Abstract
Porcine epidemic diarrhea coronavirus (PEDV) is currently devastating the United States pork industry by causing an 80–100% fatality rate in infected piglets. Coronavirus spike proteins mediate virus entry into cells, a process that requires the spike proteins to be proteolytically activated. It has been a conundrum which proteases activate PEDV entry. Here we systematically investigated the roles of different proteases in PEDV entry using pseudovirus entry, biochemical, and live virus infection assays. We found that the PEDV spike is activated by lysosomal cysteine proteases but not proprotein convertases or cell surface serine proteases. Extracellular trypsin activates PEDV entry when lysosomal cysteine proteases are inhibited. We further pinpointed cathepsin L and cathepsin B as the lysosomal cysteine proteases that activate the PEDV spike. These results advance our understanding of the molecular mechanism for PEDV entry and identify potential antiviral targets for curbing the spread of PEDV.
Keywords: cysteine protease, membrane fusion, protease inhibitor, proteolysis, virus entry
Introduction
Since 2013, porcine epidemic diarrhea coronavirus (PEDV)4 has swept throughout the United States, causing an 80–100% fatality rate in piglets and wiping out more than 10% of the pig population of America in less than a year (1–3). Currently there is no effective strategy available to keep the spread of PEDV in check. PEDV belongs to the α genus of the Coronaviridae family. An envelope-anchored spike protein guides coronavirus entry into host cells (4–7). Its ectodomain contains a receptor-binding subunit, S1, and a membrane-fusion subunit, S2. The spike exists in two different conformations: the prefusion conformation is a clove-shaped trimer with three individual S1 heads and a trimeric S2 stalk; the post-fusion conformation is a trimeric S2 that has been structurally rearranged to fuse the viral and host membranes (8–12). During virus entry, S1 first binds to a receptor on the host cell surface for viral attachment, and then S2 transitions to the post-fusion conformation for membrane fusion. For the spike to undergo conformational transitions, it needs to be proteolytically activated by one or more host proteases.
The host proteases mainly come from four different stages of the virus infection cycle: proprotein convertases during virus packaging in virus-producing cells, extracellular proteases after virus release from virus-producing cells and before virus entry into virus-targeted cells, cell surface proteases after virus attachment to virus-targeted cells, and lysosomal proteases after virus endocytosis in virus-targeted cells (9, 10). In addition, proprotein convertases were shown to activate the MERS-CoV spike after virus endocytosis in virus-targeted cells (13). PEDV is unique among coronaviruses in that its propagation in cell culture requires exogenous trypsin, and thus it is commonly believed that extracellular trypsin-like proteases in pig intestines are essential for cell entry of PEDV (14–16). However, these cell culture studies used live PEDV particles for cell entry and did not differentiate PEDV entry from other steps of the PEDV infection cycle, such as virus replication or release. Therefore, it remains to be a conundrum which proteases activate cell entry of PEDV.
We recently characterized the receptor usage and cell entry of PEDV (17). We found that PEDV S1 uses human and porcine aminopeptidase N as its main receptor and sugar as its co-receptor. In addition, PEDV infects both human and porcine cells. Importantly, we established a PEDV spike-mediated pseudovirus entry assay. In this assay, a replication-deficient retrovirus becomes pseudotyped with PEDV spike and is used to enter PEDV-susceptible host cells. The pseudovirus assay only concerns virus entry but not virus replication or release; hence, the pseudovirus entry assay has advantages over the live PEDV infection assay in virus entry studies. Here, using this PEDV pseudovirus entry assay along with biochemical and live PEDV infection assays, we systematically investigated which proteases process and activate PEDV spike, revealing the molecular mechanism for PEDV entry. Identification of the PEDV spike-processing proteases provided potential targets for the development of antiviral drugs to block PEDV entry.
Results
The Role of Proprotein Convertases in PEDV Pseudovirus Entry
To identify the proteases that activate PEDV entry, we examined potential spike-processing proteases from different stages of the virus infection cycle. We first analyzed whether proprotein convertases cleave PEDV spike during virus packaging. To this end, we packaged retrovirus particles pseudotyped with PEDV spike (i.e. PEDV pseudoviruses) in HEK293T cells (human embryonic kidney) and performed Western blotting analysis to detect the cleavage state of PEDV spike. Here the PEDV spike contained a C-terminal C9 tag and, hence, could be detected using an anti-C9 tag monoclonal antibody. Our result showed that PEDV spike remained intact on the pseudovirus surface (Fig. 1A). As a positive control, MERS-CoV spike, which also contained a C-terminal C9 tag, had been cleaved on the pseudovirus surface (Fig. 1A), consistent with previous observations that MERS-CoV entry could be activated by proprotein convertases during virus packaging in virus-producing cells (18, 19). Thus, proprotein convertases from virus-producing cells do not proteolytically activate PEDV spike or PEDV entry.
