Abstract
Hepatitis C virus (HCV) infection has been shown to induce autophagy but the mechanisms underpinning this process remain to be elucidated. Induction of autophagy requires the class III phosphatidylinositol 3-kinase, Vps34, which produces phosphatidylinositol 3-phosphate (PI3P) within the endoplasmic reticulum (ER) membrane. This recruits proteins with PI3P binding domains such as the double-FYVE-containing protein 1 (DFCP1). DFCP1 generates cup–shaped protrusions from the ER membrane, termed omegasomes, which provide a platform for the production of autophagosomes. Here we present data demonstrating that both Vps34 and DFCP1 are required for HCV genome replication, in the context of both a subgenomic replicon and virus infection, but did not affect virus entry or initial translation. Using live cell fluorescence microscopy we demonstrated that early during HCV infection the nascent viral genome replication complexes (identified by using non-structural protein NS5A as a marker) transiently colocalize with DFCP1-positive punctae (omegasomes), before the two structures move apart from each other. This observation is reminiscent of the transient association of LC3 and DFCP1 during omegasome formation, and therefore we propose that omegasomes are utilized by HCV to generate the double-membrane vesicles which are the hallmark of HCV replication complexes.
Introduction
Hepatitis C virus (HCV) is a positive-stranded RNA virus that establishes a chronic infection in 85 % of infected individuals, leading to long-term liver disease such as cirrhosis and hepatocellular carcinoma. The 9.6 kb genome is translated into a single polyprotein that is subsequently cleaved into 10 structural and non-structural proteins. The recent development of an infectious cell culture system for HCV, based on the genotype 2a isolate, JFH-1 (Wakita et al., 2005), has allowed the detailed analysis of the molecular mechanisms of virus replication. In common with most positive strand RNA viruses, the entire life cycle of HCV occurs within the cytoplasm of infected cells. The virus induces rearrangements of cytoplasmic membranes to form a structure called the ‘membranous web’ (Egger et al., 2002; Gosert et al., 2003) – a cluster of mainly double-membraned vesicles (DMV) that constitutes a replication factory where genome replication takes place. Despite recent advances in understanding the architecture of the membranous web (Paul et al., 2013; Romero-Brey et al., 2012), the biogenesis of this structure remains unclear. Although it is thought to derive from the endoplasmic reticulum (ER), the mechanisms by which DMV are produced from ER membranes, and the cellular factors involved in this process, have not been identified.
In this regard it is of interest that HCV, in common with other positive-strand RNA viruses, induces autophagy (Ait-Goughoulte et al., 2008; Dreux et al., 2009; Sir et al., 2008). Autophagy is a cellular process for the bulk degradation of cytoplasmic contents, either to allow them to be recycled, or to provide an energy source during times of nutrient starvation or stress (Tanida, 2011). Importantly, it is characterized by the formation of DMV, termed autophagosomes, which fuse with lysosomes to form autolysosomes, allowing the degradation of the vesicular contents. Following the induction of autophagy, the class III phosphatidylinositol 3-kinase (PI3K), Vps34, associates with a trimeric complex of Vps15-Beclin1-Atg14L, and these Vps34 complexes localize to the mitochondrial-associated ER membranes (MAM) (Hamasaki et al., 2013). Vps34 complexes produce phosphatidylinositol 3-phosphate (PI3P), resulting in PI3P-enriched regions that allow for the recruitment of proteins such as the double-FYVE-containing protein 1 (DFCP1), which binds PI3P via its FYVE domains (Ridley et al., 2001). DFCP1 accumulation gives rise to cup-like protrusions from the ER membrane, termed omegasomes, which provide a platform for the expansion of a structure termed the isolation membrane, and from which autophagosomes dissociate during their biogenesis (Axe et al., 2008). PI3K activity can be chemically inhibited by wortmannin or 3-methyladenine (3-MA) (Wu et al., 2010), both of which will block autophagy.
Positive-stranded RNA viruses such as picornaviruses utilize autophagy to generate the cytoplasmic membrane structures required for genome replication (Taylor & Kirkegaard, 2007), although autophagy has also been implicated in the immune response to pathogens; for review see Dreux & Chisari (2010). HCV has been shown to require either the induction of autophagy, or an involvement of proteins comprising the autophagy machinery, for the establishment of productive replication (Dreux et al., 2009; Shrivastava et al., 2011). However, the mechanisms by which HCV induces autophagy and interacts with the autophagy machinery remain poorly defined. In order to better understand the role of autophagy in the establishment of the HCV replication compartment, we used a combination of pharmacological inhibition of PI3K, genetic ablation of Vps34 and DFCP1, and fluorescent imaging of virus-infected cells early after infection. Our results reveal that HCV replication is dependent on an intact autophagosome biogenesis machinery, and we propose a novel model that implicates omegasomes as the sites for the generation of the double-membrane vesicles that are the site of genome replication complexes.
Results
PI3K activity is required for HCV genome replication
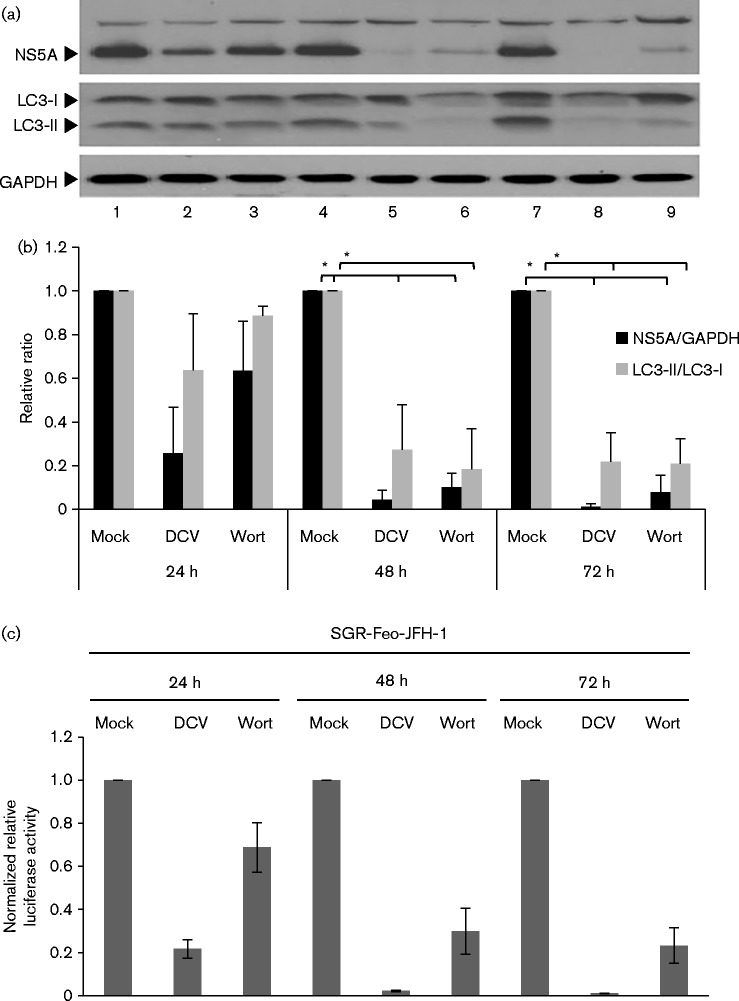
Previous work in both our lab (Street et al., 2004) and by others (He et al., 2002) has shown that HCV enhanced the activity of class I PI3K, although the role of this enhancement in virus replication was not elucidated. Whilst class I PI3K can modulate autophagy, it is the class III PI3K Vps34 that is essential for the nucleation machinery of autophagy, producing PI3P which subsequently recruits DFCP1. Therefore, to assess the significance of PI3K activity for the induction of autophagy by HCV, Huh7 cells stably harbouring a genotype 2a bicistronic JFH-1 subgenomic replicon (SGR) expressing a neomycin phosphotransferase–luciferase fusion protein (SGR-Feo-JFH-1) (Wyles et al., 2009) were treated with the well-characterized PI3K inhibitor wortmannin (Fig. 1).
