Abstract
Vitamin A homeostasis is critical to normal cellular function. Retinol-binding protein (RBP) is the sole specific carrier in the bloodstream for hydrophobic retinol, the main form in which vitamin A is transported. The integral membrane receptor STRA6 mediates cellular uptake of vitamin A by recognizing RBP-retinol to trigger release and internalization of retinol. We present the structure of zebrafish STRA6 determined to 3.9-angstrom resolution by single-particle cryo-electron microscopy. STRA6 has one intramembrane and nine transmembrane helices in an intricate dimeric assembly. Unexpectedly, calmodulin is bound tightly to STRA6 in a noncanonical arrangement. Residues involved with RBP binding map to an archlike structure that covers a deep lipophilic cleft. This cleft is open to the membrane, suggesting a possible mode for internalization of retinol through direct diffusion into the lipid bilayer.
Vitamin A is an essential nutrient for all mammals. In its retinaldehyde (retinal) form, it is vital for vision (1); as retinoic acids (RAs), it provides ligands for RAR (retinoic acid receptor) and RXR (retinoid X receptor) nuclear receptor transcription factors (2). Consequently, retinoid metabolism affects diverse biological processes (3), with disease implications from blindness to cancer (4, 5). Vitamin A deficiency is the third most common nutritional deficiency in the world, affecting millions of children and pregnant women (6).
Retinol (vitamin A alcohol) is the predominant circulating retinoid in the fasting state. In the absence of sufficient dietary vitamin A, retinol from liver stores is mobilized bound to retinol-binding protein (RBP, also called RBP4) (7), which solubilizes the hydrophobic retinol. In the bloodstream, retinol-RBP forms a complex with the transthyretin tetramer (14 kDa × 4) to prevent renal loss of RBP (21 kDa) (8). Once internalized into cells, retinol binds to cellular RBPs (CRBPs, notably CRBP1) (9, 10).
A putative cellular receptor for RBP was identified and biochemically characterized (11) and subsequently shown to be required for the transfer of retinol from RBP to CRBP (12). In 2007, Kawaguchi et al. (13) cloned this RBP receptor, which proved to be the previously named protein STRA6 (stimulated by retinoic acid 6) (14). STRA6 was predicted to be a 75-kDa multipass transmembrane (TM) protein without sequence similarity to any known transporter, channel, or receptor, and it was later experimentally demonstrated to bear nine TM segments (15). STRA6 was shown to catalyze the release of retinol from RBP, where it is tightly bound (16), and to facilitate retinol translocation across the membrane and association with CRBP1 (17). Conversion of retinol to retinyl esters for storage by enzymes, such as lecithin retinol acyltransferase (LRAT) (18), increases the efficiency of this process by increasing the amount of available apo-CRBP1 (13). In in vitro experiments, STRA6 was also shown to catalyze the exchange of retinol between holo-RBP and apo-RBP, as well as the efflux of retinol from holo-CRBP1 to apo-RBP (17). Noy and colleagues have suggested a distinct and somewhat controversial (19, 20) functional model for STRA6 (21), in which the movement of retinol is unidirectional from holo-RBP to CRBP1 and coupled to activation of a JAK-STAT (Janus kinase–signal transducers and activators of transcription) cascade, thus regulating gene expression (21, 22).
STRA6 is expressed with particular abundance in the eye, where it localizes predominantly in the basolateral membrane of the retinal pigment epithelium, and in the female reproductive organs and placenta (13, 14). Mutations in the human STRA6 gene have been linked to Matthew-Wood syndrome (MWS), which presents with ocular defects ranging from mild microphthalmia to anophthalmia, as well as with an array of other developmental abnormalities including cardiac and pulmonary defects and cognitive deficits (23). Consistent with an important role of STRA6 in maintaining vitamin A homoeostasis in the eye, a zebrafish stra6 gene knockdown model also exhibited severe ocular malformations during development (24). Stra6 knockout mice have a less severe phenotype, exhibiting defects in the visual cycle (25, 26) and impaired insulin signaling (27).
To gain insight into the mechanistic basis for STRA6 function, we determined the structure of STRA6 from zebrafish to 3.9 Å resolution by single-particle cryo-electron microscopy (cryo-EM). The protein is a dimer, and each protomer is associated at the cytoplasmic side with a molecule of calmodulin (CaM), which is unexpected given that there are no previous reports of CaM involvement with STRA6. The structure features a large lipophilic cleft on the extracellular side, to which RBP is expected to bind with induction of retinol release. Although additional pathways are possible, a window in the hydrophobic cleft is open to the bilayer, suggesting diffusion as a possible mechanism for translocation of retinol directly into the cell membrane, from which it may be internalized by CRBP1.
Structure determination by single-particle cryo-EM
We screened nine STRA6 orthologs from different species for expression and stability in nonionic detergents as green fluorescent protein (GFP) fusions in transiently transfected human embryonic kidney (HEK) 293 cells. Of these, the protein from zebrafish (75.4 kDa, 670 amino acids) performed best as judged by Western blot and fluorescence size exclusion chromatography (FSEC). We confirmed that recombinant zebrafish STRA6 is active with an HEK293 cell–based retinol uptake assay (13), which used recombinant zebrafish RBP in complex with retinol, and with cotransfection with species-matched CRBP1 and LRAT (Fig. 1A). Protein production was scaled up using baculovirus-infected Sf9 insect cells. Purified STRA6 failed to yield diffraction-quality crystals, despite the monodispersity of the sample (fig. S1A) and its thermal stability in detergents (fig. S1B).
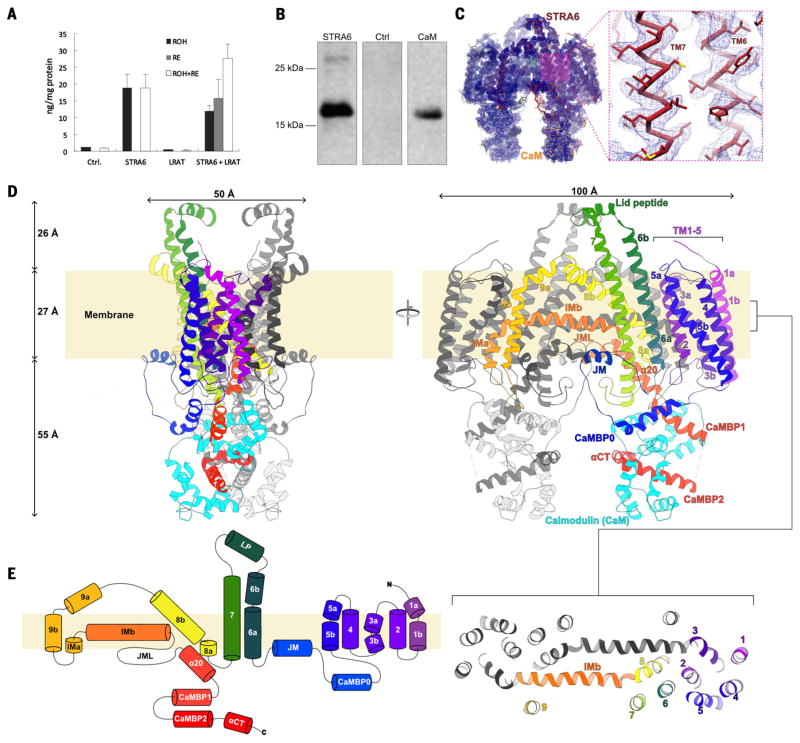
Fig. 1. Function and architecture of zebrafish STRA6 in complex with CaM.
(A) Recombinant zebrafish STRA6 exhibits retinol uptake activity. The results of a cell-based retinol uptake assay are shown as a bar graph; the assay used HEK293T cells that either were stably expressing CRBP1 (Ctrl) or stably expressing CRBP1 and transiently transfected with expression constructs for STRA6, LRAT, or STRA6 and LRAT. The amount of retinol (black bars), the amount of retinyl esters (gray bars), and total retinoid content (white bars) were determined using high-performance liquid chromatography (HPLC). Error bars represent SEM. (B) Zebrafish STRA6 is associated with CaM in a functional context. Shown here is a Western blot probed with an antibody against human CaM on the results from a metal-affinity chromatography–based purification of polyhistidine-tagged zebrafish STRA6, expressed by transient transfection in HEK293T cells. The left lane shows the results of the purification, the middle lane shows purification results for the same cell line transfected with a control plasmid, and the right lane has purified recombinant human CaM loaded as a control. (C) A Cα trace of the STRA6-CaM complex is shown in the density map (blue mesh), contoured at 6x the root mean square. STRA6 is colored dark red and CaM is colored yellow. An inset shows the local quality of the density map in the TM6-TM7 region, with side chains in stick representation. (D) Structure of the STRA6 dimer in complex with CaM. A ribbon representation of the Cα trace of the STRA6 dimer associated with CaM is shown in orthogonal views from within the plane of the membrane. One STRA6 protomer is shown in spectral coloring from violet (N terminus) to red (C terminus). The associated CaM is in cyan. The approximate location of the membrane, estimated using the PPM server (70), is depicted as a cream-colored rectangle behind the proteins. The inset below the right panel depicts the arrangement of TM helices in STRA6 as a slab of the TM region. (E) Schematic representation of the connectivity and structural elements of STRA6, shown using the same color scheme as in (D). The start and end residue number of each helical element is marked. CaMBP0, CaMBP1, and CaMBP2 are the three helical segments of STRA6, which interact with CaM.
SDS–polyacrylamide gel electrophoresis (PAGE) from purified STRA6 preparations always showed the presence of a lower-molecular-weight species (fig. S1C), identified as CaM by N-terminal sequencing, mass spectrometry, and immunoblotting from both mammalian cells, where STRA6 is functional for retinol uptake (Fig. 1, A and B), and insect cells, which were used to produce protein for structure determination (fig. S1D). The sequence of Spodoptera frugiperda CaM, the protein copurified from Sf9 cells, is not available; however, S. littoralis CaM (UniProt ID E3UJZ) is 98% identical to human and zebrafish CaM, which are 100% identical to one another. Removal of CaM was attempted by application of high concentrations of Ca2+ chelators and stringent high-ionic-strength washes of the isolated membrane fraction, but without success, which is consistent with a specific, high-affinity interaction. A molecular weight estimate for the STRA6-CaM complex determined by SEC coupled to light-scattering and refractive index measurements (fig. S1F) indicated that the detergent-solubilized complex contained ~180 kDa of protein and thus was potentially within the tractable molecular weight range of cryo-EM. To this end, the complex was reconstituted in amphipol for structure determination by this technique.
