Abstract
The acquisition of beige adipocyte features by white fat cells corresponds to protection against obesity-induced metabolic diseases in humans and animal models of type 2 diabetes. In adipose tissue, expression of the E2 small ubiquitin-like modifier ligase ubiquitin carrier protein 9 (Ubc9) is positively correlated with markers of insulin resistance and corresponds with impaired browning of human white adipocytes. However, the molecular regulation of Ubc9 expression in adipocytes and other cells remains unclear. In this study, we demonstrate that the mRNA and protein expression of Ubc9 are regulated by the microRNA miRNA-30a (miR-30a) in human subcutaneous adipocytes. Ubc9 and miR-30a exhibit inverse expression in adipose tissue, with miR-30a robustly elevated in brown fat. Depletion of Ubc9 by siRNA or enforced expression of a miR-30a mimic augments mitochondrial volume and respiration in human white adipocytes, reflecting features of brown fat cells. Furthermore, Ubc9 depletion induces a brown fat gene program in human subcutaneous adipocytes. Induction of the beige-selective gene program corresponds to stabilization of the PR domain-containing 16 (PRDM16) protein, an obligate transcriptional regulator of the brown/beige fat metabolic program in white adipocytes that interacts with Ubc9. Taken together, our data demonstrate a previously unappreciated molecular axis that controls browning of human white adipocytes.
Keywords: adipose tissue metabolism, bioenergetics, gene transcription, microRNA (miRNA), mitochondria, posttranscriptional regulation, respiration, tissue-specific transcription factor, transcription coregulator, transcription target gene
Introduction
White adipose tissue (WAT)2 mediates whole-body metabolic homeostasis by providing flexible energy storage in the form of lipids. Visceral WAT expansion and ectopic lipid accumulation in the liver, heart, and skeletal muscle are significant risk factors for insulin resistance, type 2 diabetes mellitus (T2DM), and cardiovascular disease (1). Conversely, lipid deposition in subcutaneous WAT is correlated with protection against metabolic dysfunction (2). Thus, factors that promote the sequestration of energy in subcutaneous fat may improve metabolic health.
Many studies demonstrate that subcutaneous WAT can metabolize stored energy in brown-like (“beige”) adipocytes to contribute to overall energy expenditure (3). These beige adipocytes express uncoupling protein 1 (UCP1) and display increased mitochondrial function and oxygen consumption, indicating increased lipid catabolism. Enhancing the thermogenic gene programs in subcutaneous WAT mediated by beige adipocytes protects against T2DM and associated comorbidities, including hepatic steatosis (4). Other evidence has firmly established that human brown fat volume is inversely correlated with body mass index (5) and whole-body energy expenditure (6), supporting the notion that increasing brown fat activity in humans may combat insulin resistance and T2DM. Furthermore, recent studies report that human BAT displays a molecular characteristic resembling mouse beige fat (7–9), illuminating the importance of better understanding beige adipocyte development. Therefore, strategies for augmenting human brown and beige fat function are widely sought as therapeutic approaches for T2DM and its associated metabolic sequelae.
PGC1α, KLF11, EBF2, PRDM16, and other factors have been shown to enable white adipocytes to express a brown adipose tissue (BAT) gene program (10). However, PRDM16 is the only factor that is both necessary and sufficient for browning of white fat (4, 11, 12). Pharmacological stabilization of PRDM16 protein downstream of browning agents (11) is required to maximally induce the brown fat gene program in subcutaneous adipocytes. At the molecular level, increased accumulation of PRDM16 protein through reduced ubiquitin-proteasome-mediated protein degradation acts in concert with full agonist activation of PPARγ to directly stimulate the brown adipocyte gene cassette in white adipocytes. The molecular mechanism of PRDM16 protein accumulation remains elusive.
We recently identified the E2 SUMO ligase Ubc9 as a factor that represses acquisition of brown fat features in human adipocytes (13). Ubc9 expression is increased in WAT isolated from mice and humans exhibiting insulin resistance, which motivated our studies to investigate the regulation of Ubc9 in human adipocytes and whether endogenous modes of suppression play a role in browning of white fat. In this study, we report that the microRNA miR-30a-5p regulates Ubc9 expression to regulate browning of human white fat cells. We show that miR-30a selectively targets Ubc9 in human adipocytes, leading to elevated mitochondrial activity and acquisition of brown fat features. Finally, depletion of Ubc9 by siRNA or miR-30a expression stabilizes PRDM16 protein levels and promotes genomic occupancy of PPARγ on regulatory regions of PRDM16-dependent brown adipocyte genes. Overall, our results demonstrate that the Ubc9/miR-30a axis plays a critical role in browning of human white fat cells and potentially represents a node to therapeutically stimulate human beige adipocyte recruitment for the treatment of metabolic diseases.
Results
miR-30a Regulates Ubc9 Expression in Human Adipocytes
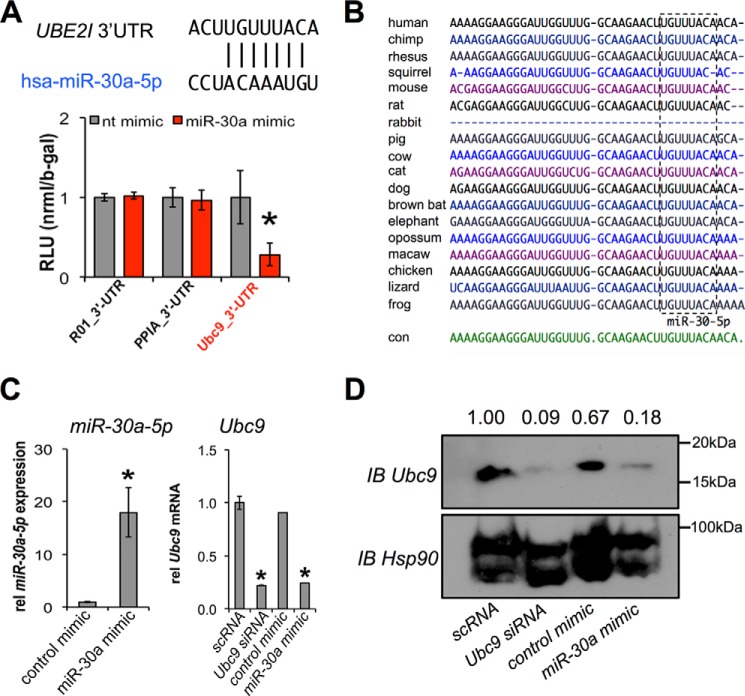
Our previous studies established Ubc9 as a regulator of the brown adipocyte gene program in human adipocytes (13). Analysis of regions upstream of the Ubc9 gene failed to identify any typical modes of transcriptional regulation. To investigate the potential posttranscriptional regulation of Ubc9, we used TargetScan (14) and starBase (15) to identify microRNAs (miRNAs) that might interact with the 3′ UTR of the human Ubc9 (UBE2I) gene. In line with previous findings (16), TargetScan and starBase analyses scored miR-30a-5p (miR-30a) as the top miRNA targeting the Ubc9 3′ UTR. We then demonstrated miR-30a binding to the Ubc9 3′ UTR by transiently co-expressing various luciferase reporter fusions of Ubc9 and miRNA mimics in human adipocytes. Results from co-transfection experiments indicated that the relative luciferase activity in Ubc9 3′ UTR-expressing cells was significantly inhibited by miR-30a, whereas other control 3′ UTR fusions that do not contain relevant miRNA binding sites were unaffected (Fig. 1A). TargetScan analysis (14) showed that miR-30a binding to the Ubc9 3′ UTR was evolutionarily conserved (Fig. 1B), which strengthened the possibility of a functional interaction. Consistent with these findings, miR-30a overexpression in human adipocytes effectively decreased endogenous Ubc9 mRNA (Fig. 1C) and protein expression (Fig. 1D). These results demonstrate that miR-30a directly targets and represses Ubc9 in adipocytes.