Fig. 1.
PI3K activity is required for HCV replication. (a) SGR-Feo-JFH-1-harbouring cells were mock treated (lanes 1, 4, 7) or treated with either daclatasvir (DCV) (100 pM) (lanes 2, 5, 8) or wortmannin (Wort; 2 μM) (lanes 3, 6, 9) for 24 h, 48 h and 72 h, respectively, prior to lysis for analysis by Western blot with the antibodies indicated. (b) Densitometry analysis of the Western blot expression data in (a). The ratios of NS5A : GADPH (black bars) or LC3-II : LC3-I (grey bars) are shown. *P < 0.05. (c). Luciferase activity is presented as values normalized to the mock-treated cells at each time point. Data represent the mean ± se (n = 3).
This analysis showed that SGR-Feo-JFH-1-harbouring cells treated with wortmannin (Fig. 1a, lanes 3, 6 and 9) exhibited a decrease in NS5A protein levels, when compared with mock-treated cells over this time period (lanes 1, 4 and 7 – quantified by densitometry, Fig. 1b). The HCV replication inhibitor daclatasvir (DCV) (Gao et al., 2010) was used as a positive control and also demonstrated a decrease in NS5A protein levels (lanes 2, 5 and 8). Wortmannin-treated cells showed a progressive loss of LC3-II over time when compared with the corresponding mock-treated controls, which is in agreement with inhibition of Vps34 class III PI3K activity. In cells treated with DCV, LC3B-II and NS5A levels decreased concurrently. These observations were corroborated by luciferase assay, which functions as an indirect measurement of levels of HCV RNA. Both DCV and wortmannin treatment resulted in a time-dependent decrease in luciferase activity that corresponded to the Western blot data (Fig. 1c). The decrease in SGR replication was not due to a cytotoxic effect of prolonged wortmannin treatment, as confirmed by MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assay (data not shown). These observations confirm that class III PI3K activity is required both for HCV genomic replication and the induction of autophagy by the virus. We therefore investigated the potential involvement of both Vps34 and DFCP1 in the HCV life cycle.
The autophagosome biogenesis machinery is required for HCV genome replication
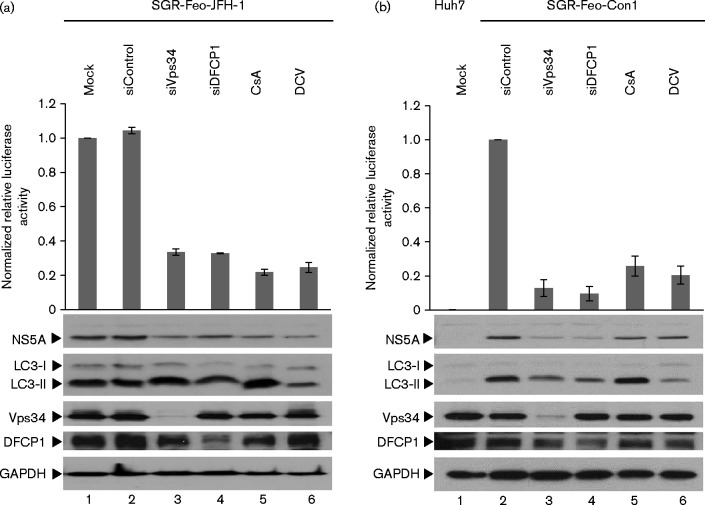
A number of previous studies have demonstrated that autophagy-related proteins are required for efficient HCV RNA replication – siRNA-mediated silencing of LC3, ATG4B, ATG7 or Beclin-1 expression results in a reduction of virus RNA accumulation (Dreux et al., 2009; Shrivastava et al., 2011; Sir et al., 2008), whilst Vps34 has been implicated in being recruited by NS4B to mediate HCV-induced autophagy (Su et al., 2011). We further investigated the role of both Vps34 and DFCP1 in HCV replication by siRNA-mediated silencing in SGR-Feo-JFH-1-harbouring cells. These cells were mock transfected or transfected with control-, Vps34- or DFCP1-targeted siRNA, and analysed at 72 h post-transfection by both Western blot and luciferase assay. As shown in Fig. 2(a), expression of both Vps34 (lane 3) and to a lesser extent DFCP1 (lane 4) could be efficiently silenced. This silencing led to an inhibition of both luciferase activity and NS5A expression (Fig. 2a), confirming the requirement for these two proteins in HCV genomic replication. As positive controls, the SGR-Feo-JFH-1-harbouring cells were treated with two well-characterized inhibitors of HCV genome replication, either cyclosporin A (CsA) (Gallay, 2009) or DCV. Both inhibitors effectively blocked HCV RNA replication as judged by reductions in both luciferase and NS5A expression (lanes 5 and 6 in Fig. 2a), and, as previously observed, CsA treatment stimulated autophagy, presumably as a result of inhibition of protein folding (Mohl et al., 2012). Interestingly, the Vps34 siRNA-mediated knockdown did not reduce LC3-II abundance (lane 3), suggesting that autophagy in Huh7 cells might be driven by both canonical and non-canonical pathways (Codogno et al., 2012). DFCP1 siRNA-mediated knockdown resulted in a decrease in LC3-II abundance (lane 4), confirming its role in autophagosome biogenesis.
Fig. 2.
Silencing of Vps34 or DFCP1 inhibits HCV SGR replication. (a) SGR-Feo-JFH-1-harbouring cells were mock transfected (lane 1), or transfected with control siRNA (lane 2) or siRNAs against Vps34 or DFCP1 (lanes 3 and 4) using RNAiMAX. At 72 h post-transfection, cells were lysed for luciferase assay or Western blot analysis with the antibodies indicated. (b) Parental Huh7 cells (lane 1) or SGR-Feo-Con1-harbouring cells (lanes 2–5) were transfected and processed as in (a) above. In both experiments, as positive controls, cells were treated with CsA (lane 5) or DCV (lane 6). Luciferase activity is presented as values normalized to the mock treated cells at each time point. Data represent the mean ± se (n = 3).
To confirm that the roles of Vps34 and DFCP1 were not restricted to the JFH-1 genotype 2a isolate, we performed the same experiment in Huh7 cells harbouring a genotype 1b (Con1) SGR (SGR-Feo-Con1) (Fig. 2b). Again silencing of either Vps34 or DFCP1 resulted in a significant decrease in luciferase and NS5A expression (lanes 3 and 4), as did treatment with CsA or DCV (lanes 5 and 6). Interestingly, in comparison with the SGR-Feo-JFH-1 data, siRNA-mediated silencing of either Vps34 or DFCP1 resulted in a similar decrease of LC3-II abundance in the SGR-Feo-Con1-harbouring cells, suggesting that replication of genotype 1b is possibly more dependent on these proteins than is replication of JFH-1. Alternatively, it is possible that this reflects a genotype-specific difference in the induction of autophagy by HCV. It is also noteworthy that silencing of Vps34 resulted in a modest reduction in DFCP1 levels for both SGR lines (lanes 3), implying that DFCP1 expression might be regulated in response to induction of autophagy. Taken together, these observations demonstrated that genome replication of both genotypes 1b and 2a HCV required Vps34 and DFCP1.
