Abstract
Tissue-specific NK cells are abundant in the pregnant uterus and interact with invading placental trophoblast cells that transform the maternal arteries to increase the fetoplacental blood supply. Genetic case-control studies have implicated killer cell Ig-like receptor (KIR) genes and their HLA ligands in pregnancy disorders characterized by failure of trophoblast arterial transformation. Activating KIR2DS1 or KIR2DS5 (when located in the centromeric region as in Africans) lower the risk of disorders when there is a fetal HLA-C allele carrying a C2 epitope. In this study, we investigated another activating KIR, KIR2DS4, and provide genetic evidence for a similar effect when carried with KIR2DS1. KIR2DS4 is expressed by ∼45% of uterine NK (uNK) cells. Similarly to KIR2DS1, triggering of KIR2DS4 on uNK cells led to secretion of GM-CSF and other chemokines, known to promote placental trophoblast invasion. Additionally, XCL1 and CCL1, identified in a screen of 120 different cytokines, were consistently secreted upon activation of KIR2DS4 on uNK cells. Inhibitory KIR2DL5A, carried in linkage disequilibrium with KIR2DS1, is expressed by peripheral blood NK cells but not by uNK cells, highlighting the unique phenotype of uNK cells compared with peripheral blood NK cells. That KIR2DS4, KIR2DS1, and some alleles of KIR2DS5 contribute to successful pregnancy suggests that activation of uNK cells by KIR binding to HLA-C is a generic mechanism promoting trophoblast invasion into the decidua.
Introduction
Natural killer cells use a combination of activating and inhibitory receptors to recognize viruses and cancerous cells (1). That the same receptors are also used to recognize fetal cells by tissue-specific uterine NK (uNK) cells (2) indicates two strong contrasting evolutionary pressures, that is, disease resistance and successful reproduction, with both showing evidence of balancing selection (3, 4). NK cells in the pregnant uterus, decidual NK (dNK) cells, are different phenotypically and functionally from peripheral blood NK (pbNK) cells (5–10). Evidence from genetic and functional studies suggests that dNK cells regulate trophoblast transformation of the uterine spiral arteries necessary for increasing the blood supply to the fetoplacental unit until the end of gestation (11–14).
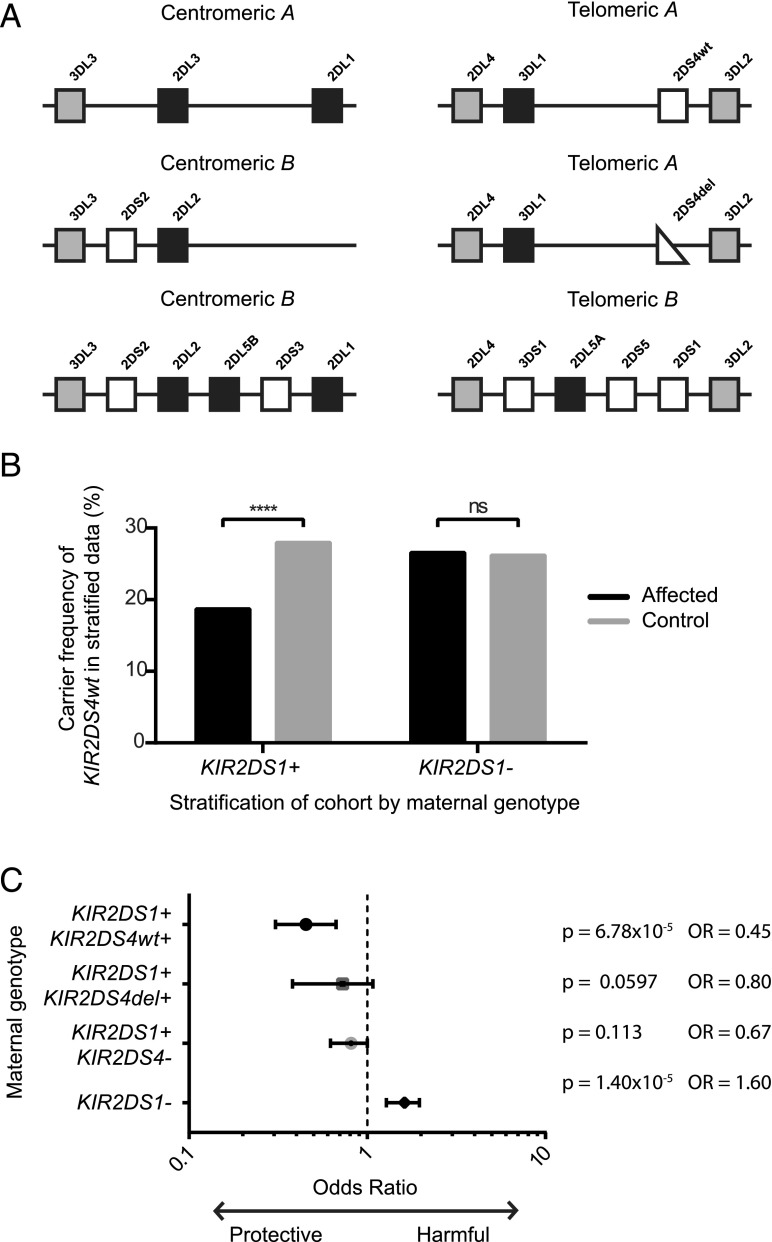
The NK cell receptors particularly implicated in reproductive health are the highly polymorphic killer cell Ig-like receptor (KIR) family (15). A KIR genotype is made up of two KIR haplotypes that can differ by both gene content and allelic variation. The genes in these haplotypes are so densely clustered on chromosome 19 that they are generally inherited as haplotypic centromeric and telomeric blocks (16, 17) (Fig. 1A). The dominant ligands for KIR are HLA-C allotypes. All individuals have KIRs that will bind to HLA-C allotypes as two groups depending on the C1 or C2 epitope that they bear. There is an increased risk of pregnancy disorders with certain inhibitory maternal KIR and fetal HLA-C combinations. Case-control genetic studies of Europeans have shown that pregnancy disorders that result from defective placentation with inadequate trophoblast arterial transformation (e.g., pre-eclampsia, fetal growth restriction, and recurrent miscarriage) are linked to an absence of the telomeric B (Tel-B) KIR region in the mother (Fig. 1A) and the presence of paternal C2 in the fetus (13, 18, 19). In contrast, pregnancies resulting in babies with increased birth weights are also associated with the presence of a paternal C2 allele in the fetus, but with a maternal Tel-B KIR region (20). The tight linkage disequilibrium (LD) of KIRs makes it difficult to determine through genetic studies alone which gene is responsible, so functional studies are required to complement this work.
FIGURE 1.
KIR2DS4wt in epistasis with KIR2DS1 is associated with a lower risk of pregnancy disorders. (A) The LD blocks that make up >94% of European KIR genotypes (17). An individual’s KIR genotype contains two haplotypes, each with one centromeric (left) and one telomeric (right) block. These blocks contain activating (white) and inhibitory (black) genes in LD. Framework genes (gray) are found in all haplotypes. The three most common telomeric blocks contain either KIR2DS4wt, KIR2DS4del, or KIR2DS1. (B) Women were stratified according to the presence or absence of the protective gene KIR2DS1, as a Breslow–Day test indicated epistasis between KIR2DS1 and KIR2DS4wt. The carrier frequency of KIR2DS4wt was then compared between women with affected pregnancies and healthy control pregnancies within each subgroup. The presence of KIR2DS4wt was protective (Cochran–Mantel–Haenszel test p = 5.7 × 10−4, OR = 0.59). (C) Then, within the women carrying KIR2DS1, the double-positive KIR2DS1+KIR2DS4wt+ are the most protected (p = 6.78 × 10−5, OR = 0.45).
Of the KIRs in the Tel-B region, activating KIR2DS1 is the most likely candidate for enhancing placentation, because it can bind to C2 allotypes. The inhibitory counterpart, KIR2DL1, also binds strongly to C2 allotypes, is present in the centromeric A and some centromeric B (Cen-B) regions, and is carried by ∼98% of individuals. Therefore, in the absence of KIR2DS1 (55–60% of Europeans), the dominant effect of paternal trophoblast C2 allotypes interacting with dNK cells is inhibition. Ligation of KIR2DS1 on dNK cells induces production of cytokines and chemokines, such as GM-CSF, which can induce trophoblast migration (12). Thus, our current model of pregnancy indicates that when C2 allotypes derived from the father are expressed by trophoblast, KIR2DS1 activates dNK cells to secrete cytokines that encourage deeper invasion of the uterus by trophoblast and promote spiral artery remodeling and a better blood supply for the fetus (2). In the absence of KIR2DS1, insufficient activation of dNK cells results in poor trophoblast invasion, placental stress, growth restriction of the fetus, and pre-eclampsia.
