Abstract
Background
Combination chemotherapy with Western anti-tuberculosis (TB) drugs is the mainstay of TB treatment. Chinese herbal medicines with either heat clearing and detoxifying effects or nourishing Yin and reducing fire effects have been used to treat TB based on the Traditional Chinese Medicine (TCM) syndromes of TB patients. This study analyzed the expression profiles of long non-coding RNAs (lncRNAs) and mRNAs in TB patients with different TCM syndromes.
Methods
TB patients were classified as pulmonary Yin deficiency (PYD) syndrome, hyperactivity of fire due to Yin deficiency (HFYD) syndrome, and deficiency of Qi and Yin (DQY) syndrome. Total RNA from 44 TB patients and healthy controls was extracted and hybridized with a human lncRNA microarray containing 30586 lncRNAs and 26109 mRNAs probes. Bioinformatics analyses, including gene ontology (GO) and pathways, were performed. Related clinical data were also analyzed.
Results
Differentially expressed mRNAs and lncRNAs were identified (fold change >2, and P < 0.05) in PYD (634 mRNAs and 566 lncRNAs), HFYD (47 mRNAs and 55 lncRNAs), and DQY (63 mRNAs and 60 lncRNAs) patients. The most enriched pathways were the hippo signaling pathway (P = 0.000164) and the protein digestion and absorption pathway (P = 5.89017E-05). Clinical analyses revealed that the lipid indexes of TB patients were abnormal and that the triglyceride concentration was significantly higher in DQY patients (P = 0.0252). Our study is the first to acquire the microarray expression profiles of lncRNAs and mRNAs and analyze pathway enrichment in PYD, HFYD, and DQY patients with TB.
Conclusions
Our analyses of the expression profiles of lncRNAs and mRNAs may represent a novel method to explore the biological essence of TCM syndromes of TB.
Electronic supplementary material
The online version of this article (doi:10.1186/s12906-016-1436-y) contains supplementary material, which is available to authorized users.
Keywords: Pulmonary tuberculosis, TCM syndrome, mRNAs, lncRNAs
Background
Pulmonary tuberculosis (TB) caused by Mycobacterium tuberculosis (Mtb) infection is a leading cause of death. Nine million new TB patients and 1.5 million TB deaths occurred globally in 2013 [1]. TB remains a public threat to human health in China. Combination chemotherapy with anti-TB drugs (isoniazid, rifampicin, pyrazinamide and ethambutol for 2 months and isoniazid and rifampicin for 4 months) is the mainstay of TB treatment [2]. Most TB cases are cured using routine anti-TB therapy, but some TB patients may develop severe side effects [3–5] or drug-resistant TB [4, 6]. The adverse effects of anti-TB drugs vary greatly among individuals [2, 7], and these effects are closely related to disease progression and the immune status of the patient [8]. Individualized treatments that strengthen the body’s immune system and enhance the efficacy and reduce the toxicity of anti-TB drugs are a new method of treating TB [2, 8].
With more than 3000 years of clinical practice, Traditional Chinese Medicine (TCM) is a fully institutionalized medical system in China [9] and has been used to treat TB for at least 500 years [10]. TCM enables individualized health care [11, 12]. Diagnoses are based on the integrity of the body and TCM syndrome differentiation, and different patients receive different prescriptions [11, 12]. The TCM syndrome is the temporary state of the patient’s comprehensive response and is the premise for treatment [13]. Disease progression and the extent of damage are generally assessed by inspection, auscultation, olfaction, interrogation, and palpation in TCM [14, 15]. Patients with the same disease can undergo different TCM syndromes, thus providing an opportunity for personalized medicine [14–16].
TB patients have been classified into three main TCM syndromes: pulmonary Yin deficiency syndrome (PYD), hyperactivity of fire due to Yin deficiency syndrome (HFYD), and deficiency of Qi and Yin syndrome (DQY) [17]. Modern medical studies have shown that the integration of Chinese and Western medicine based on the TCM syndromes of TB patients can enhance the efficacy and reduce the side effects of anti-TB drugs and improve the immune response [18–20]. For example, Chinese herbs with heat-clearing and detoxifying effects or nourishing Yin and lowering fire effects, such as Astragalus membranaceus and Radix Paeoniae Rubra (Chishao), have been used to treat TB [19–21]. Extracts from Astragalus membranaceus greatly improve the phagocytosis of mycobacteria [19, 21]. Extracts from Radix Paeoniae Rubra elevate the level of interleukin-8 [20] and drive the recruitment of T lymphocytes and neutrophils at infection sites to increase the bacteriostatic function of neutrophils [22, 23]. Extracts from Prunella vulgaris L. and Radix Sophorae Flavescentis have been shown to strengthen cell-mediated immunity in a rat model of multidrug-resistant TB [24].
However, TCM syndrome classification depends heavily on the clinical experience of TCM practitioners, and relevant fundamental experimental studies are lacking [15, 25]. The current study used Arraystar Human LncRNA Microarray technology to investigate the differential expression profiles of mRNAs and lncRNAs in PYD, HFYD, and DQY patients with pulmonary TB. The pathway enrichment of differentially expressed mRNAs and clinical indexes were also analyzed using bioinformatics methods.
Methods
Patients and control subjects
A total of 292 pretreated TB patients (aged 18 to 75 years), including 92 PYD cases, 124 HFYD cases, and 76 DQY cases, from Shaoxing Municipal Hospital (Shaoxing, Zhejiang, China) were included in the current study. All recruited TB patients were diagnosed according to the diagnostic criteria of the Ministry of Health, China, and met one of the following diagnostic criteria: positive sputum culture or smear; typical active TB findings on chest X-ray and CT scan; or pulmonary pathological lesions diagnosed as TB. TB cases with other diseases, such as hepatitis B, diabetes, extra-pulmonary TB, AIDS, and immune inhibitor users, were excluded. TB patients were classified into PYD, HFYD, DQY syndromes according to the ‘Standard of disease diagnosis and curative effect of Traditional Chinese Medicine’ [18]. A total of 115 healthy blood donors (aged 18 to 75 years) from Zhejiang Hospital (Zhejiang, China) with no history of TB, hepatitis B, AIDS, or other diseases were also included in the study.
Plasma samples were collected in heparin lithium-anticoagulant tubes and centrifuged at 3000 rpm at 4 °C for 10 min. Samples were dispensed into sterile centrifuge tubes and stored at −80 °C. Data such as lipoprotein-a, apolipoprotein-A1, apolipoprotein-B, total cholesterol (TC), high-density lipoprotein (HDL), low-density lipoprotein (LDL), and triglycerides (TG) levels were recorded for PYD, HFYD, and DQY cases and healthy controls. Differences were analyzed by one-way ANOVA followed by Tukey’s post-hoc test, χ 2 test, or unpaired t-test using GraphPad Prism 5 (GraphPad Software, Inc., USA) and one-sample t-test after taking the logarithm using SPSS 16.0.
Chemicals and reagents
TRIzol® reagent was purchased from Invitrogen Life Technologies, and the RNeasy Mini Kit was obtained from Qiagen (Valencia, CA, USA). The Quick Amp Labeling Kit (One-Color), gene expression hybridization kit, gene expression wash buffer, and microarray scanner were obtained from Agilent (California, USA). The magnetic stir plate was obtained from Corning Incorporated (New York, USA).
RNA isolation
Eleven PYD cases, 11 HFYD cases, 11 DQY cases, and 11 healthy controls were randomly chosen for the following experiments. Each experimental group was divided into three biological repeats. Plasma (200 μL) from each specimen was used to extract total RNA with TRIzol reagent (Invitrogen Life Technologies), and total RNA was eluted in 85 μL of RNase-free water. An RNeasy Mini Kit (Qiagen p/n 74104) was used to purify total RNA according to the manufacturer’s instructions. RNA quantity and concentration were evaluated using a NanoDrop ND-1000 spectrophotometer at an absorbance ratio of A260/A280. The nucleic acid was considered pure when the absorbance ratio was 1.8–2.0.