We further validated these observations in the context of infection of Huh7 cells with HCV. To do this we utilized a chimeric virus that expressed Renilla luciferase (J6/JFH-1-Rluc) (Jones et al., 2007) – thus allowing us to interrogate effects of Vps34 or DFCP1 silencing on either early or late events in the virus life cycle. Renilla luciferase activity measured at 4 h post-infection (p.i.) would give a measure of entry and initial translation of the incoming viral genome (prior to genome replication), whereas later measurements would assess effects on genome replication. Huh7 cells were transfected with siRNA for 72 h., infected with J6/JFH-1-Rluc virus at an m.o.i. of 0.5 f.f.u. per cell and analysed at 4 and 48 h p.i. by both Western blot and luciferase assay. As shown in Fig. 3, at 4 h p.i., under conditions of efficient silencing of Vps34 or DFCP1 expression, there was no significant decrease in luciferase values (compare lane 2 with 3/4), suggesting that the silencing of Vps34 and DFCP1 did not interfere with viral entry or initial translation. Interestingly, treatment of cells with inhibitors of HCV genome replication, either CsA or DCV, resulted in significant reductions in Renilla luciferase values at 4 h p.i. (lanes 5 and 6). This observation suggests that these compounds had effects on either entry or initial viral translation, in addition to direct inhibition of viral genome replication. This could be explained in the case of CsA when the unfolded protein response could result in a global attenuation of protein translation; in addition, the mechanism of action of DCV at early stages in the virus life cycle has been recently reported (Berger et al., 2014) and proposed to involve disruption of membranous web formation.
Fig. 3.
Silencing of Vps34 or DFCP1 inhibits virus replication. Huh7 cells were mock transfected (lanes 1, 7), or transfected with control siRNA (lanes 2, 8), or siRNAs against Vps34 (lanes 3, 9) or DFCP1 (lanes 4, 10) using RNAiMAX. At 72 h post-transfection, cells were infected with a chimeric J6/JFH-1Rluc reporter virus (m.o.i. of 0.5 f.f.u. per cell). At 4 or 48 h p.i., cells were lysed for luciferase assay or Western blot analysis with the antibodies indicated. As positive controls, cells were treated with CsA (lanes 5, 11) or DCV (lanes 6, 12). Luciferase activity is presented as values normalized to the control siRNA-transfected cells at each time point. Data represent the mean ± se (n = 3).
At 48 h p.i., virus replication was only robustly established in cells transfected with a control siRNA, as judged by expression of both E2 and NS5A (lane 8). Silencing of either Vps34 or DFCP1 resulted in an almost complete abrogation of virus replication (lanes 9 and 10), similar to the controls treated with either CsA or DCV. These data confirm that both Vps34 and DFCP1 are required for the establishment of HCV genome replication.
Validation of an mCherry–DFCP1 expression construct for live cell imaging
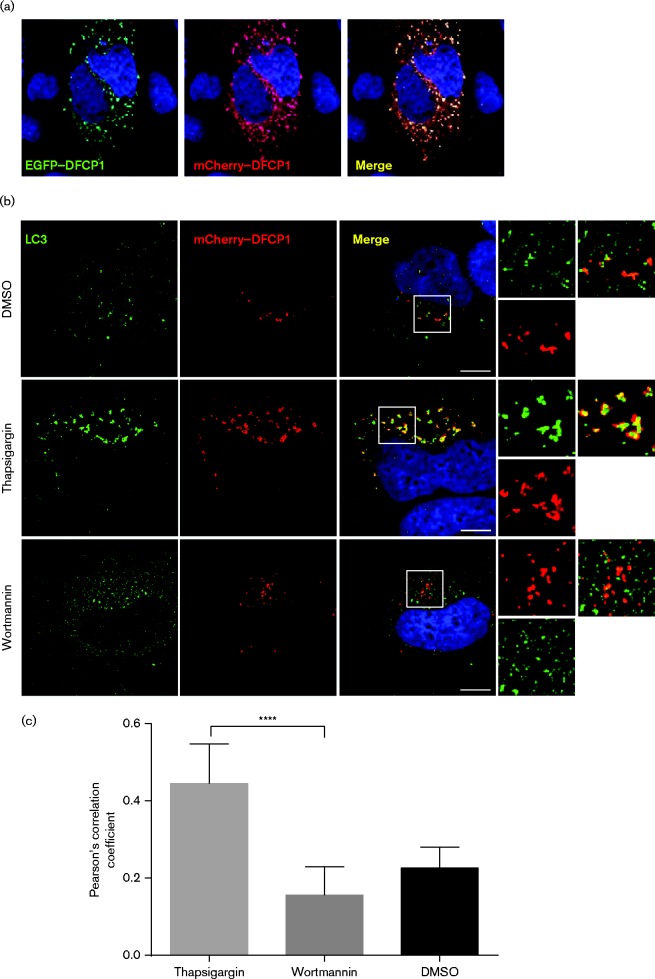
DFCP1 is recruited to localized concentrations of PI3P (generated by Vps34) and is involved in formation of omegasomes which provide a platform for the formation of isolation membranes, the progenitors of autophagosomes (Axe et al., 2008). The observation that DFCP1 was required for HCV genome replication suggested that replication complexes might associate with omegasomes at some stage during the virus life cycle. However, we reasoned that as omegasomes are transient structures the association between the replication complex and DFCP1 might also be short-lived. To assess this we wished to use live cell imaging of cells infected with an HCV derivative containing an NS5A–GFP fusion, and to facilitate this we modified an existing EGFP–DFCP1 expression construct (Axe et al., 2008) to generate an mCherry–DFCP1 fusion expression plasmid and validated its use in the context of autophagosome biogenesis. To this end, Huh7 cells were transfected with both the mCherry–DFCP1 and EGFP–DFCP1 expression constructs and imaged by fluorescence microscopy. As shown in Fig. 4(a), the distribution of the two fusion proteins overlapped completely, indicating that the mCherry–DFCP1 retained the subcellular localization of the previously characterized EGFP–DFCP1, and therefore validating its use in our experiments.
Fig. 4.
Validation of mCherry–DFCP1 as a marker of early autophagosome formation. (a) Huh7 cells were transfected with expression plasmids for EGFP–DFCP1 (Axe et al., 2008) and mCherry–DFCP1. (b) Huh7 cells were transfected with an mCherry–DFCP1 expression plasmid. Forty-eight hours post-transfection, cells were treated with vehicle (DMSO), thapsigargin (3 μM) or wortmannin (1 μM) for 3 h prior to being fixed and stained with an antibody to LC3 (green) and DAPI. Bars, 10 μm. (c) Colocalization analysis of LC3 puncta with mCherry–DFCP1 from cells in (b). The mean Pearson correlation coefficient was derived for each treatment from 6, 8 and 8 cells respectively. Data represent the mean ± sd. ****P < 0.0001.
To further validate the mCherry–DFCP1 construct, it was transfected into Huh7 cells that were then treated with thapsigargin (Tg) to induce autophagy, as previously demonstrated (Mohl et al., 2012). Cells were then fixed and labelled with an LC3 antibody for indirect immunofluorescence. Tg treatment induced autophagy, which was monitored by the appearance of abundant LC3 puncta (green) (Fig. 4b). As expected, we observed significant colocalization of LC3 with mCherry–DFCP1, as confirmed by quantification of the overlapping fluorescence signals (Fig. 4c). In contrast, following wortmannin treatment, LC3 distribution was more diffuse, as in untreated cells, and mCherry–DFCP1 did not significantly colocalize with LC3 (Fig. 4b, c). These data validated the use of the mCherry–DFCP1 construct – demonstrating that it localized to cytoplasmic puncta in a PI3K-dependent fashion and also that it colocalized with LC3 following induction of autophagy.
