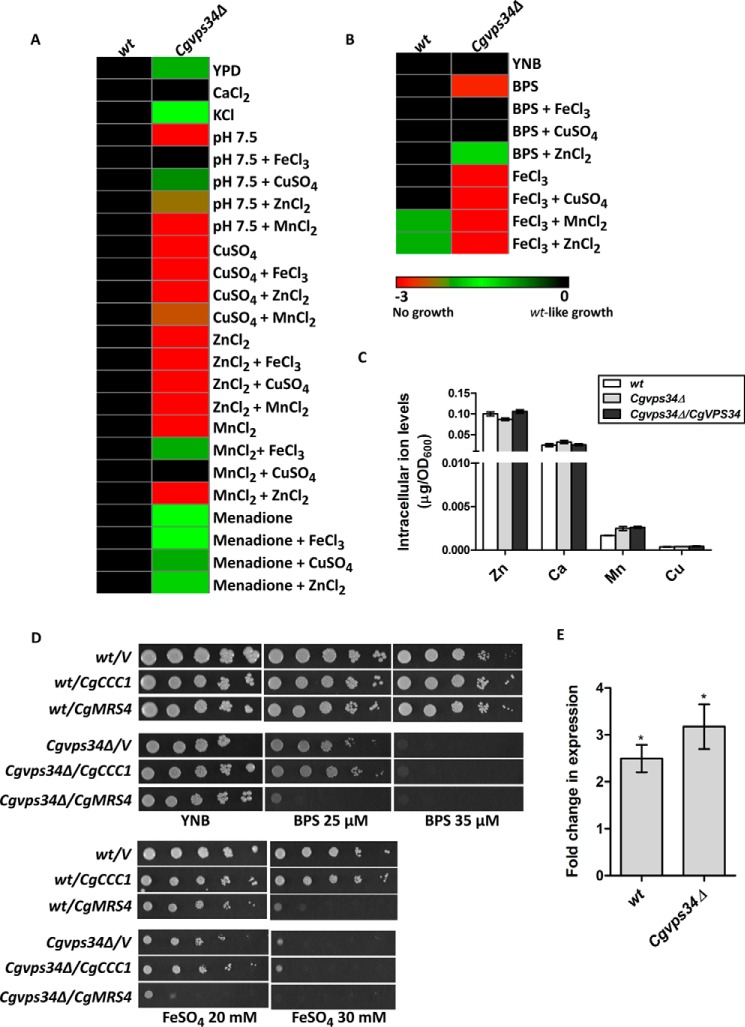
The yeast vacuole is a storage and detoxifying organelle for several metal ions including copper, manganese, iron, and zinc, and protect the cell from excess ion toxicity (22). The Cgvps34Δ mutant has previously been reported to display an enlarged vacuole and reduced levels of mature form of the vacuolar hydrolase, carboxypeptidase Y (12). To examine whether elevated susceptibility of the mutant to high-iron is due to perturbed ion storage function of the vacuole, we first evaluated growth of the Cgvps34Δ mutant in medium containing increased concentrations of calcium, potassium, copper, manganese, and zinc ions. The Cgvps34Δ mutant displayed sensitivity to high concentrations of potassium, copper, manganese, and zinc ions, but not to calcium ions under normal conditions (Fig. 5A). Of note, growth of C. glabrata mutants (Cgstv1 and Cgvma13) disrupted for the vacuolar proton-transporting V-type ATPase remained unaffected in the presence of elevated extracellular concentrations of potassium and ferric ions (29). These data indicate that despite elevated metal ion susceptibility, growth attenuation of the Cgvps34Δ mutant in the high-iron environment could not solely be due to the dysfunctional vacuole.
FIGURE 5.
The Cgvps34Δ mutant exhibits elevated susceptibility to metal ions. A, heat map illustrating growth differences in the presence of surplus metal ions. Indicated C. glabrata strains were grown overnight in YPD medium, A600 was normalized to 1.0, and cultures were serially 10-fold diluted in PBS. 3 μl of cell suspension were spotted on the indicated medium, and plates were incubated at 30 °C. After 2–5 days, yeast growth was recorded and color coded, and is indicated on the bottom. These scores reflect appearance of colonies after spotting 3 μl of six serial 10-fold culture dilutions, and are average of a minimum of three serial dilution spotting experiments. Rows correspond to various media conditions and columns to C. glabrata strains. Susceptibility of the Cgvps34Δ mutant was checked toward CaCl2 (100 mm), KCl (1.5 m), pH 7.5, CuSO4 (10 mm), ZnCl2 (3 mm), MnCl2 (3 mm), and menadione (100 μm). Metal ion concentrations used for rescue of mutant sensitivity were FeCl3 (100 μm), CuSO4 (100 μm), ZnCl2 (100 μm), and MnCl2 (100 μm). B, heat map illustrating the growth defect rescue of the Cgvps34Δ mutant under iron-limited conditions. The experiment was performed as described above. Susceptibility of the Cgvps34Δ mutant to BPS (35 μm) and FeCl3 (5 mm) was checked in the YNB medium. Metal ion concentrations used for rescue of mutant sensitivity were FeCl3 (100 μm), CuSO4 (100 μm), ZnCl2 (100 μm), and MnCl2 (100 μm). C, intracellular concentrations of zinc, calcium, manganese, and copper ions in the indicated YPD medium-grown C. glabrata cells. Cells (A600 = 50) were harvested, extracellular cations were removed with multiple washes and the ion concentration was measured by ICP-AES. Data (mean ± S.E., n = 3–6) are presented as ion (μg) per A600 cells. D, serial dilution spotting growth analysis of the indicated C. glabrata strains. Plate images were captured after 2–5 days of incubation at 30 °C. E, qPCR analysis of CgCCC1 in log-phase, CAA medium-grown wt and Cgvps34Δ cells overexpressing CgCCC1 from the CgPDC1 promoter. Cells were grown in the CAA medium for 6 h, RNA was harvested, and transcript levels of CgCCC1 were measured by qPCR. The qPCR data, normalized against the CgACT1 mRNA control, are shown as fold-change in expression in the indicated strains expressing CgCCC1 compared with the respective strains carrying plasmid alone. *, p < 0.05, paired two-tailed Student's t test.
Acquisition of several of metal ions including iron, zinc, manganese, and cadmium in S. cerevisiae is mediated by the low-affinity iron transporter, Fet4 (27, 30). To probe the possibility that elevated metal ion sensitivity of the Cgvps34Δ mutant to copper, iron, and manganese ions is due to increased activity of either CgFet4 and/or a yet to be identified nonspecific ion transporter, we supplemented medium with two different metal ions, and examined for growth restoration assuming a competition between cations for nonspecific ion transporter-dependent uptake (Fig. 5, A and B). However, addition of iron, zinc, copper, and manganese to the medium could not rescue growth defects associated with high concentrations of other metal ions suggesting that these cations are not competing for transport (Fig. 5, A and B). The only exception in this assay was the copper-dependent growth recovery in the excess-manganese medium (Fig. 5A). The molecular basis underlying this observation is yet to be deciphered. In aggregate, these results are reflective of perturbed ion homeostasis in the Cgvps34Δ mutant.
Next, to correlate the ion sensitivity phenotypes of the Cgvps34Δ mutant with intracellular cation levels, we measured the intracellular concentration of zinc, calcium, manganese, and copper ions in log-phase wt and Cgvps34Δ cells via inductively coupled plasma mass spectrometry (Fig. 5C). We observed no appreciable statistically significant differences in the content of zinc, calcium, and manganese ions between wt and the mutant (Fig. 5C). Of note, intracellular copper levels in both wt and Cgvps34Δ cells appeared to be below the detection limit of the assay (Fig. 5C). Together, these data indicate that although the Cgvps34Δ mutant exhibits sensitivity to multiple metal ions, intracellular levels of only iron are elevated in the mutant. It is possible that perturbed iron metabolism indirectly affects the uptake/homeostasis of other cations in the Cgvps34Δ mutant.
Copper and iron homeostasis are tightly linked, and the high-affinity iron uptake system is regulated by copper availability in the budding yeast S. cerevisiae (27). Furthermore, alkalinization of the medium is known to restrict the availability of metal ions (31, 32). Hence, we also checked if other phenotypes of the Cgvps34Δ mutant, viz. elevated susceptibility to alkaline pH, oxidative stress, and iron limitation, could be rescued by addition of other metal ions (Fig. 5, A and B). We found that addition of extra iron had no effect on the growth of the Cgvps34Δ mutant in the presence of oxidative stressor, menadione, but led to robust growth in the BPS-containing medium and the pH 7.5 medium (Fig. 5, A and B). Similarly, attenuated growth of the Cgvps34Δ mutant was restored by copper and zinc supplementation of the iron-limited medium (Fig. 5B). Of note, copper and zinc addition could also partially and mildly, respectively, rescue the growth defect in the pH 7.5 medium, whereas their supplementation failed to confer any growth advantage to the Cgvps34Δ mutant under oxidative stress conditions (Fig. 5, A and B). Together, these data reinforce the earlier findings that attenuated growth of the Cgvps34Δ mutant under iron-limiting and alkaline pH conditions is due to perturbed iron and copper metabolism.
Overexpression of the Vacuolar Transporter CgCcc1 Does Not Rescue Iron Toxicity in the Cgvps34Δ Mutant
To examine if surplus iron toxicity in the Cgvps34Δ mutant is due to defects in the inter-organelle iron transport, we overexpressed the vacuolar (CgCcc1) iron transporter, which imports iron into the vacuole from the cytosol, in wt and Cgvps34Δ cells (Fig. 5D). We observed that overexpression of CgCcc1 had no effect on the growth of wt and Cgvps34Δ cells in the iron-poor as well as in the iron-rich medium (Fig. 5D), thereby, ruling out misdistribution of iron between the cytosol and the vacuole as a cause for iron homeostasis defect of the Cgvps34Δ mutant. To check if the CgCCC1 gene was expressed at elevated levels, we examined its transcript levels in both wt and Cgvps34Δ cells by qPCR and found it to be 2.5–3.0-fold higher (Fig. 5E). Furthermore, to decipher the role of the mitochondria in surplus iron-mediated growth attenuation, we overexpressed CgMrs4 transporter, which imports iron into the mitochondria from the cytosol, in wt and Cgvps34Δ cells. We found that CgMrs4 expression rendered wt cells unable to grow in the iron-rich medium (Fig. 5D), which could either be due to the high mitochondrial iron level-associated toxicity or diminished cytosolic iron content. Interestingly, CgMrs4 expression aggravated the growth defect of Cgvps34Δ cells in the iron-limited medium and also made mutant cells hypersensitive to elevated iron content in the medium (Fig. 5D). There are three possible explanations for these results. First, the Cgvps34Δ mutant requires a certain threshold concentration of cytosolic iron to sustain growth in the iron-limited medium. Second, the Cgvps34Δ mutant is unable to acquire adequate iron from the low-iron environment. Third, accumulation of mitochondrial iron beyond a certain limit is toxic to Cgvps34Δ cells. Further investigations are required to test these hypotheses. Importantly, it is worth noting that CgMrs4 overexpression-mediated growth attenuation of the Cgvps34Δ mutant in the high-iron environment is in accordance with the high mitochondrial iron levels in the mutant.
Together, these data indicate that defective trafficking of iron to the vacuole may not account for attenuated growth of the Cgvps34Δ mutant under iron-limited and iron-surplus environmental conditions. Based on our metal ion supplementation studies, we speculate that perturbed ion homeostasis in the Cgvps34Δ mutant is likely to be because of the impaired functioning of the vacuole and/or enhanced ion uptake, whereas the deregulated iron metabolism is probably linked with altered mitochondrial functions.