FIGURE 1.
Ubc9 is a direct target of miR-30a. A, TargetScan and starBase predict that miR-30a binds the 3′ UTR of UBE2I (Ubc9). Plasmids with negative control (R01), PPIA, and Ubc9 3′ UTR luciferase fusions were co-transfected with a miR-30a mimic or control mimic in human adipocytes cells (n = 3; *, p < 0.05). RLU, relative light units; nt, non-targeting. B, the miR-30a putative binding site in the 3′ UTR of Ubc9 is conserved among species commonly queried in TargetScan, as shown by the sequence homology (box). con, control. C, human adipocytes were transfected with scrambled (scRNA) siRNA or Ubc9 siRNA and compared with expression of a control mimic or miR-30a mimic. qPCR was used to measure miR-30a-5p and Ubc9 relative (rel) gene expression normalized to TATA binding protein or sno412 RNA (n = 2; mean ± S.E.; *, p < 0.05). D, immunoblot (IB) analysis was used to measure the effects of siRNA and miRNA transient transfection on the protein expression of Ubc9.
miR-30a Inhibits Ubc9 Expression in Human Adipocytes
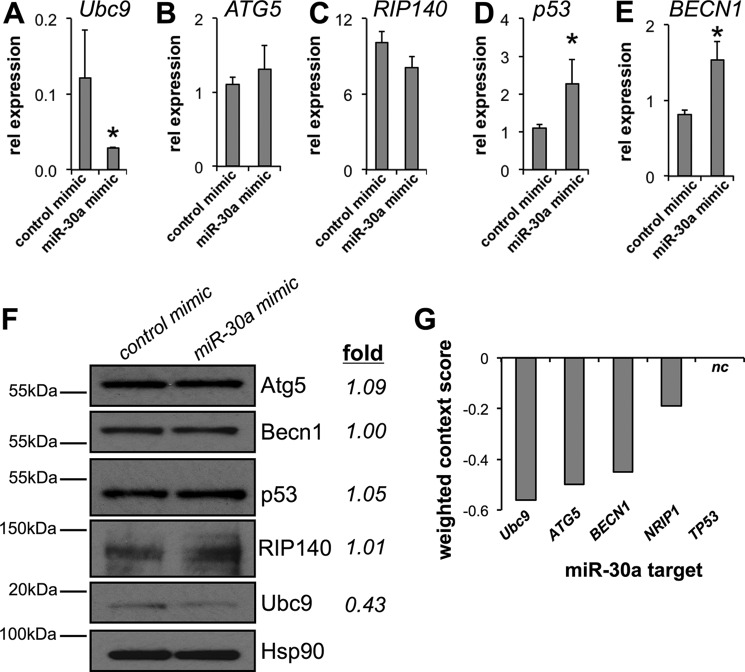
In addition to Ubc9 (16), the published target spectrum of miR-30a includes ATG5 (17), BECN1 (18), RIP140 (19), and p53 (20, 21). To experimentally determine the selectivity of miR-30a gene targeting, we transiently expressed a control or miR-30a mimic in human adipocytes. Expression of the miR-30a mimic significantly reduced Ubc9 mRNA (Fig. 2A), whereas ATG5 (Fig. 2B) and RIP140 (Fig. 2C) remained unchanged. Contrary to other studies, increased p53 (Fig. 2D) and BECN1 (Fig. 2E) gene expression was observed when miR-30a was overexpressed in human adipocytes. Immunoblotting demonstrated that Ubc9 protein was specifically reduced, whereas products of other candidate target genes were not affected by miR-30a overexpression (Fig. 2F). Lastly, the cumulative weighted context++ score (14) established that the Ubc9/miR-30a interaction was more favorable relative to other targets. The sum of these results reinforce the notion that miR-30a uniquely and directly inhibits Ubc9 expression in human adipocytes.
FIGURE 2.
miR-30a inhibits Ubc9 expression in human white adipocytes. A–E, the gene expression levels of published miR-30a targets were analyzed in human adipocytes transfected with control or miR-30a mimics for 48 h. mRNA levels of Ubc9 (A), ATG5 (B), RIP140 (C), p53 (D), and BECN1 (E) were analyzed by qPCR. rel, relative. F, immunoblotting verified exclusive suppression of Ubc9 expression using samples analyzed by qPCR. *, p < 0.05 relative to cells transfected with control mimic (n ≥ 3 independent experiments). G, TargetScan cumulative weighted context++ scores of published miR-30a targets suggest that the miR-30/Ubc9 interaction is favorable relative to other targets. More negative values indicate a stronger interaction potential. nc, not-called.