In a similar Ugandan case-control study, we found no protective effect for pre-eclampsia of the Tel-B region, including KIR2DS1 (carried by ∼20% of control women). Instead, certain alleles of an activating KIR, KIR2DS5, present in Cen-B were more frequent in controls compared with pre-eclamptic pregnancies (21). KIR2DS5 is always located in the Tel-B region in non-African populations and is carried in tight LD with KIR2DS1. It thus could contribute to the protective effect of Tel-B in Europeans, but whether it is expressed or binds C2 allotypes is still controversial. In addition to KIR2DS1 and KIR2DS5, KIR2DL5A is also present in Tel-B and remains an enigmatic KIR in terms of ligands and functions (22).
Other activating KIRs that might recognize ligands on trophoblast and influence pregnancy outcome include KIR3DS1 and KIR2DS2–4. KIR3DS1, in LD with KIR2DS1, binds HLA-B allotypes carrying the Bw4 motif (23), but HLA-B molecules are never expressed by trophoblast (24, 25). KIR2DS2 is predicted to bind the C1 motif through homology with KIR2DL2/3; the presence of fetal C1 alone is always neutral in our genetic case control studies. KIR2DS3 is not expressed at the cell surface (26). This leaves KIR2DS4, present in the telomeric A (Tel-A) region, that occurs either as a truncated (KIR2DS4del) (alleles *003/004/006 are carried by ∼80% of Europeans) or full-length (KIR2DS4wt) form (allele *001 is carried by ∼35% of Europeans). KIR2DS4del has a 22-bp deletion that introduces a frameshift mutation that results in a soluble protein with only one intact Ig-like domain (27). Whereas KIR2DS4wt has been reported to bind some HLA-C alleles carrying both the C1 and C2 epitopes, soluble KIR2DS4del does not bind HLA class I molecules (28). We previously found a negative association of KIR2DS4del with pregnancy outcome, but no positive effect of KIR2DS4wt (13).
In this study, to investigate the role of KIR other than KIR2DS1 in successful pregnancy, we have studied the expression and function of KIR2DS4 and KIR2DL5 on dNK cells. From this we demonstrate that activation of dNK cells is a general mechanism that is beneficial to pregnancy.
Materials and Methods
Primary tissue
Tissue and matched peripheral blood samples were obtained from women undergoing elective terminations in the first trimester of pregnancy; blood was also obtained from healthy volunteers. Both sets of patients gave informed consent. Ethical approval for the use of these tissues was obtained from the Cambridge Local Research Ethics Committee (REC 04/Q0108/23). Leukocytes and placental samples were isolated as previously described (29).
Cell lines
Cell lines transfected with cDNA for single KIR were used to test Ab specificities. KIR2DL1+, KIR2DL3+, KIR2DS1+, KIR2DS2+, KIR2DS4+ (30), or KIR3DS1+ (31) BWZ cells were the gift of Eric Vivier. KIR2DL2+, KIR2DS5+, KIR3DL1+ (31), or KIR3DL3+ (32) BA/F3 cells were the gift of Chiwen Chang, as was cDNA for KIR2DL5 used to transiently transfect HEK293T cells. KIR2DL4+ Jurkat cells were the gift of Kerry Campbell. Paul Norman supplied cDNA of KIR3DL2 for transient transfection into HEK293T cells.
Flow cytometry
dNK cells were gated on as live, CD9+CD56+ cells. pbNK cells were gated on as live CD56+CD3− cells. The following Abs were used: Live/Dead discriminator (Life Technologies), CD9 (SN4 or M-L13 from eBioscience or BD Biosciences, respectively), CD56 (HCD56 from BioLegend), and CD3 (SK7) from BD Biosciences. Fibroblasts and macrophages were identified using CD10 (HI10a from BioLegend) and CD14 Abs (MφP9 and HCD14 from BD Pharmingen and BioLegend), respectively. The following Abs were used to stain KIRs: UPR1 (KIR2DL5) from BioLegend and Carlos Vilches (33); 179315 (KIR2DS4), 143211 (KIR2DL1), and 181703 (KIR2DL4) from R&D Systems; FES172 (KIR2DS4) and EB6 (KIR2DL1/S1) from Beckman Coulter; CHL (KIR2DL2/3/S2) from BD Pharmingen; DX9 (KIR3DL1) from BioLegend; NKVFS1 (KIR2DL1/2/3/S1/2/4) from Abcam; 5.133 (KIR3DL2) from Marco Colonna (34); and FLAG Abs from Sigma-Aldrich. Intracellular staining was performed according to the manufacturers’ instructions with Abs against Ki647 (BD Pharmingen), CCL3 (R&D Systems), and GM-CSF (BD Biosciences).
Functional assays
Purified NK cells (CD56 positive selection using magnetic beads; Miltenyi Biotec) or mixed decidual mononuclear cells were stimulated with plate-bound anti-KIR2DS4 (179315) Abs or an isotype control for 12–48 h. After this time supernatants were removed (spun at 500 × g for 5 min to remove cellular contaminants) or stimulated cells were mechanically dislodged. Supernatants were analyzed using a chip-based fluorescence-linked immunosorbent assay (human cytokine Ab array G series 1000; RayBiotech) or a standard ELISA for CCL1 and XCL1 (DuoSets; R&D Systems). Cells activated cells in the presence of monensin and brefeldin A for 5 h were analyzed for surface expression of CD107a (H4A3; BD Pharmingen) or the intracellular cytokines listed above.
Immunohistochemistry
Paraffin sections of decidual implantation sites were heat treated in 0.1 M citrate buffer for 20 min at 99.5°C. Slides were left in hot buffer for a further 20 min for Ag unmasking. Anti-XCR1 (191704 from R&D Systems) was stained in TBS with 0.1% Tween 20 for 45 min. The staining was detected with goat anti-rabbit IgG-biotin and avidin-biotin-HRP complexes (Vector Laboratories).
Genetic typing
The case-control cohort analyzed in this study has previously been described (13). KIR and HLA-C1/2 genetic typing of new patient samples was performed as in this previous study. Two-digit HLA-C typing was performed by the Tissue Typing facility at Addenbrookes Hospital, Cambridge, U.K.
Statistical analysis
Statistical tests were carried out using the computational site http://vassarstats.net/, the statistical packages within GraphPad Prism v6 (GraphPad Software, La Jolla, CA), the Real Statistics Resource Pack for Excel 2010 (http://www.real-statistics.com/), and PLINK (version 1.07; http://pngu.mgh.harvard.edu/purcell/plink/) (35). The product rule was calculated by multiplying the observed frequency (Obs) of individual receptors (Obs [A] x Obs [B]) to generate the expected frequency of double-positive receptors (expected frequency [AB]). The following genetics tests were performed: a χ2 test and Fisher exact test with two-tailed mid-p adjustment, a Breslow–Day test, and a Cochran–Mantel–Haenszel test.
Results
KIR2DS4wt in epistasis with KIR2DS1 is associated with a lower risk of pre-eclampsia
KIR2DS4wt, the full-length form of activating KIR2DS4, is potentially important in pregnancy as it can bind to some HLA-C allotypes (28, 36). Indeed, we previously found in a case-control cohort of European women that KIR2DS4del associates with increased risk of pregnancy disorders (13). The presence of KIR2DS4wt was neutral in this analysis. However, we only considered presence/absence of this gene and did not consider the effect of both KIR telomeric regions that make up the women’s genotypes. There are three possible regions: Tel-A containing KIR2DS4wt; Tel-A containing KIR2DS4del; and Tel-B containing KIR2DS1 (Fig. 1A) that provides a strong protective effect (13). In this study, therefore, we reanalyzed this dataset for the effect of KIR2DS4wt, now controlling for the clear protective effect of KIR2DS1. Indeed, the presence/absence of KIR2DS1 does alter the effect of KIR2DS4wt, indicative of epistasis (Breslow–Day test, p = 0.003). KIR2DS4wt is protective compared with KIR2DS4del in KIR2DS1+ women (p = 5.7 × 10−4, odds ratio [OR] = 0.59) (Fig. 1B). This effect is not found in the absence of KIR2DS1 (p = 0.83, OR = 1.0). This indicates that women who carry both KIR2DS4wt and KIR2DS1 are further protected against disorders of pregnancy affecting placentation (p = 6.8 × 10−5, OR = 0.45) (Fig. 1C). Because of the similar functions and overlapping ligands of KIR2DS1 and KIR2DS4, it is likely that the epistasis detected at the statistical level reflects a biological interaction.
