Abstract
DNA polymerase theta (pol θ) is encoded in the genomes of many eukaryotes, though not in fungi. Pol θ is encoded by the POLQ gene in mammalian cells. The C-terminal third of the protein is a family A DNA polymerase with additional insertion elements relative to prokaryotic homologs. The N-terminal third is a helicase-like domain with DNA-dependent ATPase activity. Pol θ is important in the repair of genomic double-strand breaks (DSBs) from many sources. These include breaks formed by ionizing radiation and topoisomerase inhibitors, breaks arising at stalled DNA replication forks, breaks introduced during diversification steps of the mammalian immune system, and DSB induced by CRISPR-Cas9. Pol θ participates in a route of DSB repair termed “alternative end-joining” (altEJ). AltEJ is independent of the DNA binding Ku protein complex and requires DNA end resection. Pol θ is able to mediate joining of two resected 3’ ends harboring DNA sequence microhomology. “Signatures” of Pol θ action during altEJ are the frequent utilization of longer microhomologies, and the insertion of additional sequences at joining sites. The mechanism of end-joining employs the ability of Pol θ to tightly grasp a 3’ terminus through unique contacts in the active site, allowing extension from minimally paired primers. Pol θ is involved in controlling the frequency of chromosome translocations and preserves genome integrity by limiting large deletions. It may also play a backup role in DNA base excision repair. POLQ is a member of a cluster of similarly upregulated genes that are strongly correlated with poor clinical outcome for breast cancer, ovarian cancer and other cancer types. Inhibition of pol θ is a compelling approach for combination therapy of radiosensitization.
Keywords: DNA polymerase, alternative end-joining, MMEJ, DNA double strand breaks, DNA synthesis, synthetic lethality
INTRODUCTION
DNA polymerases act not only in genomic DNA replication but in various pathways of DNA repair and genome maintenance. In mammalian cells, there are ~16 known DNA polymerases that function in semiconservative DNA replication, (pols α, δ, ε), base excision repair (pol β), mitochondrial DNA replication and repair (pol γ and Primpol), non-homologous end-joining and immunological diversity (pols λ, μ, pol θ and terminal-deoxynucleotidyl transferase), and DNA damage tolerance by translesion synthesis (η, ι, κ, ζ, and Rev1). Some of these DNA polymerases have roles in more than one pathway of DNA processing [1, 2].
In mammalian cells pol θ is encoded by the POLQ gene (Polq in the mouse). The initial discovery and molecular cloning of POLQ orthologs (starting with the Drosophila Mus308 gene) and the development of mouse models for Polq disruption was covered in earlier reviews [3, 4]. Pol θ orthologs are large enzymes (290 kDa in mammalian cells) present only in multicellular organisms. They have a distinctive domain configuration, with an N-terminal helicase-like region linked to a C-terminal DNA polymerase via a central region of mostly unknown function [3, 5, 6] (Figure 1). Mammalian POLQ is broadly expressed in normal tissues.
Figure 1.
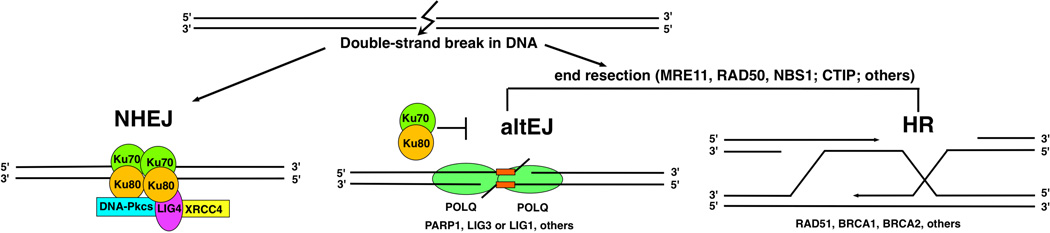
A double-strand break can be repaired by cNHEJ with minimal end processing. If the break ends are resected to produce 3’ single-stranded tails, an altEJ pathway can be invoked (involving pol θ). Alternatively, homologous recombination (HR) can take place, depending on availability of a copy of the damaged gene.
Genes with similarity to POLQ and Mus308 are present in multicellular eukaryotes, plants, and protists, but interestingly not in fungi [3, 4]. In each case, the gene products function in a DNA end-joining repair pathway. The primary strategies for DSB repair are “end-joining”, via mechanisms that process and rejoin the ends of a DSB, and homologous recombination (HR) pathways which employ an undamaged copy of the DNA [7–9] (Figure 1). “Classical” non-homologous end-joining (NHEJ) relies on DNA-end binding mediated by the Ku70-Ku80 complex (the XRCC6 and XRCC5 gene products), in concert with the DNA-dependent protein kinase (DNA-PK, PRKDC). The Ku complex and other factors inhibit processing of DNA termini, and so a majority of double-strand breaks in mammalian cells will be repaired through cNHEJ [10, 11]. If breaks are not repaired by NHEJ, the 5’ terminal strands of the broken DNA ends are resected by nucleases to generate single-stranded DNA (ssDNA) tails with 3’ ends [12, 13] (Fig 1). Resection is an essential intermediate in HR and some DSBs are channeled to repair by this pathway, particularly in S-phase cells [11, 14, 15]. A subset of DSB will be handled by alternative end-joining pathways in situations where the DNA end is not compatible with processing by cNHEJ, or if core components of the cNHEJ machinery are absent or unavailable. Alternative-end joining of DSBs can occur throughout the cell cycle in mammalian cells [9].
In general, altEJ is defined as a means for repair of DSB that is exclusive of Ku-dependent, classically defined NHEJ [16], and dependent on factors (CtIP, MRN, EXO1, etc.) that resect double-strand breaks to generate extended 3’ ssDNA tails [12, 13] (Figure 1). The biological consequences of this end-joining is manifested in different ways in different organisms [4, 17].
FUNCTIONS OF POL θ
Pol θ and double-strand break repair
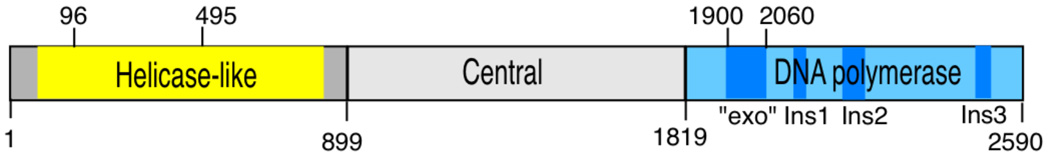
A major function for pol θ is in the defense against double-strand breaks. A defect in pol θ can lead to double-strand break-mediated genomic instability. Such instability manifests itself in different ways depending on the biological setting. The chaos1 (chromosome aberration occurring spontaneously 1) mouse was derived from a screen for animals exhibiting increased spontaneous frequencies of micronuclei (MN) in peripheral blood reticulocytes [18]. MN arise from chromosome fragments (due to chromosome breakage) or rogue chromosomes not packaged into the nucleus at the time of cell division [19, 20]. The chaos1 mutation changes Ser to Pro at amino acid 1932 in mouse pol θ, near the beginning of the polymerase domain (Figure 2) [21]. The severe effect of this amino acid change may arise from instability in the protein that prevents efficient expression [22]. Increased MN are also displayed in other cell types from Polq−/− mice [22, 23]. The frequency of MN in Polq−/− reticulocytes is further elevated by exposure of the mice to ionizing radiation (IR) or MMC [18, 21, 23].
Figure 2.
Tripartite domain structure of human pol θ. The most conserved region of the helicase-like domain is highlighted in yellow. The DNA polymerase domain (blue) includes a nonfunctional exonuclease domain (“exo”), and three insertion loops designated ins1, ins2, and ins3. A central region (grey) connecting the two domains is predicted to be largely disordered.
Polq−/− bone marrow stromal cells are selectively hypersensitive to agents that introduce DSB, including IR, the drug bleomycin, and topoisomerase inhibitors [22, 23]. A Polq-null mutant of mouse CH12F3 B cells is also more sensitive than normal cells to IR and etoposide [24]. In an siRNA-based screen for human genes affecting IR sensitivity, depletion of pol θ caused an increase in IR-induced γH2AX foci and sensitized SQ20B and HeLa cells to IR [25]. The pol θ homolog in the green algae Chlamydomonas reinhardtii was found in a screen for mutants specifically sensitivity to Zeocin (a DSB inducing agent related to bleomycin) [26].
POLQ-related synthetic lethality and epistasis
The initial demonstration of Polq-related synthetic impairment was the discovery of a marked developmental disadvantage in Atm−/− Polq−/− double mutant mice. The few surviving mice displayed DNA damage-related stress, including increased chromosomal instability and decreased body weight [21]. As a master regulator of stress responses, loss of ATM disrupts pathways that have such sequelae, including reduction of HR activity in some instances. Direct inhibition of ATM activation with the drug KU55933 increased radiosensitivity in wild type but not in Polq−/− cells, indicating that the synthetic lethality of Atm and Polq may involve more than ATM kinase activity alone [23].