Expression of miR-30a and Ubc9 Is Inversely Correlated in BAT and WAT
WAT functions primarily to store energy, whereas BAT catabolizes free fatty acids and dissipates biochemical energy as heat. Although the precise developmental origin of each tissue is undefined in humans, studies performed in mice suggest that BAT and WAT arise from different lineages (10). Previous data suggested that miR-30a is one of 19 miRNAs enriched in mouse BAT (22), which motivated our analysis of miR-30a and Ubc9 expression in WAT, BAT, and skeletal muscle. Ubc9 was more highly expressed in WAT than in BAT, with miR-30a exhibiting the opposite pattern of expression (Fig. 3A). BAT also showed higher miR-30a expression compared with skeletal muscle (Fig. 3B). Summarizing both broad profiling experiments, miR-30a showed ∼12-fold higher expression in BAT compared with either WAT or skeletal muscle. The results in WAT and BAT suggest that Ubc9 and miR-30a expression may be reciprocally regulated in the context of fat cell function.
FIGURE 3.
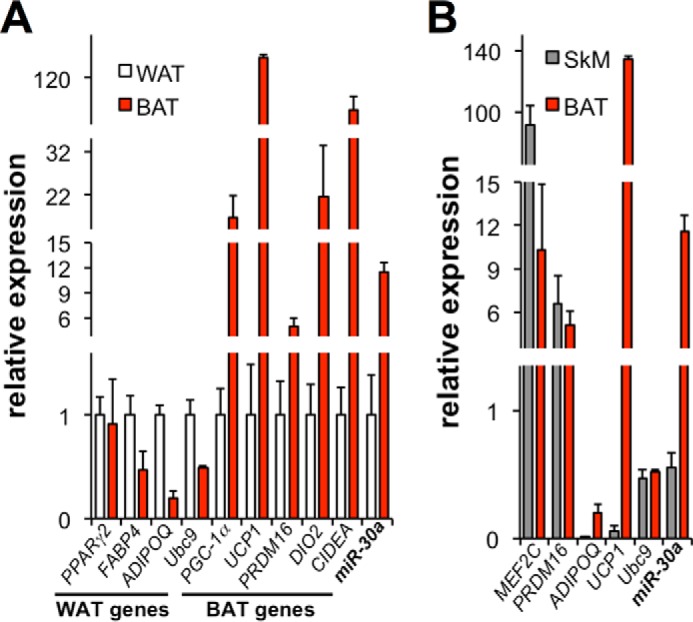
Reciprocal expression of Ubc9 and miR-30a in adipose tissues. A and B, qPCR was used to determine the expression of Ubc9, miR-30a, and marker genes from WAT and BAT (A) and skeletal muscle (SkM, B). Expression levels were normalized to TATA binding protein or U6 sno RNA (n ≥ 4 mice/group).
Manipulation of Ubc9 Expression by siRNA or a miR-30a Mimic Promotes Increased Mitochondrial Respiration
Under diverse stimuli, subcutaneous human white fat depots demonstrate substantial metabolic flexibility and are capable of acquiring features of brown fat, including increased mitochondrial biogenesis and oxidative metabolism (3). Ubc9 inhibits the acquisition of brown fat features, including mitochondriogenesis and fatty acid oxidation (13). Based on the inverse correlation between miR-30a and Ubc9 expression in adipose tissue and our recent studies positioning Ubc9 as a regulator of oxidative metabolism in human adipocytes, we hypothesized that miR-30a blocks Ubc9 expression to stimulate mitochondrial activity.
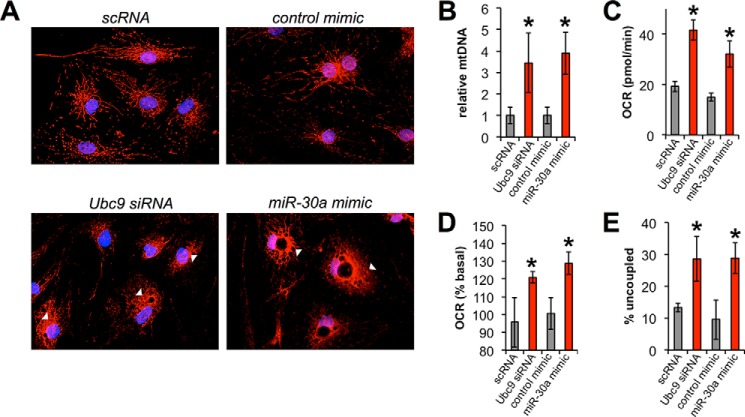
We tested our hypothesis by performing morphological and phenotypic analyses of mature human adipocytes transfected with Ubc9 siRNA, a miR-30a mimic, or appropriate controls in the presence of the browning agent rosiglitazone. We labeled mitochondria with MitoTracker to determine the effect of miR30a/Ubc9 signaling on mitochondrial content and morphology. Adipocytes transfected with Ubc9 siRNA or a miR-30a mimic showed more intense MitoTracker staining with less reticulated structure compared with controls (Fig. 4A, arrowheads). Analysis of MitoTracker staining by flow cytometry also revealed increased mitochondrial labeling in cells transfected with Ubc9 siRNA or a miR-30a mimic relative to control transfections (data not shown). To determine the effect of Ubc9 on cellular mitochondrial content, we measured total mitochondrial DNA in adipocytes following Ubc9 knockdown or miR-30a overexpression. Intriguingly, Ubc9 knockdown in human adipocytes using siRNA or a miR-30a mimic increased mtDNA content more than 3-fold (Fig. 4B). Consistent with increased mitochondrial biogenesis, knockdown of Ubc9 expression by siRNA or miR-30a mimic enhanced basal cellular respiration in adipocytes, as measured by oxygen consumption (Fig. 4C).
FIGURE 4.
The Ubc9/miR-30a axis remodels mitochondrial respiration in human white adipocytes. A, mitochondria (MitoTracker, red) and nuclei (DAPI, blue) were labeled in mature human adipocytes transfected with Ubc9 siRNA or a miR-30a mimic. Arrowheads indicate alterations in mitochondrial morphology between controls, Ubc9 siRNA, and miR-30a mimic transfections. B, mitochondrial DNA (ND6) was analyzed by qPCR from samples in A. *, p < 0.05 relative to control-transfected cells. C, basal respiration (as OCR) was measured in mature human adipocytes transfected with Ubc9 siRNA or a miR-30a mimic. D, respiration rate was measured after forskolin treatment to induce uncoupling. The percent change in OCR was normalized to baseline rates. E, percent uncoupling was calculated by subtracting the difference between oligomycin and rotenone. In this case, the OCR before oligomycin injection was set as 100%. Respiration data are presented as mean ± S.E.; n = 4 independent experiments; *, p < 0.05 relative to control transfections.