Initial inspection of the two-dimensional (2D) class averages from cryo-EM images revealed a distinctly twofold symmetric assembly, with two “legs” protruding from an ellipsoidal TM region enveloped by an amphipol shell (fig. S2). After 3D classification (fig. S3), based on an initial reference volume generated by common-lines methods (28), the refinement of a set of 56,615 particles produced a cryo-EM density map with an overall resolution of 3.9 Å (fig. S2D and table S1). Inspection of the map allowed unambiguous determination of the TM topology and the location of the N and C lobes of CaM (Fig. 1C and fig. S4, A and B). The quality of the density (fig. S5) was sufficient to allow us to generate an atomic model for 581 of the 670 STRA6 residues (26 to 630; fig. S6) and 147 of the 151 CaM residues (fig. S7 and movie S1).
Architecture of the STRA6-CaM complex
The structure shows that STRA6 is a dimer and that it is associated with CaM, in agreement with our biochemical data (fig. S1, C to F). STRA6 is folded as an intricately associated symmetric dimer, comprising 18 TM helices (nine per protomer) plus two long horizontal intramembrane (IM) helices that interact at the central dimer interface (Movie 1; Fig. 1, D and E; and fig. S4C). The N terminus of STRA6 is extracellular, leading into the first five TM helices, which form an N-terminal helical bundle domain (NTD; Fig. 1, D and E, and fig. S4D). The NTD is separated from the rest of the molecule by a long intracellular loop that connects TM5 to TM6, which contains the first (CaMBP0) of three cytoplasmic helical segments that associate with CaM, as well as a juxtamembrane (JM) helix (Fig. 1, D and E). Two long TM helices (TM6 and TM7) follow centrally, extending more than 25 Å above the extracellular membrane surface in an archlike extension and connected by a short helix-containing loop (lid peptide) proximal to the twofold axis of dimer symmetry (Fig. 1, D and E).
Movie 1. Architecture of STRA6 in complex with CaM.
A ribbon representation of the Cα trace of the STRA6 dimer associated with CaM is shown from within the plane of the membrane, rotating about the dyad axis. One STRA6 protomer is shown in spectral coloring from violet (N terminus) to red (C terminus). The associated CaM is in cyan. The other protomers are shown in black and gray.
TM8 spans the membrane at an angle and docks into the dimer-mate protomer in a TM helix–swapped arrangement that involves TM9 (Fig. 1, D and E). In total, 4811 Å2 of surface area is buried at the dimer interface. Both TM8 and TM9 are kinked sharply after two helical turns. After TM9, the polypeptide chain returns to the body of its own protomer, first through a short vertical IM helix (IMa) and then through a long horizontal one (IMb) (Fig. 1). TM3a of the dimer mate interacts at the IMa-IMb boundary (Fig. 1D and fig. S4C). IMb helices are in antiparallel contact at the dimer interface, approximately at the level of the interface between membrane leaflets (Fig. 1, D and E).
The chain emerges into the cytoplasm after an irregular but well-ordered JM loop (JML) through inclined helix α20, which is intimately associated with the cytoplasmic extensions of TM6 and TM7. Two C-terminal CaM-binding helices (CaMBP1 and CaMBP2) follow, linked by a disordered segment (Fig. 1, D and E). A C-terminal helical extension (αCT) completes the chain after a proline-induced kink (P620). The two CaM molecules, one for each STRA6 protomer, constitute a large portion of the intracellular domain of the STRA6-CaM complex, with each STRA6-CaM interface burying 3124 Å2. In the dimer, these cytoplasmic regions appear as two “legs” that protrude over 50 Å from the membrane surface into the cytoplasm (Fig. 1, C and D).
In essence, the STRA6-CaM architecture can be separated into three distinct regions (fig. S5): (i) the NTD plus the linker containing CaMBP0 and the JM helix; (ii) the central domain (CD), consisting of TMs 6 to 9, the IM, and α20; and (iii) the C-terminal tail, containing CaMBP1 and CaMBP2 and the associated CaM molecule.
STRA6-CaM interaction
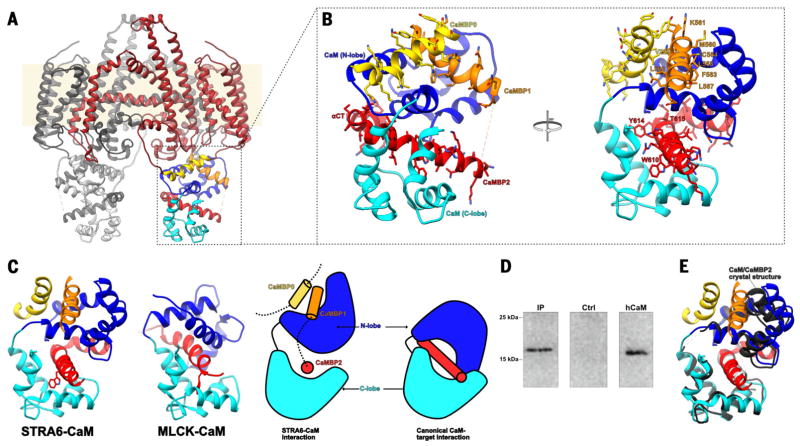
CaM binds to the cytoplasmic side of STRA6 in an unconventional arrangement (Fig. 2A and fig. S9). Overall, the intracellular region of each STRA6 protomer folds as a compact assembly in which CaM is intimately associated with three distinct STRA6 segments (Fig. 2B). The CaM structure that we observed suggests, on the basis of lobe-specific comparisons with previously solved structures of Ca2+-CaM and apo-CaM (fig. S9), that Ca2+ is bound. Ca2+ binding to CaM results in conformational changes that expose two hydrophobic binding surfaces, one in the N lobe and the other in the C lobe (29). Typically, these wrap around a single amphipathic helix (Fig. 2C), although noncanonical CaM complexes have been reported (30). In STRA6, the CaMBP1 helix, which is part of the C-terminal extension, binds exclusively to the main hydrophobic cleft in the N lobe of CaM (Fig. 2, B and C; fig. S10; and movie S2). CaMBP0, which is part of the long TM5-TM6 loop, binds to the N lobe as well, at a surface position between CaM helices 1 and 4, and it interacts additionally with CaMBP1 in a helix-helix crossing mode (Fig. 2, B and C; fig. S10; and movie S2). CaMBP2 interacts with the hydrophobic groove of the CaM C lobe in a manner similar to the canonical 1–5 interaction of the myosin light-chain kinase (MLCK) peptide with the C lobe of CaM (31) (Fig. 2C and fig. S9B), and it links the two lobes of CaM together through interactions with the polar outer face of the CaM N lobe (Fig. 2, B and C; fig. S10; and movie S2).
Fig. 2. The interactions between STRA6 and CaM.
(A) View of the STRA6-CaM complex, shown as a Cα trace in ribbon representation, with STRA6 in dark red except for CaMBP0 (yellow), CaMBP1 (orange), and CaMBP2 (red). The N and C lobes of CaM are shown in dark blue and cyan, respectively. (B) A close-up view of the main interactions between STRA6 and CaM, represented and colored as in (A). All STRA6-CaM–interacting residues are depicted in stick representation and labeled. (C) Comparison between STRA6-CaM and MLCK-CaM (canonical) binding modes in ribbon representation (left) and as a schematic (right), using the same color scheme as in (A). (D) Endogenous zebrafish CaM can be immunoprecipitated using an antibody against zebrafish STRA6. Shown here is an immunoblot probed for CaM. The left lane shows immunoprecipitation from zebrafish tissue performed using the antibody against STRA6, the middle lane shows immunoprecipitation from zebrafish tissue performed using an antibody against an unrelated protein, and the right lane shows recombinant human CaM (10 ng). (E) Superposition of the structures of the Ca2+–hCaM-CaMBP2 complex (black and gray) determined by x-ray crystallography, with the equivalent region from the STRA6-CaM complex, depicted as in (B). The side chains of CaMBP2 are depicted in stick representation. Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr.
To verify that the STRA6-CaM complex is formed under physiological conditions, we performed STRA6 immunoprecipitation experiments on zebrafish tissue by using a monoclonal antibody directed against recombinant zebrafish STRA6 (Fig. 2D and fig. S1E). Furthermore, we performed isothermal titration calorimetry experiments with synthetic CaMBP peptides and recombinant human CaM, which showed that the three main peptides are capable of binding to CaM in solution, with affinities ranging from low micromolar (CaMBP0) to subnanomolar (CaMBP2) (Table 1 and fig. S12). Lastly, to visualize the molecular details of the interaction, we determined the 1.7 Å crystal structure of human Ca2+-loaded CaM in complex with a synthetic CaMBP2 peptide (table S2 and fig. S11). This structure is very similar to that of CaM as observed in the cryo-EM (Fig. 2E), confirming the sequence assignment in this region of the STRA6-CaM cryo-EM structure and providing additional high-resolution structural information for this part of the STRA6-CaM interface (fig. S11). The adoption of a noncanonical arrangement of CaM in the complex with CaMBP2 demonstrates that this segment alone is sufficient to specify the noncanonical conformation of CaM.
Table 1. Isothermal titration calorimetry analysis of the association of CaM with STRA6 peptides.
