Abstract
For effective metabolic engineering, a toolbox of genetic components that enables predictable control of gene expression is needed. Here we present a systematic study of promoters and ribosome binding sites in the unicellular cyanobacterium Synechocystis sp. PCC 6803. A set of metal ion inducible promoters from Synechocystis were compared to commonly used constitutive promoters, by measuring fluorescence of a reporter protein in a standardized setting to allow for accurate comparisons of promoter activity. The most versatile and useful promoter was found to be PnrsB, which from a relatively silent expression could be induced almost 40-fold, nearly up to the activity of the strong psbA2 promoter. By varying the concentrations of the two metal ion inducers Ni2+ and Co2+, expression from the promoter was highly tunable, results that were reproduced with PnrsB driving ethanol production. The activities of several ribosomal binding sites were also measured, and tested in parallel in Synechocystis and Escherichia coli. The results of the study add useful information to the Synechocystis genetic toolbox for biotechnological applications.
Cyanobacteria have gained increasing attention as sustainable converters of CO2 and H2O into valuable products using solar energy1. However, engineering of cyanobacteria for production of such compounds often requires the introduction and expression of multiple genes in a well-controlled manner. This, in turn, requires well characterized and tightly regulated promoters. Having well controlled promoters is also important in cases where an engineered pathway is producing a toxic intermediate or product, which may lead to genetic instability and loss of expression if the pathway is constitutively expressed.
There are many different promoters to choose from for use in the unicellular cyanobacterium Synechocystis sp. PCC 6803 (from here on referred to as Synechocystis). Strong native promoters such as the psbA2 and rbcL promoters, the recently described “super strong” Pcpc5602 or the J23-series of promoters3 can all be useful when an “always on” constitutive expression of genes is possible or desired. However, for applications such as induced cell lysis, use of the CRISPR-Cas9 system or stable integration and expression of genes with a high metabolic burden, inducible promoters are required. Currently, there is a shortage of useful, well characterized inducible promoters for Synechocystis. Many inducible systems commonly used in Escherichia coli do not function as well in Synechocystis due to differences in cellular properties or growth requirements. Examples include the case of LacI not completely releasing the repressing of Ptrc or Ptac promoters, preventing high induction levels4,5 and problems when using the light sensitive anhydrotetracycline to induce expression from the Tet-promoter during phototrophic growth6. The green light inducible promoter PcpcG2 system described in Abe et al.7 could be useful for specialized growth conditions, but will be inherently difficult to work with during growth under normal white light.
An alternative set of regulated promoters are metal ion inducible promoters that have already been used for several applications8,9,10,11,12,13. In Synechocystis, a set of genes responsible for Ni2+, Co2+, and Zn2+ tolerance are all grouped together in a gene cluster14. The nrsBACD operon encodes a set of transporters which provide protection against Ni2+, and is induced by both Ni2+ and Co2+. Directly upstream of nrsBACD is the nrsRS operon, encoding a two-component signal transduction system where NrsS senses Ni2+ ions and phosphorylates NrsR which activates the transcription of nrsBACD and nrsRS15. nrsD is also transcribed from a promoter between nrsC and nrsD, which is repressed by InrS in the absence of Ni2+ 16.
The gene responsible for Co2+ resistance is coaT, encoding a cobalt exporting ATPase, the expression of which is induced by both Co2+ and Zn2+ ions14. In the presence of Co2+, it is transcriptionally activated by CoaR, encoded upstream of coaT17. In the absence of Co2+, CoaR acts as a repressor of coaT. Zn2+ resistance is mediated by ZiaA which is repressed by ZiaR in the absence of Zn2+. ziaR is transcribed upstream of ziaA together with sll0793, which codes for a membrane bound protein likely affiliated with Zn2+ transport18.
A frequent biotechnological application for cyanobacteria is metabolic engineering for production of a desired compound. In those utilizations, the metabolic burden placed on the cell, in the amount of resources needed to make the product and the heterologous enzymes, as well as the depletion of substrate for other pathways, can lead to severe growth reduction and genetic instability19,20. Examples include the production of mannitol where mutants commonly reverted to the wt phenotype21, the production of isobutanol where one of the enzymes commonly reduced its function22 and the production of lactic acid where an NADH:NADPH transhydrogenase could not be kept stable in the wt23. Partly because of the issue with genetic instability, metabolic studies have utilized inducible promoters. In cyanobacterial strains which have a functional induction of Ptrc and Plac such as Synechococcus elongatus PCC 7942, those are commonly used24. For Synechocystis production, PnirA have been used for production of carotenoids25, PnrsB for manoyl oxide production12 and PcoaT, PpetE, and a version of the Lac promoter were used for ethylene production13.
For biotechnological applications, promoters should be easy to use in genetic constructs, have a low leakiness, a strong induction and allow for fine-tuning the levels between those extremes. The aim of this work was to identify and characterize practical promoters and a set of ribosomal binding sites (RBS) for biotechnological purposes in Synechocystis, and to do so in a systematic way, using standardized conditions and genetic setting to allow for careful analysis and comparisons. We did not aim to investigate the functional elements or initiation dynamics of each promoter. Earlier studies have examined the transcriptional activation of some of the metal inducible promoters26,27. Here we expand on that work by providing systematic, useful data quantitatively comparing these promoters to each other and to some commonly used promoters, with fluorescent protein levels as well as formation of a model product, ethanol, as reporter.
Results and Discussion
Choice and cloning of promoters
For this study, we chose to characterize the promoters PnrsB, PnrsD, PnrsS, PcoaT and PziaA, all of which drive the expression of nickel, cobalt and zinc metal efflux pumps in the same gene cluster, and the Cu2+ inducible plastocyanin promoter PpetE which has been frequently used for expression in Synechocystis28. For comparison, we also included a selected set of other native promoters from Synechocystis: the strong promoter driving expression of the D1 subunit of Photosystem II; PpsbA2, in three versions differing by sequence length (PpsbA2S, PpsbA2M, and PpsbA2L, see Supplementary Fig. 1 for sequence details), the photosystem I subunit Ia promoter PpsaA29, the RubisCO large subunit promoter PrbcL1A4, and RNase P subunit B promoter PrnpB. All the promoters were PCR amplified from Synechocystis genomic DNA and cloned into the self-replicating vector pPMQAK14 in front of the strong ribosomal binding site RBS*28, followed by the gene encoding Enhanced yellow fluorescent protein (EYFP)30, and the terminator BBa_001531. We used an RBS calculator (https://salislab.net/software/forward)32 to predict if the presence of the native RBS on the promoters sequences would start translation prematurely, instead of at the synthetic RBS we added directly upstream of EYFP. For the promoter sequences where the native RBS were predicted to still be functional, 9 base pairs were truncated from the 3′ end of those sequences. For PrnpB, the native promoter of an RNA gene which does not have an RBS, we used the entire promoter sequence up to the start of the gene, and an RBS was added. The complete sequence of all promoters used in this study can be found in Supplementary Table 1.