NS5A puncta do not stably colocalize with DFCP1 during infection with HCV
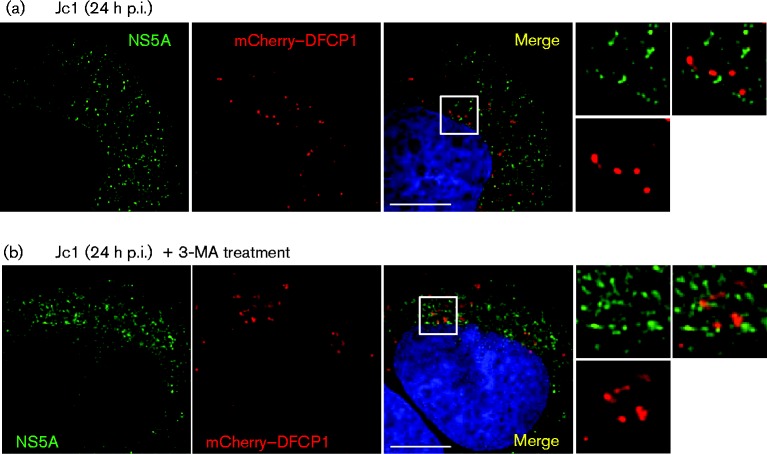
We next asked whether NS5A (as a marker for replication complexes) colocalized with DFCP1 in HCV-infected cells. Huh7 cells were therefore transfected with the mCherry–DFCP1 expression construct and subsequently infected with the chimeric Jc1 virus (Pietschmann et al., 2006) at an m.o.i. of 0.5 f.f.u. per cell. At 24 h p.i., cells were fixed and stained with an NS5A antibody for indirect immunofluorescence (Fig. 5). The distribution of mCherry–DFCP1 in Jc1-infected cells was similar to that observed in uninfected cells treated with Tg (Fig. 4), consistent with the induction of autophagy following virus infection. However, we observed no colocalization of the abundant NS5A puncta with mCherry–DFCP1. Neither did we see any colocalization of NS5A and DFCP1 when the cells were treated with an alternative PI3K inhibitor, 3-MA (Fig. 5), although interestingly this did not block the virus-induced autophagy, which we previously showed (Fig. 1) to be at least partially non-responsive to PI3K inhibition.
Fig. 5.
HCV RNA replication complexes do not colocalize with DFCP1. (a) Huh7 cells were transfected with a mCherry–DFCP1 expression plasmid and at 48 h post-transfection cells were infected with Jc1 (0.5 f.f.u. per cell). At 24 h p.i., cells were fixed and stained with antibodies to NS5A (green). (b) Infected cells (as above) were treated with 3-MA for 3 h prior to fixing. Bars, 10 μm.
A transient colocalization of NS5A and DFCP1 can be observed by live cell imaging
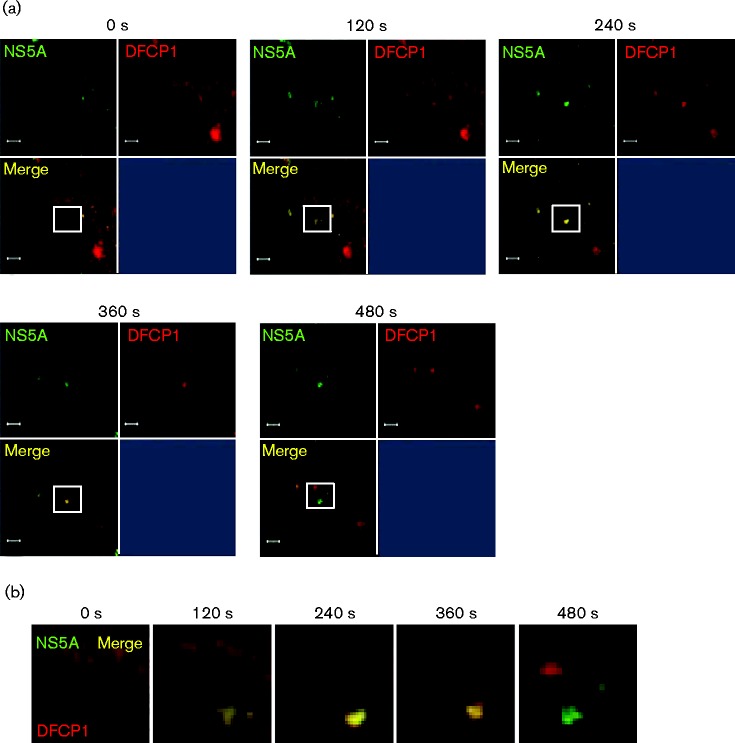
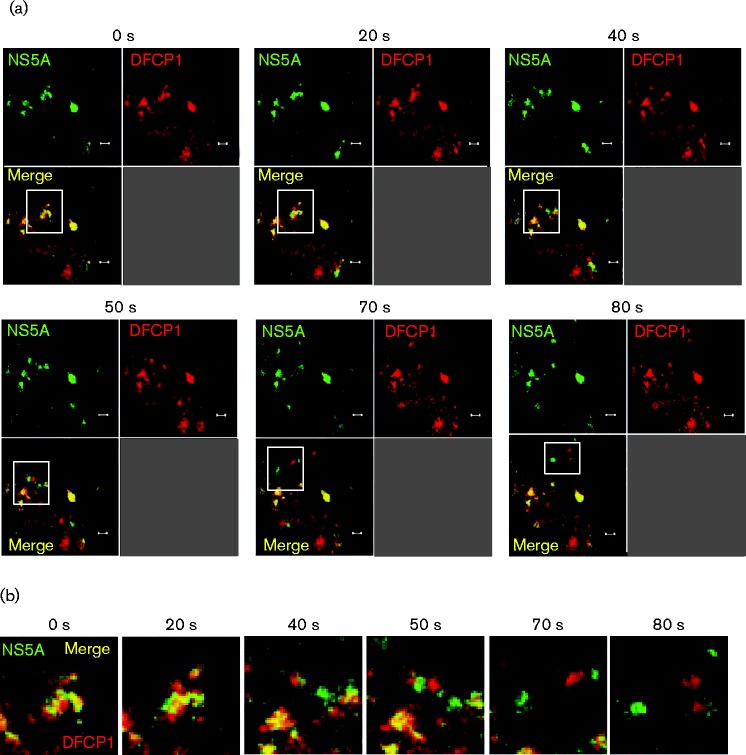
The lack of NS5A–DFCP1 colocalization was consistent with the hypothesis that the establishment of replication complexes might require DFCP1, but that once replication complexes had formed there was no further requirement for DFCP1. This raised the possibility that transient interactions between nascent replication complexes and the autophagic machinery might occur and so to test this hypothesis we proceeded to use live cell imaging: Huh7 cells were transfected with the mCherry–DFCP1 expression construct and at 48 h post-transfection cells were infected with a Jc1 derivative expressing an NS5A–GFP fusion (Jc1–GFP) (Schaller et al., 2007) at an m.o.i. of 0.5 f.f.u. per cell. Twenty-four hours post-infection, live cells were then analysed by confocal microscopy to visualize both NS5A–GFP and mCherry–DFCP1. Videos of the resulting datasets were generated and montages of time-lapse exposures are shown in Figs 6 and 7. Fig. 6(a) shows an example of the progressive accumulation of both NS5A (green) and DFCP1 (red) into a distinct punctate structure (yellow), seen first at 120 s, and the subsequent dissociation of these two into distinct structures. This observation was reminiscent of the situation with LC3 and DFCP1 previously reported to be involved in the formation of omegasomes and autophagosomes (Axe et al., 2008). Fig. 6(b) shows an enlargement of the white boxed areas from Fig. 6(a), clearly demonstrating the NS5A/DFCP1 puncta formation and separation. Fig. 7(a, b) shows a distinct example whereby a cluster of juxtaposed NS5A/DFCP1 puncta dissociate into distinct structures towards the end of the image series and traffic away from the site of colocalization. These data are consistent with a rapid and transient association of replication complexes (here represented by NS5A) with DFCP1, and furthermore provide strong evidence that omegasome formation is involved in the generation of these replication complexes.
