Abstract
Rationale: Respiratory syncytial virus (RSV) is the leading cause of acute lower respiratory tract infections and hospitalizations in infants worldwide. Known risk factors, however, incompletely explain the variability of RSV disease severity, especially among healthy children. We postulate that the severity of RSV infection is influenced by modulation of the host immune response by the local bacterial ecosystem.
Objectives: To assess whether specific nasopharyngeal microbiota (clusters) are associated with distinct host transcriptome profiles and disease severity in children less than 2 years of age with RSV infection.
Methods: We characterized the nasopharyngeal microbiota profiles of young children with mild and severe RSV disease and healthy children by 16S-rRNA sequencing. In parallel, using multivariable models, we analyzed whole-blood transcriptome profiles to study the relationship between microbial community composition, the RSV-induced host transcriptional response, and clinical disease severity.
Measurements and Main Results: We identified five nasopharyngeal microbiota clusters characterized by enrichment of either Haemophilus influenzae, Streptococcus, Corynebacterium, Moraxella, or Staphylococcus aureus. RSV infection and RSV hospitalization were positively associated with H. influenzae and Streptococcus and negatively associated with S. aureus abundance, independent of age. Children with RSV showed overexpression of IFN-related genes, independent of the microbiota cluster. In addition, transcriptome profiles of children with RSV infection and H. influenzae– and Streptococcus-dominated microbiota were characterized by greater overexpression of genes linked to Toll-like receptor and by neutrophil and macrophage activation and signaling.
Conclusions: Our data suggest that interactions between RSV and nasopharyngeal microbiota might modulate the host immune response, potentially affecting clinical disease severity.
Keywords: nasopharynx, microbiota, respiratory syncytial virus, disease severity, transcriptome profiling
At a Glance Commentary
Scientific Knowledge on the Subject
Respiratory syncytial virus (RSV) disease severity varies significantly among previously healthy children, which cannot be exclusively explained by currently known risk factors, including young age. Recent literature suggests that bacterial communities in the respiratory tract might affect RSV-induced host immune responses, thereby potentially modulating disease severity; however, a detailed assessment of this hypothesis in the target population for RSV infection is lacking.
What This Study Adds to the Field
Specific nasopharyngeal microbiota clusters enriched for Haemophilus influenzae and Streptococcus were associated with an exaggerated inflammatory host immune response in children with RSV infection. This immune response was characterized, among others, by enhanced Toll-like receptor signaling and increased expression of neutrophil- and macrophage-related transcripts and clinically with more severe RSV disease.
Globally, respiratory syncytial virus (RSV) is the most frequent viral cause of acute lower respiratory infections in children younger than 5 years of age. In addition, RSV is responsible for significant morbidity worldwide and mortality in infants in the developing world (1, 2).
Most children experience a primary RSV infection before 2 years of age (3), yet only 2 to 3% require hospitalization (1, 4). Medical comorbidities and young age increase the risk for severe RSV infection (4–6). Nevertheless, the majority of infants who are hospitalized with RSV infection are previously healthy and have no predisposing risk factors for severe disease (4, 7). Disease severity in these infants has been linked to a dysregulated host immune response, characterized among others by inadequate cytokine responses (8–11) and neutrophil influx in the respiratory tract (12, 13).
Besides the direct virus–host interaction, certain bacterial members of the respiratory tract microbiome might influence host responses to RSV, therewith modulating inflammation and possibly disease severity, yet few studies have addressed this hypothesis in the clinical setting. Recent reports, however, suggest that the composition of the nasopharyngeal microbiome affects the overall risk of developing respiratory tract infections (14) and is associated with the severity of acute respiratory symptoms (15). We characterized the nasopharyngeal microbiota using 16S-rRNA–based sequencing and analyzed whole-blood RNA transcriptional profiles in outpatients with RSV and infants hospitalized with an RSV infection, as well as healthy control subjects. We sought to define the nasopharyngeal microbiota profiles in infants with RSV disease and their relationship with host immune responses and disease severity.
Methods
Study Population
From 2010 to 2014 we conducted a prospective observational study during four consecutive RSV seasons at Nationwide Children’s Hospital, Columbus, Ohio. Previously healthy children less than 2 years of age with a first episode of RSV infection were enrolled either at the outpatient clinics (“outpatients”) or within a median of 24 hours (interquartile range [IQR], 17–39 h) of admission in the pediatric ward or the pediatric intensive care unit (PICU) (“inpatients”). Asymptomatic healthy control subjects were enrolled during routine primary care visits or elective surgery not involving the respiratory tract. For study criteria, see the Methods section of the online supplement. In addition to the need for hospitalization, RSV disease severity was assessed using a clinical disease severity score and by the need for supplemental oxygen, PICU admission, and length of stay (16).
Sample Collection, Storage, and Processing
At enrollment, we obtained from both patients and control subjects a blood sample for white blood cell count with differential and transcriptome analysis, a nasopharyngeal bacterial swab for bacterial quantitative polymerase chain reaction (PCR) and microbiome analysis, and a nasal wash for RSV quantitation. Sample collection, processing, and storage were performed as previously described (11, 17, 18) and summarized in the online supplement Methods.
Bacterial High-Throughput Sequencing and Bioinformatic Processing
Nasopharyngeal bacterial DNA was isolated as described previously (19, 20). A PCR amplicon library was generated by amplification of the V5 to V7 region of the 16S-rRNA gene (21). Quality filtering, clustering of sequences in operational taxonomic units (OTUs), and taxonomic annotation were performed using QIIME version 1.8 (online supplement Methods) (22). Data have been deposited in the National Center for Biotechnology Information GenBank database (accession number: SRP069222).
Host Gene Expression Profiling
RNA was extracted from whole-blood samples and hybridized onto Illumina HT12-V4 beadchips. Data import, background subtraction, and data normalization were performed as previously described (16, 23). Because our dataset included samples from two microarray batches, we applied an empirical Bayes (EB) method (ComBat, sva R-package) (24) to adjust for nonbiological variation between batches. Data are deposited in the National Center for Biotechnology Information Gene Expression Omnibus (accession number: GSE77087).
Statistical Analysis
To assess the relationship between nasopharyngeal microbiota and host characteristics, including age, clinical phenotype (healthy control, outpatient, or inpatient), and antibiotic treatment, all patient samples were subjected to a similarity-based, unsupervised hierarchical clustering approach. Major and minor classifier taxa for the resulting clusters were identified by random forest analysis. Subsequently, we applied multivariable linear models to study the relationship between host characteristics and relative abundance of these classifier taxa. Adjusted effect sizes and 95% confidence intervals were calculated for each predictor.