Transcriptional Response to Changes in the Environmental Iron Content Is Not Impaired in the Cgvps34Δ Mutant
Iron limitation- and iron excess-mediated growth inhibition of the Cgvps34Δ mutant suggested that mutant cells may be deficient in responding to variations in the environmental iron concentration. Hence, we examined transcriptional response of the Cgvps34Δ mutant to an increase and a decrease in the external iron levels. C. glabrata cells are known to up-regulate and down-regulate expression of the high affinity iron transport system in response to iron-limited and iron-excess conditions, respectively (19). To determine transcript levels of iron-responsive genes, we performed qPCR analysis on cells grown either in the YNB medium or the YNB medium supplemented with BPS/ferric chloride for 2 h (Fig. 6A). We observed that although CgFTR1, CgFET3, and CgSIT1 genes, compared with wt cells, were expressed at low basal levels in the Cgvps34Δ mutant (Fig. 2, B and C), their transcript levels increased and decreased in response to iron depletion and iron surplus conditions, respectively (Fig. 6A). Of note, this response was similar to that of wt cells (Fig. 6A) indicating that both wt and Cgvps34Δ mutant cells are able to sense and mount an appropriate transcriptional response upon low- and high-iron stress.
FIGURE 6.
Cellular response to environmental iron concentration is not impaired in the Cgvps34Δ mutant. A, qPCR analysis of CgFTR1, CgFET3, and CgSIT1 gene expression in wt and Cgvps34Δ mutant upon 2 h growth in the YNB medium (Y) lacking or containing 50 μm BPS (B) or 500 μm ferric chloride (F). Data (mean ± S.E., n = 3–6) were normalized to an internal CgACT1 mRNA control, and represent fold-change in expression upon BPS and ferric chloride treatment compared with YNB-grown cultures. Paired, two-tailed, Student's t test was used (*, p < 0.05; **, p < 0.005; ***, p < 0.0001). B and C, representative immunoblots of CgSlt2 (B) and CgHog1 (C) phosphorylation indicated in C. glabrata cells grown for 2 h in YNB medium (Y) and YNB medium containing either 50 μm BPS (B) or 500 μm ferric chloride (F) at 30 °C. CgGapdh was used as a loading control. The ImageJ densitometry software was used to quantify intensity of individual bands in three independent Western blots. CgSlt2 and CgHog1 phosphorylation signals in each lane were normalized to the corresponding CgGapdh signal, and data (± S.E.) are presented as fold-change in phosphorylation levels in treated samples compared with untreated (YNB-grown, considered as 1.0) samples for each strain below the blot.
C. glabrata cells are also known to respond to a low-iron environment by activating the HOG-MAPK pathway and to a high-iron environment by activating the HOG as well as the protein kinase C (PKC)-mediated cell wall integrity pathway (19). To determine the status of these two pathways in a varied iron environment, we assessed the phosphorylation levels of CgSlt2 and CgHog1, the terminal MAPKs of PKC and HOG signaling cascades, respectively, in wt and Cgvps34Δ cells. As reported previously (12), constitutive activation of the PKC signaling pathway was observed in log-phase Cgvps34Δ cells (Fig. 6B). Furthermore, we found a similar increase in phosphorylated forms of CgSlt2 and CgHog1 between wt and the mutant under iron-limited and iron-excess conditions (Fig. 6B). In accordance, overexpression of neither CgHog1 nor CgSlt2 kinase could rescue the attenuated growth of the Cgvps34Δ mutant in iron-limited and iron-surplus media (data not shown). Altogether, these findings suggest that the Cgvps34Δ mutant is fully proficient in reconfiguring its transcriptional regulatory and MAPK signal transduction networks in response to iron depletion and iron-excess stress.
CgFet3 Is Required for Plasma Membrane Localization of the CgFtr1 Transporter
The above mentioned findings raised the question of why, despite mounting an appropriate transcriptional and stress signaling response, Cgvps34Δ cells could not grow in low- and high-iron medium. To address this, we decided to inspect the functioning of the iron transport machinery in the Cgvps34Δ mutant. The high-affinity iron uptake system in C. glabrata is composed of an iron permease (CgFtr1) and a copper ferroxidase (CgFet3), which are assumed to form a complex (10). To gain a better understanding of localization and activity of CgFtr1 and CgFet3 in C. glabrata, we first performed topology analysis of CgFtr1 (408 amino acids) and CgFet3 (635 amino acids) using the online tool harrier.nagahama-i-bio.ac.jp/sosui/sosui_submit.html, which revealed the presence of seven and two transmembrane helices in CgFtr1 and CgFet3, respectively. Each transmembrane domain was about 23 amino acids in length. The transmembrane domains in CgFtr1 were connected through 3 cytosolic and 3 extracellular loops with amino and carboxyl termini facing the cell wall and the cytosol, respectively (Fig. 7A). In contrast, CgFet3 consists of two membrane-spanning helices with the N-terminal catalytic domain toward the exterior of the cell (Fig. 7A). Overall, the predicted topology of CgFtr1 and CgFet3 is similar to that of their counterparts in S. cerevisiae.
FIGURE 7.
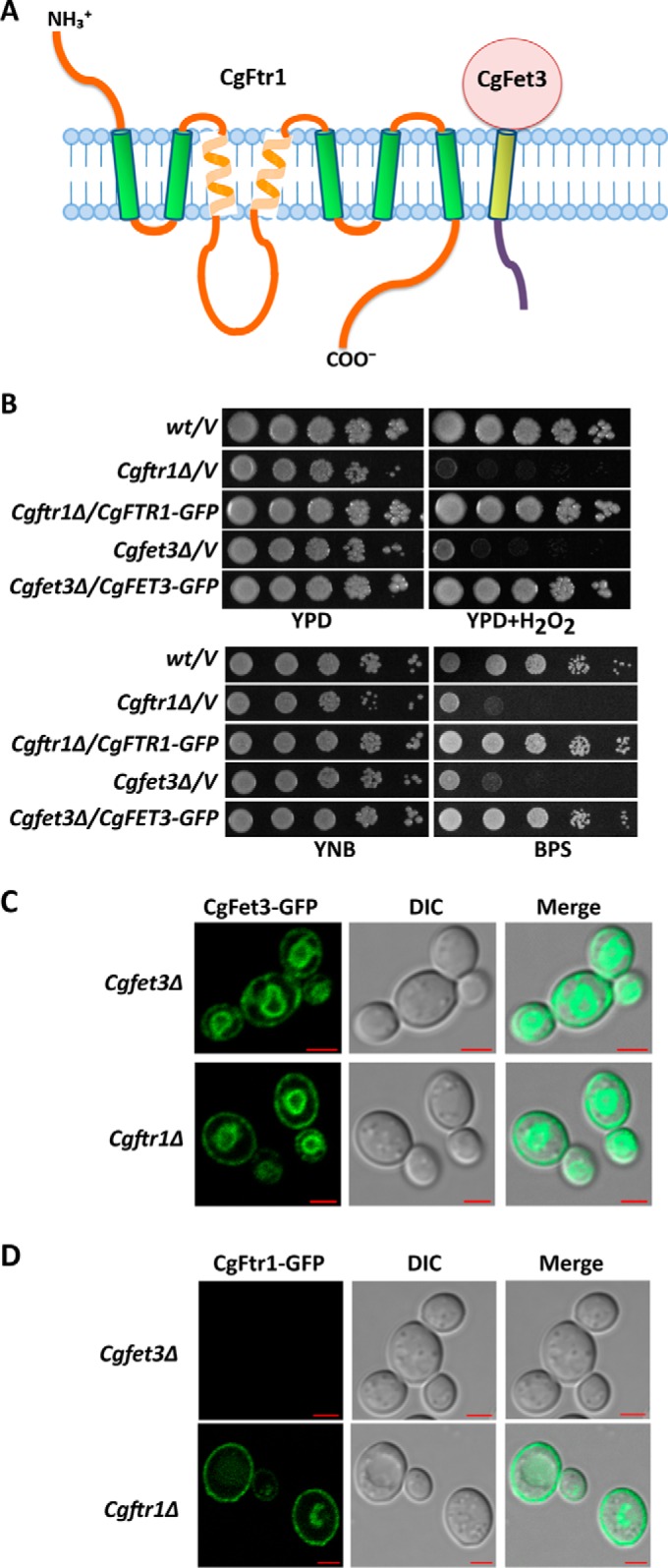
CgFet3 is required for CgFtr1 expression and localization. A, schematic illustration of membrane topology of CgFtr1 as predicted by the SOSUI online tool. The NH3+ and COO− indicate the amino and carboxyl termini, respectively, of CgFtr1. B, serial dilution spotting growth analysis of the indicated C. glabrata strains. Hydrogen peroxide and BPS were added to final concentrations of 20 mm and 25 μm, respectively. Plate images were captured after 2–4 days of incubation at 30 °C. C and D, representative confocal images of CAA medium-grown log-phase Cgftr1Δ and Cgfet3Δ cells expressing CgFet3-GFP (C) and CgFtr1-GFP (D). Cgftr1Δ and Cgfet3Δ cells carrying CgFtr1-GFP and CgFet3-GFP, respectively, were used as controls that display plasma membrane localization of the respective GFP fusion proteins. Bar, 2 μm.
The Ftr1 permease and Fet3 ferroxidase in S. cerevisiae are co-trafficked to and from the cell membrane (33). To examine localization of C. glabrata Ftr1 and Fet3 proteins, we first generated CgFtr1-GFP and CgFet3-GFP fusion proteins by inserting GFP (green fluorescent protein) at the C terminus of CgFtr1 and CgFet3. Cgftr1Δ and Cgfet3Δ mutants are known to display reduced growth in the iron-limited medium and in the presence of the oxidative stressor, hydrogen peroxide (10). Expression of CgFtr1-GFP and CgFet3-GFP in the respective mutants could rescue attenuated growth in the BPS-containing medium as well as in the medium containing hydrogen peroxide (Fig. 7B) suggesting that C-terminal GFP fusion proteins of CgFtr1 and CgFet3 are being expressed as well as are functional.
Next, we examined the requirement for CgFtr1 for proper localization of CgFet3 and vice versa (Fig. 7, C and D), to gain a better understanding of transport of CgFtr1 and CgFet3 to the plasma membrane. We found CgFet3 to be located on the cell membrane as well as on an intracellular organelle in the Cgftr1Δ mutant (Fig. 7C) suggesting that CgFet3 transport to the plasma membrane does not require CgFtr1. In contrast, no fluorescence signal was detectable in the Cgfet3Δ mutant expressing the CgFtr1-GFP protein (Fig. 7D) indicating that lack of CgFet3 probably leads to mislocalization and degradation of CgFtr1.
To investigate the effect of CgVPS34 disruption on activity of the CgFet3 enzyme, we measured the cell surface-associated oxidase activity, using the p-phenylenediamine dihydrochloride as a substrate, in wt and Cgvps34Δ cells. As shown in Fig. 8, oxidase activity in the Cgvps34Δ mutant was found to be 2.3-fold higher compared with that in the wt cells. The Cgfet3Δ mutant was used as a control in this assay, and expectedly, it displayed 60% lower oxidase activity compared with wt cells. Ectopic expression of CgVPS34 restored the cell surface-associated oxidase activity in the Cgvps34Δ mutant to wt levels (Fig. 8). It is worth noting here that although elevated oxidase activity of CgFet3 is in contrary to lower CgFET3 transcript levels in the Cgvps34Δ mutant, it does partly explain the higher intracellular iron content of Cgvps34Δ cells. Furthermore, these data raise the question whether enhanced activity of CgFet3 is due to the defect in recycling of the enzyme complex from the cell membrane.
FIGURE 8.