Fig. 6.
Transient association of nascent HCV RNA replication complexes and DFCP1 imaged by live cell microscopy. Huh7 cells were transfected with a mCherry–DFCP1 expression plasmid and at 48 h post-transfection cells were infected with Jc1–GFP (0.5 f.f.u. per cell). At 24 h p.i. live cell imaging was performed and movies of infected cells captured. (a) Representative images captured at the time points indicated. Bars, 2 μm. (b) A montage of the images indicated in the white squares in (a), captured every 120 s.
Fig. 7.
Transient association of nascent HCV RNA replication complexes and DFCP1 imaged by live cell microscopy. Huh7 cells were transfected with a mCherry–DFCP1 expression plasmid and at 48 h post-transfection cells were infected with Jc1–GFP (0.5 f.f.u. per cell). At 24 h p.i., live cell imaging was performed and movies of infected cells captured. (a) Representative images captured at the time points indicated. Bars, 2 μm. (b) A montage of the images indicated in the white squares in (a), captured at the times indicated.
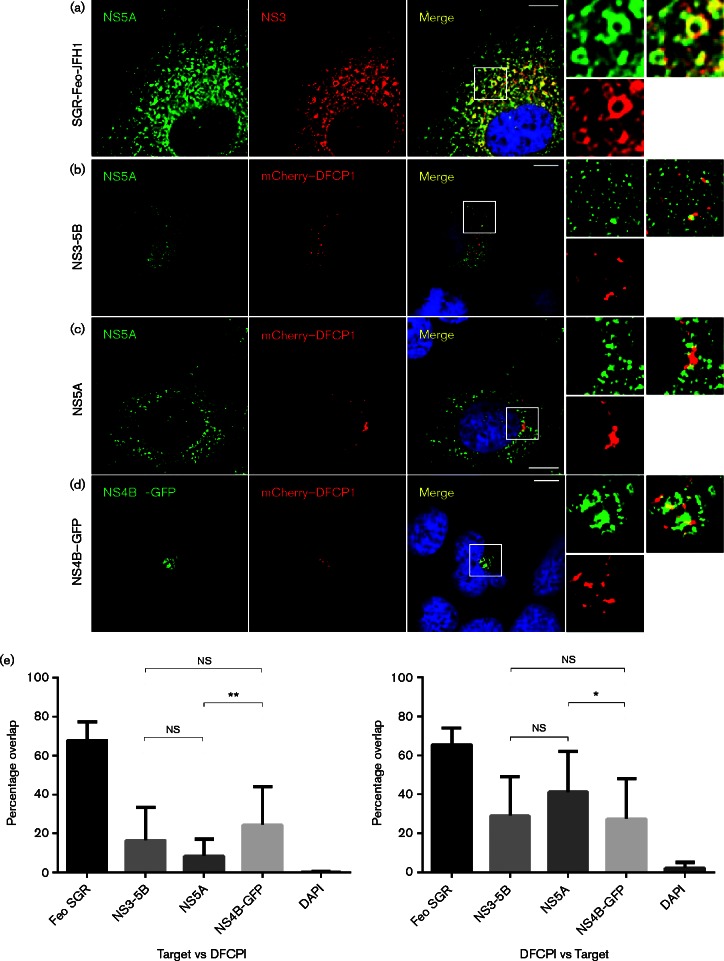
We then reasoned that the transient colocalization of replication complexes and DFCP1 might be an intrinsic function of one of the non-structural proteins. NS4B and NS5A were the most likely candidates as both have been shown to individually generate DMV or single-membrane vesicles (SMV) (Paul et al., 2013; Romero-Brey et al., 2012). Additionally, recent evidence has pointed to a role for NS4B in the induction of autophagy and accumulation of autophagosomes (Su et al., 2011; Wang et al., 2015). Furthermore it was conceivable that subsequent formation of active replication complexes then did not require DFCP1 and the colocalization was disrupted. To test this hypothesis we co-transfected Huh7 cells with plasmids expressing a JFH-1 NS3-5B polyprotein from a human cytomegalovirus (CMV) promoter (Ross-Thriepland et al., 2013) and mCherry–DFCP1 (Fig. 8). Both NS5A and mCherry–DFCP1 exhibited intense punctate staining over a background of a less bright reticular distribution, with NS5A showing a more dispersed pattern throughout the cytoplasm. Quantitative analysis of the colocalization revealed that, in comparison with the positive control (NS5A-NS3 colocalization in SGR-Feo-JFH1 cells), there was a low but significant level of colocalization of NS5A and DFCP1. This analysis (Fig. 8e) showed that approximately 20 % of NS5A-positive punctae were also DFCP1-positive, whereas ∼30 % of DFCP1 punctae were NS5A-positive. Similar observations were made in Huh7 cells transfected with plasmids expressing either JFH-1 NS5A or NS4B–GFP with mCherry–DFCP1. We conclude that both NS4B and NS5A transiently associate with the autophagic machinery (exemplified by DFCP1), but once active HCV replication complexes are formed these associations are disrupted.
Fig. 8.
Partial colocalization of NS4B or NS5A expressed alone or in the context of the remaining replicase proteins with DFCP1. (a) SGR-Feo-JFH1-harbouring cells were fixed and stained with antibodies to NS5A and NS3. Huh7 cells were co-transfected with expression plasmids for mCherry–DFCP1 and (b) JFH-1 NS3-5B, (c) NS5A or (d) NS4B–GFP for 48 h. In (b) and (c) cells were fixed and stained with a sheep polyclonal antiserum to NS5A (green). Bars, 10 μm. (e) Colocalization analysis of images from (a–d). Mean Manders’ overlap coefficients were derived from >10 cells for each condition. Data represent the mean ± sd. *P < 0.05. **P < 0.005.
Discussion
In this study we demonstrated that early events in the generation of autophagosomes play a key role in the biogenesis of the membranous compartment required for HCV genome replication. Specifically, our data point to roles for the class III PI3K, Vps34, and the double FYVE-domain containing protein, DFCP1 (which specifically binds to PI3P lipids), in this process. This conclusion stems from the following lines of evidence.
Pharmacological inhibition demonstrated that class III PI3K activity was required for HCV genome replication.
Silencing of Vps34 and DFCP1 expression by siRNA strongly inhibited HCV genome replication both in the context of a SGR (genotypes 2a and 1b) and during virus infection, whilst having no effect on virus entry or the translation of incoming viral RNA.
Live cell imaging revealed evidence for transient colocalization of NS5A with DFCP1 during virus infection, before the two proteins dissociated into distinct structures.
Our data complement the recent, elegant study of Romero-Brey et al. (2012), who showed that the membranous web in HCV-infected cells comprised predominantly DMVs and derived from the ER. In particular, they identified DMVs as protrusions from the ER membrane. The potential parallels between this process and the formation of autophagosomes are striking, particularly with regard to the role of DFCP1 in mediating the formation of omegasomes at ER membranes (Axe et al., 2008). Romero-Brey et al. (2012) also highlighted the similarities between the morphology of the membrane rearrangements seen in HCV infection and those of unrelated viruses such as the coronaviruses and arteriviruses. Further to this, another recent study (Cottam et al., 2011) demonstrated that the nsp6 protein of various coronaviruses (including the SARS coronavirus), or the nsp5-7 protein of the arterivirus, porcine reproductive and respiratory syndrome virus (PPRSV), located to the ER where they recruited Vps34 and DFCP1, leading to omegasome formation. Our data complement this study and we propose therefore that the similarities between HCV and coronavirus or arterivirus membrane rearrangements may, at least in part, be explained by a common mechanism of biogenesis of these membrane structures.