To establish a role for Ubc9 or miR-30a in mitochondrial uncoupling and thermogenesis, we measured the oxygen consumption rate (OCR) in response to cAMP elevation (23). Adipocytes transfected with Ubc9 siRNA or miR-30a mimic showed a 20% greater response to forskolin compared with nonspecific miRNA or siRNA controls (Fig. 4D). After subtraction of the non-mitochondrial OCR from the oligomycin-insensitive OCR, the percentage of uncoupling was significantly increased under conditions of Ubc9 siRNA or miR-30a overexpression (Fig. 4E). In sum, Ubc9 inhibition by siRNA or miR-30a transfection increases mitochondrial content and cellular respiratory capacity.
The PPARγ/PRDM16 Thermogenic Gene Program Is Regulated by the Ubc9/miR-30a Axis in Human Adipocytes
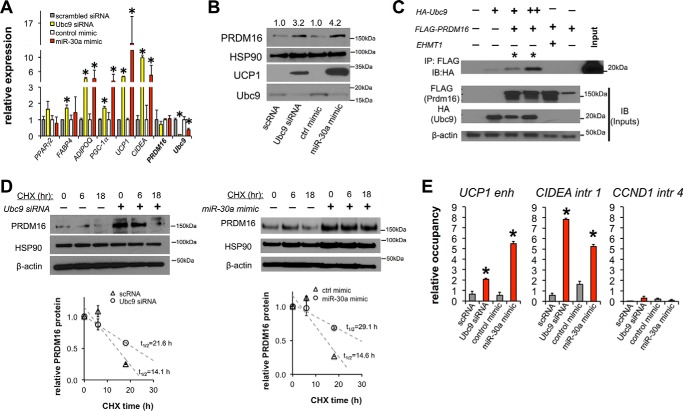
Based on the observation that Ubc9 siRNA or miR-30a mimic transfection increased mitochondrial activity relative to control conditions, we hypothesized that the Ubc9/miR-30a axis would induce thermogenic gene expression in human adipocytes. To test this hypothesis, we transfected mature human adipocytes with Ubc9 siRNA or a miR-30a mimic. Ubc9 knockdown or miR-30a mimic (Fig. 5A) transfections produced a similar effect on thermogenic genes, including significant induction of genes preferentially expressed in BAT (UCP1, PGC-1α, and CIDEA). Notably, induction of UCP1 was selectively enhanced over classic white adipocyte genes such as FABP4, suggesting that the effects of Ubc9 siRNA or miR-30a mimic were not due to primary effects on adipogenesis. Subsequent immunoblot analysis confirmed that Ubc9 knockdown by siRNA or miR-30a mimic increased both PRDM16 and UCP1 protein levels (Fig. 5B). Previous data established that PRMD16 and PPARγ interact to promote expression of thermogenic genes in subcutaneous adipocytes (11, 24). These data, combined with our recent observation that Ubc9 interacts with PPARγ (13), suggested that Ubc9 may interact with PRDM16 as part of a complex. As shown in Fig. 5C, immunoprecipitation of FLAG-PRDM16 and immunoblotting for HA-Ubc9 showed co-precipitation of PRDM16 with Ubc9, confirming that these proteins associate in a common complex.
FIGURE 5.
PRDM16 stabilization and browning of human white adipocytes are regulated by the Ubc9/miR-30a axis. Ubc9 siRNA, miR-30a, or transfection controls were introduced into mature human adipocytes. A, the effects of Ubc9 siRNA and miR-30a overexpression in human adipocytes were characterized by qPCR analysis of PPARγ2, FABP4, ADIPOQ, PGC1α, UCP1, CIDEA, PRDM16, and Ubc9 mRNA levels (n ≥ 3 independent experiments; *, p < 0.05 relative to control transfections). B, expression levels of Ubc9, PRDM16, and UCP1 were analyzed by immunoblotting for human adipocytes transfected with Ubc9 siRNA or a miR-30a mimic. C, HEK293T cells were transfected with HA-Ubc9 and FLAG-PRDM16. Lysates were co-immunoprecipitated (IP) with anti-FLAG antibodies, and the precipitates were analyzed by immunoblotting (IB). The interaction between EHMT1 and PRDM16 was a positive control for PRDM16 binding partners. D, PRDM16 protein levels in a cycloheximide (CHX) chase experiment were analyzed by immunoblotting (top panel) and quantified by densitometric analysis of PRDM16 protein degradation rates normalized to HSP90 expression (bottom panel, n = 2 independent experiments). E, PPARγ ChIP was performed in human adipocytes transfected with Ubc9 siRNA, miR-30a, or transfection controls. qPCR was used to analyze genomic occupancy using primers flanking PPARγ binding sites in the UCP1 enhancer (enh), and CIDEA intron (intr) 1 regions. An intronic region of Cyclin D1 served as a negative control (n = 2 independent experiments; *, p < 0.05 relative to control transfections). All data are expressed as mean ± S.E.
Ubc9 knockdown or miR-30a-mediated repression of Ubc9 increased previously described (11) PRDM16-dependent brown adipocyte gene expression (UCP1, PGC-1α, and CIDEA) independent of increased PRDM16 mRNA (Fig. 5A). These data, coupled with higher PRDM16 protein levels (Fig. 5B), suggested that Ubc9 depletion may result in stabilization of PRDM16. To confirm increased PRDM16 protein half-life, we performed cycloheximide pulse-chase experiments and found that Ubc9 siRNA or the miR-30a mimic extended the half-life of PRDM16 by at least 6 h (Fig. 5D).
Agonists that maximally stimulate the master regulator of brown and white adipocytes, PPARγ, increase the half-life of PRDM16 as an obligatory step to selectively potentiate the BAT gene program in subcutaneous fat cells (11). To activate BAT genes in white fat cells, PRDM16 interacts with PPARγ at enhancer regions critical for brown adipocyte identity (24, 25). We investigated whether the increased stability of PRDM16 corresponded with increased occupancy of PPARγ on the enhancer region upstream of UCP1 (26, 27) and a predicted CIDEA intronic region (28) in human adipocytes transfected with Ubc9 siRNA or a miR-30a mimic. ChIP-qPCR revealed that PPARγ occupancy on the UCP1 enhancer and intron 1 of CIDEA was significantly increased following knockdown of Ubc9 by siRNA or miR-30a (Fig. 5E). Together, our findings demonstrate that the depletion of Ubc9 stimulates PPARγ activity (13) and stabilizes PRDM16, corresponding to activation of the BAT energy metabolism program in human adipocytes.