Determination of fluorescent reporter levels from promoter constructs
Promoter constructs were transferred into Synechocystis by conjugation and an empty vector control was included. The concentrations of metal ions used to induce the promoters were chosen based on a balance between having high enough concentration to get a high induction and low enough to not stress the cells. For Synechocystis, the half growth-inhibitory concentration (IC50) of Ni2+ was previously reported to be 27 μM, 8 μM for Co2+, between 8 and16 μM for Zn2+, and 2 μM for Cu2+ 27. Our initial designation of standard metal inducer concentration was 5 μM for Ni2+ and 4 μM for Zn2+, as per the recommendations of Blasi et al.27. The Co2+ concentration used was 6 μM, and 0.5 μM for Cu2+, as in Guerrero et al.13. The BG11 medium used to cultivate Synechocystis contains trace amounts of some of the metal ions used to induce expression33. There is no Ni2+ added in BG11, but it does contains 0.17 μM Co2+, 0.77 μM Zn2+, and 0.32 μM Cu2+, which potentially could induce the expression of some of the promoters. However, the purpose of our study was to investigate how these promoters can be used in a biotechnological application, where an absence of certain metals could affect the function of proteins responsible for vital processes in the cell, and a strict requirement for keeping the growth medium free from some metals would be impractical and likely not economically feasible. Therefore, all experiments were done using complete BG11, including the trace metals, except in certain control experiments as indicated below.
Synechocystis cells carrying the respective promoter constructs were used to inoculate fresh cultures, which were grown for two days before induction of expression by addition of the various metal ions. The cells were cultivated for an additional two days, and then fluorescence from expressed EYFP was determined (Fig. 1a).
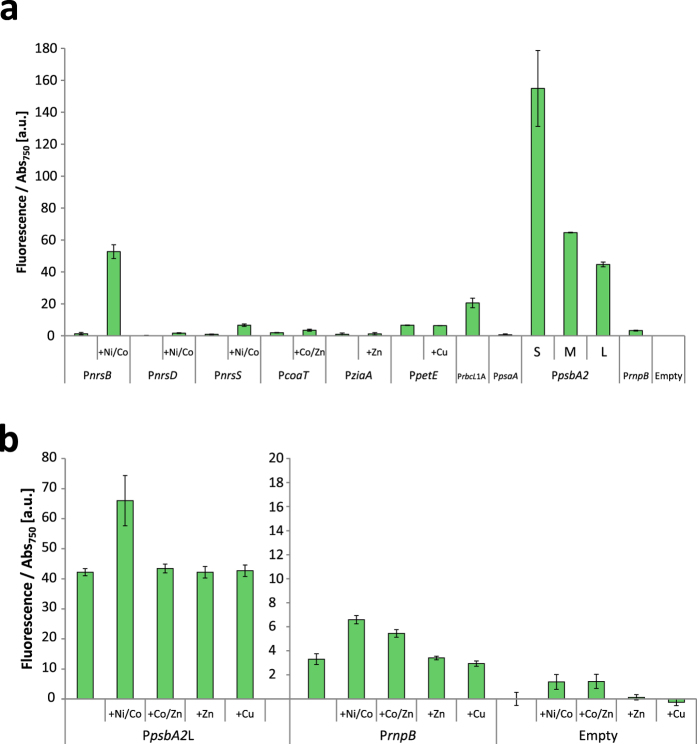
Figure 1. Promoter activities.
(a) Activity of promoters in Synechocystis, as measured by EYFP fluorescence. (b) Effect of metal induction on fluorescence for control strains. Two y-axis scales are used to better visualize low expressed samples. Promoter activities were measured as EYFP fluorescence per Abs750 two days after induction by addition of metal ions. The fluorescence of the un-induced empty vector control strain was subtracted from each sample. Metal ion concentrations used for induction were 5 μM Ni2+, 6 μM Co2+, 4 μM Zn2+ and 0.5 μM Cu2+. Error bars represent SD (n = 4).
The Ni2+ and Co2+ induced promoter PnrsB showed a low leakiness and a high induction rate, going from half of the levels of the reference promoter PrnpB to a 39-fold increase of expression levels. The other two promoters from the nickel tolerance system, PnrsD and PnrsS, were induced fourteen and seven times, respectively, but had a lower maximal expression than PnrsB. The Co2+ induced promoter PcoaT showed a low expression and low induction by its inducer metals Co2+ and Zn2+, while the Zn2+ dependent promoter PziaA had very weak activity both in the presence and absence of added Zn2+. The petE promoter showed no change in expression upon addition of Cu2+, likely due to it already being fully induced by the 0.32 μM Cu2+ that is present in BG11.
For the strong photosystem II promoter PpsbA2, the length of native sequence included in the promoter had a large effect on resulting expression levels, with the short version having more than three times higher expression than the long one. A difference between the promoters is that PpsbA2S lacks the beginning of an antisense RNA which has a role in stabilizing the psbA2 mRNA34. However, due to the stabilization effect of the antisense RNA, one would expect higher expression with the full sequence intact, and that would also require a promoter sequence within the EYFP gene to promote the transcription of the antisense RNA, which to our knowledge there is not. Another study identified repressor binding sites in PpsbA2, deletion of which resulted in increased expression of the promoter35. All of those binding sites reported in that study are still present on the three versions of PpsbA2 used in this study, but our results suggest a possibility of additional regulatory sites even further upstream.
Although the PpsbA2 is one of the strongest native promoters, expressing one of the most abundant gene transcript in the cell36, and being commonly used for heterologous chemical production37,38,39, it is still weaker than the synthetic Ptrc promoter. That is important to consider when choosing a promoter since the limitation for production is commonly enzyme availability, something that has been seen for several products, including lactic acid40,41, ethanol42, β phellandrene43, and ethylene44. By normalizing the PpsbA2 expression observed here to PrnpB and comparing with another study where Ptrc and PrnpB were measured6, the inferred differences in expression are 1.8, 4.2 and 6.1 times weaker for PpsbA2S, PpsbA2M and PpsbA2L respectively compared to Ptrc1O.
Curiously, even though the photosystem I promoter PpsaA has been widely used for transgenic expression in Synechocystis, we could only detect a minimal expression from this promoter in our construct. We used the short A4 sequence version of PpsaA from Muramatsu and Hihara29. A possible cause for the lack of expression is that the combination of that promoter with the EYFP and RBS sequences used created an unfavorable 5′UTR, preventing efficient expression45.
Metal-ion effect on fluorescence signal in control strains
As a control experiment, we wanted to investigate whether there is an effect of addition of metals on accumulation of fluorescent protein, even when expressed from the native constitutive promoters. For these experiments, we chose PpsbA2L and PrnpB as reference promoters. PpsbA2L was picked for its similar strength to the induced levels of PnrsB, and PrnpB because it is considered to be constitutive under standard cultivation conditions46. The addition of metals to the reference strains increased fluorescence for some of the metal combinations, up to twice the levels of the non-metal induced cultures, but much lower than the effect on fluorescence from the PnrsB construct induced with Ni2+ (Fig. 1b). To our knowledge, neither PpsbA2 nor PrnpB are directly metal inducible, and even the empty vector strain that did not contain the EYFP gene slightly increased in fluorescence, indicating that the effect was unspecific.
It is known that the fluorescent signal will increase if the cells grow slower due to slower dilution of EYFP proteins, which is the main mechanism of EYFP removal47. This effect could be somewhat diminished by addition of a protein degradation tag on EYFP, targeting it for continuous degradation4. However, this option was not explored in this paper, since our aim was primarily to study the usefulness of the investigated promoters for expression of proteins, not the dynamics of expression. Instead, due to this unspecific increase in fluorescence, we decided that for all further comparisons we would treat control promoters and metal induced promoters with the same induction conditions, so as to not over-estimate the specific effect of induction.