A major sensitization to IR by simultaneous inactivation of Polq and homologous recombination activity was first found in Drosophila [27]. Mutant larvae with a spn-A (Rad51) mutation are radiosensitive, and addition of a mus308-mutation results in synergistic IR hypersensitivity [27]. Inactivation of FANCD2, involved in HR repair for some lesions, is incompatible with simultaneous inactivation of pol θ in mice [28]. Knockdown of POLQ is also reported to inhibit the viability of BRCA1 mutant cells [29]. A Polq mutant in chicken DT40 cells, was reported to have greatly reduced levels of homologous recombination repair of I-Sce-I-mediated DSBs [30]. However, such a role is not compatible with the other results that we summarize here.
There is evidence that poly-(ADP-ribose)polymerase, especially PARP1, participates in an altEJ pathway [31–34]. It is therefore of interest to determine whether pol θ and PARP1 work together in altEJ, but this is not yet resolved. PARP1 inhibitors have enhanced toxicity in BRCA1 or BRCA2-defective cells. This is sometimes attributed to inhibition of a backup break restitution process that uses components of BER, but an alternative is that PARP1 inhibition impairs an end-joining pathway that is important to maintain normal genomic replication [35]. A linkage between PARP and pol θ action was suggested in studies of Polq-defective cells, which are sensitive to the topoisomerase I inhibitor camptothecin. The PARP inhibitor olaparib confers minimal additional sensitization of Polq−/− cells to camptothecin [22]. On the other hand, POLQ depletion from FANCD2-defective human cells or mouse tumor cells enhanced their PARP inhibitor sensitivity, implying that pol θ and PARP are not entirely in the same pathway [28]. Pol θ localizes to tracks of DNA breaks formed by laser radiation [36], as does PARP1, and it was suggested that PARP1 recruits POLQ to promote altEJ [29]. However, more clarification is needed because localization of tagged pol θ to microirradiated tracks in HeLa cells was reduced both by PARP inhibitor (where PARP is retained on DNA) and by siPARP1 (which decreases total PARP) [29].
Pol θ and “alternative end-joining” of DNA double-strand breaks
Pol θ is a component of an end-joining pathway for DSB, which appears to be the main route by which pol θ defends against the genomic toxicity of IR and other DNA damaging agents. The relevant end joining pathway is variously referred to as alternative end joining (altEJ), microhomology-mediated end-joining (MMEJ), synthesis-dependent end-joining (SD-MMEJ), or theta-mediated end-joining (TMEJ). It is referred to here as altEJ to emphasize its distinction from the “canonical” non-homologous end-joining (cNHEJ) pathway that involves the Ku70-Ku80 proteins. The role of pol θ in altEJ was discovered initially in studies of Drosophila. In vivo break joining assays of DSBs induced by I-SceI nuclease showed a role of Mus308 in a synthesis-dependent end joining process [37], independent of Ku70 and Lig4.
The preferred substrates and outcomes of altEJ are being clarified by assays with extrachromosomal substrates transfected into mammalian cells. Pol θ-dependent altEJ is the main process responsible for the joining of substrates with “pre-resected” tails (e.g., substrates with long 3’-ssDNA tails) [22]. Pol θ-dependent altEJ is dominant in Ku-deficient cells, or in situations where end-resection is unregulated or excessive [38]. Inactivation of both Ku-dependent NHEJ and pol θ-dependent altEJ is not well-tolerated by mouse cells [38].
The occurrence of “microhomologies” (> 1–2 bp or greater) at sites of break rejoining is considered a defining signature of altEJ. In the mouse, pol θ-dependent altEJ accounts for most repair associated with microhomologies, and is made efficient by coupling removal of nonhomologous tails with a microhomology search and microhomology-primed synthesis across broken ends [38]. In Drosophila, joining events in mus308 mutants show decreased utilization of longer micro homologies at repair junctions [27].
A second signatureof pol θ-dependent altEJ is the production of templated DNA insertions at some sites of DSB joining. Such insertions occur during Mus308-dependent repair of directed double-strand breaks in Drosophila [27] and in C. elegans [39]. DNA joining during retrohoming of linear group II intron RNAs in Drosophila also results in the formation of insertions dependent on pol θ [40]. In the mouse, such insertions have been observed during class switch recombination (CSR) of immunoglobulin genes. AltEJ is used as a backup pathway for joining of double-strand break intermediates in CSR [24]. The insertions are dependent on POLQ [22]. Pol θ-dependent insertions in mammalian cells are typically 2 to 30 bp long, are largely templated [22] and can arise from sequences directly adjacent to the resected ends, or from sequences many kb distant [22], or even sequences arising from another chromosome [38].
A significant fraction of insertions observed upon joining of CRISPR-Cas9 induced breaks are also POLQ-dependent [29, 41]. These insertions can be explained by initiation of synthesis at microhomologies by POLQ (using nearby or distant sites as a template) [39, 40] sometimes with cycles of slippage and reinitiation [22].
Unprotected telomeres are normally joined by NHEJ, but can be joined by altEJ as a backup in a POLQ-dependent manner [29]. Other evidence pointing to a role for POLQ in guarding against major changes in genomic DNA replication was found by examining the consequences of replication through endogenous G-quadruplex (G4) forming sequences. The FANCJ homolog dog1 in C. elegans normally provides a major defense against G4-induced instability. In the absence of dog1, C. elegans Pol θ is important in preventing large catastrophic deletions. When Pol θ is operational, only small deletions are formed, and at their joining sites they show the microhomology and occasional insertion signature of pol θ action [39]. Pol θ is also important for preventing large deletions when other TLS polymerases are absent from the genome [38, 42].
Possible modulation of HR by Polq status
Drosophila pol θ does not participate in homology-directed repair and its action is independent of Rad51 [27]. Polq-mediated end-joining typically does not compete with either cNHEJ or HR repair. However, it is engaged more frequently in NHEJ deficient cells [38]. Cellular Rad51 “foci”, which may represent RAD51 loading on resected ends, were found to increase about 1.5-fold by siRNA suppression of POLQ in U2OS cells [28] or mouse MEFs [29] after exposure to 4 Gy of IR. As measured by an I-SceI based assay in U2OS cells, HR was reported to increase about 2-fold after siRNA suppression of Polq. It was suggested that this might be related to a function of the helicase-like domain in modulating Rad51 loading [28], but further investigation is required to test this idea.
Mutants of POLQ homologs in Arabidopsis (TEBICHI), C. elegans (polq-1), and Drosophila (Mus308) are hypersensitive to ICL-inducing agents [3], whereas Polq-defective mammalian cells are not appreciably hypersensitive to such agents. These different consequences of defects in POLQ-dependent altEJ may arise because organisms differ in the priority of engaging DNA repair pathways. In proliferating mammalian cells, ICLs are usually dealt with by a Fanconi anemia-related pathway that gives rise to enzymatically-induced double-strand breaks that are channeled into HR [43]. In Drosophila and some other organisms, an altEJ-dependent pathway may be more important for resolving ICL-associated double-strand breaks.
Chromosome translocations
Chromosome translocations can arise in cells when either altEJ or cNHEJ are inactivated, indicating that either process is capable of joining broken chromosome ends [44]. The Myc-IgH translocation in mice (a model for the oncogenic Burkitt lymphoma translocation) is initiated by action of the AIDCA-encoded enzyme, and is increased by ~4-fold in Polq-defective mice [22]. CRISPR-Cas9 initiated translocations have been examined in mouse pluripotent stem cells derived by oncogene transfection of MEFs. After CRISPR-Cas9 cleavage of the Rosa26 locus on mouse chr 6 and the H3f3b locus on Chr 11, the translocation frequency was decreased by about 4 fold in Polq-defective cells [29]. As expected, insertion sequences at translocation breakpoints were absent in Polq-defective cells. In contrast, examining the same translocation in MEF cells showed no significant change in Polq−/− single mutants, a 2 fold enhancement in Ku70−/− mutants, and about 3 fold enhancement in a Polq−/− Ku70−/− double mutant [38].
Functions at DNA replication forks
There are indications that pol θ action can be closely coordinated with DNA replication. As described below, POLQ gene expression correlates with expression of genes involved in cell cycle control and DNA replication. The enhanced micronuclei phenotype of POLQ disruption is phenocopied by the chaos3 mutation, isolated in the same screen as chaos1. Chaos3 is a hypomorphic allele of Mcm4, a component of the helicase operating during semiconservative DNA replication [45]. The pol θ homolog TEBICHI is important for normal Arabidopsis plant development and contributes to alleviating DNA replication stress [46, 47]. In C. elegans, pol θ limits extensive deletions at DNA replication fork barriers, but generates small indels, templated by DNA adjacent to the excision site [48, 49]. Furthermore, pol θ suppression in the human RKO cell line modestly shifts DNA replication timing throughout the genome [50]. It is possible that the influence of pol θ on replication timing is a result of a continuous function for pol θ in repairing endogenous damage arising when DNA replication forks stall in particular sequences (for example G4 DNA). A parallel similar situation that shifts replication timing is seen with pol ζ, which has a role in preventing DSB during genomic replication [51]. There is a direct association of pol θ with the origin replication complex protein ORC1 [50], and this may be important in recruiting pol θ to rescue broken replication forks. Consistent with this, stalled DNA replication forks recover less well in pol θ-defective cells [28].