Discussion
Regulation of the transcriptional programs operant in BAT and BAT-like tissues has received significant attention because of the discovery of these depots in adult humans (5, 29, 30) and the well established roles of BAT in protection against obesity and diabetes in rodent models of human disease (31, 32). Subcutaneous white fat cells exposed to cold, β-adrenergic receptor stimuli, or PPARγ agonists acquire features of brown adipocytes, including UCP1 expression and increased mitochondrial biogenesis (11, 12, 33–36). Subcutaneous adipocytes that combine efficient lipid storage and catabolism represent an uncharacterized therapeutic opportunity to protect against T2DM and its associated cardiovascular sequelae. Despite obvious therapeutic potential, translational efforts to leverage browning for metabolic therapies are hindered by a lack of understanding of the mechanisms that specifically “brown” white fat. In this study, we show that inhibiting Ubc9 via siRNA or miR-30a overexpression promotes white adipocyte browning by driving a PRDM16-dependent gene profile associated with energy expenditure. Thus, inhibiting Ubc9 promotes the expression of genes associated with energy expenditure and browning of white fat (13).
In this study, we sought to understand the regulation of Ubc9 protein and mRNA expression in adipocytes. Analysis of the proximal promoter region showed no relevant adipocyte-specific transactivator binding sites upstream of the Ubc9 gene as a means of logical regulation. Indeed, the Ubc9 promoter region is devoid of TATA boxes (37). These findings indicate that Ubc9 expression in adipocytes may be regulated by mechanisms independent of classical transcriptional programs. We used two comprehensive online miRNA analysis prediction tools, TargetScan (14) and starBase (15), to nominate miRNAs that target Ubc9. In starBase, which uses sequencing of small RNAs bound to mRNAs inside the RNA-induced silencing complex, the putative miR-30a-Ubc9 interaction was the most significantly anti-correlated cognate miRNA-mRNA pair for Ubc9. miR-30 family members share the same seed sequence and exhibit broad tissue expression (38, 39), which implies a vast target gene spectrum. Our in vitro analyses showed that miR-30a binds the Ubc9 3′ UTR, leading to both Ubc9 protein and mRNA suppression in human adipocytes. Additional analysis showed that miR-30a expression in human adipocytes inhibits expression of Ubc9 but not other published targets, including ATG5 (17), BECN1 (18), RIP140 (19), and p53 (20, 21). The reason for these discrepancies with the published literature remain unknown and are likely related to tissue-specific functions of miRNAs and other biochemical contexts that influence miRNA-mRNA interactions (40, 41). Other studies support a role for miR-30 in human adipocytes (42), but further studies are required to identify the full spectrum of miR-30a targets and the molecular regulation by transcription factors required for adipocyte identity. Nonetheless, the selectivity of miR-30a for Ubc9 suggests a distinct function in human adipocytes.
In this study, the expression of Ubc9 and miR-30a was inversely correlated in WAT and BAT, but no relationship was observed in skeletal muscle. Such a tissue-specific counter-regulatory balance between Ubc9 and miR-30a may direct white-to-beige adipocyte conversion. When Ubc9 is depleted by siRNA or miR-30a, expression of genes associated with energy expenditure and BAT was increased, which translated to remodeled mitochondrial structures necessary for higher respiration rates (35, 43). This result reflects the BAT-like gene expression profile, suggesting that inhibition of Ubc9 by miR-30a browns human subcutaneous adipocytes.
Our work implicates miR-30a as a coordinating molecule that modulates an integrated gene program to elevate oxidative respiration and brown adipocyte features. One other factor, PRDM16, is essential for browning of white fat and systemic homeostasis (4, 12). Stabilization of PRDM16 protein downstream of browning agents (11) is required to maximally induce the brown fat program in subcutaneous adipocytes. Recent studies indicate that PRDM16 recruits transcriptional machinery to enhancer regions bound by PPARγ to license brown adipocyte genes. Our results suggest a mechanism whereby abating the Ubc9 interaction with PRDM16 allows selective expression of WAT and BAT transcriptional programs.
In line with these studies, we discovered that the browning effects of Ubc9 depletion by siRNA or miR-30a are tightly associated with increased accumulation of PRDM16 protein and occupancy of PPARγ on regulatory regions of genes synergistically regulated by PRDM16 and PPARγ. Ubc9 is required for SUMOylation that alters cellular localization, protein-protein interactions, and/or protein stability. Recent work established that PRDM16 is SUMOylated (44) in protein complexes containing Ubc9 and known repressive factors, including CtBP1/2 (45). These data motivate future studies aimed at identifying the mechanistic and physiologic implications of PRDM16 SUMOylation in adipocytes.
In investigating the regulation of Ubc9 in human adipocytes, our results have revealed another layer that regulates browning of white fat. Additional characterization of how the PPARγ-PRDM16-Ubc9 complex is regulated by miR-30a or other factors may uncover a specific transcriptional module that directs white and brown fat identity in subcutaneous adipocytes. Further functional characterization of the signaling networks bracketing the Ubc9/miR-30a axis in animal models is expected to lead to identification of new therapeutic targets and strategies against obesity and related metabolic disorders.
Materials and Methods
Cell Culture and Differentiation
Subcutaneous primary human preadipocytes were provided by Zen-Bio Inc. Human preadipocytes were maintained in DMEM/F12 with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin (growth medium). Confluent cells were differentiated using growth medium supplemented with 100 nm human insulin, 0.250 mm 3-isobutyl-1-methylxanthine, 500 nm dexamethasone, and 3 μm rosiglitazone. Based on a previous microarray analysis (46), adipocytes were considered mature at day 8. After transfections, white-to-brown adipocyte transition was stimulated by treating cells with rosiglitazone for 4 days.
RNA Extraction and qPCR Analysis
Total RNA was extracted from cells using the RNeasy kit. To measure relative mRNA expression, qPCR was performed with TaqMan reagents using a StepOne real-time PCR system (Life Technologies). Invariant controls included TATA binding protein, 18S rRNA, RNU48, and sno412. The TaqMan and Roche Universal Probe (UPL) gene expression assays are detailed in supplemental Tables 1 and 2.
Animals
Animal experiments were performed according to procedures approved by the Institutional Animal Care and Use Committee at Baylor College of Medicine. Only male C57BL/6J mice were used for analysis of gene expression.
siRNA and miRNA Transfection
Mature adipocytes were transfected with Ubc9 siRNA (Qiagen), mismatched siRNA control (Dharmacon), a miR-30a-5p mimic (Active Motif), or a control mimic (Active Motif) at a final concentration of 20 nm using Dharmafect transfection reagent (Dharmacon). After transfection, cells were incubated for 48 h at 5% CO2 and 37 °C.