Expression at different inducer concentrations
To test the range of induction of PnrsB, PcoaT and PpetE, and to see how much each inducer contributed to the promoter activity, the cells were subjected to a range of concentrations of their inducer metal ions. In this experiment, the BG11 medium used was modified to not include the inducing metal ion(s). Starting from Co2+-free BG11, there was an increase of the fluorescence in cultures with the PnrsB-driven EYFP construct with increasing concentrations of Ni2+ which plateaued at 5 μM (Fig. 2a). Addition of both inducers, Ni2+ and Co2+, gave a response similar to when only Ni2+ was used, and induction with only Co2+ gave 3.5 times lower expression than with 5 μM Ni2+. Both for PcoaT and PpetE, using Co2+ or Cu2+ higher than the initial level of induction did not lead to higher expression, but removal of the inducer metals from BG11 cut the expression in half, compared to complete BG11.
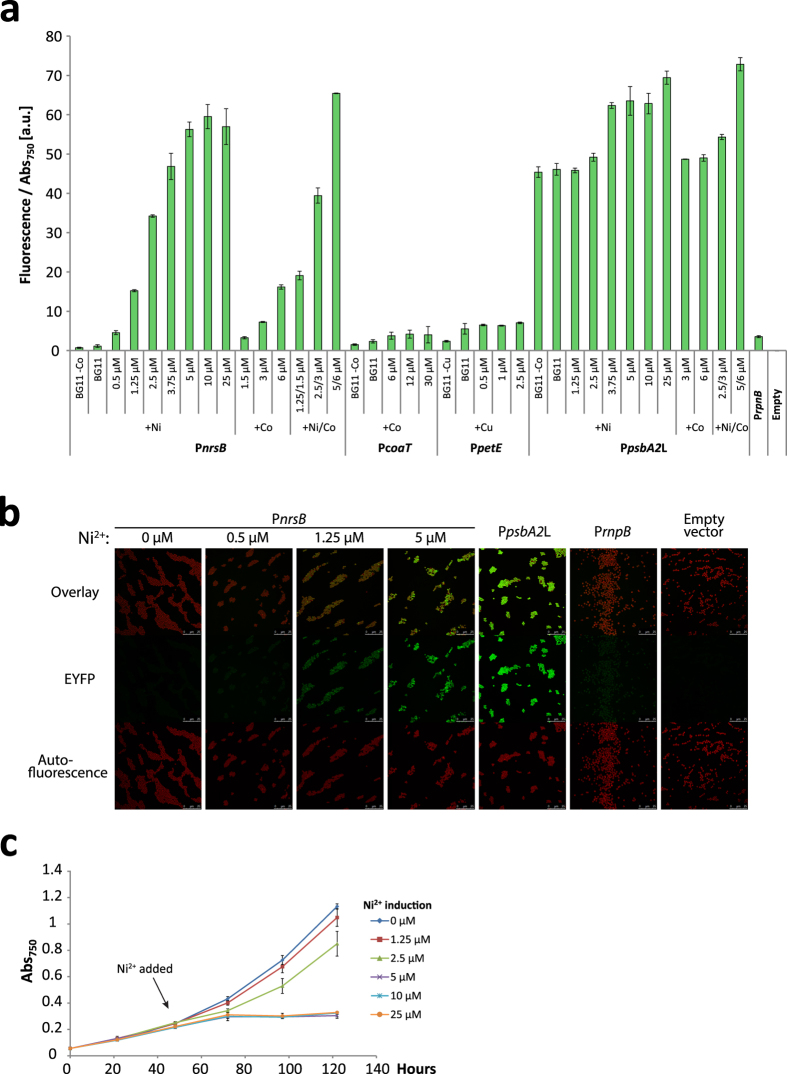
Figure 2. Metal ion induction effects on promoter activity and cell growth.
(a) Promoter activities measured as EYFP fluorescence per Abs750 two days after induction. The fluorescence of the empty vector control strain was subtracted from each sample. PrnpB and the empty vector strain were grown in BG11. Error bars represent SD (n = 4). (b) Fluorescent microscopy image of cells expressing EYFP driven by PnrsB at different Ni2+ induction strengths, PpsbA2L, PrnpB or the empty vector. Bottom panels: chlorophyll autofluorescence, middle panels: EYFP fluorescence, top panel: overlay of EYFP and chlorophyll signals. (c) Growth of wild type Synechocystis with different amounts of Ni2+. Cells were grown at 12 μmol photons m−2 s−1 and Ni2+ was added after 48 h. Error bars represent SD (n = 3).
To determine if the lower expression of PnrsB at reduced Ni2+ induction concentrations was due to a lower expression level in every cell in the culture or a binary reduction where the culture becomes a mixture of on and off cells, laser scanning confocal microscopy was used. Cells with the PnrsB promoter at induction concentrations 0 μM, 0.5 μM, 1.25 μM and 5 μM showed a clear and continuous increase in EYFP fluorescence, spread out across all cells (Fig. 2b). This indicates that at lower inducer concentrations, less transcription occurs at the cell level, and not only on the population level.
As described above, we based our initial selection of inducer concentration on literature data. To see at which concentration nickel had detrimental effects on growth in our cultures, wild type Synechocystis cells were grown for 48 h and then subjected to different amounts of Ni2+. Already at 5 μM, there was a large reduction of growth while at 2.5 μM, the effect on growth was less severe (Fig. 2c). Due to the relatively high induction of PnrsB (Fig. 2a) and low toxicity of Ni2+ at 2.5 μM, we decided to use that as the standard concentration for the following experiments.
Measurement of promoter activity over time
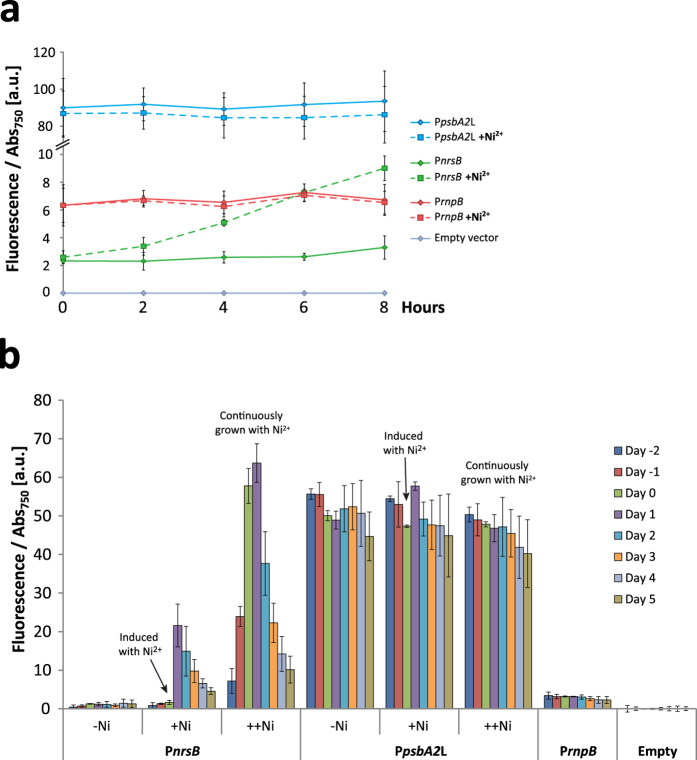
Unlike most inducible promoter systems, use of metal inducible promoters has an additional complication in that the inducer molecule is being actively pumped out of the cells. Therefore, the concentration of metal ions inside the cells might change as more efflux pumps are induced, even though the metal ion concentration in the medium remains constant. To determine how the induction of PnrsB changes over time, fluorescence was measured over the course of a day and the course of a week. When the response of PnrsB to Ni2+ was measured over eight hours, the induction had led to a detectable increased expression already after 2 hours (Fig. 3a). To see how expression varied over a longer time period, the EYFP accumulation across eight days of growth was assayed in strains growing without induction, with induction on the second day, and with Ni2+ continuously present in the medium (Fig. 3b). In the PnrsB strain, the highest amount of fluorescence was observed when reaching the early stationary phase (~OD750 1.3, “Day 1” in Fig. 3b) after which the fluorescence went rapidly down, something not seen in the PpsbA2 strain. This peak in fluorescence in the PnrsB strain could possibly be caused by a lowered concentration of Ni2+ transporters in each cell due to cell division during the rapid growth occurring the days before the peak, increasing the intracellular Ni2+ concentration. It was also seen that continuous growth with nickel gave a higher maximal induction than when induced after 2 days.
