Summary
Cancer progression depends on both cell-intrinsic processes and interactions between different cell types. However, large scale assessment of cell type composition and molecular profiles of individual cell types within tumors remains challenging. To address this, we developed Epigenomic Deconvolution (EDec), an in silico method that infers cell type composition of complex tissues as well as DNA methylation and gene transcription profiles of constituent cell types. By applying EDec to The Cancer Genome Atlas (TCGA) breast tumors we detect changes in immune cell infiltration related to patient prognosis, and a striking change in stromal fibroblast to adipocyte ratio across breast cancer subtypes. We further show that a less adipose stroma tends to display lower levels of mitochondrial activity and to be associated with cancerous cells with higher levels of oxidative metabolism. These findings highlight the role of stromal composition in the metabolic coupling between distinct cell types within tumors.
eTOC blurb
Onuchic et al. develop an in silico deconvolution technique (EDec) that can accurately estimate cell type composition and molecular profiles of constituent cell types in the context of breast tumors. Application to breast cancers from TCGA data reveals association between stromal composition and the metabolic phonotype of breast tumors. Explore consortium data at the Cell Press IHEC webportal at www.cell.com/consortium/IHEC.
Introduction
Molecular profiling of breast tumors has led to their categorization into different subtypes with distinct risks and underlying biology. Of particular interest is the classification into 5 intrinsic subtypes, which can be performed using the PAM50 classifier (Parker et al., 2009). However, most molecular profiling studies to date have been performed on bulk tissue samples, ignoring the complexity of the breast tissue, with its multiple cell types and the interactions between them. Valuable evidence for the significance of heterotypic interactions comes from the study of cell type composition of tumors, as exemplified by the prognostic value of immune cell infiltration (Coussens et al., 2013; Liu et al., 2014) and of epigenomic (Hu et al., 2005) and transcriptomic (Finak et al., 2008) perturbations within stromal cells (Tlsty and Coussens, 2006). Laser capture microdissection (LCM), cell sorting, and other physical methods to isolate cell types from solid tumors for molecular profiling are technically challenging, and severely limit throughput (Debey et al., 2004). A number of methods for in silico deconvolution have been developed to address this problem using as input gene expression profiles (Aran et al., 2015; Gentles et al., 2015; Houseman and Ince, 2014; Kuhn et al., 2011; Li and Xie, 2013; Newman et al., 2015; Shen-Orr et al., 2010; Venet et al., 2001; Yoshihara et al., 2013; Zhong et al., 2013) and, more recently, DNA methylation profiles (Houseman et al., 2012, 2014, 2016; Zheng et al., 2014; Zou et al., 2014; Rahmani et al., 2016) of tissue homogenates. However, the ability of these methods to infer cell type composition of solid tumors and interpret the states of constituent cell types is limited, thus hampering the study of cellular states and cellular interactions within the tumor microenvironment.
To address this gap, we developed EDec, a deconvolution method based on a heuristic for constrained matrix factorization using quadratic programming. The deconvolution is based on cell-type specific patterns of DNA methylation. Such patterns are acquired during normal cellular differentiation, maintained through cell division, and serve as chemically stable cellular markers. We reasoned that methylation profiles would be more amenable to deconvolution than gene expression due to their linearity, measurement within the complete (0–1) dynamic range, and technology-independence (including both bisulfite sequencing and array platforms).
Previous methylation-based deconvolution methods either make direct use of reference methylation profiles of constituent cell types (Houseman et al., 2012) or ignore such references (Houseman et al., 2014, 2016; Rahmani et al., 2016; Zou et al., 2014). Highly accurate reference methylation profiles, essential for reference-based deconvolution approaches, are unavailable for many solid tissues, arguing for a reference-free approach. However, reference methylation profiles from representative cell lines are available and can provide valuable information if used to improve inference while minimizing bias. Toward this goal, EDec uses relevant reference information in indirect ways to minimize bias. First, it uses references to identify sets of loci that are likely to exhibit variation in methylation levels across constituent cell types of a given tissue (feature selection), while taking a reference-free approach to the deconvolution problem itself. Second, it identifies constituent cell types by comparing their deconvoluted molecular profiles to reference profiles.