KIR2DS4 is expressed by a large proportion of both pbNK and dNK cells
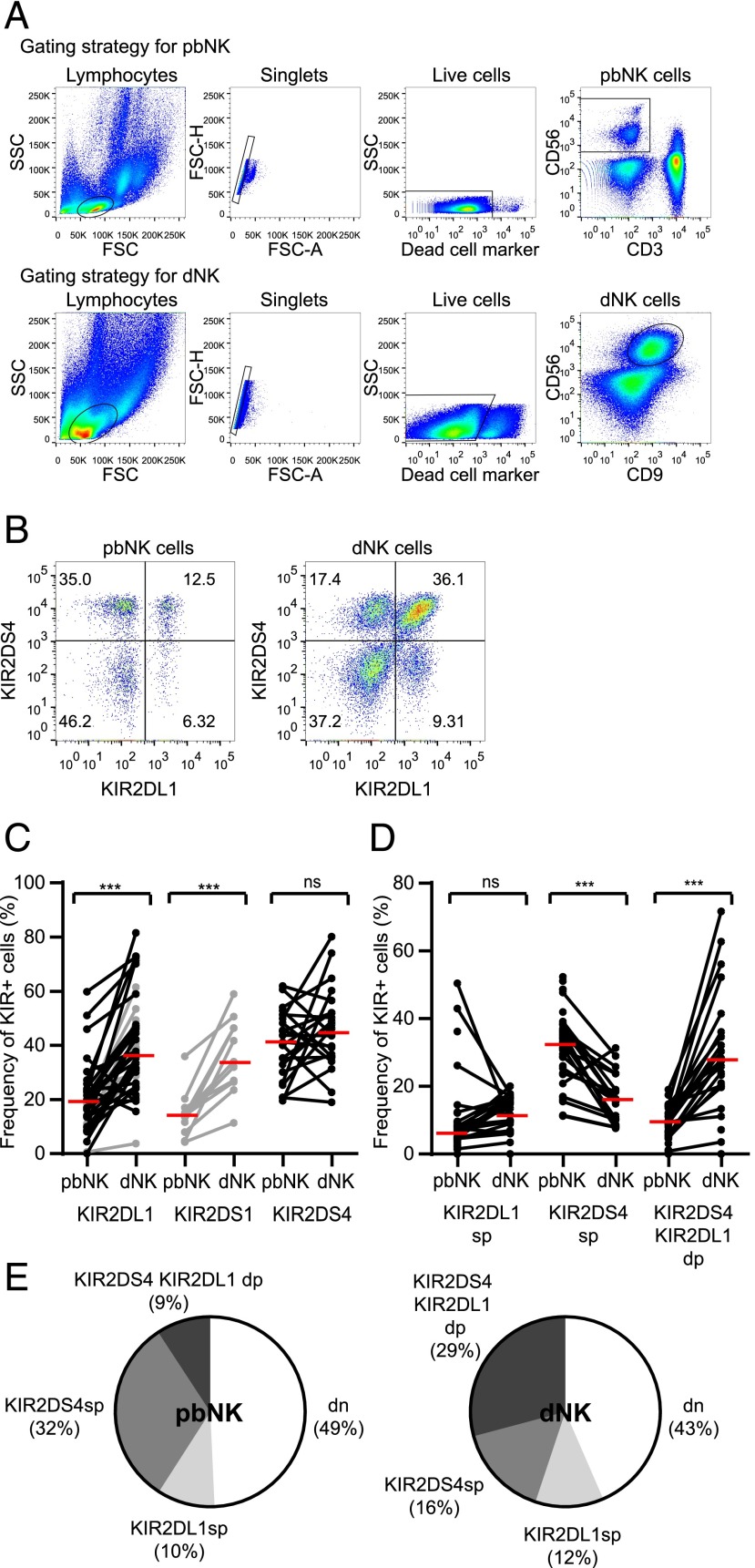
Two mAbs (FES172 and 179315) were tested to confirm specificity against KIR2DS4 on cell lines expressing single KIR (Supplemental Fig. 1). The frequency of KIR2DS4+CD56+ cells is high in both dNK and pbNK cell populations (Fig. 2A–C). In contrast, both KIR2DS1 and KIR2DL1 have an increased frequency of expression in dNK cells compared with pbNK cells (12, 37, 38) and so, in accordance with the product rule, there is a higher frequency of dNK cells coexpressing these KIRs than for pbNK cells (12). This means that the proportion of cells coexpressing KIR2DS4 and other KIRs is probably different for dNK and pbNK cells. We chose to look at the distribution of KIR2DS4 relative to KIR2DL1, because KIR2DL1 is carried by almost all donors, allowing us to analyze KIR coexpression with sufficient statistical power. KIR2DL1 is also critical to our model of pregnancy disorders, as it is strongly inhibitory for HLA-C allotypes bearing C2 epitopes. Our findings (Fig. 2D, 2E) show that in pbNK cells, most KIR2DS4+ cells lack KIR2DL1 (Fig. 2E, mid gray segment), but in dNK cells, most KIR2DS4+ cells coexpress KIR2DL1 (Fig. 2E, dark gray segment). This increased coexpression obeys the product rule (Supplemental Fig. 2A), suggesting it reflects the combined frequency of KIR2DL1 and KIR2DS4. In line with this, Ki-67 staining shows that KIR+ dNK cells proliferate more than KIR− cells, but there is no preferential proliferation by KIR2DS4+KIR2DL1+ cells compared with single-positive cells (Supplemental Fig. 2B). Therefore, in donors carrying KIR2DS4wt, a large proportion of dNK cells coexpresses KIR2DS4 with other KIRs that have the potential to modulate its function.
FIGURE 2.
KIR2DS4 is expressed by a large proportion of both pbNK and dNK cells. (A) Flow cytometry plots from a typical donor showing the gating strategy for pbNK and dNK cells. (B) Flow cytometry plots showing KIR2DS4 and KIR2DL1 staining on pbNK and dNK cells from a representative donor. The percentage of cells in each quadrant is shown. (C) The proportion of KIR2DS4+ cells was compared for pbNK and dNK cells in matched donors (n = 22). The proportion of KIR2DL1+ (n = 41) and KIR2DS1+ (n = 11) NK cells is shown for comparison only. Data points for KIR2DS1 and some KIR2DL1 (shown in gray) are already published (12) and are reproduced with permission from the Journal of Clinical Investigation. (D and E) The proportion of NK cells from each KIR+ subset (single-positive [sp], double-positive [dp], or double-negative [dn] for the receptors) was compared for pbNK and dNK cells. (D) Values for individual donors. Black lines represent donors, red lines represent the median, ***p < 0.001 by Wilcoxon signed rank test. (E) The mean values for each subset are displayed as a pie chart.
Using KIR Fc-fusion proteins, KIR2DS4 binds and responds to certain HLA-C alleles carrying both C1 and C2 epitopes (28, 36). Binding of KIR2DS4 on dNK cells to trophoblast HLA-C ligands might affect the frequency of KIR2DS4+ cells, but we find no difference in the proportion of dNK cells expressing KIR2DS4 when the mother or fetus carries its ligands (Supplemental Fig. 3A, 3B). There is a suggestion that allogeneic ligands affect KIR2DS4 expression, as there are fewer dNK cells expressing KIR2DS4 when the fetus alone carries a ligand, compared with the mother alone (Supplemental Fig. 3C). Given that KIR2DS4wt is protective in genetic case-control studies only in the presence of KIR2DS1, and that both are mutually exclusive on a KIR haplotype, protected individuals must have one copy of each gene. Therefore, we analyzed the effect of KIR2DS4wt copy number on frequency of expression: as two copies, as one copy in the presence of KIR2DS4del, or as one copy in the presence of KIR2DS1. KIR2DS4 frequency on dNK cells is similar in these different genetic backgrounds, suggesting that an altered frequency of KIR2DS4+ dNK cells in KIR2DS1+KIR2DS4+ individuals is not the mechanism by which KIR2DS4 provides protection against pregnancy disorders (Supplemental Fig. 3D).