Figure 3. Promoter driven EYFP signal change over time.
Promoter activity measured as EYFP fluorescence per Abs750. (a) All strains were induced with 2.5 μM at 0 hours and measured every second hour. (b) The fluorescence of strains with and without induction was measured for eight days. The PnrsB and PpsbA2L strains were either grown in BG11 (−Ni), with addition of 2.5 μM Ni2+ at day 0 (+Ni) or grown continuously with 2.5 μM Ni2+ (++Ni). PrnpB and the empty vector strain were grown in BG11 without Ni2+. The fluorescence of the empty vector control strain was subtracted from each sample for each day or hour. Error bars represent SD (n = 4).
One way to prevent the export of the inducer would be to integrate the genes to be expressed in the nrsBACD operon itself, thereby have them be expressed by the native PnrsB while at the same time knocking out the nickel export system. An nrsBACD knock out strain would have a reduced tolerance to Ni2+ 14, but might also allow for induction at lower metal concentrations. However, this option was not explored within the scope of this paper.
Ethanol production with PnrsB
To evaluate the potential usefulness and capacity of the investigated promoters for actual biotechnology applications, and to get a measure of the total promoter expression output across several days, we made a new construct with PnrsB driving the expression of genes for ethanol production48. Expression patterns from reporter molecules and metabolic products can differ, hence the need to measure PnrsB activity using a product based reporter system49. Since ethanol readily diffuses across the cell membrane and accumulates in the media, we can use ethanol accumulation as a reporter to compare the effect of different promoters for the expression. Pyruvate decarboxylase (pdc) from Zymomonas mobilis and the native Synechocystis alcohol dehydrogenase (slr1192)42 were cloned in a new PnrsB-driven construct, which was introduced into Synechocystis. The resulting strain was grown with different concentrations of Ni2+, and ethanol accumulation and growth were measured. Over the course of 8 days, the highest ethanol accumulation in the medium reached 65 mg/L (Fig. 4a).
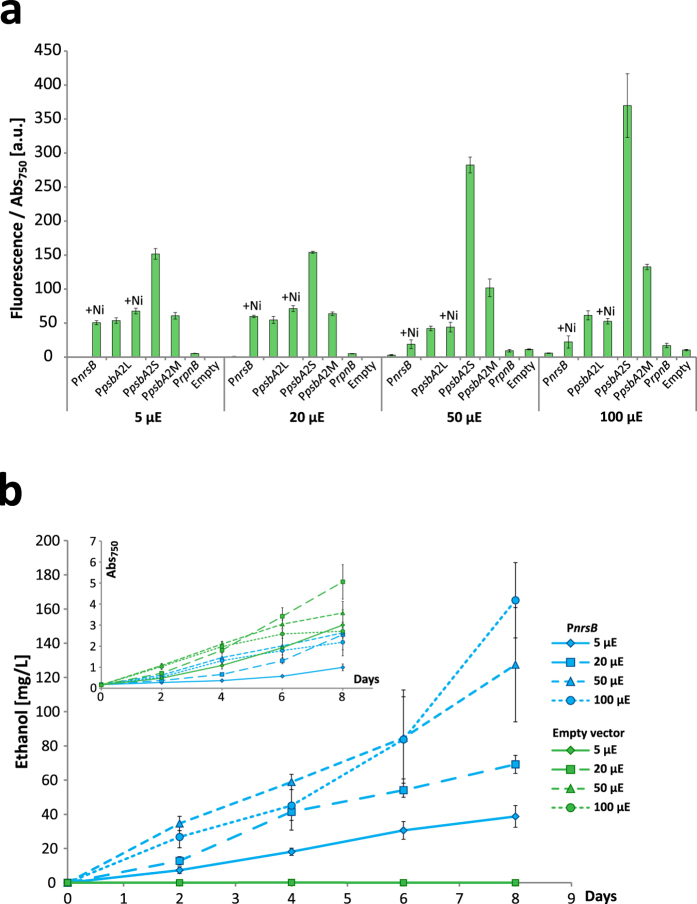
Figure 4. Ethanol production and growth with varying amount of Ni2+ induction.
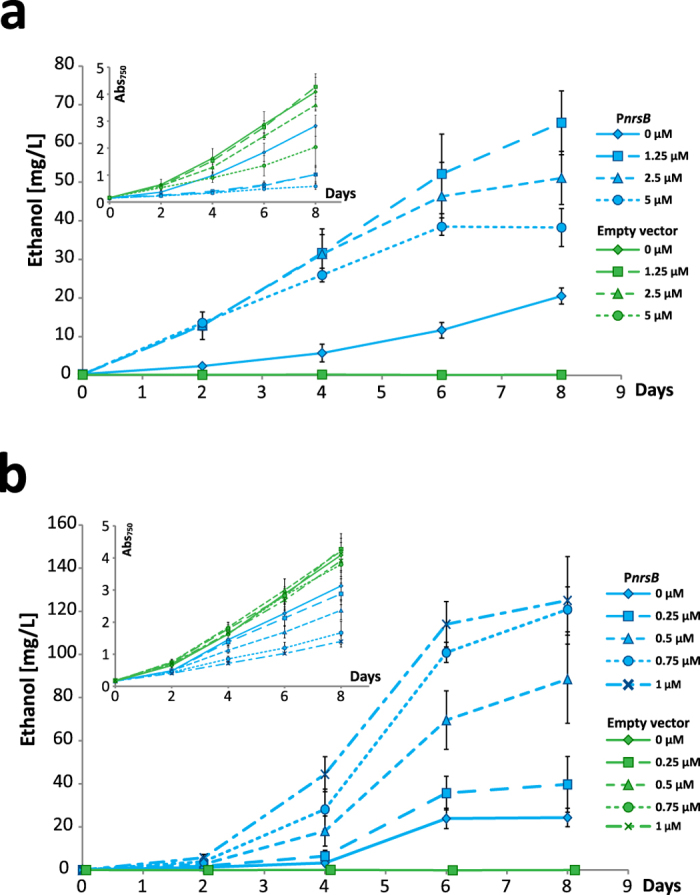
A strain with PnrsB driven expression of pdc and slr1192 and the empty vector strain were induced with a range of Ni2+ concentrations, from (a) 0–5 μM Ni2+ or (b) 0–1 μM. Error bars represent SD (n = 4).