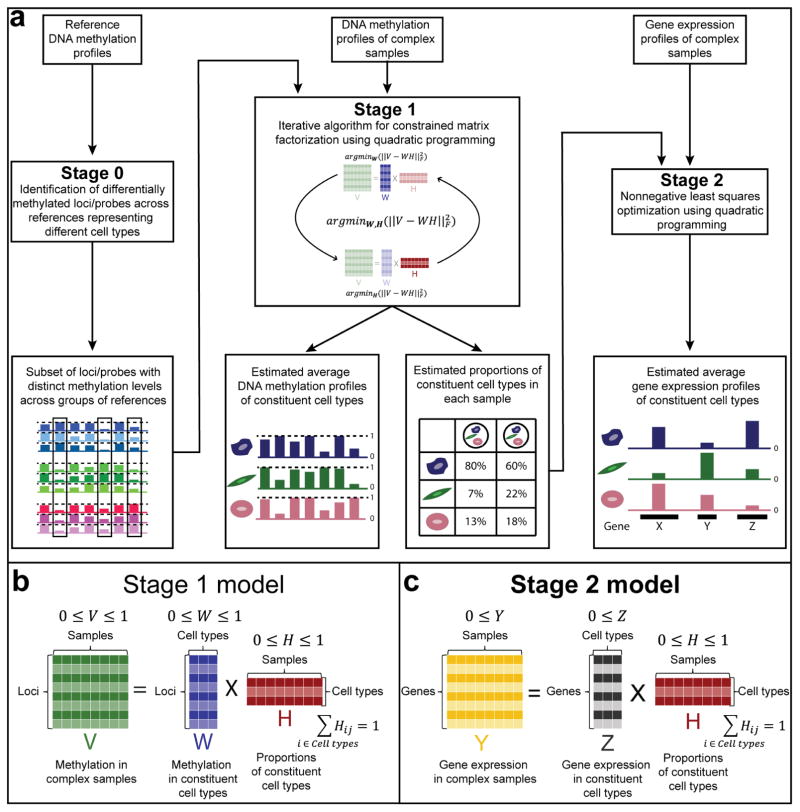
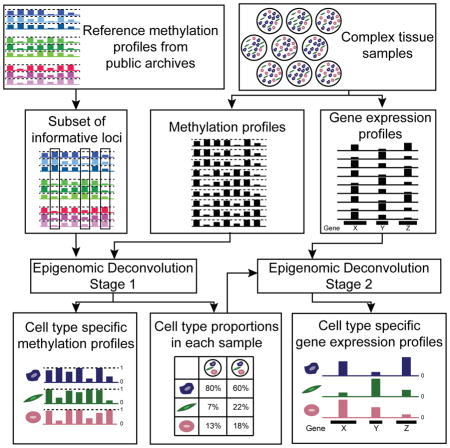
EDec consists of three stages (0,1, and 2, Figure 1). Starting with methylation profiles of tumor homogenates over loci selected based on reference methylation profiles (Figure 1a - Stage 0), EDec estimates both cell type proportions and methylation profiles of constituent cell types using a reference-free approach (Figure 1a - Stage 1) similar to previous reference-free techniques (Gaujoux and Seoighe, 2011; Houseman et al., 2016). The proportion estimates are then used as a “key” to deconvolute gene expression profiles of constituent cell types (Figure 1a - Stage 2).
Figure 1. Description of the EDec method.
(a) The EDec method has 2 main stages (Stages 1 and 2), preceded by a preparation stage (Stage 0). In Stage 0, a set of reference methylation profiles is used to select a set of genomic loci or array probes with distinct methylation levels across groups of references representing different constituent cell types. Methylation profiles of complex tissue samples over the set of loci/probes selected in Stage 0 are used as the input for the Stage 1 of the EDec method. In Stage 1, EDec estimates both the average methylation profiles of constituent cell types and the proportions of constituent cell types in each input sample using an iterative algorithm for constrained matrix factorization using quadratic programming. Stage 2 of EDec takes as input the gene expression profiles of the same tissue samples profiled for DNA methylation, as well as the proportions of constituent cell types for those samples, estimated in Stage 1, and outputs the gene expression profiles of constituent cell types. (b) Representation of the model associated with Stage 1 of EDec method. (c) Representation of the model used for gene expression deconvolution in Stage 2 of the EDec method.
EDec proof of concept experiments were performed using both Illumina methylation arrays and RainDance Technologies’ ThunderStorm BS-seq (Komori et al., 2011; Paul et al., 2014) targeted bisulfite sequencing. The method is validated using both computer simulations and profiling experiments on prepared cell mixtures. By applying EDec to the breast cancer datasets generated by The Cancer Genome Atlas (TCGA) (The Cancer Genome Atlas Network, 2012) we predict cellular proportions and methylation states of constituent cell types within breast tumors as well as infer changes in gene expression within each constituent cell type. Such predictions were largely confirmed by comparisons with cell type composition estimates based on H&E staining, and by comparison against gene expression profiles of specific cell types isolated through LCM. We show that cancerous epithelial cells exhibit methylomes distinct from those of normal epithelium. EDec also replicates the previously reported association between increased immune cell infiltration in triple negative breast cancer and better prognosis (Adams et al., 2014). We further detect expression changes that are highly consistent with known hallmarks of cancer, and with known roles of specific cell types within breast cancer. Lastly, we observe that the degree of stromal adiposity across breast cancer subtypes predicts the pattern of metabolic coupling observed between cancer epithelium and stroma.
Results
Epigenomic Deconvolution Method
The first stage of EDec (Figure 1a - Stage 1) performs constrained matrix factorization to find cell type specific methylation profiles and constituent cell type proportions that minimize the Euclidian distance between their linear combination and the original matrix of tissue methylation profiles (Figure 1b). The minimization algorithm involves an iterative procedure that, in each round, alternates between estimating constituent cell type proportions and methylation profiles by solving constrained least squares problems through quadratic programming. The minimization problem is made tractable by the constraints that methylation measurements (beta values) and cell type proportions are numbers in the [0,1] interval, and that cell type proportions within a sample add up to one. These constraints restrict the space of possible solutions, thus making it possible for the local iterative search to reproducibly find a global minimum and an accurate solution. One key requirement for EDec is that cell type proportions vary across samples. A second requirement is that there must be significant differences across constituent cell type methylation profiles. The latter requirement can be met by providing EDec with loci expected to vary in methylation levels across constituent cell types (Figure 1a - Stage 0).