FIGURE 1.
Proprotein convertases do not activate PEDV pseudovirus entry. A, Western blotting analysis of PEDV spike in pseudovirus particles. Retroviruses pseudotyped with PEDV spike (i.e. PEDV pseudoviruses) were produced in HEK293T cells and then subjected to Western blotting analysis using antibody against its C-terminal C9 tag. B and C, Huh-7 cells (B) or PK-15 cells (C) were preincubated with proprotein convertase inhibitor (PPCi, Dec-RVKR-CMK) at the indicated concentrations and then transduced by PEDV pseudoviruses. Empty vector-packaged pseudoviruses (Mock) were used as a negative control. D, as a positive control, Huh-7 cells were transduced by retroviruses pseudotyped with MERS-CoV spike (i.e. MERS-CoV pseudoviruses). The pseudovirus entry efficiency was characterized as luciferase activity accompanying the entry. The pseudovirus entry in target cells without any inhibitor treatment was taken as 100%. Error bars indicate S.E. (n = 5).
We also examined whether proprotein convertases from virus-targeted cells cleave PEDV spike during virus endocytosis. Our result showed that a proprotein convertase inhibitor, Dec-RVKR-CMK, did not affect PEDV pseudovirus entry into Huh-7 cells (human liver) or PK-15 cells (porcine kidney) (Fig. 1, B and C). As a positive control, MERS-CoV pseudoviruses demonstrated decreased entry into Huh-7 cells in the presence of the proprotein convertase inhibitor (Fig. 1D), consistent with previous observations that MERS-CoV entry could be activated by proprotein convertases after virus endocytosis in virus-targeted cells (13). Thus, proprotein convertases from virus-targeted cells do not proteolytically activate PEDV spike or PEDV entry either.
The Role of Cell Surface Proteases in PEDV Pseudovirus Entry
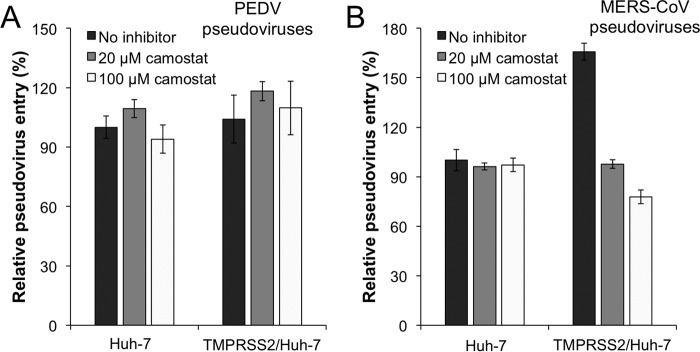
Next we investigated whether cell surface proteases activate PEDV entry. Previous studies demonstrated that Huh-7 cells do not express the cell surface serine protease TMPRSS2 (19, 20). Here we showed that PEDV pseudoviruses entered Huh-7 cells efficiently, indicating that PEDV entry does not require activation by TMPRSS2 (Fig. 2A). Furthermore, exogenously expressing TMPRSS2 in Huh-7 cells did not enhance PEDV pseudovirus entry (Fig. 2A). Additionally, a TMPRSS2 inhibitor, camostat, had no impact on PEDV pseudovirus entry into Huh-7 cells exogenously expressing or not expressing TMPRSS2 (Fig. 2A). These results all suggest that TMPRSS2 does not activate PEDV entry into host cells. As a positive control, MERS-CoV pseudovirus entry was enhanced in Huh-7 cells exogenously expressing TMPRSS2 (Fig. 2, A and B). Moreover, the enhanced MERS-CoV pseudovirus entry in Huh-7 cells exogenously expressing TMPRSS2 could be reversed by camostat (Fig. 2B). These results were consistent with previous observations that MERS-CoV entry could be activated by TMPRSS2 (21, 22). As a negative control, camostat did not affect MERS-CoV pseudovirus entry into Huh-7 cells not expressing TMPRSS2 (Fig. 2B). Therefore, we can rule out the role of cell surface serine proteases in processing PEDV spike and activating PEDV entry.
FIGURE 2.
Cell surface serine proteases do not activate PEDV pseudovirus entry. A and B, Huh-7 cells transiently transfected with the empty pCAGGS vector or TMPRSS2 in the pCAGGS vector were preincubated with camostat (a cell surface serine proteases inhibitor) at the indicated concentrations and transduced by PEDV pseudoviruses (A) or MERS-CoV pseudoviruses (B). The pseudovirus entry in empty pCAGGS vector-transfected Huh-7 cells without any inhibitor treatment was taken as 100%. Error bars indicate S.E. (n = 5).