Base excision repair
Pol θ may serve as a backup polymerase for base excision repair (BER). In mammalian cells, most single-nucleotide BER is catalyzed by pol β [36, 52]. The C-terminal region of human pol θ displays a weak 5’-deoxyribose-phosphate lyase (dRp-lyase) activity that could facilitate BER [53]. In chicken DT40 cells, Polq and Polq Polb mutants are more sensitive to MMS than either of the single mutants. Extracts from the Polq mutant DT40 cell line appeared to have a reduced capacity for both short and long patch BER [36]. Pol θ may be a backup enzyme for BER in the C. elegans, which lacks pol β, although at least one additional DNA polymerase also appears to function in BER in the nematode [54].
BIOCHEMICAL PROPERTIES AND STRUCTURE
Polymerase activity
Human pol θ has been studied as a recombinant full-length protein produced from a baculovirus vector in insect cells [5, 6], and as active constructs of the C-terminal polymerase domain [55, 56]. The full-length protein is active on substrates including oligonucleotide primer-templates, hairpin primer-templates, activated calf thymus DNA and poly (dA)-oligo (dT) [5, 6]. Recombinant pol θ is relatively resistant to aphidicolin, an inhibitor of eukaryotic replicative DNA polymerases (pols α,δ,ε) [5, 57–59]. Pol θ is sensitive to dideoxynucleoside triphosphate (ddNTP), consistent with the presence of a tyrosine residue in motif 4, which facilitates ddNTP incorporation [5, 60].
Fidelity
The A family of polymerases typically comprises high-fidelity polymerases, yet pol θ is remarkably error prone. In a primer extension assay to measure DNA polymerase fidelity, pol θ frequently misincorporated a G or T across from a T in the template [5, 6]. Human pol θ generated single base errors in a gap-filling reaction at a 10 to 100-fold higher rate than the other A family DNA polymerases pol γ and pol ν [61]. The enzyme generates single-base substitutions at an average rate of 2.4 × 10−3, comparable in rate to the inaccurate family Y human pol κ (5.8×10−3) [62]. Pol θ adds single nucleotides in homopolymeric runs at unusually high rates during gap filling, exceeding 1% in certain sequence contexts. Such +1 frameshifts indicate a propensity for slippage of the template relative to the primer. Even though it harbors an exonuclease-like domain, purified pol θ lacks a 3’ → 5’ exonuclease activity. It may associate in mammalian cells with a separate 3’ → 5’ proofreading activity [63].
Extension of single-stranded DNA by error-prone templated synthesis
Pol θ has a unique ability to add nucleotides to the 3’ ends of single-stranded DNA [64], primed by minimal pairing with other available DNA molecules. Under physiological conditions, pol θ does not have terminal transferase-like activity [64]. Synthesis by pol θ in this context is consistent with the unusually efficient ability of the polymerase to extend from mismatched DNA termini [6, 65], and its tendency towards primer-template slippage [61]. These properties of pol θ provide a mechanistic explanation for its contribution to altEJ. In vivo studies are giving insight as to the preferred structures for POLQ-catalyzed extension [22].
In vitro, joining experiments have been done only with a C-terminal fragment of pol θ [55, 66]. It was shown that the C-terminal fragment of pol θ performs end joining of partially resected DNAs with a 3’ overhang, provided that microhomology extends to more than 2 bp [66]. Further experiments showed that human pol θ, and no other DNA polymerase, promotes MMEJ in vitro and in vivo. It will be of interest to examine the action of pol θ and DNA ligases at double-strand breaks with 3’-single-stranded overhangs that closely mimic the resected ends of a DNA double-strand break.
Bypass of DNA damage
Pol θ can efficiently incorporate an A residue opposite an abasic (AP) sites and possesses the rare ability to extend past an AP site, a lesion that usually constitutes a formidable block to DNA polymerases [67, 68]. It remains to be determined whether this AP site bypass activity is a function of pol θ in vivo. Thymine glycol (Tg) is a common product of reactive oxygen species-mediated damage to DNA. Pol θ and Pol ν can efficiently bypass both enantiomers (5R,5S) of Tg, whereas pol η extends efficiently only from the 5R-Tg [6, 69, 70]. Pol θ cannot insert a base opposite the common UV-radiation induced cyclobutane pyrimidine dimers or (6-4) photoproducts. However, if pol ι [71] is used to incorporate bases opposite a 6-4PP, pol θ can extend the poorly matched primer-terminus [6, 65]. Pol θ is also able to efficiently extend mismatched A:G, A:T, and A:C termini [65]. The ability of pol θ to bypass Tg, AP sites, or other lesions might be useful during joining of 3’ tails arising following IR-induced DNA damage, where multiply damaged sites of DNA will occur surrounding a DNA break.
Because POLQ has a relatively high error rate and can bypass AP sites, several investigations explored the possibility of a function for POLQ in somatic hypermutation of immunoglobulin genes but on balance, it appears that POLQ does not participate in the major events leading to somatic hypermutation [72].
Structural basis of extension of poorly matched termini by the Pol domain
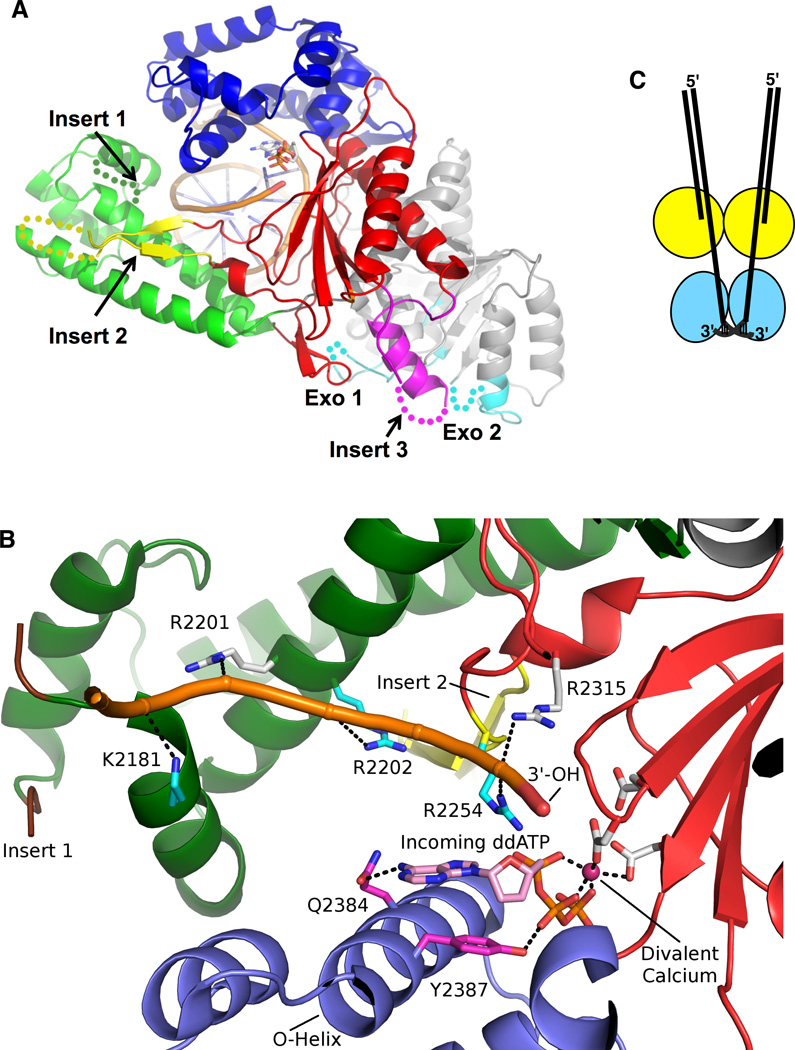
The ability of pol θ to extend from mismatched, poorly matched, or unmatched termini is beginning to be understood by biochemical and structural examination of the DNA polymerase domain. A fragment of pol θ (residues 1792–2590) encompassing the pol domain and a portion of the central domain (Figure 2) is active as a DNA polymerase when produced as a recombinant protein in E. coli. Shorter fragments are much less active, or inactive [52, 55] indicating that this region of the central domain is required for efficient polymerase activity or proper folding. The active fragment is able to bypass AP sites and Tg adducts, showing that these activities are independent of the N-terminal helicase-like domain. Several conserved insertion loops, absent from bacterial homologs, intervene within the family A polymerase fold of pol θ. Two insertion loops in the vestigial exonuclease domain (exo1 and exo2) and three in the polymerase catalytic domain (inserts 1, 2, and 3) are unique to pol θ (Figure 3A), and specific deletions of these inserts have been analyzed with respect to activity. Insert 1, which is located at the tip of the thumb, comprises ~22 amino acids between the first and second conserved motifs in that subdomain and its sequence is highly conserved throughout vertebrates [6]. In other A-family DNA polymerases, the thumb region influences DNA binding and frameshift fidelity. Deletions in the thumb region of E. coli Pol I can cause errors in processivity and an increased use in misaligned primer/template complex [60, 73, 74]. Elimination of Insert 1 of the active C-terminal pol θ fragment reduced processivity of the enzyme but had little, if any, bearing on the translesion synthesis properties of the enzyme [55]. Insert 2 is 52 amino acids long, falls between the second and third motifs and has the least sequence conservation among the inserts whereas insert 3 (33 amino acids) is located between the fifth and sixth motifs (Fig. 2A and B). Removal of either inserts 2 or 3 reduced activity on undamaged DNA and eliminated the ability of POLQ to bypass AP sites or Tg lesions [55]. Deletion of residues 2264–2315, which includes much of insert 2, was shown to abrogate POLQ’s ability to extend single-stranded oligonucleotides [64].