KIR2DS4 activation on dNK cells induces cytokine responses
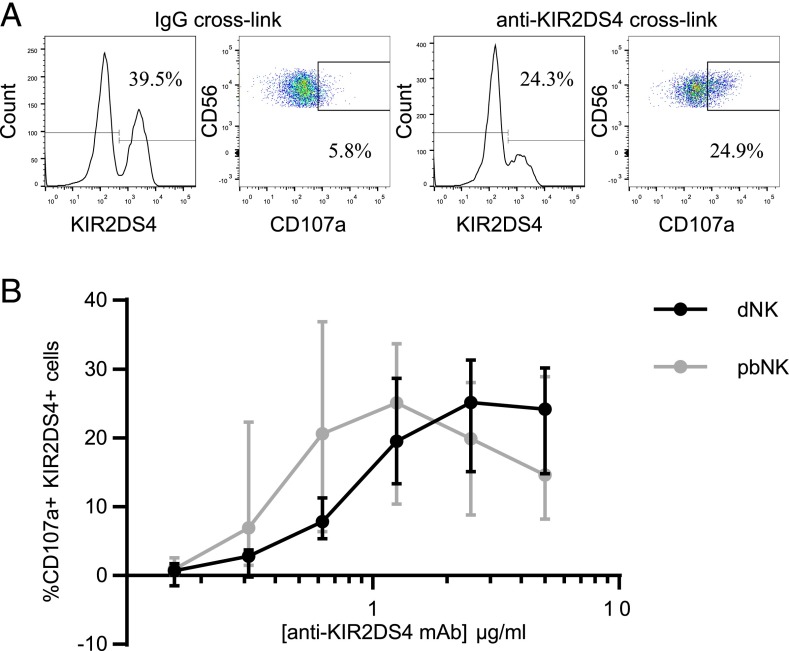
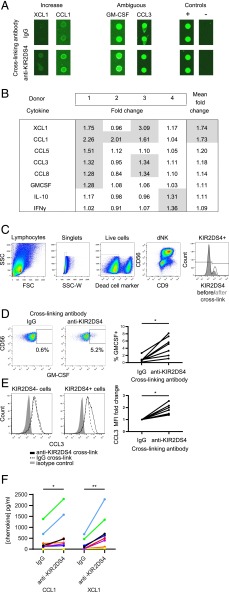
HLA-C ligands for KIR2DS4 are shared with other NK receptors on dNK cells. To investigate the functional consequences specific to activation of KIR2DS4 alone, we used cross-linking with a specific mAb. Decidual NK cells are poor killers, as measured by chromium-release assays (6, 9, 39), but CD107a degranulation does occur in the presence of low-dose IL-15 (40) and offers a reproducible assay to quantify dNK cell activation. We find that degranulation of both pbNK and dNK cells occurs in response to increasing concentrations of anti-KIR2DS4 (Scheirer–Ray–Hare modification of Kruskal–Wallis test, effect of mAb concentration p = 4.1 × 10−10) (Fig. 3). Because cytokine responses are more physiologically relevant to human pregnancy than is degranulation (12, 41), we next analyzed the cytokines produced following KIR2DS4 stimulation of dNK cells using a semiquantitative screen of 120 cytokines (Supplemental Table I). Mixed decidual mononuclear cells were cocultured in wells coated with anti-KIR2DS4 or control IgG Ab so that contact with stromal cells is maintained, as this improves viability. We identified eight candidates that were upregulated >1.25-fold in at least one out of four donors tested (Fig. 4A, 4B). After cross-linking with anti-KIR2DS4, flow cytometry (GM-CSF and CCL3) or ELISA (XCL1 and CCL1) assays were used to validate four of these eight cytokines (Fig. 4C–F). The percentage of dNK cells positive for intracellular GM-CSF and the median fluorescence intensity for CCL3 increases (p < 0.05) (Fig. 4C–E) and secretion assayed by ELISA for both XCL1 (p < 0.01) and CCL1 (p < 0.05) is augmented (Fig. 4F). In summary, stimulation of KIR2DS4 on dNK cells triggers the release of cytokines, many of which are related to the cytokines upregulated at the mRNA level by dNK cells upon KIR2DS1 activation (XCL2, CCL3L3, GM-CSF, IFNG) (12), although to our knowledge this is the first time XCL1 and CCL1 have been identified as secreted by dNK cells in response to activating signals.
FIGURE 3.
KIR2DS4 is functional on dNK cells. pbNK and dNK cells from KIR2DS4+ donors were incubated in wells coated with anti-KIR2DS4 or an isotype control for 5 h in the presence of monensin. (A) dNK cells from a represenative donor, gated as in Fig. 2, are shown stained for KIR2DS4 and CD107a following activation with plate-bound Ab (anti-KIR2DS4 or an isotype control). (B) The percentage of KIR2DS4+ NK cells positive for CD107a upon activation was calculated by subtracting the percentage CD107a+ when cells were cross-linked with IgG. The extent of degranulation for a range of Ab concentrations is shown. pbNK and dNK cells were not from the same donor. Scheirer–Ray–Hare modification of Kruskal–Wallis test, effect of concentration p = 4.1 × 10−10; effect of cell type p = 0.24; effect of interaction p = 0.87. Bars represent medians and interquartile ranges.
FIGURE 4.
Cytokine secretion by dNK cells in response to KIR2DS4 activation. (A and B) A semiquantitative fluorescent chip-based sandwich ELISA was used to screen for 120 cytokines in supernatants taken from mixed decidual leukocytes of KIR2DS4+ donors (see Supplemental Table I). Leukocytes were cultured on Ab-coated plastic for 12–24 h, where the only cells to express KIR2DS4 were the dNK cells. Fluorescent spots for cytokines of interest are highlighted in (A). The cropped regions of interest are taken from different chips and different donors. They are grouped according to whether they show a >1.5-fold increase in secretion on average across all donors (Increase); secretion that was already high within the isotype control stimulation, so the screen was insensitive (Ambiguous); and control spots (Control). (B) The cytokines that were upregulated >1.25-fold upon KIR2DS4 activation in at least one of four donors tested are listed in the table. The mean fold change across all four donors is shown to the right. Values >1.25-fold are highlighted in gray. (C–E) Mixed decidual leukocytes were cultured on plastic coated with either anti-KIR2DS4 Ab or an isotype control (IgG2a) in the presence of monensin and brefeldin A. After 5 h, cells were fixed and live CD56+CD9+KIR2DS4+ dNK cells were identified by flow cytometry (C). Although KIR2DS4 expression reduced upon cross-linking (C), most retained KIR2DS4 expression. These KIR2DS4+ dNK cells were assessed for intracellular cytokines: (D) GM-CSF (n = 7) and (E) CCL3 (n = 7). (F) When Abs for flow cytometry were not available, purified dNK cells were cultured on Ab-coated plastic for 12–48 h and the production of CCL1 and XCL1 (n = 8) was detected in supernatants by commercial sandwich ELISA. Results are color coded according to donor. *p < 0.05, **p < 0.01 by Wilcoxon signed rank test.
