Abstract
Lysine demethylation of proteins such as histones is catalyzed by several classes of enzymes, including the FAD-dependent amine oxidases KDM1A/B. The KDM1 family is homologous to the mitochondrial monoamine oxidases MAO-A/B and produces hydrogen peroxide in the nucleus as a byproduct of demethylation. Here, we show KDM1A is highly thiol-reactive in vitro and in cellular models. Enzyme activity is potently and reversibly inhibited by the drug disulfiram and by hydrogen peroxide. Hydrogen peroxide produced by KDM1A catalysis reduces thiol labeling and inactivates demethylase activity over time. MALDI-TOF mass spectrometry indicates that hydrogen peroxide blocks labeling of cysteine 600, which we propose forms an intramolecular disulfide with cysteine 618 to negatively regulate the catalytic activity of KDM1A. This activity-dependent regulation is unique among histone-modifying enzymes but consistent with redox sensitivity of epigenetic regulators.
Keywords: chemical biology, disulfide, drug screening, enzyme inactivation, flavoprotein, histone demethylase, hydrogen peroxide, reactive oxygen species (ROS), redox regulation, thiol
Introduction
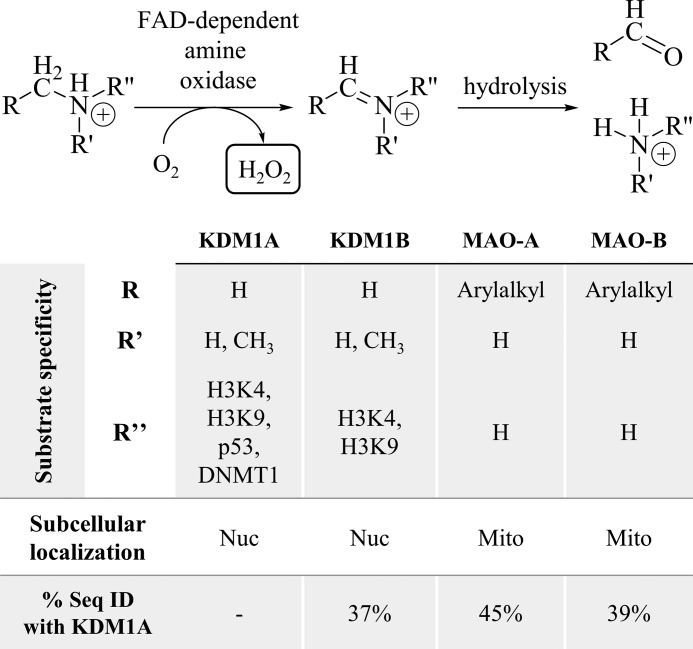
Post-translational N-methylation of lysine residues in proteins such as histones plays a widespread role in normal and pathological biological processes, including transcriptional regulation, chromatin remodeling, cell signaling, and assembly of protein complexes, among other functions. Lysines can be mono-, di-, or tri-methylated, with each modification resulting in different functional outcomes depending on the degree of methylation and its context (1). Oxidative removal of histone lysine N-methyl groups is catalyzed by two mechanistically distinct classes of lysine-specific demethylases (KDMs)3 (2). Most identified demethylases are JmjC domain-containing, iron (II) and α-ketoglutarate-dependent oxygenases (KDM subfamilies 2–8). These enzymes hydroxylate the N-methyl group of their substrates producing a hemi-aminal intermediate that fragments to the demethylated lysine and formaldehyde, a catalytic mechanism capable of demethylating mono-, di-, or tri-methylated substrates (3). In contrast, the amine oxidase domain-containing demethylases utilize a flavin adenine dinucleotide (FAD) cofactor (KDM subfamily 1). These enzymes, KDM1A and KDM1B, couple the reduction of FAD to FADH2 with the oxidation of the C-N methylamine bond to a hydrolytically labile iminium ion, a mechanism only compatible with mono- and di-methylated substrates (Fig. 1) (4–6). The KDM1 family features a catalytic amine oxidase (AO) domain homologous to the mitochondrial monoamine oxidases MAO-A and MAO-B, which have been pharmacological targets for antidepressants and neuroprotective agents for decades due to their function to catalyze the oxidative deamination of neurotransmitters (7).
FIGURE 1.
FAD-dependent amine oxidases generate H2O2 to deaminate their substrates. The nuclear (nuc) amine oxidases KDM1A/B remove methyl groups from lysine residues on histone H3, as well as other nuclear proteins. The mitochondrial (mito) amine oxidases MAO-A/B catabolize small molecule amines such as neurotransmitters. All four enzymes share considerable structural homology and sequence identity (% seq ID) in their amine oxidase catalytic domains.
All KDMs require molecular oxygen to demethylate their substrates. KDM subfamilies 2–8 generate a reactive Fe(IV) oxo species, which inserts an oxygen atom into a substrate C-H bond, whereas the KDM1 family uses molecular oxygen to re-oxidize their flavin cofactor to the quinone species (3). Beyond the mechanistic requirement for oxygen, most JmjC domain-containing KDMs are inducible in hypoxic conditions and many are direct targets of the hypoxia inducible factor HIF-1 (8). The role of the KDM1 family in hypoxia is less well-established, but KDM1A has been shown to mediate hypoxia-induced histone lysine demethylation at the BRCA1 and RAD51 promoters in MCF-7 breast cancer cells and the MLH1 promoter in RKO colon cancer cells (9, 10). In addition to their requirement for molecular oxygen, all KDMs liberate the former N-methyl group as formaldehyde. The fate of formaldehyde and general mechanisms that protect KDMs from cross-linking damage remain unclear. However, KDM1A has been shown to bind tetrahydrofolate, which serves as the formaldehyde acceptor for the enzymes dimethylglycine dehydrogenase and sarcosine dehydrogenase and may play a similar role for KDM1A (11).
A key feature that distinguishes class 1 KDMs from class 2–8 KDMs is the production of hydrogen peroxide (H2O2), which is generated when oxygen accepts electrons to re-oxidize the FADH2 cofactor. Production of H2O2 in the nucleus is significant as it is known to cause mutagenic changes to DNA, including formation of 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxoG). This modified nucleobase is repaired by the base excision repair (BER) pathway (12), and recruitment of BER machinery to the 8-oxoG lesions caused by KDM1A catalytic activity has been linked to estrogen-, Myc-, and androgen-induced transcriptional activation (13–15). These examples suggest a critical role for H2O2 generation in the mechanism of KDM1A distinct from other classes of histone lysine demethylases. To the best of our knowledge, the importance of H2O2 in KDM1B-catalyzed lysine demethylation has yet to be described. In this work, we describe an additional role for H2O2 in KDM1A biochemistry. We show that KDM1A is inactivated by cysteine oxidation, and that catalytically generated H2O2 negatively regulates demethylase activity. We propose a mechanism where KDM1A utilizes a thiol/disulfide switch to sense H2O2, a unique auto-oxidation mechanism among histone modifiers but consistent with general mechanisms of redox sensing by epigenetic enzymes.
Results
KDM1A Is Reversibly Inhibited by Thiol-reactive Compounds
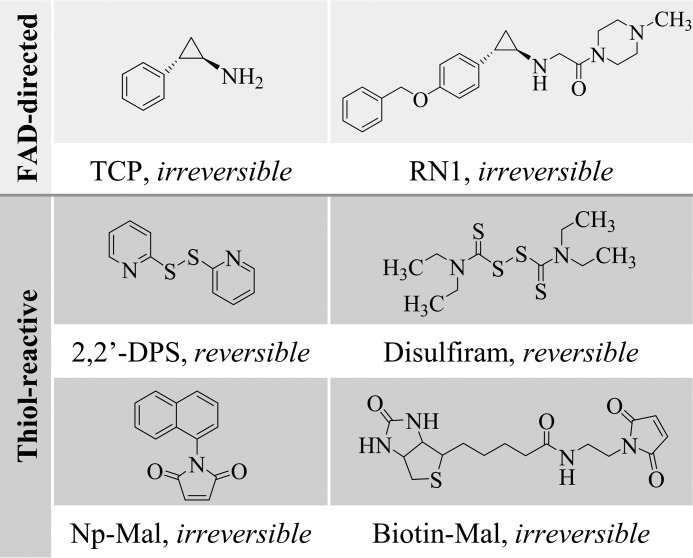
Tranylcypromine (TCP; Parnate) inhibits FAD-dependent amine oxidases by forming covalent cofactor adducts (7). TCP derivatives, such as RN1, exploit the same mechanism of action but with improved selectivity for KDM1A over other enzyme family members such as the monoamine oxidases (Fig. 2) (16). Using these compounds as positive controls, a high-throughput screen followed by detailed secondary assays was used to identify novel KDM1A inhibitors. Notably, many of the small molecules initially identified as potent, structurally novel KDM1A inhibitors were found to have potential for reactivity with thiol groups. Thiol-reactive small molecules are known to be promiscuous enzyme inhibitors, and are generally disfavored for therapeutic development due to metabolic liability (17). However, given our interest in the nuclear redox activity of KDM1A and emerging evidence for dynamic cysteine metabolism in other epigenetic regulators, including histone deacetylases (HDACs) that are known to complex with KDM1A, we sought to further characterize KDM1A's thiol-reactivity. Two mechanistic classes of thiol-reactive compounds were utilized: the FDA-approved drug disulfiram and the oxidizing agent 2,2′-dithiodipyridine (2,2′-DPS) form reversible disulfide bonds, whereas maleimides such as N-naphthylmaleimide (Np-Mal) or biotinylated maleimide (Biotin-Mal) form essentially irreversible sulfur-carbon thioether bonds and are selective for thiols at physiological pH (Fig. 2, note that thiol-maleimide adducts can be reversed with extended incubation in reducing environments (18)). Both classes of small molecules inhibit KDM1A activity as measured by a high-throughput mass spectrometry assay (Fig. 3A). Enzyme inactivation was measured after a 10-min pre-incubation with compound followed by a 20-min demethylation reaction; however, all compounds studied here are expected to form covalent adducts and so relative potencies may exhibit time-dependence. Under these conditions, all thiol-reactive inhibitors were measured to have apparent IC50 values of less than 1 μm.