Similar to how tissue methylation profiles are modelled, tissue gene expression profiles can also be modeled by the linear combination of the expression profiles of its constituent cell types. However, due to the less constrained nature of gene expression measurements ([0,∞]) vs. methylation measurements ([0,1]), the same reference-free approach used in Stage 1 is not as effective for gene expression deconvolution. Therefore, instead of using that approach, when both DNA methylation and gene expression profiles are available for the same set of samples (e.g., from the same tissue homogenate), EDec-Stage 2 uses the cell proportions estimated in Stage 1 as a fixed input when estimating the average gene expression profiles of constituent cell types through a constrained least squares fit using quadratic programming with solutions constrained to [0,∞] (Figure 1a - Stage 2 and Figure 1c).
Validation using in silico mixtures of methylation profiles derived from breast cancer-related cell lines
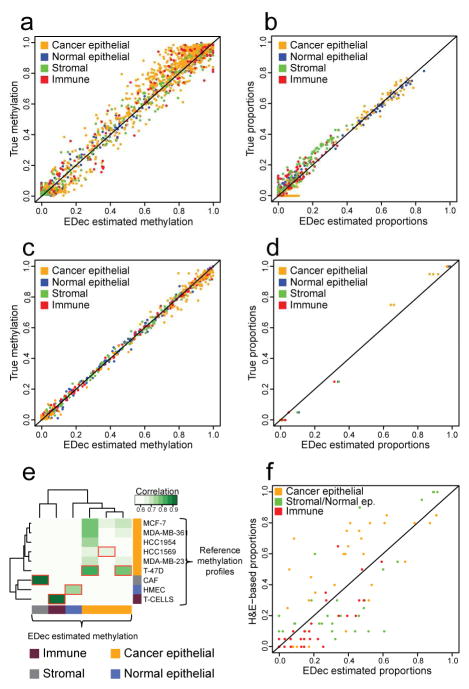
We first validated the core EDec algorithm (Stage 1) on simulated mixtures of experimentally derived DNA methylation profiles (9 cell lines: 6 breast cancer, 1 normal breast epithelial, 1 immune, and 1 cancer associated fibroblast (CAF)). Among the 1,000 target genomic regions included in this breast cancer methylation-focused panel (Supplemental Table 2), 149 exhibited particularly distinct methylation patterns across different breast cell types (based on reference epigenomes) (Kundaje et al., 2015), and were used in EDec Stage 1. The simulation dataset consisted of 100 mixtures, each composed of 4 cell types (one breast cancer cell line, one normal mammary epithelial cell type, one stromal cell type, and one immune cell type). About half of the simulated mixtures contained on average higher levels of breast cancer (60%) and immune cell types (20%), representing distributions observed in tumor samples such as those in the TCGA dataset. To simulate the presence of different breast cancer subtypes, different simulated mixtures had a different cancerous epithelium constituent. Specifically, the breast cancer cell type for each mixture was chosen randomly from the set of 6 breast cancer cell lines. Simulated normal breast contained higher than average levels of normal epithelial (60%) and stromal cell types (30%). To better represent real samples, random noise was introduced into the methylation profiles across all samples (see Extended Experimental Procedures). We applied EDec to this dataset assuming 9 different cell types in the model (6 possible breast cancer cell lines, one normal epithelial, one stromal, and one immune). EDec accurately estimated DNA methylation profiles (r = 0.982, Figure 2a) and proportions (r = 0.983) for all constituent cell types (Figure 2b).
Figure 2. EDec validation on simulated mixtures, experimental mixtures, and solid tumors.
(a) Estimated versus true methylation levels for each constituent cell type and locus involved in the simulated mixtures dataset. (b) Estimated versus true proportions for each constituent cell type in each of the samples involved in the simulated mixtures dataset. (c) Estimated versus true methylation levels for each constituent cell type and locus profiled in the experimental mixtures dataset. (d) Estimated versus true proportions for each constituent cell type in each of the samples profiled in the experimental mixtures dataset. (e) Heat map representing the level of correlation between the estimated methylation profiles from the application of EDec to the targeted bisulfite sequencing dataset and the reference methylation profiles. Red boxes indicate the highest level of correlation for each estimated methylation profile. The estimated methylation profiles were labeled as cancer epithelial, normal epithelial, immune, or stromal based on what reference methylation profile was most correlated to each of them. (f) Proportion of constituent cell types estimated by EDec for samples in the targeted bisulfite sequencing dataset versus pathologist estimated proportions (H&E staining). Color key for all panels: orange (MCF-7), blue (HMEC), green (CAF), and red (T-cell).
Validation on cell line mixtures profiled by targeted bisulfite sequencing
We next validated EDec on cellular mixtures prepared in vitro. Specifically, we profiled 10 samples using targeted bisulfite sequencing and applied EDec using the set of 149 loci selected in EDec-Stage 0. Four of the 10 samples were pure cells lines, including: MCF-7, HMEC (Human Mammary Epithelial Cells), a CAF cell line, and CD8+ cytotoxic T-cells. The other six samples consisted of three pairwise combinations (MCF-7/HMEC, MCF-7/T-cells and MCF-7/CAF), each in two proportions (75%:25%, and 95%:5%). There was a strong concordance between the EDec estimated and the true proportions (r = 0.996, Figure 2c). In addition, the estimated methylation profiles for the 4 different cellular fractions closely matched the methylation profiles of cells used to create the mixtures (r = 0.998, Figure 2d).