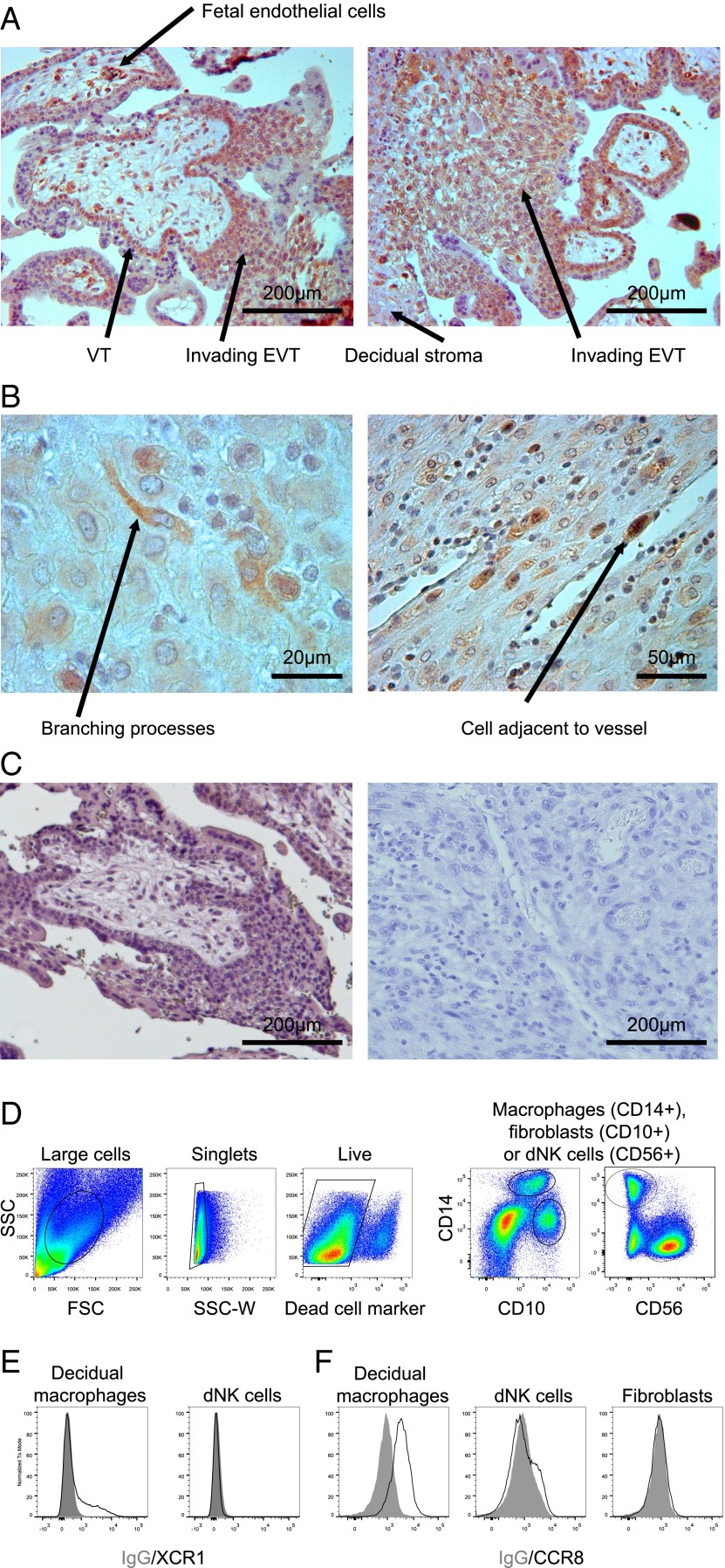
Trophoblast and maternal decidual cells express receptors for newly identified cytokines produced by activated dNK cells
Recently we have shown that GM-CSF induces migration of human primary trophoblast cells (12). CCL3 production by decidual and trophoblast cells may attract NK cells, as well as monocytes and T cells, which all bear receptors for this cytokine (42, 43). Receptors for chemokines XCL1 and CCL1 have not been described on cells at the site of placentation. We therefore stained sections of decidua and placenta and cell isolates by flow cytometry for these receptors. Several cell types within the placenta, including fetal endothelial cells, villous trophoblast, and invasive EVT express XCR1, the receptor for XCL1 (Fig. 5). Within the dNK cell–rich decidua, XCR1 is found on cells with branching processes (Fig. 5B), identified by flow cytometry as a small proportion of the CD14+ macrophage population (Fig. 5D, 5E). CCR8, the receptor for CCL1, is expressed by all decidual macrophages and a small proportion of dNK cells (Fig. 5F).
FIGURE 5.
Receptors for XCL1 and CCL1 on placenta and in the pregnant uterus. (A-C) Immunohistochemical localization of XCR1, the receptor for XCL1, with DAB substrate and Carazzi's hematoxylin nuclear counterstain. (A) XCR1 was identified on trophoblast invading the pregnant uterus. (B) Within the maternal compartment, XCR1 was identified on individual cells with branching processes often adjacent to vessels. (C) Isotype control staining of trophoblast (left panel) invading the uterus (right panel). (D) Chemokine receptor expression on live CD14+ decidual macrophages was confirmed by flow cytometry for (E) XCR1 (n = 6) and (F) CCR8 (n = 5). A population of cells that did not express the receptor (dNK cells for XCR1 and fibroblasts for CCR8, because some dNK cells express low amounts of CCR8) is shown for comparison. EVT, extravillous trophoblast; VT, villous trophoblast.
KIR2DL5, the only inhibitory receptor in the Tel-B region, is not expressed on the surface of dNK cells
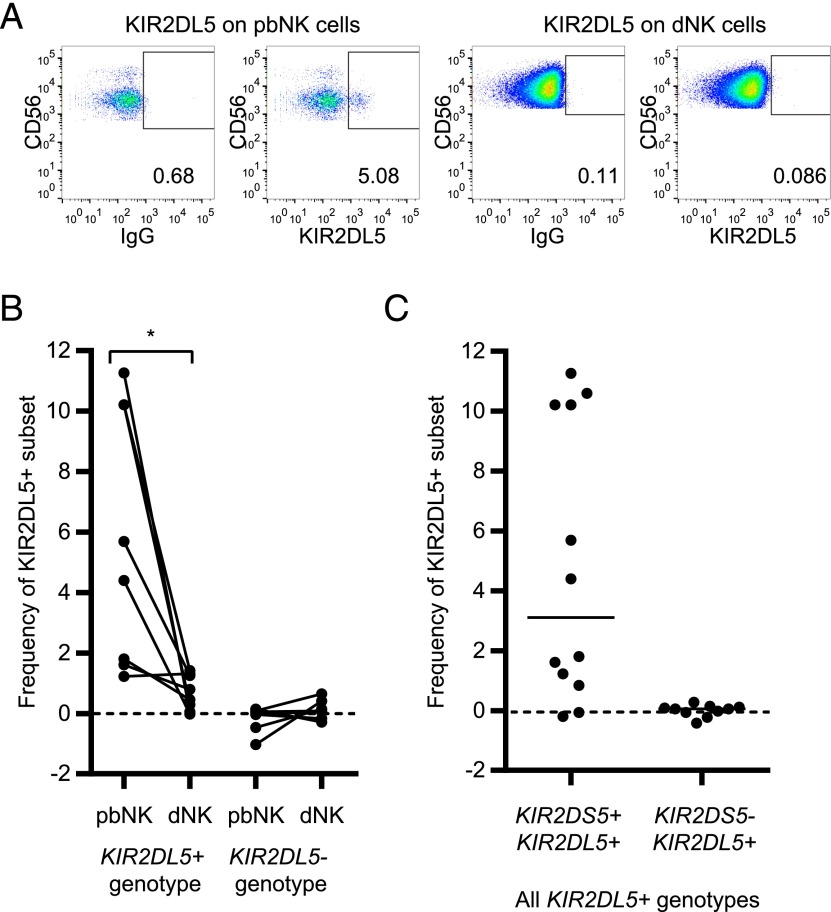
In the Tel-B region, KIR2DS1 is in LD with KIR2DL5A, which codes for an orphan inhibitory receptor. To determine whether KIR2DL5A affects dNK cell activation and pregnancy outcome, we first looked for expression of KIR2DL5A in dNK cells. KIR2DL5 alleles are also found in the Cen-B region, where they are known as KIR2DL5B (Fig. 1A). To distinguish between these alleles we used Ab UPR1, which binds the most common KIR2DL5A allele in Europeans, KIR2DL5A*001 (∼30% Europeans) (33), but not KIR2DL5A*005 (∼8% Europeans) or KIR2DL5B (∼20–40% Europeans) (22). UPR1 binds KIR2DL5 and not other KIRs, which we confirmed using KIR-negative cell lines transfected with single KIR genes (Supplemental Fig. 1). In donors where there were detectable KIR2DL5+ pbNK cells, there was no surface expression of KIR2DL5 on dNK cells (Fig. 6A, 6B). The donors who expressed KIR2DL5 in blood always also carry KIR2DS5 (Fig. 6C), which is in LD with KIR2DS1 and KIR2DL5*001 in the Tel-B haplotype in Europeans, suggesting we might only detect Tel-B KIR2DL5*001 and not other KIR2DL5 allotypes. The absence of KIR2DL5 surface expression on dNK cells means it is unlikely to be affecting the activity of dNK cells, and so the protective effect of the Tel-B region is due to activating KIR alone.