Figure 3.
A. Crystal structure of the polymerase domain of human pol θ.
The polymerase domain harbors the canonical fingers (colored in blue), palm (red), and thumb (green) subdomains as well as an exonuclease-like domain devoid of proofreading activity (shown in grey). The crystal structure revealed that the polymerase domain comprises five unique insertions: three in the polymerase domain (insertions 1–3) and two in the exonuclease-like domain (exo1 and exo2). Disordered segments not visible in the crystal structure are indicated with dotted lines.
B. Features of pol θ that grasp the primer-terminus. In this view of the ternary complex of the pol θ DNA polymerase domain, five Lys and Arg residues make specific contacts with phosphate residues (shown as nubs) in the primer DNA strand (orange). Two of these contacts are conserved in all A-family polymerases (shown with white carbons), and three of them are unique residues in pol θ (cyan carbons), with Arg2254 emerging from the distinctive Insert 2 (yellow). Additional unique contacts are made by Gln2384 and Tyr2387, which contact the major groove side of the base and the phosphate of the incoming nucleotide, respectively [from data in [56]]. For the sake of clarity, only the backbone of the primer strand is shown. Color coding of the protein subdomains is as in Figure 3A.
C. Possible mechanism for alignment of terminal microhomologies, based on the dimerization of the pol θ helicase-like domain (POLQ-HLD) as observed in crystal structures [78]. Connectivity between the polymerase and POLQ-HLD, not shown here, could be in cis or trans.
Crystal structures of the polymerase domain (aa 1792–2590) of human pol θ have been obtained [56]. One structure depicts insertion of ddATP opposite an abasic site analog during translesion DNA synthesis. The second structure describes a cognate complex with ddGTP. In addition to upstream contacts to the primer DNA strand mediated by the specialized thumb, pol θ establishes unique contacts to the primer terminal phosphate via insert 2 (Figure 3B). In this way, pol θ uniquely grasps the primer to bypass DNA lesions, and to extend poorly base-paired DNA termini to mediate alternative end-joining of DNA double-strand breaks. Mutation of the arginine contacting the 3’ end of the primer into a valine (corresponding residue in bacterial family A polymerases) yields an enzyme variant R2254V that is impeded in its ability to bypass AP sites [56]. In the vestigial exonuclease-like domain (aa 1819–2090) the catalytic carboxylates are mutated, explaining the lack of 3’-5’ exonuclease activity [56].
Pol ν is a 900 amino acid protein in human cells, harboring a polymerase fold related to that of Mus308 and pol θ [70, 75, 76]. Pol ν and pol θ are the only A-family polymerases in the nucleus and both differ markedly from the prototypical members of this family in that they are remarkably error-prone. Crystal structures of human pol ν bound to primer/template DNA revealed a polymerase catalytic core that resembles that of its bacterial orthologs, such as E. coli or Bacillus Pol I [77]. The N-terminal exonuclease-like domain is more divergent: the catalytic carboxylate residues are absent and a loop blocks access to the potential active site preventing any 3’-5’ exonuclease activity. Pol ν has been shown to incorporate dTTP preferentially regardless of the nature of the template base [76, 77]. This unique ability appears to be due to a lysine residue in the O helix, K679 [76, 77], which resides near the nascent base pair. This residue is conserved among pol ν protein sequences, but not in pol θ, in which the corresponding residue is a glutamine. Similarly to pol θ, pol ν harbors 3 insertion elements in its polymerase domain, albeit shorter. Insert 2 appears to play a role in pol ν ’s ability to loop out the primer strand [77].
Helicase-like domain
Pol θ is the only polymerase known to include a helicase domain. The helicase-like domain of pol θ (POLQ-HLD) has 7 conserved motifs of the superfamily II (SF2) DNA and RNA helicase family [5]. POLQ-HLD exhibits some single-stranded DNA-dependent ATPase activity, though lower in comparison to that of HELQ. No overt helicase activity of POLQ-HLD has been reported [5, 78], and it is possible that the enzyme only displays helicase activity with an as yet untested substrate or requires accessory factors.
The DNA polymerase activity of pol θ is necessary to prevent cell death and chromosome breaks (micronuclei) caused by a double-strand break-inducing agent. Disruption of the ATPase activity in POLQ-HLD did not, however, alter the correcting function of pol θ addition to knockout cells [22]. No activity has yet been shown for POLQ-HLD, other than DNA-dependent ATPase function [5]. The crystal structures of the helicase-like domain of pol θ was recently reported, unliganded (aa 1–894) or in complex with ADP or a non-hydrolyzable ATP analog (aa 67–894) [78]. As with the C-terminal polymerase domain of pol θ, the helicase-like domain relies on unique inserts to accomplish new functions: additional helices in domain 4 contribute to the formation of a tetrameric interface. Comparison with an archaeal Hel308 [79] helicase revealed key differences between the two proteins: The beta hairpin that functions in strand separation in Hel308 is much shorter in POLQ-HLD. The latter also lacks residues critical for ratcheting in Hel308 [79].
POLQ-HLD is a tetramer (a dimer of dimers) in solution and in crystallo, suggesting that at minimum the enzyme acts as a dimer [78]. A hypothesis put forward for how two pol θ molecules could work together is that each molecule operates on either side of a DNA break. POLQ-HLD may participate in the microhomology annealing step thus preparing the annealed substrate for further processing by the polymerase domain [78] (Figure 3B).
Central region
The POLQ-HLD and the C-terminal polymerase domain are separated by a long region of mostly unknown function. Based on disorder prediction algorithms this domain is presumed to be largely flexible and disordered. The central region is longer in vertebrates than in plants or invertebrates [22]. A recent paper identified three possible RAD51 binding sites in pol θ [28], one towards the end of POLQ-HLD (aa 861–865) and two in the central region (aa 1,297–1,303, and 1,315–1,319) though the latter two sites are not conserved in evolutionarily-based alignments.
POLQ AND CANCER
POLQ expression and cancer outcome
POLQ is one of a group of genes where higher levels of expression confer a survival advantage for tumors. An early analysis of patients from Japan compared expression of POLQ mRNA in tumor tissue and matched control tissue from the same individuals. Higher relative POLQ expression was found in stomach, lung, and colon cancers [80]. Division of colon cancers into two groups based on POLQ expression showed that the group expressing higher levels of POLQ had poorer survival than the lower expressing group by an average of about 24 months. A study of colorectal cancer patients from France found that higher expression of a group of 47 DNA replication-related genes (including POLQ) in tumors was significantly correlated with poorer patient survival [81]. Patients having higher expression of a small set of genes (POLQ and several DNA replication initiation genes) had shorter survival times [81]. An analysis focusing on human breast cancers found that of 14 nuclear DNA polymerase genes, only POLQ expression was significantly higher in the cancer tissues in comparison to normal tissues. When the data were divided into high and low POLQ-expressing groups, higher POLQ expression in breast cancers was correlated with poorer survival outcomes [82]. Another report of POLQ expression in early breast cancer found the highest POLQ expression in ER negative and high-grade tumors; higher expression was correlated with shorter relapse-free survival times [83]. POLQ is one of the genes frequently upregulated in a group of oral squamous cell carcinomas from Brazil [84]. In a study of early to mid-stage non small-cell lung cancers, higher expression of a five-gene prognostic signature including POLQ was associated with poor outcome [85]. Similarly, analysis of TCGA data on POLQ gene expression in ovarian carcinoma shows that its expression correlates with tumor grade [28].
These correlations are of interest, but it is important to note that POLQ is representative of a cluster of genes, with similar expression patterns, that correlate with cancer subtype and outcome. Relevant to breast cancer, POLQ is present in a 76 gene signature for prediction of distant metastasis in lymph node-negative primary tumors [86], in a 97 gene set predicting higher risk of recurrence in histologic grade 2 tumors [87], and in a more broad “374 gene signature set” where high expression correlates with poor recurrence-free survival in diverse histopathological types [88] Analysis of chronic myeloid leukemia samples identified the 20 most correlated gene expression probes where overexpression was associated with non-responsiveness to imatinib (Gleevec) treatment; POLQ was in this gene set [89].