H, enthalpy; S, entropy; T, absolute temperature (kelvin); G, Gibbs free energy; KD, dissociation constant.
| Peptide (site) | N (sites) | ΔH (kcal/mol) | TΔS (kcal/mol) | ΔG (kcal/mol) | KD (nM) |
|---|---|---|---|---|---|
| CaMBP0* | 1.01 ± 0.07 | −7.8 ± 0.8 | 0.02 ± 0.1 | −7.8 ± 0.7 | 2.56 × 103 ± 722 |
| CaMBP1* | 0.99 ± 0.02 | −7.3 ± 0.1 | 0.10 ± 0.02 | −7.4 ± 0.1 | 1.68 × 103 ± 140 |
| CaMBP2* | 0.90 ± 0.02 | −13.9 ± 0.2 | −0.11 ± 0.03 | −13.7 ± 0.1 | 0.9 ± 0.3 |
| CaMBP1.2† (high) | 0.88 ± 0.02 | −39.4 ± 1.6 | −3.34 ± 0.2 | −36.1 ± 1.4 | 9.5 ± 1.2 |
| CaMBP1.2† (low) | 0.98 ± 0.09 | −7.6 ± 1.0 | 0.06 ± 0.1 | −7.6 ± 1.0 | 1.93 × 103 ± 281 |
The STRA6 peptides were designed on the basis of the STRA6-CaM complex structure and are defined as follows: CaMBP0, residues 222 to 237; CaMBP1, residues 554 to 571; and CaMBP2, residues 600 to 626. Experiments were performed at 37°C in 20 mM HEPES (pH 7.4), 50 mM NaCl, 5 mM MgCl2, and 0.5 mM TCEP, using 30 injections at 10 μl per injection.
This thermogram was fit using a two-site model (termed “high” and “low,” referring to affinity) because of the endothermic region of the thermogram that was observed as the system approached saturation.
The cryo-EM analysis shows the STRA6-CaM complex to be rigid; no conformations differing substantially in the STRA6-CaM arrangement are identifiable by maximum-likelihood 3D classification of the data set (fig. S3). All STRA6 residues interacting with CaM, as well as residues forming the CaMBP1/CaMBP0 interface, are conserved across all species (fig. S7 and movie S2). The disordered CaMBP1-CaMBP2 linker is conserved in sequence for its central and acidic motif, EEGIQLV in zebrafish STRA6, with the Glu-Gly segment invariant across STRA6 orthologs (shown in bold in fig. S7). Lastly, two human STRA6 polymorphisms in CaMBP2, R655C and T644M (equivalent to R626 and T615 in zebrafish, respectively), are strongly associated with MWS (table S3) (32).
Surface features of the STRA6 dimer
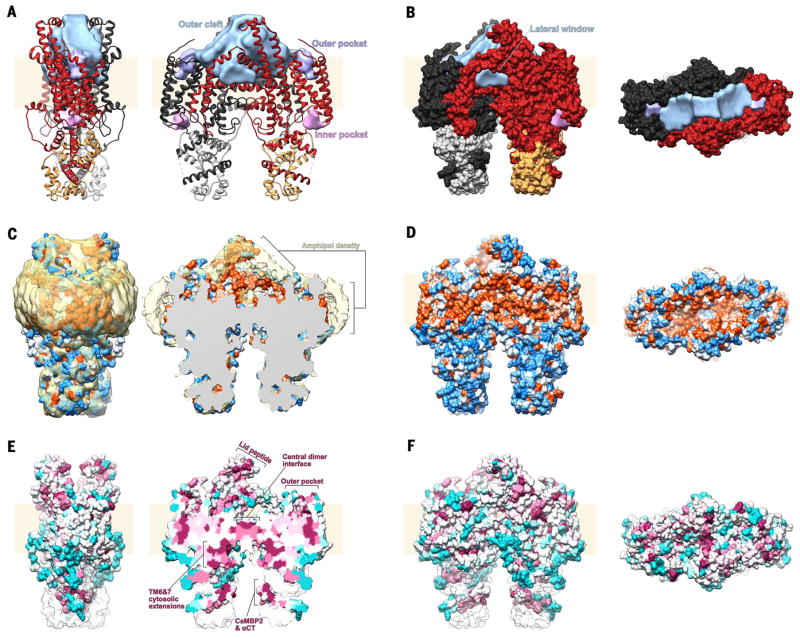
The archlike TM6–lid peptide–TM7 extensions, together with outer portions of the TM8 and TM9 helices, define the sides of a large extracellular cleft (19,174 Å3 in volume; Materials and methods), which we refer to as the outer cleft. This cleft is shaped like an arrowhead, and it extends from a “floor” of dyad-related IM helices at the mid-bilayer level to just under a “ceiling” of lid peptides ~25 Å above the membrane surface (Fig. 3A). The outer cleft is bounded along the short edges by the NTD, which protrudes only slightly above the membrane, defining large openings to the extracellular space at either side of the lid peptides. Toward the base of the outer cleft, TM8, TM9, and the IM helix define a triangular “lateral window” that is open to the lipid bilayer (Fig. 3B). Many of the residues lining the cleft are conserved (fig. S13A) and hydrophobic (Fig. 3, C and D). There is a clear shell of density corresponding to the polar part of the amphipol belt surrounding the TM region (Fig. 3C), as has been observed in other cryo-EM structures of membrane proteins (33, 34). Specific to STRA6, however, a similar shell of density is also located at surfaces bounding the outer cleft (Fig. 3C), suggesting that the hydrophobic alkyl tails of the amphipol occupy the hydrophobic cleft of the purified protein, and that consequently this cleft would be lipid-filled in a native context.
Fig. 3. Surface features of STRA6.
(A) Ribbon representation of the STRA6-CaM complex in two orthogonal views along the plane of the membrane, with one STRA6 protomer shown in dark red and one CaM in gold; the other protomers are shown in black and gray. Surface features are labeled and shown in light blue (outer cleft), purple (outer pocket), and pink (inner pocket). Volumes were calculated using the Voss Volume Voxelator server (63) with an outer probe radius of 10 Å and an inner probe radius of 3 Å. (B) Spacefill representation of the STRA6-CaM complex in two views along (left) and from above (right) the plane of the membrane, colored as in (A). (C) Spacefill representation of the STRA6-CaM complex shown as in (A), colored by hydrophobicity according to the Kyte-Doolittle scale (71), ranging from blue (−4.5, most polar) to white (0.0) to orange (4.5, most hydrophobic). The amphipol layer surrounding the molecule is shown in yellow. The image on the right is a slice from the middle of the outer cleft. (D) Spacefill representation of the STRA6-CaM complex shown as in (B), colored by hydrophobicity as in (C). (E) Spacefill representation of the STRA6-CaM complex shown as in (C), colored by conservation from cyan (least conserved) to maroon (most conserved). (F) Spacefill representation of the STRA6-CaM complex shown as in (B) and (D) and colored as in (E).
Also on the extracellular side, but on the periphery of STRA6, a deep outer pocket (782 Å3 in volume) is formed within the NTD (Fig. 3A). It is somewhat polar in nature (Fig. 3, C and D) and lined by two histidine residues (H41 and H160) along TM1 and TM4 and two tyrosine residues (Y130 and Y131) along TM3, all of which are conserved (fig. S13, B and C). A smaller pocket on the intracellular side of the NTD (Fig. 3A) is lined by histidine (H145 and H86) and tyrosine (Y150 and Y200) residues, although these are less conserved (fig. S13, B and D).
Molecular details of the outer cleft and pockets
The outer cleft is separated from the cytosol by a two-layer structure at the level of the cytoplasmic leaflet of the membrane (Figs. 3A and 4A). The top layer, corresponding to the floor of the outer cleft, is formed by the antiparallel dyad-related IM helices (Figs. 3A and 4A). The bottom layer is formed by the two JMLs, which interact symmetrically with one another and each with the IM helices of the apposed protomers (Figs. 3A and 4A). The two layers are connected by interactions between conserved residues R511 in the IM helix and D539 in the JML. Disruption of this salt bridge by mutation of an equivalent residue in human STRA6 (D560H) results in MWS (35).
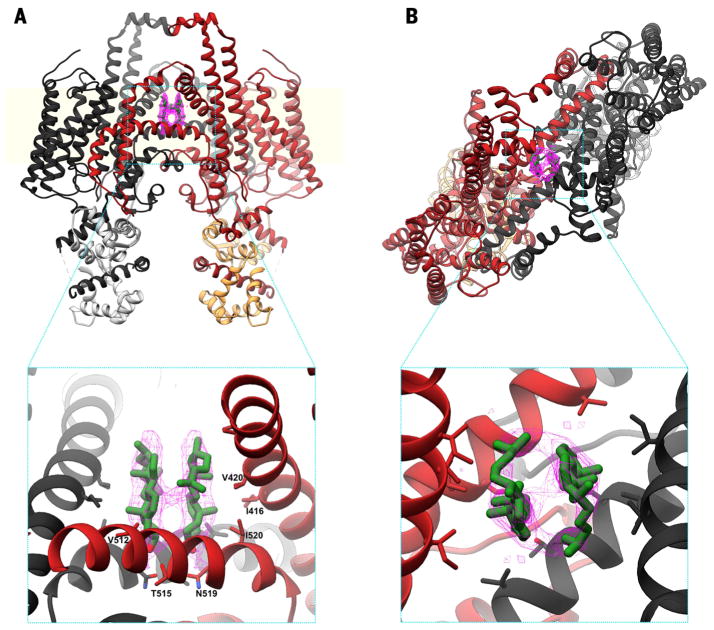
Fig. 4. Putative cholesterol binding sites in the STRA6 outer cleft.
(A) Ribbon representation of the STRA6-CaM complex, shown along the plane of the membrane and colored as in Fig. 3A. Putative cholesterol molecules are shown in green, with the corresponding density in pink, as a mesh. (B) View from above the plane of the membrane, shown and colored as in (A). The two insets show molecular details of the interactions between the putative cholesterols and residues in the STRA6 outer cleft; key interacting residues are labeled.