In contrast, a number of other viruses have previously been reported to usurp the autophagic machinery in distinct ways to facilitate genome replication. Poliovirus was the first such virus and it has been documented that the viral 2BC and 3A proteins colocalize with LC3 and the late endosomal marker LAMP-1, leading to the proposal that these proteins recruit modified LC3 to form a complete autophagosome-like vesicle which functions as a platform for RNA replication (Taylor & Kirkegaard, 2007). A distinct example is dengue virus (DENV), a virus in the Flavivirus genus and therefore more closely related to HCV. DENV infection has been shown to induce autophagy which promotes viral replication (Lee et al., 2008). Furthermore, it was observed that the DENV NS1 protein and dsRNA (as markers of replication complexes) colocalized with LC3, the ribosomal protein L28 and the mannose 6-phosphate receptor, suggesting that the DENV replication/translation machinery is present on amphisomes, which result from the fusion of autophagosomes and endosomes (Panyasrivanit et al., 2009). The differences between HCV and DENV are intriguing, given their close phylogenetic relationship; further to this the membrane architecture of DENV replication complexes is distinct from that of HCV (Welsch et al., 2009), implying that the biogenesis of these structures is via distinct mechanisms.
We (Mohl et al., 2012), and others (Sir et al., 2008), have previously shown that the five non-structural proteins (NS3-5B) encoded by the SGR are necessary for the induction of autophagy by HCV. The roles of the individual proteins in this process remains unclear, although it has been shown that both NS4B and NS5B have been shown to physically interact with components of the autophagic machinery – NS4B with Rab5 and Vps34 (Su et al., 2011), and NS5B with Atg5 (Guévin et al., 2010). We have performed co-immunoprecipitation experiments but these have not yielded any biochemical evidence for a physical interaction between DFCP1 and either NS4B or NS5A. However, we cannot rule out the possibility that the transient colocalization of NS4B/NS5A and DFCP1 suggests that any interaction would be both weak and short-lived, thus hampering a biochemical analysis. It would be of interest to investigate if other proteins known to be involved in the early stages of autophagosome formation, such as the WIPI proteins (Dooley et al., 2014), which, like DFCP1, are also PI3K effectors. Such experiments are ongoing in our laboratory.
Further to the involvement of DFCP1, our data demonstrate a novel role for the class III PI3K Vps34 in HCV genome replication which has not previously been documented. Although Su et al. (2011) showed that 3-MA treatment or siRNA silencing of Vps34 reduced levels of HCV-mediated LC3 lipidation, another study (Sir et al., 2012) reported these treatments had no effect. Importantly, neither study showed any data pertaining to effects on virus replication. However, indirect support for a role of Vps34 comes from a report that HCV infection upregulated Beclin1 expression, which could favour the formation of a Vps34-Vps15-Beclin1-Atg14L complex (Shrivastava et al., 2011), together with the observation that Beclin1 silencing reduced HCV genome replication (Dreux et al., 2009).
Collectively, these results are consistent with a model for the formation of the DMVs required for HCV genome replication, this is presented in Fig S1 (available in the online Supplementary Material). We propose that the replication complex (NS3-5B) interacts with the early autophagic machinery (Vps34 and associated proteins) on the ER membrane. Vps34 activity is then required to produce a concentration of PI3P on the ER membrane which in turn recruits DFCP1. DFCP1 then promotes the formation of omegasomes and the replication complexes are recruited on to these nascent structures. The mechanisms by which autophagosomes are generated from omegasomes are not entirely clear. In a simplified model (Simonsen & Stenmark, 2008), omegasomes bud from the ER membrane to become autophagosomes. Our model would predict that as the omegasomes bud from the ER membrane, instead of becoming autophagosomes their progression is subverted to the formation of the DMVs which ultimately comprise the membranous web. However, a more complex model (Hayashi-Nishino et al., 2009) suggests that the isolation membrane is generated from the tip of the omegasome, engulfing cytoplasmic components and then separating away from the omegasome. Of note, the timescale of this process as observed in this study (up to 10 min from formation of the NS5A–DFCP1 puncta to their eventual dissociation – see Figs 6 and 7), is consistent with the timescale observed previously for the transient association of GFP–DFCP1 and RFP–LC3 during autophagosome formation (also approximately 10 min) (Axe et al., 2008). The difference in size between autophagosomes ( < 1.5 μm) and DMVs (100–150 nm) (Romero-Brey et al., 2012) is also consistent with the hypothesis that DMVs derive from autophagosomes.
Clearly our observations raise additional questions, such as how are replication complexes transferred from the ER membrane on to the omegasomes and subsequently into the DMVs? Intriguingly, we (Mohl et al., 2012), and others (Dreux et al., 2009), do not see colocalization of NS5A (as a marker for replication complexes) with LC3, suggesting that the replication complex might be reconfiguring autophagosome membranes, and possibly displacing some of the proteins normally found in that compartment. Again, this is a controversial area with other studies providing evidence for colocalization of LC3 with NS5A and NS5B (Sir et al., 2012); clearly this issue also needs to be clarified. The complex mechanics of membrane rearrangements during the morphogenesis of both autophagosomes and the HCV replication factory remain obscure.
Methods
Cell culture
Huh7 and 7.5 cells were cultured in Dulbecco's modified Eagle's Medium (DMEM), supplemented with 10 % (v/v) FBS, 100 U penicillin ml− 1, 100 μg streptomycin ml− 1, 2 mM l-glutamine and 1 % (v/v) non-essential amino acids (Gibco) at 37 °C, 5 % CO2, in a humidified incubator. Huh7 cells stably harbouring subgenomic replicons were maintained in G418 (Melford) at 500 μg ml− 1.
Virus assays
For virus infection, medium removed from Huh7.5 cells previously transfected with the appropriate in vitro transcribed virus RNA was clarified by centrifugation at 1200 g for 5 min. Huh7 cells were seeded onto a 96-well microtitre plate prior to titration of virus by focus forming assay as previously described (Mohl et al., 2012). During subsequent experiments, cells were routinely infected at an m.o.i. of 0.5 focus forming units (f.f.u.) per cell.
Construction of mCherry–DFCP1
The mCherry–DFCP1 construct was generated by excising the DFCP1 coding sequence from a GFP–DFCP1 construct (a kind gift from Nicholas Ktistakis, Babraham Institute, Cambridge; Axe et al., 2008) using XhoI/BamHI and cloning into an mCherry expression plasmid (Addgene).
DNA transfections
Prior to transfections, cells were incubated with serum-free medium. Transfection mixtures comprised 2 μg DNA ml− 1, 10 μg polyethylenimine (PEI) ml− 1, in Optimem (GIBCO), or 0.25 μg DNA ml− 1, 0.5 μl FuGENE6 transfection reagent (Promega) in Optimem, and were equilibrated at room temperature for 45 min prior to inoculation into the appropriate wells and incubation at 37 °C. Eight hours post-transfection, the transfection medium was replaced by fresh, complete, DMEM.
siRNA transfections
For siRNA experiments, 7.5 × 104 cells had previously been seeded 24 h before transfection. Cells were transfected with siRNA (75 pmol) using Lipofectamine RNAiMAX (Invitrogen) in Optimem (GIBCO) for 24 h. Transfection medium was subsequently replaced by fresh DMEM as previously described. Silencing was allowed to progress for 72 h in replicon-harbouring cells, and 120 h in virus-infected cells, post-transfection, before the cells were harvested for analysis using either Glasgow lysis buffer (GLB) or passive lysis buffer (PLB; Promega).