To study the association between nasopharyngeal microbiota and host gene expression in infants with RSV infection, we used a stepwise approach that was initiated by unsupervised analysis of either microbiota or host gene expression data. (1) For the microbiota-driven approach, we first reran the clustering analysis as described above for RSV-infected children with paired blood microarray samples. Differentially expressed genes between each microbiota cluster and healthy control subjects were identified using multivariable linear models (limma) (25), after adjusting for age and sex. Genes were functionally annotated using modular analysis (26, 27) and Ingenuity Pathway Analysis (IPA) software (online supplement Methods). (2) For the gene expression–driven approach, we first decomposed the host transcriptome dataset of patients with RSV by principal component analysis, extracting the gene principal components (gPCs) required to explain more than 50% of the variance in the dataset. Next, we fitted multivariable linear models and calculated adjusted effect sizes to assess the association between gPCs and the relative abundance of the classifier taxa, adjusting for age (28). For each gPC, the 5% transcripts with the highest impact were subjected to DAVID pathway enrichment analysis (29). Subsequently, we analyzed differences in fold-change expression of genes involved in significantly enriched host immune response pathways for each microbiota cluster. We used the Benjamini–Hochberg method to correct for multiple testing. All statistical analyses were performed in R version 3.2.2 and IBM SPSS Statistics version 21 (online supplement Methods).
Results
Baseline Characteristics of the Study Population
A total of 132 children, 106 with RSV infection (84 inpatients and 22 outpatients) and 26 healthy control subjects, were enrolled and included in the primary analyses. Baseline characteristics of the study participants are described in Table 1. Overall, children hospitalized with RSV infection were younger than outpatients with RSV and healthy control subjects. In addition, we observed that inpatients were treated with antibiotics more frequently than outpatients (46.4 vs. 18.2%, respectively; P < 0.0005). In most inpatients (33 of 39; 84.6%) antibiotic treatment was initiated less than or equal to 2 days before sampling.
Table 1.
Demographics and Clinical Parameters Stratified by Healthy Control Subjects and Outpatients/Inpatients with Respiratory Syncytial Virus Infection (N = 132)
| Healthy (n = 26) | Outpatients (n = 22) | Inpatients (n = 84) | P Value | |
|---|---|---|---|---|
| Demographic characteristics | ||||
| Age, mo | 6.6 (2.0–10.2) | 7.2 (5.0–12.2) | 2.6 (1.4–4.9) | <0.0005* |
| Male sex | 19 (73.1) | 12 (54.5) | 50 (59.5) | 0.36† |
| Breastfeeding | 5/17 (29.4) | 2/22 (9.1) | 19/82 (23.2) | 0.25‡ |
| Antibiotics | 0 (0.0) | 4 (18.2) | 39 (46.4) | <0.0005† |
| Days of antibiotics before sampling | N/A | § | 1.0 (1.0–2.0) | N/A |
| Days of symptoms at enrollment | N/A | 3.5 (2.0–5.5) | 5.0 (4.0–6.0) | 0.03|| |
| Disease severity | ||||
| Length of stay, d | N/A | N/A | 3.2 (2.0–5.5) | N/A |
| O2 requirement | N/A | 0 (0.0) | 71 (84.5) | <0.0005‡ |
| CDSS | ||||
| Mild | N/A | 21 (95.5) | 18 (21.4) | |
| Moderate | N/A | 1 (4.5) | 41 (48.8) | |
| Severe | N/A | 0 (0.0) | 25 (29.8) | <0.0005† |
| PICU admission | N/A | N/A | 35 (41.7) | N/A |
| White blood cells/μl¶ | 7.9 (6.9–9.4) | 10.0 (7.3–11.8) | 11.8 (9.0–14.7) | <0.0005* |
| Neutrophils, % | 21.0 (13.5–27.5) | 26.5 (19.3–45.8) | 35.5 (27.8–49.3) | <0.0005* |
| Lymphocytes, % | 67.0 (59.8–76.5) | 57.0 (35.0–70.5) | 50.5 (39.0–61.0) | <0.0005* |
| Monocytes, % | 7.0 (5.0–11.0) | 12.0 (8.8–15.8) | 12.0 (8.0–16.0) | 0.001* |
Definition of abbreviations: CDSS = clinical disease severity score, which ranks from 0–15 and is stratified in: mild (0–5), moderate (6–10), and severe (11–15) respiratory syncytial virus disease; N/A = not applicable; PICU = pediatric intensive care unit.
Data reported are median (interquartile range) or n (%). Statistically significant P values (P < 0.05) are shown in boldface.
Statistically significant differences between groups were calculated using Kruskal–Wallis test for continuous variables.
Statistically significant differences between groups were calculated using chi-square test for categorical data.
Statistically significant differences between groups were calculated using Fisher’s exact test for categorical data.
Four children received antibiotic treatment, 2, 10, 12, and 21 d before sampling.
Statistically significant differences between groups were calculated using Mann-Whitney test for continuous variables.
On twenty samples, no white blood cell count and differential was performed (2, 4, and 14 samples of healthy children, outpatients, and inpatients, respectively).
Characterization of the Microbiota Analysis
Using 16S-rRNA–based sequencing, we obtained a total of 646,727 high-quality sequences (median 4,778; IQR, 4,050–5,516 sequences per sample) from the total set of nasopharyngeal samples. These sequences were binned in 482 OTUs representing 21 phyla including 122 genera. The nasopharyngeal bacterial community composition was characterized by high relative abundance of Proteobacteria (43.4%), Firmicutes (40.9%), Actinobacteria (12.4%), and Bacteroidetes (2.6%), accounting for 99.3% of all sequences. The presence and abundance of the highest-ranking OTUs, including Haemophilus influenzae, Streptococcus (pneumoniae), Moraxella (catarrhalis), or Staphylococcus aureus, was verified using quantitative PCR and BLASTN (online supplement Results; Table E1).
Unsupervised Clustering Analysis of Subjects on the Basis of Nasopharyngeal Microbiota Composition
Samples from children with RSV and healthy control subjects were hierarchically clustered based on similarities in their nasopharyngeal microbiota composition, which was visualized using a dendrogram (Figure 1A). Using clustering indices, we identified 11 clusters; 5 of these clusters contained more than three samples (n = 123) and were therefore considered for subsequent analyses. We identified five OTUs that were most discriminative for the classification of subjects in these clusters (“[major] classifier taxa”): H. influenzae, Streptococcus, Corynebacterium, Moraxella, and S. aureus. In addition, we distinguished five minor classifier taxa, including several streptococcal species (i.e., Streptococcus rank 28, 30, and 46), Dolosigranulum and Moraxella rank 19 (online supplement Methods; Figure E1). Healthy control subjects and patients with RSV (both outpatients and inpatients) were distributed unevenly over the clusters (Fisher’s exact test P = 0.052), with greater numbers of patients with RSV than healthy control subjects in the H. influenzae– and Streptococcus-dominated clusters (Table 2). Within the inpatient cohort, we did not observe significant differences in disease severity, as defined by length of hospital stay, clinical disease severity score, PICU admission, or oxygen requirement between clusters. However, rates of PICU admission and length of stay tended to be lower in children within the Moraxella cluster than in children included in the other clusters (18% vs. 40–60% and 2.0 vs. 3.3–3.9 days, respectively; Table 3).
Figure 1.