The two IM helices form mostly hydrophobic contacts, except for a conserved asparagine (N519) and threonine (T515) pair, arranged around the twofold axis (Fig. 4A). The density maps show two lipid-like densities extending into the outer cleft from these polar residues (Fig. 4 and fig. S5). These densities are unlikely to correspond to retinol, given that no retinol was detectable in the purified complex by spectroscopy. Although not conclusive, the shape of the density is consistent with cholesterol (Fig. 4), with each sterol oriented so that its hydroxyl group interacts with the N519-T515 pair. The two-layer structure is closed, and thus conformational changes effecting the separation of the symmetrically related IM helices and JMLs would be required to permit retinol transfer along this route. However, there is no obvious barrier to retinol diffusion from the outer cleft into the lipid bilayer via the lateral window (Figs. 3B and 4A).
Discussion
The unanticipated constitutive association of CaM with STRA6 occurs in vivo, as demonstrated by immunoprecipitation experiments performed at physiological concentrations (Fig. 2D and fig. S1E). CaM is a binding partner and functional regulator for diverse membrane protein families including ion channels, transporters, and enzymes (30). CaM adopts an unusual conformation in which it binds to three STRA6 helical segments (Fig. 2). There is one instance of an EF hand–containing protein with similarly disposed domains, namely, the activation domains of apicomplexan Ca2+-activated, Ca2+-dependent protein kinases (36) (fig. S9A). A comparable arrangement has not been observed for CaM itself, although distinct noncanonical CaM-target complexes have been characterized, such as, for example, the structure of the complex of CaM with the anthrax edema factor toxin (37). We determined the 1.7 Å–resolution crystal structure of Ca2+-CaM in complex with CaMBP2 (Fig. 2E and fig. S11), the STRA6 peptide with the highest affinity (0.9 nM). Notably, CaMBP2 alone induces the same noncanonical conformation of Ca2+-CaM as observed in the cryo-EM structure of STRA6-CaM. The seeming obligate nature of the CaM-STRA6 interaction explains previous observations that the C-terminal tail of the receptor is structurally and functionally indispensable (15). Several MWS mutations are located in this region (table S3), and random mutagenesis of STRA6 identified multiple mutations in the C-terminal tail that eliminate surface expression and prevent correct folding of the receptor (38). The role of CaM—and its regulation by Ca2+—is enigmatic because no direct link between Ca2+ and retinol transport has been identified. Nevertheless, this interaction is conserved and structurally essential.
The CaM-interacting regions, as well as several other structural elements of STRA6, are conserved in RBPR2, a STRA6-like protein expressed primarily in the liver and intestine (39) (fig. S14), suggesting a similar architecture with potential functional differences in these specialized retinol storage and uptake tissues. In humans, the NTD of RBPR2 is expressed as a separate polypeptide (39), supporting our assignment of the NTD as a separable domain (fig. S5). More distantly related lipocalin receptors such as LMBR1 also have nine predicted TM helices and have been suggested to form dimers (40), and consequently we anticipate they may adopt a STRA6-like fold.
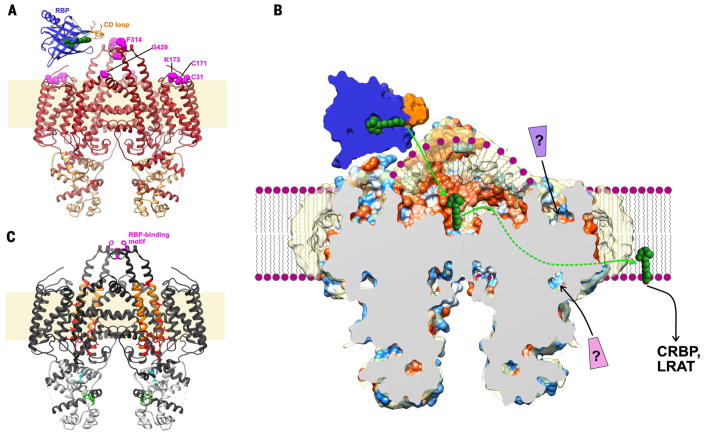
The most notable feature of STRA6 is the large extracellular-facing hydrophobic cleft, which spans from the mid-bilayer to the lid peptides at the apex of the TM6–lid peptide–TM7 arch, 25 Å above the membrane surface (Fig. 3A). Human STRA6 residues that were implicated previously in RBP interactions (314 to 320, zebrafish numbering) by functional mutagenesis studies (38) all map to the lid peptide (Fig. 5A), thus putting constraints on the position of RBP in its interaction with STRA6. Furthermore, insertion of a Myc tag at the apex of the TM8-TM9 loop has been shown to impair RBP binding while not affecting surface expression of STRA6 (15). Mutants that disrupt a predicted C31-C171 disulfide bond within the NTD also show reduced RBP binding (15). The regions with impaired RBP binding, together with prior work on RBP itself that showed dependence on its CD loop for interaction with the lid peptide (11, 41, 42), allow us to place constraints on the location of the RBP-binding site (Fig. 5A). On the basis of this orientation and the hydrophobic nature of both retinol and the STRA6 outer cleft, it is reasonable to postulate that RBP binding to the lid peptide weakens RBP affinity for retinol, leading to deposition of retinol in the STRA6 outer cleft. It remains to be determined whether the catalysis of retinol release is primarily due to induced structural changes in the retinol exit pathway of RBP or to the enforced proximity of the retinol to the surface of the lipid bilayer.
Fig. 5. Possible mechanism for STRA6-mediated retinol uptake.
(A) Ribbon representation of the STRA6-CaM complex, viewed from the plane of the membrane, with STRA6 colored in red and CaM in gold. Residues previously shown to be important for RBP binding are shown as magenta spheres. Green spheres, retinol. (B) Schematic of STRA6-mediated retinol release from RBP into the outer cleft and translocation to the lipid bilayer (shown as purple spheres and wavy lines) through the lateral window. Question marks indicate putative ligands binding to the NTD; green and white arrows show two potential retinol exit pathways, via the lateral window and the central dimer interface, respectively; and coloring of the STRA6 surface is as in Fig. 3C (by hydrophobicity). (C) Previous work mapped onto the STRA6 structure, shown as follows: the RBP-binding motif in the lid peptide in magenta (38); the region probed by acute chemical modification of single cysteine point mutants (48) in orange, and residues in this region, modification of which severely affects retinol uptake from RBP, in red; the putative CRBP1 interacting motif (45), located in CaMBP0, in cyan; and the putative STAT5 binding motif (22), located in CaMBP2, in green.
The outer cleft is hydrophobic, it extends down about halfway through the bilayer, and it is exposed to the membrane via the lateral window (Fig. 3B). Furthermore, the outer cleft is covered by a shell of density, presumably from the polar backbone of the amphipol (Fig. 3C). These observations suggest that the outer cleft is occupied in vivo by bulk lipid rather than aqueous solvent. Such an environment could provide a low-energy pathway for retinol to partition into the lipid bilayer. The lipid composition within the outer cleft remains to be determined; however, broadly speaking, we envision two likely possibilities. In one, the content of the outer cleft could be similar to that of the surrounding membrane, namely, a distorted monolayer of phospholipids (Fig. 5B). Alternatively, it could be enriched in specific components such as cholesterol, perhaps similar to what is observed in lipid rafts (43).
Consistent with the above, we observed several distinct densities within this cleft, two of which we have tentatively assigned as cholesterol (Fig. 4). These could either originate from the expression host or represent hydrolysis products of the cholesteryl hemisuccinate used to stabilize the protein during purification (fig. S1B). The two putative cholesterol molecules are bound by their hydroxyl groups to the conserved T515-N519 site at the floor of the outer cleft. The fact that retinol, like cholesterol, is also hydrophobic with an apical hydroxyl group raises the possibility that the observed positions of the putative cholesterol molecules may also represent binding sites for retinol.
As noted in the previous section, the outer cleft is separated from the cytosol by a two-layer protein barrier at the level of the cytoplasmic leaflet of the membrane (Figs. 3 and 4). Because there is no evidence of a pore through which retinol could translocate via this central structure, a conformational change would be required. However, the structure suggests an alternative passageway for retinol by direct diffusion into the membrane through the lateral window (Fig. 5C). This hypothesis is consistent with the observation of (i) CRBP- and LRAT-independent STRA6-mediated retinol uptake from holo-RBP, (ii) STRA6-independent uptake of retinol from the membrane by CRBP1, and (iii) CRABP- and STRA6-dependent internalization of RA from a RA-RBP complex (44). Furthermore, retinol in the plasma membrane inhibits STRA6-dependent uptake of retinol from holo-RBP (17), raising the possibility of a mechanism of exchange between membrane-and RBP-bound retinol. The lateral window could provide a key element for such a mechanism.
The evidence for direct association of CRBP with STRA6 is ambiguous (19, 45). Residues I232 and L233, which have been implicated in CRBP-binding (45), are located in our structure within CaMBP0, where they interact directly with CaM (Fig. 5C). Although the proposed mechanism of retinol uptake through the lateral window does not require direct binding of CRBP1 to STRA6, it does not preclude this interaction, which could also play a regulatory role. The STRA6 structure also suggests likely sites of interaction with other cytoplasmic partners. CaMBP1 and CaMBP2 are connected by a linker, that is disordered in the structure and that contains a conserved sequence motif (EEGIQLV; fig. S7) similar to the box2 motif of cytokine receptors, which is known to mediate recruitment of JAK family members through their FERM and SH2 domains (46, 47). Furthermore, two residues (Y614 and T615) that have been implicated in JAK-dependent regulation of STRA6 (45, 48) are located on CaMBP2 (Fig. 5D). These two residues are not solvent-accessible in our current structure, implying that a conformational change of either CaM or STRA6 would be required for their modification.
A previous study involving scanning cysteine mutagenesis of TM6 and TM7 identified a number of modifications that severely inhibited retinol uptake (48). Our structure shows that these residues are clustered near the cytoplasmic ends of these helices and that most are buried (Fig. 5D), precluding direct involvement in retinol translocation absent a substantial conformational change.