Western blotting
The polyclonal sheep anti-NS5A serum was described previously (Macdonald et al., 2003). Other antibodies were obtained from either Cell Signalling Technologies (Vps34) or Abcam (GAPDH, LC3 and DFCP1). AP33 (Genentech) was used to detect the HCV envelope glycoprotein E2. For Western blotting, cells were lysed in GLB (Macdonald et al., 2003) – comprising 10 mM PIPES-NaOH, pH 7.2, 1 % Triton X-100, 120 mM KCl, 30 mM NaCl, 5 mM MgCl2, 10 % glycerol, 0.1 % SDS and protease inhibitors. Lysates were incubated on ice for 30 min prior to centrifugation at 2800 g for 5 min. Protein concentrations were determined by bicinchoninic acid (BCA) assay. Equal amounts of protein (10 μg) were resolved by 12 % or 15 % SDS-PAGE. Proteins were transferred to a PVDF membrane (Millipore) and blocked for 1 h in 1 × TBS containing 5 % BSA or 10 % skimmed milk powder. Membranes were probed with appropriate primary antibodies overnight at 4 °C followed by horseradish-peroxidase-conjugated secondary antibodies (Sigma), and Western blots visualized using an in-house enhanced chemiluminescence (ECL) reagent.
Luciferase assay
Luciferase activity was measured by lysing 2 × 105 cells in 150 μl PLB (Promega). Lysates were incubated on ice for 30 min prior to centrifugation at 2800 g for 5 min. Fifty microlitres of cell lysate was dispensed into a white-bottomed, 96-well plate (Greiner), which was read in a BMG Labtech optical plate reader following the addition of 50 μl of either Luciferase Assay Reagent or Stop & Glo (Promega).
Indirect immunofluorescence microscopy
Huh7 cells (2 × 104) that had previously been seeded onto coverslips in 24-well tissue culture plates were prepared by washing the coverslips three times in PBS before fixing in 4 % paraformaldehyde (PFA) for 10 min. PFA was removed and the coverslips were washed two times in PBS. For permeabilization, 0.2 % Triton X-100 in PBS was added to the wells and incubated at room temperature (RT) for 10 min. Permeabilization solution was removed and the coverslips washed three times in PBS. Antibodies were diluted in 1 × PBS and incubated for 1 h at RT. Following antibody removal, unbound antibody was removed with three 1 × PBS washes. Coverslips were drained of excess fluid and mounted on slides using Vector Shield (Vector Laboratories) or ProLong Gold (Life Technologies) and sealed with nail varnish. Coverslips were imaged using a Deltavision Deconvolution Microscope for replicon or transfected cells.
Live cell microscopy
Huh7 cells (5 × 104) were seeded onto 35 mm glass-bottom dishes (Ibidi) 24 h prior to transfection with mCherry–DFCP1 plasmid using PEI. Eight hours post-transfection, medium was replaced by fresh DMEM, and at 48 h post-transfection, the transfected cells were infected with Jc1-GFP virus. 24 h p.i., virus-containing medium was replaced with phenol-red-free DMEM medium. Cells were imaged using an LSM 780 confocal microscope (Zeiss) under BSL3 containment conditions. Co-infected/transfected cells were identified and imaged. The image series collected were coalesced into videos at 12 frames per second.
Colocalization analysis
Microscopy images were analysed in ImageJ (NIH) with colocalization studies conducted using the JACoP plugin (Bolte & Cordelières, 2006). Statistical significance was calculated using Welch's unpaired t-test in Prism.
Acknowledgements
We thank Charles Rice (The Rockefeller University, New York) for the Huh7.5 cells and J6/JFH-1Rluc construct, Takaji Wakita (National Institute of Infectious Diseases, Tokyo) for the pJFH-1 construct and SGR-neo-JFH-1 replicon construct, David Wyles (UCSD) for the SGR-Feo-JFH1 and SGR-Feo-Con1 replicon constructs, Ralf Bartenschlager (University of Heidelberg) for the Jc1 and Jc1–GFP virus constructs and Nicholas Ktistakis (Babraham Institute, Cambridge) for the EGFP–DFCP1 construct. We are grateful to Douglas Ross-Thriepland and Philip Tedbury for helpful advice during this project. This work was funded by a Wellcome Trust Investigator Award to M. H. (grant number 096670). C. B. is a PhD student funded by a Wellcome Trust Four Year PhD scheme entitled The Molecular Basis of Biological Mechanisms (grant number 099759), J. M. is a Royal Society University Research Fellow (grant number UF100419), the Zeiss LSM780 confocal microscope used for live cell imaging of HCV infected cells was funded by a Royal Society equipment grant (grant number RG110306).
Supplementary Data
Supplementary Data
References
- Ait-Goughoulte M., Kanda T., Meyer K., Ryerse J. S., Ray R. B., Ray R. (2008). Hepatitis C virus genotype 1a growth and induction of autophagy J Virol 822241–2249 10.1128/JVI.02093-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axe E. L., Walker S. A., Manifava M., Chandra P., Roderick H. L., Habermann A., Griffiths G., Ktistakis N. T. (2008). Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum J Cell Biol 182685–701 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger C., Romero-Brey I., Radujkovic D., Terreux R., Zayas M., D. Paul, Harak C., Hoppe S., Gao M., other authors (2014). Daclatasvir-like inhibitors of NS5A block early biogenesis of hepatitis C virus-induced membranous replication factories, independent of RNA replication Gastroenterology 1471094–1105, e25 10.1053/j.gastro.2014.07.019. [DOI] [PubMed] [Google Scholar]
- Bolte S., Cordelières F. P. (2006). A guided tour into subcellular colocalization analysis in light microscopy J Microsc 224213–232 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- Codogno P., Mehrpour M., Proikas-Cezanne T. (2012). Canonical and non-canonical autophagy: variations on a common theme of self-eating? Nat Rev Mol Cell Biol 137–12. [DOI] [PubMed] [Google Scholar]
- Cottam E. M., Maier H. J., Manifava M., Vaux L. C., Chandra-Schoenfelder P., Gerner W., Britton P., Ktistakis N. T., Wileman T. (2011). Coronavirus nsp6 proteins generate autophagosomes from the endoplasmic reticulum via an omegasome intermediate Autophagy 71335–1347 10.4161/auto.7.11.16642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley H. C., Razi M., Polson H. E., Girardin S. E., Wilson M. I., Tooze S. A. (2014). WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12-5-16L1 Mol Cell 55238–252 10.1016/j.molcel.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreux M., Chisari F. V. (2010). Viruses and the autophagy machinery Cell Cycle 91295–1307 10.4161/cc.9.7.11109. [DOI] [PubMed] [Google Scholar]
- Dreux M., Gastaminza P., Wieland S. F., Chisari F. V. (2009). The autophagy machinery is required to initiate hepatitis C virus replication Proc Natl Acad Sci U S A 10614046–14051 10.1073/pnas.0907344106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger D., Wölk B., Gosert R., Bianchi L., Blum H. E., Moradpour D., Bienz K. (2002). Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex J Virol 765974–5984 10.1128/JVI.76.12.5974-5984.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallay P. A. (2009). Cyclophilin inhibitors Clin Liver Dis 13403–417 10.1016/j.cld.2009.05.002. [DOI] [PubMed] [Google Scholar]