Validation on breast tumor samples profiled by targeted bisulfite sequencing
We next generated DNA methylation profiles for 31 breast tumors and 8 normal breast samples using targeted bisulfite sequencing. We applied EDec assuming six constituent cell types (see Extended Experimental Procedures), and asked how similar the estimated methylation profiles were to a set of external reference methylation profiles (Figure 2e). Three of the six estimated methylation profiles were most similar to one of the reference breast cancer cell lines. The three remaining profiles had particularly high correlation with the methylation profiles of either CD8+ cytotoxic T-cells, CAF cell line, or the HMEC cell line. This indicates that EDec identifies three components that explain the diversity of cancerous epithelial cells in those samples, while the other three components correspond to an immune fraction, a fibroblast/stromal fraction, and a normal epithelial fraction.
To further validate EDec, clinical pathologist evaluations of cell type composition were obtained for 29 of the 39 samples based on H&E staining. The pathologist estimated proportions for cancerous epithelial, normal epithelial, stromal, and immune fractions. Since the EDec method had proportion estimates for three different cancer epithelial fractions, we combined the proportions for those three fractions to make the two techniques comparable. Despite observing good consistency for the cancer epithelial and immune fractions, we observed low correlation for the normal epithelial and stromal fractions. We reasoned that the low correlation may be explained by extensive epithelial-mesenchymal transitions that may blur the boundary between epithelial and stromal cells. We therefore modified the analysis by combining proportion estimates of normal epithelial and stromal components and examined concordance of EDec and H&E proportion estimates for three fractions (cancerous epithelial, normal epithelial/stromal, and immune). The estimates were highly concordant for all three cell type fractions (r = 0.74, p-value < 10−15, Figure 2f). The highest correlation was for the immune fraction (0.78) and the lowest for cancerous epithelial fraction (0.67). The concordance between these two techniques indicates that EDec’s estimates of proportions and methylation profiles correspond to real cell types, and are not just general components that explain variability in the methylation dataset.
Deconvolution of breast tumors from the TCGA collection confirms the role of immune response in tumor progression
We next applied EDec to deconvolute DNA methylation profiles of 1061 breast tumors and 123 adjacent normal breast samples generated using Infinium HumanMethylation arrays by TCGA (The Cancer Genome Atlas Network, 2012). We selected 391 informative loci (EDec - Stage 0) from 45 reference DNA methylation profiles gathered from the NCBI GEO archive for the following four relevant cell types: cancer epithelial (25), normal epithelial (3), stromal (9), and immune (8). (Figure 3a) (see Extended Experimental Procedures).
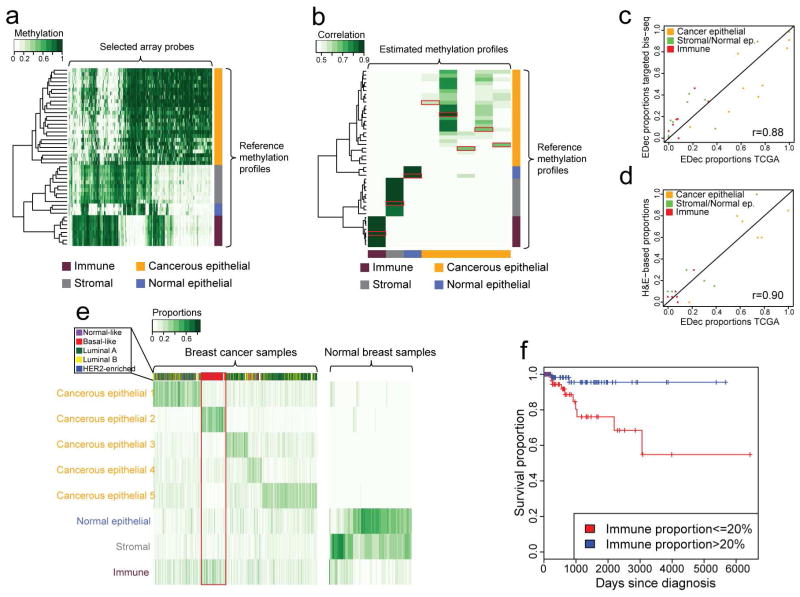
Figure 3. Analysis of DNA methylation profiles of breast tumors samples from the TCGA collection using EDec.