Luciferase Reporter Assays
We used replication-deficient adenoviruses coupled with poly-lysine (46, 47) to express 3′ UTR luciferase fusions (Active Motif) in human adipocytes. miRNA binding to control regions or the 3′ UTR of Ubc9 was determined using LightSwitch assay reagents (Active Motif).
Antibodies
The following antibodies were used for immunoblotting: Ubc9 (Genetex), HSP90 (Cell Signaling Technology), polyclonal UCP1 (Abcam), PRDM16 (Abcam), RIP140 (Santa Cruz Biotechnology), ATG5 (Genetex), p53 (Santa Cruz Biotechnology), Beclin-1 (Genetex), and β-actin (Sigma).
Immunoblotting
Cells were collected by scraping and lysed in RIPA buffer supplemented with the appropriate protease and phosphatase inhibitors. Immunoblot analysis was performed with whole cell lysates run on 4–12% BisTris NuPage (Millipore) gels and transferred onto Immobilon-P transfer membranes (Millipore), followed by antibody incubation. Immunoreactive bands were visualized by chemiluminescence.
Co-immunoprecipitation
HeLa cells were transfected using Lipofectamine 2000 (Life Technologies) in 10-cm dishes when cells were ∼80% confluent. Cells were transfected with expression plasmids for FLAG-PRDM16 and HA-Ubc9 or FLAG-PRDM16 and HA-EHMT1. After 24 h, cells were lysed with RIPA buffer plus protease and phosphatase inhibitors. The lysate was incubated with HA (Cell Signaling Technology) followed by protein A beads. Protein A beads were washed with RIPA buffer followed by elution in Laemmli buffer (Bio-Rad) and immunoblotted using FLAG or HA antibodies.
ChIP
ChIP was performed using the ChIP-IT high sensitivity kit (Active Motif) according to the instructions of the manufacturer. Chromatin fragmentation was performed using a Bioruptor water bath sonicator (Diagenode). Sheared protein-DNA complexes were immunoprecipitated with anti-PPARγ (Santa Cruz Biotechnology, H-100) or rabbit IgG control (Santa Cruz Biotechnology). For ChIP-qPCR, enrichment was measured using SYBR Green (Applied Biosystems). Primer sequences can be found in Supplemental Table 3.
Cellular Respiration
Respiration was measured in human adipocytes using an XF24 analyzer (Seahorse Bioscience). Preadipocytes were plated into V7-PS plates and differentiated before siRNA or miRNA transfection. After transfection, the medium was replaced with 37 °C unbuffered DMEM containing 4.5 g/liter glucose, sodium pyruvate (1 mmol/liter), and l-glutamine (2 mmol/liter). Basal respiration was defined before sequential addition of forskolin, oligomycin, rotenone, and antimycin A. In select experiments, the OCR purely attributed to mitochondria was determined by subtracting the rotenone/antimycin A-insensitive OCR.
Mitochondrial DNA Analysis
Mitochondrial DNA content was determined by qPCR. Total DNA was isolated using the DNeasy kit (Qiagen). Real-time qPCR was performed on a StepOne real-time qPCR system (Life Technologies) using Platinum SYBR Green qPCR Supermix-UDG (Life Technologies). Reactions were prepared according to the recommendations of the manufacturer in a total volume of 25 μl. mtDNA primers were designed using the human mitochondrial genome sequence within the NADH dehydrogenase subunit 6 (ND6) gene. 18S rRNA was used as the invariant control.
Mitochondrial Labeling
Mitochondria were labeled using MitoTracker CMX-ROS (Life Technologies). Live cells were pulsed with 500 nm MitoTracker for 15 min. Mitochondrial labeling was followed by cell fixation in 4% paraformaldehyde. Ammonium chloride was used to quench autofluorescence derived from residual paraformaldehyde. DAPI (Sigma) was used for nucleus labeling. Imaging was performed with the DeltaVision core image restoration microscope (Applied Precision).
Statistical Analyses
The data presented were acquired from a minimum of two independent experiments performed on multiple days unless otherwise indicated. Statistical significance was assessed by unpaired Student's t test. All tests were carried out at a 95% confidence interval.
Author Contributions
E. H. K. and S. M. H. conceived the hypothesis, designed the study, and performed the majority of the experiments. Y. C. and S. K. performed experiments. E. H. K. and S. M. H. wrote the manuscript. B. Y. contributed reagents and tools. D. A. B., M. P. H., B. Y., B. H., Y. C., S. K., and S. E. M. contributed to discussion and edited the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
This work was supported by National Institutes of Health Grants K01DK096093 and R03DK105006, American Heart Association Beginning Grant-in-Aid 15BGIA25850025, the Baylor College of Medicine Bridge to Independence program, and the Alkek Center for Molecular Discovery (to S. M. H.). This work was also supported by the Caroline Weiss Law Foundation, National Institutes of Health Grant R21CA205257, and the Prostate Cancer Research Foundation (to S. E. M.). Additional support was provided by National Institutes of Health Grants F30CA196108 (to D. A. B.), R01GM033976 (to A. R. M.), K01DK081446 (to B. H.), and R01DK97441 (to S.K.); American Cancer Society Grant RSG-13-061-01-TBE (to B. H.), and Baylor College of Medicine Diabetes and Endocrinology Research Center Grant P30-DK079638. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Tables 1–3.
- WAT
- white adipose tissue
- T2DM
- type 2 diabetes mellitus
- BAT
- brown adipose tissue
- PPARγ
- peroxisome proliferator-activated receptor γ
- SUMO
- small ubiquitin-like modifier
- miRNA
- microRNA
- OCR
- oxygen consumption rate
- qPCR
- quantitative PCR
- RIPA
- radioimmune precipitation assay.