FIGURE 2.
Chemical structures of FAD-directed and thiol-reactive KDM1A inhibitors. Thiol-reactive probes form disulfide bonds that are readily reversed in reducing environments, or essentially irreversible thioether bonds.
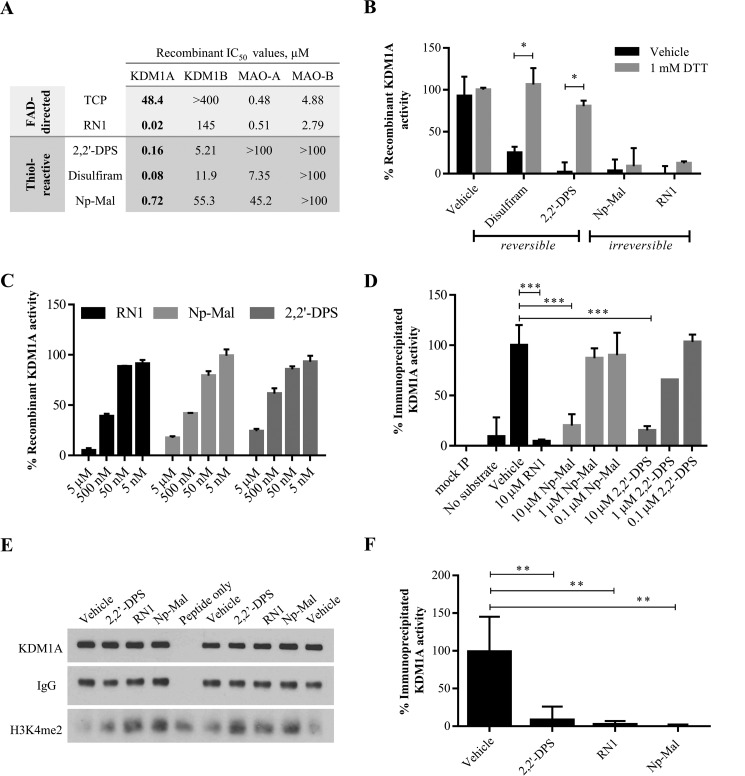
FIGURE 3.
KDM1A is reversibly inhibited by thiol-reactive small molecules. A, in vitro enzymatic assays with recombinant GST-tagged KDM1A/B or MAO-A/B reveal potent inhibition of KDM1A by reversible and essentially irreversible thiol-reactive small molecules. Apparent IC50 values reported in μm for 20 min (KDM1A) and 60 min (KDM1B, MAO-A/B) reactions. B, inhibition of KDM1A by disulfide forming thiol-reactive small molecules (disulfiram, 2,2′-DPS) is reversible by addition of DTT. Covalent modification by Np-Mal or RN1 is not reversible by DTT. Compounds (1 μm) were pre-incubated with KDM1A for 10 min prior to reduction, and demethylation of H3K4me2 starting material was detected by LC-MS. Error bars indicate S.D.; *, p < 0.05 by 2-tailed t test with correction for multiple comparisons. C, 6×His-tagged KDM1A is inhibited by thiol-reactive inhibitors. Enzyme activity was monitored by detection of catalytically generated H2O2 with an HRP-coupled assay. D–F, thiol-reactive compounds inhibit activity of full-length KDM1A immunoprecipitated from HeLa cells. D, thiol-reactive inhibitors reduce catalytically-generated H2O2 produced by immunoprecipitated KDM1A as measured by HRP-coupled detection. Error bars indicate S.D. in n = 2 replicate reactions; ***, p < 0.001 by 1-way ANOVA with correction for multiple comparisons. E, Western blot analysis of the depletion of H3K4me2 starting material by immunoprecipitated KDM1A. For both cell-free assays, immunoprecipitated KDM1A was pre-incubated with inhibitor for 10 min prior to demethylation of H3K4me2 peptide substrate for 1 h. F, quantification of Western blot data of immunoprecipitated KDM1A inhibition by thiol-reactive compounds (10 μm) in n = 4 assays from biological duplicates. Error bars indicate S.D.; **, p < 0.01 by one-way ANOVA with correction for multiple comparisons.
Based on this panel of inhibitors, we suspected that covalent modification of one or more cysteine residues on KDM1A resulted in enzyme inactivation. Consistent with this mechanism, inhibition by disulfide-forming compounds (10 μm, 10-min pre-incubation) was reversed following addition of an excess of the thiol reducing agent DTT (1 mm, Fig. 3B). In contrast, the thioether-forming Np-Mal and FAD-adduct forming RN1 were irreversible under these conditions. We and others have previously described the use of HRP-coupled assays to detect catalytically generated H2O2 and to profile KDM1A inhibition (16, 19). Using this assay, thiol-reactive inhibitors were confirmed to inhibit H2O2 production by KDM1A (Fig. 3C). Notably, only the LC-MS-based assay was compatible with reducing agents such as DTT and was thus critical to these and subsequent studies. Both MAO-A and MAO-B are known to be modified and inhibited by high concentrations of thiol-reactive compounds in a process competitive with substrate binding (20). To determine the relative thiol-reactivity among homologous FAD-dependent amine oxidases, a panel of mass spectrometry assays to monitor KDM1A/B activity and enzymatically coupled assays for MAO-A/B activity were conducted. Even with longer reaction durations (60 min for KDM1B/MAO-A/B versus 20 min for KDM1A), thiol-reactive inhibitors were found to be 30–90-fold more potent against recombinant KDM1A versus the second-most inhibited enzyme (Fig. 3A). In particular, MAO-B was not inhibited by thiol-reactive compounds under the conditions tested, consistent with hours-long treatment required for cysteine modification (20).
The recombinant KDM1A used in this study contains all 9 cysteine residues encoded in the full-length protein and was purified with an N-terminal GST tag. To ensure consistency among reactions, recombinant KDM1A was pre-reduced with immobilized TCEP resin then desalted with buffer exchange immediately prior to each assay. Although the GST tag served as a useful surrogate marker for total recombinant protein in several Western blot-based experiments, as no commercially available antibodies were found that detected the recombinant protein, GST itself can be modified by high concentrations of thiol-reactive compounds (21). We thus confirmed that His6-tagged KDM1A is also inhibited by thiol-reactive compounds (Fig. 3C), but noted that this enzyme had significantly lower demethylase activity than the GST-tagged protein. In addition, full-length KDM1A was immunoprecipitated from HeLa cells. Immunoprecipitated enzyme activity was monitored by the HRP-coupled assay (Fig. 3D) or by Western blotting to detect demethylation of a synthetic 21-mer peptide substrate (Fig. 3, E and F). Hydrogen peroxide production and peptide demethylation were both inhibited by the disulfide-forming 2,2′-DPS, the thioether-forming Np-Mal, and the FAD-directed inhibitor RN1, suggesting that full-length KDM1A is also inhibited by thiol-reactive compounds.
Cys-600 Is Modified by Biotin-Mal
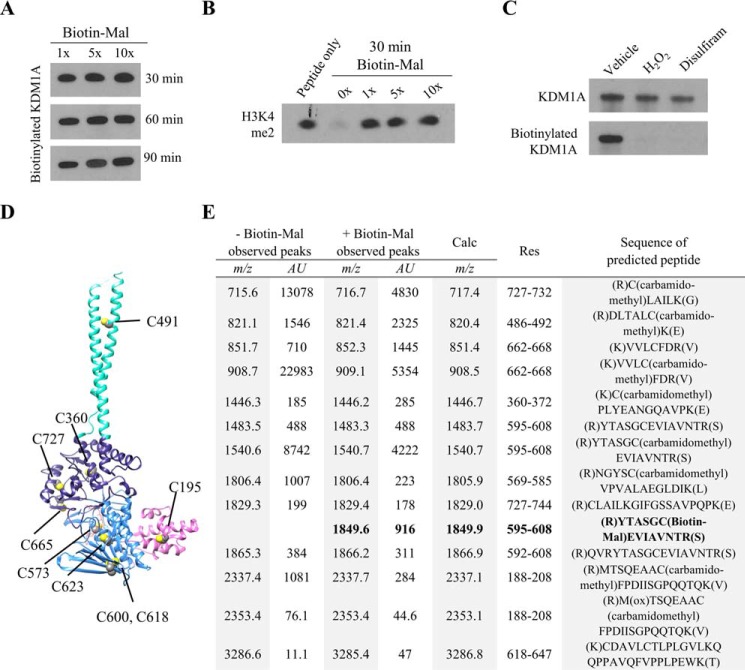
To gain further insight into the mechanism and possible significance of KDM1A thiol-reactivity, we sought to identify which cysteine residue, or residues, were modified. To this end, pre-reduced, recombinant KDM1A was labeled with a 1-, 5-, or 10-fold excess of biotinylated maleimide (Biotin-Mal, Fig. 2). Even after extended reaction durations, equivalent levels of Biotin-Mal labeling were observed across all conditions by blotting with a streptavidin-HRP conjugate (Fig. 4A). Furthermore, treatment with one equivalent of Biotin-Mal resulted in complete inhibition of recombinant KDM1A (Fig. 4B). Taken together, these results suggest that a single cysteine residue on recombinant KDM1A is modified by Biotin-Mal labeling and is sufficient to inhibit enzyme activity. Pre-treatment of recombinant KDM1A with thiol-reactive compounds such as disulfiram followed by buffer exchange blocked Biotin-Mal labeling (Fig. 4C), consistent with covalent thiol modification as a general mechanism of inhibition.