- Gao M., Nettles R. E., Belema M., Snyder L. B., Nguyen V. N., Fridell R. A., Serrano-Wu M. H., Langley D. R., Sun J. H., other authors (2010). Chemical genetics strategy identifies an HCV NS5A inhibitor with a potent clinical effect Nature 46596–100 10.1038/nature08960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosert R., Egger D., Lohmann V., Bartenschlager R., Blum H. E., Bienz K., Moradpour D. (2003). Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons J Virol 775487–5492 10.1128/JVI.77.9.5487-5492.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guévin C., Manna D., Bélanger C., Konan K. V., Mak P., Labonté P. (2010). Autophagy protein ATG5 interacts transiently with the hepatitis C virus RNA polymerase (NS5B) early during infection Virology 4051–7 10.1016/j.virol.2010.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki M., Furuta N., Matsuda A., Nezu A., Yamamoto A., Fujita N., Oomori H., Noda T., Haraguchi T., other authors (2013). Autophagosomes form at ER-mitochondria contact sites Nature 495389–393 10.1038/nature11910. [DOI] [PubMed] [Google Scholar]
- Hayashi-Nishino M., Fujita N., Noda T., Yamaguchi A., Yoshimori T., Yamamoto A. (2009). A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation Nat Cell Biol 111433–1437 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- He Y., Nakao H., Tan S. L., Polyak S. J., Neddermann P., Vijaysri S., Jacobs B. L., Katze M. G. (2002). Subversion of cell signaling pathways by hepatitis C virus nonstructural 5A protein via interaction with Grb2 and P85 phosphatidylinositol 3-kinase J Virol 769207–9217 10.1128/JVI.76.18.9207-9217.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C. T., Murray C. L., Eastman D. K., Tassello J., Rice C. M. (2007). Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus J Virol 818374–8383 10.1128/JVI.00690-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. R., Lei H. Y., Liu M. T., Wang J. R., Chen S. H., Jiang-Shieh Y. F., Lin Y. S., Yeh T. M., Liu C. C., Liu H. S. (2008). Autophagic machinery activated by dengue virus enhances virus replication Virology 374240–248 10.1016/j.virol.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald A., Crowder K., Street A., McCormick C., Saksela K., Harris M. (2003). The hepatitis C virus NS5A protein inhibits activating protein-1 function by perturbing Ras-ERK pathway signalling J Biol Chem 27817775–17784 10.1074/jbc.M210900200. [DOI] [PubMed] [Google Scholar]
- Mohl B. P., Tedbury P. R., Griffin S., Harris M. (2012). Hepatitis C virus-induced autophagy is independent of the unfolded protein response J Virol 8610724–10732 10.1128/JVI.01667-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panyasrivanit M., Khakpoor A., Wikan N., Smith D. R. (2009). Co-localization of constituents of the dengue virus translation and replication machinery with amphisomes J Gen Virol 90448–456 10.1099/vir.0.005355-0. [DOI] [PubMed] [Google Scholar]
- Paul D., Hoppe S., Saher G., Krijnse-Locker J., Bartenschlager R. (2013). Morphological and biochemical characterization of the membranous hepatitis C virus replication compartment J Virol 8710612–10627 10.1128/JVI.01370-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietschmann T., Kaul A., Koutsoudakis G., Shavinskaya A., Kallis S., Steinmann E., Abid K., Negro F., Dreux M., other authors (2006). Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras Proc Natl Acad Sci U S A 1037408–7413 10.1073/pnas.0504877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley S. H., Ktistakis N., Davidson K., Anderson K. E., Manifava M., Ellson C. D., Lipp P., Bootman M., Coadwell J., other authors (2001). FENS-1 and DFCP1 are FYVE domain-containing proteins with distinct functions in the endosomal and Golgi compartments J Cell Sci 1143991–4000. [DOI] [PubMed] [Google Scholar]
- Romero-Brey I., Merz A., Chiramel A., Lee J. Y., Chlanda P., Haselman U., Santarella-Mellwig R., Habermann A., Hoppe S., other authors (2012). Three-dimensional architecture and biogenesis of membrane structures associated with hepatitis C virus replication PLoS Pathog 8e1003056. 10.1371/journal.ppat.1003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Thriepland D., Amako Y., Harris M. (2013). The C terminus of NS5A domain II is a key determinant of hepatitis C virus genome replication, but is not required for virion assembly and release J Gen Virol 941009–1018 10.1099/vir.0.050633-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller T., Appel N., Koutsoudakis G., Kallis S., Lohmann V., Pietschmann T., Bartenschlager R. (2007). Analysis of hepatitis C virus superinfection exclusion by using novel fluorochrome gene-tagged viral genomes J Virol 814591–4603 10.1128/JVI.02144-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastava S., Raychoudhuri A., Steele R., Ray R., Ray R. B. (2011). Knockdown of autophagy enhances the innate immune response in hepatitis C virus-infected hepatocytes Hepatology 53406–414 10.1002/hep.24073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen A., Stenmark H. (2008). Self-eating from an ER-associated cup J Cell Biol 182621–622 10.1083/jcb.200807061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sir D., Chen W. L., Choi J., Wakita T., Yen T. S., Ou J. H. (2008). Induction of incomplete autophagic response by hepatitis C virus via the unfolded protein response Hepatology 481054–1061 10.1002/hep.22464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sir D., Kuo C. F., Tian Y., Liu H. M., Huang E. J., Jung J. U., Machida K., Ou J. H. (2012). Replication of hepatitis C virus RNA on autophagosomal membranes J Biol Chem 28718036–18043 10.1074/jbc.M111.320085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street A., Macdonald A., Crowder K., Harris M. (2004). The hepatitis C virus NS5A protein activates a phosphoinositide 3-kinase-dependent survival signaling cascade J Biol Chem 27912232–12241 10.1074/jbc.M312245200. [DOI] [PubMed] [Google Scholar]
- Su W. C., Chao T. C., Huang Y. L., Weng S. C., Jeng K. S., Lai M. M. (2011). Rab5 and class III phosphoinositide 3-kinase Vps34 are involved in hepatitis C virus NS4B-induced autophagy J Virol 8510561–10571 10.1128/JVI.00173-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I. (2011). Autophagy basics Microbiol Immunol 551–11 10.1111/j.1348-0421.2010.00271.x. [DOI] [PubMed] [Google Scholar]
- Taylor M. P., Kirkegaard K. (2007). Modification of cellular autophagy protein LC3 by poliovirus J Virol 8112543–12553 10.1128/JVI.00755-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakita T., Pietschmann T., Kato T., Date T., Miyamoto M., Zhao Z., Murthy K., Habermann A., Kräusslich H. G., other authors (2005). Production of infectious hepatitis C virus in tissue culture from a cloned viral genome Nat Med 11791–796 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Tian Y., Ou J. H. (2015). HCV induces the expression of Rubicon and UVRAG to temporally regulate the maturation of autophagosomes and viral replication PLoS Pathog 11e1004764. 10.1371/journal.ppat.1004764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsch S., Miller S., Romero-Brey I., Merz A., Bleck C. K., Walther P., Fuller S. D., Antony C., Krijnse-Locker J., Bartenschlager R. (2009). Composition and three-dimensional architecture of the dengue virus replication and assembly sites Cell Host Microbe 5365–375 10.1016/j.chom.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y. T., Tan H. L., Shui G., Bauvy C., Huang Q., Wenk M. R., Ong C. N., Codogno P., Shen H. M. (2010). Dual role of 3-methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide 3-kinase J Biol Chem 28510850–10861 10.1074/jbc.M109.080796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyles D. L., Kaihara K. A., Korba B. E., Schooley R. T., Beadle J. R., Hostetler K. Y. (2009). The octadecyloxyethyl ester of (S)-9-[3-hydroxy-2-(phosphonomethoxy) propyl]adenine is a potent and selective inhibitor of hepatitis C virus replication in genotype 1A, 1B, and 2A replicons Antimicrob Agents Chemother 532660–2662 10.1128/AAC.01546-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data