DNA microarray
The Human LncRNA Microarray V3.0 (Arraystar Co. USA) allows the global profiling of human lncRNAs and protein-coding transcripts. An estimated 30,586 lncRNAs were constructed using the most highly respected public transcriptome databases, including Refseq, Gencode, and UCSC known genes, and the lncRNA microarray can detect 26,109 coding transcripts. A specific exon or splice junction probe accurately identified each transcript. Negative probes and positive probes (housekeeping genes) were also printed onto the array for hybridization quality control [26].
RNA labeling and array hybridization
Total RNA (1 μg) from each group was amplified and transcribed into cyanine 3-labeled cRNA according to the instructions for the Quick Amp Labeling Kit, One-Color (Agilent). The labeled cRNAs were purified, and the concentration and specific activity (pmol Cy3/μg cRNA) were measured using a NanoDrop ND-1000 spectrophotometer. Hybridization was performed using an Agilent Gene Expression Hybridization Kit according to the manufacturer’s guidelines. Briefly, final 1× blocking agent and 1× fragmentation buffer were added to the labeled cRNA and incubated at 60 °C to fragment RNA for 30 min. GE Hybridization Buffer HI-RPM was mixed with the samples to stop the fragmentation reaction. A gasket slide was loaded into the Agilent SureHyb chamber before the hybridization samples were dispensed into the gasket well, and the Human LncRNA Array V3.0 slide was assembled. The slides were hybridized in a hybridization oven at 65 °C for 17 h, washed with Gene Expression Wash Buffer, fixed and immediately scanned in an Agilent Microarray Scanner (Agilent p/n G2565BA) [27].
Data analysis
The array images were analyzed using Agilent Feature Extraction (version 11.0.1.1) software, and subsequent quantile normalization and further data analyses were performed in the GeneSpring GX v11.5.1 package (Agilent Technologies). LncRNAs and mRNAs flagged as Present or Marginal (“All Targets Value”) in at least three of 12 samples were chosen for further data analyses to remove transcripts with unreliable expression. Significantly differentially expressed lncRNAs and mRNAs between the two groups were identified using Volcano Plot filtering, and the expression patterns were analyzed using hierarchical clustering [28].
lncRNA classification and pathway analysis
To explore potential functional relationship between lncRNAs and related coding genes, significantly expressed ncRNAs were classified into different subgroups, including enhancer lncRNAs near coding genes, enhancer lncRNA profiling, homeobox transcription factor (HOX) cluster profiling, long intergenic noncoding RNAs (lincRNAs) near coding genes, and lincRNA profiling. Pathway analyses of differentially expressed mRNAs were performed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. Gene ontology (GO) was analyzed online (http://www.geneontology.org) to determine the broad attributes of genes and gene products, which were classified into three domains: biological process, cellular component, and molecular function. The overlap between differentially expressed genes and GO annotation was also analyzed using Fisher’s exact test [28].
Results
Clinical characteristics of TB cases with different TCM syndromes
PYD patients exhibited the following clinical symptoms and signs: tussiculation; scant sticky and white sputum or blood-stained sputum; dry mouth and pharynx; red tongue with thin fur; and weak and rapid pulse (Fig. 1a).
Fig. 1.
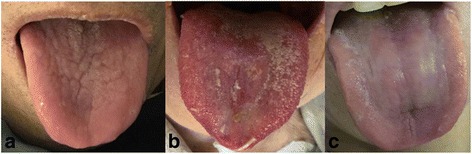
Tongue manifestations of TB patients. a PYD syndrome of TB (red tongue with thin fur). b HFYD syndrome of TB (red or dark red tongue with thin yellow or eroded fur). c DQY syndrome of TB (red and tender tongue with thin fur)
HFYD patients exhibited the following clinical characteristics: cough and breathlessness; hemoptysis; scant sticky sputum with white or yellow color; dry mouth and pharynx; red cheeks in the afternoon; tidal fever; steaming sensation in the bone; night sweats; red or dark red tongue with thin yellow or eroded fur; and weak and rapid pulse (Fig. 1b).
DQY patients exhibited the following clinical symptoms: cough with shortness of breath; clear and thin sputum; hemoptysis; physical and mental fatigue; spontaneous perspiration and night sweats; abdominal distension; anorexia; loose stool; red, tender tongue with thin fur; and weak and rapid pulse (Fig. 1c).
Statistical analysis of clinical indexes in TB cases
Statistical analyses of clinical indexes were performed between 92 PYD cases (aged 40.13 ± 16.16 years), 124 HFYD cases (aged 40.85 ± 15.85 years), 76 DQY cases (aged 43.95 ± 13.49 years), and 115 healthy controls (aged 39.30 ± 9.60 years). There were no significant differences in gender or age between the four groups. However, one-way ANOVA showed that the cholesterol levels of TB patients were significantly different compared to the healthy controls. The TC values in PYD cases, HFYD cases, DQY cases, and healthy controls were 3.83 ± 1.06 mmol/L, 3.85 ± 0.80 mmol/L, 3.79 ± 0.87 mmol/L, and 4.65 ± 0.52 mmol/L, respectively (P < 0.0001). HDL values were 1.05 ± 0.36 mmol/L, 1.10 ± 0.40 mmol/L, 1.04 ± 0.35 mmol/L, and 1.44 ± 0.26 mmol/L, respectively (P < 0.0001). LDL values were 2.74 ± 0.52 mmol/L, 2.31 ± 0.66 mmol/L, 2.30 ± 0.72 mmol/L, and 2.73 ± 0.52 mmol/L, respectively (P = 0.0002). TG values were 1.08 ± 0.57 mmol/L, 0.97 ± 0.41 mmol/L, 1.20 ± 0.89 mmol/L, and 0.95 ± 0.33 mmol/L, respectively (P = 0.0085). The TG value was significantly higher in DQY cases than in PYD and DQY cases (P = 0.0231, Table 1). One-sample t-tests were also performed to analyze differences between TB patients with different TCM syndromes and normal reference ranges after taking the logarithm. Various significant differences between TB patients with different TCM syndromes and normal reference values of blood fat indexes were observed (Additional file 1).
Table 1.