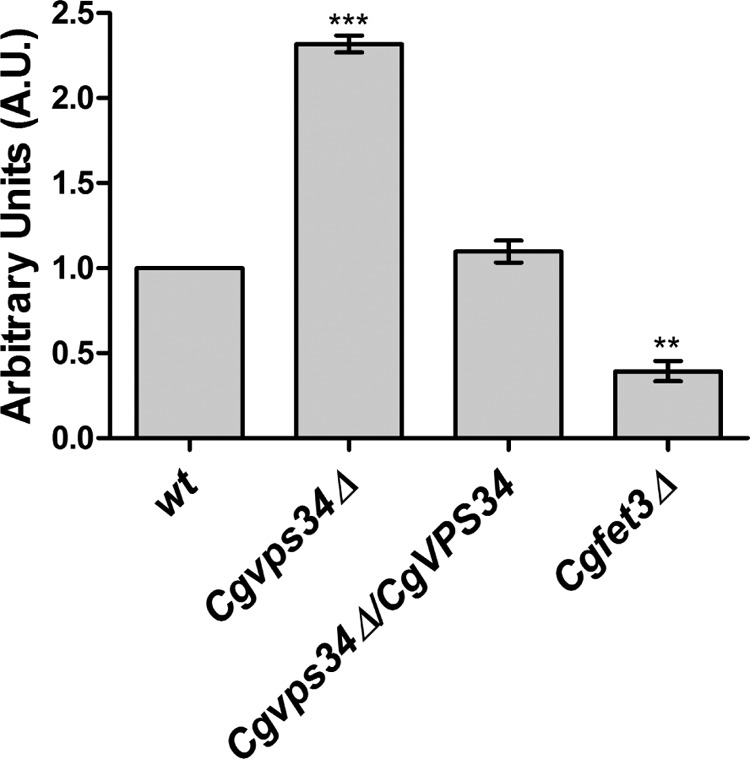
The Cgvps34Δ mutant displays increased oxidase activity. As indicated, overnight YPD medium-grown C. glabrata strains were inoculated in the YPD medium. After 6 h of incubation at 30 °C, cells were collected, washed, normalized to 1.0 A600, and were incubated with 0.2% p-PD. The rate of p-PD oxidation was recorded spectrophotometrically at 570 nm, and expressed in arbitrary units. The oxidase activity of wt cells was considered as 1.0. The Cgfet3Δ mutant was used as a control, which displayed 40% of wt oxidase activity. Paired, two-tailed, Student's t test was used (**, p < 0.005; ***, p < 0.0001).
Retrograde Transport of the CgFtr1 Transporter Is Defective in the Cgvps34Δ Mutant
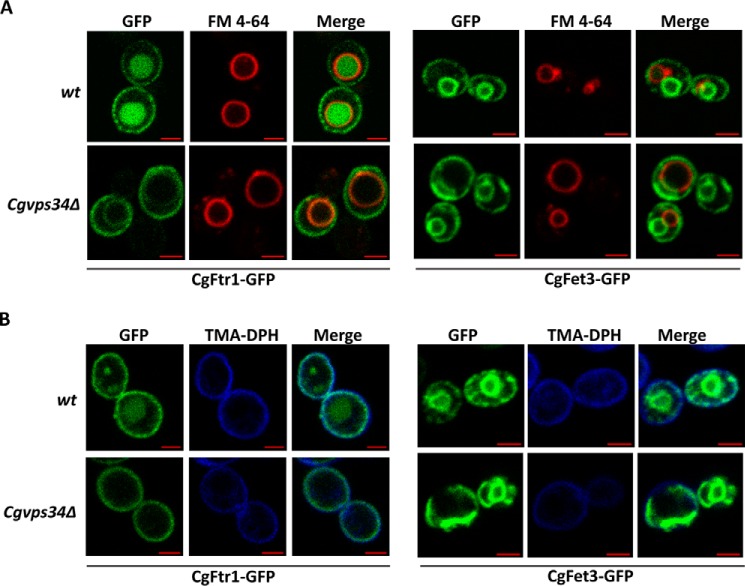
Because CgVps34 has previously been implicated in regulated trafficking of cellular proteins and processing of Epa1 adhesin from the cell surface (12), we next checked if higher CgFet3 oxidase activity in the Cgvps34Δ mutant could be due to impaired retrograde transport of CgFtr1 permease and CgFet3 ferroxidase proteins from the cell membrane. We expressed CgFtr1-GFP and CgFet3-GFP in wt and Cgvps34Δ cells and examined their localization by confocal microscopy. Under regular iron log phase conditions, we found CgFtr1 to localize to both the plasma membrane and vacuole in wt cells (Fig. 9A). In contrast, the vacuolar localization of CgFtr1 was markedly diminished in Cgvps34Δ cells (Fig. 9A). Intriguingly, CgFet3-GFP was primarily located on the plasma membrane and the membrane of an intracellular organelle in both wt and Cgvps34Δ cells under log-phase conditions (Fig. 9A). To verify the plasma membrane localization of CgFtr1-GFP and CgFet3-GFP, wt and Cgvps34Δ cells were labeled with TMA-DPH (trimethylamino-diphenyl-1,3,5-hexatriene), which marks solely the plasma membrane (34). As shown in Fig. 9B, the green fluorescence signal of CgFtr1-GFP and CgFet3-GFP overlapped with that of the TMA-DPH-stained cell membrane indicative of plasma membrane localization of CgFtr1 and CgFet3 proteins in wt and Cgvps34Δ cells.
FIGURE 9.
CgFtr1-GFP and CgFet3-GFP are expressed at the plasma membrane in the Cgvps34Δ mutant. A, CAA medium-grown, log-phase wt and Cgvps34Δ cells expressing either CgFtr1-GFP or CgFet3-GFP were preloaded with FM4-64 (16 μm) for 1 h prior to cell collection for slide preparation. Images were captured using the Zeiss LSM 700 META confocal microscope. Bar, 2 μm. B, CAA medium-grown, log-phase wt and Cgvps34Δ cells expressing either CgFtr1-GFP or CgFet3-GFP were labeled with TMA-DPH (1.0 μm) for 10 min at 25 °C in the dark. Cells were washed with TE buffer and imaged using the Zeiss LSM 700 META confocal microscope. Bar, 2 μm.
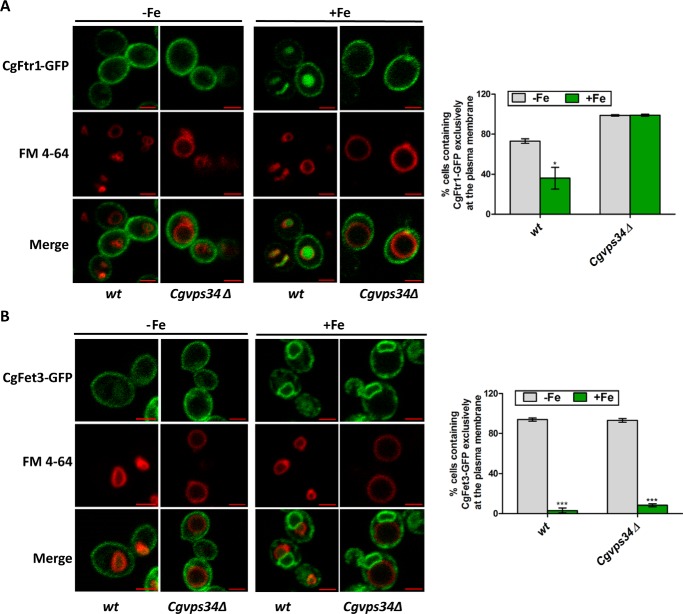
Furthermore, in response to iron limitation, CgFtr1-GFP did not localize to the vacuole as cellular fluorescence was limited only to the plasma membrane in both wt and Cgvps34Δ cells (Fig. 10A). Contrarily, the vacuolar localization and the cell membrane localization of CgFtr1-GFP was enhanced and diminished, respectively, in wt cells upon growth in the iron-surplus medium (Fig. 10A). Of note, localization of CgFtr1-GFP in the Cgvps34Δ mutant did not change in response to iron-repleted conditions, and remained primarily confined to the plasma membrane in the vast majority (95%) of cells (Fig. 10A). These data indicate that Cgvps34Δ cells are deficient in the retrograde transport of CgFtr1-GFP from the plasma membrane in the high-iron environment, which could partly account for elevated susceptibility of the Cgvps34Δ mutant to the surplus iron.
FIGURE 10.
Retrograde trafficking of CgFtr1 is impeded in the Cgvps34Δ mutant. A and B, overnight CAA medium-grown, wt and Cgvps34Δ cells expressing CgFtr1-GFP (A) and CgFet3-GFP (B) were inoculated in the CAA medium containing 50 μm BPS. After 12 h incubation at 30 °C, cells were collected, washed with CAA, and inoculated to the CAA medium containing either 50 μm BPS (−Fe) or 500 μm ferrous ammonium sulfate and 1 mm sodium ascorbate (+Fe). Post 2 h growth at 30 °C, cells were imaged using the Zeiss LSM 700 META confocal microscope. Bar, 2 μm. For each strain, a minimum of 160 cells displaying green fluorescence were counted, and data are presented as percentage of cells with CgFtr1-GFP (A) and CgFet3-GFP (B) at the plasma membrane on the right side of panels. Unpaired, two-tailed, Student's t test was used (*, p < 0.05; ***, p < 0.0001).
Similar to CgFtr1-GFP, ∼90% of wt and Cgvps34Δ cells exhibited plasma membrane localization of CgFet3-GFP under iron-limiting conditions (Fig. 10B). Furthermore, in the iron-excess medium, plasma membrane targeting of CgFet3-GFP was drastically reduced as only 1–5% of wt and Cgvps34Δ cells contained CgFet3-GFP exclusively on the cell membrane (Fig. 10B). CgFet3-GFP was primarily confined to the intracellular organelle membrane in wt and Cgvps34Δ cells upon growth in the iron-rich medium (Fig. 10B), thereby, precluding involvement of CgVps34 in the retrograde transport of CgFet3-GFP.
Four key findings emerge from these localization studies. First, CgFet3 is required for membrane localization of CgFtr1, whereas CgVps34 is dispensable for trafficking of CgFtr1 and CgFet3 to the cell membrane. Second, environmental iron content determines the recycling of CgFtr1 and CgFet3 from the cell membrane with high-iron resulting in relocation to intracellular organelles, thereby, setting the stage for either recycling or degradation of CgFtr1 and CgFet3 proteins. Third, retrograde transport of CgFtr1 and CgFet3 probably occur independently of each other. Last, transport of CgFtr1 to the vacuole in response to surplus iron requires PI3-kinase.
The Cgvps34Δ Mutant Is Defective in Mouse Colonization in Vivo
CgVps34 has previously been shown to be essential for survival in human and mice macrophages and in murine model of systemic candidiasis (12). C. glabrata wild-type cells are able to undergo intracellular proliferation, whereas the Cgvps34Δ mutant is killed in human THP-1 macrophages (12, 35). As one of the macrophage defense mechanisms is to restrict iron availability to engulfed pathogens to control their proliferation (36), we, next, sought to examine whether the incapacity of the Cgvps34Δ mutant to survive the intracellular milieu of macrophages is because of its deregulated iron metabolism and probably intrinsic high-iron requirement. For this, we first checked the intracellular behavior of wt and Cgvps34Δ cells pre-grown either in the iron-limited or iron-surplus medium. Proliferation of wt and survival of Cgvps34Δ cells in macrophages remained unaffected by prior iron chelation or supplementation (data not shown) ruling out any effect of internal iron reserves on the intracellular replication. Of note, these data are inconsistent with an earlier study reporting increased intracellular survival of C. glabrata cells, which were grown in the ferrichrome-containing iron-rich medium prior to macrophage infection (18). This discordance could be due to the nature of the iron source used in this (ferric chloride) and the earlier study (ferrichrome). Furthermore, although the macrophage environment is supposed to be a low-iron environment, macrophage-internalized C. glabrata wild-type cells are known to down-regulate the expression of high-affinity iron transport genes after 10 h of co-incubation (35) raising the possibility that macrophage internal milieu either is an iron-rich environment, or requirement of the high-affinity iron uptake system is less at later infection stages. To decipher the iron status of macrophages, we checked the localization of CgFtr1-GFP in internalized C. glabrata wt cells after 6 h co-incubation with macrophages. As shown in Fig. 11A, CgFtr1-GFP was present exclusively on the cell membrane in macrophage-ingested wt and Cgvps34Δ cells indicating that C. glabrata cells encounter iron limitation in early stages of macrophage infection. These results also corroborate earlier findings that iron limitation-induced cell membrane localization of CgFtr1 is not impaired in Cgvps34Δ cells.