The cultures grown with 1.25 μM Ni2+ had the highest ethanol production, which was somewhat surprising, since the EYFP data suggested that higher induction than that would increase expression further, and in a previous study, it was shown that ethanol production could be increased several times upon gene duplication of slr1192 and pdc, effectively increasing the expression levels of the enzymes42. However, a clear decrease in growth could be observed in the ethanol producing strain upon addition of Ni2+, an effect not seen in the empty vector control. When measuring the ethanol at lower induction levels, from 0–1 μM Ni2+, the production increased with increasing amounts of Ni2+ while the growth rate decreased (Fig. 4b). It may be that at higher concentrations of the inducer, the combined stresses of Ni2+ exposure and ethanol production counteract any increase in product formation which could result from enhanced protein expression levels per cell. At lower levels of induction, the effects of Ni2+ are not as severe, with higher yield as a result.
Attempts to make an ethanol producing strain of Synechocystis where the expression was driven by PpsbA2, in order to compare it with the PnrsB driven ethanol production, were unsuccessful. When tested in E. coli, the PpsbA2-driven construct was leading to ethanol production, but in Synechocystis, the ethanol producing capability with this construct was rapidly lost (data not shown). We were unable to get a stable ethanol producing strain for any of the three PpsbA2 variants, in agreement with a previous study50, while the PnrsB strain lost its production after too long exposure to Ni2+. This effect may likely be attributed to genetic instability of the ethanol production construct: in experiments where expression levels were higher, a larger proportion of cells in the culture lost their ability to generate ethanol, due to selective pressure on the cells. This has been reported previously as a problem for continuous ethanol production51.
The genetic instability of ethanol production highlights the importance of inducible promoters for genetically stable production strains. For complex multi-step pathway products, PnrsB could serve as a gatekeeper to prevent genetic instability, controlling expression of the first enzymes in the pathway while a strong constitutive promoter expresses the rest, thereby only completing the full pathway when Ni2+ is added.
Effect of light on PnrsB activity
In a previous study where we used PnrsB for the production of the high value compound manoyl oxide, we observed a decrease in production at high light, something we speculated could be due to reduced function of PnrsB at higher light12. In order to investigate this effect, we performed experiments to determine the influence of light intensity on EYFP expression as well as ethanol production. For PnrsB-driven EYFP expression, increasing the light from 20 μE to 50 μE did reduce the induced fluorescence with more than half, with similar reduction at 100 μE (Fig. 5a). The short and medium length sequence of PpsbA2 increased EYFP expression as the light increased, consistent with literature52. For the longer version of the PpsbA2 promoter, expression did not increase with higher light intensity, for reasons as of yet unknown. When PnrsB was driving the expression of ethanol producing genes, induced with 2.5 μM Ni2+, cultures growing at 50 and 100 μE accumulated more ethanol after eight days than cultures grown at 20 μE, even if normalized per biomass (Fig. 5b). This inconsistence with the fluorescence data may be due to increased substrate availability for ethanol production under higher light conditions, leading to more product being formed even if the enzyme expression levels are lower than at lower light.
Figure 5. Effect of light intensity on PnrsB activity.
(a) Promoter activity measured as EYFP fluorescence per Abs750. Cells were grown at different light intensities and fluorescence was measured two days after induction with 2.5 μM Ni2+. The fluorescence of the empty vector control strain at 5 μE was subtracted from each sample. (b) A strain with PnrsB driven expression of pdc and slr1192 and the empty vector strain were grown at different light intensities, and ethanol and growth was measured. μE = μmol photons m−2 s−1. Error bars represent SD (n = 4).
Promoter activity in E. coli
For cloning purposes, it can be beneficial to have the promoter turned off in the cloning host, thus preventing selection pressure to mutate detrimental genes. Therefore, we investigated the Synechocsytis promoters tested in this paper for activity in E. coli. The promoters J23101, J23110 and J23119 from the Registry of standard biological parts31 were used as references for the strength of expression. None of the Synechocystis promoters had any discernable activity except PpsbA2 (Fig. 6). The transcriptional machinery of Synechocystis has several differences compared to the one in E. coli, e.g. a split β´ subunit in the RNA polymerase and a different sigma factor composition53, explaining the large differences in expression pattern between the two strains. Of the PpsbA2 strains, the long version had the strongest expression in E. coli and the medium length the lowest, a third of the long one. In contrast, expression form PpsbA2 in Synechocystis was strongest with the short version while the long version had the weakest expression (Fig. 1a).
Figure 6. Promoter activities in E. coli.
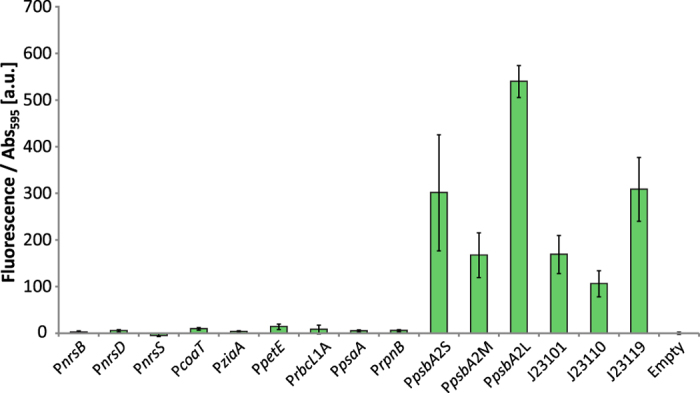
Promoter activities measured as fluorescence per Abs595. The fluorescence of the empty vector control strain was subtracted from each sample. Error bars represent SD (n = 6).
Characterization of ribosome binding sites
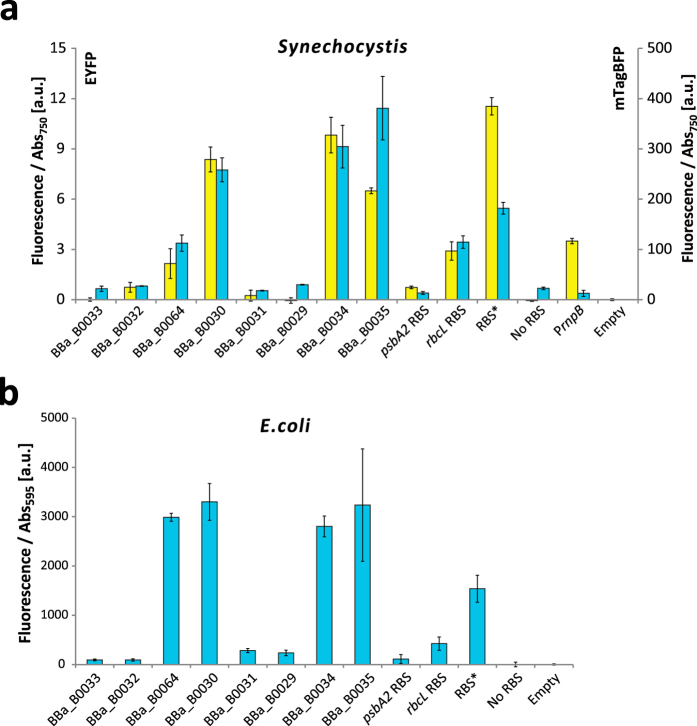
Other than promoters, the choice of ribosomal binding site can have a large impact on product formation in metabolic engineering studies44,49. Also, having a library of different strengths of RBSs available can be useful for constructing operons with variable strength of expression of each gene. Therefore, we selected 11 RBS sequences to characterize, eight from the BioBrick Registry of standard biological parts31, the two native RBSs upstream of the psbA2 and rbcL genes, and the synthetic RBS*28, and compared their expression in Synechocystis (see Supplementary Table 2 for sequence details). Because the activity of RBSs can vary highly depending on sequence context45, each RBS was measured using two constructs designed to have high sequence dissimilarities in the upstream and downstream sequences from the RBS, one expressing EYFP driven by PpetE, and the other mTagBFP driven by PpsbA2S54. The eight RBSs from the BioBrick registry had a large variation in expression strength and showed a similar relative activity for both fluorescent markers, indicating that the genetic context did not influence expression in major ways for the two sequences used to test the RBSs in this study (Fig. 7a), although it should be noted that the pattern might not hold for other sequences.The exceptions were BBa_B0035 and RBS*, which both changed in relative strengths, depending on if the RBSs where measured in the EYFP constructs or the mTagBFP constructs. Both BBa_B0029 and BBa_B0031 had weak expression even though they have been reported to have levels comparable to BBa_B0030 in E. coli31.