FIGURE 4.
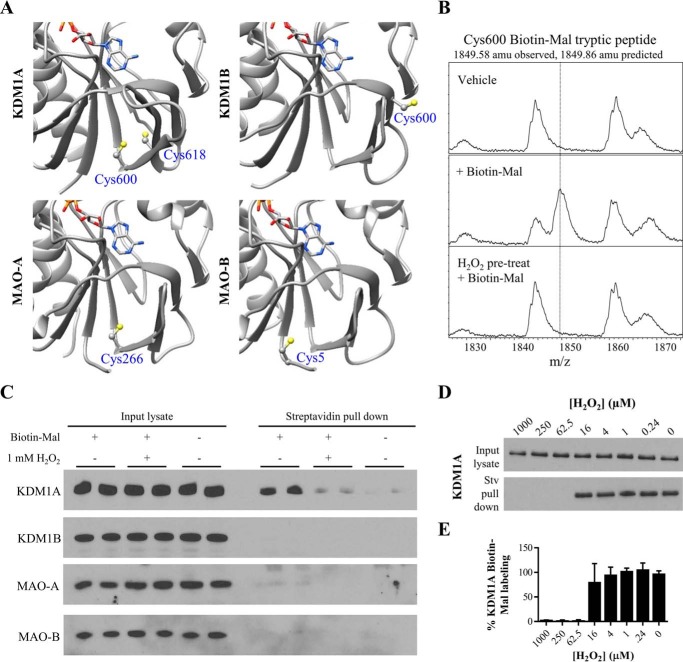
MALDI-TOF analysis of KDM1A tryptic digests identifies Cys-600 as a site of Biotin-Mal labeling. A, recombinant KDM1A (5 μm) was pre-reduced, desalted with buffer exchange, and labeled with various equivalents of Biotin-Mal for 30, 60, or 90 min. The extent of thiol labeling was monitored by blotting with a streptavidin-HRP conjugate. B, inhibition of recombinant KDM1A by Biotin-Mal labeling was determined by Western blot analysis of the depletion of H3K4me2 starting material. Recombinant KDM1A was pre-incubated with Biotin-Mal for 30 min prior to demethylation of H3K4me2 peptide substrate for 1 h. C, pre-treatment of recombinant KDM1A with H2O2 or disulfiram followed by desalting with buffer exchange blocks labeling with Biotin-Mal, as measured by blotting with a streptavidin-HRP conjugate. D, crystal structure (PDB ID: 2HKO) indicating the locations of the nine cysteine residues of KDM1A. Cys-195 is in the SWIRM domain (pink), Cys-573, -600, -618, and -623 are in the FAD-binding amine oxidase domain (light blue), Cys-360, -665, and -727 are in the substrate-binding amine oxidase domain (dark blue), and Cys-491 is in the tower domain (teal). E, tabular summary of cysteine-containing tryptic KDM1A peptides identified by MALDI-TOF. Recombinant KDM1A (5 μm) was pre-reduced and desalted with buffer exchange, then labeled with 10 μm Biotin-Mal for 20 min prior to in-gel alkylation and digestion.
To determine which of the nine cysteine residues was modified (Fig. 4D), samples of recombinant KDM1A were labeled with a 2-fold excess of Biotin-Mal or vehicle control, subjected to in-gel trypsin digestion, and the resulting tryptic digest peptides were measured with MALDI-TOF mass spectrometry. Under optimized conditions, 73% sequence coverage was obtained including peaks corresponding to all cysteine-containing peptides (Fig. 4E). Consistent with the observed labeling stoichiometry, only Cys-600 was labeled by Biotin-Mal. This residue is conserved in all mammalian forms of KDM1A and is located in the FAD-binding portion of the amine oxidase domain (Fig. 4D). The corresponding cysteine residues in MAO-A and MAO-B are the only to be modified by a related biotinylated maleimide (Cys-266 of clorgyline-inactivated MAO-A and Cys-5 of pargyline-inactivated MAO-B, respectively (20)), suggesting that the reactivity of this cysteine residue may be conserved.
Cys-600 Forms a Putative Disulfide with Cys-618
Examination of the crystal structures of KDM1A revealed the presence of Cys-618 proximal to Cys-600 (Fig. 5A). Comparison of KDM1A/B and MAO-A/B indicates that only KDM1A contains this pair of cysteines in FAD-binding amine oxidase domain. Although the Cys-600/Cys-618 sulfur atoms were not oriented toward one another in the solved structures, the average S-S distance was ∼4.8 Å and the average Cb-Cb distance was ∼5.1 Å in a sampling of available KDM1A structures (data not shown), raising the possibility of intramolecular disulfide formation (22).
FIGURE 5.
KDM1A forms a putative intramolecular disulfide bond. A, crystal structures of the FAD-binding amine oxidase domains of KDM1A/B and MAO-A/B indicate a unique pair of proximal cysteine residues in KDM1A which may be capable of disulfide bond formation (respective PDB accession codes: 2HKO, 4GUU, 2BXR, and 2XFU, only residues in the amine oxidase domains are displayed). Cys-600 is ∼5 Å away from Cys-618 in the crystal structure of KDM1A, and this pair of cysteines abuts the Rossmann fold responsible for FAD cofactor binding. B, MALDI-TOF analysis of KDM1A tryptic digests reveals Biotin-Mal labeling of Cys-600 is blocked when KDM1A is pre-treated with H2O2. C, KDM1A is readily labeled with 10 μm Biotin-Mal in SH-SY5Y cell lysate and pulled down on streptavidin-agarose beads, and labeling is blocked by pulse pre-treatment (10 min) of intact cells with 1 mm H2O2. Other FAD-dependent amine oxidases do not appear to be as thiol-reactive. D, dose response curve of pulse pre-treatment (10 min) of intact SH-SY5Y cells with H2O2. E, quantification of KDM1A Biotin-Mal labeling, n = 2 assays from biological duplicates. Error bars indicate S.D.
Production of H2O2 in the nucleus is a distinguishing feature of KDM1A/B biochemistry and may have important functional consequences; furthermore, proximal cysteine thiols can be reversibly oxidized by H2O2 to form disulfide bonds. On this basis, we were motivated to test the reactivity of KDM1A with H2O2. When oxidized, cysteine thiols are no longer nucleophilic and are thus do not readily react with electrophilic maleimides (23). Consistent with this mechanism, treatment of recombinant KDM1A with 1 mm H2O2, followed by buffer exchange, blocked Biotin-Mal labeling as detected by blotting with a streptavidin-peroxidase conjugate (Fig. 4C). Furthermore, MALDI-TOF analysis of tryptic peptides confirmed that pre-treatment of recombinant KDM1A with H2O2 blocked Biotin-Mal labeling of Cys-600 (Fig. 5B). Efforts to directly detect the disulfide-linked tryptic peptide by MALDI-TOF were unsuccessful, possibly due to disulfide scrambling.
To test if cysteine oxidation might be relevant to full-length, cellular KDM1A, intact SH-SY5Y cells were pulse treated with 1 mm H2O2 for 10 min, then collected and washed with PBS prior to lysis in buffer containing Biotin-Mal. The lysis buffer used for this experiment was degassed to minimize artifactual oxidation and adjusted to a relatively low pH (6.5) to bias the selectivity of maleimide labeling toward cysteine thiol nucleophiles (23). Under these conditions, pre-treatment with H2O2 reduced Biotin-Mal labeling of KDM1A to near background levels (Fig. 5C). These data indicate that cellular KDM1A is thiol-reactive and sensitive to H2O2. SH-SY5Y cells were selected as a model system due to their expression of all four homologous FAD-dependent amine oxidases. Under these conditions, KDM1A appears to be relatively more thiol-reactive than its homologues KDM1B and MAOA/B, consistent with the in vitro activity observed with recombinant enzymes (Fig. 3A). However, these data do not exclude the possibility that KDM1B and MAO-A/B were not detected due to Biotin-Mal modification preventing epitope recognition by their respective antibodies.
The intracellular steady-state concentration of H2O2 is reported to be in the nanomolar to low micromolar range (23). To determine the apparent IC50 value of cellular KDM1A oxidation, SH-SY5Y cells were pulse-treated with a dose-response curve of H2O2, followed by lysis with Biotin-Mal labeling and pull-down on streptavidin-agarose beads. Oxidation of native KDM1A in intact cells blocked Biotin-Mal labeling within the range of 16 to 62.5 μm H2O2 (Fig. 5, D and E). Although these concentrations exceed physiological relevance, the concentration of H2O2 that diffused into the cell nucleus under these experimental conditions may be lower than the concentration applied exogenously to cells. Thus, this experiment likely underestimates the peroxide sensitivity of KDM1A.