Nasopharyngeal microbiome composition in young children with respiratory syncytial virus (RSV) infection and healthy control (HC) subjects and associations with host characteristics. (A) Dendrogram visualizing an average linkage hierarchical clustering of individuals on the basis of the Bray–Curtis dissimilarity matrix. The branches of the tree structure were colored according to health status (HC subjects, green; patients with RSV, yellow). Information on age and disease severity is depicted adjacent to the branch ends. Stacked bar charts show the relative abundance of the 15 highest-ranked operational taxonomic units (OTUs) and of residual bacteria specified by phylum. OTUs are color coded according to phylum: Firmicutes, red; Proteobacteria, blue; Actinobacteria, yellow; and Bacteroidetes, green. On the basis of clustering indices, an optimal number of 11 clusters was identified, 5 of which comprised more than three study participant samples. Classifier taxa for these five clusters (color-coded horizontal panels) were: Streptococcus (STR), light red; Haemophilus influenzae (HPH), dark blue; Corynebacterium (COR), yellow; Moraxella (MOR), light blue; and Staphylococcus aureus (STA), dark red. Gray panels mark individuals not included in any of these five clusters. (B) A nonmetric multidimensional scaling (NMDS) plot was used to visualize the associations between nasopharyngeal microbiota clusters (see color coding in A) and host characteristics: age group (round arrowheads), health status (arrowheads, bold text), and antibiotic treatment (arrowheads, bold italic text), depicting both the individual nasopharyngeal microbiota composition (data points [n = 132] and ellipses [SD of data points] colored according to cluster) and the 10 highest-ranked OTUs. We observed that RSV hospitalization was related to STR- and HPH-enriched profiles. Additionally, we observed an association between outpatients, older age, and MOR enrichment, whereas STA-dominated profiles were observed primarily in young, healthy infants. The orthogonal orientation (∼90°angle) of the vectors associated with health status and age imply that these host characteristics are not highly correlated, which was verified by permutational multivariate analysis of variance (unadjusted R2 severity, 5.7%; age, 6.7%; and interaction severity–age, 5.9%). AB = antibiotic treatment; Inp = inpatients; Outp = outpatients.
Table 2.
Demographics of Healthy Control Subjects and Respiratory Syncytial Virus Cases Stratified by Nasopharyngeal Microbiome Profile
| Cluster (N = 123)* |
||||||
|---|---|---|---|---|---|---|
| HPH (n = 19) | STR (n = 32) | COR (n = 24) | MOR (n = 29) | STA (n = 19) | P Value | |
| Health status | ||||||
| Healthy | 1 (5.3) | 3 (9.4) | 7 (29.2) | 7 (24.1) | 6 (31.6) | |
| Outpatient | 4 (21.1) | 4 (12.5) | 2 (8.3) | 11 (37.9) | 1 (5.3) | |
| Inpatient | 14 (73.7) | 25 (78.1) | 15 (62.5) | 11 (37.9) | 12 (63.2) | 0.007† |
| Demographic parameters | ||||||
| Age, mo | 6.7 (3.3–8.9) | 2.5 (1.6–6.7) | 5.8 (2.4–11.7) | 5.1 (3.3–9.4) | 1.2 (0.7–2.4) | <0.0005‡ |
| Breastfeeding | 4/18 (22.2) | 2/30 (6.7) | 3/20 (15.0) | 4/27 (14.8) | 11/18 (61.1) | 0.0005† |
| Antibiotics | 11 (57.9) | 13 (40.6) | 8 (33.3) | 5 (17.2) | 3 (15.8) | 0.017§ |
| WBC/μl|| | 11.9 (9.5–17.5) | 9.4 (6.7–12.7) | 10.8 (9.3–15.1) | 10.1 (7.4–13) | 9.1 (7.9–10.9) | 0.122‡ |
| Neutrophils, % | 40.0 (34.5–58.5) | 34.0 (22.5–49.5) | 29.0 (19.5–51.0) | 25.0 (16.0–42.3) | 24.5 (19.0–32.0) | 0.006‡ |
| Lymphocytes, % | 40.0 (29.0–51.5) | 50.0 (38.5–66.0) | 57.5 (39.0–67.3) | 61.0 (47.0–72.0) | 60.0 (54.5–63.3) | 0.003‡ |
| Monocytes, % | 12.0 (8.5–15.0) | 13.0 (8.5–17.5) | 8.5 (5.0–10.3) | 9.5 (6.0–14.0) | 11.5 (6.8–13.8) | 0.044‡ |
Definition of abbreviations: COR = Corynebacterium; HPH = Haemophilus influenzae; MOR = Moraxella; STA = Staphylococcus aureus; STR = Streptococcus; WBC = white blood cells.
Data reported are median (interquartile range) or n (%). Statistically significant P values (P < 0.05) are shown in boldface. Clusters were enriched for H. influenzae, Streptococcus, Corynebacterium, Moraxella, or S. aureus.
Nine individuals could not be assigned to a large (>3 individuals) microbiota cluster.
Statistically significant differences between groups were calculated using Fisher’s exact test for categorical data.
Statistically significant differences between groups were calculated using Kruskal–Wallis test for continuous variables.
Statistically significant differences between groups were calculated using chi-square test for categorical data.
On 19 samples, no white blood cell count and differential was performed (2, 3, 2, 7, and 5 samples included in the HPH, STR, COR, MOR, and STA clusters, respectively).
Table 3.
Measures of Severity in Hospitalized Respiratory Syncytial Virus Cases Stratified by Nasopharyngeal Microbiota Profile
| Cluster (N = 77)* |
||||||
|---|---|---|---|---|---|---|
| HPH (n = 14) | STR (n = 25) | COR (n = 15) | MOR (n = 11) | STA (n = 12) | P Value | |
| Demographic characteristics | ||||||
| Age, mo | 5.6 (1.5–7.6) | 2.2 (1.4–3.3) | 4.1 (2.2–9.1) | 3.5 (1.8–5.1) | 1.2 (0.6–2.2) | 0.002† |
| Male sex | 8 (57.1) | 10 (40.0) | 12 (80.0) | 5 (45.5) | 9 (75.0) | 0.08‡ |
| Breastfeeding | 4/14 (28.6) | 2/25 (8.0) | 2/14 (14.3) | 1/10 (10) | 8/12 (66.7) | 0.002‡ |
| Antibiotics | 10 (71.4) | 12 (48.0) | 8 (53.3) | 3 (27.3) | 3 (25.0) | 0.11§ |
| Disease severity | ||||||
| LOS | 3.9 (1.9–12.1) | 3.8 (2.3–6.3) | 3.3 (2.7–4.9) | 2.0 (1.4–3.2) | 3.6 (2.0–5.5) | 0.10† |
| O2 required | 12 (85.7) | 24 (96.0) | 13 (86.7) | 8 (72.7) | 9 (75.0) | 0.22‡ |
| CDSS | 7.0 (4.8–10.3) | 9.0 (6.0–12.0) | 13.0 (8.0–13.0) | 7.0 (5.0–8.0) | 6.5 (5.0–10.8) | 0.06† |
| PICU | 6 (42.9) | 10 (40.0) | 9 (60.0) | 2 (18.2) | 6 (50) | 0.31‡ |
Definition of abbreviations: CDSS = clinical disease severity score; COR = Corynebacterium; HPH = Haemophilus influenzae; LOS = length of hospital stay; MOR = Moraxella; PICU = pediatric intensive care unit; STA = Staphylococcus aureus; STR = Streptococcus.