The Role of Lysosomal Cysteine Proteases in PEDV Pseudovirus Entry
Next we examined whether lysosomal cysteine proteases activate PEDV entry. To this end, we carried out PEDV pseudovirus entry into Huh-7 or PK-15 cells in the presence of a lysosomal acidification inhibitor, bafilomycin A1, or a lysosomal cysteine protease inhibitor, E-64d. We found that both inhibitors significantly reduced PEDV pseudovirus entry into host cells in a dose-dependent manner (Fig. 3, A and B). As a control, only bafilomycin A1, but not E-64d, significantly reduced VSV pseudovirus entry into Huh-7 and PK-15 cells (Fig. 3, C and D). The result from the control experiment is consistent with previous reports that VSV entry into host cells depends on endocytosis but not lysosomal cysteine proteases (23, 24). The control experiment also showed that the inhibitors did not have nonspecific cytotoxic effects on target cells. Thus, lysosomal cysteine proteases play a critical role in PEDV entry.
FIGURE 3.

Lysosomal cysteine proteases activate PEDV pseudovirus entry. A and B, Huh-7 cells (A) or PK-15 cells (B) were preincubated with bafilomycin A1 (Baf-A1, a lysosomal acidification inhibitor) or E-64d (a lysosomal cysteine protease inhibitor) at the indicated concentrations and then transduced by PEDV pseudoviruses. C and D, retroviruses pseudotyped with VSV envelop glycoprotein (i.e. VSV pseudoviruses) were used as a control. The pseudovirus entry in target cells without any inhibitor treatment was taken as 100%. Error bars indicate S.E. (n = 5).
We went further to pinpoint the specific lysosomal cysteine proteases that cleave PEDV spike and activate PEDV entry. We focused on cathepsin L and cathepsin B because both of these cathepsins have been identified previously to process the spike proteins from other coronaviruses, including severe acute respiratory syndrome and MERS coronaviruses (19, 24–27). To identify the role of cathepsin L and cathepsin B in PEDV entry, we carried out PEDV pseudovirus entry in the presence of inhibitors that are specific for cathepsin L (i.e. inhibitor Z-FY-CHO) or cathepsin B (i.e. CA-074 Me), respectively. The result showed that both inhibitors dramatically reduced PEDV pseudovirus entry into Huh-7 and PK-15 cells (Fig. 4, A and B).
FIGURE 4.

Cathepsin L and cathepsin B activate PEDV pseudovirus entry. A and B, before being infected by PEDV pseudoviruses, Huh-7 cells (A) or PK-15 cells (B) were preincubated with 50 μm cathepsin L inhibitor (i.e. Z-FY-CHO) or 50 μm cathepsin B inhibitor (i.e. CA-074 Me). After pseudovirus attachment to target cells, unbound pseudovirus particles were removed, and bound pseudovirus particles were either treated or not treated with 40 μg/ml exogenous trypsin. PEDV pseudovirus entry in the absence of inhibitor or exogenous trypsin was taken as 100% in each cell line. PEDV pseudovirus entry in the absence of inhibitor and in the presence of trypsin is shown separately in Fig. 6. Error bars indicate S.E. (n = 4). C, PEDV spike was transiently expressed in HEK293T cells, the cells were lysed through sonication, and the expressed spike protein was subsequently subjected to cathepsin L or cathepsin B cleavage at gradient concentrations at pH 5.6. The spike was detected using an antibody against its C-terminal C9 tag.
To provide direct biochemical evidence that cathepsin L and cathepsin B cleave PEDV spike, we expressed PEDV spike in HEK293T cells, lysed the cells, and treated the cell-expressed PEDV spike with recombinant cathepsin L and cathepsin B, respectively, at pH 5.6 (i.e. the working pH level for cathepsins). We then detected the cleavage state of the cell-expressed PEDV spike using Western blotting analysis. Our result showed that, at relatively low concentrations (e.g. 1 μg/ml), cathepsin L cleaved PEDV spike to S2 (Fig. 4C). At higher concentrations (e.g. 4 μg/ml), cathepsin L further cleaved PEDV S2. On the other hand, at relative low concentrations (e.g. 1 μg/ml), cathepsin B did not cleave PEDV spike efficiently. At higher concentrations (e.g. 10 μg/ml), cathepsin B cleaved PEDV spike to S2 but failed to further cleave PEDV S2 (Fig. 4C). Hence, PEDV spike is more sensitive to the cleavage of cathepsin L than to the cleavage of cathepsin B. In sum, host lysosomal cysteine proteases, particularly cathepsin L and cathepsin B, process PEDV spike and activate PEDV entry.
Recognizing that lysosomal cysteine proteases differ in their expression levels among different tissues (28), we selected IPI-21 cells (porcine small intestine) for repeating PEDV pseudovirus entry in the presence of lysosomal cysteine proteases (Fig. 5). The porcine small intestine is the major target organ for PEDV infections (29–31). Our result showed that PEDV pseudovirus entry into IPI-21 cells could be inhibited by the lysosomal acidification inhibitor bafilomycin A1, the lysosomal cysteine protease inhibitor E-64d, and cathepsin-l- and cathepsin-B-specific inhibitors. Therefore, lysosomal cysteine proteases activate PEDV entry into cells from porcine small intestine.