FIGURE 11.
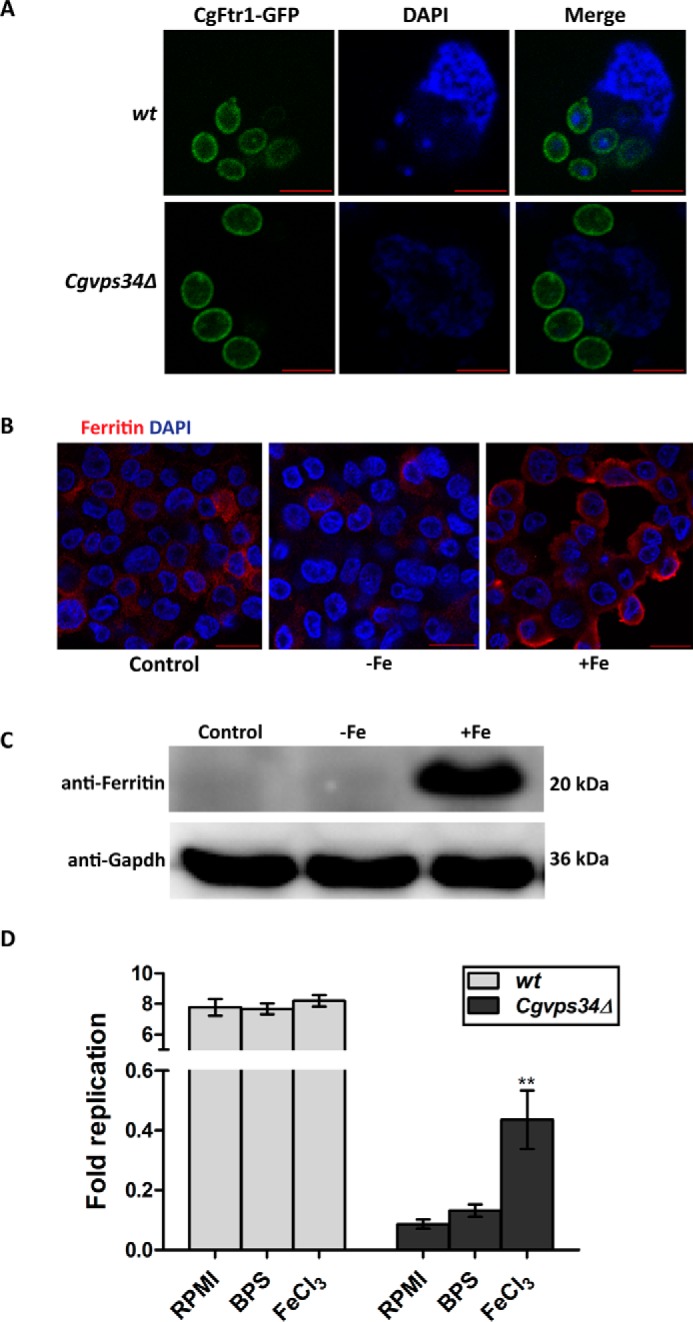
Macrophage-internalized C. glabrata cells express CgFtr1 on the plasma membrane. A, representative confocal images showing localization of CgFtr1-GFP on the plasma membrane in C. glabrata wt and Cgvps34Δ cells recovered from THP-1 macrophages after 6 h of infection. DAPI was used to stain nuclei. Bar, 5 μm. B, representative confocal images depicting immunofluorescence staining of ferritin (in red) in THP-1 cells grown in RPMI medium (control), serum-free RPMI medium containing 50 μm BPS (−Fe), and RPMI medium plus 50 μm ferric chloride (+Fe). Nuclei were stained with DAPI (in blue). Bar, 20 μm. C, a representative immunoblot illustrating ferritin levels in THP-1 macrophages grown in RPMI medium (control), serum-free RPMI medium containing 50 μm BPS (−Fe), and RPMI medium plus 50 μm ferric chloride (+Fe). THP-1 macrophages were lysed and extracts containing 70 μg of protein were resolved on 15% SDS-PAGE and immunoblotted with anti-ferritin antibody. Gapdh was used as a loading control. D, intracellular survival of wt and Cgvps34Δ cells in THP-1 macrophages as determined by the cfu-based assay. THP-1 cells were treated with phorbol 12-myristate 13-acetate (16 nm) for 12 h and recovered in RPMI plus 10% FBS medium. After 12 h, THP-1 macrophages were cultured in RPMI complete medium (RPMI plus 10% FBS; RPMI), the iron surplus medium (RPMI complete and 50 μm ferric chloride; FeCl3), or the iron-limited medium (serum-free RPMI containing 50 μm BPS; BPS). After 24 h growth in varied iron concentrations, THP-1 cells were washed, incubated in the serum-free RPMI medium, and infected either with wt or Cgvps34Δ cells to a multiplicity of infection of 0.1. Extracellular yeast cells were washed three times with PBS and internalized cells were collected by lysing macrophages in water 2 and 24 h post-infection. Yeast colonies were counted after 2 days of plating appropriate lysate dilutions. Fold-replication reflect the ratio of the number of intracellular C. glabrata cells at 24 h to the number at 2 h after macrophage internalization. Data represent mean ± S.E. (n = 5). **, p < 0.005; two-tailed unpaired Student's t test was used.
Next, to examine the effect of varied extracellular iron concentration in the medium upon survival of the Cgvps34Δ mutant in macrophages, we infected wt and Cgvps34Δ cells to THP-1 macrophages cultured in the iron-restricted (serum-free RPMI medium containing BPS) and iron-supplemented medium (serum and ferric chloride-containing RPMI medium) as described previously (37). Iron in macrophages is stored in the form of an iron storage protein ferritin, which is composed of 24 subunits that are arranged in an octahedral symmetry (38). To check if macrophage culturing in the aforementioned media alters intracellular levels of ferritin, we performed immunofluorescence as well as Western blot analysis using the anti-ferritin antibody. As shown in Fig. 11B, THP-1 cells grown in regular RPMI medium and RPMI medium containing BPS displayed very low levels of ferritin expression, whereas surplus iron medium-cultured THP-1 cells exhibited intense ferritin staining. Similarly, whereas ferritin protein levels were not detected in RPMI- and RPMI plus BPS medium-cultured THP-1 cells, growth in the ferric chloride-supplemented medium led to a significant increase in intracellular levels of ferritin (Fig. 11C). Together, these results indicate that RPMI plus BPS and RPMI plus ferric chloride media mirror low and high environmental iron conditions, respectively. Notably, no appreciable differences in macrophage viability were observed upon culturing in iron-depleted and iron-repleted conditions, as checked by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay (data not shown).
Furthermore, intracellular proliferation of wt cells remained unaffected by external iron concentrations indicating that iron availability is not the limiting factor for intracellular replication of C. glabrata wt cells (Fig. 11D). Intriguingly, whereas survival rate of the Cgvps34Δ mutant remained unaffected in iron-restricted macrophages, a 5-fold higher survival was observed in iron-supplemented macrophages compared with the RPMI medium-grown macrophages (Fig. 11D). These data suggest that higher extracellular iron levels promote the survival of Cgvps34Δ cells in macrophages, which could be attributed, in part, to better growth of the Cgvps34Δ mutant in iron-rich macrophages.
Iron availability is known to modulate the ability of the bacterial pathogen Staphylococcus aureus to form biofilms (39). Hence, to investigate the effect of extracellular iron on the biofilm-forming ability of C. glabrata, we measured the capacity of wt and Cgvps34Δ cells to form biofilm under normal, iron-depleted and iron-repleted conditions. We found the biofilm-forming capacity of Cgvps34Δ cells to be 10-fold lower compared with that of wt cells in the normal growth medium (Fig. 12A). Environmental iron content has previously been reported to regulate adherence of C. glabrata to Lec2 epithelial cells, with surplus iron medium-grown cells exhibiting 3-fold higher adherence to Lec2 cells compared with regular iron medium-cultured cells (19). Consistent with these results, the ability of C. glabrata wt cells to form biofilms was 60% higher in the high-iron environment (Fig. 12B). Similarly, a 2-fold increase in biofilm formation was observed in the Cgvps34Δ mutant under surplus iron conditions (Fig. 12B). Of note, the BPS medium-grown wt and Cgvps34Δ cells formed biofilms on polystyrene plates to the same extent as the corresponding RPMI medium-grown cells (Fig. 12B). These data demonstrate that CgVPS34 disruption led to abrogation of two important virulence traits of C. glabrata, and this abolishment may contribute to the reduced virulence of the Cgvps34Δ mutant.
FIGURE 12.
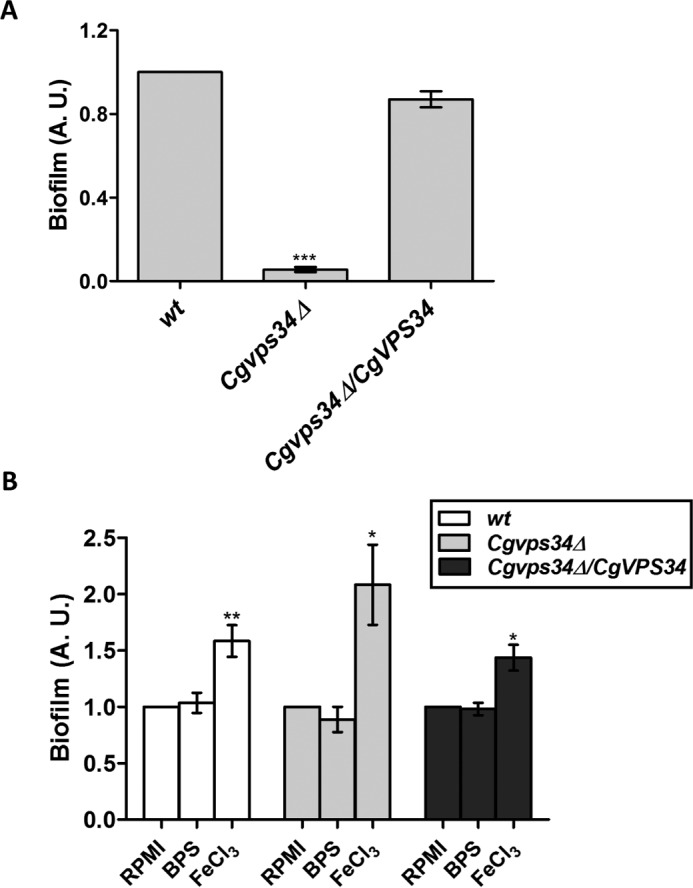
Deletion of CgVPS34 significantly attenuates biofilm-forming capacity of C. glabrata cells. A, the indicated C. glabrata strains were grown in RPMI medium containing 10% FBS for 48 h in a 24-well polystyrene plate followed by crystal violet (0.4% in 20% (v/v) ethanol solution) staining for 45 min. One ml of 95% ethanol was used for destaining, and the amount of crystal violet stain in ethanol was measured by recording the absorbance at 595 nm. Data (mean ± S.E.; n = 3) reflect the number of adherent mutant cells relative to wt cells (set to 1.0). ***, p < 0.0001; two-tailed paired Student's t test was used. B, biofilm forming ability of the indicated strains was determined as mentioned above, but with one modification that cells were also grown in the RPMI medium containing 100 μm BPS (BPS) and RPMI medium containing 500 μm ferric chloride and 10% FBS (FeCl3). Data (mean ± S.E.; n = 6) reflect the number of adherent cells after treatment relative to untreated RPMI-grown cells (set to 1.0). *, p < 0.05; **, p < 0.005; two-tailed paired Student's t test was used.