Data reported are median (interquartile range) or n (%). Statistically significant P values (P < 0.05) are shown in boldface. Clusters were enriched for H. influenzae, Streptococcus, Corynebacterium, Moraxella, or S. aureus.
Excluding 26 healthy control subjects, 22 outpatients, and 7 children who could not be assigned to a large (>3 individuals) microbiota cluster.
Statistically significant differences between groups were calculated using Kruskal–Wallis for continuous variables.
Statistically significant differences between groups were calculated using Fisher’s exact test for categorical data.
Statistically significant differences between groups were calculated using chi-square test for categorical data.
Next, we visualized the interactions between microbiota composition per individual stratified by cluster in relation to the most abundant bacterial community members (including classifier taxa) and host-specific variables, including disease severity, age, and antibiotic treatment, using nonmetric multidimensional scaling (Figure 1B) and assessed these interactions using multivariable linear regression models (Figure 2). We observed that the abundance of H. influenzae and Streptococcus OTUs was positively associated with RSV hospitalization (P = 0.005 and P = 0.004, respectively), whereas S. aureus abundance was inversely associated with the need for hospitalization (P = 0.025), irrespective of age. In addition, high abundance of Moraxella was observed most often in outpatients (P = 0.034). Multivariable linear regression analyses within the RSV cohort demonstrated no effect of antibiotic use on the relative abundance of classifier species after adjusting for age (P > 0.05; Table E2).
Figure 2.
Independent associations between disease severity, age, and the relative abundance of classifier taxa. Multivariable linear regression modeling was used to assess the independent association between disease severity, defined as the need for hospitalization, and age (independent variables), and the relative abundance of classifier taxa (Haemophilus influenzae, Streptococcus, Corynebacterium, Moraxella, and Staphylococcus aureus; outcome variable). Adjusted effect sizes (aESs) are visualized by a forest plot (outpatients, gray squares; inpatients, white squares; age, black squares) and 95% confidence intervals (CIs) (bars). Benjamini-Hochberg–corrected P values (q values) were calculated. Background colors indicate the classifier species (Streptococcus, light red; H. influenzae, dark blue; Corynebacterium, yellow; Moraxella, light blue; and S. aureus, dark red). We observed that higher abundance of H. influenzae and Streptococcus was positively associated (i.e., aES > 0) with hospitalization, whereas S. aureus enrichment was negatively associated (i.e., aES < 0) with being an inpatient and was observed more often in young infants.
Host Transcriptome Profiles Stratified by Microbiota Clusters: Modular Analysis
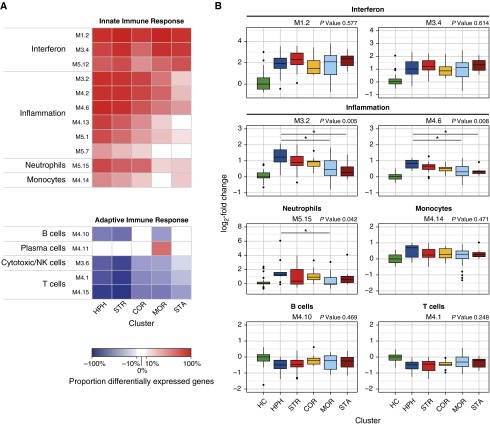
Paired nasopharyngeal and whole-blood samples were available for 104 of 132 study subjects. Baseline characteristics of this subcohort and hierarchical clustering of patients with RSV were similar to those of the whole cohort (Tables E3 and E4, Figure E2). We sought to test whether host transcriptional profiles in patients with RSV infection could be partially influenced by the nasopharyngeal microbiota. To this end, transcriptional profiles of patients with RSV stratified by microbiota cluster (Streptococcus, n = 12; H. influenzae, n = 19; Corynebacterium, n = 9; Moraxella, n = 26; and S. aureus, n = 8) and healthy control subjects (n = 23) were analyzed through gene-wise linear models (limma). Using nonstringent statistical filtering, these models identified 8,841 differentially expressed transcripts across all pairwise comparisons (Figure E3), adjusted for age and sex. Modular analysis of these transcripts showed increased modular expression (i.e., the cumulative proportion of over- and underexpressed genes) of IFN-related modules ranging from 43 to 100% in patients with RSV regardless of the microbiota profiles. However, expression of inflammation modules differed between clusters and were higher in the H. influenzae– and Streptococcus-dominated clusters (modular expression, 38–94% and 26–89%, respectively) compared with the S. aureus cluster (−1 to 39%). Moreover, overexpression of neutrophil-related genes (module M5.15) varied from 75% in the H. influenzae cluster to 21% in the S. aureus cluster (Figure 3A). In contrast, B cell, T cell, and cytotoxic/natural killer cell modules were overall underexpressed, which was particularly evident for the H. influenzae– and Streptococcus-enriched clusters (modular expression, −87% to −50%), compared with the other clusters (−51% to −3%). Last, we also observed differences in median expression values of inflammation and neutrophil module–associated genes between the different clusters (Figure 3B).
Figure 3.
Transcriptional modular repertoire analysis stratified by nasopharyngeal microbiome cluster. (A) Heat map depicting the difference between the proportion of over- and underexpressed genes (modular expression; Figure E3) per cluster compared with healthy control (HC) subjects on a per-module basis. The color gradient indicates the proportion of differentially expressed genes per module (modular expression) ranging from −100% (blue) to 100% (red). Modular expression between −10% and 10% is shown in white and reflects no differences compared with HC subjects. Although IFN-related modules (M1.2, M3.4, M5.12) were similarly overexpressed in all clusters, we observed increased expression of inflammation (M3.2, M4.2, M4.6, M4.13, M5.1, M5.7), neutrophil (M.5.15), and monocyte/macrophage modules (M4.14) in the Haemophilus influenzae (HPH) and Streptococcus (STR) clusters. Contrarily, the shared adaptive immune response to respiratory syncytial virus was underexpressed and distinguished by particularly strong suppression of B cell– (M4.10) and T/natural killer (NK) cell–related modules (M3.6, M3.1, M4.15) in the STR cluster. (B) Box plots showing the per-cluster median fold-change gene expression relative to the HC baseline. A representative selection of modules is shown. Median fold-change expression values of inflammation (M3.2 and M4.6) and neutrophil modules (M5.15) in the HPH-enriched cluster were significantly higher than that in the Moraxella (and Staphylococcus aureus) clusters. Statistically significant differences in median fold change between clusters was assessed using a Kruskal–Wallis test (P values shown) with Nemenyi post hoc test (bars; *P < 0.05). COR = Corynebacterium; MOR = Moraxella; STA = Staphylococcus aureus.