Characteristics of TB cases with PYD, HFYD and DQY syndromes
| Healthy Controls (N = 115) | PYD (N = 92) | HFYD (N = 124) | DQY (N = 76) | P value | |
|---|---|---|---|---|---|
| Age, age range (Mean ± SD) | 39.30 ± 9.60 | 40.13 ± 16.16 | 40.85 ± 15.85 | 43.95 ± 13.49 | 0.1409a |
| Gender (female: male) | 58/57 | 40/52 | 50/74 | 28/48 | 0.6558b |
| Abnormal chest radiograph (X, CT), no. (%) | ND | 92(100) | 124(100) | 76(100) | / |
| Positive sputum smears, no. (%) | ND | 77(84.00) | 100(81) | 56(74.00) | / |
| Lipoprotein a (mg/L) | ND | 261.41 ± 279.31 | 196.81 ± 189.58 | 264.72 ± 242.90 | / |
| Apolipoprotein A1 (apoA1) (g/L) | ND | 1.05 ± 0.24 | 1.11 ± 0.28 | 1.07 ± 0.22 | / |
| Apolipoprotein B (apoB) (g/L) | ND | 0.78 ± 0.24 | 0.81 ± 0.20 | 0.76 ± 0.17 | / |
| Total cholesterol (TC) (mmol/L) | 4.65 ± 0.52 | 3.83 ± 1.06 | 3.85 ± 0.80 | 3.79 ± 0.87 | <0.0001*** a |
| High-density lipoprotein (HDL) (mmol/L) | 1.44 ± 0.26 | 1.05 ± 0.36 | 1.10 ± 0.40 | 1.04 ± 0.35 | <0.0001*** a |
| Low-density lipoprotein (LDL) (mmol/L) | 2.74 ± 0.52 | 2.43 ± 1.20 | 2.31 ± 0.66 | 2.30 ± 0.72 | 0.0002*** a |
| Triglyceride (TG) (mmol/L) | 0.95 ± 0.33 | 1.08 ± 0.57 | 0.97 ± 0.41 | 1.20 ± 0.89 | 0.0085** a |
| 1.02 ± 0.49 | 1.20 ± 0.89 | 0.0252* c |
There were no significant differences in gender and age between healthy controls and TB cases with PYD, HFYD and DQY syndromes. However, the cholesterol levels of TB patients, such as TC, HDL, LDL, and TG, were significantly different from those of healthy controls
a P-value between healthy controls and TB cases with PYD, HFYD and DQY syndromes for one-way ANOVA followed by Tukey’s post-hoc test
b P-value between healthy controls and TB cases with PYD, HFYD and DQY syndromes for the χ2 test
c P-value between HFYD cases and other TB patients for the unpaired t-test
N number of subjects, ND not determined. *P < 0.05. **P < 0.01. *** P < 0.001
lncRNA microarray profiling of TB cases with different TCM syndromes
The distribution of samples for microarray detection is shown in Additional file 2. Microarray profiling of 30,586 lncRNAs was analyzed using Arraystar Human LncRNA Microarray V3.0, and lncRNAs with fold changes >2.00 and P < 0.05 were considered significantly different. A total of 566 differentially expressed lncRNAs were identified in PYD cases, including 347 up-regulated and 219 down-regulated lncRNAs. Fifty-five differentially expressed lncRNAs were identified in HFYD cases, including 31 up-regulated and 24 down-regulated lncRNAs. Sixty differentially expressed lncRNAs were identified in DQY cases, including 35 up-regulated and 25 down-regulated lncRNAs. Most of the differentially expressed lncRNAs were intergenic lncRNAs (61.34%), natural antisense lncRNAs (14.63%), or intronic antisense lncRNAs (11.49%). The remainder were exon sense-overlapping lncRNAs, intron sense-overlapping lncRNAs, and bidirectional lncRNAs. Figure 2 shows the volcano plots and hierarchical clustering of the differentially expressed lncRNAs.
Fig. 2.
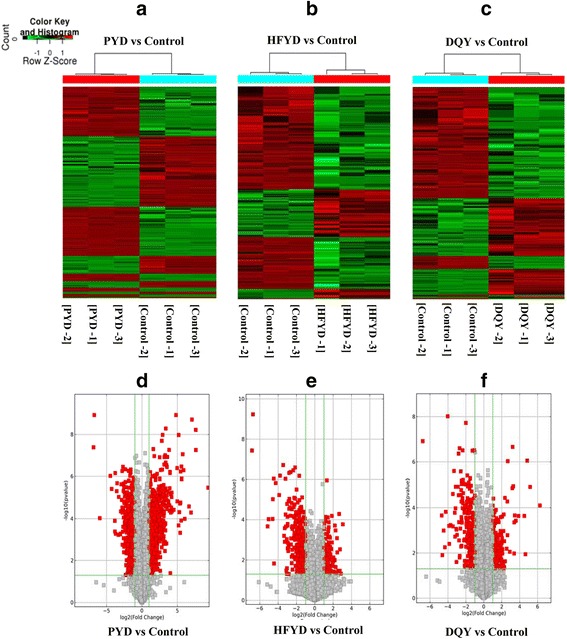
lncRNA volcano plots and hierarchical clustering dendrogram of TB patients with distinctive TCM syndromes. a-c lncRNA volcano plots (fold change >2.0 and P < 0.05). The vertical lines correspond to 2.0-fold up and down (log2 ratio), and the horizontal line represents a P-value of 0.05. Each point in the plot represents a different transcript. Transcripts are isolated based on statistical significance and magnitude of change. Red squares represent significantly differentially expressed lncRNAs. d–f Hierarchical clustering dendrogram representing distinguishable lncRNA expression profiling of samples. PYD: pulmonary Yin deficiency syndrome; HFYD: hyperactivity of fire due to Yin deficiency syndrome; DQY: deficiency of Qi and Yin syndrome
mRNA microarray profiling of TB cases with different TCM syndromes
Microarray profiling of 26,109 mRNAs was also performed using Arraystar Human LncRNA Microarray V3.0. A total of 634 differentially expressed mRNAs were identified in PYD patients, including 404 up-regulated and 230 down-regulated mRNAs. Forty-seven differentially expressed mRNAs were identified in HFYD patients, including 28 up-regulated and 19 down-regulated mRNAs. Sixty-three differentially expressed mRNAs were identified in DQY patients, including 44 up-regulated and 19 down-regulated mRNAs. The volcano plots and hierarchical clustering of differentially expressed mRNAs are shown in Fig. 3.
Fig. 3.
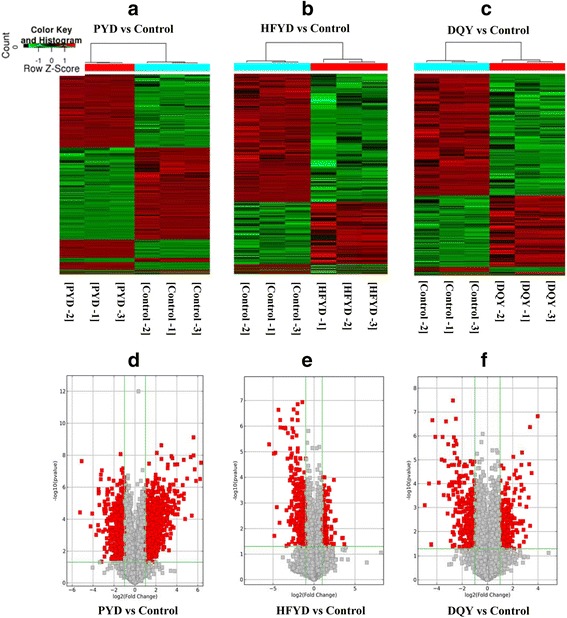
mRNA volcano plots and hierarchical clustering dendrogram of TB patients with distinctive TCM syndromes. a–c mRNA volcano plots (fold change >2.0 and P < 0.05). The vertical lines correspond to 2.0-fold up and down (log2 ratio), and the horizontal line represents a P-value of 0.05. Each point in the plot represents a different transcript. Transcripts are isolated based on statistical significance and magnitude of change. Red squares represent significantly differentially expressed mRNAs. d–f Hierarchical clustering dendrogram representing distinguishable mRNA expression profiling of the samples. PYD: pulmonary Yin deficiency syndrome; HFYD: hyperactivity of fire due to Yin deficiency syndrome; DQY: deficiency of Qi and Yin syndrome
Biological analysis
GO analysis was performed to determine the functions of genes and gene products involved in biological processes, cellular components and molecular functions. Fisher’s exact test was performed to determine the overlap between the differentially expressed list and the GO annotation. The significance of GO term enrichment among the differentially expressed genes was shown using P values. The highest enriched GO terms among differentially expressed transcripts between TB syndromes were cellular process (GO: 0009987; Ontology: biological process, P = 4.23124E-05) (Fig. 4a), cytoplasm (GO: 0005737; Ontology: cellular component, P = 0.000754106) (Fig. 4b), and protein binding (GO: 0005515; Ontology: molecular function, P = 0.001293641) (Fig. 4c).