Figure 7. Comparative strengths of ribosomal binding sites.
(a) Activity of RBSs in Synechocystis, measured using EYFP (yellow bars) and mTagBFP (blue bars). The eight RBSs from the BioBrick library are sorted left to right with increasing strength based on values from the literature31. All EYFP constructs were expressed by PpetE and mTagBFP construct by PpsbA2S except the “PrnpB” strain which were expressing EYFP and mTagBFP using PrnpB and RBS*. (b) Activity of RBSs in E. coli using mTagBFP fluorescence. RBS activity was measured by fluorescence per Abs750 for Synechocystis or Abs595 for E. coli and the fluorescence of the empty vector control strain was subtracted from each sample. Error bars represent SD (n = 4 for Synechocystis, n = 6 for E. coli).
All mTagBFP expressing RBS-constructs were also tested in E. coli, to see whether RBSs have comparable relative strengths in both organisms. The results showed that the relative strengths of the RBSs to each other were similar when expressed in E. coli as in Synechocystis (Fig. 7b), consistent with the fact that the Synechocystis and E. coli ribosomes have the same core anti-Shine-Dalgarno sequence (5′-CCUCC-3′)55. This is in sharp contrast to the differences in promoter function, where nine out of ten Synechocystis promoters tested in this study were not functional in E. coli. This experimentally verified RBS study may prove useful for balancing gene expressions in operons, especially since predicted RBS strengths may not correlate well with the actual strengths56.
Conclusions
In this report, we investigated the activity of a set of native promoters and a library of RBSs in Synechocystis, and provide systematic, comparative data on their respective output levels and tuneability. Of the tested promoters, we found that the nickel inducible nrsB promoter was the most versatile with a strong, well-regulated expression that can be fine-tuned by varying the concentration of the inducer. We also demonstrated that these characteristics can be utilized for expression of a production pathway, by using PnrsB for inducible ethanol formation. Drawbacks in using PnrsB include a reduced expression at higher light, an efflux of the inducer which might lead to a variable intracellular concentration, and the toxicity of Ni2+ at higher concentrations. However, despite those shortcomings, we believe it is a viable choice for many applications that require inducible expression.
It is important to note that promoter and RBS activity can vary based on genetic context45, and thus, depending on the specific genes expressed, the expression strength may be different. Nevertheless, the characterization performed in this study provides useful information on the relative strength of the promoters and RBSs, and enhances our understanding of the induction pattern of the promoters we have examined here.
Methods
Vector construction
The promoter sequences from Supplementary Table 1 were amplified by PCR using Synechocystis genomic DNA as template with primers flanked by EcoRI and SpeI sites. Using 3A assembly57, promoter parts were ligated together with a BioBrick containing RBS*, EYFP and a terminator (BBa_B0015) and then ligated into the broad-host-range shuttle vector pPMQAK14. For PpsbA2M and PpsbA2S, the RBS was included on the primers and thus, no BioBrick scar was created between promoter and RBS. For the ethanol construct, pdc was amplified from Zymomonas mobilis, slr1192 and PnrsB from Synechocystis, and the parts ligated into pPMQAK1 using 3A assembly. Correctly assembled vectors were verified by sequencing. The vectors were transferred to Synechocystis by triparental mating as described previously58, using the pRL443 conjugative plasmid. Isolation of correct transformants was verified by PCR amplification of extracted DNA, to confirm presence of the desired constructs.
For the RBS constructs, PpetE or PpsbA2 were combined with the RBSs, EYFP or mTagBFP respectively, and terminator BBa_B0015 using 3A assembly into pPMQAK1. The “No RBS” constructs was made without RBS, directly linking promoter to the fluorescent protein gene. The EYFP constructs contained the sequence TCTAGA between the RBS and EYFP while the mTagBFP constructs contained that sequence between PpsbA2 and RBS and TACTAG between RBS and mTagBFP. The vectors were conjugated into Synechocystis in the same way as for the promoter constructs.
Cultivation conditions
Escherichia coli strain DH5α (Invitrogen) was used for subcloning and cells were grown in LB supplemented with 50 μg mL−1 kanamycin. Synechocystis PCC 6803 cultures were grown at 30 °C and 20 μmol photons m−2 s−1 except when otherwise noted, in BG11 medium with addition of 10 μg mL−1 kanamycin in 6-well plates containing 4–6 mL of media. For the experiment where fluorescence was measured over eight days, cultures were grown in 25 ml BG11 in 100 ml Erlenmeyer-flasks.
Fluorescence measurements
Fresh Synechocystis cells were seeded to an Abs750 of 0.15 and grown for two days. Solutions of metal salts (NiCl2 × 6 H2O (Merck), ZnSO4 × 7 H2O (Merck), Co(NO3)2 × 6 H2O (Merck), or CuSO4 × 5 H2O (Riedel-de Haën)) were then added to the cultures to obtain the appropriate concentrations of metal ions. After another two days, fluorescence and absorbance was measured with a Plate Chameleon V Microplate Reader (Hidex) using Microtest 96-well Optilux Black Assay Plates (BD Falcon) at 485 nm excitation and 535 nm emission for EYFP and 390 nm excitation and 460 nm emission for mTagBFP measurements. Optical density at 750 nm for Synechocystis and 595 nm for E. coli was used as a measure of the number of cells in the cultures. For each promoter or RBS strain, cells was grown in duplicates and measured with three technical replicates, and each experiment was repeated twice. Standard deviations were calculated from the four biological replicates. For data analysis, the background was subtracted from absorbance and fluorescence measurements using a blank BG11 sample. Fluorescence was then divided by the optical density to get a value representing average fluorescence per cell. The fluorescence per cell value of cells containing the empty vector was subtracted from all samples.
For activity tests in E. coli, cultures were grown overnight in LB, then 2 μl was added to 198 μl of M9 media59 in a 96-well plate. Cells were grown for 6 hours in triplicates and then fluorescence was measured in the same way as described above.