FIGURE 5.
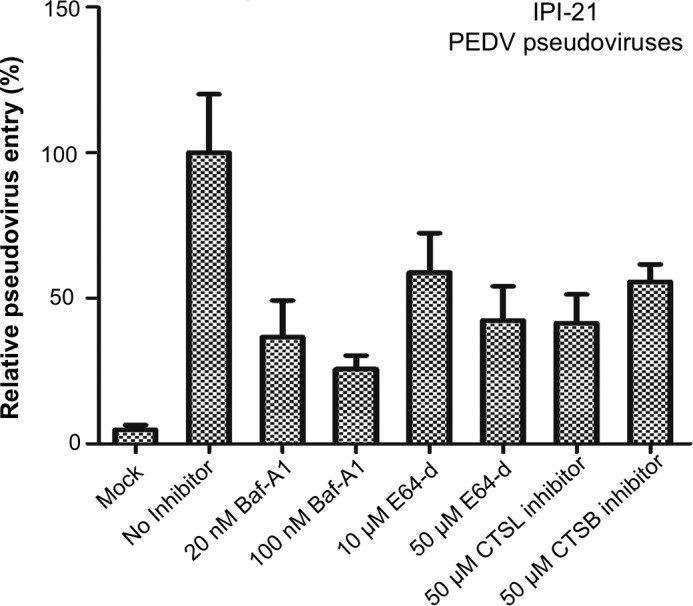
Lysosomal cysteine proteases activate PEDV pseudovirus entry into porcine small intestine cells. IPI-21 cells from porcine small intestine, which is the primary target organ for PEDV, were preincubated with Baf-A1, E-64d, cathepsin L (CTSL) inhibitor (i.e. Z-FY-CHO), and cathepsin B (CTSB) inhibitor (i.e. CA-074 Me) at the indicated concentrations and then transduced by PEDV pseudoviruses. Empty pcDNA vector-packaged pseudoviruses (Mock) were used as a negative control. The pseudovirus entry mediated by PEDV spike in the absence of any inhibitor was taken as 100%. Error bars indicate S.E. (n = 4).
The Role of Extracellular Proteases in PEDV Pseudovirus Entry
We also tackled the confounding role of the extracellular protease trypsin in PEDV entry. Previous studies showed that exogenous trypsin could activate the entry of severe acute respiratory syndrome and MERS-CoV into host cells after the viruses had already been attached to host cells (19, 26, 32). Hence, we added trypsin after PEDV pseudoviruses had been attached to Huh-7 or PK-15 cells. Our result revealed that trypsin slightly reduced PEDV pseudovirus entry into Huh7 and PK-15 cells (Fig. 6, A and B). On the other hand, in the presence of a cathepsin L or cathepsin B inhibitor, the dramatically reduced PEDV pseudovirus entry into host cells could be partially rescued by extracellular trypsin (Fig. 4, A and B). Taken together, extracellular trypsin has the potential to process PEDV spike when lysosomal cysteine proteases are inhibited or unavailable; however, when available, lysosomal cysteine proteases play a major role in PEDV entry into host cells.
FIGURE 6.

Extracellular protease trypsin serves a backup role in activating PEDV pseudovirus entry. A and B, after PEDV pseudoviruses were incubated with Huh-7 cells (A) or PK-15 cells (B), unbound PEDV pseudovirus particles were removed, and bound PEDV pseudovirus particles were either treated or not treated with exogenous trypsin at the indicated concentrations. Empty pcDNA vector-packaged pseudoviruses (Mock) were used as a negative control. The pseudovirus entry mediated by PEDV spike in the absence of exogenous trypsin was taken as 100%. Error bars indicate S.E. (n = 4).
The Role of Lysosomal Cysteine Proteases in Live PEDV Entry
Last we investigated the role of lysosomal cysteine proteases in live PEDV infection in cell culture (Fig. 7). Without trypsin, PEDV replicated inefficiently in Vero CCL81 cells (monkey kidney) but still at a detectable level. PEDV replication in Vero CCL81 cells was reduced to nearly undetectable levels by the lysosomal cysteine protease inhibitor E-64d, cathepsin L inhibitor, or cathepsin B inhibitor. These results are consistent with the pseudovirus entry assay, confirming that lysosomal cysteine proteases play critical roles in PEDV entry into host cells.
FIGURE 7.
Lysosomal cysteine proteases activate live PEDV entry into cells. Vero CCL81 cells were pretreated with one of the lysosomal cysteine protease inhibitors (E64d, cathepsin L, or cathepsin B) before they were infected by live PEDV. 24 h post-infection, cells were chemically fixed and incubated with FITC-labeled mouse anti-PEDV N protein antibody. PEDV-positive cells were observed using a fluorescence microscope.