Lastly, to probe into the effect of abated biofilm-forming capacity of Cgvps34Δ cells on their murine organ colonization potential, we performed a kinetic study of BALB/c mice infection in the disseminated candidiasis model. Female BALB/c mice were infected either with wt or Cgvps34Δ mutant cells, three target organs (kidneys, liver, and spleen) were harvested 1, 3, and 5 days after infection, and the fungal burden was analyzed by the cfu-based assay. At day 1 post-infection, we observed 10-, 10- and 100-fold lower cfus in liver, spleen, and kidneys, respectively, of Cgvps34Δ-infected mice compared with fungal load in wt-infected mice (Fig. 13, A–C). On day 3 post-infection, yeast cfus in the Cgvps34Δ-infected mice were further decreased by 70–100-fold (Fig. 13, A–C). Notably, no significant decrease in fungal burden in kidneys, liver, and spleen of wt-infected mice was observed between days 1 and 3 (Fig. 13, A–C). After 5 days of infection, very few yeast cfus were recovered from organs of the Cgvps34Δ-infected mice (Fig. 13, A–C). In contrast, wt-infected mice still had 104-105 C. glabrata cells in their kidneys, liver, and spleen (Fig. 13, A–C). Altogether, these data indicate that attenuated virulence of the Cgvps34Δ mutant in the systemic murine candidiasis model is probably a combined effect of lower colonization of host cells, and increased clearance by the host immune system.
FIGURE 13.
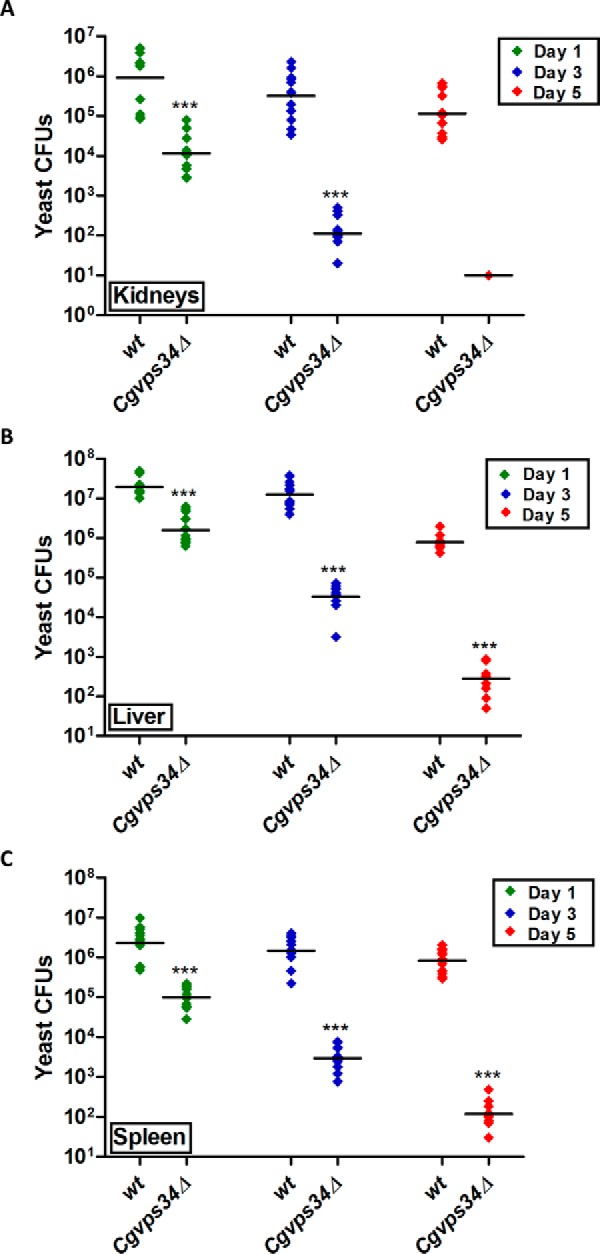
The Cgvps34Δ mutant displays reduced colonization in a murine model of systemic candidiasis. YPD medium-grown wt or Cgvps34Δ cells (4 × 107) were injected into the tail vein of 6–8-week-old, female BALB/c mice, and mice were sacrificed at the indicated days post-infection. Diamonds represent the cfu recovered from target organs, kidneys (A), liver (B), and spleen (C), of the individual mouse. The cfu geometric mean (n = 8–14) for each strain is depicted by a horizontal line. Statistically significant differences in cfu are marked (***, p < 0.0001; Mann-Whitney test). Of note, of 10 mice infected with the Cgvps34Δ mutant, we could recover yeast cfu from kidneys of only one animal 5 days post-inoculation.
Discussion
The ability to survive varied iron environmental niches is pivotal to the survival of opportunistic pathogenic microbes (40). C. glabrata, a successful pathogenic yeast, relies on both high-affinity iron uptake and host-specific iron uptake systems to acquire iron from host niches (10), and the strategies of C. glabrata cells to survive and proliferate in low- and high-iron environments are beginning to be unfolded. Here, we report an essential role for the phosphoinositide 3-kinase CgVps34 in the maintenance of iron homeostasis. Our results suggest that CgVps34-mediated iron homeostasis promotes survival of C. glabrata cells in both iron-deficient and iron-sufficient conditions (Fig. 3D). Furthermore, we demonstrate for the first time that retrograde trafficking of the CgFtr1 transporter in response to high-iron is disrupted in the Cgvps34Δ mutant (Fig. 10A), probably resulting in the incessant iron uptake and impaired iron homeostasis. Although disruption of CgVps34 results in aberrant functioning of both the mitochondria and the vacuole in C. glabrata, our data indicate a more prominent contribution of the mitochondrial iron metabolic machinery to CgVps34-mediated cellular iron homeostasis. Iron deficiency is closely linked with respiratory incompetence, and yeast mutants with defects in vacuolar functions and/or biogenesis are also sensitive to metal ions (23, 24, 41). These effects have been attributed either to ion storage defects and/or expression of less selective divalent metal cation transporters due to deficient assembly of the high-affinity transport system (24, 42, 43). Three pieces of evidence suggest that metal dyshomeostasis in the Cgvps34Δ mutant is unlikely to be solely due to impaired vacuolar functions. First, metal ion sensitivity profile of the Cgvps34Δ mutant appears to be distinct (Fig. 5, A and B) from the one reported for vacuolar ATPase-defective mutants (29). Second, there was no appreciable competition among metal ions for the nonspecific transporter-dependent uptake (Fig. 5A). Third, overexpression of the vacuolar iron importer CgCcc1 affected mutant growth in neither the iron-poor nor iron-rich medium (Fig. 5D). Furthermore, iron toxicity associated with mitochondrial importer CgMrs4 overexpression in the wild-type strain (Fig. 5D) indicates that C. glabrata cells probably mobilize extra iron to the mitochondria.
The high-affinity iron uptake system in C. glabrata is known to be largely under the transcriptional control (10), however, our data highlight posttranslational regulation of the iron transporter CgFtr1, which is reflected in high-iron induced internalization and possible degradation of CgFtr1 (Fig. 10A). Similar to S. cerevisiae (27, 33, 44), the high-affinity iron uptake system is composed of a transmembrane transporter CgFtr1 and the cell membrane-localized ferroxidase CgFet3 in C. glabrata (10). The multicopper oxidase Fet3 in S. cerevisiae is involved in oxidation of ferrous ion to ferric ion, prior to ferric ion transport by Ftr1 across the membrane (33, 44, 45). Targeting of both Ftr1 and Fet3 to the plasma membrane requires the presence of the other protein (33). Furthermore, proper assembly and localization of the Fet3-Ftr1 transport system in S. cerevisiae is a prerequisite to acquire iron from the iron-restricted medium (46, 47). Of note, high environmental iron results in internalization and degradation of the Ftr1-Fet3 complex (46). In contrast, low environmental iron induces diversion of the degradative pathway-destined Fet3-Ftr1 complex into the endosomal recycling pathway for targeting back to the cell membrane via a specific retromer complex in S. cerevisiae (46–49).
Our data suggest the existence of a different mechanism for the assembly and transport of the CgFtr1-CgFet3 system in C. glabrata, as CgFet3 localization at the plasma membrane was not affected in the Cgftr1Δ mutant (Fig. 7C). Similarly, PI3-kinase is required for retrograde trafficking of CgFtr1, but not of CgFet3 in response to high-iron (Fig. 14), indicating that CgFtr1 and CgFet3 are unlikely to be co-trafficked to, and from the cell membrane. It is also worth noting that Vps34 in S. cerevisiae has not been implicated in retrieval of CgFtr1-CgFet3 from the cell membrane (48–50). Consistent with this, the C. glabrata Vps30 protein (12), unlike its ortholog in S. cerevisiae, which is an integral constituent of PI3K complexes (50), is not required for intracellular protein trafficking (12). These findings reflect functional diversification between the S. cerevisiae and C. glabrata PI3K. Furthermore, as the C. glabrata genome harbors two other ORFs, CAGL0J08481g and CAGL0K12738g, coding for putative oxidoreductase and ferroxidase, respectively, the possibility that CgFtr1 forms a complex with one of these oxidases for its transport and/or function in the absence of CgFet3 (CAGL0F06413p) cannot be precluded. It should be noted, however, that cell surface localization of CgFet3 in C. glabrata wild-type cells, and attenuated growth of the Cgfet3Δ mutant in the iron-limited medium renders this possibility unlikely.
FIGURE 14.
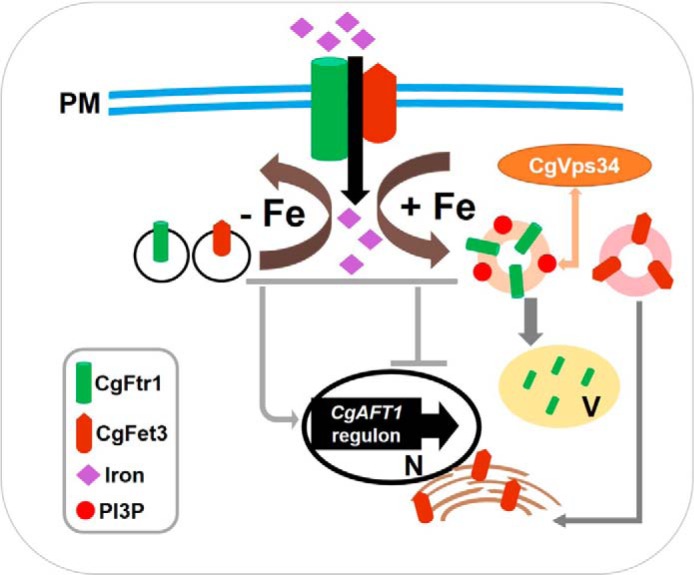
A schematic model depicting the role of CgVps34 in regulation of CgFtr1 trafficking and iron homeostasis. Components of the high-affinity iron uptake system, CgFtr1 and CgFet3, shuttle between the plasma membrane, and intracellular organelles. Although they function at the cell surface in an iron-restricted environment, they are sent back to intracellular organelles under iron-rich conditions for degradation and/or recycling. The phosphoinositide 3-kinase CgVps34, which phosphorylates inositol phosphate and produces PI3P, is essential for retrograde trafficking of CgFtr1 from the cell membrane in response to an increase in the environmental iron content. PM, plasma membrane; V, vacuole; N, nucleus; PI3P, phosphatidylinositol 3-phosphate.