(a) Heat map representing the methylation levels over the chosen set of array probes for the reference methylation profiles. (b) Heat map representing the correlation between the methylation profiles estimated by EDec and the reference methylation profiles. Red boxes indicate the highest correlation for each estimated methylation profile. (c) Scatterplot of EDec cell type proportion estimates for 9 TCGA samples based on targeted bisulfite sequencing (y-axis) and microarray (x-axis). (d) Scatterplot between EDec and pathologist (H & E) estimates of proportions of constituent cell types for a subset (six samples) of the TCGA dataset for which H&E staining-based estimates were available. (e) EDec estimated proportions of constituent cell types for samples in the TCGA dataset. Side bar represents separation of TCGA cancers samples into PAM50 expression subtypes. The red box highlights the samples best explained by the cancerous epithelial 2 profile which are almost exclusively classified as basal-like. (f) Kaplan-Meier plot indicating the significant difference in prognosis (p-value < 0.01) for patients within the group of samples best explained by the cancer epithelial 2 profile (red box in panel F; basal-like) with high versus low estimated immune cell type proportion. See also Figures S1 and S2.
EDec-Stage 1 (Figure 1a) was then applied to the TCGA DNA methylation data over the 391 probes, assuming 4–15 constituent cell types. Reference methylation profiles (20) were added to the TCGA dataset to improve stability of convergence (Extended Experimental Procedures and Figure S5). Based on model reproducibility and goodness of fit (see Extended Experimental Procedures), we chose the model with 8 cell types for all further analyses. We generated heat maps of correlations between the 8 EDec-estimated methylation profiles and each GEO reference methylation profile (Figure 3b). The correlations suggest that EDec identified methylation profiles corresponding to one immune, one stromal, one normal epithelial, and 5 different cancerous epithelial components.
DNA methylation profiles were also generated for nine of the TCGA samples using targeted bisulfite sequencing. This allowed us to compare EDec estimated proportions for those samples based on sequencing data, in the context of 39 breast tissue samples profiled by bisulfite sequencing, versus those estimates based on arrays in the context of 1184 TCGA samples (Figure 3c). Estimated proportions were highly correlated (r = 0.88), suggesting that EDec operates independently of the methylation profiling method. EDec and pathologist (H&E staining) proportion estimates were also consistent (r = 0.90) (Figure 3d).
Consistent with expectations, EDec predicts normal breast samples to have negligible proportions of cancerous epithelial cells, while in breast tumors those cell types are generally the ones with highest proportions (Figure 3e). We also observe that the cancerous cell fraction of the different breast cancer samples is explained by a different combination of the five cancerous epithelial components, with one of them typically being dominant. Grouping tumor samples based on the dominant cancer epithelial component showed some concordance with their PAM50 classification (Parker et al., 2009). In particular, basal-like samples were nearly all in the same EDec-defined group (Figure 3e - red box). We further investigated methylation heterogeneity of the epithelial fraction over the 391 chosen probes within and between tumor subtypes (Extended Experimental Procedures and Figure S1). Luminal B tumors had the most heterogeneous profiles, while normal breast samples had the most homogeneous epithelial profile. Despite having an intermediary level of heterogeneity, Basal-like tumors exhibited epithelial methylation profiles highly distinct from the other breast tumor subtypes.
We also found that tumor subtypes differ significantly in the degree of infiltration by either immune or stromal cells (Figure S2). Normal-like samples contained the highest median stromal proportion (18%) and Luminal B tumors the lowest (4%). Basal-like tumors displayed the highest median degree of immune cell infiltration (21%), while Luminal B tumors again had the lowest (7%). Normal breast tissue samples displayed a much higher median proportion of stromal cells (37%) than breast tumors (8%).
We next investigated whether the predicted immune proportion was associated with survival of basal-like breast cancer patients. Indeed, patients with greater than 20% immune cell infiltration survived significantly longer (p < 0.01) than those with less than 20% (Figure 3f), consistent with previous microscopy-based evaluation of immune cell infiltration (Adams et al., 2014). We also investigated whether immune infiltration levels in adjacent normal tissue were related to immune infiltration levels in the matched tumor sample. No such correlation was observed, indicating that immune infiltration of tumors is not dependent on the amount of immune cells in the surrounding normal tissue (Figure S2).
Deconvolution of RNA-seq profiles of breast tumors from the TCGA collection reveals cell-type specific tumorigenic perturbations with the tumor microenvironment
Given the availability of both mRNA-sequencing and DNA methylation profiles for the TCGA breast samples, we applied EDec-Stage 2 to estimate gene expression profiles of constituent cell types. EDec-Stage 2 was independently applied to 6 subsets of the 1,114 TCGA expression profiles, corresponding to the five PAM50 subtypes (Luminal A (523 samples), Luminal B (207), HER2-enriched (78), Basal-like (173), Normal-like (33)) (Parker et al., 2009), plus normal breast tissue samples (100). We combined the eight EDec Stage 1-estimated proportions (Figure 3e) into the following three cell type fractions: epithelial (including 5 cancer epithelial and 1 normal epithelial), stromal, and immune. Proportion estimates for those three cell types were then used in EDec Stage-2 to estimate expression profiles of epithelial, stromal, and immune cell types for each PAM50 subtype and normal breast.
EDec predicts epithelial specific expression of ESR1, PGR, and FOXA1 in Luminal A and Luminal B subtypes (Figure 4a), consistent with previous reports (Toss and Cristofanilli, 2015). Due to poor model fit, as indicated by large error bars, cell-type specific expression could not be established for a number of genes, ERBB2 within HER2-enriched tumors being the most conspicuous example. The poor fit of the model for that gene is due to its exceedingly high variance in expression within epithelial cells of this tumor type (Figure S3). We can show through simulations (Extended Experimental Procedures) that this effect is mitigated by increasing the number of input breast cancer samples. We note that the large estimated standard error provides a clear signal that cell-type specific expression cannot be established for specific genes, thus preventing erroneous conclusion suggested by high mean values.