Discussion
Our study has elucidated a long-standing puzzle regarding which proteases activate PEDV entry into host cells. Previous studies identified the extracellular protease trypsin as required for PEDV infection in cell culture, which led to the conclusion that intestinal proteases are essential for PEDV entry (14–16). However, these previous studies all used live PEDV virus particles and therefore were unable to differentiate between PEDV entry and other steps in the PEDV infection cycle, such as virus replication or release. Indeed, an electron microscopic study showed that PEDV release is a limiting step in the PEDV infection cycle and that trypsin is required for PEDV release (33). To separate PEDV entry from other steps of the PEDV infection cycle, we performed a PEDV pseudovirus entry assay in which PEDV spike-packaged pseudovirus particles can only enter host cells but cannot replicate or be released. Thus the PEDV pseudovirus entry assay provides a simplified system for studying PEDV entry (17). Using this assay, we showed that PEDV entry does not depend on proprotein convertases or cell surface proteases. Instead, PEDV entry is activated by lysosomal cysteine proteases. Using both pseudovirus entry and direct biochemical assays, we further identified cathepsin L and cathepsin B as the specific lysosomal cysteine proteases that can process PEDV spike. We obtained the result using several cell lines, including IPI-21 cells from porcine small intestine, the major target organ for PEDV infections. Hence, our finding is relevant for pig infection and development of antivirals. Further, we confirmed this result using a live PEDV infection assay in cell culture. Our study also elucidated the puzzling role of extracellular trypsin in PEDV entry. When cathepsins are available, trypsin is not as efficient as cathepsins in activating PEDV entry. However, when cathepsins are inhibited or unavailable, trypsin can fill in the role of cathepsins by activating PEDV entry. Because this study focuses on PEDV entry, the role of trypsin in other steps of the PEDV infection cycle, such as PEDV release, remains to be investigated using other research approaches. Nevertheless, our study has laid out a blueprint for systematically examining the roles of proteases in virus entry and provided insight into the puzzling molecular mechanism for PEDV entry.
PEDV is currently sweeping through the pig population of America with little hindrance because neither vaccines nor antiviral drugs are available to curb its spread. Our study suggests that cysteine protease inhibitors such as MDL 28170 can serve as a class of antiviral agents that potentially block PEDV infections (34). Moreover, our finding suggests that cysteine protease inhibitors alone may not be sufficient to block PEDV entry because trypsin can have a backup role in activating PEDV entry. Instead, combinational use of cysteine protease inhibitors and trypsin inhibitors may be more effective to block PEDV entry and treat infected pigs.
Materials and Methods
Cell Lines and Plasmids
HEK293T (human embryonic kidney), PK-15 (porcine kidney), and Vero CCL81 (monkey kidney) cells were obtained from the American Type Culture Collection. IPI-21 (porcine small intestine) cells were purchased from Sigma-Aldrich. Huh-7 (human hepatoma) cells were kindly provided by Dr. Charles M. Rice at Rockefeller University. These cell lines were cultured in DMEM supplemented with 10% FBS, 2 mm l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin (Life Technologies).
The genes of MERS-CoV spike (GenBank accession no. AFS88936.1) and PEDV spike (GenBank accession no. AGO58924.1) were each cloned into the pcDNA3.1(+) vector (Life Technologies) with a C-terminal C9 tag. The genes of cathepsin L (GenBank accession no. CAA30981.1) and cathepsin B (GenBank accession no. AAA52129.1) were synthesized from GenScript and were each cloned into the pFastBac1 vector (Life Technologies) with a C-terminal His6 tag. The plasmid of human TMPRSS2 was kindly provided by Dr. Tom Gallagher at Loyola University Medical Center.
Protein Expression and Purification
Cathepsin L and cathepsin B were expressed as an inactive proform in insect cells and purified as described previously (19). Briefly, the full-length proteases containing a C-terminal His6 tag were expressed in Sf9 insect cells using the Bac-to-Bac expression system (Life Technologies), secreted to cell culture medium, and purified using a HiTrap Chelating HP column and Superdex 200 gel filtration column (GE Healthcare), sequentially. Purified pro-cathepsin L and pro-cathepsin B were autoactivated as described previously (35). Briefly, purified pro-cathepsin L or pro-cathepsin B were diluted 10-fold using 100 mm sodium acetate (pH 4.0) and incubated at 37 °C for 1 h. The activated mature proteases were further purified on a Superdex 200 gel filtration column.