Fig. 4.
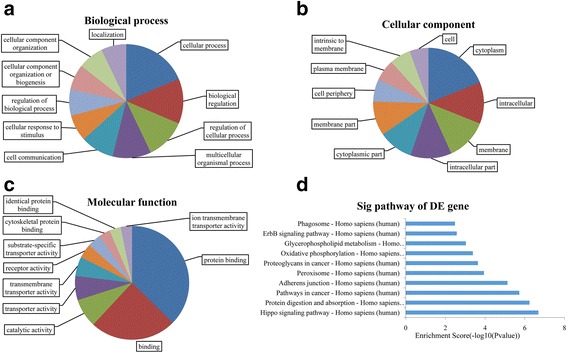
Gene ontology and pathway analyses of differentially expressed transcripts
Pathway analysis indicated that 61 pathways were associated with differentially expressed transcripts. The most enriched pathway was “Hippo signaling pathway-Homo sapiens (human)” (P = 0.000164), which was composed of 19 differentially expressed genes, and “Protein digestion and absorption-Homo sapiens (human)” (P = 5.89017E-05), which was also composed of 19 differentially expressed genes (Fig. 4d). Seven transcripts (COL4A6, PGA3, PGA4, PGA5, SLC1A5, SLC7A8, and SLC9A3) involved in pathways of protein digestion and absorption were up-regulated in PYD patients compared to HFYD and DQY patients. Sixteen transcripts linked to oxidative phosphorylation pathways were up-regulated in HFYD patients compared to the healthy controls, and two of these transcripts (CYC1 and PPA2) were up-regulated in HFYD patients compared to PYD and DQY patients. However, two transcripts (ACOT1 and PTPLA) related to the fatty acid elongation pathway were up-regulated in DQY patients compared to the healthy controls. We screened 20 differentially expressed mRNAs and lncRNAs in different TCM syndromes of TB based on the fold changes and the results of statistical analyses (Tables 2, 3 and 4).
Table 2.
Differentially expressed lncRNAs and mRNAs in TB cases with PYD syndrome
| Seqname | GeneSymbol | P-value | Absolute Fold Change | Associated Gene |
||||
|---|---|---|---|---|---|---|---|---|
| A vs. control | A vs. B | A vs. C | A vs. control | A vs. B | A vs. C | |||
| lncRNA | ||||||||
| ENST00000536029 | RP11-392P7.8 | 3.41E-07 | 8.50E-04 | 1.35E-04 | 47.87 | 44.37 | 44.85 | GPRC5D |
| ENST00000543515 | RP11-392P7.8 | 4.86E-05 | 1.72E-05 | 1.43E-04 | 39.01 | 44.01 | 31.53 | GPRC5D |
| NR_036524 | MICA | 1.70E-05 | 1.20E-04 | 4.19E-05 | 44.00 | 36.07 | 29.72 | MICA |
| ENST00000467458 | RP11-796I2.2 | 1.48E-05 | 4.18E-07 | 1.91E-05 | 31.88 | 49.41 | 43.14 | PRKAG2 |
| uc002mej.1 | AK056073 | 2.89E-06 | 1.40E-07 | 1.16E-07 | 5.28 | 5.46 | 5.52 | ACSBG2 |
| TCONS_00002486 | XLOC_001036 | 5.51E-06 | 1.81E-06 | 7.44E-04 | −5.57 | −5.62 | −4.58 | KCNN3 |
| DA401339 | / | 1.76E-04 | 8.76E-04 | 4.92E-04 | −3.65 | −4.94 | −5.47 | POU3F2 |
| NR_036569 | PILRB | 1.73E-05 | 3.32E-05 | 1.42E-04 | −4.32 | −3.77 | −4.53 | PILRB |
| ENST00000458682 | LINC00202 | 1.72E-03 | 1.94E-03 | 3.23E-03 | −5.12 | −5.48 | −4.45 | / |
| ENST00000517670 | RP11-363E6.3 | 5.20E-05 | 9.24E-03 | 6.41E-04 | −2.66 | −2.75 | −2.74 | FABP5 |
| mRNA | ||||||||
| NM_033641 | COL4A6 | 6.59E-04 | 1.23E-02 | 3.81E-05 | 3.04 | 3.63 | 2.76 | / |
| NM_001079807 | PGA3 | 1.32E-05 | 1.62E-03 | 9.73E-06 | 2.71 | 2.41 | 2.86 | / |
| NM_001079808 | PGA4 | 7.14E-06 | 2.51E-03 | 9.10E-05 | 5.07 | 5.30 | 4.12 | / |
| NM_014224 | PGA5 | 2.76E-06 | 2.84E-04 | 1.83E-02 | 2.88 | 4.08 | 2.81 | / |
| ENST00000412532 | SLC1A5 | 5.62E-03 | 2.18E-03 | 8.24E-04 | 4.78 | 11.40 | 8.89 | / |
| NM_012244 | SLC7A8 | 6.75E-05 | 2.77E-03 | 1.52E-03 | 4.45 | 4.81 | 3.47 | / |
| NM_004174 | SLC9A3 | 5.38E-06 | 5.64E-06 | 5.33E-06 | 4.85 | 4.81 | 4.86 | / |
| NM_199235 | COLEC11 | 3.00E-05 | 6.87E-04 | 5.33E-03 | −6.77 | −2.80 | −4.07 | / |
| NM_020664 | DECR2 | 1.37E-05 | 7.59E-05 | 7.84E-04 | −3.54 | −3.61 | −3.76 | / |
| NM_018248 | NEIL3 | 2.06E-04 | 8.97E-05 | 9.04E-05 | −2.97 | −2.97 | −3.28 | / |
Twenty of the most significantly differentially expressed lncRNAs and mRNAs in TB cases with PYD syndrome compared to healthy controls and TB cases with HFYD syndrome and DQY syndrome
Fold change >2: up-regulated; fold change < -2: down-regulated
A: PYD syndrome; B: HFYD syndrome; C: DQY syndrome
Table 3.