References
- 1. Samuel V. T., and Shulman G. I. (2016) The pathogenesis of insulin resistance: integrating signaling pathways and substrate flux. J. Clin. Invest. 126, 12–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Després J. P. (2012) Body fat distribution and risk of cardiovascular disease an update. Circulation 126, 1301–1313 [DOI] [PubMed] [Google Scholar]
- 3. Sidossis L., and Kajimura S. (2015) Brown and beige fat in humans: thermogenic adipocytes that control energy and glucose homeostasis. J. Clin. Invest. 125, 478–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cohen P., Levy J. D., Zhang Y., Frontini A., Kolodin D. P., Svensson K. J., Lo J. C., Zeng X., Ye L., Khandekar M. J., Wu J., Gunawardana S. C., Banks A. S., Camporez J. P., Jurczak M. J., et al. (2014) Ablation of PRDM16 and beige adipose causes metabolic dysfunction and a subcutaneous to visceral fat switch. Cell 156, 304–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cypess A. M., Lehman S., Williams G., Tal I., Rodman D., Goldfine A. B., Kuo F. C., Palmer E. L., Tseng Y. H., Doria A., Kolodny G. M., and Kahn C. R. (2009) Identification and importance of brown adipose tissue in adult humans. N. Engl. J. Med. 360, 1509–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chondronikola M., Volpi E., Børsheim E., Porter C., Annamalai P., Enerbäck S., Lidell M. E., Saraf M. K., Labbe S. M., Hurren N. M., Yfanti C., Chao T., Andersen C. R., Cesani F., Hawkins H., and Sidossis L. S. (2014) Brown adipose tissue improves whole-body glucose homeostasis and insulin sensitivity in humans. Diabetes 63, 4089–4099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sharp L. Z., Shinoda K., Ohno H., Scheel D. W., Tomoda E., Ruiz L., Hu H., Wang L., Pavlova Z., Gilsanz V., and Kajimura S. (2012) Human BAT possesses molecular signatures that resemble beige/brite cells. PLoS ONE 7, e49452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shinoda K., Luijten I. H., Hasegawa Y., Hong H., Sonne S. B., Kim M., Xue R., Chondronikola M., Cypess A. M., Tseng Y. H., Nedergaard J., Sidossis L. S., and Kajimura S. (2015) Genetic and functional characterization of clonally derived adult human brown adipocytes. Nat. Med. 21, 389–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wu J., Boström P., Sparks L. M., Ye L., Choi J. H., Giang A. H., Khandekar M., Virtanen K. A., Nuutila P., Schaart G., Huang K., Tu H., van Marken Lichtenbelt W. D., Hoeks J., Enerbäck S., et al. (2012) Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell 150, 366–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Seale P. (2015) Transcriptional regulatory circuits controlling brown fat development and activation. Diabetes 64, 2369–2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ohno H., Shinoda K., Spiegelman B. M., and Kajimura S. (2012) PPARγ agonists induce a white-to-brown fat conversion through stabilization of PRDM16 protein. Cell Metab. 15, 395–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Seale P., Conroe H. M., Estall J., Kajimura S., Frontini A., Ishibashi J., Cohen P., Cinti S., and Spiegelman B. M. (2011) Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. J. Clin. Invest. 121, 96–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hartig S. M., Bader D. A., Abadie K. V., Motamed M., Hamilton M. P., Long W., York B., Mueller M., Wagner M., Trauner M., Chan L., Bajaj M., Moore D. D., Mancini M. A., and McGuire S. E. (2015) Ubc9 impairs activation of the brown bat energy metabolism program in human white adipocytes. Mol. Endocrinol. 29, 1320–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Agarwal V., Bell G. W., Nam J. W., and Bartel D. P. (2015) Predicting effective microRNA target sites in mammalian mRNAs. eLife 4, e05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li J. H., Liu S., Zhou H., Qu L. H., and Yang J. H. (2014) starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 42, D92–D97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wu F., Zhu S., Ding Y., Beck W. T., and Mo Y. Y. (2009) MicroRNA-mediated regulation of Ubc9 expression in cancer cells. Clin. Cancer Res. 15, 1550–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yu Y., Yang L., Zhao M., Zhu S., Kang R., Vernon P., Tang D., and Cao L. (2012) Targeting microRNA-30a-mediated autophagy enhances imatinib activity against human chronic myeloid leukemia cells. Leukemia 26, 1752–1760 [DOI] [PubMed] [Google Scholar]
- 18. Zou Z., Wu L., Ding H., Wang Y., Zhang Y., Chen X., Chen X., Zhang C. Y., Zhang Q., and Zen K. (2012) MicroRNA-30a sensitizes tumor cells to cis-platinum via suppressing beclin 1-mediated autophagy. J. Biol. Chem. 287, 4148–4156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hu F., Wang M., Xiao T., Yin B., He L., Meng W., Dong M., and Liu F. (2015) miR-30 promotes thermogenesis and the development of beige fat by targeting RIP140. Diabetes 64, 2056–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Forini F., Kusmic C., Nicolini G., Mariani L., Zucchi R., Matteucci M., Iervasi G., and Pitto L. (2014) Triiodothyronine prevents cardiac ischemia/reperfusion mitochondrial impairment and cell loss by regulating miR30a/p53 axis. Endocrinology 155, 4581–4590 [DOI] [PubMed] [Google Scholar]
- 21. Li J., Donath S., Li Y., Qin D., Prabhakar B. S., and Li P. (2010) miR-30 regulates mitochondrial fission through targeting p53 and the dynamin-related protein-1 pathway. PLoS Genet. 6, e1000795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sun L., Xie H., Mori M. A., Alexander R., Yuan B., Hattangadi S. M., Liu Q., Kahn C. R., and Lodish H. F. (2011) Mir-193b-365 is essential for brown fat differentiation. Nat. Cell Biol. 13, 958–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yehuda-Shnaidman E., Buehrer B., Pi J., Kumar N., and Collins S. (2010) Acute stimulation of white adipocyte respiration by PKA-induced lipolysis. Diabetes 59, 2474–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Seale P., Bjork B., Yang W., Kajimura S., Chin S., Kuang S., Scimè A., Devarakonda S., Conroe H. M., Erdjument-Bromage H., Tempst P., Rudnicki M. A., Beier D. R., and Spiegelman B. M. (2008) PRDM16 controls a brown fat/skeletal muscle switch. Nature 454, 961–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Harms M. J., Lim H. W., Ho Y., Shapira S. N., Ishibashi J., Rajakumari S., Steger D. J., Lazar M. A., Won K. J., and Seale P. (2015) PRDM16 binds MED1 and controls chromatin architecture to determine a brown fat transcriptional program. Genes Dev. 29, 298–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bordicchia M., Liu D., Amri E. Z., Ailhaud G., Dessì-Fulgheri P., Zhang C., Takahashi N., Sarzani R., and Collins S. (2012) Cardiac natriuretic peptides act via p38 MAPK to induce the brown fat thermogenic program in mouse and human adipocytes. J. Clin. Invest. 