The N-terminal TM1-TM5 bundle may represent a regulatory module, which could potentially act as a sensor for an unidentified ligand. The NTD contains conserved polar pockets on both sides of the membrane (Fig. 3A and fig. S13), which could constitute ligand-binding sites. Free retinoids are known to stimulate RBP-dependent, STRA6-mediated retinol uptake activity (49), making them plausible candidates for such a role. Interestingly, TM3 packs against the IMa helix of the adjacent protomer. Because IMa is contiguous with the IMb helix that lines the outer cavity floor, it is conceivable that a ligand-induced change in NTD conformation could affect the efficiency of STRA6-mediated retinol release from holo-RBP and/or translocation.
Overall, the structure of STRA6 presented here is consistent with a model of retinol release from RBP into the lipid-filled outer cleft and its direct transfer into the membrane by diffusion through the lateral window (Fig. 5C). The release and transfer of ligands into specialized lipidic environments could represent a general mechanism for protein-mediated uptake of hydrophobic molecules.
Materials and methods
Expression constructs and small-scale expression screening
Nine STRA6 orthologs (human, mouse, chicken, zebrafish, dog, horse, bovine, rat, pig) were cloned into pFM1.2 (50) with a GFP and decahistidine tag engineered to express as a fusion at either and both the N- or C- terminus of the proteins. The constructs were transiently transfected into adherent HEK293T cells using lipofectamine (Invitrogen). The transfected cells were harvested after incubation for 72 hours at 37°C. The harvested cell pellets were lysed and solubilized for 1 hour on ice in a buffer containing n-dodecyl-β-D-maltopyranoside (βDDM). Samples were ultracentrifuged to remove debris, protein concentration determined by Bradford assay (BioRad) (51), and 20 μl of supernatant was loaded on a SDS-PAGE for Western blot, which was then probed with anti-GFP antibody (Invitrogen) and detected using Western Blot Luminol reagent (Santa Cruz). The expression levels and stability of the constructs were further assayed by FSEC. 10 ml of Freestyle 293-F cell culture (Invitrogen) was transiently transfected with each construct at a density of 1 × 106 cells per ml using polyethyleneimine (PEI) (Polysciences), grown in shaker flasks for 72 hours at 37°C and harvested by centrifugation. Cell pellets were treated following the same protocol as described above, and the supernatant was injected onto a Superdex 200 Increase 10/300 GL (GE Healthcare Life Sciences) SEC column connected to a Prominence UFLC system (Shimadzu) fitted with an RF-10AXL fluorescence detector (Shimadzu).
Large-scale protein expression and membrane preparation
STRA6 from zebrafish (Danio rerio) was cloned into pIEX/Bac-1 plasmid modified to include a C-terminal decahistidine tag and thrombin cleavage site and transfected together with baculovirus bacmid DNA (Sapphire Baculovirus DNA and Transfection kit, Allele Biotech) into Spodoptera frugiperda Sf9 cells for generation of recombinant baculovirus. The recombinant virus was amplified to high titer following standard procedures. Sf9 cells were infected at a density of 2 × 106 cells per ml with high titer virus at an approximate multiplicity of infection (MOI) of 1, grown in shaker flasks for 72 hours at 27°C and harvested by centrifugation. Cell pellets were resuspended in low salt buffer consisting of 10 mM HEPES pH 7.5, 10 mM KCl, 10 mM MgCl2, 0.5 mM PMSF, EDTA-free complete protease inhibitor cocktail (Roche), DNase, and RNase and lysed by gentle brane fractions were isolated by ultracentrifugation, resuspended by homogenization and washed a minimum of two times, until the supernatant following ultracentrifugation was clear, in high salt buffer (10 mM HEPES pH 7.5, 10 mM KCl, 1 M NaCl, 10 mM MgCl2, 0.5 mM PMSF, EDTA-free complete protease inhibitor tablet DNase, RNase). Extensively washed membranes were then resuspended again by homogenization in buffer containing 30 mM HEPES, pH 7.5, 200 mM NaCl, 0.5 mM PMSF, protease inhibitor cocktail III (Millipore) and stored at −80°C until use.
Protein purification
The membrane fraction was thawed on ice and solubilized by the addition of detergent—lauryl maltose neopentyl glycol (LMNG) or βDDM with or without cholesteryl hemisuccinate (CHS) in a 10 to 1 ratio—to a final concentration of 1% (w/v) detergent and 2 mg/ml protein, and gently agitated for 1.5 hours at 4°C. Insoluble matter was removed by ultracentrifugation, the supernatant was transferred to fresh tubes and imidazole added to a final concentration of 40 mM. The sample was then added to pre-equilibrated Ni2+-NTA resin (Qiagen) and the mixture allowed to gently rotate overnight at 4°C. After transferring the mixture to a column, the resin was washed with 10 column volumes of buffer containing 60 mM imidazole, and the protein was eluted with buffer containing 200 mM imidazole. The eluted protein was loaded on a Superdex 200 Increase 10/300 GL SEC column (GE Healthcare Life Sciences) pre-equilibrated with buffer containing 20 mM HEPES pH 7.0, 150 mM NaCl, and detergent (LMNG or βDDM) at twice the critical micellar concentration (CMC). Typically, final yields ranged from 0.7 to 1mg purified protein per liter of Sf9 cells.
Light-scattering and refractive index measurements
Recombinant STRA6 was purified by metal affinity chromatography in buffer containing 20 mM HEPES, pH 7.0, 200 mM NaCl, 0.025% (w/v) βDDM, 0.0025% (w/v) CHS and subjected to SEC coupled to UV, dynamic light-scattering, and refractive index detectors to determine its molecular weight (52). SEC was performed on a FPLC (SEC-MALS) system attached to a DAWN HELEOS Multi Angle Light Scattering (MALS) detector (Wyatt technology). Light-scattering data were collected for multiple scans, and molecular masses and relative contributions to size were determined using ASTRA software (Wyatt technology).
Thermostability of STRA6 in detergent
STRA6 was extracted from purified membranes by addition of βDDM or LMNG with or without CHS. After an initial purification step by metal affinity chromatography, each sample was injected on a small-scale analytical SEC column (Superdex 200 5/150GL, GE Life Sciences) to determine conditions that yielded a sharp monodisperse peak. These optimized conditions were subsequently purified protein was desalted using a PD miditrap G-25 column (GE Healthcare) and separate aliquots heated for 10 min over a range of temperatures from 25° to 70°C. After centrifugation to remove aggregated or precipitated protein, the supernatant was injected on an analytical SEC column in buffer containing 20 mM HEPES pH 7.0, 150 mM NaCl, and detergent with or without CHS. Thermal denaturation curves were constructed by measuring the height of the SEC peak measured from samples at each of the different temperatures (fig. S1D). The height of the SEC peak for sample maintained at 4°C was used as control for normalization. The thermal denaturation curves were fitted with a Boltzmann sigmoidal equation using SigmaPlot (SYSTAT Software).
Protein preparation for cryo-EM
STRA6 was purified in buffer containing LMNG and CHS in a 10:1 ratio (w/w). After elution from Ni2+-NTA resin, the protein was desalted and reconstituted into amphipol A8-35 (Anatrace) at a ratio of 1:3 by weight. The mixture was incubated for 4 to 5 hours with gentle agitation at 4°C and the detergent was removed by the addition of Bio-Beads (Bio-Rad) and overnight incubation at 4°C. The reconstituted protein was then loaded on a Superdex 200 Increase 10/300 GL SEC column (GE Healthcare Life Sciences) equilibrated in buffer without detergent, to remove free amphipol and residual detergent. The eluted protein from the peak fractions (at ~0.6 mg/ml) was used without further concentration for cryo-electron microscopy as described below.
Zebrafish immunoprecipitations
Zebrafish heads (3 days post fertilization) were resuspended in lysis buffer consisting of 10 mM HEPES pH 7.5, 10 mM KCl, 10 mM MgCl2, 0.5 mM PMSF, EDTA-free complete protease inhibitor cocktail (Roche) and triturated using a syringe attached with a 25 gauge needle. Membranes were isolated by ultracentrifugation and solubilized at 1.5 mg/ml protein concentration in βDDM for 2 hours, and the insoluble material was cleared by ultracentrifugation. For each point, a volume corresponding to ~500 mg of solubilized protein was diluted to a final volume of 1 ml with 100 μl of supernatant from a hybridoma culture, 10 μl of a 50:50 pre-equilibrated mixture of protein A–coupled and protein G–coupled Sepharose resin (GE healthcare), and buffer containing 0.1% βDDM. Assays were incubated overnight at 4°C under gentle rotation, washed three times with 1 ml of 20 mM HEPES pH 7.5, 150 mM NaCl, 0.1% βDDM, and eluted with 50 μl of 100 mM glycine pH 2.5, 0.1% βDDM. 45 μl of the sample was recovered and the pH neutralized by transferring to a fresh tube containing 5 μl of 1.5 M Tris-HCl pH 8.8.