Differentially expressed lncRNAs and mRNAs in TB cases with HFYD syndrome
| Seqname | GeneSymbol | P-value | Absolute Fold Change | Associated Gene |
||||
|---|---|---|---|---|---|---|---|---|
| B vs. control | B vs. A | B vs. C | B vs. control | B vs. A | B vs. C | |||
| lncRNA | ||||||||
| TCONS_00008415 | XLOC_003867 | 4.72E-02 | 1.71E-02 | 3.09E-02 | 7.24 | 14.65 | 10.20 | / |
| NR_024499 | FMR1-AS1 | 3.45E-02 | 2.96E-02 | 3.62E-02 | 6.64 | 8.06 | 7.25 | FMR1 |
| ENST00000510610 | RP11-706C16.8 | 1.95E-04 | 1.72E-05 | 2.80E-05 | 4.57 | 6.58 | 6.39 | LY6D |
| ENST00000521600 | PVT1 | 8.03E-03 | 1.69E-03 | 1.88E-02 | 4.11 | 5.83 | 4.95 | / |
| ENST00000448365 | AC078842.4 | 6.88E-03 | 3.39E-03 | 2.75E-03 | 3.09 | 3.25 | 4.11 | PTN |
| uc021pjm.1 | HM358976 | 2.11E-04 | 5.85E-08 | 4.50E-05 | −5.46 | −4.76 | −5.48 | / |
| ENST00000441029 | RP4-553 F4.2 | 4.27E-05 | 9.12E-04 | 4.26E-04 | −3.49 | −3.27 | −4.65 | ZNF341 |
| ENST00000444112 | AC005808.3 | 4.42E-03 | 2.13E-03 | 2.58E-02 | −2.66 | −3.30 | −3.08 | ZNF217 |
| TCONS_00004246 | XLOC_002087 | 7.60E-05 | 1.39E-03 | 3.01E-04 | −4.72 | −3.12 | −3.88 | / |
| ENST00000434072 | GNG12-AS1 | 1.72E-03 | 1.54E-04 | 1.60E-03 | −2.96 | −6.46 | −2.62 | WLS |
| mRNA | ||||||||
| NM_001916 | CYC1 | 1.26E-04 | 1.33E-03 | 1.67E-04 | 5.39 | 5.61 | 7.44 | / |
| NM_176869 | PPA2 | 1.44E-02 | 1.67E-02 | 4.48E-02 | 2.70 | 2.02 | 2.06 | / |
| ENST00000418434 | CASP5 | 2.61E-02 | 3.75E-02 | 1.67E-02 | 4.29 | 3.82 | 5.21 | / |
| NM_182609 | ZNF677 | 8.52E-03 | 2.47E-02 | 2.56E-03 | 4.35 | 3.61 | 5.60 | / |
| NM_001135086 | PRSS41 | 3.90E-02 | 1.15E-02 | 4.07E-02 | 4.34 | 9.42 | 4.54 | / |
| NM_207174 | ABCG1 | 4.65E-03 | 2.59E-03 | 3.68E-02 | 6.35 | 8.32 | 4.40 | / |
| NM_001101 | ACTB | 1.95E-03 | 3.10E-04 | 1.99E-03 | −2.45 | −4.22 | −3.09 | / |
| NM_001135575 | C6orf228 | 1.23E-03 | 1.10E-03 | 3.89E-03 | −6.66 | −7.04 | −5.13 | / |
| NM_001166415 | EHHADH | 3.06E-02 | 2.25E-02 | 1.93E-02 | −2.79 | −2.87 | −3.15 | / |
| NM_152496 | MANEAL | 2.40E-02 | 1.28E-02 | 1.36E-02 | −5.78 | −8.54 | −8.88 | / |
Twenty of the most significantly differentially expressed lncRNAs and mRNA in TB cases with HFYD syndrome compared to healthy controls and TB cases with PYD syndrome and DQY syndrome
Fold change >2: up-regulated; fold change < -2: down-regulated
A: PYD syndrome; B: HFYD syndrome; C: DQY syndrome
Table 4.
Differentially expressed lncRNAs and mRNAs in TB cases with DQY syndrome
| Seqname | Gene Symbol | s-value | Absolute Fold Change | Associated Gene |
||||
|---|---|---|---|---|---|---|---|---|
| C vs. control | C vs. A | C vs. B | C vs. control | C vs. A | C vs. B | |||
| lncRNA | ||||||||
| ENST00000412526 | LINC00161 | 1.07E-03 | 1.72E-06 | 7.63E-04 | 4.59 | 6.66 | 4.13 | / |
| uc004erg.1 | BC028211 | 2.34E-02 | 4.18E-02 | 1.53E-02 | 5.21 | 4.40 | 7.54 | SLC25A5 |
| NR_036516 | C17orf62 | 1.13E-05 | 1.19E-05 | 2.03E-04 | 9.79 | 9.70 | 7.45 | C17orf62 |
| ENST00000434601 | LINC00422 | 1.05E-02 | 1.62E-02 | 2.06E-02 | 10.46 | 7.46 | 6.49 | / |
| ENST00000522460 | RP1-84O15.2 | 8.86E-03 | 9.02E-03 | 9.01E-03 | 7.49 | 7.42 | 7.42 | / |
| ENST00000452110 | PI4KAP1 | 1.63E-02 | 8.34E-05 | 8.64E-03 | −2.42 | −8.92 | 5.27 | / |
| BM728564 | / | 1.88E-03 | 5.54E-03 | 4.47E-02 | −9.20 | −5.69 | 3.54 | / |
| ENST00000528696 | CTD-3076O17.1 | 1.88E-03 | 4.12E-02 | 1.57E-02 | −2.15 | −4.07 | −2.09 | ADAMTS17 |
| ENST00000582033 | CTD-2124B20.2 | 9.98E-03 | 1.21E-04 | 1.86E-04 | −2.81 | −4.08 | −2.39 | / |
| TCONS_00012029 | XLOC_005561 | 9.25E-05 | 3.03E-02 | 1.68E-04 | −2.90 | −3.51 | −2.29 | / |
| mRNA | ||||||||
| NM_013322 | SNX10 | 2.73E-02 | 2.72E-02 | 2.99E-02 | 7.36 | 7.39 | 6.99 | / |
| NM_004436 | ENSA | 9.27E-04 | 9.47E-04 | 4.01E-03 | 8.45 | 8.36 | 6.76 | / |
| NM_153182 | MINA | 5.81E-03 | 5.91E-03 | 5.90E-03 | 10.37 | 10.27 | 10.28 | / |
| NM_014178 | STXBP6 | 1.50E-07 | 1.83E-07 | 1.59E-07 | 15.71 | 15.56 | 15.56 | / |
| NM_183375 | PRSS48 | 3.58E-05 | 1.94E-06 | 1.27E-06 | 11.26 | 11.92 | 12.46 | / |
| NM_001037161 | ACOT1 | 2.39E-02 | 7.52E-02 | 3.50E-01 | 2.15 | 1.68 | 1.67 | / |
| NM_014241 | PTPLA | 2.25E-02 | 7.65E-01 | 5.54E-02 | 3.11 | 1.05 | 2.17 | / |
| NM_001040443 | PHF11 | 1.82E-02 | 1.01E-02 | 2.71E-02 | −2.71 | −3.85 | −2.93 | / |
| NM_001099455 | CPPED1 | 1.21E-03 | 2.01E-03 | 2.32E-02 | −5.78 | −4.58 | −2.25 | / |
| NM_080627 | SOGA1 | 7.61E-04 | 2.65E-04 | 4.96E-04 | −2.48 | −4.33 | −3.32 | / |
Twenty of the most significantly differentially expressed lncRNAs and mRNA in TB cases with DQY syndrome compared to healthy control s and TB cases with PYD syndrome and HFYD syndrome
Fold change >2: up-regulated; fold change < -2: down-regulated
A: PYD syndrome; B: HFYD syndrome; C: DQY syndrome
Discussion
TCM states that disease occurs when Yin and Yang are unbalanced or the flow of Qi and blood is disturbed [15]. The major causes of TB are infection by Mtb and Yin deficiency [29]. Consumption of lung-Yin in the early stage of TB causes pulmonary Yin deficiency syndrome (PYD). Hyperactivity of liver-fire occurs with the development of lung-Yin consumption and leads to hyperactivity of fire due to Yin deficiency syndrome (HFYD). Harmony of Qi and blood is disturbed in some chronic TB patients and may cause a deficiency of Qi and Yin syndrome (DQY) [29, 30]. There is a lack of research on the subtle changes between different TCM syndromes, which is the major challenge in the interpretation of the theories of TCM using traditional methods [15, 17, 29]. We previously investigated the proteomic profiles of TB cases with TCM syndromes using SELDI-TOF MS and iTRAQ-2DLC-MS/MS and identified several differentially expressed serum proteins in PYD, HFYD, and DQY cases [10, 31]. Thus, subtle changes between different TCM syndromes of TB were reflected in serum proteomics, suggesting that subtle changes in PYD, HFYD and DQY cases may also be reflected in transcriptomics.
In the current study, more differentially expressed mRNAs and lncRNAs were detected in PYD patients than HFYD and DQY patients. A total of 634 mRNAs and 566 lncRNAs were differentially expressed in PYD patients. However, 47 mRNAs and 55 lncRNAs were differentially expressed in HFYD patients, and 63 mRNAs and 60 lncRNAs were differentially expressed in DQY patients. These results indicate that more abnormal gene expression occurred in PYD patients.