Cysteine thiols can be reversibly oxidized by H2O2 to form sulfenic acids, which may further react with nearby thiols to form disulfide bonds. Several tool compounds have been developed to detect sulfenic acids based on their reactivity with 1,3-diones such as dimedone (23). Efforts to trap and detect sulfenic acid formation in H2O2-treated recombinant KDM1A or in SH-SY5Y cell lysate with dimedone or a biotinylated derivative (BP1) were unsuccessful in both Western blotting analysis and mass spectrometry experiments (data not shown), suggesting that this oxidized intermediate may be rapidly converted to a disulfide species. Under harsh conditions, thiols are oxidized by H2O2 to form sulfinic and sulfonic acids, modifications that are essentially irreversible by reducing agents such as DTT (23). No differences were observed between oxidized and control tryptic peptide MALDI-TOF spectra in the absence of Biotin-Mal labeling, suggesting that any oxidation caused by H2O2 was reversed during the reduction step of the in-gel digestion protocol. While other mechanisms cannot be excluded, these data are fully consistent with the structurally guided hypothesis of intramolecular disulfide bond formation between Cys-600 and Cys-618 of KDM1A.
Activity-dependent Oxidation of KDM1A
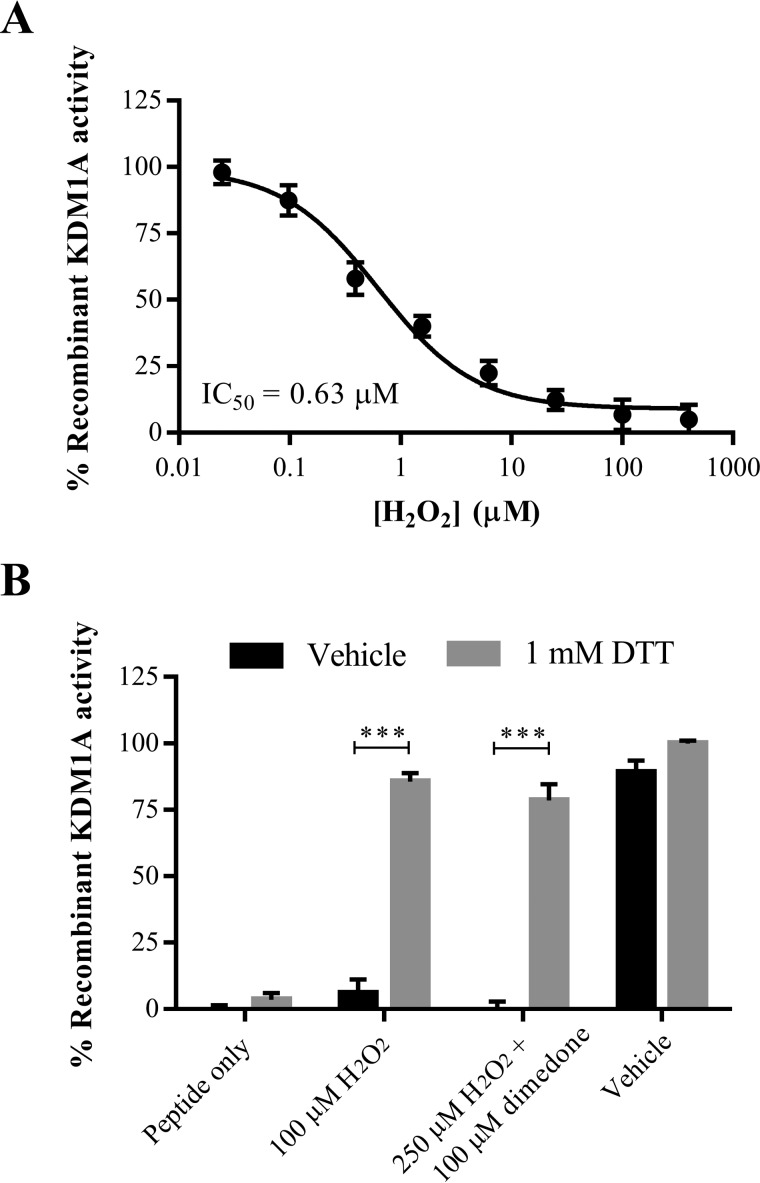
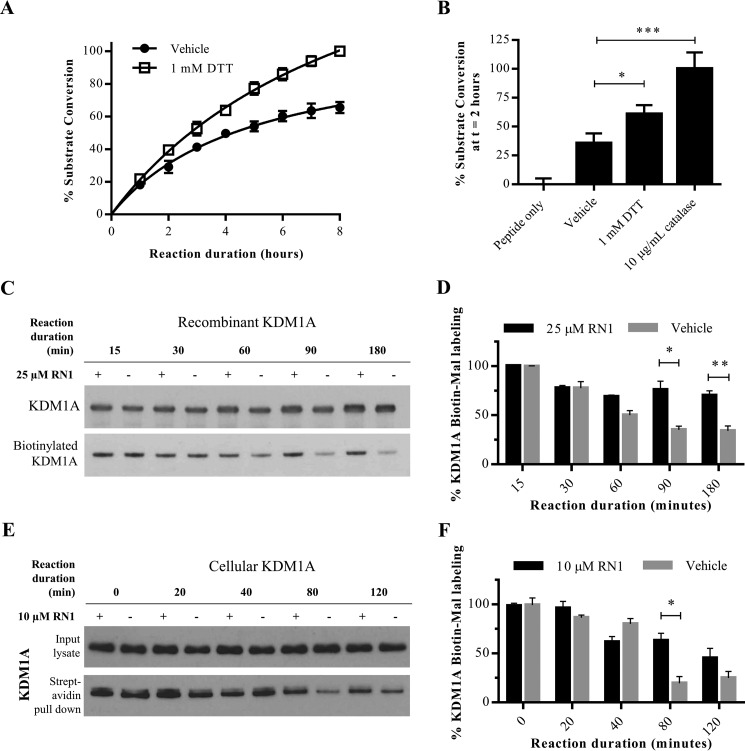
Modification of KDM1A with thiol-reactive small molecules results in enzyme inhibition, and H2O2 pre-treatment blocks these modifications via putative disulfide bond formation. We thus reasoned that H2O2 itself could inhibit KDM1A's enzyme activity. Indeed, exogenously applied H2O2 was found to inhibit the catalytic activity of recombinant KDM1A with an apparent IC50 value around 630 nm in an LC-MS assay, and inhibition was fully reversed by addition of the reducing agent DTT (Fig. 6, A and B). Consistent with lack of BP1 labeling and with rapid disulfide bond formation, pre-treatment of recombinant KDM1A with the sulfenic acid-trapping reagent dimedone had no effect on the reversibility of enzyme inhibition (Fig. 6B). Concentrations of H2O2 required for complete inhibition of KDM1A were an order of magnitude greater than the enzyme concentration (200 nm). We thus postulated that the H2O2 produced by KDM1A as a byproduct of catalysis might negatively regulate enzyme activity by cysteine oxidation after several rounds of substrate conversion. To test this hypothesis, recombinant KDM1A was treated with a large excess of synthetic peptide substrate (1,000-fold) and substrate demethylation was monitored by LC-MS over time. The demethylase activity of vehicle-treated KDM1A plateaued after several hours, whereas catalysis continues unabated in the presence of DTT (Fig. 7A).
FIGURE 6.
Recombinant KDM1A is inhibited by hydrogen peroxide. A, exogenously applied H2O2 inhibits recombinant KDM1A with an apparent IC50 value of 630 nm, error bars indicate S.D. B, inhibition of recombinant KDM1A by H2O2 or dimedone plus H2O2 is reversible by the addition of DTT. KDM1A was pre-incubated with dimedone (100 μm) or vehicle for 10 min, then treated with H2O2 (100 μm or 250 μm) for 10 min prior to reduction. Error bars indicate S.D.; ***, p < 0.001 by 2-tailed t test with correction for multiple comparisons. For both A and B, demethylation of H3K4me2 starting material was detected by LC-MS after 1-h reaction.
FIGURE 7.
Activity-dependent regulation of KDM1A. A, demethylation of H3K4me2 peptide substrate by recombinant KDM1A is enhanced in the presence of DTT (1 mm), or B, the enzyme catalase (10 μg/ml). For both A and B, demethylation of H3K4me2 starting material was detected by LC-MS. Error bars indicate S.D.; *, p < 0.05; ***, p < 0.001 by one-way ANOVA with correction for multiple comparisons. C, labeling of recombinant KDM1A with Biotin-Mal (25 μm) is reduced with extended demethylation reaction durations. Addition of the FAD-directed inhibitor RN1 (25 μm) blocks the time-dependent reduction in labeling. D, quantification of Biotin-Mal labeling as measured by blotting with a streptavidin-HRP conjugate and normalized to total recombinant KDM1A over three replicate experiments. Error bars indicate S.D.; *, p < 0.05; **, p < 0.01, by 2-tailed t test with correction for multiple comparisons. E, labeling of KDM1A in SH-SY5Y cells by Biotin-Mal (25 μm) is reduced with extended incubation in PBS. Addition of the FAD-directed inhibitor RN1 (25 μm) blocks the time-dependent reduction in labeling. F, quantification of Biotin-Mal labeling as measured by pull down with streptavidin-agarose beads and Western conjugate and normalized to total recombinant KDM1A over three replicate experiments. Error bars indicate S.D.; *, p < 0.05 by 2-tailed t test with correction for multiple comparisons.