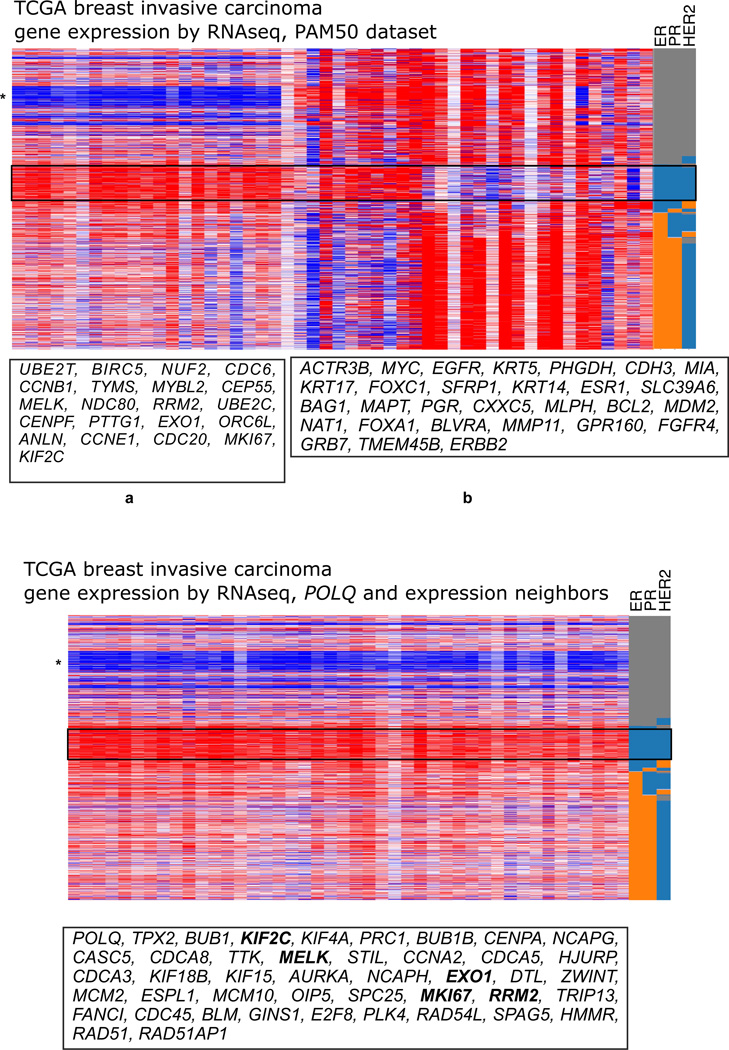
As an example of POLQ expression in relation to similarly expressed genes, we consider an example in breast invasive carcinoma. In Figure 4, gene expression heatmaps were generated from TCGA data (cancergenome.nih.gov). The top panel was produced using an established set of 50 genes, the PAM50 set, effective in subtyping breast cancers on the basis of gene expression [90]. As shown in the top panel of Figure 4, triple negative breast cancers have a distinct pattern of gene expression. Note the pattern in the group of genes at the left box marked “a” in the PAM50 set. In the bottom panel, the expression of POLQ and its 43 most similar gene expression neighbors were analyzed with the same set of breast cancer data. The gene expression pattern for POLQ and its expression neighbors closely matches the pattern for the set of PAM50 “a” set. Indeed, five of the PAM50 “a” genes are also POLQ nearest expression neighbors (Figure 4). Many of the genes in the POLQ expression neighbors set, including POLQ itself and AURKA [91] would serve as surrogates for similarly expressed genes in the PAM50 dataset, and mark the same set of triple-negative breast cancers; inclusion of POLQ in such analyses is not essential to predict outcome. It does appear that high expression of the group of genes that includes POLQ may promote cancer progression by increasing resistance to endogenous replication stress in a concerted manner [85]. Many of the POLQ expression neighbors are involved in DNA replication, cell cycle control, and DNA repair. It is notable that POLQ is the only DNA polymerase that emerges from this type of analysis, suggesting that it indeed confers a selective advantage to tumors. It remains to be determined whether all members of this group of genes contribute to such an advantage. Another open question is how the expression of these genes is co-regulated at the transcriptional level.
Figure 4. POLQ is expressed in a cluster of genes predictive of breast cancer subtypes.
The heatmaps show gene expression (high, dark red; low, dark blue) derived from the UCSC cancer genomics website (https://genome-cancer.ucsc.edu/proj/site/hgHeatmap/). Here, data were analyzed using the dataset “TCGA breast invasive carcinoma gene expression by RNAseq (IlluminaHiSeq), pancan normalized • N=1215”. The data are organized according to estrogen receptor (ER), progesterone receptor (PR), and HER2 amplification status (HER2) in the columns at the right. The orange color shows positive status, blue is negative, and grey shows undetermined status. Each row is a different breast cancer case. The area of the heatmaps containing triple-negative breast cancers is boxed with a solid line. The gene subset “a” in the top heatmap has a distinctive pattern of exceptionally high expression in triple-negative cancers (red boxed area) and exceptionally low expression in another set of cancers (blue starred region). The heatmap was produced with the subtype predictor PAM50 set of genes [90], with each column representing a gene in the order shown in the list. The bottom heatmap used POLQ and its 43 nearest expression neighbors where expression information is available in the target TCGA dataset. The expression neighbors were determined with the multi-experiment matrix tool (MEM, http://biit.cs.ut.ee/mem/index.cgi, output filtered with the text search “breast”) by querying AFFY44 human microarrays. Each column represents a gene in the order shown in the list. The heatmap pattern for the POLQ expression neighbors genes closely resembles the pattern for the subset of PAM50 genes shown at the left in the top panel, and five of the genes overlap between these sets as indicated in bold.
Polymorphic variants and cancer
Many studies have searched for single nucleotide polymorphism (SNP) associations with cancer risk or outcomes, but these must be interpreted cautiously until functional data are obtained [92]. One comparison found that a variant of SNP rs587553 in the promoter region of POLQ was more common in hereditary cases of breast cancer than in sporadic cases [93]. It was suggested that the SNP variation might affect binding of the YY1 transcription factor [93]. This is unlikely, as YY1 binding consensus sequences (http://jaspar.genereg.net) do not overlap with rs587553. A SNP variant rs7632907 associated with increased risk of esophageal squamous cell carcinoma was proposed to be relevant to POLQ [94], but we note that this SNP is in the 3’ untranslated region of an adjacent gene, STXBP5L, and is not part of the POLQ transcript.
Pol θ as a target for cancer therapy
Because suppression of POLQ results in sensitivity of cells to ionizing radiation and some DSB-inducing drugs, suppression of POLQ might be a useful adjuvant to these DNA damaging therapies [3, 25, 82]. Cells of at least some cancers may be susceptible because they appear to thrive on higher levels of POLQ expression. Because HR-defective cells are more sensitive to POLQ ablation, HR defective tumors might be especially vulnerable. POLQ includes at least two enzymatic activities that could be targeted by small molecule inhibitors. Further consideration of dual inhibition of POLQ and PARP1 is also warranted.
It would be clinically beneficial if tumor cells, but not normal cells, could be sensitized by POLQ suppression, but it remains to be seen whether this is generally the case. Suppression of POLQ by siRNA is variable and no general rules have emerged to control radiation sensitivity [25]. It is encouraging that despite the radiation sensitivity of mouse cell lines, the hematopoietic system of Polq−/− mice seems resilient to IR [23]. Approaches using POLQ suppression on tumors should also be alert to the fact that loss of POLQ may also lead to damaging genetic alterations in tumor cells, such as potentially oncogenic chromosome translocations or regional genomic deletions.
Acknowledgments
These studies were funded by National Institutes of Health grants R01 CA052040 (S.D.) and CA097175 (R.D.W.) and grant RP130297 from the Cancer Prevention and Research Institute of Texas (R.D.W.) and the Grady F. Saunders Ph.D. Distinguished Research Professorship (R.D.W.). We thank Karl E. Zahn and Andrew W. Malaby for generating figures 3A and 3B. We appreciate the helpful comments on the manuscript provided by Kei-ichi Takata and Junya Tomida.
Abbreviations used
- DSB
double-strand break
- IR
ionizing radiation
- altEJ
alternative end-joining
- NHEJ
non-homologous end-joining
- MN
micronuclei
- pol θ
DNA polymerase theta
Footnotes
Conflict of Interest Statement
The authors declare that they have no financial, personal or professional competing interests that could be construed to have influenced this paper.