What is intriguing is why the Cgvps34Δ mutant is susceptible to both high- and low-iron stresses. Growth attenuation in the high-iron environment could be attributed, in part, to the inability to degrade the high-affinity iron transporter CgFtr1. Although it remains to be experimentally demonstrated, the current data (high mitochondrial iron content, elevated mitochondrial aconitase activity, increased ATP levels, and increased expression of genes coding for iron-utilizing enzymes), reflective of the iron-rich intracellular environment of the Cgvps34Δ mutant, indicate that the mutant requires high-iron to keep the basic cellular processes running efficiently, as iron consumption of the mitochondria is probably high. Mitochondria, the key producer of cellular energy currency ATP, plays a pivotal role in iron homeostasis primarily through generation and utilization of Fe-S clusters (28). Of note, inadequate iron or copper is known to impede the assembly of metal-containing systems involved in oxidative phosphorylation, thereby, decelerating cellular respiration (25). Hence, it is possible that an increased need for energy contributes to the attenuated growth of Cgvps34Δ cells in the iron-poor medium. Alternatively, sensitivity of the Cgvps34Δ mutant to low-iron stress could be a side effect of altered nutrient acquisition and requirement, as global defects in protein trafficking may perturb transport of several critical nutrients including metal ions.
Surplus iron-mediated toxicity is lethal, and two terminal MAPKs, CgHog1 and CgSlt2, of the high osmolarity glycerol and the protein kinase C (PKC)-mediated cell wall integrity pathway, respectively, have been implicated in the survival of high-iron stress in C. glabrata (19). Our data identified PI3-kinase to be a novel and a critical component of the iron homeostatic machinery. Extending this analysis further, we have also screened the C. glabrata deletion strain library (51) for growth reduction in the high-iron medium, and identified five (CgKdx1, CgMkk2, CgRom2, CgSlg1, and CgRlm1) and four (CgPbs2, CgSho1, CgSte20, and CgSte50) putative components of PKC and HOG pathways, respectively, in addition to the PI3K regulatory subunit CgVps15, and six constituents (CgBcy1, CgCch1, CgCmp2, CgCnb1, CgCrz1, and CgEcm7) of the calcineurin signaling pathway to be required for survival of high-iron stress (data not shown). The precise role these signaling pathways play, and the cross-talk between proteins of these cascades, if any, remain to be determined. However, these data point to the existence of a complex regulatory system, composed of at least four stress signaling cascades, to ensure iron homeostasis in the iron-rich environment in C. glabrata. Of note, Candida spp. are likely to encounter high-iron levels in the gastrointestinal tract (52).
The biofilm formation attribute is considered a major contributor to nosocomial C. glabrata infections (53). It has been reported earlier that compared with 104-105 wild-type cells still inhabiting the BALB/c mouse organs, the Cgvps34Δ mutant is almost completely cleared 7 days post-infection in the systemic model (12). Multiple processes including adherence to host tissue, cell proliferation, and secretion of extracellular matrix components are required for establishment of a mature biofilm (54). Here, we show that Cgvps34Δ cells are both poor biofilm formers (Fig. 12A) and mouse organ colonizers (Fig. 13). Intriguingly, in the presence of surplus iron, biofilm formation capacity of wild-type as well as Cgvps34Δ cells is significantly enhanced (Fig. 12B) indicating that high iron-induced biofilm formation can partially bypass the normal requirement for PI3K. Future investigations will be focused on examining the architecture of Cgvps34Δ biofilms made under normal and iron-sufficient conditions, and study their impact on survival in iron-poor and iron-rich host niches. Taken together, our data demonstrate that CgVps34 plays a pivotal role in regulation of iron homeostasis, biofilm-forming capacity, and organ colonization in vivo, and is indispensable for high iron-induced retrograde trafficking of CgFtr1 from the cell membrane.
Experimental Procedures
Strains, Media, and Growth Conditions
C. glabrata wild-type (wt) and mutant strains were derivatives of BG2 strain and routinely grown in rich YPD medium, CAA medium, or synthetically defined YNB medium at 30 °C with constant shaking at 200 rpm. Bacterial parental strains and strains carrying plasmids were grown in LB medium and LB medium containing ampicillin, respectively, at 37 °C with constant shaking at 200 rpm. To prepare media of different pH, YNB/YPD medium was buffered with HEPES and the desired pH was obtained by adding HCl or NaOH. Logarithmic (log)-phase cells were obtained by reinoculating the overnight grown culture in fresh medium for 4 h. To prepare iron-depleted and iron-surplus medium, BPS and ferric chloride (FeCl3) were added to the YNB medium to final concentrations of 50 and 500 μm, respectively. For gene expression analysis of iron uptake genes and activation of MAPK pathway proteins, log-phase grown cells were grown in iron-depleted and iron-surplus media for 2 h. For metal ion sensitivity studies, strain growth on solid medium was recorded, and results were presented as heat map using the Matrix2png program.
For CgFtr1-GFP and CgFet3-GFP localization studies, iron starvation was induced by culturing overnight CAA medium-grown cells in the BPS (50 μm)-supplemented CAA medium for 12 h, as described previously (49). For iron shock assays, iron-starved cells were washed once with CAA medium, suspended in CAA medium containing sodium ascorbate (1 mm) and ferrous ammonium sulfate (500 μm), and incubated at 30 °C for 2 h. Strains and plasmids used in this study are listed in Tables 1 and 2.
TABLE 1.
Strains used in the study
| Yeast strain | Genotype | Reference |
|---|---|---|
| YRK19 | ura3Δ::Tn903 G418R BG14 | 58 |
| YRK20 | URA3 BG462 | 59 |
| YRK662 | ura3Δ::Tn903 G418R Cgvps15Δ::nat1 | 12 |
| YRK695 | URA3 Cgvps15Δ::nat1 | 12 |
| YRK707 | ura3Δ::Tn903 G418R Cgvps34Δ::nat1 | 12 |
| YRK709 | URA3 Cgvps34Δ::nat1 | 12 |
| YRK714 | URA3 Cgftr1Δ::nat1 | 10 |
| YRK750 | URA3 Cgfet3Δ::nat1 | 10 |
| YRK843 | ura3Δ::Tn903 G418R Cgvps15Δ::nat1/pRK942 | 12 |
| YRK845 | ura3Δ::Tn903 G418R Cgvps34Δ::nat1/pRK984 | 12 |
| YRK875 | ura3Δ::Tn903 G418R Cgfet3Δ::nat1 | 10 |
| YRK931 | ura3Δ::Tn903 G418R Cgftr1Δ::nat1 | 10 |
| YRK964 | URA3 Cghog1Δ::nat1 | 10 |
| YRK1150 | ura3Δ::Tn903 G418R Cgvps34::nat1/pRK1037 | 12 |
| YRK1178 | ura3Δ::Tn903 G418R/pRK999 | Lab collection |
| YRK1251 | ura3Δ::Tn903 G418R/pRK1056 | Lab collection |
| YRK1255 | ura3Δ::Tn903 G418R/pRK1058 | Lab collection |
| YRK1328 | ura3Δ::Tn903 G418R Cgvps34Δ::nat1/pRK1018 | This study |
| YRK1385 | ura3Δ::Tn903 G418R/pRK1018 | This study |
| YRK1387 | ura3Δ::Tn903 G418R/pRK1100 | This study |
| YRK1389 | ura3Δ::Tn903 G418R/pRK1102 | This study |
| YRK1393 | ura3Δ::Tn903 G418R Cgvps34Δ::nat1/pRK1100 | This study |
| YEK1395 | ura3Δ::Tn903 G418R Cgvps34Δ::nat1/pRK1102 | This study |
| YRK1421 | ura3Δ::Tn903 G418R Cgftr1Δ::nat1/pRK1018 | This study |
| YRK1423 | ura3Δ::Tn903 G418R Cgftr1Δ::nat1/pRK1100 | This study |
| YRK1425 | ura3Δ::Tn903 G418R Cgfet3Δ::nat1/pRK1018 | This study |
| YRK1427 | ura3Δ::Tn903 G418R Cgfet3Δ::nat1/pRK1102 | This study |
| YRK1482 | ura3Δ::Tn903 G418R Cgvps34Δ::nat1/pRK1056 | This study |
| YRK1486 | ura3Δ::Tn903 G418R Cgvps34Δ::nat1/pRK1058 | This study |
| YRK1488 | ura3Δ::Tn903 G418R Cgvps34Δ::nat1/pRK999 | This study |
| YRK1589 | ura3Δ::Tn903 G418R Cgfet3Δ::nat1/pRK1100 | This study |
| YRK1591 | ura3Δ::Tn903 G418R Cgftr1Δ::nat1/pRK1102 | This study |
TABLE 2.
Plasmids used in the study
| Plasmid | Description | Reference |
|---|---|---|
| pRK74 | A CEN-ARS plasmid (pGRB2.2) of C. glabrata carrying S. cerevisiae URA3 as a selection marker. MCS sites are flanked by S. cerevisiae PGK1 promoter at one end and by 3′ UTR of HIS3 at the other end | 60 |
| pRK942 | CgVPS15 ORF (3.4 kb) cloned in XmaI-XhoI sites in the pRK74 plasmid | 12 |
| pRK984 | CgVPS34 ORF (2.4 kb) cloned in SalI-XmaI sites in the pRK74 plasmid | 12 |
| pRK999 | A CEN-ARS plasmid (Addgene-ID 45323) of C. glabrata carrying S. cerevisiae URA3 as a selection marker. MCS sites are flanked by S. cerevisiae PDC1 promoter at one end and by 3′ UTR of HIS3 at the other end | 61 |
| pRK1018 | A CEN-ARS plasmid (Addgene-ID 45343) of C. glabrata carrying S. cerevisiae URA3 as a selection marker. MCS sites are flanked by S. cerevisiae PGK1 promoter at one end and by 3′ UTR of HIS3 at the other end. yEGFP is introduced at the C terminus | 61 |
| pRK1037 | CgVPS34N750K (2.4 kb) cloned in SalI-XmaI sites in the pRK74 plasmid | 12 |
| pRK1056 | CgMRS4 ORF (0.92 kb) cloned in XmaI-XhoI sites in the pRK999 plasmid | Lab collection |
| pRK1058 | CgCCC1 ORF (0.92 kb) cloned in BamHI-SalI sites in the pRK999 plasmid | Lab collection |
| pRK1100 | CgFTR1 ORF (1.22 kb) cloned in XbaI-XmaI sites in the pRK1018 plasmid | This study |
| pRK1102 | CgFET3 ORF (1.90 kb) cloned in XbaI-XmaI sites in the pRK1018 plasmid | This study |
RNA Sequencing Analysis
The RNA sequencing experiment was performed with two biological replicates. Briefly, YPD-grown log-phase wt and Cgvps34Δ cells were collected and washed twice with chilled diethyl pyrocarbonate-treated water. RNA was extracted using the acid-phenol method, and RNA samples were sent to Genotypic Technology's genomic facility, Bangalore, India. Library preparation was performed at Genotypic Technology's Genomics facility following Illumina TruSeq RNA library protocol outlined in “TruSeq RNA Sample Preparation Guide” (part 15008136; Rev. A; Nov. 2010). Briefly, 1 μg of total RNA was subjected to poly(A) purification of mRNA. Purified mRNA was fragmented for 2 min at 94 °C in the presence of divalent cations and reverse transcribed using the SuperScript III reverse transcriptase and random hexamers. Second strand cDNA was synthesized in the presence of DNA polymerase I and RNase H. Illumina Adapters were ligated to cDNA molecules, and the library was amplified using 8 cycles of PCR for enrichment of adapter-ligated fragments. Furthermore, 76-bp paired end reads were produced with Illumina NextSeq500 and quality-checked using FastQC. Raw reads were used to identify differentially expressed genes (greater than 1.5-fold change and a p value of ≤0.05) with the help of CUFFLINKS (55) using the Candida genome database as the reference genome. The raw RNA-Seq datasets have been deposited in the NCBI-SRA database (submission: SUB1954295; SRA: SRP090201; BioProject: PRJNA343588).
Measurement of Intracellular Metal Ion Levels
Intracellular metal ion levels were measured using inductively coupled plasma-mass spectrometry analysis. Briefly, YPD-grown log phase cells (total A600 = 50) were harvested and washed twice with MilliQ water. Washed cells were suspended in 500 μl of 3% nitric acid and lysed at 96 °C for 16 h. The cell debris was removed by centrifugation, and intracellular metal ion levels were determined in the supernatant at the National Centre for Compositional Characterization of Material, Bhabha Atomic Research Centre, Hyderabad.