FIGURE 6.
KIR2DL5 is detected by flow cytometry on the surface of pbNK cells, but not dNK cells. (A) Flow cytometry plots from a typical donor showing matched pbNK and dNK cells, gated as in Fig. 2, stained for KIR2DL5 (mAb UPR1) or an isotype control (IgG1). (B) The frequency of the KIR2DL5+ population was defined as percentage UPR1+ minus percentage IgG1+. The frequency of this population was measured for all pbNK and dNK cells from donors where there was UPR1+ staining in blood (n = 8). For comparison, staining of KIR2DL5− donors is shown alongside (n = 7). Each line represents one donor. *p < 0.05 by Wilcoxon signed rank test. (C) All donors that carried KIR2DL5 according to sequence-specific primer–PCR were assessed for KIR2DL5 staining of pbNK, but a positive KIR2DL5+ subset was only seen as part of the genotype that carried KIR2DS5 alongside KIR2DL5. Lines show the median. Each dot represents one donor (n = 22).
Discussion
We have shown that KIR2DS4wt is associated with lower risk of pregnancy disorders in the presence of KIR2DS1, representing a synergistic interaction. KIR2DS4wt has been linked to higher viral load and increased transmission of HIV infection (44–47), as well as with clinical outcomes in arthritis (48–50), cancer (51), and allogeneic cell transplantation (52). Because KIRs are in tight LD and there are confounding effects of genes on alternative haplotypes, it can be difficult to determine which particular KIR has a role in disease (53). In the present study, we have ruled out the effect of alternative haplotypes by stratifying the cohort according to the presence of Tel-B. A clear biological rationale for a particular KIR’s involvement can also help distinguish the effect of KIRs in tight LD. KIR2DS4wt is in LD with KIR3DL1 alleles, but in the context of pregnancy, the ligand for KIR3DL1, HLA-Bw4, is not expressed by trophoblast. HLA-Bw4 can be expressed by stromal cells, so it is possible it modulates uNK cell activity. Nevertheless, KIR2DS4, known to bind to certain HLA-C allotypes expressed on trophoblast, is the likely candidate for the protective effect.
To explain how a particular KIR could affect trophoblast migration, the candidate KIR needs to be expressed by NK cells in contact with EVT in the decidua. KIR2DS4 is expressed by a large proportion of dNK cells, and its frequency of expression follows the product rule of coexpression with other KIRs. Coexpression of KIRs is relevant because the balance of activating and inhibitory signals within the cell determines activation of NK cells. Similar to KIR2DL1, KIR2DS1 has increased frequency of expression in dNK cells compared with pbNK cells. KIR2DS4wt could swing the balance in favor of activation when coexpressed with KIR2DS1 or in the context of activating cytokines, but it may fail to activate dNK cells in the absence of another activating receptor. Indeed, KIR2DS1 may also require the presence of another activating receptor to have measurable effects on population genetics, but unlike KIR2DS4wt, KIR2DS1 is in LD with two other activating receptors in Europeans. There is precedence for co-operation of activating KIRs from pregnancy and allogeneic hematopoietic stem cell transplantation, where cumulative Cen-B and Tel-B haplotypes that carry multiple activating KIR contribute to increasing beneficial effects (18, 54). Indeed, our finding that certain centromeric alleles of another different activating KIR, KIR2DS5, is protective against pre-eclampsia in Ugandans (21) supports this model. There is still limited evidence that KIR2DS4 responds to HLA-C molecules, but our preliminary findings suggest that the size of the KIR2DS4+ dNK cell subset is smaller when KIR2DS4 ligands are present in the fetus, but not the mother. This observation supports the hypothesis that KIR2DS4 does bind these HLA-C ligands on trophoblast.
Why should KIR2DS4 act as a coreceptor in this way, requiring the presence of another activating receptor? The mechanism of this co-operation remains unclear, but we can exclude some factors. First, we have shown that the frequency of expression of KIR2DS4wt on dNK cells is unaffected by the presence of Tel-B or Tel-A on the women’s other haplotype. Therefore, higher frequency of expression of KIR2DS4wt in the presence of KIR2DS1 cannot be the mechanism by which epistasis is achieved. Similarly, there is only one prevalent allele of KIR2DS1 and functional KIR2DS4 among Europeans (KIR2DS1*002 and KIR2DS4*001), so allelic variation on particular haplotypes is unlikely to affect the association in our European case-control cohorts. One reason for the dependence of KIR2DS4wt on the presence of KIR2DS1 could be the nature of its interaction with HLA-C molecules. Although there are functional responses of KIR2DS1+ NK cells upon interaction with HLA-C alleles carrying C2 epitopes ex vivo (12, 55, 56), similar responses of KIR2DS4+ NK cells have only been demonstrated for HLA-C*0401 (36) and HLA-A*1102 (28). The interaction of KIR2DS4 with HLA-C could be of lower avidity than that of KIR2DS1; KIR2DS4 recognition of HLA-C allotypes might be peptide-dependent, as has been shown for KIR3DS1 (23); or KIR2DS4 may be interacting with open conformers of HLA molecules (57) expressed by trophoblast. All these factors could affect the way KIR2DS4 binds to HLA-C molecules on trophoblast.
Specialized functions for pbNK and dNK cells are likely to have arisen because of the conflicting demands of disease resistance and reproductive success (3). When trying to assess the impact of KIRs in health and disease, it is necessary, therefore, to study these receptors in the species and tissue of interest. Upon triggering of KIR2DS4 with specific Abs, dNK cells degranulate and secrete cytokines, such as GM-CSF, that are known to have direct effects on trophoblast migration, and other cytokines (XCL1, CCL1, and CCL3) that have the potential to directly impact trophoblast and other cells in the decidua, including decidual macrophages. Recently, KIR2DS4 has been highlighted for promoting trogocytosis (58), a process that has been implicated in dNK cell acquisition of HLA-G from trophoblast (59). There may be several mechanisms, therefore, by which triggering of dNK cells could aid placentation.
The view that immune cells must be suppressed for successful pregnancy, both locally in the uterus and systemically, originated with Medawar (60) and the birth of transplant biology. There is now mounting evidence that for uNK cells this is not correct. We show that activation of dNK cells through KIR2DS4wt provides help to trophoblast migration and the establishment of pregnancy. Perhaps KIR2DS5 in the Cen-B region may protect Africans from pre-eclampsia in the same way (21). Moreover, we find here that inhibitory receptor KIR2DL5A in the protective Tel-B region is not expressed by dNK cells, suggesting it does not affect pregnancy outcome. Taken together, these data support a model of generic activation of dNK cells counteracting strong inhibition by KIR2DL1 and benefitting pregnancy.
Supplementary Material
Acknowledgments
We thank all the donors and research nurses for providing samples. We thank Carlos Vilches, Hugo Hilton, Paul Norman, Peter Parham, Eric Vivier, Chiwen Chang, John Trowsdale, Kerry Campbell, Michela Falco, and Massimo Vitale for reagents relating to this study, as well as Sarah Peacock and Craig Taylor of the Tissue Typing facility at Addenbrookes Hospital Cambridge. We thank Daniel Davis for helpful comments on the manuscript.
This work was supported by the Wellcome Trust, the Centre for Trophoblast Research, the British Heart Foundation, and the Cambridge Philosophical Society.
The online version of this article contains supplemental material.