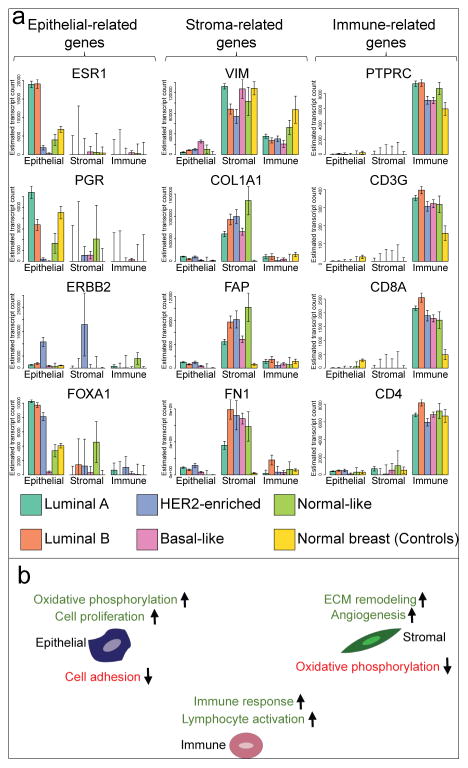
Figure 4. Cell type specific gene expression.
(a) Bar-plots represent the estimated expression profiles of 12 different genes within constituent cell types for each of the breast cancer intrinsic subtypes, as well as for the set of normal breast (control) samples. (b) Summary of main enriched gene sets among up- or down-regulated genes between cancer and normal breast in each cell type. See also Figures S3 and S4.
EDec predicts stroma-specific expression of vimentin (VIM), a general mesenchymal cell marker (Kalluri and Zeisberg, 2006), in normal breast and in all tumor subtypes. Conversely, the stroma-specific expression of COL1A1, FAP, and FN1 is observed in tumors, but not in normal breast (Figure 4a). That observation is consistent with the activation of such genes in CAFs, the main constituent of the tumor stroma (Kalluri and Zeisberg, 2006).
EDec correctly predicts immune-specific expression of immune cell markers (PTPRC, CD3G, CD8A, and CD4) in every group of samples (Figure 4a). Note that the CD8+ T-cell marker CD8A shows significantly higher expression in breast cancers than in normal breasts, consistent with observations that the immune components of breast tumors contains a larger proportion of CD8+ T-cells compared to the immune component of normal breasts.
We next compared gene expression profiles of the three tumor-constituent cell types against the profiles of their normal control counterparts. A gene set enrichment analysis (Huang et al., 2009a, 2009b) was then performed on the resulting sets of differentially expressed genes. Figure 4b summarizes the top gene set enrichments for genes up- or down-regulated in tumor cells compared the normal controls (for full set of gene set enrichments see Supplemental Table 1). The terms found to be enriched in each of the sets of differentially expressed genes are consistent with known hallmarks of cancer (sustaining proliferative signaling, activating invasion and metastasis, inducing angiogenesis, deregulating cellular energetics, avoiding immune destruction, etc.) and with the known roles of each cell type within breast tumors (e.g., “extracellular matrix remodeling” genes up-regulated specifically in stromal cells and “sustaining inflammation in tumor” category in immune cells) (Hanahan and Weinberg, 2011).
We next sought to further validate EDec-Stage 2 predictions of differentially expressed genes against a previously published dataset, in which gene expression profiling was performed on epithelial and stromal components of matched invasive carcinomas and adjacent normal tissue, after LCM (Ma et al., 2009). Despite the fact that the study did not separate out the immune component, focused on the fibrous portion of the stroma (both in normal breast and breast cancer), and used microarrays to profile expression, we still observe significant overlaps between the differentially expressed genes predicted by EDec and those observed in the LCM dataset (Figure S4). Consistency is observed both for expression differences in epithelial and stromal components.