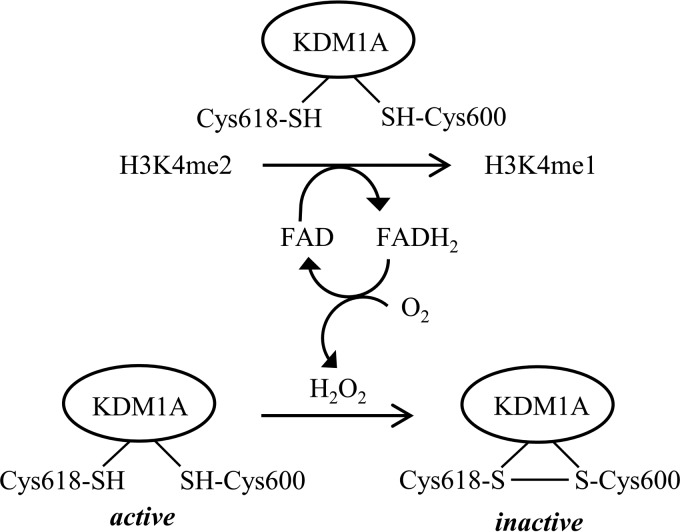
In addition to H2O2, molecular oxygen also oxidizes thiols to disulfide bonds and would be predicted to reduce KDM1A activity over time. To distinguish between these two oxidants, the enzyme catalase was added to decompose H2O2 before its concentration was sufficient to inhibit KDM1A. Addition of catalase to the KDM1A reaction with excess substrate significantly increased substrate demethylation, suggesting that H2O2 is the relevant inactivating oxidant (Fig. 7B). Finally, we tested if KDM1A catalytic activity results in decreased thiol-reactivity over time. Recombinant KDM1A or native KDM1A in intact SH-SY5Y cells was treated with vehicle or inactivated with the FAD-directed inhibitor RN1. Recombinant KDM1A was then mixed with an excess of substrate, whereas cellular KDM1A demethylated its endogenous histone and non-histone substrates. Thiol-reactivity was probed by Biotin-Mal labeling at various time intervals. Initial labeling efficiencies were comparable, indicating that RN1 inhibition is non-competitive with thiol labeling (Fig. 7, C–F). For recombinant KDM1A, an initial drop in labeling efficiency was seen for both active and inactive forms of KDM1A, which may reflect oxidation by O2. However, the labeling efficiency of RN1-inactivated KDM1A plateaus, whereas labeling of the catalytically-active protein diminishes over time (Fig. 7, C and D). Biotin-Mal labeling gradually decreased over time in the cellular assay, but KDM1A labeling was significantly greater when catalytic activity was blocked by RN1 (Fig. 7, E and F). Under these conditions, we propose that KDM1A is feedback inhibited by the H2O2 byproduct of catalysis via a thiol/disulfide switch (Fig. 8).
FIGURE 8.
Working model of KDM1A activity-dependent regulation by a thiol/disulfide redox switch.
Discussion
Epigenetic machinery is regulated by the cellular redox state through several mechanisms, including fluctuations in the availability of metabolites utilized as substrates and cofactors, interaction with redox-sensitive transcription factors, modification by redox-sensitive enzymes, and direct modification by ROS/reactive nitrogen species (RNS) (24). The evolution of two mechanistically-distinct classes of histone lysine demethylases points toward important differences in their regulation, including by their enzymatic chemistries. Enzymes such as flavoproteins that produce ROS are rare in the nucleus, and oxidation of DNA by the H2O2 generated from KDM1A catalysis has been implicated in transcriptional regulation by BER (13–15). Given this context, we were intrigued to discover that KDM1A is highly thiol-reactive both in vitro and in cellular models. Our data indicate that KDM1A enzymatic activity is potently and rapidly inhibited by multiple classes of thiol-reactive compounds, including reversible inhibition by H2O2. Pre-treatment with H2O2 blocks labeling of Cys-600 by Biotin-Mal, implicating intramolecular disulfide bond formation with Cys-618 as regulating KDM1A activity. Notably, this pair of cysteines abuts the conserved Rossmann fold responsible for co-factor binding, suggesting that cross-strand disulfide bond formation may allosterically regulate FAD accessibility or redox potential (22, 25, 26).
Definitive assignment of cysteine reactivity and disulfide bond formation is challenging. Thiol-labeling of MAO-A and MAO-B is substantially reduced by competition with enzyme substrate, indicating that enzyme conformations vary significantly in their cysteine reactivity and/or accessibility (20). Further complicating this analysis is the observation that TCP may form unstable adducts with MAO cysteine residues (27). Our data suggest that KDM1A thiol reactivity is non-competitive with RN1 inactivation, although other FAD-directed inhibitors remain to be tested. Prior to determination of its crystal structure, MAO-A was proposed to contain a redox-sensitive active-site disulfide bond (28). The solved structure of MAO-A later revealed a disulfide linkage between Cys-321 and Cys-323 (PDB ID: 2BXR), although these residues are not near the active site and are not conserved in KDM1A (29). Subsequent electrospray mass spectrometry experiments provided no evidence for intramolecular MAO-A disulfide bond formation, raising the possibility of artifactual oxidation during crystallization (20). Crystallography experiments on oxidized KDM1A may provide additional evidence for disulfide bond formation and clarify the mechanism of enzyme inactivation.
KDM1A is an attractive therapeutic target due to its elevated expression in many cancers and possible role in neurologic disease and viral pathogenesis, and numerous assays exist to profile inhibition by novel compounds (30). Our results as well as others' reports indicate that additional screening strategies are needed to overcome the predominant mode of inhibition via thiol-reactivity and general redox sensitivity to find more tractable, drug-like small molecule inhibitors (31). One potential avenue to circumvent unwanted thiol reactivity is to mutate candidate cysteine residues. This approach has led to mixed results with the monoamine oxidases; for example, site-directed mutagenesis of MAO-A Cys-374 and the corresponding MAO-B Cys-365 to serine resulted in loss of enzyme activity, whereas cysteine to alanine mutants remained active (32). Despite highly similar overall folds, the rate of thiol reactivity is vastly greater for MAO-A than MAO-B, and appears to be greater still for KDM1A, suggesting that subtleties in enzyme structure modulate reactivity. In light of these observations, we favored a chemical tool-based approach to study KDM1A cysteine reactivity.
We found that both KDM1A and KDM1B are inhibited by the FDA-approved drug disulfiram. Disulfiram has been used clinically as an aldehyde dehydrogenase inhibitor, where it inhibits catalysis by modification of active-site cysteines. A class 2 KDM, JMJD2A, has also been reported to be inhibited by disulfiram via disruption of its cysteine-rich zinc-binding site (33). KDM1B contains a unique N-terminal zinc-finger domain consisting of a C4H2C2-type zinc finger and a CW-type zinc finger. Mutation of zinc-finger cysteine residues results in subtle conformational alterations that impair FAD binding and demethylase activity (34). We speculate that disulfiram inhibits KDM1B by a similar mechanism involving cysteine modification and ejection of zinc, but further work is required to test this mode of inactivation. As others have reported, the enzymatic activity of recombinant KDM1B was substantially lower than recombinant KDM1A in vitro, so further efforts will be necessary to determine if KDM1B is negatively regulated by its own H2O2 production (35).
We found that the H2O2 generated by multiple rounds of KDM1A catalysis was linked with diminished enzyme activity in vitro and reduced Biotin-Mal labeling in cells, suggesting that auto-oxidation negatively regulates enzyme activity. To the best of our knowledge, this is the first example of an activity-dependent thiol/disulfide switch among epigenetic enzymes. The physiological relevance of this mechanism remains to be tested, but this chemistry fits within an emerging picture of cysteine-mediated redox regulation of chromatin biology. ROS-generating stimuli result in intramolecular disulfide bond formation between Cys-667 and Cys-669 of HDAC4, promoting nuclear exportation (36). Peroxide-sensitive, regulatory intermolecular disulfides have been proposed to form between the transcription factor FoxO and the p300/CBP acetyltransferase (37). Recently, the Sin3A-associated protein 30-like (SAP30L), a key protein in Sin3A repressive complexes, was found to form two intramolecular disulfide bonds with concomitant release of zinc from a C3H type zinc finger upon H2O2 treatment (38). In addition to cysteine-mediated regulation by ROS, critical cysteine residues on the histone deacetylase HDAC2, a component of KDM1A repressive complexes, appears to be regulated by RNS. Brain-derived neurotrophic factor (BDNF) triggers NO synthesis and induces S-nitrosylation of histone deacetylase 2 (HDAC2) at Cys-262 and Cys-274 in neurons, resulting not in altered enzymatic activity but rather in release from chromatin (39). These same cysteine residues and their homologs on HDAC1 and 3 are carbonylated upon exposure to cigarette smoke and other alkylating agents, resulting in enzyme inactivation (40). These findings collectively point to multiple mechanisms by which cysteine modification regulates chromatin-modifying complexes, and raise the possibility that KDM1A-generated H2O2 may oxidize additional epigenetic regulators.
Our proposed mechanism of thiol/disulfide regulation of KDM1A raises several questions which remain to be addressed in a cellular context. What is the relevant reducing agent that opposes KDM1A oxidation? Notably, the nuclear glutathione and thioredoxin-1 systems responsible for reduction of many protein cysteines are controlled independently from their cytoplasmic counterparts, and are not in redox equilibrium with one another (41). Does negative feedback regulation terminate bursts of highly localized KDM1A activity, such as that resulting in 8-oxoG modification of target genes? Finally, does KDM1A oxidize any of its non-histone substrates? For example, exogenous H2O2 induces methylation of the transcription factor Sp1, a modification that strengthens its interaction with HDAC1 and that is sustained by KDM1A inhibition with pargyline or by KDM1A knock down (42). The H2O2 generated by KDM1A may play multiple roles in the biochemistry of this enzyme and its co-regulators.