Ethanol measurements
Cells were seeded to Abs750 0.15 in 25 ml BG11 with 50 μg mL−1 kanamycin and 2.5 μM Ni2+ except when otherwise noted. Samples were taken every other day and Abs750 and ethanol accumulation was measured. Cultures were grown with four replicates for eight days at 20 μmol photons m−2 s−1 except when otherwise noted. For measuring ethanol, samples from cultures were pelleted and 1 μl of the supernatant was injected in a Clarus 580 Perkin Elmer FID gas chromatograph (GC) with a packed column (1.8 m × 2 mm i.d., Cat No. N9305013-ZW5531, Perkin Elmer). Injection was done through a packed injector, the carrier gas was N2 at 20 ml min−1. The GC program was: 130 °C for 5.5 min, then ramp to 230 °C at 45 °C min−1 and held for 5 min. The area of the ethanol peak at 4.96 min was converted into mg/L by using a correlation factor determined by measuring a standard of pure ethanol mixed with BG11 (R2 > 0.99).
Microscopy
Microscopy imaging was carried out on a Leica TCS SP5 confocal microscope. EYFP was excited at 514 nm using laser light and emission was collected between 527 and 546 nm. Autofluorescence was excited using the same wavelength and emission was collected between 605 and 710 nm. Gain and offset were identical between each sample to ensure equal treatment.
Additional Information
How to cite this article: Englund, E. et al. Evaluation of promoters and ribosome binding sites for biotechnological applications in the unicellular cyanobacterium Synechocystis sp. PCC 6803. Sci. Rep. 6, 36640; doi: 10.1038/srep36640 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work was supported by the Swedish Energy Agency (grant no 38334-1) and a scholarship from the Chinese Service Center for Scholarly Exchange to Feiyan Liang (201304910361). The BioBrick containing RBS*, EYFP and B0015 as well the Synechocystis strains PrnpB and Empty vector were a gift from Hsin-Ho Huang. The standard curve for ethanol quantitation was made by Isabel Moreno de Palma.
Footnotes
Author Contributions E.E. conceived the project, designed and conducted experiments and wrote the paper together with P.L. and F.L. constructed and tested the PpsbA2M and PpsbA2S constructs. P.L. provided advice on experiment design and co-wrote the paper with E.E. All authors approved the manuscript before submission.
References
- Savakis P. & Hellingwerf K. J. Engineering cyanobacteria for direct biofuel production from CO2. Curr. Opin. Biotechnol. 33, 8–14 (2015). [DOI] [PubMed] [Google Scholar]
- Zhou J. et al. Discovery of a super-strong promoter enables efficient production of heterologous proteins in cyanobacteria. Sci. Rep. 4, 4500 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camsund D., Heidorn T. & Lindblad P. Design and analysis of LacI-repressed promoters and DNA-looping in a cyanobacterium. J. Biol. Eng. 8, doi: 10.1186/1754-1611-1188-1184 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H.-H., Camsund D., Lindblad P. & Heidorn T. Design and characterization of molecular tools for a Synthetic Biology approach towards developing cyanobacterial biotechnology. Nucleic Acids Res. 38, 2577–2593 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albers S. C., Gallegos V. A. & Peebles C. A. Engineering of genetic control tools in Synechocystis sp. PCC 6803 using rational design techniques. J Biotechnol 216, 36–46 (2015). [DOI] [PubMed] [Google Scholar]
- Huang H.-H. & Lindblad P. Wide-dynamic-range promoters engineered for cyanobacteria. J. Biol. Eng. 7, doi: 10.1186/1754-1611-1187-1110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe K. et al. Engineering of a green light inducible gene expression system in Synechocystis sp. PCC6803. Microb. Biotechnol. 7, 177–183 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peca L., Kós P. B., Máté Z., Farsang A. & Vass I. Construction of bioluminescent cyanobacterial reporter strains for detection of nickel, cobalt and zinc. FEMS Microbiol. Lett. 289, 258–264 (2008). [DOI] [PubMed] [Google Scholar]
- Liu X. & Curtiss R. Nickel-inducible lysis system in Synechocystis sp. PCC 6803. Proc. Natl. Acad. Sci. USA 106, 21550–21554 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheah Y. E., Albers S. C. & Peebles C. A. M. A novel counter-selection method for markerless genetic modification in Synechocystis sp. PCC 6803. Biotechnol. Prog. 29, 23–30 (2013). [DOI] [PubMed] [Google Scholar]
- Čelešnik H. et al. Biosafety of biotechnologically important microalgae: intrinsic suicide switch implementation in cyanobacterium Synechocystis sp. PCC 6803. Biol. open, bio. 017129 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund E., Andersen-Ranberg J., Miao R., Hamberger B. & Lindberg P. Metabolic engineering of Synechocystis sp. PCC 6803 for production of the plant diterpenoid manoyl oxide. ACS Synth. Biol. 4, 1270–1278 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero F., Carbonell V., Cossu M., Correddu D. & Jones P. R. Ethylene synthesis and regulated expression of recombinant protein in Synechocystis sp. PCC 6803. PLoS One 7, e50470, doi: 10.1371/journal.pone.0050470 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Domínguez M., Lopez-Maury L., Florencio F. J. & Reyes J. C. A gene cluster involved in metal homeostasis in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 182, 1507–1514 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Maury L., García-Domínguez M., Florencio F. J. & Reyes J. C. A two-component signal transduction system involved in nickel sensing in the cyanobacterium Synechocystis sp. PCC 6803. Mol. Microbiol. 43, 247–256 (2002). [DOI] [PubMed] [Google Scholar]
- Foster A. W., Patterson C. J., Pernil R., Hess C. R. & Robinson N. J. Cytosolic Ni(II) sensor in cyanobacterium: Nickel detection follows nickel affinity across four families of metal sensors. J. Biol. Chem. 287, 12142–12151, doi: 10.1074/jbc.M111.338301 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford J. C., Cavet J. S. & Robinson N. J. Cobalt-dependent transcriptional switching by a dual-effector MerR-like protein regulates a cobalt-exporting variant CPx-type ATPase. J. Biol. Chem. 274, 25827–25832 (1999). [DOI] [PubMed] [Google Scholar]
- Thelwell C., Robinson N. J. & Turner-Cavet J. S. An SmtB-like repressor from Synechocystis PCC 6803 regulates a zinc exporter. Proc. Natl. Acad. Sci. USA 95, 10728–10733 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G. et al. Metabolic burden: Cornerstones in synthetic biology and metabolic engineering applications. Trends Biotechnol. 34, 652–664 (2016). [DOI] [PubMed] [Google Scholar]
- Jones P. R. Genetic instability in cyanobacteria - an elephant in the room? Front. Bioeng. Biotechnol. 2, 12, doi: 10.3389/fbioe.2014.00012 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen J. H. & Frigaard N. U. Engineering of photosynthetic mannitol biosynthesis from CO2 in a cyanobacterium. Metab. Eng. 21, 60–70 (2014). [DOI] [PubMed] [Google Scholar]
- Kusakabe T. et al. Engineering a synthetic pathway in cyanobacteria for isopropanol production directly from carbon dioxide and light. Metab. Eng. 20, 101–108 (2013). [DOI] [PubMed] [Google Scholar]
- Angermayr S. A., Paszota M. & Hellingwerf K. J. Engineering a cyanobacterial cell factory for production of lactic acid. Appl. Environ. Microbiol. 78, 7098–7106 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atsumi S., Higashide W. & Liao J. C. Direct photosynthetic recycling of carbon dioxide to isobutyraldehyde. Nat. Biotechnol. 27, 1177–1180 (2009). [DOI] [PubMed] [Google Scholar]
- Qi Q. et al. Application of the Synechococcus nirA promoter to establish an inducible expression system for engineering the Synechocystis tocopherol pathway. Appl. Environ. Microbiol. 71, 5678–5684 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peca L., Kos P. & Vass I. Characterization of the activity of heavy metal-responsive promoters in the cyanobacterium Synechocystis PCC 6803. Acta Biol. Hung. 58, 11–22 (2007). [DOI] [PubMed] [Google Scholar]
- Blasi B., Peca L., Vass I. & Kos P. B. Characterization of stress responses of heavy metal and metalloid inducible promoters in Synechocystis PCC 6803. J. Microbiol. Biotechnol. 22, 166–169 (2012). [DOI] [PubMed] [Google Scholar]
- Heidorn T. et al. In Methods Enzymol. Vol. 497 (ed Christopher A. Voigt) 539–579 (Academic Press, 2011). [Google Scholar]
- Muramatsu M. & Hihara Y. Characterization of high-light-responsive promoters of the psaAB genes in Synechocystis sp. PCC 6803. Plant Cell Physiol. 47, 878–890 (2006). [DOI] [PubMed] [Google Scholar]
- Miyawaki A. et al. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature 388, 882–887 (1997). [DOI] [PubMed] [Google Scholar]
- iGEM Registry. Registry of Standard Biological Parts. http://parts.igem.org/ (Date of access:11/10/2016).