- Cen-B
- centromeric B
- dNK
- decidual NK
- KIR
- killer-cell Ig-like receptor
- LD
- linkage disequilibrium
- Obs
- observed frequency
- OR
- odds ratio
- pbNK
- peripheral blood NK
- Tel-A
- telomeric A
- Tel-B
- telomeric B
- uNK
- uterine NK.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Lanier L. L. 2005. NK cell recognition. Annu. Rev. Immunol. 23: 225–274. [DOI] [PubMed] [Google Scholar]
- 2.Moffett-King A. 2002. Natural killer cells and pregnancy. Nat. Rev. Immunol. 2: 656–663. [DOI] [PubMed] [Google Scholar]
- 3.Parham P., Moffett A. 2013. Variable NK cell receptors and their MHC class I ligands in immunity, reproduction and human evolution. Nat. Rev. Immunol. 13: 133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moffett A., Colucci F. 2015. Co-evolution of NK receptors and HLA ligands in humans is driven by reproduction. Immunol. Rev. 267: 283–297. [DOI] [PubMed] [Google Scholar]
- 5.Koopman L. A., Kopcow H. D., Rybalov B., Boyson J. E., Orange J. S., Schatz F., Masch R., Lockwood C. J., Schachter A. D., Park P. J., Strominger J. L. 2003. Human decidual natural killer cells are a unique NK cell subset with immunomodulatory potential. J. Exp. Med. 198: 1201–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kopcow H. D., Allan D. S. J., Chen X., Rybalov B., Andzelm M. M., Ge B., Strominger J. L. 2005. Human decidual NK cells form immature activating synapses and are not cytotoxic. Proc. Natl. Acad. Sci. USA 102: 15563–15568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharkey A. M., Xiong S., Kennedy P. R., Gardner L., Farrell L. E., Chazara O., Ivarsson M. A., Hiby S. E., Colucci F., Moffett A. 2015. Tissue-specific education of decidual NK cells. J. Immunol. 195: 3026–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferry B. L., Starkey P. M., Sargent I. L., Watt G. M., Jackson M., Redman C. W. 1990. Cell populations in the human early pregnancy decidua: natural killer activity and response to interleukin-2 of CD56-positive large granular lymphocytes. Immunology 70: 446–452. [PMC free article] [PubMed] [Google Scholar]
- 9.King A., Birkby C., Loke Y. W. 1989. Early human decidual cells exhibit NK activity against the K562 cell line but not against first trimester trophoblast. Cell. Immunol. 118: 337–344. [DOI] [PubMed] [Google Scholar]
- 10.Ritson A., Bulmer J. N. 1989. Isolation and functional studies of granulated lymphocytes in first trimester human decidua. Clin. Exp. Immunol. 77: 263–268. [PMC free article] [PubMed] [Google Scholar]
- 11.Ashkar A. A., Di Santo J. P., Croy B. A. 2000. Interferon γ contributes to initiation of uterine vascular modification, decidual integrity, and uterine natural killer cell maturation during normal murine pregnancy. J. Exp. Med. 192: 259–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiong S., Sharkey A. M., Kennedy P. R., Gardner L., Farrell L. E., Chazara O., Bauer J., Hiby S. E., Colucci F., Moffett A. 2013. Maternal uterine NK cell-activating receptor KIR2DS1 enhances placentation. J. Clin. Invest. 123: 4264–4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hiby S. E., Apps R., Sharkey A. M., Farrell L. E., Gardner L., Mulder A., Claas F. H., Walker J. J., Redman C. W., Morgan L., et al. 2010. Maternal activating KIRs protect against human reproductive failure mediated by fetal HLA-C2. [Published erratum appears in 2011 J. Clin. Invest. 121:455.] J. Clin. Invest. 120: 4102–4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kieckbusch J., Gaynor L. M., Moffett A., Colucci F. 2014. MHC-dependent inhibition of uterine NK cells impedes fetal growth and decidual vascular remodelling. Nat. Commun. 5: 3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khakoo S. I., Carrington M. 2006. KIR and disease: a model system or system of models? Immunol. Rev. 214: 186–201. [DOI] [PubMed] [Google Scholar]
- 16.Robinson J., Mistry K., Mcwilliam H., Lopez R., Marsh S. G. E. 2009. IPD–the Immuno Polymorphism Database. Nucleic Acids Res. 38: D863–D869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang W., Johnson C., Jayaraman J., Simecek N., Noble J., Moffatt M. F., Cookson W. O., Trowsdale J., Traherne J. A. 2012. Copy number variation leads to considerable diversity for B but not A haplotypes of the human KIR genes encoding NK cell receptors. Genome Res. 22: 1845–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hiby S. E., Walker J. J., O’shaughnessy K. M., Redman C. W. G., Carrington M., Trowsdale J., Moffett A. 2004. Combinations of maternal KIR and fetal HLA-C genes influence the risk of preeclampsia and reproductive success. J. Exp. Med. 200: 957–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hiby S. E., Regan L., Lo W., Farrell L., Carrington M., Moffett A. 2008. Association of maternal killer-cell immunoglobulin-like receptors and parental HLA-C genotypes with recurrent miscarriage. Hum. Reprod. 23: 972–976. [DOI] [PubMed] [Google Scholar]
- 20.Hiby S. E., Apps R., Chazara O., Farrell L. E., Magnus P., Trogstad L., Gjessing H. K., Carrington M., Moffett A. 2014. Maternal KIR in combination with paternal HLA-C2 regulate human birth weight. J. Immunol. 192: 5069–5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakimuli A., Chazara O., Hiby S. E., Farrell L., Tukwasibwe S., Jayaraman J., Traherne J. A., Trowsdale J., Colucci F., Lougee E., et al. 2015. A KIR B centromeric region present in Africans but not Europeans protects pregnant women from pre-eclampsia. Proc. Natl. Acad. Sci. USA 112: 845–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cisneros E., Moraru M., Gómez-Lozano N., López-Botet M., Vilches C. 2012. KIR2DL5: an orphan inhibitory receptor displaying complex patterns of polymorphism and expression. Front. Immunol. 3: 289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Connor G. M., Vivian J. P., Gostick E., Pymm P., Lafont B. A., Price D. A., Rossjohn J., Brooks A. G., McVicar D. W. 2015. Peptide-dependent recognition of HLA-B*57:01 by KIR3DS1. J. Virol. 89: 5213–5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.King A., Burrows T. D., Hiby S. E., Bowen J. M., Joseph S., Verma S., Lim P. B., Gardner L., Le Bouteiller P., Ziegler A., et al. 2000. Surface expression of HLA-C antigen by human extravillous trophoblast. Placenta 21: 376–387. [DOI] [PubMed] [Google Scholar]
- 25.Apps R., Murphy S. P., Fernando R., Gardner L., Ahad T., Moffett A. 2009. Human leucocyte antigen (HLA) expression of primary trophoblast cells and placental cell lines, determined using single antigen beads to characterize allotype specificities of anti-HLA antibodies. Immunology 127: 26–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.VandenBussche C. J., Mulrooney T. J., Frazier W. R., Dakshanamurthy S., Hurley C. K. 2009. Dramatically reduced surface expression of NK cell receptor KIR2DS3 is attributed to multiple residues throughout the molecule. Genes Immun. 10: 162–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsu K. C., Liu X.-R., Selvakumar A., Mickelson E., O’Reilly R. J., Dupont B. 2002. Killer Ig-like receptor haplotype analysis by gene content: evidence for genomic diversity with a minimum of six basic framework haplotypes, each with multiple subsets. J. Immunol. 169: 5118–5129. [DOI] [PubMed] [Google Scholar]
- 28.Graef T., Moesta A. K., Norman P. J., Abi-Rached L., Vago L., Older Aguilar A. M., Gleimer M., Hammond J. A., Guethlein L. A., Bushnell D. A., et al. 2009. KIR2DS4 is a product of gene conversion with KIR3DL2 that introduced specificity for HLA-A*11 while diminishing avidity for HLA-C. J. Exp. Med. 206: 2557–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Male V., Trundley A., Gardner L., Northfield J., Chang C., Apps R., Moffett A. 2010. Natural killer cells in human pregnancy. In Natural Killer Cell Protocols. Methods in Molecular Biology, Vol. 612 Campbell K. S., ed. Humana Press, Totowa, NJ, p. 447–463. [DOI] [PubMed] [Google Scholar]