References
- 1.Hübscher U, Maga G, Spadari S. Eukaryotic DNA polymerases. Annu Rev Biochem. 2002;71:133–163. doi: 10.1146/annurev.biochem.71.090501.150041. [DOI] [PubMed] [Google Scholar]
- 2.Lange SS, Takata K, Wood RD. DNA polymerases and cancer. Nat Rev Cancer. 2011;11:96–110. doi: 10.1038/nrc2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yousefzadeh M, Wood R. DNA polymerase POLQ and cellular defense against DNA damage. DNA Repair. 2013;12:1–9. doi: 10.1016/j.dnarep.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beagan K, McVey M. Linking DNA polymerase theta structure and function in health and disease. Cell Mol Life Sci. 2016;73:603–615. doi: 10.1007/s00018-015-2078-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seki M, Marini F, Wood RD. POLQ (Pol θ), a DNA polymerase and DNA-dependent ATPase in human cells. Nucleic Acids Res. 2003;31:6117–6126. doi: 10.1093/nar/gkg814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seki M, Masutani C, Yang LW, Schuffert A, Iwai S, Bahar I, Wood RD. High-efficiency bypass of DNA damage by human DNA polymerase Q. EMBO J. 2004;23:4484–4494. doi: 10.1038/sj.emboj.7600424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kass EM, Jasin M. Collaboration and competition between DNA double-strand break repair pathways. FEBS Lett. 2010;584:3703–3708. doi: 10.1016/j.febslet.2010.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rassool FV, Tomkinson AE. Targeting abnormal DNA double strand break repair in cancer. Cell Mol Life Sci. 2010;67:3699–3710. doi: 10.1007/s00018-010-0493-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thompson LH. Recognition, signaling, and repair of DNA double-strand breaks produced by ionizing radiation in mammalian cells: The molecular choreography. Mutation Research. 2012;751:158–246. doi: 10.1016/j.mrrev.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 10.Shibata A, Conrad S, Birraux J, Geuting V, Barton O, Ismail A, Kakarougkas A, Meek K, Taucher-Scholz G, Lobrich M, Jeggo PA. Factors determining DNA double-strand break repair pathway choice in G2 phase. EMBO J. 2011;30:1079–1092. doi: 10.1038/emboj.2011.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kakarougkas A, Jeggo PA. DNA DSB repair pathway choice: an orchestrated handover mechanism. Br J Radiol. 2014;87:20130685. doi: 10.1259/bjr.20130685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bennardo N, Cheng A, Huang N, Stark JM. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genetics. 2008;4:e1000110. doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee K, Lee SE. Saccharomyces cerevisiae Sae2- and Tel1-dependent single-strand DNA formation at DNA break promotes microhomology-mediated end joining. Genetics. 2007;176:2003–2014. doi: 10.1534/genetics.107.076539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hartlerode A, Odate S, Shim I, Brown J, Scully R. Cell cycle-dependent induction of homologous recombination by a tightly regulated I-SceI fusion protein. PLoS One. 2011;6:e16501. doi: 10.1371/journal.pone.0016501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karanam K, Kafri R, Loewer A, Lahav G. Quantitative live cell imaging reveals a gradual shift between DNA repair mechanisms and a maximal use of HR in mid S phase. Mol Cell. 2012;47:320–329. doi: 10.1016/j.molcel.2012.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deriano L, Roth DB. Modernizing the nonhomologous end-joining repertoire: alternative and classical NHEJ share the stage. Annual review of genetics. 2013;47:433–455. doi: 10.1146/annurev-genet-110711-155540. [DOI] [PubMed] [Google Scholar]
- 17.Sfeir A, Symington LS. Microhomology-Mediated End Joining: A Back-up Survival Mechanism or Dedicated Pathway? Trends Biochem Sci. 2015;40:701–714. doi: 10.1016/j.tibs.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shima N, Hartford SA, Duffy T, Wilson LA, Schimenti KJ, Schimenti JC. Phenotype-based identification of mouse chromosome instability mutants. Genetics. 2003;163:1031–1040. doi: 10.1093/genetics/163.3.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nusse M, Miller BM, Viaggi S, Grawe J. Analysis of the DNA content distribution of micronuclei using flow sorting and fluorescent in situ hybridization with a centromeric DNA probe. Mutagenesis. 1996;11:405–413. doi: 10.1093/mutage/11.4.405. [DOI] [PubMed] [Google Scholar]
- 20.Dertinger SD, Torous DK, Tometsko KR. Simple and reliable enumeration of micronucleated reticulocytes with a single-laser flow cytometer. Mutat Res. 1996;371:283–292. doi: 10.1016/s0165-1218(96)90117-2. [DOI] [PubMed] [Google Scholar]
- 21.Shima N, Munroe RJ, Schimenti JC. The mouse genomic instability mutation chaos1 is an allele of Polq that exhibits genetic interaction with Atm. Mol Cell Biol. 2004;24:10381–10389. doi: 10.1128/MCB.24.23.10381-10389.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yousefzadeh MJ, Wyatt DW, Takata K, Mu Y, Hensley SC, Tomida J, Bylund GO, Doublié S, Johansson E, Ramsden DA, McBride KM, Wood RD. Mechanism of suppression of chromosomal instability by DNA polymerase POLQ. PLoS Genetics. 2014;10:e1004654. doi: 10.1371/journal.pgen.1004654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goff JP, Shields DS, Seki M, Choi S, Epperly MW, Dixon T, Wang H, Bakkenist CJ, Dertinger SD, Torous DK, Wittschieben J, Wood RD, Greenberger JS. Lack of DNA polymerase θ (POLQ) radiosensitizes bone marrow stromal cells in vitro and increases reticulocyte micronuclei after total-body irradiation. Radiat Res. 2009;172:165–174. doi: 10.1667/RR1598.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, Gao X, Wang JY. Comparison of two POLQ mutants reveals that a polymerase-inactive POLQ retains significant function in tolerance to etoposide and gamma-irradiation in mouse B cells. Genes Cells. 2011;16:973–983. doi: 10.1111/j.1365-2443.2011.01550.x. [DOI] [PubMed] [Google Scholar]
- 25.Higgins GS, Prevo R, Lee YF, Helleday T, Muschel RJ, Taylor S, Yoshimura M, Hickson ID, Bernhard EJ, McKenna WG. A small interfering RNA screen of genes involved in DNA repair identifies tumor-specific radiosensitization by POLQ knockdown. Cancer Res. 2010;70:2984–2993. doi: 10.1158/0008-5472.CAN-09-4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Plecenikova A, Slaninova M, Riha K. Characterization of DNA Repair Deficient Strains of Chlamydomonas reinhardtii Generated by Insertional Mutagenesis. PLoS One. 2014;9:e105482. doi: 10.1371/journal.pone.0105482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan SH, Yu AM, McVey M. Dual Roles for DNA Polymerase θ in Alternative End-Joining Repair of Double-Strand Breaks in Drosophila. PLoS Genet. 2010;6:e1001005. doi: 10.1371/journal.pgen.1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ceccaldi R, Liu JC, Amunugama R, Hajdu I, Primack B, Petalcorin MI, O'Connor KW, Konstantinopoulos PA, Elledge SJ, Boulton SJ, Yusufzai T, D'Andrea AD. Homologous-recombination-deficient tumours are dependent on Poltheta-mediated repair. Nature. 2015;518:258–262. doi: 10.1038/nature14184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mateos-Gomez PA, Gong F, Nair N, Miller KM, Lazzerini-Denchi E, Sfeir A. Mammalian polymerase θ promotes alternative NHEJ and suppresses recombination. Nature. 2015;518:254–257. doi: 10.1038/nature14157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kohzaki M, Nishihara K, Hirota K, Sonoda E, Yoshimura M, Ekino S, Butler JE, Watanabe M, Halazonetis TD, Takeda S. DNA polymerases nu and θ are required for efficient immunoglobulin V gene diversification in chicken. J Cell Biol. 2010;189:1117–1127. doi: 10.1083/jcb.200912012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Howard SM, Yanez DA, Stark JM. DNA damage response factors from diverse pathways, including DNA crosslink repair, mediate alternative end joining. PLoS Genet. 2015;11:e1004943. doi: 10.1371/journal.pgen.1004943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mansour WY, Borgmann K, Petersen C, Dikomey E, Dahm-Daphi J. The absence of Ku but not defects in classical non-homologous end-joining is required to trigger PARP1-dependent end-joining. DNA Repair (Amst) 2013;12:1134–1142. doi: 10.1016/j.dnarep.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 33.Audebert M, Salles B, Calsou P. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J Biol Chem. 2004;279:55117–55126. doi: 10.1074/jbc.M404524200. [DOI] [PubMed] [Google Scholar]