Experimental Procedures
Inhibitors and Reagents
TCP, disulfiram, 2,2′-DPS, and dimedone were purchased from Sigma Aldrich. Np-Mal was purchased from ChemBridge (catalogue no. 51332107). Biotin-Mal was purchased from Anaspec (catalogue no. AS-60643). RN1 was synthesized as previously described (16). Inhibitors were used without further purification and were stored as 25 mm stock solutions in DMSO at −20 °C. Dilutions in aqueous buffer were prepared immediately prior to use. Hydrogen peroxide (Sigma Aldrich, cat# 216763) was stored as a 37% solution at 4 °C. DTT (Sigma Aldrich) and 2-iodoacetamide (Alfa Aesar) were stored as powders at 4 °C, and solutions were prepared immediately before use.
Recombinant Enzymes and Substrates
Recombinant KDM1A (GenBankTM Accession No. NM_015013, human amino acids 158-end with N-terminal GST tag, BPS Biosciences, catalogue no. 50100, lot no. 91006) was expressed in Escherichia coli and purified as previously described (16, 19). KDM1B (GenBankTM Accession No. XM_005248926.1, human full-length with N-terminal GST tag, Active Motif catalogue no. 31470, lot no. 21814001) was expressed in Sf9 insect cells and purified by the manufacturer. The 21-mer peptide corresponding to the histone H3 dimethylated lysine 4 tail (NH2-ARTK(me2)QTARKSTGGKAPRKQKA-COOH, abbreviated H3K4me2) was synthesized by the Massachusetts Institute of Technology Biopolymers Laboratory and the purity was confirmed by HPLC and MALDI-TOF mass spectrometry (expected mass = 2297.71, observed ion = 2298.30). Monoamine oxidase A (human, Sigma Aldrich, catalogue no. M7316, lot no. 047K1130) and monoamine oxidase B (human, Sigma Aldrich, catalogue no. M7441, lot no. 021M2162) were expressed in BTI insect cells and purified by the manufacturer.
Cell Lines
HeLa and SH-SY5Y cells were maintained in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (Gibco) and 100 units/ml penicillin-streptomycin (Gibco) at 37 °C and 5% CO2. Cells were collected at near confluence by brief (1–2 min) treatment with 0.05% trypsin-EDTA (Gibco). Trypsin was quenched with 5 volumes of medium, cells were pelleted by centrifugation for 5 min at 1,000 rpm, then media was aspirated, and the cells were washed 2× with 5 volumes of PBS prior to subsequent experimentation.
Recombinant Amine Oxidase Assays (Fig. 3A)
KDM1A (30 nm) was pre-incubated with inhibitors for 10 min in 50 mm pH 7.4 sodium phosphate buffer, followed by addition of H3K4me2 peptide substrate (5 μm) in a total reaction volume of 30 μl. The demethylation reaction was quenched with 1% formic acid after 20 min, then detection of substrate conversion to H3K4me1 and H3K4me0 was accomplished on an Agilent RF300 mass spectrometry system with RapidFire chromatography in line with a triple stage quadrapole mass spectrometer (RapidFire MS) as previously described (16). KDM1B (60 nm) was pre-incubated with inhibitors for 10 min in 50 mm pH 8.1 sodium phosphate buffer followed by addition of H3K4me2 peptide substrate (5 μm) in a total reaction volume of 15 μl. The demethylation reaction was quenched with 1% formic acid after 1 h, then detection of substrate conversion to H3K4me1 and H3K4me0 was accomplished by LC-MS using an Agilent 6310 ion trap mass spectrometer with an ESI source connected to an Agilent 1200 series HPLC with isocratic elution of 5% acetonitrile (ACN) in 0.1% formic acid (FA) at a flow rate of 0.5 ml/min through an Agilent Eclipse XBD-C8 reversed-phase column. Percent enzyme activity was calculated from ratio of H3K4me1 and H3K4me0 peak areas to the H3K4me2 peak area in inhibited wells relative to control wells. MAO-A and MAO-B were pre-incubated with compounds for 10 min, then enzyme activity was assayed using the MAO-Glo assay kit (Promega #V1401) according to the manufacturer's protocol in Proxiplate 384 Plus plates (Perkin Elmer) with a miniaturization of the final assay volume to 20 μl as previously described (16). Reactions were quenched after 1 h by adding reconstituted luciferin detection reagent, and fluorescence at 570 nm was measured on a Perkin Elmer Wallac Envision 2103 Multilabel plate reader. Percent enzyme activity was calculated from the fluorescence readings of inhibited wells relative to control wells. For all enzymes, the compound concentration resulting in 50% inhibition (apparent IC50) was determined by 3-parameter non-linear regression of the plot of log[inhibitor] versus enzyme activity in GraphPad Prism 6 (GraphPad Software, Inc.).
Recombinant KDM1A Activity Assays
With the exception of the inhibition data in Fig. 3, A and C, KDM1A assays were run as follows: recombinant KDM1A was pre-reduced for 15 min on ice with immobilized tris(2-carboxyethyl) phosphine (TCEP) resin (Thermo Scientific), which had been equilibrated in assay buffer, 50 mm, pH 7.4, sodium phosphate. Following reduction, the buffer was exchanged using Zeba spin desalting columns (7K molecular weight cut-off, Thermo Scientific) that had been pre-equilibrated with assay buffer. Demethylation reactions were quenched with 1% formic acid and detection of substrate conversion to H3K4me1 and H3K4me0 was accomplished by LC-MS as described for the KDM1B assay. For dithiothreitol (DTT) reversibility experiments and inhibition by H2O2, 1.5 μm KDM1A was reduced with an equal volume of TCEP resin, desalted, then diluted to a final [KDM1A] of 200 nm. KDM1A was pre-incubated with inhibitors for 10 min, followed by addition of 1 mm DTT or vehicle control. After a 10-min reduction, H3K4me2 substrate was added to a final concentration of 10 μm in a final volume of 30 μl, and the reaction was allowed to proceed for 1 h. For feedback inhibition experiments, 5 μm recombinant KDM1A was reduced with an equal volume of TCEP beads, desalted, then diluted to a final [KDM1A] of 400 nm. DTT (final concentration of 1 mm), catalase (Sigma Aldrich, catalogue no. C1345, final concentration of 10 μg/ml) or vehicle control were added, followed by H3K4me2 substrate (final concentration of 500 μm) in total reaction volume of 10 μl. The demethylation reactions were quenched at various time points as indicated. For Fig. 3C, 6×His-KDM1A (30 nm) was pre-incubated with inhibitors for 10 min in 50 mm, pH 7.4, sodium phosphate buffer with 0.01% Brij-35 detergent (Calbiochem) in 384-well black non-sterile plates (Corning). Following addition of H3K4me2 substrate (10 μm) for a final volume of 30 μl, the reaction proceeded for 20 min at room temperature, during which time the detection reagent was prepared. HRP (Sigma, no. P2088, 5KU) and ADHP (ABD Bioquest no. 11000, in DMSO) were diluted in assay buffer, then 30 μl was added to each well for a final concentration of 0.06 units of HRP and 40 μm ADHP. The plates were immediately read on a PerkinElmer Wallac Envision 2103 Multilabel plate reader (excitation filter: 485 nm; emission filter: 595 nm).
Immunoprecipitation Assay
HeLa cells were lysed with sonication at 4 °C (Fischer FB120, 50% power for 30 s, alternating 1 s on and 2 s off) in ice-cold PBS containing 0.15% Ipegal CA-630 (Sigma Aldrich) and 1× protease inhibitors (Roche, complete, EDTA-free, prepared just prior to use). Lysate was pre-cleared for 1 h, nutated with anti-KDM1A antibody (Abcam, catalogue no. 129195, lot no. YI120618DS) for 1 h, then immunoprecipitated overnight at 4 °C at a ratio of 12 million HeLa cells per 50 μl of packed protein-agarose A beads (Roche) per 5 μl of antibody per tube. KDM1A-bound beads were pooled, washed three times with 50 mm phosphate buffer, pH 7.4 with 0.1% Brij-35 (Calbiochem), then divided into wells of a 384-well plate or into Eppendorf tubes (25 μl of 1:1 bead slurry per reaction). Immunoprecipitated KDM1A was pre-incubated with inhibitors (10 μm) or vehicle control for 10 min before addition of 10 μm H3K4me2 substrate for a final reaction volume of 30 μl. Reactions were incubated at 37 °C for 1 h. Reactions were run in technical replicates, and the entire procedure was repeated twice. For Fig. 3D, 30 μl of HRP and ADHP detection reagent were added to each well as described above, and the plates were immediately read on a PerkinElmer Wallac Envision 2103 Multilabel plate reader (excitation filter: 485 nm; emission filter: 595 nm). For Fig. 3E, samples were quenched by addition of sample loading buffer + DTT. Proteins were separated on pre-cast 4–12% Bis-Tris NuPAGE gels (Invitrogen) with MES running buffer (Invitrogen) for 30 min at 200 V, transferred to PVDF membrane (Millipore), blocked with 5% nonfat milk in TBST then co-probed overnight with 1:1,000 anti-KDM1A (Abcam, catalogue no. 129195, lot no. YI120618DS or Cell Signaling, catalogue no. 2184S, lot no. 1) and 1:1,000 anti-H3K4me2 (Cell Signaling, catalogue no. 9725S, lot no. 1) at 4 °C. Specificity of the H3K4me2 antibody for the recombinant H3K4me2 substrate starting material was confirmed with a substrate-only control and an unmethylated peptide substrate control (data not shown). Detection was achieved with an HRP-linked anti-rabbit secondary antibody and ECL substrate (Thermo Scientific) and captured on autoradiography film (LabScientific). Films were scanned and band intensities from exposures within a linear range were quantified in ImageJ (NIH). Percent enzyme activity was calculated from the remaining H3K4me2 starting material of inhibited lanes relative to control lanes.