122, 1022–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kozak U. C., Kopecky J., Teisinger J., Enerbäck S., Boyer B., and Kozak L. P. (1994) An upstream enhancer regulating brown fat specific expression of the mitochondrial uncoupling protein gene. Mol. Cell Biol. 14, 59–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mikkelsen T. S., Xu Z., Zhang X., Wang L., Gimble J. M., Lander E. S., and Rosen E. D. (2010) Comparative epigenomic analysis of murine and human adipogenesis. Cell 143, 156–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. van Marken Lichtenbelt W. D., Vanhommerig J. W., Smulders N. M., Drossaerts J. M., Kemerink G. J., Bouvy N. D., Schrauwen P., and Teule G. J. (2009) Cold-activated brown adipose tissue in healthy men. N. Engl. J. Med. 360, 1500–1508 [DOI] [PubMed] [Google Scholar]
- 30. Virtanen K. A., Lidell M. E., Orava J., Heglind M., Westergren R., Niemi T., Taittonen M., Laine J., Savisto N. J., Enerbäck S., and Nuutila P. (2009) Functional brown adipose tissue in healthy adults. N. Engl. J. Med. 360, 1518–1525 [DOI] [PubMed] [Google Scholar]
- 31. Hamann A., Flier J. S., and Lowell B. B. (1996) Decreased brown fat markedly enhances susceptibility to diet-induced obesity, diabetes, and hyperlipidemia. Endocrinology 137, 21–29 [DOI] [PubMed] [Google Scholar]
- 32. Lowell B. B., S-Susulic V., Hamann A., Lawitts J. A., Himms-Hagen J., Boyer B. B., Kozak L. P., and Flier J. S. (1993) Development of obesity in transgenic mice after genetic ablation of brown adipose tissue. Nature 366, 740–742 [DOI] [PubMed] [Google Scholar]
- 33. Petrovic N., Walden T. B., Shabalina I. G., Timmons J. A., Cannon B., and Nedergaard J. (2010) Chronic peroxisome proliferator-activated receptor-γ (PPARγ) activation of epididymally derived white adipocyte cultures reveals a population of thermogenically competent, UCP1-containing adipocytes molecularly distinct from classic brown adipocytes. J. Biol. Chem. 285, 7153–7164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tiraby C., Tavernier G., Lefort C., Larrouy D., Bouillaud F., Ricquier D., and Langin D. (2003) Acquirement of brown fat cell features by human adipocytes. J. Biol. Chem. 278, 33370–33376 [DOI] [PubMed] [Google Scholar]
- 35. Wilson-Fritch L., Burkart A., Bell G., Mendelson K., Leszyk J., Nicoloro S., Czech M., and Corvera S. (2003) Mitochondrial biogenesis and remodeling during adipogenesis and in response to the insulin sensitizer rosiglitazone. Mol. Cell Biol. 23, 1085–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ye L., Wu J., Cohen P., Kazak L., Khandekar M. J., Jedrychowski M. P., Zeng X., Gygi S. P., and Spiegelman B. M. (2013) Fat cells directly sense temperature to activate thermogenesis. Proc. Natl. Acad. Sci. U.S.A. 110, 12480–12485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ying S., Dünnebier T., Si J., and Hamann U. (2013) Estrogen receptor α and nuclear factor Y coordinately regulate the transcription of the SUMO-conjugating UBC9 gene in MCF-7 breast cancer cells. PloS ONE 8, e75695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hamilton M. P., Rajapakshe K., Hartig S. M., Reva B., McLellan M. D., Kandoth C., Ding L., Zack T. I., Gunaratne P. H., Wheeler D. A., Coarfa C., and McGuire S. E. (2013) Identification of a pan-cancer oncogenic microRNA superfamily anchored by a central core seed motif. Nat. Commun. 4, 2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Civelek M., Hagopian R., Pan C., Che N., Yang W. P., Kayne P. S., Saleem N. K., Cederberg H., Kuusisto J., Gargalovic P. S., Kirchgessner T. G., Laakso M., and Lusis A. J. (2013) Genetic regulation of human adipose tissue microRNA expression and its consequences for metabolic traits. Hum. Mol. Genet. 22, 3023–3037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Preusse M., Theis F. J., and Mueller N. S. (2016) miTALOS v2: analyzing tissue specific microRNA function. PloS ONE 11, e0151771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vidigal J. A., and Ventura A. (2015) The biological functions of miRNAs: lessons from in vivo studies. Trends Cell Biol. 25, 137–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zaragosi L. E., Wdziekonski B., Brigand K. L., Villageois P., Mari B., Waldmann R., Dani C., and Barbry P. (2011) Small RNA sequencing reveals miR-642a-3p as a novel adipocyte-specific microRNA and miR-30 as a key regulator of human adipogenesis. Genome Biol. 12, R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Keuper M., Jastroch M., Yi C. X., Fischer-Posovszky P., Wabitsch M., Tschöp M. H., and Hofmann S. M. (2014) Spare mitochondrial respiratory capacity permits human adipocytes to maintain ATP homeostasis under hypoglycemic conditions. FASEB J. 28, 761–770 [DOI] [PubMed] [Google Scholar]
- 44. Nishikata I., Nakahata S., Saito Y., Kaneda K., Ichihara E., Yamakawa N., and Morishita K. (2011) Sumoylation of MEL1S at lysine 568 and its interaction with CtBP facilitates its repressor activity and the blockade of G-CSF-induced myeloid differentiation. Oncogene 30, 4194–4207 [DOI] [PubMed] [Google Scholar]
- 45. Kajimura S., Seale P., Tomaru T., Erdjument-Bromage H., Cooper M. P., Ruas J. L., Chin S., Tempst P., Lazar M. A., and Spiegelman B. M. (2008) Regulation of the brown and white fat gene programs through a PRDM16/CtBP transcriptional complex. Genes Dev. 22, 1397–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hartig S. M., He B., Newberg J. Y., Ochsner S. A., Loose D. S., Lanz R. B., McKenna N. J., Buehrer B. M., McGuire S. E., Marcelli M., and Mancini M. A. (2012) Feed-forward inhibition of androgen receptor activity by glucocorticoid action in human adipocytes. Chem. Biol. 19, 1126–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Allgood V. E., Zhang Y., O'Malley B. W., and Weigel N. L. (1997) Analysis of chicken progesterone receptor function and phosphorylation using an adenovirus-mediated procedure for high-efficiency DNA transfer. Biochemistry 36, 224–232 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.