Host Transcriptome Profiles Stratified by Microbiota Clusters: Pathway Analysis
To confirm the significance and robustness of the observed differences in gene expression between the different microbiota clusters, we applied multivariable linear modeling using stringent statistical filtering (limma), adjusting for multiple testing, age, and sex. Despite that numbers of patients were comparable among groups, we found that the H. influenzae and Streptococcus clusters had a higher number of differentially expressed transcripts (1,315 and 1,435 transcripts, respectively) than the three other microbiota clusters (range, 207–651 transcripts; Figure 4A). Using IPA, we confirmed the findings derived from the modular analysis and dissected the common response to RSV independent of the microbiota cluster, as well as the specific host response elicited by each of the microbiota clusters. By comparing the differentially expressed genes shared by patients in all or all but one cluster, we identified a shared component of the RSV signature that was independent of the patient’s microbiota profile and was characterized by overexpression of genes involved in IFN signaling (207 transcripts; Figures 4B and 4C). In addition, we analyzed the differentially expressed transcripts associated with the H. influenzae– and Streptococcus-enriched clusters (which were associated with clinical RSV disease severity; Figure 2) versus the Corynebacterium, Moraxella, and S. aureus clusters. IPA of the genes expressed in the Streptococcus and/or H. influenzae cluster (1,257 transcripts; Figure 4B and 4D) showed strong overexpression of pathways associated with macrophage and neutrophil activation and/or signaling (IL-8 and IL-17A signaling, among others) and bacteria and virus pattern recognition receptors. Contrarily, genes exclusively associated with the Corynebacterium, Moraxella, and/or S. aureus cluster (Figure 4B) were not related to specific (immune-related) pathways (q = 0.48 for each of the 10 highest-ranking pathways; Figure 4E). These results suggest an additive effect of the presence of Streptococcus and H. influenzae on the RSV-induced host immune response, characterized by stronger activation of inflammatory pathways, which in turn might be related to a more severe RSV disease phenotype.
Figure 4.
Identification and functional annotations of respiratory syncytial virus (RSV) and microbiota-specific differentially expressed genes between patients with RSV and healthy control (HC) subjects. (A) Heat maps depict differentially expressed genes between HC subjects and each microbiota cluster (limma, stringent filter: P < 0.01, log2-fold change > 1.25, Benjamini–Hochberg multiple test correction), adjusted for age and sex. Normalized expression is indicated as overexpressed (red) or underexpressed (blue) compared with the median expression of HC subjects (yellow). Heat maps were scaled according to number of samples and transcripts per comparison. The highest number of differentially expressed transcripts was observed in the Streptococcus (STR; 1,435 genes) and Haemophilus influenzae (HPH) clusters (1,315 genes) compared with the other microbiota clusters. (B) Venn diagram showing the intersection between differentially expressed genes derived from pairwise comparisons between patients with RSV stratified by microbiome profiles and HC subjects (A). We observed a considerable overlap between the STR and HPH clusters (464 genes). In contrast, the Corynebacterium (COR, 323 genes), Moraxella (MOR, 651 genes), and Staphylococcus aureus clusters (STA, 207 genes) showed a small number of differentially expressed genes versus HC subjects. Using ingenuity pathway analysis, shared and cluster-specific genes were extracted and functionally annotated: (C) genes for each of the clusters were intersected and those shared between all or all but one clusters (209 genes; common RSV signature); (D) genes shared between or unique to the STR and HPH clusters (1,257 genes; shared STR/HPH signature); and (E) genes shared between COR, MOR, and STA, but not the STR and HPH clusters (258 genes; shared COR/MOR/STA signature), were extracted. For each gene list of interest, the 10 strongest associated over-/underexpressed pathways were visualized; Benjamini-Hochberg–corrected P values (q values) associated with a specific pathway were calculated using Fisher’s exact test and visualized on a log10 scale. (C) We hypothesized that the genes shared between all or four out of five clusters would represent the common RSV signature, as this feature is shared between all clusters. Indeed, we observed these genes were strongly associated with IFN signaling (q value = 8.1 × 10−8). (D) Genes shared between or unique to the HPH and STR clusters were involved (q value ranging from 6.5 × 10−4 to 5.9 × 10−6) in the activation of neutrophils (N-formylmethionyl-leucyl-phenylalanine [fMLP], triggering receptor expressed on myeloid cells [TREM]-1, IL-8, and IL-17A signaling) and macrophages (inducible nitric oxide synthase [iNOS] signaling and production of nitric oxide and reactive oxygen species [ROS]). In addition, genes related to the pattern recognition of bacteria and viruses were up-regulated, including Toll-like receptor (TLR) genes TLR4, TLR6, and TLR8. (E) Genes shared between COR, MOR, and STA, but not the STR and HPH clusters, were not significantly associated with any of the 10 highest-ranking canonical pathways (q = 0.48). EIF = eukaryotic initiation factor; mTOR = mechanistic target of rapamycin; PRRs = pattern recognition receptors; trx = transcripts.
Unsupervised Gene Expression Data Analysis
To confirm the findings derived from the microbiota-driven analyses, we used a second analytical approach in which we decomposed the host transcriptome data in an unsupervised manner through principal component analysis. Within the RSV patient cohort, we identified seven gPCs that were visualized simultaneously with the microbial community composition of patients, classifier species, and metadata in a nonmetric multidimensional scaling plot (Figure 5A). Multivariable linear regression modeling identified four gPCs (gPC1, gPC3, gPC6, and gPC7) that were significantly associated with the relative abundance of the major classifier taxa, adjusted for age and multiple testing (Figure 5B). Among others, we found that gPC1 was positively associated with Streptococcus and H. influenzae abundance and gPC6 and gPC7 with S. aureus abundance (Figure 5B, online supplement Results). Of the top 5% of transcripts contributing to gPC1, DAVID pathway enrichment analysis identified 37 genes involved in immune response pathways. Of these 37 genes, 24 were cytokine related and mostly involved in neutrophil recruitment (IL-8 receptor, IL-17A), neutrophil activation (oncostatin M [OSM], IL-6 family cytokine), and neutrophil and macrophage differentiation (Figure 5C), and 13 genes were related to Toll-like receptor (TLR) signaling, including TLR2, TLR4, TLR5, and TLR8 genes (Figure 5C, online supplement Results, Table E5). None of the genes linked to gPC6/gPC7 and S. aureus were involved in enriched immune response signaling pathways (data not shown). Fold-change expression of genes related to TLR signaling and neutrophil activation/recruitment was significantly higher within the H. influenzae and the Streptococcus clusters than in the other microbiota clusters (Figure E4), confirming our previous findings using supervised analyses. For associations between other gPCs and microbiota and functional annotations see online supplement Results and Table E6.
Figure 5.