- 30.Schleinitz N., Cognet C., Guia S., Laugier-Anfossi F., Baratin M., Pouget J., Pelissier J.-F., Harle J.-R., Vivier E., Figarella-Branger D. 2008. Expression of the CD85j (leukocyte Ig-like receptor 1, Ig-like transcript 2) receptor for class I major histocompatibility complex molecules in idiopathic inflammatory myopathies. Arthritis Rheum. 58: 3216–3223. [DOI] [PubMed] [Google Scholar]
- 31.Trundley A., Frebel H., Jones D., Chang C., Trowsdale J. 2007. Allelic expression patterns of KIR3DS1 and 3DL1 using the Z27 and DX9 antibodies. Eur. J. Immunol. 37: 780–787. [DOI] [PubMed] [Google Scholar]
- 32.Trundley A. E., Hiby S. E., Chang C., Sharkey A. M., Santourlidis S., Uhrberg M., Trowsdale J., Moffett A. 2006. Molecular characterization of KIR3DL3. Immunogenetics 57: 904–916. [DOI] [PubMed] [Google Scholar]
- 33.Estefanía E., Flores R., Gómez-Lozano N., Aguilar H., López-Botet M., Vilches C. 2007. Human KIR2DL5 is an inhibitory receptor expressed on the surface of NK and T lymphocyte subsets. J. Immunol. 178: 4402–4410. [DOI] [PubMed] [Google Scholar]
- 34.Döhring C., Scheidegger D., Samaridis J., Cella M., Colonna M. 1996. A human killer inhibitory receptor specific for HLA-A1,2. J. Immunol. 156: 3098–3101. [PubMed] [Google Scholar]
- 35.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A. R., Bender D., Maller J., Sklar P., de Bakker P. I. W., Daly M. J., Sham P. C. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katz G., Markel G., Mizrahi S., Arnon T. I., Mandelboim O. 2001. Recognition of HLA-Cw4 but not HLA-Cw6 by the NK cell receptor killer cell Ig-like receptor two-domain short tail number 4. J. Immunol. 166: 7260–7267. [DOI] [PubMed] [Google Scholar]
- 37.Sharkey A., Gardner L., Hiby S. 2008. Killer Ig-like receptor expression in uterine NK cells is biased toward recognition of HLA-C and alters with gestational age. J. Immunol. 181: 39–46. [DOI] [PubMed] [Google Scholar]
- 38.Male V., Sharkey A., Masters L., Kennedy P. R., Farrell L. E., Moffett A. 2011. The effect of pregnancy on the uterine NK cell KIR repertoire. Eur. J. Immunol. 41: 3017–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verma S., Hiby S. E., Loke Y. W., King A. 2000. Human decidual natural killer cells express the receptor for and respond to the cytokine interleukin 15. Biol. Reprod. 62: 959–968. [DOI] [PubMed] [Google Scholar]
- 40.Apps R., Sharkey A., Gardner L., Male V., Kennedy P., Masters L., Farrell L., Jones D., Thomas R., Moffett A. 2011. Ex vivo functional responses to HLA-G differ between blood and decidual NK cells. Mol. Hum. Reprod. 17: 577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hanna J., Goldman-Wohl D., Hamani Y., Avraham I., Greenfield C., Natanson-Yaron S., Prus D., Cohen-Daniel L., Arnon T. I., Manaster I., et al. 2006. Decidual NK cells regulate key developmental processes at the human fetal-maternal interface. Nat. Med. 12: 1065–1074. [DOI] [PubMed] [Google Scholar]
- 42.Red-Horse K., Drake P. M., Gunn M. D., Fisher S. J. 2001. Chemokine ligand and receptor expression in the pregnant uterus: reciprocal patterns in complementary cell subsets suggest functional roles. Am. J. Pathol. 159: 2199–2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Drake P. M., Gunn M. D., Charo I. F., Tsou C. L., Zhou Y., Huang L., Fisher S. J. 2001. Human placental cytotrophoblasts attract monocytes and CD56bright natural killer cells via the actions of monocyte inflammatory protein 1α. J. Exp. Med. 193: 1199–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hong H. A., Paximadis M., Gray G. E., Kuhn L., Tiemessen C. T. 2013. KIR2DS4 allelic variants: differential effects on in utero and intrapartum HIV-1 mother-to-child transmission. Clin. Immunol. 149: 498–508. [DOI] [PubMed] [Google Scholar]
- 45.Merino A., Malhotra R., Morton M., Mulenga J., Allen S., Hunter E., Tang J., Kaslow R. A. 2011. Impact of a functional KIR2DS4 allele on heterosexual HIV-1 transmission among discordant Zambian couples. J. Infect. Dis. 203: 487–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Merino A. M., Dugast A. S., Wilson C. M., Goepfert P. A., Alter G., Kaslow R. A., Tang J. 2014. KIR2DS4 promotes HIV-1 pathogenesis: new evidence from analyses of immunogenetic data and natural killer cell function. PLoS One 9: e99353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Olvera A., Pérez-Álvarez S., Ibarrondo J., Ganoza C., Lama J., Lucchetti A., Cate S., Hildebrand W., Bernard N., Gomez G., et al. 2015. The HLA-C*04:01/KIR2DS4 gene combination and 1 HLA alleles with high population frequency drive rate of HIV disease progression. AIDS 29: 507–517. [DOI] [PubMed] [Google Scholar]
- 48.Zhou J., Tang X., Ding Y., An Y., Zhao X. 2013. Natural killer cell activity and frequency of killer cell immunoglobulin-like receptors in children with different forms of juvenile idiopathic arthritis. Pediatr. Allergy Immunol. 24: 691–696. [DOI] [PubMed] [Google Scholar]
- 49.Yen J.-H., Lin C.-H., Tsai W.-C., Wu C.-C., Ou T.-T., Hu C.-J., Liu H.-W. 2006. Killer cell immunoglobulin-like receptor gene’s repertoire in rheumatoid arthritis. Scand. J. Rheumatol. 35: 124–127. [DOI] [PubMed] [Google Scholar]
- 50.Majorczyk E., Pawlik A., Gendosz D., Kuśnierczyk P. 2014. Presence of the full-length KIR2DS4 gene reduces the chance of rheumatoid arthritis patients to respond to methotrexate treatment. BMC Musculoskelet. Disord. 15: 256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Giebel S., Boratyn-Nowicka A., Karabon L., Jedynak A., Pamula-Pilat J., Tecza K., Kula D., Kowal M., Frydecka I., Grzybowska E. 2014. Associations between genes for killer immunoglobulin-like receptors and their ligands in patients with epithelial ovarian cancer. Hum. Immunol. 75: 508–513. [DOI] [PubMed] [Google Scholar]
- 52.Bao X. J., Hou L. H., Sun A. N., Qiu Q. C., Yuan X. N., Chen M. H., Chen Z. X., He J. 2010. The impact of KIR2DS4 alleles and the expression of KIR in the development of acute GVHD after unrelated allogeneic hematopoietic SCT. Bone Marrow Transplant. 45: 1435–1441. [DOI] [PubMed] [Google Scholar]
- 53.Hammond J. A., Carrington M., Khakoo S. I. 2016. A vision of KIR variation at super resolution. Immunology 148: 249–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cooley S., Weisdorf D. J., Guethlein L. A., Klein J. P., Wang T., Le C. T., Marsh S. G., Geraghty D., Spellman S., Haagenson M. D., et al. 2010. Donor selection for natural killer cell receptor genes leads to superior survival after unrelated transplantation for acute myelogenous leukemia. Blood 116: 2411–2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pittari G., Liu X.-R., Selvakumar A., Zhao Z., Merino E., Huse M., Chewning J. H., Hsu K. C., Dupont B. 2013. NK cell tolerance of self-specific activating receptor KIR2DS1 in individuals with cognate HLA-C2 ligand. J. Immunol. 190: 4650–4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stewart C. A., Laugier-Anfossi F., Vély F., Saulquin X., Riedmuller J., Tisserant A., Gauthier L., Romagné F., Ferracci G., Arosa F. A., et al. 2005. Recognition of peptide-MHC class I complexes by activating killer immunoglobulin-like receptors. Proc. Natl. Acad. Sci. USA 102: 13224–13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goodridge J. P., Burian A., Lee N., Geraghty D. E. 2013. HLA-F and MHC class I open conformers are ligands for NK cell Ig-like receptors. J. Immunol. 191: 3553–3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pesce S., Carlomagno S., Moretta A., Sivori S., Marcenaro E. 2015. Uptake of CCR7 by KIR2DS4+ NK cells is induced upon recognition of certain HLA-C alleles. J. Immunol. Res. 2015: 754373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tilburgs T., Evans J. H., Crespo Â. C., Strominger J. L. 2015. The HLA-G cycle provides for both NK tolerance and immunity at the maternal–fetal interface. Proc. Natl. Acad. Sci. USA 112: 13312–13317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Medawar P. 1953. Some immunological and endocrinological problems raised by the evolution of viviparity in vertebrates. Symp. Soc. Exp. Biol. 7: 320–338. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.