Grid preparation and data collection
3 μl of the central fraction of the peak (0.68 mg/ml) was applied to the surface of a holey gold grid, prepared in-house from Quantifoil R1.3 carbon grids according to the procedure described by Russo and colleagues (54). The grid was blotted on both sides using Whatman ashless filter paper for 3.5 to 4 s at a blot force of 3 in a Vitrobot Mark IV (FEI) at 100% humidity and 4°C, then plunged in cooled liquid ethane after a wait time of 30 s. 2599 micrographs were collected, as dose fractionated stacks (71 frames with a total nominal dose of 100 e−/Å2) on a F30 Polara microscope (FEI) operated in electron counting mode, at a nominal magnification of 31000×, with a calibrated pixel size of 1.255 Å at the specimen plane. A single stack was collected from each hole (fig. S2G). Pixel size was calibrated by measuring real space correlation of a known crystal structure [Protein Data Bank (PDB) ID 2XOA; N-terminal 3 domains of RyR1] to maps calculated with a range of voxel sizes for a ryanodine receptor (RyR1) data set collected on the same microscope at the same nominal magnification. After gain correction, frame alignment and dose correction were performed in UNBLUR (55), and CTF estimation in CTFFIND4 (56) (fig. S2F). 2196 micrographs remained after screening based on visual inspection of the aligned averages and power spectra. 3000 particles were picked manually in RELION 1.3, extracted and subjected to 2D classification. Six classes representative of the most common views of the molecule were used (with application of a 30 Å lowpass filter) to generate templates for autopicking as implemented in relion_autopick (57). ~300,000 autopicked particles were subjected to extensive 2D classification to eliminate poor-quality particles and nonprotein contaminants. 20 high-quality 2D class averages were used to generate candidate initial models as references for refinement using EMAN2 (e2initialmodel.py) (28). After an initial trial refinement of the whole set of particles in RELION 1.3 without imposition of symmetry (in which secondary structural elements were clearly visible), 3D classification (K = 4, T = 4) was performed without the imposition of symmetry, and was used to identify a class of 70,000 particles with improved homogeneity (fig. S3), and with evident C2 symmetry. Refinement of this class with application of C2 symmetry yielded a map with final resolution of 4.18 Å according to the Fourier shell correlation criterion using the “gold standard” protocol (58). Inspection of the density map revealed features consistent with the estimated resolution, including clearly evident helical pitch throughout the map, and well-defined density for bulky side chains (fig. S5). At this point, it was evident that the reference model was of the wrong hand (the alpha helices were left-handed), and so the correct hand was obtained by reflecting the map about the X-Y plane using EMAN2 (e2proc3d.py). After the initial reconstruction, 3D classification was rerun with C2 symmetry enforced (K = 4, T = 4), allowing identification of a further improved class of 56,614 particles, which were corrected for the motion of individual particles using alignmparts_lmbfgs with an estimated dose of 1.25 e− per frame. Refinement of this improved set of polished particles using the 3D-auto-refine procedure of RELION 1.3 yielded a reconstruction with an estimated resolution of 3.9 Å. The mask used for Fourier shell correlation calculations was generated by creating a 10 Å low pass filtered map from the atomic model using the molmap command of UCSF Chimera, and using relion_mask_create to generate a soft-edged mask from this map. The map was low pass filtered at 3.5Å and a negative B-factor of −113.68 was applied to enhance visibility of high-resolution features using relion_ postprocess. Analysis of the orientational distribution of particles contributing to the final reconstruction demonstrates that STRA6 appears to adopt a preferred orientation under the conditions used for data collection, with the view parallel to the membrane so that both CaM “legs” are visible being the most common (fig. S2A).
Model building and refinement
Model building was initiated with the placement of the N and C lobes of CaM. Both apo-CaM and Ca2+-bound CaM structures were tested; Ca2+-bound CaM structures showed an excellent fit to the density map for both lobes, whereas no apo-CaM structure matched the density. Initial orientations of the N and C lobes (derived from the structure of human Ca2+-CaM, PDB ID 1CDL, and mutated in silico to match the sequence of Spodoptera littoralis CaM, UniProt ID E3UJZ8) were obtained using the Jiggle-fit routine of COOT (59). Initial model building of STRA6 was informed by previous experimental studies of the transmembrane topology (15), computational predictions of secondary structure and disordered regions using the Xtalpred server (60), and the spacing of bulky hydrophobic residues in the sequence, which are especially recognizable in the density map. Model building was performed manually in COOT (59), alternating with cycles of automated real-space refinement in phenix. real_space_refine (61). All structural figures were prepared using UCSF Chimera (62). Analysis of clefts and pockets was performed using the Voss Volume Voxelator (3V) server with an outer probe radius of 10 Å and an inner probe radius of 3 Å (63). Sequence alignments were performed using MUSCLE (64) and all sequence alignment figures were prepared using Jalview (65).
Cross validation and model-map FSC calculations
Potential effects of overfitting in the final refined model were assessed by the following procedure. Two maps were calculated from random half sets of particles using relion_reconstruct; post-processing was performed with the same parameters as for the original full map using relion_postprocess. One of these maps (the “refine” half-map) was used to refine the final model, with all atoms randomly displaced by 0.2 Å (phenix.pdbtools sites.shake=0.2) to reduce the influence of overfitting on the initial model. The final model was used to generate a “model map” using phenix.model_map, which, after both maps (the model map and the test/refine/full maps) were masked using the same soft mask as used for resolution calculations, was used for FSC calculations performed with EMAN2.11 (e2proc3d.py–calcfsc). The FSC curves calculated thusly are presented in fig. S6B.
Mammalian cell culture and generation of stable lines
HEK293 cells were maintained at 37°C, in a humidified environment enriched with 5% CO2. HEK293 cells were grown in DMEM (Gibco) supplemented with 10% FBS (Gibco), penicillin, streptomycin, and L-glutamine (Pen/Strep/L-Glu; Gibco). Zebrafish CRBP1 was cloned into pFM1.2 and RBP was cloned into pFM1.2R (50) with a hexahistidine tag appended to the C terminus. Each plasmid was mixed with plasmid carrying a puromycin resistance gene (1:5) before cotransfection using lipofectamine (Invitrogen) or PEI (Polysciences) into 293 GnTi− cells (66). Stable integrants were selected by addition of 5 μg/ml puromycin to the growth medium 24 hours after transfection. Production cell lines from fluorescent colonies were selected either by FACS sorting in Autoclone mode and subsequent visual inspection of the resulting colonies, to identify single, highly fluorescent colonies, or by manual picking of the most intense fluorescent colonies after antibiotic selection (50). Expression levels of selected colonies were checked by small-scale purification tests—performed by metal-affinity chromatography—to select the best lines for expansion.
Zebrafish RBP expression and purification
The stable mammalian cell line secreting RBP was maintained in DMEM supplemented with 10% FBS, Pen/Strep/L-Glu, and 5 μg/ml puromycin. Media was harvested from scaled-up cultures as monolayers in 225-cm2 culture flasks (Costar) after 10 days. After centrifugation to remove cell debris, the media was adjusted to 30 mM HEPES, pH 7.5, 50 mM NaCl, and 5 mM imidazole for purification by metal affinity chromatography. After binding to equilibrated Ni2+-NTA resin for 30 min, the slurry was transferred to a column, washed with 15 column volumes of buffer containing 10 mM imidazole, and the protein was eluted with buffer containing 200 mM imidazole. The eluted protein was desalted using PD MidiTrap G-25 (GE Healthcare) and purity was assessed by SDS-PAGE.
Holo-RBP preparation
Purified zebrafish RBP was precipitated in ice-chilled 100% ethanol to remove endogenous retinol. After ultracentrifugation, the precipitated RBP was resuspended in denaturing buffer (50 mM Tris pH 8.0, 6.6 M guanidine hydrochloride). After incubation at room temperature for 30 min, the sample (1 ml at 5 mg/ml) was slowly added drop-wise to a 10 ml ice-cold refolding buffer (0.4 M L-arginine, 20 mM HEPES pH 7.5, 100 mM NaCl) containing 0.1 mM all-trans retinol (Sigma). All the experimental steps were carried out in the dark and samples were protected from light at all times from this point onwards. The refolding mixture was gently agitated overnight at 4°C. After centrifugation to remove aggregates, the sample was concentrated to 1 ml, loaded on a desalting column (PD MidiTrap G-25, GE Healthcare) and further purified by SEC (Superdex 200 10/300, AKTA, GE column) to remove free retinol. The concentration of eluted fractions was determined by Bradford assay (BioRad) (51). The success of retinol incorporation was assessed by measuring the absorption spectrum of the protein samples (Beckman Coulter DU730) at 260 to 400 nm and confirmed by HPLC (details described below in cell-based retinol uptake assay section).
Cell-based retinol uptake assay
Parental HEK293 cells or a cell line stably expressing zebrafish CRBP1 were seeded in six well plates at 0.5 × 106 cells per well the evening before the experiment and expression constructs transfected in triplicates. 72 hours after transfection, the media was replaced with serum free DMEM and incubated for an additional 16 hours in standard cell-maintenance conditions. Holo-RBP was added to a final concentration of 1 μM to the medium, and cells were maintained as such for 6 hours in standard cell-maintenance conditions. To detect retinol uptake, holo-RBP–containing media was removed from the cells, and these were gently washed three times with PBS and harvested by centrifugation at 800 × g. Cell pellets were resuspended with 300 μl of PBS, frozen on dry ice and stored at −70°C. Cells were lysed by repeated freeze-thaw cycles. 20 μl of the cell suspension was used to determine protein concentration by Bradford assay (51) (BioRad), and the remaining 280 μl was extracted with hexane followed by loading on HPLC run in reverse phase. Retinol and retinyl ester peaks were separated on a 4.6 × 250 mm Denali C18 column (Grace, Deerfield, IL) preceded by a C18 guard column (PerkinElmer, Waltham, MA) using acetonitrile, methanol, and methylene chloride (70:15:15, v/v) as the mobile phase flowing at 1.8 ml/min. Quantitative analysis was performed by comparing retention times and spectral data of experimental compounds with those of authentic standards.
Peptides
All peptides were synthesized and purchased from Bio-Synthesis. Peptide sequences corresponding to regions in zebrafish STRA6 were as follows: CaMBP0. residues 222 to 237 (EDLSSSYY RDYVKKILKKKK); CaMBP1, residues 554 to 571 (SQSHPVMKAFCGLLLQSS); CaMBP2, residues 600 to 626 (VSNAKRARAHWQLLYTLVNNPSLVGSR); CaMBP1_2, residues 554 to 571 and 601 to 626 separated by a flexible 8-resiude linker (SQSHPVMKAFCGLLLQSSGGGEGGGGSNAKRARAHWQLLYTLVNNPSLVGS R). The peptides were protected using N-terminal acetylation and C-terminal amidation, and purity was confirmed by HPLC and amino acid analysis.