Nasopharyngeal microbiome composition in infants with respiratory syncytial virus (RSV) infection and associations with host characteristics and gene principal components (gPCs). (A) A nonmetric multidimensional scaling (NMDS) plot was used to jointly visualize individual microbiota profiles (data points [n = 74] and ellipses [SD of data points] are colored according to cluster), the 10 most-abundant nasopharyngeal microbiota, host characteristics (age and hospitalization status), and gPCs extracted by principal component analysis from the host transcriptome dataset. (B) Multivariable linear regression models were fitted to investigate the association between classifier taxa (explanatory variables) and gPCs (outcome variables) within the RSV cohort (n = 74), corrected for age and multiple testing (Benjamini–Hochberg correction). We used a forest plot to visualize adjusted effect size (aES) (colored squares) and 95% confidence intervals (CIs) (bars). Only gPCs with significant results (defined as q < 0.15, depicted in bold) are shown. We verified the positive associations between Streptococcus (STR) and Haemophilus influenzae (HPH) and gPC1 and between Staphylococcus aureus (STA) enrichment and gPC6 and gPC7, and the negative association between Corynebacterium (COR) abundance and gPC3 observed using NMDS (A). Color coding: STR, light red; HPH, dark blue; COR, yellow; Moraxella, light blue; and STA, dark red. (C) Heat map depicts the log2-fold change gene expression of gPC1 genes identified using DAVID pathway enrichment analyses for each individual, stratified by health status/microbiota cluster. Pathways were selected based on a cluster enrichment score of greater than 1 and a Benjamini-Hochberg–corrected P value < 0.05. Genes covered by more than one probe are shown as [gene abbreviation].1 or [gene abbreviation].2 (e.g., MAPK14.1). Normalized expression is indicated as overexpressed (red) or underexpressed (blue) compared with the median expression of healthy control subjects (yellow). Explanations of the abbreviations of the depicted genes are given in Table E5. HC = healthy controls; Inp = inpatients; MOR = Moraxella; Outp = outpatients; TLR = Toll-like receptor.
Discussion
In this article, we provided evidence that RSV infection is associated with the presence of specific nasopharyngeal microbiota, distinguished by enrichment of H. influenzae and Streptococcus. Microbiota profiles enriched for either of these two bacterial pathogens were significantly associated with enhanced clinical disease severity, as defined by the need for hospitalization, independent of age. We observed that infants within H. influenzae– and Streptococcus-enriched clusters mounted a distinct host inflammatory response characterized by overexpression of genes related to TLR signaling and neutrophil recruitment and activation, as compared with children with another microbiota composition, particularly those in the S. aureus cluster.
To date, studies addressing the interactions between potentially pathogenic bacteria inhabiting the upper respiratory tract and RSV have been largely focused on the synergistic relationship between S. pneumoniae and RSV, which appears to be of bidirectional nature. RSV infection appears to predispose to invasive pneumococcal disease, as evidenced by epidemiological (30, 31), in vitro (32, 33), and in vivo studies (34). Alternatively, the reverse causative mechanism, where pneumococcal carriage predisposes to RSV hospitalization, might also be plausible, given observations in which vaccination with pneumococcal conjugate vaccine leads to a reduction in colonization with the most prevalent pneumococcal serotypes and decreased rates of RSV hospitalizations (30, 35). Similarly, cooccurrence of H. influenzae and RSV (36) may be explained by both RSV predisposing to H. influenzae colonization (37) and H. influenzae affecting the susceptibility to (38) and/or severity of RSV infection (39), suggesting that these associations are not mutually exclusive.
Our data suggest that the presence of specific microbes colonizing the upper respiratory tract mucosa is associated with modulation of the systemic host immune response during RSV infection. A similar phenomenon was previously reported for the interaction between the resident microbiome and influenza virus. In those studies, bacterial TLR agonists, including lipopolysaccharide, seemed responsible for immune modulation during coinfections (40, 41). Alternatively, up-regulation of adhesion molecules may augment a proinflammatory signal, which is supported by a recent study in human primary airway epithelial cells, showing a synergistic effect of nontypeable H. influenzae followed by RSV inoculation on intercellular adhesion molecule (ICAM)-1 expression and subsequent increased production of IL-6 and IL-8 (39). Indeed, we previously showed that children colonized with gram-negative bacteria had increased serum IL-6 and IL-8 concentrations compared with those colonized with gram-positive bacteria or respiratory flora (18). In agreement with those findings, we observed that differentially expressed genes that were unique and/or shared between infants with an H. influenzae– and Streptococcus–enriched nasopharyngeal microbiome composition, representing individuals with a more severe RSV disease phenotype, were related to OSM (IL-6 cytokine group), IL-8, and IL-17A signaling, which all contribute to neutrophil recruitment and activation (42–45). Altogether, these data support the hypothesis that microbial cosignaling might be involved in a proinflammatory response leading to enhanced neutrophil recruitment and activation, resulting in more severe RSV disease. The fact that the type and magnitude of RSV-induced IFN responses were similar across patients, independent of their microbiota composition, substantiates these conclusions.
Strengths of our study include the unique and relatively large cohort of well-characterized young children from whom simultaneous nasopharyngeal and whole-blood samples were obtained and comprehensively analyzed. We confirmed that the infant nasopharyngeal bacterial communities are capable of adopting five distinct community compositions (14, 15), underscoring the representativeness of the study cohort. In addition, our study is the first to assess the interrelation between the upper respiratory tract microbiome and the host systemic transcriptome immune response in young children with RSV infection and to document how these interactions might influence the clinical disease phenotype. This was accomplished using two separate, data-driven analytical approaches, which contributed to the robustness of the observations.
Our study also has limitations. First, healthy control subjects and patients with RSV not requiring hospitalization were older than inpatients with RSV, which is inherent to this disease. Nevertheless, we adjusted for age throughout the study using multivariable regression models. A second confounder could have been the influence of antibiotic treatment on microbiota composition; however, we did not observe a major independent effect of antibiotics on the microbial community composition, plausibly because antibiotic treatment was initiated at a median time of 1 day before sampling. Additionally, there was a trend for the effect of antibiotic use in the opposite direction of the one we would expect (i.e., antibiotic treatment was positively associated with increased H. influenzae and Streptococcus abundance, not negatively). Third, except for the need for hospitalization, we lacked the statistical power to make solid statements on specific associations between microbiota clusters and other parameters of disease severity within the RSV inpatient cohort. Last, the cross-sectional study design precludes statements on cause–effect relationships between microbiota, host immune response, and RSV disease severity. Further studies to elucidate the cause–effect mechanisms underlying the observed associations are needed.
We conclude that a nasopharyngeal microbiota composition characterized by H. influenzae and Streptococcus, as opposed to S. aureus, is associated with a distinct host inflammatory immune response and enhanced disease severity as defined by more frequent need for RSV hospitalization. Altogether, our data suggest that viral–bacterial interactions within the ecological respiratory niche modulate the systemic host immune response to RSV, thereby potentially influencing disease phenotypes.
Acknowledgments
Acknowledgment
The authors thank Gail Arthur, Michael Lawson, Paula Davies, and Grace Wentzel for their efforts in enrolling study subjects; Phuong Nguyen, Esperanza Aguiano, and Nicole Baldwin for their help with RNA processing and hybridization; Sara Mertz for her technical assistance; and Marinus J. C. Eijkemans for his input on the statistical analysis. They also thank all the participating children and their families.
Footnotes
Supported in part by the Netherlands Organization for Scientific Research through NWO-VIDI grant 91715359 and ZonMW grant 91209010, and Wilhelmina Children’s Hospital intramural funds (D.B.); the National Institute of Allergy and Infectious Diseases grants AI089987 and AI112524 (O.R. and A.M.); Nationwide Children’s Hospital intramural funds grant 299814 (A.M.); and the European Society for Pediatric Infectious Diseases (ESPID Fellowship Award), the Finnish Medical Foundation, the Foundation for Pediatric Research, and Maud Kuistila Memorial Foundation (S.H.). The sponsors had no role in study design, data collection, data analysis, data interpretation, decision to publish, or preparation of the manuscript.