Pseudovirus Entry into Human and Pig Cells
Retroviruses pseudotyped with PEDV spike, MERS-CoV spike, or VSV envelope glycoprotein were generated as described previously (17, 19). Briefly, HEK293T cells were co-transfected with a plasmid carrying an Env-defective, luciferase-expressing HIV, type 1 genome (pNL4–3.luc.R-E-) and a plasmid encoding PEDV spike or MERS-CoV spike using Lipofectamine 3000 reagent (Life Technologies) according to the instructions of the manufacturer. Supernatants containing pseudoviruses were harvested 72 h after transfection and centrifuged at 1200 × g for 10 min to remove cell debris. Pseudoviruses were concentrated using Amicon Ultra centrifugal filter units with a 100-kDa molecular weight cutoff and then aliquoted and frozen at −80 °C for later use.
Retroviruses pseudotyped with PEDV spike, MERS-CoV spike, VSV envelope glycoprotein, or empty vector (mock) were used to transduce Huh-7 cells, Huh-7 cells transiently expressing human TMPRSS2, PK-15 cells, or IPI-21 cells in 96-well plates. For trypsin processing of pseudoviruses, the initial DMEM was removed after pseudovirus attachment for 2 h and subsequently replaced with serum-free DMEM with 0, 10, or 40 μg/ml l-1-tosylamido-2-phenylethyl chloromethyl ketone-treated trypsin (Sigma-Aldrich). After incubating with trypsin at 37 °C for 10 min, DMEM supplemented with 75 μg/ml soybean trypsin inhibitor (Sigma-Aldrich) was added to neutralize trypsin. Cells were incubated at 37 °C for another 12 h, and the medium was replaced with fresh DMEM. 48 h later, cells were washed with PBS and lysed. Aliquots of cell lysates were transferred to an Optiplate-96 plate (PerkinElmer Life Sciences), and luciferase substrate (Promega) was added. Relative luciferase units were measured using an EnSpire plate reader (PerkinElmer Life Sciences).
Inhibition of Pseudovirus Entry into Human and Pig Cells Using Inhibitors
Inhibition of pseudovirus entry using various protease inhibitors was carried out as described previously (18). Briefly, target cells were preincubated with 10 or 50 μm proprotein convertase inhibitor Dec-RVKR-CMK (Enzo Life Sciences), 20 or 100 μm camostat mesylate (Sigma-Aldrich), 20 or 100 nm bafilomycin A1 (Sigma-Aldrich), 10 or 50 μm E-64d (Sigma-Aldrich), 50 μm cathepsin L inhibitor Z-FY-CHO (Santa Cruz Biotechnology), or 50 μm cathepsin B inhibitor CA-074 Me (Santa Cruz Biotechnology) at 37 °C for 1 h. The cells were subsequently transduced by retroviruses pseudotyped with PEDV spike, MERS-CoV spike, or VSV envelope glycoprotein. The cells were incubated at 37 °C for 6–8 h, and then the medium was replaced with fresh DMEM. 48 h later, the cells were lysed and measured for luciferase activity.
Western Blotting Analysis of Spike Cleavage by Proprotein Convertases
PEDV and MERS-CoV pseudoviruses were packaged in HEK293T cells, lysed, and then subjected to Western blotting analysis. The C9-tagged spikes were detected using an anti-C9 tag monoclonal antibody (Santa Cruz Biotechnology).
Western Blotting Analysis of Spike Cleavage by Lysosomal Cysteine Proteases
HEK293T cells were transfected with plasmids encoding PEDV spike or MERS-CoV spike. 48 h after transfection, the cells were harvested, washed with PBS, and lysed by sonication. Cell lysates were then incubated with activated cathepsin L or cathepsin B at gradient concentrations at pH 5.6 and 37 °C for 30 min and subjected to Western blotting analysis. The C9-tagged spikes were detected using an anti-C9 tag monoclonal antibody (Santa Cruz Biotechnology).
Inhibition of Live PEDV Entry into Host Cells Using Inhibitors
Vero CCL81 cells were washed three times with DMEM and pretreated with 50 μm E-64d, 50 μm cathepsin L inhibitor, or 50 μm cathepsin B inhibitor in DMEM. After 1 h, cells were infected with PEDV strain Ohio VBS2 at a multiplicity of infection of 0.5 as described previously (17). 2 h after infection, cells were washed with DMEM three times to remove unbound PEDV particles. DMEM was supplemented with 0.018% (w/v) tryptose phosphate broth (Sigma) and 0.02% yeast extract (Sigma), and the respective inhibitor at the above concentration was then added. 24 h after infection, cells were washed twice with PBS and fixed with 4.0% (v/v) paraformaldehyde and 0.2% (v/v) glutaraldehyde in 0.1 m potassium phosphate buffer (pH 7.4) at 22 °C for 15 min. They were then washed three times with PBS. After permeabilization with 0.1% Triton X-100 in PBS for 15 min, the cells were washed with PBS and blocked with PBS containing 2% bovine serum albumin at room temperature for 1 h. The cells were then incubated with FITC-labeled mouse anti-PEDV N protein antibody (Medgene Labs, Brookings, SD) in 0.2% BSA in PBS at dilution of 1:100 at 4 °C overnight. Cells were examined under an Olympus fluorescence microscope system.