- Salis H. M., Mirsky E. A. & Voigt C. A. Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotech. 27, 946–950 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanier R. Y., Kunisawa R., Mandel M. & Cohen-Bazire G. Purification and properties of unicellular blue-green algae (order Chroococcales). Bacteriol. Rev. 35, 171–205 (1971). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai I. et al. Positive regulation of psbA gene expression by cis-encoded antisense RNAs in Synechocystis sp. PCC 6803. Plant Physiol. 160, 1000–1010 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson J., Salih G. F., Ghebramedhin H. & Jansson C. Deletion mutagenesis of the 5′ psbA2 region in Synechocystis 6803: Identification of a putative cis element involved in photoregulation. Mol. Cell Biol. Res. Commun. 3, 292–298 (2000). [DOI] [PubMed] [Google Scholar]
- Kopf M. et al. Comparative analysis of the primary transcriptome of Synechocystis sp. PCC 6803. DNA Res. 21, 527–539 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagarde D., Beuf L. & Vermaas W. Increased production of zeaxanthin and other pigments by application of genetic engineering techniques to Synechocystis sp. strain PCC 6803. Appl. Environ. Microbiol. 66, 64–72 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dexter J. & Fu P. Metabolic engineering of cyanobacteria for ethanol production. Energy Environ. Sci. 2, 857–864, doi: 10.1039/B811937F (2009). [DOI] [Google Scholar]
- Lindberg P., Park S. & Melis A. Engineering a platform for photosynthetic isoprene production in cyanobacteria, using Synechocystis as the model organism. Metab. Eng. 12, 70–79 (2010). [DOI] [PubMed] [Google Scholar]
- Angermayr S. A. et al. Exploring metabolic engineering design principles for the photosynthetic production of lactic acid by Synechocystis sp. PCC6803. Biotechnol. Biofuels 7, 99, doi: 10.1186/1754-6834-7-99 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angermayr S. A. & Hellingwerf K. J. On the use of metabolic control analysis in the optimization of cyanobacterial biosolar cell factories. J. Phys. Chem. B 117, 11169–11175 (2013). [DOI] [PubMed] [Google Scholar]
- Gao Z., Zhao H., Li Z., Tan X. & Lu X. Photosynthetic production of ethanol from carbon dioxide in genetically engineered cyanobacteria. Energy Environ. Sci. 5, 9857–9865 (2012). [Google Scholar]
- Formighieri C. & Melis A. Regulation of beta-phellandrene synthase gene expression, recombinant protein accumulation, and monoterpene hydrocarbons production in Synechocystis transformants. Planta 240, 309–324 (2014). [DOI] [PubMed] [Google Scholar]
- Xiong W. et al. The plasticity of cyanobacterial metabolism supports direct CO2 conversion to ethylene. Nat. Plants 1, doi: 10.1038/nplants.2015.53 (2015). [DOI] [Google Scholar]
- Mutalik V. K. et al. Quantitative estimation of activity and quality for collections of functional genetic elements. Nat. Methods 10, 347–353 (2013). [DOI] [PubMed] [Google Scholar]
- Alfonso M., Perewoska I. & Kirilovsky D. Redox control of psbA gene expression in the cyanobacterium Synechocystis PCC 6803. Involvement of the cytochrome b6/f complex. Plant Physiol. 122, 505–516 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leveau J. H. & Lindow S. E. Predictive and interpretive simulation of green fluorescent protein expression in reporter bacteria. J. Bacteriol. 183, 6752–6762 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng M.-D. & Coleman J. R. Ethanol synthesis by genetic engineering in cyanobacteria. Appl. Environ. Microbiol. 65, 523–528 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver J. W., Machado I. M., Yoneda H. & Atsumi S. Combinatorial optimization of cyanobacterial 2,3-butanediol production. Metab. Eng. 22, 76–82 (2014). [DOI] [PubMed] [Google Scholar]
- Yoshikawa K., Hirasawa T. & Shimizu H. Effect of malic enzyme on ethanol production by Synechocystis sp. PCC 6803. J. Biosci. Bioeng. 119, 82–84 (2015). [DOI] [PubMed] [Google Scholar]
- Dexter J., Armshaw P., Sheahan C. & Pembroke J. The state of autotrophic ethanol production in cyanobacteria. J. Appl. Microbiol. 119, 11–24 (2015). [DOI] [PubMed] [Google Scholar]
- Mohamed A. & Jansson C. Influence of light on accumulation of photosynthesis-specific transcripts in the cyanobacterium Synechocystis 6803. Plant Mol. Biol. 13, 693–700 (1989). [DOI] [PubMed] [Google Scholar]
- Imamura S. & Asayama M. Sigma factors for cyanobacterial transcription. Gene Regul. Syst. Bio. 3, 65–87 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subach O. M. et al. Conversion of red fluorescent protein into a bright blue probe. Chem. Biol. 15, 1116–1124 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Campbell A. & Karlin S. Correlations between Shine-Dalgarno sequences and gene features such as predicted expression levels and operon structures. J. Bacteriol. 184, 5733–5745 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markley A. L., Begemann M. B., Clarke R. E., Gordon G. C. & Pfleger B. F. Synthetic biology toolbox for controlling gene expression in the cyanobacterium Synechococcus sp. strain PCC 7002. ACS Synth. Biol. 4, 595–603 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty R., Endy D. & Knight T. Engineering BioBrick vectors from BioBrick parts. J. Biol. Eng. 2, doi: 10.1186/1754-1611-2-5 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhai J. & Wolk C. P. In Methods Enzymol. Vol. 167 (eds. Packer L. & Glazer A. N.) 747–754 (Academic Press, 1988). [DOI] [PubMed] [Google Scholar]
- Harwood C. R. & Cutting S. M. In Molecular biological methods for Bacillus (eds Harwood C. R. & Cutting S. M.) (Wiley, 1990). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.