Determination of Mitochondrial Aconitase Activity and Mitochondrial Iron Levels
Mitochondrial extract preparation and aconitase activity measurement were performed as described previously (56, 57). Briefly, cells were grown to exponential phase for 6 h in YPD medium, washed with water, and collected. The crude mitochondrial extracts were prepared by lysing cells with glass beads, and protein amounts in extracts were quantified using the BCA kit. 5 μg of protein samples diluted in KH2PO4 buffer (25 mm, pH 7.2) containing Triton X-100 (0.05%) were incubated with isocitrate dehydrogenase (1 unit ml−1), NADP (0.2 mm), sodium citrate (1 mm), and MnCl2 (0.6 mm). The rate of NADP reduction by isocitrate dehydrogenase, as a readout of aconitase activity, was measured spectrophotometrically at 340 nm for 40 min. For estimation of iron levels in the mitochondria, crude mitochondrial fractions were subjected to a four-step sucrose gradient centrifugation (60, 32, 23, and 15% sucrose gradient), and pure mitochondrial fractions were collected at the 32–60% sucrose gradient interface. Isolated pure mitochondrial samples (200 μg of protein) were incubated with Tris-Cl (100 mm, pH 7.4) containing SDS (0.6%), dithionite (20 mm), and BPS (10 mm) for 5 min at 20 °C. The membrane debris was removed by centrifugation, and the absorption spectrum of BPS-bound iron was recorded between 500 and 700 nm wavelength. The difference between A540 (BPS-iron specific) and A700 (nonspecific) was used to determine the iron levels.
p-Phenylenediamine Oxidase Activity Assay
YPD-grown log-phase cells were collected and washed twice with MilliQ water. Cells corresponding to 1.0 A600 were incubated with 0.2% p-phenylenediamine (p-PD) in the assay buffer (100 mm sodium acetate, pH 5.3) at room temperature for 15 min, and increase in the absorbance, reflective of p-PD oxidation, was recorded spectrophotometrically at 570 nm.
Cloning of C. glabrata ORFs
CgFTR1 (CAGL0I06743g, 1.2 kb) and CgFET3 (CAGL0F06413g, 1.9 kb) ORFs were PCR amplified from genomic DNA of the wt strain using the Phusion High-Fidelity DNA polymerase, and cloned downstream of the PGK1 promoter in the XmaI and XbaI restriction sites in the pGRB2.3 plasmid containing GFP at the C terminus. The functionality of GFP fusion proteins was verified by mutant complementation assays. Sequence of primers used are available upon request.
Microscopy
For microscopic observations, the point scanning confocal system (Zeiss LSM 700) equipped with ×63 and ×100/1.44 NA objective was used. For staining of the mitochondria in C. glabrata, MitoTracker Green FM stain was used. YPD-grown log-phase cells were collected, washed, and suspended in HEPES buffer (10 mm, pH 7.4) containing glucose (5%) and MitoTracker Green FM (100 nm). After 30 min incubation in dark at room temperature, cells were pelleted down, washed, suspended in HEPES buffer, and visualized under the confocal microscope.
For localization studies of CgFtr1-GFP in wt and the Cgvps34Δ mutant in iron-starved and iron shock conditions, confocal microscopy was performed on live cells. For ferritin staining, phorbol 12-myristate 13-acetate-activated THP-1 cells were treated with BPS and FeCl3 in a 4-chambered glass slide for 24 h. Cells were washed twice with PBS and incubated with formaldehyde (3.7%) for 15 min at room temperature. Cells were again washed with PBS, permeabilized with Triton-X (0.1%) for 5 min, and incubated in blocking buffer (2% bovine serum albumin in PBS). After 1 h, slides were probed with anti-ferritin primary antibody followed by anti-rabbit Alexa Fluor-568 secondary antibody. For DAPI staining, Vectashield mounting medium containing DAPI was used and cells were visualized under the confocal microscope. All imaging was done at room temperature. Image acquisition and analysis were done using the ZEN software.
Biofilm Formation
YPD-grown log-phase C. glabrata cells were collected and washed twice with sterile PBS. Cells (0.5 A600) were seeded in a 24-well polystyrene plate and incubated at 37 °C for 90 min with constant shaking at 70 rpm. Two gentle PBS washes were given to each well and incubated in three different media, (i) serum-free RPMI 1640 containing BPS (100 μm); (ii) RPMI 1640 with FBS (10%), and (iii) RPMI 1640 with FBS (10%) and FeCl3 (500 μm), was continued for another 48 h with replacement of 500 μl of spent medium with the respective fresh RPMI medium at 24 h. Unbound C. glabrata cells were removed with PBS washes, plates were air-dried, and wells containing adherent, biofilm-forming yeast cells were stained with crystal violet solution (0.4% (w/v) in 20% ethanol). After 45 min, C. glabrata biofilms were washed four times with MilliQ water, destained with 95% ethanol, and absorbance of the destaining solution was recorded at 595 nm. Absorbance values of wells without C. glabrata cells were subtracted from those of yeast-containing wells, and the biofilm ratio was calculated by dividing the mutant absorbance units by those of wt cells.
Immunoblot Analysis
Western blot analysis was performed with C. glabrata cells and THP-1 cells following standard protocols. To detect CgSlt2 phosphorylation, CgHog1 phosphorylation, and CgGapdh levels, log-phase cells grown in iron-depleted and iron-surplus media for 2 h were collected and washed. The protein was extracted using the homogenizing buffer (50 mm Tris, pH 7.5, 2 mm EDTA) containing 1 mm phenylmethylsulfonyl fluoride, 10 mm sodium fluoride, 1 mm sodium orthovanadate, and protease inhibitor mixture by glass bead lysis method. Furthermore, an equal amount of protein was loaded on 10% SDS-PAGE and blots were probed with anti-phospho-p44/42 MAP kinase (Thr202/Tyr204; Cell Signaling Technology number 4370S), anti-phospho-p38 MAP kinase (Thr180/Tyr182; Cell Signaling Technology number 4511S), and anti-Gapdh (Abcam number ab22555) antibodies.
To detect ferritin levels in THP-1 cells, cells were treated with BPS and FeCl3 for 24 h as described previously (37). Next, cells were lysed in NETN buffer (20 mm Tris-HCl, pH 8.0, 100 mm NaCl, 1 mm EDTA, 0.5% Nonidet P-40) containing 50 mm β-glycerophosphate, 10 mm NaF, and 1× protease inhibitor on ice for 30 min. An equal amount of proteins was loaded on 15% SDS-PAGE and blots were probed with anti-ferritin (Abcam number ab7332) and anti-Gapdh (Abcam number ab22555) antibodies. ImageJ software (National Institutes of Health, Bethesda, MD) was used to quantify the band intensities and all data were normalized to CgGapdh protein levels.
Mice Infection Assay
Mice infection experiments were performed at the CDFD animal facility, VIMTA Labs Limited, Hyderabad, India, in accordance with guidelines of the Committee for the Purpose of Control and Supervision of Experiments on Animals, Government of India. The protocol was approved by the Institutional Animal Ethics Committee of the Vimta Labs Ltd. Procedures used in this protocol were designed to minimize animal suffering. Groups of 6–8-week-old female BALB/c mice were injected with YPD medium-grown C. glabrata cells (4 × 107; 100 μl of PBS cell suspension) through the tail vein, sacrificed 1, 3, and 5 days post-infection and three organs (kidneys, liver, and spleen) were harvested. Fungal burden in murine organs was determined by plating appropriate dilutions of tissue homogenates on YPD medium supplemented with penicillin and streptomycin antibiotics.
Other Procedures
qPCR, THP-1 macrophage infection, intracellular ROS determination, ATP level determination, and serial dilution spotting assay were performed as described previously (12, 29, 35).
Statistical Analysis
Statistical analysis was performed using the GraphPad Prism software. Organ fungal burdens were analyzed using the non-parametric Mann-Whitney test. Intergroup comparisons were made using the two-tailed Student's t test.
Author Contributions
V. S. and R. K. conceived and designed the study. V. S. and R. P. acquired and analyzed the data. V. S. and R. K. wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Kavela Sridhar and Jayant Pundalikrao Hole, and V. Joy Prashant and A. Lakshmi Annapurna, for their help with animal experiments, and confocal microscopy, respectively. We are grateful to Brendan Cormack for the gift of C. glabrata deletion strain library, and Vivek K. Srivastava for the mutant screen.
This work was supported in part by Department of Biotechnology, Government of India Grants BT/PR5145/MED/29/470/2012, BT/PR7388/MED/29/650/2012, and BT/PR7123/BRB/10/1149/2012 (to R. K.). The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Table S1.
- PI3P
- phosphatidylinositol 3-phosphate
- ROS
- reactive oxygen species
- HOG
- high osmolarity glycerol
- GO
- gene ontology
- qRT
- quantitative RT
- ICP-AES
- inductively coupled plasma atomic emission spectrometry
- BPS
- bathophenanthrolinedisulfonic acid
- TMA-DPH
- trimethylamino-diphenyl-1,3,5-hexatriene
- p-PD
- p-phenylenediamine.