- 34.Wang M, Wu W, Wu W, Rosidi B, Zhang L, Wang H, Iliakis G. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 2006;34:6170–6182. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Helleday T. The underlying mechanism for the PARP and BRCA synthetic lethality: clearing up the misunderstandings. Mol Oncol. 2011;5:387–393. doi: 10.1016/j.molonc.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshimura M, Kohzaki M, Nakamura J, Asagoshi K, Sonoda E, Hou E, Prasad R, Wilson SH, Tano K, Yasui A, Lan L, Seki M, Wood RD, Arakawa H, Buerstedde JM, Hochegger H, Okada T, Hiraoka M, Takeda S. Vertebrate POLQ and POL beta cooperate in base excision repair of oxidative DNA damage. Mol Cell. 2006;24:115–125. doi: 10.1016/j.molcel.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu AM, McVey M. Synthesis-dependent microhomology-mediated end joining accounts for multiple types of repair junctions. Nucleic Acids Res. 2010;38:5706–5717. doi: 10.1093/nar/gkq379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wyatt DW, Feng W, Conlin MP, Yousefzadeh MJ, Roberts SA, Wood RD, Gupta GP, Ramsden DA. Essential roles for Polymerase θ mediated end joining in repair of chromosome breaks, submitted. 2016 doi: 10.1016/j.molcel.2016.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koole W, van Schendel R, Karambelas AE, van Heteren JT, Okihara KL, Tijsterman M. A Polymerase θ-dependent repair pathway suppresses extensive genomic instability at endogenous G4 DNA sites. Nat Commun. 2014;5:3216. doi: 10.1038/ncomms4216. [DOI] [PubMed] [Google Scholar]
- 40.White TB, Lambowitz AM. The retrohoming of linear group II intron RNAs in Drosophila melanogaster occurs by both DNA ligase 4-dependent and -independent mechanisms. PLoS Genet. 2012;8:e1002534. doi: 10.1371/journal.pgen.1002534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Schendel R, Roerink SF, Portegijs V, van den Heuvel S, Tijsterman M. Polymerase θ is a key driver of genome evolution and of CRISPR/Cas9-mediated mutagenesis. Nat Commun. 2015;6:7394. doi: 10.1038/ncomms8394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roerink SF, van Schendel R, Tijsterman M. Polymerase θ-mediated end joining of replication-associated DNA breaks in C. elegans. Genome Res. 2014;24:954–962. doi: 10.1101/gr.170431.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kottemann MC, Smogorzewska A. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature. 2013;493:356–363. doi: 10.1038/nature11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei PC, Chang AN, Kao J, Du Z, Meyers RM, Alt FW, Schwer B. Long Neural Genes Harbor Recurrent DNA Break Clusters in Neural Stem/Progenitor Cells. Cell. 2016;164:644–655. doi: 10.1016/j.cell.2015.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shima N, Alcaraz A, Liachko I, Buske TR, Andrews CA, Munroe RJ, Hartford SA, Tye BK, Schimenti JC. A viable allele of Mcm4 causes chromosome instability and mammary adenocarcinomas in mice. Nat Genet. 2007;39:93–98. doi: 10.1038/ng1936. [DOI] [PubMed] [Google Scholar]
- 46.Inagaki S, Suzuki T, Ohto MA, Urawa H, Horiuchi T, Nakamura K, Morikami A. Arabidopsis TEBICHI, with helicase and DNA polymerase domains, is required for regulated cell division and differentiation in meristems. Plant Cell. 2006;18:879–892. doi: 10.1105/tpc.105.036798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Inagaki H, Ohye T, Kogo H, Kato T, Bolor H, Taniguchi M, Shaikh TH, Emanuel BS, Kurahashi H. Chromosomal instability mediated by non-B DNA: cruciform conformation and not DNA sequence is responsible for recurrent translocation in humans. Genome Res. 2009;19:191–198. doi: 10.1101/gr.079244.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koole W, van Schendel R, Karambelas AE, van Heteren JT, Okihara KL, Tijsterman M. A Polymerase Theta-dependent repair pathway suppresses extensive genomic instability at endogenous G4 DNA sites. Nat Commun. 2014;5:3216. doi: 10.1038/ncomms4216. [DOI] [PubMed] [Google Scholar]
- 49.Roerink SF, van Schendel R, Tijsterman M. Polymerase theta-mediated end joining of replication-associated DNA breaks in C. elegans. Genome Res. 2014;24:954–962. doi: 10.1101/gr.170431.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fernandez-Vidal A, Guitton-Sert L, Cadoret JC, Drac M, Schwob E, Baldacci G, Cazaux C, Hoffmann JS. A role for DNA polymerase theta in the timing of DNA replication. Nat Commun. 2014;5:4285. doi: 10.1038/ncomms5285. [DOI] [PubMed] [Google Scholar]
- 51.Seplyarskiy VB, Bazykin GA, Soldatov RA. Polymerase zeta Activity Is Linked to Replication Timing in Humans: Evidence from Mutational Signatures. Mol Biol Evol. 2015;32:3158–3172. doi: 10.1093/molbev/msv184. [DOI] [PubMed] [Google Scholar]
- 52.Prasad R, Longley MJ, Sharief FS, Hou EW, Copeland WC, Wilson SH. Human DNA polymerase θ possesses 5'-dRP lyase activity and functions in single-nucleotide base excision repair in vitro. Nucleic Acids Res. 2009;37:1868–1877. doi: 10.1093/nar/gkp035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prasad R, Longley MJ, Sharief FS, Hou EW, Copeland WC, Wilson SH. Human DNA polymerase theta possesses 5'-dRP lyase activity and functions in single-nucleotide base excision repair in vitro. Nucleic Acids Res. 2009;37:1868–1877. doi: 10.1093/nar/gkp035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Asagoshi K, Lehmann W, Braithwaite EK, Santana-Santos L, Prasad R, Freedman JH, Van Houten B, Wilson SH. Single-nucleotide base excision repair DNA polymerase activity in C. elegans in the absence of DNA polymerase beta. Nucleic Acids Res. 2012;40:670–681. doi: 10.1093/nar/gkr727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hogg M, Seki M, Wood RD, Doublié S, Wallace SS. Lesion bypass activity of DNA polymerase θ (POLQ) is an intrinsic property of the pol domain and depends on unique sequence inserts. J Mol Biol. 2011;405:642–652. doi: 10.1016/j.jmb.2010.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zahn KE, Averill AM, Aller P, Wood RD, Doublié S. Human DNA polymerase θ grasps the primer terminus to mediate DNA repair. Nat Struct Mol Biol. 2015;22:304–311. doi: 10.1038/nsmb.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sheaff R, Ilsley D, Kuchta R. Mechanism of DNA polymerase alpha inhibition by aphidicolin. Biochemistry. 1991;30:8590–8597. doi: 10.1021/bi00099a014. [DOI] [PubMed] [Google Scholar]
- 58.Dresler SL, Gowans BJ, Robinson-Hill RM, Hunting DJ. Involvement of DNA polymerase delta in DNA repair synthesis in human fibroblasts at late times after ultraviolet irradiation. Biochemistry. 1988;27:6379–6383. doi: 10.1021/bi00417a028. [DOI] [PubMed] [Google Scholar]
- 59.Cheng CH, Kuchta RD. DNA polymerase epsilon: aphidicolin inhibition and the relationship between polymerase and exonuclease activity. Biochemistry. 1993;32:8568–8574. doi: 10.1021/bi00084a025. [DOI] [PubMed] [Google Scholar]
- 60.Doublie S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 61.Arana ME, Seki M, Wood RD, Rogozin IB, Kunkel TA. Low-fidelity DNA synthesis by human DNA polymerase θ. Nucleic Acids Res. 2008;36:3847–3856. doi: 10.1093/nar/gkn310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goodman MF. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu Rev Biochem. 2002;71:17–50. doi: 10.1146/annurev.biochem.71.083101.124707. [DOI] [PubMed] [Google Scholar]
- 63.Maga G, Shevelev I, Ramadan K, Spadari S, Hubscher U. DNA polymerase θ purified from human cells is a high-fidelity enzyme. J Mol Biol. 2002;319:359–369. doi: 10.1016/S0022-2836(02)00325-X. [DOI] [PubMed] [Google Scholar]