A total of 19 mRNAs involved in the pathway of protein digestion and absorption were differentially expressed in PYD cases compared to the healthy controls, and seven transcripts (COL4A6, PGA3, PGA4, PGA5, SLC1A5, SLC7A8, and SLC9A3) were also up-regulated in PYD cases compared to HFYD and DQY cases. COL4A6 encodes the alpha-6 chain of type IV collagen of basal membranes and is related to the prognosis of esophageal squamous cell carcinoma [32]. PGA3, PGA4, and PGA5 encode pepsinogen A (PGA), and the differential expression of PGA3, PGA4, and PGA5 is related to the pre-neoplastic nature in patients with Barrett’s esophagus [33]. SLC7A8 and SLC9A3 are solute carrier family genes, and the product of SLC7A8 participates in the transport of amino acids [34]. SLC9A3 encodes Na(+)/H(+) exchanger (NHE3), which is down-regulated in patients with ulcerative colitis [35]. TCM considers the spleen the ‘mother organ’ to the lungs, and thus the spleen may be similarly affected by conditions that affect the ‘child organ’ [36]. PYD syndrome often leads to spleen deficiency, and the clinical symptoms of PYD patients with spleen deficiency were anorexia, poor appetite, and loose stools [36]. Patients with spleen deficiency are generally characterized by digestive system disorders [37]. Therefore, we suspect that differentially expressed mRNAs (COL4A6, PGA3, PGA4, PGA5, SLC1A5, SLC7A8, and SLC9A3) involved in the protein digestion and absorption pathway may be related to the spleen deficiency in PYD patients.
The expression of 16 mRNAs involved in the pathway of oxidative phosphorylation was significantly increased in HFYD patients compared to the healthy controls, and two mRNAs (CYC1, PPA2) were also up-regulated. CYC1 encodes the cytochrome c1 subunit of respiratory chain complex III, which mediates the transfer of electrons from cytochrome b to cytochrome c during oxidative phosphorylation [38–40]. The PPA2 gene encodes mitochondrial pyrophosphatase 2, which catalyzes the hydrolysis of pyrophosphate to generate inorganic phosphate in cellular enzymatic reactions [41]. PPA2 is required for the maintenance of mitochondrial DNA and the synthesis of DNA, RNA, cAMP, and cGMP [41]. Oxidative phosphorylation is the major source of ATP and energy production, and mitochondria are the primary site of oxidative phosphorylation reactions [42]. Pulmonary TB is a typical consumptive disease with symptoms of weight loss, energy expenditure, and fat reduction [43]. Therefore, increased expression of CYC1 and PPA2 mRNAs in the pathway of oxidative phosphorylation may be linked to the energy consumption observed in HFYD syndrome.
Two mRNAs (ACOT1, PTPLA) involved in the fatty acid elongation pathway were up-regulated in DQY patients compared to the healthy controls. ACOT1 encodes acyl-CoA thioesterase 1, which hydrolyzes acyl-CoAs into free fatty acids and CoASH, thereby regulating intracellular levels of free fatty acids and CoASH [44]. PTPLA (also known as HACD1) encodes 3-hydroxyacyl-CoA dehydratase 1, which affects the third step (dehydration) in the elongation of very long chain fatty acids and is necessary for muscle function [45]. Clinical analyses revealed that cholesterol levels (TG, TC, HDL, LDL) differed significantly between TB patients and healthy controls, and the value of TG was significantly increased in DQY patients compared to PYD and DQY patients (Table 1). The hydrolysis of triglyceride (fat) from TB patients is necessary for the survival and virulence of Mycobacterium tuberculosis [46, 47]. Therefore, the up-regulated ACOT1 and PTPLA mRNAs linked to the fatty acid elongation pathway may be associated with the abnormality of blood fats in DQY syndrome.
Differentially expressed lncRNAs were identified in PYD, HFYD, and DQY patients. Ten differentially expressed lncRNAs for each TCM syndrome of TB were screened based on fold changes. G-protein coupled receptor family C group 5 member D, MHC class I polypeptide-related sequence A isoform 2,5’-AMP-activated protein kinase subunit gamma-2 isoform a, and paired immunoglobulin-like type 2 receptor beta precursor were the associated proteins of differentially expressed lncRNAs in PYD cases. Lymphocyte antigen 6D precursor, pleiotrophin precursor, and zinc finger protein were the associated proteins of differentially expressed lncRNAs in HFYD patients. ADP/ATP translocase 2 and a disintegrin and metalloproteinase with thrombospondin motifs 17 preproprotein were the associated proteins of differentially expressed lncRNAs in DQY patients. However, the functions of most lncRNAs have not been reported. Therefore, the differentially expressed lncRNAs in PYD, HFYD, and DQY patients and their functional relationship with the subtle changes between different TCM syndromes of TB requires further investigation.
Conclusion
This study revealed significantly altered lncRNA and mRNA expression profiles in the PYD, HFYD and DQY syndromes of TB. The pathway enrichment of differentially expressed transcripts was also analyzed using bioinformatics methods. The enhanced expression of mRNAs involved in the protein digestion and absorption pathway in PYD patients may be related to the spleen deficiency in PYD syndrome. The increased expression of CYC1 and PPA2 mRNAs in the oxidative phosphorylation pathway may be linked to the energy consumption in HFYD syndrome. The up-regulated ACOT1 and PTPLA mRNAs linked to the fatty acid elongation pathway may be associated with the abnormality of blood fats in DQY syndrome. These results indicated that the expression profile analysis of lncRNAs and mRNAs may be a novel method to explore the biological essence of TCM syndromes. However, the functional roles of lncRNAs and mRNAs in different TCM syndromes of TB require further investigation.
Acknowledgments
The authors thank all the blood donors from Shaoxing Municipal Hospital (Shaoxing, China) and Zhejiang Hospital (Zhejiang, China) and Jun-Liang Jiang and Yong-Liang Ma for transporting blood samples.
Funding
This work was supported by grants from the National Basic Research Program of China (No.2014CB543002), National Special Sci-Tech Projects (No. 2012ZX10005001-006), and National Natural Science Foundation of China (No.81573709, No.81273882). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
The datasets analyzed in the current study are available in Refseq, Gencode, and the UCSC known genes repository. The URLs are http://www.ncbi.nlm.nih.gov/refseq/, http://www.gencodegenes.org/, and http://www.genome.ucsc.edu/FAQ/FAQdownloads.html, respectively.
Authors’ contributions
JCL designed the experiments. LLW diagnosed and differentiated all the TB patients, and LYS collected blood samples and clinical data. TTJ performed the experiments and analyzed the results with ZLC and CW. All authors discussed the results, and JCL and TTJ wrote the manuscript. All authors read and approved the final manuscript.
Competing of interests
The authors declare that they have no competing interests.
Consent for publication
Written informed consent was obtained from all study participants for publication of individual age, gender, tongue manifestation images, and other clinical details. A copy of the written consent form is available for review by the Editor of this journal.
Ethics approval and consent to participate
The Medical Ethics Committee of Zhejiang University (China) approved this study, and written informed consent was obtained from all donors prior to blood collection.