Purification of human CaM
The vector for human CaM was generously supplied by L. M. Amzel at Johns Hopkins University in a pET24 plasmid without an affinity tag, which was transformed and expressed in Escherichia coli strain BL21(DE3) using 0.5 mM IPTG at 37°C. Bacterial cell pellets were resuspended in Buffer A (50 mM Tris, pH 7.5, 1 mM DTT) and lysed using a combination of lysozyme (0.1 mg/ml) and sonication. The lysate was clarified via centrifugation at 15,000 rpm. The resulting protein solution was extensively dialyzed against Buffer B [50 mM Tris, pH 7.5, 10 mM β-mercaptoethanol (β-ME)] and applied to a diethylaminoethanol (DEAE) sepharose column (Sigma-Aldrich) for initial purification. The protein was eluted from the DEAE column using a linear gradient of 10 to 35% Buffer C (50 mM Tris, pH 7.5, 10 mM β-ME, 1 M NaCl). After pooling the protein fractions, the sample was dialyzed extensively against Buffer D (10 mM Tris, pH 7.5, 500 mM NaCl, 10 mM CaCl2, and 0.25 mM DTT) and applied to a Phenyl Sepharose column (Sigma-Aldrich). The protein was eluted from the column using a single step of 100% Buffer E (10 mM Tris, pH 7.5, 500 mM NaCl, 10 mM EDTA, and 0.25 mM DTT). As a final purification step, the pooled Phenyl Sepharose fractions were concentrated to <5 ml and loaded onto a Superdex 200 (S200-PG) SEC column (GE Healthcare) and eluted using Buffer F {20 mM HEPES, pH 7.4, 50 mM NaCl, 5 mM MgCl2, 1 mM TCEP [tris(2-carboxyethyl)phosphine], and 0.02% NaN3} as mobile phase. Throughout the purification process, the fractions containing the protein were identified using a combination of Bradford Assay and SDS-PAGE, and the final protein sample purity was assessed using SDS-PAGE and native PAGE (>99%). Pure protein was concentrated to 20 mg/ml, flash-frozen, and stored in 0.5 ml aliquots at –80°C.
Isothermal titration calorimetry
All isothermal titration calorimetry experiments were performed using a VP-ITC system obtained from MicroCal (Northampton, MA). Samples were extensively dialyzed against Buffer G (20 mM HEPES pH 7.4, 50 mM NaCl, 10 mM CaCl2, 5 mM MgCl2, and 0.5 mM TCEP), and experiments were performed at 37°C. Initial experiments had CaM in the syringe at a concentration of 150 to 500 μM with each STRA6 peptide at a concentration of 15 to 50 μM in the reaction cell. A series of 30 injections of 10 μl of protein solution was performed at 5-min intervals. The reverse titrations with CaM in the reaction cell at concentrations of 15 to 20 μM and peptide in the syringe at concentrations of 150 to 200 μM were also performed for confirmation of the initial results. The data were processed and analyzed with the MicroCal Origin 7 software and corrected by the heat of injection from the basal heat remaining after saturation and confirmed by titration into buffer only as a control. For CaMBP0 and CaMBP1, a one-site binding mode was used to fit the data, using a nonlinear least-squares algorithm, whereas CaMBP2 was fit using a two-site binding mode with a Levenberg-Marquardt nonlinear regression model, due to the endothermic interval before saturation (or initial binding interval in the reversed titration).
X-ray crystallography on CaM-CaMBP2
Crystallization experiments were performed using sitting-drop vapor diffusion methods as follows: CaM-CaMBP2 crystals were set in drops of 1.0 μl protein solution containing 20 mg/ml CaM, 1.2 mM CaMBP2, 20 mM HEPES pH 7.4, 50 mM NaCl, 10 mM CaCl2, 5 mM MgCl2, and 0.5 mM TCEP with 0.8% (v/v) DMSO with 1.0 μl mother liquor containing 0.1 M imidazole pH 5.5, 28% (v/v) polyethylene glycol monomethyl ether 550. Crystals formed over the course of 1 to 10 days at a temperature of 281 K before they were harvested. No additional cryo-protection was used before flash-cooling into liquid nitrogen. Diffraction data were collected remotely at the Northeastern Collaborative Access Team using beamline 24ID-E at the Advanced Photon Source (Argonne National Laboratory) at 100 K using an ADSC Q315 (315 mm × 315 mm) detector. A 1.74 Å data set was obtained at a wavelength of 0.97919 Å with 1.0° oscillation for each frame and processed then integrated using Mosflm (67) with a space group of C121. Diffraction statistics are summarized in table S2. Model building was started using molecular replacement with a search model derived from the CaM C lobe of a previously determined structure (PDB accession code 1ZUZ) using the PHENIX (61) AUTOMR (68) function. The models were completed via manual building within COOT (59) with peptide, ligands, and water incorporation guided by the m|Fo| - D|Fc| omit maps and iteratively refined with the PHENIX. REFINE (69) function. All structure refinement statistics can be found in table S2.
Supplementary Material
Acknowledgments
This work was supported by a NIH National Institute of General Medical Sciences initiative to the New York Consortium on Membrane Protein Structure (U54 GM095315). O.B.C. was supported by a Charles H. Revson Senior Fellowship. We thank G. Sciara and G. Ahlsen for sharing with us their expertise for the light-scattering experiments; F. Insaidoo, L. A. White, I. Ubarretxena, Y. Gomez, S. Brenner-Morton, and A. Nemes for their contributions during the initial stages of this work; B. Kloss, R. Bruni, and R. Kalathur from the New York Structural Biology Center for their many contributions to the project; R. Grassucci for assistance with data collection; A. des Georges, A. Marks, and R. Axel for advice and helpful discussions; and L. Hamberger for assistance with running the Mancia laboratory. The CaM-CaMBP2 structure has been deposited in the PDB under accession code 5K8Q. The STRA6-CaM coordinates have been deposited in the PDB under accession code 5SY1, and the map has been deposited in the Electron Microscopy Data Bank under accession code EMD-8315.
Footnotes
REFERENCES AND NOTES
- 1.Palczewski K. Chemistry and biology of vision. J Biol Chem. 2012;287:1612–1619. doi: 10.1074/jbc.R111.301150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al Tanoury Z, Piskunov A, Rochette-Egly C. Vitamin A and retinoid signaling: Genomic and nongenomic effects. J Lipid Res. 2013;54:1761–1775. doi: 10.1194/jlr.R030833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goodman DS. Vitamin A and retinoids in health and disease. N Engl J Med. 1984;310:1023–1031. doi: 10.1056/NEJM198404193101605. [DOI] [PubMed] [Google Scholar]
- 4.Shirakami Y, Lee SA, Clugston RD, Blaner WS. Hepatic metabolism of retinoids and disease associations. Biochim Biophys Acta. 2012;1821:124–136. doi: 10.1016/j.bbalip.2011.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.di Masi A, et al. Retinoic acid receptors: From molecular mechanisms to cancer therapy. Mol Aspects Med. 2015;41:1–115. doi: 10.1016/j.mam.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 6.West KP., Jr Extent of vitamin A deficiency among preschool children and women of reproductive age. J Nutr. 2002;132:2857S–2866S. doi: 10.1093/jn/132.9.2857S. [DOI] [PubMed] [Google Scholar]
- 7.Soprano DR, Blaner WS. In: The Retinoids: Biology, Chemistry, and Medicine. Sporn MB, Roberts AB, Goodman DS, editors. Raven Press; 1994. pp. 257–282. [Google Scholar]
- 8.Vieira M, Saraiva MJ. Transthyretin: A multifaceted protein. Biomol Concepts. 2014;5:45–54. doi: 10.1515/bmc-2013-0038. [DOI] [PubMed] [Google Scholar]
- 9.Ross AC. Cellular metabolism and activation of retinoids: Roles of cellular retinoid-binding proteins. FASEB J. 1993;7:317–327. doi: 10.1096/fasebj.7.2.8440409. [DOI] [PubMed] [Google Scholar]
- 10.Li Y, Wongsiriroj N, Blaner WS. The multifaceted nature of retinoid transport and metabolism. Hepatobiliary Surg Nutr. 2014;3:126–139. doi: 10.3978/j.issn.2304-3881.2014.05.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sivaprasadarao A, Boudjelal M, Findlay JB. Solubilization and purification of the retinol-binding protein receptor from human placental membranes. Biochem J. 1994;302:245–251. doi: 10.1042/bj3020245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sundaram M, Sivaprasadarao A, DeSousa MM, Findlay JB. The transfer of retinol from serum retinol-binding protein to cellular retinol-binding protein is mediated by a membrane receptor. J Biol Chem. 1998;273:3336–3342. doi: 10.1074/jbc.273.6.3336. [DOI] [PubMed] [Google Scholar]
- 13.Kawaguchi R, et al. A membrane receptor for retinol binding protein mediates cellular uptake of vitamin A. Science. 2007;315:820–825. doi: 10.1126/science.1136244. [DOI] [PubMed] [Google Scholar]
- 14.Bouillet P, et al. Developmental expression pattern of Stra6, a retinoic acid-responsive gene encoding a new type of membrane protein. Mech Dev. 1997;63:173–186. doi: 10.1016/S0925-4773(97)00039-7. [DOI] [PubMed] [Google Scholar]
- 15.Kawaguchi R, Yu J, Wiita P, Ter-Stepanian M, Sun H. Mapping the membrane topology and extracellular ligand binding domains of the retinol binding protein receptor. Biochemistry. 2008;47:5387–5395. doi: 10.1021/bi8002082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cogan U, Kopelman M, Mokady S, Shinitzky M. Binding affinities of retinol and related compounds to retinol binding proteins. Eur J Biochem. 1976;65:71–78. doi: 10.1111/j.1432-1033.1976.tb10390.x. [DOI] [PubMed] [Google Scholar]
- 17.Kawaguchi R, Zhong M, Kassai M, Ter-Stepanian M, Sun H. STRA6-catalyzed vitamin A influx, efflux, and exchange. J Membr Biol. 2012;245:731–745. doi: 10.1007/s00232-012-9463-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruiz A, et al. Molecular and biochemical characterization of lecithin retinol acyltransferase. J Biol Chem. 1999;274:3834–3841. doi: 10.1074/jbc.274.6.3834. [DOI] [PubMed] [Google Scholar]