Recombinant KDM1A Biotin-Mal Labeling Assays
For stoichiometry experiments, 5 μm recombinant KDM1A was reduced with 2 equivalent volumes of TCEP resin, desalted with buffer exchange, then split into 3 aliquots, which were treated with 1× (5 μm), 5× (25 μm), or 10× (50 μm) biotin-maleimide (Biotin-Mal). Aliquots from each sample were removed after 30, 60, or 90 min of labeling and quenched with 40 mm DTT for 10 min, followed by addition of sample loading buffer for analysis by Western blotting. To determine the demethylase activity of Biotin-Mal labeled KDM1A, an aliquot was removed after 30 min of labeling and diluted to 100 nm in 50 mm phosphate buffer, pH 7.4, containing 10 μm H3K4me2 substrate for a final reaction volume of 30 μl. Detection of H3K4me2 was achieved with Western blotting as described above. For differential labeling experiments, 3 μm recombinant KDM1A was reduced with 2 equivalent volumes of TCEP resin, desalted with buffer exchange, then split into 3 aliquots, which were treated with 1 mm H2O2, 25 μm disulfiram, or vehicle control for 20 min. Each aliquot was then desalted with buffer exchange and labeled with Biotin-Mal at a final concentration of 10 μm for 20 min. Excess maleimide was quenched with 40 mm DTT for 10 min, followed by addition of sample loading buffer for analysis by Western blotting. Biotinylation was detected with a streptavidin-HRP conjugate (Millipore, catalogue no. 18-152), then the membrane was stripped and re-probed for total recombinant protein with 1:10,000 anti-GST (Cell Signaling, catalogue no 2625P, lot no 7) as no commercially-available KDM1A antibodies tested were able to recognize the recombinant protein. For activity-dependent labeling, 5 μm recombinant KDM1A was reduced with an equal volume of TCEP beads, desalted, then diluted to a final [KDM1A] of 400 nm. Recombinant KDM1A was pre-treated with 25 μm RN1 or vehicle control for 10 min before addition of H3K4me2 substrate (final concentration of 500 μm) in total reaction volume of 10 μl. At the time points indicated, 25 μm Biotin-Mal was added for 10 min, then excess maleimide was quenched with 40 mm DTT for 10 min followed by addition of sample loading buffer. Biotinylated and total protein was detected as described above and band intensities were quantified in ImageJ. Biotinylated KDM1A was normalized to total recombinant KDM1A for statistical analyses.
Matrix-assisted Laser Desorption/Ionization (MALDI) Time-of-Flight (TOF) Mass Spectrometry
A solution of 5 μm recombinant KDM1A was reduced with 1 equivalent volume of TCEP resin, desalted with buffer exchange, then split into 2 aliquots, which were treated with 1 mm H2O2 or vehicle control for 20 min. Each sample was desalted with buffer exchange, then split into two aliquots and treated with 10 μm Biotin-Mal or vehicle control for 20 min. Excess maleimide was quenched with 40 mm DTT for 10 min, followed by addition of sample loading buffer. A total of 10 μg of recombinant protein sample was loaded per lane of a pre-cast 4–12% Bis-Tris NuPAGE gel (Invitrogen). The gel was stained with Coomassie Brilliant Blue R-250 staining solution (Bio-Rad) then destained overnight with 50:40:10 methanol/water/acetic acid. Bands were excised, washed with 50% ACN in 50 mm pH 8 ammonium bicarbonate, reduced with 100 μl of 10 mm DTT for 30 min at 80 °C, then alkylated with 55 mm iodoacetamide for 20 min in the dark. Excised bands were washed again then incubated overnight with 10 ng/μl sequencing grade modified porcine trypsin (Promega) in 50 mm pH 8 ammonium bicarbonate at 37 °C. Peptides were extracted in 1% trifluoroacetic acid (TFA), mixed with an equal volume of 70 mg/ml 2,5-dihydroxybenzoic acid matrix (Sigma) in 7:3 ACN/0.1% TFA, then deposited on an MTP 384 polished steel BC target plate (Bruker) and analyzed with an autoflex speed LRF MALDI-TOF mass spectrometer (Bruker). The acquired spectra were manually referenced against a virtual digest (UCSF Protein Prospector MS-Digest) including variable oxidation states and missed cut sites. Samples were deposited in triplicate, and the experiment was repeated twice.
Streptavidin Pull-down Assays
Lysis buffer (50 mm pH 6.5 sodium phosphate with 1% Ipegal CA-630 and 1× protease inhibitors), was degassed by sonication under vacuum for 20 min, then chilled on ice. For differential labeling experiments, SH-SY5Y cells were collected, washed with PBS, then pulse-treated with H2O2 or vehicle control for 20 min. For activity-dependent labeling, SH-SY5Y cells were treated with 10 μm RN1 or DMSO vehicle control for the durations indicated. After compound treatment, cells were pelleted and washed with PBS. Ice-cold lysis buffer was supplemented with 10 μm Biotin-Mal or vehicle control immediately prior to use, and cells were lysed at a density of 2 million SH-SY5Y cells/ml with sonication. After 20 min of labeling, input samples were reserved and 100 μl of pre-equilibrated 1:1 streptavidin-agarose slurry (Life Technologies catalogue no. 951) was added per ml of lysate and nutated for 1 h. The strepativin-agarose beads were washed three times in unsupplemented lysis buffer, then an equal volume of sample loading buffer + DTT was added to each bead slurry and samples were boiled prior to analysis by Western blot. Technical replicates were run on adjacent lanes, and the experiment was repeated twice. Membranes were probed for KDM1A (1:5000, Cell Signaling, catalogue no. 2184S, lot no. 1), KDM1B (1:5000, Abcam catalogue no. ab52001, lot no. GR81275-1), MAO-A (1:1000, Santa Cruz, catalogue no. sc-20156, lot no. D2710) or MAO-B (1:1000, Abcam catalogue no. ab125010, lot no. GR188691-2).
Statistical Analyses
Statistical analysis of enzyme activity and blot densitometry data were performed using unpaired, two-tailed Student's t tests with post hoc Holm-Sidak correction for multiple comparisons or 1-way ANOVA with post hoc Dunnett correction for multiple comparisons to determine p values, as indicated in each figure legend. Statistical testing and graph preparation were performed using GraphPad Prism 6 (GraphPad Software, Inc.). All error bars indicate standard deviation (S.D.). A p value of <0.05 was considered significant. Relative degree of significance is indicated in figures by the number of asterisks and is defined in each figure legend.
Author Contributions
E. L. R. designed, performed, and analyzed the experiments and wrote the paper. J. M. H. and S. J. H. conceived and coordinated the study. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
The members of the Haggarty and Hooker laboratories are thanked for many helpful discussions. Kyle Strom and Norman Lee of Boston University assisted with the MALDI-TOF experiments.
This project was supported by National Institute of Mental Health Grant R01MH095088. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors declare that they have no conflicts of interest with the contents of this article.
- KDM
- lysine-specific demethylase
- FAD
- flavin adenine dinucleotide
- BER
- base excision repair
- TCP
- tranylcypromine
- HDAC
- histone deacetylase
- AO
- amine oxidase.