Switch from adipose to fibrous stroma supports oxidative metabolism in cancerous cells
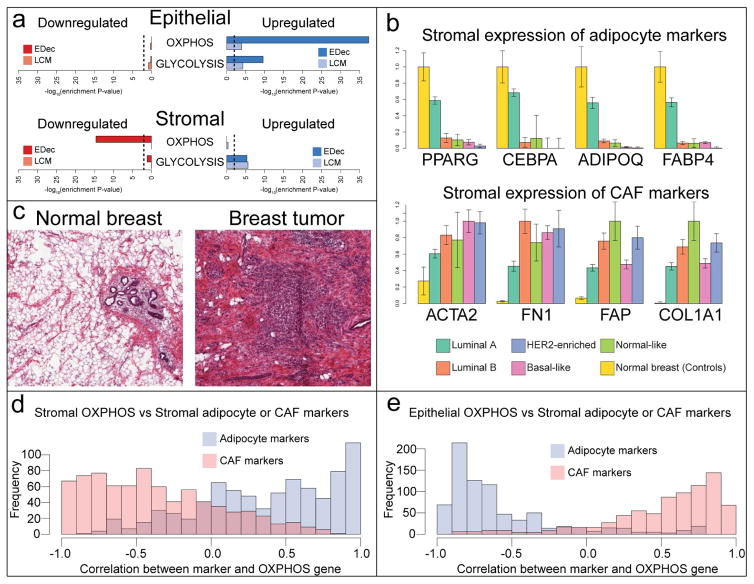
Tumor cells are often more glycolytic than their normal counterparts even in the presence of oxygen. This phenomenon is known as the Warburg effect (Wallace, 2005) and is thought to occur due to the higher anabolic needs of highly proliferative tumor cells (Vander Heiden et al., 2009). Consistent with this phenomenon, we observe enrichment for glycolysis genes among those upregulated in cancer epithelium compared to normal epithelium (Figure 5a). However, contrary to the reduction in mitochondrial activity in cancerous cells predicted by the Warburg effect, we observe strong enrichment for genes involved in oxidative phosphorylation (OXPHOS) among those upregulated in cancer epithelium compared to normal epithelium (Figure 5a). Further, upregulation both glycolysis and OXPHOS genes can be confirmed in comparisons of gene expression profiles of tumor versus normal breast epithelium after LCM (Figure 5a).
Figure 5. Switch from adipose to fibrous stroma influences the metabolic phenotype of the tumor.
(a) Enrichment of either OXPHOS or GLYCOLYSIS gene sets (hallmark gene sets MSigDB (Liberzon et al., 2015)) among those up- or down-regulated in epithelial or stromal cells of breast cancer. Cell type specific differential expression analysis was performed with either by applying EDec to TCGA dataset, or in the LCM dataset. Dashed lines represent a p-value of 0.01. (b) Estimated stromal expression of either adipocyte or CAF markers across breast cancer subtypes. (c) Representative H&E staining images of matched tumor and normal breast samples from TCGA (TCGA-BH-A0B2). (d) Histogram of correlations between stromal expression of OXPHOS genes and stromal expression of marker genes of either adipocyte or CAF across breast cancer subtypes. (e) Histogram of correlations between epithelial expression of OXPHOS genes and stromal expression of marker genes of either adipocyte or CAF across breast cancer subtypes.
The upregulation of both glycolytic and oxidative pathways in cancer cells comes with a demand for nutrients and oxygen, which can be met both by increased angiogenesis and potentially by the support of other cells in the microenvironment. The previously proposed reverse Warburg effect model (Martinez-Outschoorn et al., 2015, 2014; Pavlides et al., 2009) postulates that tumor cells can induce shutdown of oxidative metabolism in the surrounding stromal cells, causing them to reduce oxygen consumption and to secrete high energy metabolites produced through glycolysis. Those metabolites may then be taken up by cancerous cells to fuel their own oxidative metabolism. Consistent with that model, we observe enrichment for OXPHOS genes among those downregulated in tumor stroma, and for glycolysis genes among those upregulated in the tumor stroma (Figure 5a).
Given that adipocytes have higher rates of mitochondrial activity than fibroblasts (Hofmann et al., 2012; Wilson-Fritch et al., 2003), the observed downregulation of OXPHOS genes in the tumor stroma may reflect the change in stromal composition, from a more adipose (oxidative) stroma in normal breast to a more fibrous (glycolytic) stroma in breast tumors. To determine whether such change indeed occurs we examined expression levels of adipocyte (PPARG, CEBPA, ADIPOQ, FABP4) or CAF (ACTA2, FN1, FAP, COL1A1) markers in the stroma of normal breast and different breast tumor subtypes (Figure 5b). Adipocyte markers are highly expressed in the stroma of normal breast and Luminal A tumors, with negligible expression in other tumor subtypes. CAF markers, in contrast, seem to display the opposite pattern of expression. Such observations are consistent with fibrosis in breast tumors, and with the higher incidence of tumors with adipose stroma among those of the Luminal A subtype (Jung et al., 2015). The change in stromal adipocyte content between normal breast and breast tumor is also apparent in H&E staining slides gathered from matched tumor/normal samples from TCGA (Figure 5c). In the LCM dataset, only the fibrous portion of the stroma was selected for analysis both in normal breast and in breast tumors. Therefore, consistent with the idea that the observed changes in stromal OXPHOS gene expression result from a change from adipose to fibrous stroma, no change in expression of those genes is observed in the LCM dataset (Figure 5a).
We next asked whether the change from adipose to fibrous stroma was associated with a change from oxidative to glycolytic stroma. To examine this, we analyzed the correlation between the expression of either adipocyte or CAF markers in the stroma and the stromal expression of OXPHOS genes across breast cancer subtypes. We observed that, as expected, the stromal expression of most OXPHOS genes had a strong positive correlation with the stromal expression of adipocyte markers, while the expression of CAF markers in the stroma was negatively correlated with OXPHOS genes (Figure 5d).