- 19.Zhong M, Kawaguchi R, Kassai M, Sun H. Apo-RBP, holo-RBP, and insulin resistance. Mol Cell Biol. 2014;34:2105–2106. doi: 10.1128/MCB.01425-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noy N. Reply to “Apo-RBP, Holo-RBP, and insulin resistance”. Mol Cell Biol. 2014;34:2107. doi: 10.1128/MCB.01699-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noy N. Signaling by retinol and its serum binding protein. Prostaglandins Leukot Essent Fatty Acids. 2015;93:3–7. doi: 10.1016/j.plefa.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berry DC, Jin H, Majumdar A, Noy N. Signaling by vitamin A and retinol-binding protein regulates gene expression to inhibit insulin responses. Proc Natl Acad Sci USA. 2011;108:4340–4345. doi: 10.1073/pnas.1011115108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chassaing N, et al. Phenotypic spectrum of STRA6 mutations: From Matthew-Wood syndrome to non-lethal anophthalmia. Hum Mutat. 2009;30:E673–E681. doi: 10.1002/humu.21023. [DOI] [PubMed] [Google Scholar]
- 24.Isken A, et al. RBP4 disrupts vitamin A uptake homeostasis in a STRA6-deficient animal model for Matthew-Wood syndrome. Cell Metab. 2008;7:258–268. doi: 10.1016/j.cmet.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amengual J, et al. STRA6 is critical for cellular vitamin A uptake and homeostasis. Hum Mol Genet. 2014;23:5402–5417. doi: 10.1093/hmg/ddu258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruiz A, et al. Retinoid content, visual responses, and ocular morphology are compromised in the retinas of mice lacking the retinol-binding protein receptor, STRA6. Invest Ophthalmol Vis Sci. 2012;53:3027–3039. doi: 10.1167/iovs.11-8476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berry DC, et al. The STRA6 receptor is essential for retinol-binding protein-induced insulin resistance but not for maintaining vitamin A homeostasis in tissues other than the eye. J Biol Chem. 2013;288:24528–24539. doi: 10.1074/jbc.M113.484014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang G, et al. EMAN2: An extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 29.Hoeflich KP, Ikura M. Calmodulin in action: Diversity in target recognition and activation mechanisms. Cell. 2002;108:739–742. doi: 10.1016/S0092-8674(02)00682-7. [DOI] [PubMed] [Google Scholar]
- 30.Tidow H, Nissen P. Structural diversity of calmodulin binding to its target sites. FEBS J. 2013;280:5551–5565. doi: 10.1111/febs.12296. [DOI] [PubMed] [Google Scholar]
- 31.Ikura M, et al. Solution structure of a calmodulin-target peptide complex by multidimensional NMR. Science. 1992;256:632–638. doi: 10.1126/science.1585175. [DOI] [PubMed] [Google Scholar]
- 32.Pasutto F, et al. Mutations in STRA6 cause a broad spectrum of malformations including anophthalmia, congenital heart defects, diaphragmatic hernia, alveolar capillary dysplasia, lung hypoplasia, and mental retardation. Am J Hum Genet. 2007;80:550–560. doi: 10.1086/512203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liao M, Cao E, Julius D, Cheng Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature. 2013;504:107–112. doi: 10.1038/nature12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu P, et al. Three-dimensional structure of human γ-secretase. Nature. 2014;512:166–170. doi: 10.1038/nature13567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Esch H, Jansen A, Bauters M, Froyen G, Fryns JP. Encephalopathy and bilateral cataract in a boy with an interstitial deletion of Xp22 comprising the CDKL5 and NHS genes. Am J Med Genet A. 2007;143A:364–369. doi: 10.1002/ajmg.a.31572. [DOI] [PubMed] [Google Scholar]
- 36.Wernimont AK, et al. Structures of parasitic CDPK domains point to a common mechanism of activation. Proteins. 2011;79:803–820. doi: 10.1002/prot.22919. [DOI] [PubMed] [Google Scholar]
- 37.Drum CL, et al. Structural basis for the activation of anthrax adenylyl cyclase exotoxin by calmodulin. Nature. 2002;415:396–402. doi: 10.1038/415396a. [DOI] [PubMed] [Google Scholar]
- 38.Kawaguchi R, Yu J, Wiita P, Honda J, Sun H. An essential ligand-binding domain in the membrane receptor for retinol-binding protein revealed by large-scale mutagenesis and a human polymorphism. J Biol Chem. 2008;283:15160–15168. doi: 10.1074/jbc.M801060200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alapatt P, et al. Liver retinol transporter and receptor for serum retinol-binding protein (RBP4) J Biol Chem. 2013;288:1250–1265. doi: 10.1074/jbc.M112.369132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hesselink RW, Findlay JB. Expression, characterization and ligand specificity of lipocalin-1 interacting membrane receptor (LIMR) Mol Membr Biol. 2013;30:327–337. doi: 10.3109/09687688.2013.823018. [DOI] [PubMed] [Google Scholar]
- 41.Melhus H, Båvik CO, Rask L, Peterson PA, Eriksson U. Epitope mapping of a monoclonal antibody that blocks the binding of retinol-binding protein to its receptor. Biochem Biophys Res Commun. 1995;210:105–112. doi: 10.1006/bbrc.1995.1633. [DOI] [PubMed] [Google Scholar]
- 42.Redondo C, Vouropoulou M, Evans J, Findlay JB. Identification of the retinol-binding protein (RBP) interaction site and functional state of RBPs for the membrane receptor. FASEB J. 2008;22:1043–1054. doi: 10.1096/fj.07-8939com. [DOI] [PubMed] [Google Scholar]
- 43.Edidin M. The state of lipid rafts: From model membranes to cells. Annu Rev Biophys Biomol Struct. 2003;32:257–283. doi: 10.1146/annurev.biophys.32.110601.142439. [DOI] [PubMed] [Google Scholar]
- 44.Kawaguchi R, et al. Receptor-mediated cellular uptake mechanism that couples to intracellular storage. ACS Chem Biol. 2011;6:1041–1051. doi: 10.1021/cb200178w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berry DC, O’Byrne SM, Vreeland AC, Blaner WS, Noy N. Cross talk between signaling and vitamin A transport by the retinol-binding protein receptor STRA6. Mol Cell Biol. 2012;32:3164–3175. doi: 10.1128/MCB.00505-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Usacheva A, et al. Contribution of the Box 1 and Box 2 motifs of cytokine receptors to Jak1 association and activation. J Biol Chem. 2002;277:48220–48226. doi: 10.1074/jbc.M205757200. [DOI] [PubMed] [Google Scholar]
- 47.Wallweber HJ, Tam C, Franke Y, Starovasnik MA, Lupardus PJ. Structural basis of recognition of interferon-α receptor by tyrosine kinase 2. Nat Struct Mol Biol. 2014;21:443–448. doi: 10.1038/nsmb.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhong M, Kawaguchi R, Ter-Stepanian M, Kassai H, Sun M. Vitamin A transport and the transmembrane pore in the cell-surface receptor for plasma retinol binding protein. PLOS ONE. 2013;8:e73838. doi: 10.1371/journal.pone.0073838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kawaguchi R, Zhong M, Kassai M, Ter-Stepanian M, Sun H. Differential and isomer-specific modulation of vitamin A transport and the catalytic activities of the RBP receptor by retinoids. J Membr Biol. 2013;246:647–660. doi: 10.1007/s00232-013-9578-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Assur Z, Hendrickson WA, Mancia F. Tools for coproducing multiple proteins in mammalian cells. Methods Mol Biol. 2011;801:173–187. doi: 10.1007/978-1-61779-352-3_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 52.Kendrick BS, Kerwin BA, Chang BS, Philo JS. Online size-exclusion high-performance liquid chromatography light scattering and differential refractometry methods to determine degree of polymer conjugation to proteins and protein-protein or protein-ligand association states. Anal Biochem. 2001;299:136–146. doi: 10.1006/abio.2001.5411. [DOI] [PubMed] [Google Scholar]
- 53.Mancusso R, Karpowich NK, Czyzewski BK, Wang DN. Simple screening method for improving membrane protein thermostability. Methods. 2011;55:324–329. doi: 10.1016/j.ymeth.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Russo CJ, Passmore LA. Ultrastable gold substrates for electron cryomicroscopy. Science. 2014;346:1377–1380. doi: 10.1126/science.1259530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grant T, Grigorieff N. Measuring the optimal exposure for single particle cryo-EM using a 2.6 Å reconstruction of rotavirus VP6. eLife. 2015;4:e06980. doi: 10.7554/eLife.06980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Scheres SH. Semi-automated selection of cryo-EM particles in RELION-1.3. J Struct Biol. 2015;189:114–122. doi: 10.1016/j.jsb.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scheres SH, Chen S. Prevention of overfitting in cryo-EM structure determination. Nat Methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Slabinski L, et al. XtalPred: A web server for prediction of protein crystallizability. Bioinformatics. 2007;23:3403–3405. doi: 10.1093/bioinformatics/btm477. [DOI] [PubMed] [Google Scholar]
- 61.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pettersen EF, et al. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 63.Voss NR, Gerstein M. 3V: cavity, channel and cleft volume calculator and extractor. Nucleic Acids Res. 2010;38:W555–W562. doi: 10.1093/nar/gkq395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reeves PJ, Callewaert N, Contreras R, Khorana HG. Structure and function in rhodopsin: High-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc Natl Acad Sci USA. 2002;99:13419–13424. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. iMOSFLM: A new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Afonine PV, Grosse-Kunstleve RW, Adams PD. A robust bulk-solvent correction and anisotropic scaling procedure. Acta Crystallogr D Biol Crystallogr. 2005;61:850–855. doi: 10.1107/S0907444905007894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lomize MA, Pogozheva ID, Joo H, Mosberg HI, Lomize AL. OPM database and PPM web server: Resources for positioning of proteins in membranes. Nucleic Acids Res. 2012;40:D370–D376. doi: 10.1093/nar/gkr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.