References
- 1. Pfaller M. A., Pappas P. G., and Wingard J. R. (2006) Invasive fungal pathogens: current epidemiological trends. Clin. Infect. Dis. 43, S3–S14 [Google Scholar]
- 2. Brown G. D., Denning D. W., Gow N. A., Levitz S. M., Netea M. G., and White T. C. (2012) Hidden killers: human fungal infections. Science Translat. Med. 4, 165rv113. [DOI] [PubMed] [Google Scholar]
- 3. Pfaller M. A., and Diekema D. J. (2007) Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 20, 133–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pfaller M., Neofytos D., Diekema D., Azie N., Meier-Kriesche H. U., Quan S. P., and Horn D. (2012) Epidemiology and outcomes of candidemia in 3648 patients: data from the Prospective Antifungal Therapy (PATH Alliance(R)) registry, 2004–2008. Diagn. Microbiol. Infect. Dis. 74, 323–331 [DOI] [PubMed] [Google Scholar]
- 5. Lortholary O., Renaudat C., Sitbon K., Madec Y., Denoeud-Ndam L., Wolff M., Fontanet A., Bretagne S., Dromer F., and French Mycosis Study Group (2014) Worrisome trends in incidence and mortality of candidemia in intensive care units (Paris area, 2002–2010). Intensive Care Med. 40, 1303–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fidel P. L. Jr, Vazquez J. A., and Sobel J. D. (1999) Candida glabrata: review of epidemiology, pathogenesis, and clinical disease with comparison to C. albicans. Clin. Microbiol. Rev. 12, 80–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tortorano A. M., Dho G., Prigitano A., Breda G., Grancini A., Emmi V., Cavanna C., Marino G., Morero S., Ossi C., Delvecchio G., Passera M., Cusumano V., David A., Bonaccorso G., et al. (2012) Invasive fungal infections in the intensive care unit: a multicentre, prospective, observational study in Italy (2006–2008). Mycoses 55, 73–79 [DOI] [PubMed] [Google Scholar]
- 8. Kaur R., Domergue R., Zupancic M. L., and Cormack B. P. (2005) A yeast by any other name: Candida glabrata and its interaction with the host. Curr. Opin. Microbiol. 8, 378–384 [DOI] [PubMed] [Google Scholar]
- 9. Roetzer A., Gabaldón T., and Schüller C. (2011) From Saccharomyces cerevisiae to Candida glabrata in a few easy steps: important adaptations for an opportunistic pathogen. FEMS Microbiol. Lett. 314, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Srivastava V. K., Suneetha K. J., and Kaur R. (2014) A systematic analysis reveals an essential role for high-affinity iron uptake system, haemolysin and CFEM domain-containing protein in iron homoeostasis and virulence in Candida glabrata. Biochem. J. 463, 103–114 [DOI] [PubMed] [Google Scholar]
- 11. Gabaldón T., and Carreté L. (2016) The birth of a deadly yeast: tracing the evolutionary emergence of virulence traits in Candida glabrata. FEMS Yeast Res. 16, fov110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rai M. N., Sharma V., Balusu S., and Kaur R. (2015) An essential role for phosphatidylinositol 3-kinase in the inhibition of phagosomal maturation, intracellular survival and virulence in Candida glabrata. Cell. Microbiol. 17, 269–287 [DOI] [PubMed] [Google Scholar]
- 13. Katso R., Okkenhaug K., Ahmadi K., White S., Timms J., and Waterfield M. D. (2001) Cellular function of phosphoinositide 3-kinases: implications for development, homeostasis, and cancer. Annu. Rev. Cell Dev. Biol. 17, 615–675 [DOI] [PubMed] [Google Scholar]
- 14. Jean S., and Kiger A. A. (2014) Classes of phosphoinositide 3-kinases at a glance. J. Cell Sci. 127, 923–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vanhaesebroeck B., Stephens L., and Hawkins P. (2012) PI3K signalling: the path to discovery and understanding. Nat. Rev. Mol. Cell Biol. 13, 195–203 [DOI] [PubMed] [Google Scholar]
- 16. Wang J., and Pantopoulos K. (2011) Regulation of cellular iron metabolism. Biochem. J. 434, 365–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hood M. I., and Skaar E. P. (2012) Nutritional immunity: transition metals at the pathogen-host interface. Nat. Rev. Microbiol. 10, 525–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nevitt T., and Thiele D. J. (2011) Host iron withholding demands siderophore utilization for Candida glabrata to survive macrophage killing. PLoS Pathog. 7, e1001322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Srivastava V. K., Suneetha K. J., and Kaur R. (2015) The mitogen-activated protein kinase CgHog1 is required for iron homeostasis, adherence and virulence in Candida glabrata. FEBS J. 282, 2142–2166 [DOI] [PubMed] [Google Scholar]
- 20. Ramírez M. A., and Lorenz M. C. (2007) Mutations in alternative carbon utilization pathways in Candida albicans attenuate virulence and confer pleiotropic phenotypes. Eukaryotic Cell 6, 280–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shingu-Vazquez M., and Traven A. (2011) Mitochondria and fungal pathogenesis: drug tolerance, virulence, and potential for antifungal therapy. Eukaryotic Cell 10, 1376–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Klionsky D. J., Herman P. K., and Emr S. D. (1990) The fungal vacuole: composition, function, and biogenesis. Microbiol. Rev. 54, 266–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dimmer K. S., Fritz S., Fuchs F., Messerschmitt M., Weinbach N., Neupert W., and Westermann B. (2002) Genetic basis of mitochondrial function and morphology in Saccharomyces cerevisiae. Mol. Biol. Cell 13, 847–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li S. C., and Kane P. M. (2009) The yeast lysosome-like vacuole: endpoint and crossroads. Biochim. Biophys. Acta 1793, 650–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. De Freitas J., Wintz H., Kim J. H., Poynton H., Fox T., and Vulpe C. (2003) Yeast, a model organism for iron and copper metabolism studies. Biometals 16, 185–197 [DOI] [PubMed] [Google Scholar]
- 26. Cyert M. S., and Philpott C. C. (2013) Regulation of cation balance in Saccharomyces cerevisiae. Genetics 193, 677–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sutak R., Lesuisse E., Tachezy J., and Richardson D. R. (2008) Crusade for iron: iron uptake in unicellular eukaryotes and its significance for virulence. Trends Microbiol. 16, 261–268 [DOI] [PubMed] [Google Scholar]
- 28. Lill R., Hoffmann B., Molik S., Pierik A. J., Rietzschel N., Stehling O., Uzarska M. A., Webert H., Wilbrecht C., and Mühlenhoff U. (2012) The role of mitochondria in cellular iron-sulfur protein biogenesis and iron metabolism. Biochim. Biophys. Acta 1823, 1491–1508 [DOI] [PubMed] [Google Scholar]
- 29. Bairwa G., Rasheed M., Taigwal R., Sahoo R., and Kaur R. (2014) GPI (glycosylphosphatidylinositol)-linked aspartyl proteases regulate vacuole homoeostasis in Candida glabrata. Biochem. J. 458, 323–334 [DOI] [PubMed] [Google Scholar]
- 30. Dix D. R., Bridgham J. T., Broderius M. A., Byersdorfer C. A., and Eide D. J. (1994) The FET4 gene encodes the low affinity Fe(II) transport protein of Saccharomyces cerevisiae. J. Biol. Chem. 269, 26092–26099 [PubMed] [Google Scholar]
- 31. Martínez C. E., and Motto H. L. (2000) Solubility of lead, zinc and copper added to mineral soils. Environ. Pollut. 107, 153–158 [DOI] [PubMed] [Google Scholar]
- 32. Serrano R., Bernal D., Simón E., and Ariño J. (2004) Copper and iron are the limiting factors for growth of the yeast Saccharomyces cerevisiae in an alkaline environment. J. Biol. Chem. 279, 19698–19704 [DOI] [PubMed] [Google Scholar]
- 33. Stearman R., Yuan D. S., Yamaguchi-Iwai Y., Klausner R. D., and Dancis A. (1996) A permease-oxidase complex involved in high-affinity iron uptake in yeast. Science 271, 1552–1557 [DOI] [PubMed] [Google Scholar]
- 34. Abe F., and Hiraki T. (2009) Mechanistic role of ergosterol in membrane rigidity and cycloheximide resistance in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1788, 743–752 [DOI] [PubMed] [Google Scholar]
- 35. Rai M. N., Balusu S., Gorityala N., Dandu L., and Kaur R. (2012) Functional genomic analysis of Candida glabrata-macrophage interaction: role of chromatin remodeling in virulence. PLoS Pathog. 8, e1002863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Weiss G., and Schaible U. E. (2015) Macrophage defense mechanisms against intracellular bacteria. Immunol. Rev. 264, 182–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Almeida R. S., Brunke S., Albrecht A., Thewes S., Laue M., Edwards J. E., Filler S. G., and Hube B. (2008) the hyphal-associated adhesin and invasin Als3 of Candida albicans mediates iron acquisition from host ferritin. PLoS Pathog. 4, e1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Linder M. C. (2013) Mobilization of stored iron in mammals: a review. Nutrients 5, 4022–4050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Johnson M., Cockayne A., Williams P. H., and Morrissey J. A. (2005) Iron-responsive regulation of biofilm formation in Staphylococcus aureus involves fur-dependent and fur-independent mechanisms. J. Bacteriol. 187, 8211–8215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cassat J. E., and Skaar E. P. (2013) Iron in infection and immunity. Cell Host Microbe 13, 509–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ramsay L. M., and Gadd G. M. (1997) Mutants of Saccharomyces cerevisiae defective in vacuolar function confirm a role for the vacuole in toxic metal ion detoxification. FEMS Microbiol. Lett. 152, 293–298 [DOI] [PubMed] [Google Scholar]
- 42. Van Ho A., Ward D. M., and Kaplan J. (2002) Transition metal transport in yeast. Annu. Rev. Microbiol. 56, 237–261 [DOI] [PubMed] [Google Scholar]
- 43. Kane P. M. (2007) The long physiological reach of the yeast vacuolar H+-ATPase. J. Bioenerg. Biomembr. 39, 415–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Askwith C., Eide D., Van Ho A., Bernard P. S., Li L., Davis-Kaplan S., Sipe D. M., and Kaplan J. (1994) The FET3 gene of S. cerevisiae encodes a multicopper oxidase required for ferrous iron uptake. Cell 76, 403–410 [DOI] [PubMed] [Google Scholar]
- 45. De Silva D. M., Askwith C. C., Eide D., and Kaplan J. (1995) The FET3 gene product required for high affinity iron transport in yeast is a cell surface ferroxidase. J. Biol. Chem. 270, 1098–1101 [DOI] [PubMed] [Google Scholar]
- 46. Felice M. R., De Domenico I., Li L., Ward D. M., Bartok B., Musci G., and Kaplan J. (2005) Post-transcriptional regulation of the yeast high affinity iron transport system. J. Biol. Chem. 280, 22181–22190 [DOI] [PubMed] [Google Scholar]
- 47. Singh A., Severance S., Kaur N., Wiltsie W., and Kosman D. J. (2006) Assembly, activation, and trafficking of the Fet3p.Ftr1p high affinity iron permease complex in Saccharomyces cerevisiae. J. Biol. Chem. 281, 13355–13364 [DOI] [PubMed] [Google Scholar]
- 48. Strochlic T. I., Setty T. G., Sitaram A., and Burd C. G. (2007) Grd19/Snx3p functions as a cargo-specific adapter for retromer-dependent endocytic recycling. J. Cell Biol. 177, 115–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Strochlic T. I., Schmiedekamp B. C., Lee J., Katzmann D. J., and Burd C. G. (2008) Opposing activities of the Snx3-retromer complex and ESCRT proteins mediate regulated cargo sorting at a common endosome. Mol. Biol. Cell 19, 4694–4706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Burda P., Padilla S. M., Sarkar S., and Emr S. D. (2002) Retromer function in endosome-to-Golgi retrograde transport is regulated by the yeast Vps34 PtdIns 3-kinase. J. Cell Sci. 115, 3889–3900 [DOI] [PubMed] [Google Scholar]
- 51. Schwarzmüller T., Ma B., Hiller E., Istel F., Tscherner M., Brunke S., Ames L., Firon A., Green B., Cabral V., Marcet-Houben M., Jacobsen I. D., Quintin J., Seider K., Frohner I., et al. (2014) Systematic phenotyping of a large-scale Candida glabrata deletion collection reveals novel antifungal tolerance genes. PLoS Pathog. 10, e1004211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Noble S. M. (2013) Candida albicans specializations for iron homeostasis: from commensalism to virulence. Curr. Opin. Microbiol. 16, 708–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rodrigues C. F., Silva S., and Henriques M. (2014) Candida glabrata: a review of its features and resistance. Eur. J. Clin. Microbiol. Infect. Dis. 33, 673–688 [DOI] [PubMed] [Google Scholar]
- 54. Fanning S., and Mitchell A. P. (2012) Fungal biofilms. PLoS Pathog. 8, e1002585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G., van Baren M. J., Salzberg S. L., Wold B. J., and Pachter L. (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bulteau A. L., Ikeda-Saito M., and Szweda L. I. (2003) Redox-dependent modulation of aconitase activity in intact mitochondria. Biochemistry 42, 14846–14855 [DOI] [PubMed] [Google Scholar]
- 57. Meisinger C., Pfanner N., and Truscott K. N. (2006) Isolation of yeast mitochondria. Methods Mol. Biol. 313, 33–39 [DOI] [PubMed] [Google Scholar]
- 58. Cormack B. P., Ghori N., and Falkow S. (1999) An adhesin of the yeast pathogen Candida glabrata mediating adherence to human epithelial cells. Science 285, 578–582 [DOI] [PubMed] [Google Scholar]
- 59. De Las Peñas A., Pan S. J., Castaño I., Alder J., Cregg R., and Cormack B. P. (2003) Virulence-related surface glycoproteins in the yeast pathogen Candida glabrata are encoded in subtelomeric clusters and subject to RAP1- and SIR-dependent transcriptional silencing. Genes Dev. 17, 2245–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Frieman M. B., McCaffery J. M., and Cormack B. P. (2002) Modular domain structure in the Candida glabrata adhesin Epa1p, a β1,6-glucan-cross-linked cell wall protein. Mol. Microbiol. 46, 479–492 [DOI] [PubMed] [Google Scholar]
- 61. Zordan R. E., Ren Y., Pan S. J., Rotondo G., De Las Peñas A., Iluore J., and Cormack B. P. (2013) Expression plasmids for use in Candida glabrata. G3 3, 1675–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.