Author Contributions
C. L., Y. Z., S. J., L. D., J. L., and F. L. designed the experiments and wrote the manuscript. C. L., Y. M., Y. Y., Y. Z., and J. S. performed the experiments. C. L., Y. M., Y. Y., Y. Z., J. S., Y. Z., S. J., L. D., J. L., and F. L. analyzed the data.
This work was supported by NIAID, National Institutes of Health Grants R01AI089728 (to F. L). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- PEDV
- porcine epidemic diarrhea coronavirus
- MERS-CoV
- Middle East respiratory syndrome coronavirus
- Dec-RVKR-CMK
- decanoyl-Arg-Val-Lys-Arg-chloromethylketone
- Z-FY-CHO
- Z-Phe-Tyr-aldehyde
- CA-074 Me
- CA-074 methyl ester.
References
- 1. Mole B. (2013) Deadly pig virus slips through US borders. Nature 499, 388. [DOI] [PubMed] [Google Scholar]
- 2. Stevenson G. W., Hoang H., Schwartz K. J., Burrough E. R., Sun D., Madson D., Cooper V. L., Pillatzki A., Gauger P., Schmitt B. J., Koster L. G., Killian M. L., and Yoon K. J. (2013) Emergence of porcine epidemic diarrhea virus in the United States: clinical signs, lesions, and viral genomic sequences. J. Vet. Diagn. Invest. 25, 649–654 [DOI] [PubMed] [Google Scholar]
- 3. Chen Q., Li G., Stasko J., Thomas J. T., Stensland W. R., Pillatzki A. E., Gauger P. C., Schwartz K. J., Madson D., Yoon K. J., Stevenson G. W., Burrough E. R., Harmon K. M., Main R. G., and Zhang J. (2014) Isolation and characterization of porcine epidemic diarrhea viruses associated with the 2013 disease outbreak among swine in the United States. J. Clin. Microbiol. 52, 234–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Perlman S., and Netland J. (2009) Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 7, 439–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li W., Wong S. K., Li F., Kuhn J. H., Huang I. C., Choe H., and Farzan M. (2006) Animal origins of the severe acute respiratory syndrome coronavirus: insight from ACE2-S-protein interactions. J. Virol. 80, 4211–4219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li F. (2015) Receptor recognition mechanisms of coronaviruses: a decade of structural studies. J. Virol. 89, 1954–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li F. (2016) Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 3, 237–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li F., Berardi M., Li W., Farzan M., Dormitzer P. R., and Harrison S. C. (2006) Conformational states of the severe acute respiratory syndrome coronavirus spike protein ectodomain. J. Virol. 80, 6794–6800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Belouzard S., Millet J. K., Licitra B. N., and Whittaker G. R. (2012) Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses 4, 1011–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Heald-Sargent T., and Gallagher T. (2012) Ready, set, fuse! The coronavirus spike protein and acquisition of fusion competence. Viruses 4, 557–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Walls A. C., Tortorici M. A., Bosch B. J., Frenz B., Rottier P. J., DiMaio F., Rey F. A., and Veesler D. (2016) Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer. Nature 531, 114–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kirchdoerfer R. N., Cottrell C. A., Wang N., Pallesen J., Yassine H. M., Turner H. L., Corbett K. S., Graham B. S., McLellan J. S., and Ward A. B. (2016) Pre-fusion structure of a human coronavirus spike protein. Nature 531, 118–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Millet J. K., and Whittaker G. R. (2014) Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl. Acad. Sci. U.S.A. 111, 15214–15219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hofmann M., and Wyler R. (1988) Propagation of the virus of porcine epidemic diarrhea in cell culture. J. Clin. Microbiol. 26, 2235–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Oka T., Saif L. J., Marthaler D., Esseili M. A., Meulia T., Lin C. M., Vlasova A. N., Jung K., Zhang Y., and Wang Q. (2014) Cell culture isolation and sequence analysis of genetically diverse US porcine epidemic diarrhea virus strains including a novel strain with a large deletion in the spike gene. Vet. Microbiol. 173, 258–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wicht O., Li W., Willems L., Meuleman T. J., Wubbolts R. W., van Kuppeveld F. J., Rottier P. J., and Bosch B. J. (2014) Proteolytic activation of the porcine epidemic diarrhea coronavirus spike fusion protein by trypsin in cell culture. J. Virol. 88, 7952–7961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu C., Tang J., Ma Y., Liang X., Yang Y., Peng G., Qi Q., Jiang S., Li J., Du L., and Li F. (2015) Receptor usage and cell entry of porcine epidemic diarrhea coronavirus. J. Virol. 