Abbreviations
- DQY
Deficiency of Qi and Yin
- GO
Gene ontology
- HDL
High-density lipoprotein
- HFYD
Hyperactivity of fire due to Yin deficiency
- HOX
Homeobox transcription factors
- KEGG
Kyoto encyclopedia of genes and genomes
- LDL
Low-density lipoprotein
- lincRNAs
long intergenic noncoding RNAs
- lncRNAs
Long non-coding RNAs
- Mtb
Mycobacterium tuberculosis
- PYD
Pulmonary Yin deficiency
- TB
Tuberculosis
- TC
Total cholesterol
- TCM
Traditional Chinese medicine
- TG
Triglyceride
Additional files
Clinical data for TB cases with PYD, HFYD and DQY syndromes and normal reference ranges. P values between TB cases with PYD, HFYD and DQY syndromes and the normal reference range were determined by one-sample t-test after taking the logarithm and comparison to the median. *P < 0.05. **P < 0.01. *** P < 0.001 (DOCX 17 kb)
Sample data for TB PYD, HFYD, and DQY patients and healthy controls. (DOCX 15 kb)
Contributor Information
Ting-Ting Jiang, Email: jiang_tingting1989@126.com.
Li-Liang Wei, Email: weililiang1981@163.com.
Li-Ying Shi, Email: shi-lee-ying@163.com.
Zhong-Liang Chen, Email: zlchen_m@163.com.
Chong Wang, Email: knight_880102@163.com.
Chang-Ming Liu, Email: liucm523@126.com.
Zhong-Jie Li, Email: lizhongjie@zju.edu.cn.
Ji-Cheng Li, Phone: +86-571-88208088, Email: lijichen@zju.edu.cn.
References
- 1.[Internet] World Health Organization, Fact sheet N°104. http://www.who.int/mediacentre/factsheets/fs104/en/index.html Accessed Mar 2015.
- 2.Ramachandran G, Swaminathan S. Role of pharmacogenomics in the treatment of tuberculosis: a review. Pharmacogenomics Pers Med. 2012;5:89–98. doi: 10.2147/PGPM.S15454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burman WJ, Reves RR. Hepatotoxicity from rifampin plus pyrazinamide: lessons for policymakers and messages for care providers. Am J Resp Crit CareMed. 2001;164:1112–1113. doi: 10.1164/ajrccm.164.7.2109052. [DOI] [PubMed] [Google Scholar]
- 4.Singla R, Sharma SK, Mohan A, Makharia G, Sreenivas V, Jha B, et al. Evaluation of risk factors for antituberculosis treatment induced hepatotoxicity. Indian J Med Res. 2010;132:81–86. [PubMed] [Google Scholar]
- 5.Tsuyuguchi K, Wada M. Management of adverse effects with antituberculosis chemotherapy [in Japanese] Kekkaku. 2011;86:87–99. [PubMed] [Google Scholar]
- 6.Tripathi RP, Bisht SS, Ajay A, Sharma A, Misra M, Gupt MP. Developments in chemical approaches to treat tuberculosis in the last decade. Curr Med Chem. 2012;19:488–517. doi: 10.2174/092986712798918815. [DOI] [PubMed] [Google Scholar]
- 7.Evans DA, Manley KA, Mc KV. Genetic control of isoniazid metabolism in man. Br Med J. 1960;2:485–491. doi: 10.1136/bmj.2.5197.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heyckendorf J, Olaru ID, Ruhwald M, Lange C. Getting personal perspectives on individualized treatment duration in multidrug-resistant and extensively drug-resistant tuberculosis. Am J Respir Crit Care Med. 2014;190:374–383. doi: 10.1164/rccm.201402-0363PP. [DOI] [PubMed] [Google Scholar]
- 9.Guo Z, Yu S, Guan Y, Li YY, Lu YY, Zhang H, et al. Molecular mechanisms of same TCM syndrome for different diseases and different TCM syndrome for same disease in chronic hepatitis B and liver cirrhosis. Evid Based Complement Alternat Med. 2012;2012:120350. doi: 10.1155/2012/120350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu J, Li Y, Wei L, Yang X, Xie Z, Jiang T, et al. Screening and identification of potential biomarkers and establishment of the diagnostic serum proteomic model for the traditional Chinese medicine syndromes of tuberculosis. J Ethnopharmacol. 2014;155:1322–1331. doi: 10.1016/j.jep.2014.07.025. [DOI] [PubMed] [Google Scholar]
- 11.Wang X, Zhang A, Sun H, Wang P. Systems biology technologies enable personalized traditional Chinese medicine: a systematic review. Am J Chin Med. 2012;40:1109–1122. doi: 10.1142/S0192415X12500826. [DOI] [PubMed] [Google Scholar]
- 12.Zhang A, Sun H, Wang P, Han Y, Wang X. Future perspectives of personalized medicine in traditional Chinese medicine: a systems biology approach. Complement Ther Med. 2012;20:93–99. doi: 10.1016/j.ctim.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 13.Kanawong R, Obafemi-Ajayi T, Ma T, Xu D, Li S, Duan Y. Automated tongue feature extraction for ZHENG classification in traditional Chinese medicine. Evid Based Complement Alternat Med. 2012;2012:912852. doi: 10.1155/2012/912852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheung F. TCM: made in China. Nature. 2011;480:S82–83. doi: 10.1038/480S82a. [DOI] [PubMed] [Google Scholar]
- 15.Tang JL, Liu BY, Ma KW. Traditional Chinese medicine. Lancet. 2008;372:1938–1940. doi: 10.1016/S0140-6736(08)61354-9. [DOI] [PubMed] [Google Scholar]
- 16.Dai J, Sun S, Cao H, Zheng N, Wang W, Gou X, et al. Applications of new technologies and new methods in ZHENG differentiation. Evid Based Complement Alternat Med. 2012;2012:298014. doi: 10.1155/2012/298014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang SS, Zhou J, Zhang YF, Wang L, Feng X, Wang Y. Study on the rule of TCM syndromes in pulmonary tuberculosis [in Chinese] World J Integ Tradit Western Med. 2014;9:498–500. [Google Scholar]
- 18.Liu Y, He F. Chinese medicine in the treatment of tuberculosis of the status and prospect [in Chinese] Guiding J Tradit Chin Med Pharmacy. 2010;16:100–102. [Google Scholar]
- 19.Xu HD, You CG, Zhang RL, Gao P, Wang ZR. Effects of Astragalus polysaccharides and astragalosides on the phagocytosis of Mycobacterium tuberculosis by macrophages. J Int Med Res. 2007;35:84–90. doi: 10.1177/147323000703500108. [DOI] [PubMed] [Google Scholar]
- 20.Wang L, Yang CL, Or TC, Chen G, Zhou J, Li JC, et al. Differential effects of Radix Paeoniae Rubra (Chishao) on cytokine and chemokine expression inducible by mycobacteria. Chin Med. 2011;6:14. doi: 10.1186/1749-8546-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang M, Guan X, Chi Y, Robinson N, Liu JP. Chinese herbal medicine as adjuvant treatment to chemotherapy for multidrug-resistant tuberculosis (MDR-TB): a systematic review of randomised clinical trials. Tuberculosis. 2015;95:364–372. doi: 10.1016/j.tube.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 22.Pace E, Gjomarkaj M, Melis M, Profita M, Spatafora M, Vignola AM, et al. Interleukin-8 induces lymphocyte chemotaxis into the pleural space - role of pleural macrophages. Am J Resp Crit Care. 1999;159:1592–1599. doi: 10.1164/ajrccm.159.5.9806001. [DOI] [PubMed] [Google Scholar]
- 23.Godaly G, Young DB. Mycobacterium bovis bacille calmette Guerin infection of human neutrophils induces CXCL8 secretion by MyD88-dependent TLR2 and TLR4 activation. Cell Microbiol. 2005;7:591–601. doi: 10.1111/j.1462-5822.2004.00489.x. [DOI] [PubMed] [Google Scholar]