- 64.Hogg M, Sauer-Eriksson AE, Johansson E. Promiscuous DNA synthesis by human DNA polymerase θ. Nucleic Acids Res. 2012;40:2611–2622. doi: 10.1093/nar/gkr1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Seki M, Wood RD. DNA polymerase θ (POLQ) can extend from mismatches and from bases opposite a (6-4) photoproduct. DNA Repair (Amst) 2008;7:119–127. doi: 10.1016/j.dnarep.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kent T, Chandramouly G, McDevitt SM, Ozdemir AY, Pomerantz RT. Mechanism of microhomology-mediated end-joining promoted by human DNA polymerase θ. Nat Struct Mol Biol. 2015;22:230–237. doi: 10.1038/nsmb.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hogg M, Wallace SS, Doublie S. Crystallographic snapshots of a replicative DNA polymerase encountering an abasic site. EMBO J. 2004;23:1483–1493. doi: 10.1038/sj.emboj.7600150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hogg M, Wallace SS, Doublie S. Bumps in the road: how replicative DNA polymerases see DNA damage. Curr Opin Struct Biol. 2005;15:86–93. doi: 10.1016/j.sbi.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 69.Kusumoto R, Masutani C, Iwai S, Hanaoka F. Translesion synthesis by human DNA polymerase eta across thymine glycol lesions. Biochemistry. 2002;41:6090–6099. doi: 10.1021/bi025549k. [DOI] [PubMed] [Google Scholar]
- 70.Takata KI, Shimizu T, Iwai S, Wood RD. Human DNA polymerase N (POLN) is a low-fidelity enzyme capable of error-free bypass of 5S-thymine glycol. J Biol Chem. 2006;281:23445–23455. doi: 10.1074/jbc.M604317200. [DOI] [PubMed] [Google Scholar]
- 71.Vaisman A, Lehmann AR, Woodgate R. DNA polymerases eta and iota. Adv Protein Chem. 2004;69:205–228. doi: 10.1016/S0065-3233(04)69007-3. [DOI] [PubMed] [Google Scholar]
- 72.Martomo SA, Saribasak H, Yokoi M, Hanaoka F, Gearhart PJ. Reevaluation of the role of DNA polymerase θ in somatic hypermutation of immunoglobulin genes. DNA Repair (Amst) 2008;7:1603–1608. doi: 10.1016/j.dnarep.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Minnick DT, Astatke M, Joyce CM, Kunkel TA. A thumb subdomain mutant of the large fragment of Escherichia coli DNA polymerase I with reduced DNA binding affinity, processivity, and frameshift fidelity. J Biol Chem. 1996;271:24954–24961. doi: 10.1074/jbc.271.40.24954. [DOI] [PubMed] [Google Scholar]
- 74.Cannistraro VJ, Taylor JS. DNA-thumb interactions and processivity of T7 DNA polymerase in comparison to yeast polymerase eta. J Biol Chem. 2004;279:18288–18295. doi: 10.1074/jbc.M400282200. [DOI] [PubMed] [Google Scholar]
- 75.Marini F, Kim N, Schuffert A, Wood RD. POLN, a nuclear PolA family DNA polymerase homologous to the DNA cross-link sensitivity protein Mus308. J Biol Chem. 2003;278:32014–32019. doi: 10.1074/jbc.M305646200. [DOI] [PubMed] [Google Scholar]
- 76.Takata K, Arana ME, Seki M, Kunkel TA, Wood RD. Evolutionary conservation of residues in vertebrate DNA polymerase N conferring low fidelity and bypass activity. Nucleic Acids Res. 2010;38:3233–3244. doi: 10.1093/nar/gkq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee YS, Gao Y, Yang W. How a homolog of high-fidelity replicases conducts mutagenic DNA synthesis. Nat Struct Mol Biol. 2015;22:298–303. doi: 10.1038/nsmb.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Newman JA, Cooper CD, Aitkenhead H, Gileadi O. Structure of the Helicase Domain of DNA Polymerase Theta Reveals a Possible Role in the Microhomology-Mediated End-Joining Pathway. Structure. 2015;23:2319–2330. doi: 10.1016/j.str.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Buttner K, Nehring S, Hopfner KP. Structural basis for DNA duplex separation by a superfamily-2 helicase. Nat Struct Mol Biol. 2007;14:647–652. doi: 10.1038/nsmb1246. [DOI] [PubMed] [Google Scholar]
- 80.Kawamura K, Bahar R, Seimiya M, Chiyo M, Wada A, Okada S, Hatano M, Tokuhisa T, Kimura H, Watanabe S, Honda I, Sakiyama S, Tagawa M, O-Wang J. DNA polymerase θ is preferentially expressed in lymphoid tissues and upregulated in human cancers. Int J Cancer. 2004;109:9–16. doi: 10.1002/ijc.11666. [DOI] [PubMed] [Google Scholar]
- 81.Pillaire MJ, Selves J, Gordien K, Gourraud PA, Gentil C, Danjoux M, Do C, Negre V, Bieth A, Guimbaud R, Trouche D, Pasero P, Mechali M, Hoffmann JS, Cazaux C. A 'DNA replication' signature of progression and negative outcome in colorectal cancer. Oncogene. 2010;29:876–887. doi: 10.1038/onc.2009.378. [DOI] [PubMed] [Google Scholar]
- 82.Lemée F, Bergoglio V, Fernandez-Vidal A, Machado-Silva A, Pillaire M-J, Bieth A, Gentil C, Baker L, Martin A-L, Leduc C, Lam E, Magdeleine E, Filleron T, Oumouhou N, Jordan L, Kaina B, Seki M, Grimal F, Lacroix-Triki M, Thompson A, Roche H, Bourdon J-C, Wood RD, Hoffmann J-S, Cazaux C. POLQ up-regulation is associated with poor survival in breast cancer, perturbs DNA replication and promotes genetic instability. Proc Natl Acad Sci (USA) 2010;107:13390–13395. doi: 10.1073/pnas.0910759107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Higgins GS, Harris AL, Prevo R, Helleday T, McKenna WG, Buffa FM. Overexpression Of POLQ Confers a Poor Prognosis In Early Breast Cancer Patients. Oncotarget. 2010;1:175–184. doi: 10.18632/oncotarget.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lessa RC, Campos AH, Freitas CE, Silva FR, Kowalski LP, Carvalho AL, Vettore AL. Identification of upregulated genes in oral squamous cell carcinomas. Head Neck. 2013;35:1475–1481. doi: 10.1002/hed.23169. [DOI] [PubMed] [Google Scholar]
- 85.Allera-Moreau C, Rouquette I, Lepage B, Oumouhou N, Walschaerts M, Leconte E, Schilling V, Gordien K, Brouchet L, Delisle MB, Mazieres J, Hoffmann JS, Pasero P, Cazaux C. DNA replication stress response involving PLK1, CDC6, POLQ, RAD51 and CLASPIN upregulation prognoses the outcome of early/mid-stage non-small cell lung cancer patients. Oncogenesis. 2012;1:e30. doi: 10.1038/oncsis.2012.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, Talantov D, Timmermans M, Meijer-van Gelder ME, Yu J, Jatkoe T, Berns EM, Atkins D, Foekens JA. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 87.Sotiriou C, Wirapati P, Loi S, Harris A, Fox S, Smeds J, Nordgren H, Farmer P, Praz V, Haibe-Kains B, Desmedt C, Larsimont D, Cardoso F, Peterse H, Nuyten D, Buyse M, Van de Vijver MJ, Bergh J, Piccart M, Delorenzi M. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98:262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 88.Lauss M, Kriegner A, Vierlinger K, Visne I, Yildiz A, Dilaveroglu E, Noehammer C. Consensus genes of the literature to predict breast cancer recurrence. Breast Cancer Res Treat. 2008;110:235–244. doi: 10.1007/s10549-007-9716-3. [DOI] [PubMed] [Google Scholar]
- 89.de Lavallade H, Finetti P, Carbuccia N, Khorashad JS, Charbonnier A, Foroni L, Apperley JF, Vey N, Bertucci F, Birnbaum D, Mozziconacci MJ. A gene expression signature of primary resistance to imatinib in chronic myeloid leukemia. Leuk Res. 2010;34:254–257. doi: 10.1016/j.leukres.2009.09.026. [DOI] [PubMed] [Google Scholar]
- 90.Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, Quackenbush JF, Stijleman IJ, Palazzo J, Marron JS, Nobel AB, Mardis E, Nielsen TO, Ellis MJ, Perou CM, Bernard PS. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Prat A, Parker JS, Fan C, Perou CM. PAM50 assay and the three-gene model for identifying the major and clinically relevant molecular subtypes of breast cancer. Breast Cancer Res Treat. 2012;135:301–306. doi: 10.1007/s10549-012-2143-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Clarkson SG, Wood RD. Polymorphisms in the human XPD (ERCC2) gene, DNA repair capacity and cancer susceptibility: An appraisal. DNA Repair (Amst) 2005;4:1068–1074. doi: 10.1016/j.dnarep.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 93.Brandalize APC, Schuler-Faccini L, Hoffmann JS, Caleffi M, Cazaux C, Ashton-Prolla P. A DNA repair variant in POLQ (c.-1060A >G) is associated to hereditary breast cancer patients: a case-control study. BMC Cancer. 2014;14:850. doi: 10.1186/1471-2407-14-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li WQ, Hu N, Hyland PL, Gao Y, Wang ZM, Yu K, Su H, Wang CY, Wang LM, Chanock SJ, Burdett L, Ding T, Qiao YL, Fan JH, Wang Y, Xu Y, Shi JX, Gu F, Wheeler W, Xiong XQ, Giffen C, Tucker MA, Dawsey SM, Freedman ND, Abnet CC, Goldstein AM, Taylor PR. Genetic variants in DNA repair pathway genes and risk of esophageal squamous cell carcinoma and gastric adenocarcinoma in a Chinese population. Carcinogenesis. 2013;34:1536–1542. doi: 10.1093/carcin/bgt094. [DOI] [PMC free article] [PubMed] [Google Scholar]