89, 6121–6125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang Y., Liu C., Du L., Jiang S., Shi Z., Baric R. S., and Li F. (2015) Two mutations were critical for bat-to-human transmission of Middle East respiratory syndrome coronavirus. J. Virol. 89, 9119–9123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang Y., Du L., Liu C., Wang L., Ma C., Tang J., Baric R. S., Jiang S., and Li F. (2014) Receptor usage and cell entry of bat coronavirus HKU4 provide insight into bat-to-human transmission of MERS coronavirus. Proc. Natl. Acad. Sci. U.S.A. 111, 12516–12521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bertram S., Glowacka I., Blazejewska P., Soilleux E., Allen P., Danisch S., Steffen I., Choi S. Y., Park Y., Schneider H., Schughart K., and Pöhlmann S. (2010) TMPRSS2 and TMPRSS4 facilitate trypsin-independent spread of influenza virus in Caco-2 cells. J. Virol. 84, 10016–10025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shirato K., Kawase M., and Matsuyama S. (2013) Middle East respiratory syndrome coronavirus infection mediated by the transmembrane serine protease TMPRSS2. J. Virol. 87, 12552–12561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gierer S., Bertram S., Kaup F., Wrensch F., Heurich A., Krämer-Kühl A., Welsch K., Winkler M., Meyer B., Drosten C., Dittmer U., von Hahn T., Simmons G., Hofmann H., and Pöhlmann S. (2013) The spike protein of the emerging betacoronavirus EMC uses a novel coronavirus receptor for entry, can be activated by TMPRSS2, and is targeted by neutralizing antibodies. J. Virol. 87, 5502–5511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hofmann H., Hattermann K., Marzi A., Gramberg T., Geier M., Krumbiegel M., Kuate S., Uberla K., Niedrig M., and Pöhlmann S. (2004) S protein of severe acute respiratory syndrome-associated coronavirus mediates entry into hepatoma cell lines and is targeted by neutralizing antibodies in infected patients. J. Virol. 78, 6134–6142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Qian Z., Dominguez S. R., and Holmes K. V. (2013) Role of the spike glycoprotein of human Middle East respiratory syndrome coronavirus (MERS-CoV) in virus entry and syncytia formation. PLoS ONE 8, e76469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Simmons G., Gosalia D. N., Rennekamp A. J., Reeves J. D., Diamond S. L., and Bates P. (2005) Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. U.S.A. 102, 11876–11881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Simmons G., Reeves J. D., Rennekamp A. J., Amberg S. M., Piefer A. J., and Bates P. (2004) Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc. Natl. Acad. Sci. U.S.A. 101, 4240–4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Huang I. C., Bosch B. J., Li F., Li W., Lee K. H., Ghiran S., Vasilieva N., Dermody T. S., Harrison S. C., Dormitzer P. R., Farzan M., Rottier P. J., and Choe H. (2006) SARS coronavirus, but not human coronavirus NL63, utilizes cathepsin L to infect ACE2-expressing cells. J. Biol. Chem. 281, 3198–3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bando Y., Kominami E., and Katunuma N. (1986) Purification and tissue distribution of rat cathepsin L. J. Biochem. 100, 35–42 [DOI] [PubMed] [Google Scholar]
- 29. Li W., van Kuppeveld F. J., He Q., Rottier P. J., and Bosch B. J. (2016) Cellular entry of the porcine epidemic diarrhea virus. Virus Res. 10.1016/j.virusres.2016.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ducatelle R., Coussement W., Debouck P., and Hoorens J. (1982) Pathology of experimental CV777 coronavirus enteritis in piglets: II: electron microscopic study. Vet. Pathol. 19, 57–66 [DOI] [PubMed] [Google Scholar]
- 31. Ducatelle R., Coussement W., Pensaert M. B., Debouck P., and Hoorens J. (1981) In vivo morphogenesis of a new porcine enteric coronavirus, CV 777. Arch. Virol. 68, 35–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Simmons G., Zmora P., Gierer S., Heurich A., and Pöhlmann S. (2013) Proteolytic activation of the SARS-coronavirus spike protein: cutting enzymes at the cutting edge of antiviral research. Antiviral Res. 100, 605–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shirato K., Matsuyama S., Ujike M., and Taguchi F. (2011) Role of proteases in the release of porcine epidemic diarrhea virus from infected cells. J. Virol. 85, 7872–7880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mehdi S. (1991) Cell-penetrating inhibitors of calpain. Trends Biochem. Sci. 16, 150–153 [DOI] [PubMed] [Google Scholar]
- 35. Nomura T., Fujishima A., and Fujisawa Y. (1996) Characterization and crystallization of recombinant human cathepsin L. Biochem. Biophys. Res. Commun. 228, 792–796 [DOI] [PubMed] [Google Scholar]