Author Contributions: W.A.A.d.S.P., S.H., E.A.M.S., O.R., D.B., and A.M. designed the experiments. A.M., D.B., and O.R. wrote the study protocols. M.-C.S.-A. and D.M.C. were responsible for patient recruitment and clinical data collection. E.B. was responsible for clinical data collection and conventional quantitative polymerase chain reaction data. W.A.A.d.S.P. and R.H. were responsible for sample preparation for 16S-rRNA sequencing. D.B. and W.A.A.d.S.P. were responsible for bioinformatic processing of bacterial sequences. W.A.A.d.S.P., S.H., B.S., and A.M. were responsible for microarray profiling and post-processing of data. D.C. was responsible for the microarray modular repertoire. W.A.A.d.S.P., S.H., B.S., D.B., and A.M. were responsible for statistical analyses. All authors were involved in data interpretation and drafting of the manuscript.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.201602-0220OC on May 2, 2016
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Stockman LJ, Curns AT, Anderson LJ, Fischer-Langley G. Respiratory syncytial virus-associated hospitalizations among infants and young children in the United States, 1997-2006. Pediatr Infect Dis J. 2012;31:5–9. doi: 10.1097/INF.0b013e31822e68e6. [DOI] [PubMed] [Google Scholar]
- 2.Nair H, Nokes DJ, Gessner BD, Dherani M, Madhi SA, Singleton RJ, O’Brien KL, Roca A, Wright PF, Bruce N, et al. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet. 2010;375:1545–1555. doi: 10.1016/S0140-6736(10)60206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glezen WP, Taber LH, Frank AL, Kasel JA. Risk of primary infection and reinfection with respiratory syncytial virus. Am J Dis Child. 1986;140:543–546. doi: 10.1001/archpedi.1986.02140200053026. [DOI] [PubMed] [Google Scholar]
- 4.Boyce TG, Mellen BG, Mitchel EF, Jr, Wright PF, Griffin MR. Rates of hospitalization for respiratory syncytial virus infection among children in medicaid. J Pediatr. 2000;137:865–870. doi: 10.1067/mpd.2000.110531. [DOI] [PubMed] [Google Scholar]
- 5.Hall CB, Weinberg GA, Blumkin AK, Edwards KM, Staat MA, Schultz AF, Poehling KA, Szilagyi PG, Griffin MR, Williams JV, et al. Respiratory syncytial virus-associated hospitalizations among children less than 24 months of age. Pediatrics. 2013;132:e341–e348. doi: 10.1542/peds.2013-0303. [DOI] [PubMed] [Google Scholar]
- 6.Kristensen K, Hjuler T, Ravn H, Simões EAF, Stensballe LG. Chronic diseases, chromosomal abnormalities, and congenital malformations as risk factors for respiratory syncytial virus hospitalization: a population-based cohort study. Clin Infect Dis. 2012;54:810–817. doi: 10.1093/cid/cir928. [DOI] [PubMed] [Google Scholar]
- 7.García CG, Bhore R, Soriano-Fallas A, Trost M, Chason R, Ramilo O, Mejias A. Risk factors in children hospitalized with RSV bronchiolitis versus non-RSV bronchiolitis. Pediatrics. 2010;126:e1453–e1460. doi: 10.1542/peds.2010-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smyth RL, Mobbs KJ, O’Hea U, Ashby D, Hart CA. Respiratory syncytial virus bronchiolitis: disease severity, interleukin-8, and virus genotype. Pediatr Pulmonol. 2002;33:339–346. doi: 10.1002/ppul.10080. [DOI] [PubMed] [Google Scholar]
- 9.Bennett BL, Garofalo RP, Cron SG, Hosakote YM, Atmar RL, Macias CG, Piedra PA. Immunopathogenesis of respiratory syncytial virus bronchiolitis. J Infect Dis. 2007;195:1532–1540. doi: 10.1086/515575. [DOI] [PubMed] [Google Scholar]
- 10.García C, Soriano-Fallas A, Lozano J, Leos N, Gomez AM, Ramilo O, Mejias A. Decreased innate immune cytokine responses correlate with disease severity in children with respiratory syncytial virus and human rhinovirus bronchiolitis. Pediatr Infect Dis J. 2012;31:86–89. doi: 10.1097/INF.0b013e31822dc8c1. [DOI] [PubMed] [Google Scholar]
- 11.Mella C, Suarez-Arrabal MC, Lopez S, Stephens J, Fernandez S, Hall MW, Ramilo O, Mejias A. Innate immune dysfunction is associated with enhanced disease severity in infants with severe respiratory syncytial virus bronchiolitis. J Infect Dis. 2013;207:564–573. doi: 10.1093/infdis/jis721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McNamara PS, Ritson P, Selby A, Hart CA, Smyth RL. Bronchoalveolar lavage cellularity in infants with severe respiratory syncytial virus bronchiolitis. Arch Dis Child. 2003;88:922–926. doi: 10.1136/adc.88.10.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geerdink RJ, Pillay J, Meyaard L, Bont L. Neutrophils in respiratory syncytial virus infection: a target for asthma prevention. J Allergy Clin Immunol. 2015;136:838–847. doi: 10.1016/j.jaci.2015.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Biesbroek G, Tsivtsivadze E, Sanders EAM, Montijn R, Veenhoven RH, Keijser BJF, Bogaert D. Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am J Respir Crit Care Med. 2014;190:1283–1292. doi: 10.1164/rccm.201407-1240OC. [DOI] [PubMed] [Google Scholar]
- 15.Teo SM, Mok D, Pham K, Kusel M, Serralha M, Troy N, Holt BJ, Hales BJ, Walker ML, Hollams E, et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe. 2015;17:704–715. doi: 10.1016/j.chom.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mejias A, Dimo B, Suarez NM, Garcia C, Suarez-Arrabal MC, Jartti T, Blankenship D, Jordan-Villegas A, Ardura MI, Xu Z, et al. Whole blood gene expression profiles to assess pathogenesis and disease severity in infants with respiratory syncytial virus infection. PLoS Med. 2013;10:e1001549. doi: 10.1371/journal.pmed.1001549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sheeran P, Jafri H, Carubelli C, Saavedra J, Johnson C, Krisher K, Sánchez PJ, Ramilo O. Elevated cytokine concentrations in the nasopharyngeal and tracheal secretions of children with respiratory syncytial virus disease. Pediatr Infect Dis J. 1999;18:115–122. doi: 10.1097/00006454-199902000-00007. [DOI] [PubMed] [Google Scholar]