References
- 1. Mosammaparast N., and Shi Y. (2010) Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu. Rev. Biochem. 79, 155–179 [DOI] [PubMed] [Google Scholar]
- 2. Burg J. M., Link J. E., Morgan B. S., Heller F. J., Hargrove A. E., and McCafferty D. G. (2015) KDM1 class flavin-dependent protein lysine demethylases. Peptide Science 104, 213–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Walport L. J., Hopkinson R. J., and Schofield C. J. (2012) Mechanisms of human histone and nucleic acid demethylases. Curr. Opin. Chem. Biol. 16, 525–534 [DOI] [PubMed] [Google Scholar]
- 4. Gaweska H., Henderson Pozzi M., Schmidt D. M., McCafferty D. G., and Fitzpatrick P. F. (2009) Use of pH and kinetic isotope effects to establish chemistry as rate-limiting in oxidation of a peptide substrate by LSD1. Biochemistry 48, 5440–5445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shi Y., Lan F., Matson C., Mulligan P., Whetstine J. R., Cole P. A., Casero R. A., and Shi Y. (2004) Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119, 941–953 [DOI] [PubMed] [Google Scholar]
- 6. Karytinos A., Forneris F., Profumo A., Ciossani G., Battaglioli E., Binda C., and Mattevi A. (2009) A novel mammalian flavin-dependent histone demethylase. J. Biol. Chem. 284, 17775–17782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yang M., Culhane J. C., Szewczuk L. M., Jalili P., Ball H. L., Machius M., Cole P. A., and Yu H. (2007) Structural basis for the inhibition of the LSD1 histone demethylase by the antidepressant trans-2-phenylcyclopropylamine. Biochemistry 46, 8058–8065 [DOI] [PubMed] [Google Scholar]
- 8. Melvin A., and Rocha S. (2012) Chromatin as an oxygen sensor and active player in the hypoxia response. Cell. Signal. 24, 35–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lu Y., Chu A., Turker M. S., and Glazer P. M. (2011) Hypoxia-induced epigenetic regulation and silencing of the BRCA1 promoter. Mol. Cell. Biol. 31, 3339–3350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lu Y., Wajapeyee N., Turker M. S., and Glazer P. M. (2014) Silencing of the DNA mismatch repair gene MLH1 induced by hypoxic stress in a pathway dependent on the histone demethylase LSD1. Cell Rep. 8, 501–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Luka Z., Moss F., Loukachevitch L. V., Bornhop D. J., and Wagner C. (2011) Histone demethylase LSD1 is a folate-binding protein. Biochemistry 50, 4750–4756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li J., Braganza A., and Sobol R. W. (2013) Base excision repair facilitates a functional relationship between Guanine oxidation and histone demethylation. Antioxidants Redox Signaling 18, 2429–2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Perillo B., Ombra M. N., Bertoni A., Cuozzo C., Sacchetti S., Sasso A., Chiariotti L., Malorni A., Abbondanza C., and Avvedimento E. V. (2008) DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science 319, 202–206 [DOI] [PubMed] [Google Scholar]
- 14. Amente S., Bertoni A., Morano A., Lania L., Avvedimento E., and Majello B. (2010) LSD1-mediated demethylation of histone H3 lysine 4 triggers Myc-induced transcription. Oncogene 29, 3691–3702 [DOI] [PubMed] [Google Scholar]
- 15. Yang S., Zhang J., Zhang Y., Wan X., Zhang C., Huang X., Huang W., Pu H., Pei C., Huang Y., Huang S., and Li Y. (2015) KDM1A triggers androgen-induced miRNA transcription via H3K4me2 demethylation and DNA oxidation. The Prostate 75, 936–946 [DOI] [PubMed] [Google Scholar]
- 16. Neelamegam R., Ricq E. L., Malvaez M., Patnaik D., Norton S., Carlin S. M., Hill I. T., Wood M. A., Haggarty S. J., and Hooker J. M. (2012) Brain-penetrant LSD1 inhibitors can block memory consolidation. ACS Chem. Neurosci. 3, 120–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dahlin J. L., Nissink J. W. M., Strasser J. M., Francis S., Higgins L., Zhou H., Zhang Z., and Walters M. A. (2015) PAINS in the assay: chemical mechanisms of assay interference and promiscuous enzymatic inhibition observed during a sulfhydryl-scavenging HTS. J. Med. Chem. 58, 2091–2113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Baldwin A. D., and Kiick K. L. (2011) Tunable degradation of maleimide–thiol adducts in reducing environments. Bioconjugate Chem. 22, 1946–1953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Forneris F., Binda C., Vanoni M. A., Battaglioli E., and Mattevi A. (2005) Human histone demethylase LSD1 reads the histone code. J. Biol. Chem. 280, 41360–41365 [DOI] [PubMed] [Google Scholar]
- 20. Hubálek F., Pohl J., and Edmondson D. E. (2003) Structural comparison of human monoamine oxidases A and B mass spectrometry monitoring of cysteine reactivities. J. Biol. Chem. 278, 28612–28618 [DOI] [PubMed] [Google Scholar]
- 21. Tamai K., Satoh K., Tsuchida S., Hatayama I., Maki T., and Sato K. (1990) Specific inactivation of glutathione S-transferases in class Pi by SH-modifiers. Biochem. Biophys. Res. Commun. 167, 331–338 [DOI] [PubMed] [Google Scholar]
- 22. Wouters M. A., Fan S. W., and Haworth N. L. (2010) Disulfides as redox switches: from molecular mechanisms to functional significance. Antioxidants Redox Signaling 12, 53–91 [DOI] [PubMed] [Google Scholar]
- 23. Paulsen C. E., and Carroll K. S. (2013) Cysteine-mediated redox signaling: chemistry, biology, and tools for discovery. Chem. Rev. 113, 4633–4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cyr A. R., and Domann F. E. (2011) The redox basis of epigenetic modifications: from mechanisms to functional consequences. Antioxidants Redox Signaling 15, 551–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Forneris F., Binda C., Adamo A., Battaglioli E., and Mattevi A. (2007) Structural basis of LSD1-CoREST selectivity in histone H3 recognition. J. Biol. Chem. 282, 20070–20074 [DOI] [PubMed] [Google Scholar]
- 26. Yang M., Gocke C. B., Luo X., Borek D., Tomchick D. R., Machius M., Otwinowski Z., and Yu H. (2006) Structural basis for CoREST-dependent demethylation of nucleosomes by the human LSD1 histone demethylase. Mol. Cell 23, 377–387 [DOI] [PubMed] [Google Scholar]
- 27. Silverman R. B., and Zieske P. A. (1986) Identification of the amino acid bound to the labile adduct formed during inactivation of monoamine oxidase by 1-phenylcyclopropylamine. Biochem. Biophys. Res. Commun. 135, 154–159 [DOI] [PubMed] [Google Scholar]
- 28. Sablin S. O., and Ramsay R. R. (1998) Monoamine oxidase contains a redox-active disulfide. J. Biol. Chem. 273, 14074–14076 [DOI] [PubMed] [Google Scholar]
- 29. De Colibus L., Li M., Binda C., Lustig A., Edmondson D. E., and Mattevi A. (2005) Three-dimensional structure of human monoamine oxidase A (MAO A): relation to the structures of rat MAO A and human MAO B. Proc. Natl. Acad. Sci. U.S.A. 102, 12684–12689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hayward D., and Cole P. (2016) LSD1 histone demethylase assays and inhibition. Methods Enzymol. 573, 261–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wigle T. J., Swinger K. K., Campbell J. E., Scholle M. D., Sherrill J., Admirand E. A., Boriack-Sjodin P. A., Kuntz K. W., Chesworth R., Moyer M. P., Scott M. P., and Copeland R. A. (2015) A high-throughput mass spectrometry assay coupled with redox activity testing reduces artifacts and false positives in lysine demethylase screening. J. Biomol. Screen. 20, 810–820 [DOI] [PubMed] [Google Scholar]
- 32. Vintém A. P. B., Price N. T., Silverman R. B., and Ramsay R. R. (2005) Mutation of surface cysteine 374 to alanine in monoamine oxidase A alters substrate turnover and inactivation by cyclopropylamines. Bioorganic Medicinal Chemistry 13, 3487–3495 [DOI] [PubMed] [Google Scholar]
- 33. Rose N. R., Thalhammer A., Seden P. T., Mecinović J., and Schofield C. J. (2009) Inhibition of the histone lysine demethylase JMJD2A by ejection of structural Zn (II). Chem. Commun. 42, 6376–6378 [DOI] [PubMed] [Google Scholar]
- 34. Zhang Q., Qi S., Xu M., Yu L., Tao Y., Deng Z., Wu W., Li J., Chen Z., and Wong J. (2013) Structure-function analysis reveals a novel mechanism for regulation of histone demethylase LSD2/AOF1/KDM1b. Cell Res. 23, 225–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kakizawa T., Mizukami T., Itoh Y., Hasegawa M., Sasaki R., and Suzuki T. (2016) Evaluation of phenylcyclopropylamine compounds by enzymatic assay of lysine-specific demethylase 2 in the presence of NPAC peptide. Bioorganic Medicinal Chemistry Letters 26, 1193–1195 [DOI] [PubMed] [Google Scholar]
- 36. Ago T., Liu T., Zhai P., Chen W., Li H., Molkentin J. D., Vatner S. F., and Sadoshima J. (2008) A redox-dependent pathway for regulating class II HDACs and cardiac hypertrophy. Cell 133, 978–993 [DOI] [PubMed] [Google Scholar]
- 37. Dansen T. B., Smits L. M., van Triest M. H., de Keizer P. L., van Leenen D., Koerkamp M. G., Szypowska A., Meppelink A., Brenkman A. B., Yodoi J., Holstege F. C., and Burgering B. M. (2009) Redox-sensitive cysteines bridge p300/CBP-mediated acetylation and FoxO4 activity. Nat. Chem. Biol. 5, 664–672 [DOI] [PubMed] [Google Scholar]
- 38. Laitaoja M., Tossavainen H., Pihlajamaa T., Valjakka J., Viiri K., Lohi O., Permi P., and Jänis J. (2015) Redox-dependent disulfide bond formation in SAP30L corepressor protein: Implications for structure and function. Protein Science 25, 572–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nott A., Watson P. M., Robinson J. D., Crepaldi L., and Riccio A. (2008) S-Nitrosylation of histone deacetylase 2 induces chromatin remodelling in neurons. Nature 455, 411–415 [DOI] [PubMed] [Google Scholar]
- 40. Doyle K., and Fitzpatrick F. A. (2010) Redox signaling, alkylation (carbonylation) of conserved cysteines inactivates class I histone deacetylases 1, 2, and 3 and antagonizes their transcriptional repressor function. J. Biol. Chem. 285, 17417–17424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Go Y.-M., and Jones D. P. (2010) Redox control systems in the nucleus: mechanisms and functions. Antioxidants Redox Signaling 13, 489–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chuang J.-Y., Chang W.-C., and Hung J.-J. (2011) Hydrogen peroxide induces Sp1 methylation and thereby suppresses cyclin B1 via recruitment of Suv39H1 and HDAC1 in cancer cells. Free Rad. Biol. Med. 51, 2309–2318 [DOI] [PubMed] [Google Scholar]