- 24.Lu J, Ye S, Qin R, Deng Y, Li CP. Effect of Chinese herbal medicine extracts on cell-mediated immunity in a rat model of tuberculosis induced by multiple drug-resistant bacilli. Mol Med Rep. 2013;8:227–232. doi: 10.3892/mmr.2013.1491. [DOI] [PubMed] [Google Scholar]
- 25.Lu AP, Jiang M, Zhang C, Chan K. An integrative approach of linking traditional Chinese medicine pattern classification and biomedicine diagnosis. J Ethnopharmacol. 2012;141:549–556. doi: 10.1016/j.jep.2011.08.045. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Gao SG, Liu G, Jia RN, Fan DM, Feng XS. Microarray expression profile analysis of long non-coding RNAs in human gastric cardiac adenocarcinoma. Cell Physiol Biochem. 2014;33:1225–1238. doi: 10.1159/000358692. [DOI] [PubMed] [Google Scholar]
- 27.Wang P, Lu SH, Mao HL, Bai YN, Ma TL, Cheng ZL, et al. Identification of biomarkers for the detection of early stage lung adenocarcinoma by microarray profiling of long noncoding RNAs. Lung Cancer. 2015;88:147–153. doi: 10.1016/j.lungcan.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 28.McKiernan PJ, Molloy K, Cryan SA, McElvaney NG, Greene CM. Long noncoding RNA are aberrantly expressed in vivo in the cystic fibrosis bronchial epithelium. Int J Biochem Cell B. 2014;52:184–191. doi: 10.1016/j.biocel.2014.02.022. [DOI] [PubMed] [Google Scholar]
- 29.Zeng JF, Ouyang JJ. Overview and prospect on treatment of pulmonary tuberculosis with serial prescription of TCM [in Chinese] Guiding J TCM. 2006;12:70–72. [Google Scholar]
- 30.Wang SS, Zhou J, Fang D, Zhou JM. Research on TCM law of tuberculosis and related to objects indicators [in Chinese] Clin J Chinese Med. 2011;3:13–14. [Google Scholar]
- 31.Jiang TT, Wang C, Wei LL, Yu XM, Shi LY, Xu DD, et al. Serum protein gamma-glutamyl hydrolase, Ig gamma-3 chain C region, and haptoglobin are associated with the syndromes of pulmonary tuberculosis in traditional Chinese medicine. BMC Complement Altern Med. 2015;15:243. doi: 10.1186/s12906-015-0686-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baba Y, Iyama KI, Ikeda K, Ishikawa S, Hayashi N, Miyanari N, et al. The expression of type IV collagen alpha 6 chain is related to the prognosis in patients with esophageal squamous cell carcinoma. Ann Surg Oncol. 2008;15:555–565. doi: 10.1245/s10434-007-9592-4. [DOI] [PubMed] [Google Scholar]
- 33.Pals G, Eriksson AW, Pronk JC, Frants RR, Klinkenbergknol EC, Bosma A, et al. Differential expression of pepsinogen isozymogens in a patient with barrett esophagus. Clin Genet. 1988;34:90–97. doi: 10.1111/j.1399-0004.1988.tb02842.x. [DOI] [PubMed] [Google Scholar]
- 34.Pineda M, Fernandez E, Torrents D, Estevez R, Lopez C, Camps M, et al. Identification of a membrane protein, LAT-2, that co-expresses with 4F2 heavy chain, an L-type amino acid transport activity with broad specificity for small and large zwitterionic amino acids. J Biol Chem. 1999;274:19738–19744. doi: 10.1074/jbc.274.28.19738. [DOI] [PubMed] [Google Scholar]
- 35.Fonseca-Camarillo G, Yamamoto-Furusho JK. Gene expression of solute carrier family 9 (sodium/hydrogen exchanger) 3, (SLC9A3) is downregulated in patients with ulcerative colitis. Inflamm Bowel Dis. 2012;18:1197–1198. doi: 10.1002/ibd.22968. [DOI] [PubMed] [Google Scholar]
- 36.He S, Xu JX, Zhu W, Xiang S, Tang C, Lu TT. The therapy of pulmonary tuberculosis with the relationship of lung and spleen [in Chinese] Guiding J Tradit Chin Med Pharmacy. 2013;28:7–9. [Google Scholar]
- 37.Zhang Y, Liu Y, Li XP, Li J, Li XF, Chen L, et al. Evidence-based novel changes in prevalence and symptom characteristics of spleen deficiency syndrome in persons of varied health status and different ages: across-sectional observational study. Evid Based Complement Alternat Med. 2014;2014:818235. doi: 10.1155/2014/818235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaignard P, Menezes M, Schiff M, Bayot A, Rak M, Ogier de Baulny H, et al. Mutations in CYC1, encoding cytochrome c1subunit of respiratory chain complex III, cause insulin-responsive hyperglycemia. Am J Hum Genet. 2013;93:384–389. doi: 10.1016/j.ajhg.2013.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crofts AR. The cytochrome bc (1) complex: function in the context of structure. Annu Rev Physiol. 2004;66:689–733. doi: 10.1146/annurev.physiol.66.032102.150251. [DOI] [PubMed] [Google Scholar]
- 40.Li GD, Fu D, Liang WQ, Fan L, Chen K, Shan LC, et al. CYC1 silencing sensitizes osteosarcoma cells to TRAIL-induced apoptosis. Cell Physiol Biochem. 2014;34:2070–2080. doi: 10.1159/000366402. [DOI] [PubMed] [Google Scholar]
- 41.Curbo S, Lagier-Tourenne C, Carrozzo R, Palenzuela L, Lucioli S, Hirano M, et al. Human mitochondrial pyrophosphatase: cDNA cloning and analysis of the gene in patients with mtDNA depletion syndromes. Genomics. 2006;87:410–416. doi: 10.1016/j.ygeno.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 42.Ravera S, Bartolucci M, Cuccarolo P, Litame E, Illarcio M, Calzia D, et al. Oxidative stress in myelin sheath: The other face of the extramitochondrial oxidative phosphorylation ability. Free Radic Res. 2015;49:1156–1164. doi: 10.3109/10715762.2015.1050962. [DOI] [PubMed] [Google Scholar]
- 43.Schwenk A, Hodgson L, Rayner CF, Griffin GE, Macallan DC. Leptin and energy metabolism in pulmonary tuberculosis. Am J Clin Nutr. 2003;77:392–398. doi: 10.1093/ajcn/77.2.392. [DOI] [PubMed] [Google Scholar]
- 44.Hunt MC, Rautanen A, Westin MA, Svensson LT, Alexson SE. Analysis of the mouse and human acyl-CoA thioesterase (ACOT) gene clusters shows that convergent, functional evolution results in a reduced number of human peroxisomal ACOTs. Faseb J. 2006;20:1855–1864. doi: 10.1096/fj.06-6042com. [DOI] [PubMed] [Google Scholar]
- 45.Muhammad E, Reish O, Ohno Y, Scheetz T, DeLuca A, Searby C, et al. Congenital myopathy is caused by mutation of HACD1. Hum Mol Genet. 2013;22:5229–5236. doi: 10.1093/hmg/ddt380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Munoz-Elias EJ, McKinney JD. Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat Med. 2005;11:638–644. doi: 10.1038/nm1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arora P, Goyal A, Natarajan VT, Rajakumara E, Verma P, Gupta R, et al. Mechanistic and functional insights into fatty acid activation in Mycobacterium tuberculosis. Nat Chem Biol. 2009;5:166–173. doi: 10.1038/nchembio.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analyzed in the current study are available in Refseq, Gencode, and the UCSC known genes repository. The URLs are http://www.ncbi.nlm.nih.gov/refseq/, http://www.gencodegenes.org/, and http://www.genome.ucsc.edu/FAQ/FAQdownloads.html, respectively.