- 18.Suárez-Arrabal MC, Mella C, Lopez SM, Brown NV, Hall MW, Hammond S, Shiels W, Groner J, Marcon M, Ramilo O, et al. Nasopharyngeal bacterial burden and antibiotics: influence on inflammatory markers and disease severity in infants with respiratory syncytial virus bronchiolitis. J Infect. 2015;71:458–469. doi: 10.1016/j.jinf.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 19.Biesbroek G, Sanders EAM, Roeselers G, Wang X, Caspers MPM, Trzciński K, Bogaert D, Keijser BJF. Deep sequencing analyses of low density microbial communities: working at the boundary of accurate microbiota detection. PLoS One. 2012;7:e32942. doi: 10.1371/journal.pone.0032942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wyllie AL, Chu MLJN, Schellens MHB, van Engelsdorp Gastelaars J, Jansen MD, van der Ende A, Bogaert D, Sanders EAM, Trzciński K. Streptococcus pneumoniae in saliva of Dutch primary school children. PLoS One. 2014;9:e102045. doi: 10.1371/journal.pone.0102045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bogaert D, Keijser B, Huse S, Rossen J, Veenhoven R, van Gils E, Bruin J, Montijn R, Bonten M, Sanders E. Variability and diversity of nasopharyngeal microbiota in children: a metagenomic analysis. PLoS One. 2011;6:e17035. doi: 10.1371/journal.pone.0017035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heinonen S, Jartti T, Garcia C, Oliva S, Smitherman C, Anguiano E, de Steenhuijsen Piters WAA, Vuorinen T, Ruuskanen O, Dimo B, et al. Rhinovirus detection in symptomatic and asymptomatic children: value of host transcriptome analysis. Am J Respir Crit Care Med. 2016;193:772–782. doi: 10.1164/rccm.201504-0749OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 25.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chaussabel D, Baldwin N. Democratizing systems immunology with modular transcriptional repertoire analyses. Nat Rev Immunol. 2014;14:271–280. doi: 10.1038/nri3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chaussabel D, Quinn C, Shen J, Patel P, Glaser C, Baldwin N, Stichweh D, Blankenship D, Li L, Munagala I, et al. A modular analysis framework for blood genomics studies: application to systemic lupus erythematosus. Immunity. 2008;29:150–164. doi: 10.1016/j.immuni.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, Snapper SB, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 30.Weinberger DM, Klugman KP, Steiner CA, Simonsen L, Viboud C. Association between respiratory syncytial virus activity and pneumococcal disease in infants: a time series analysis of US hospitalization data. PLoS Med. 2015;12:e1001776. doi: 10.1371/journal.pmed.1001776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stensballe LG, Hjuler T, Andersen A, Kaltoft M, Ravn H, Aaby P, Simões EAF. Hospitalization for respiratory syncytial virus infection and invasive pneumococcal disease in Danish children aged <2 years: a population-based cohort study. Clin Infect Dis. 2008;46:1165–1171. doi: 10.1086/529438. [DOI] [PubMed] [Google Scholar]
- 32.Smith CM, Sandrini S, Datta S, Freestone P, Shafeeq S, Radhakrishnan P, Williams G, Glenn SM, Kuipers OP, Hirst RA, et al. Respiratory syncytial virus increases the virulence of Streptococcus pneumoniae by binding to penicillin binding protein 1a: a new paradigm in respiratory infection. Am J Respir Crit Care Med. 2014;190:196–207. doi: 10.1164/rccm.201311-2110OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Avadhanula V, Rodriguez CA, Devincenzo JP, Wang Y, Webby RJ, Ulett GC, Adderson EE. Respiratory viruses augment the adhesion of bacterial pathogens to respiratory epithelium in a viral species- and cell type-dependent manner. J Virol. 2006;80:1629–1636. doi: 10.1128/JVI.80.4.1629-1636.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stark JM, Stark MA, Colasurdo GN, LeVine AM. Decreased bacterial clearance from the lungs of mice following primary respiratory syncytial virus infection. J Med Virol. 2006;78:829–838. doi: 10.1002/jmv.20631. [DOI] [PubMed] [Google Scholar]
- 35.Madhi SA, Klugman KP Vaccine Trialist Group. A role for Streptococcus pneumoniae in virus-associated pneumonia. Nat Med. 2004;10:811–813. doi: 10.1038/nm1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van den Bergh MR, Biesbroek G, Rossen JWA, de Steenhuijsen Piters WAA, Bosch AATM, van Gils EJM, Wang X, Boonacker CWB, Veenhoven RH, Bruin JP, et al. Associations between pathogens in the upper respiratory tract of young children: interplay between viruses and bacteria. PLoS One. 2012;7:e47711. doi: 10.1371/journal.pone.0047711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGillivary G, Mason KM, Jurcisek JA, Peeples ME, Bakaletz LO. Respiratory syncytial virus-induced dysregulation of expression of a mucosal beta-defensin augments colonization of the upper airway by non-typeable Haemophilus influenzae. Cell Microbiol. 2009;11:1399–1408. doi: 10.1111/j.1462-5822.2009.01339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vissing NH, Chawes BLK, Bisgaard H. Increased risk of pneumonia and bronchiolitis after bacterial colonization of the airways as neonates. Am J Respir Crit Care Med. 2013;188:1246–1252. doi: 10.1164/rccm.201302-0215OC. [DOI] [PubMed] [Google Scholar]
- 39.Gulraiz F, Bellinghausen C, Bruggeman CA, Stassen FR. Haemophilus influenzae increases the susceptibility and inflammatory response of airway epithelial cells to viral infections. FASEB J. 2015;29:849–858. doi: 10.1096/fj.14-254359. [DOI] [PubMed] [Google Scholar]
- 40.Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, Murray TS, Iwasaki A. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci USA. 2011;108:5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abt MC, Osborne LC, Monticelli LA, Doering TA, Alenghat T, Sonnenberg GF, Paley MA, Antenus M, Williams KL, Erikson J, et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity. 2012;37:158–170. doi: 10.1016/j.immuni.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoshino H, Laan M, Sjöstrand M, Lötvall J, Skoogh BE, Lindén A. Increased elastase and myeloperoxidase activity associated with neutrophil recruitment by IL-17 in airways in vivo. J Allergy Clin Immunol. 2000;105:143–149. doi: 10.1016/s0091-6749(00)90189-1. [DOI] [PubMed] [Google Scholar]
- 43.Roos AB, Sethi S, Nikota J, Wrona CT, Dorrington MG, Sandén C, Bauer CM, Shen P, Bowdish D, Stevenson CS, et al. IL-17A and the promotion of neutrophilia in acute exacerbation of chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2015;192:428–437. doi: 10.1164/rccm.201409-1689OC. [DOI] [PubMed] [Google Scholar]
- 44.Lu Y-J, Gross J, Bogaert D, Finn A, Bagrade L, Zhang Q, Kolls JK, Srivastava A, Lundgren A, Forte S, et al. Interleukin-17A mediates acquired immunity to pneumococcal colonization. PLoS Pathog. 2008;4:e1000159. doi: 10.1371/journal.ppat.1000159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Følsgaard NV, Schjørring S, Chawes BL, Rasmussen MA, Krogfelt KA, Brix S, Bisgaard H. Pathogenic bacteria colonizing the airways in asymptomatic neonates stimulates topical inflammatory mediator release. Am J Respir Crit Care Med. 2013;187:589–595. doi: 10.1164/rccm.201207-1297OC. [DOI] [PubMed] [Google Scholar]