The reverse Warburg effect model predicts that a glycolytic stroma associates with oxidative cancerous epithelial cells, whereas an oxidative stroma would be associated with more glycolytic tumor cells. Given that a fibrous stroma seems to be more glycolytic than an adipose one, we hypothesized that a change from adipose to fibrous stroma would associate with a change from glycolytic to oxidative cancerous epithelium. We therefore analyzed the degree of correlation between the expression of either adipocyte or CAF markers in the stroma and the expression of OXPHOS genes in the epithelial fraction across breast cancer subtypes. Stromal expression of CAF markers was indeed positively correlated with epithelial OXPHOS gene expression, while adipocyte marker expression in the stroma was negatively correlated with OXPHOS gene expression in the epithelial fraction (Figure 5e). Interestingly, the stromal expression of CAV1, a gene whose low expression in breast cancer stroma is known to associate with negative prognosis and with tumors with reverse Warburg metabolism (Martinez-Outschoorn et al., 2015, 2014; Pavlides et al., 2009), is strongly correlated with the expression of adipocyte markers in the stroma (mean r = 0.97), providing further support for the hypothesis that stromal adiposity associates negatively and the stromal fibroblast content associates positively with the reverse Warburg pattern of metabolism.
Discussion
The Epigenomic Deconvolution (EDec) method provides accurate platform-independent estimation of cell type proportions, DNA methylation profiles, and gene expression profiles of constituent cell types. By significantly relaxing the dependence on reference methylation profiles of constituent cell types compared to previous methods (Houseman et al., 2012), EDec enables deconvolution of complex tumor tissues where highly accurate references are unavailable. In contrast to reference-free methods (Houseman et al., 2016, 2014; Rahmani et al., 2016; Zheng et al., 2014; Zou et al., 2014), EDec’s indirect use of surrogate references greatly assisted in the interpretation of deconvolution results, allowing us to uncover more complex biological patterns than possible by applying other deconvolution techniques. Further, unlike previous methylation-based deconvolution methods, EDec does not require that each cell type be explained by a single component (e.g., cancerous epithelial fraction in the TCGA dataset was modeled by five different components), thus making it possible to model the full diversity of cancerous epithelial cells. Despite such methodological advances, we note that the current tissue models obtained by EDec still only approximate the full complexity of breast tumors. For example, more detailed deconvolution of individual components of the stromal and immune fractions would likely yield additional biological insights.
By addressing the confounding issue of tissue heterogeneity, EDec enables the comparison of tumors of various cell type compositions based on inferred molecular profiles of constitutive cancer epithelial cells and also the comparisons between cancer cell fractions of tumors and experimentally more tractable cell line models. EDec reveals that methylome profiles of breast cancer cells are distinct from those of normal epithelial cell types, and that they can be mapped to specific groups of cancer cell lines. We also observe that cancerous cells of basal-like tumors have particularly distinct cellular identity as indicated by their distinct methylation profiles.
By providing information about the epigenomic and transcriptomic states of both cancerous epithelial and non-epithelial tumor cells, the method enables the study of heterotypic interactions driving tumor progression. The most striking pattern that emerged from our analyses is metabolic coupling between epithelium and stroma that seems to be related to the degree of adiposity of the stroma. Specifically, upregulation of both glycolysis and OXPHOS in cancerous epithelial cells supports the idea that, despite the long-postulated Warburg effect, cancer cells in breast tumors still upregulate their energy production through OXPHOS in comparison to normal cells (Zu and Guppy, 2004). Further, the switch from adipose to fibrous stroma leads to lower stromal mitochondrial activity, which in turn seems to support up-regulation of OXPHOS in cancerous epithelial cells. Our findings therefore refine the reverse Warburg effect model (Martinez-Outschoorn et al., 2015, 2014; Pavlides et al., 2009) by showing that it may be mediated by changes in cell type composition of tumor stroma. It is tempting to speculate that the differences in stroma composition across tumor subtypes may be related to a different capacity of distinct tumor types to induce the conversion of adipocytes into fibroblasts (Bochet et al., 2013; Dirat et al., 2011), which would be more supportive of reverse Warburg metabolism. Despite these encouraging results, that are largely confirmed by expression profiling of microdissected tumors, further experiments focusing on protein and metabolite levels in different tumor cell types will be needed to conclusively confirm this model.
In conclusion, EDec reveals layers of biological information about distinct cell types within solid tumors and about their heterotypic interactions that were previously inaccessible at such large scale due to tissue heterogeneity. EDec improves on previous methods by employing a data-driven approach that makes indirect use of reference profiles of constituent cell types and adequately models the variability of methylation profiles across different cancer cells. We note that EDec is a general technique and could potentially be applied to different types of tumors and other complex non-tumor tissues. However, such applications would involve new feature selection with a set of references appropriate for that tissue, and would need to be validated. In addition to the method itself, we have also developed a “deconvoluted breast cancer” data resource for breast tumors and normal breast tissues within the TCGA collection (http://genboree.org/theCommons/projects/edec). This resource can now be further explored by the community to derive or test new hypotheses.
Experimental procedures
ThunderStorm BS-Seq assay and breast cancer target panel
A set of 1000 target regions of around 300bp in length were preselected for targeted bisulfite sequencing based on previous reports of their involvement in breast cancer biology (see Supplemental Table 2). Of the 1000 genomic regions, 149 were selected based on cell type specific methylation based on Roadmap Epigenomics reference DNA methylation profiles (Kundaje et al., 2015).
Primer pairs designed to specifically amplify each selected target region were designed by RainDance Technologies. The ThunderStorm BS-seq assay using that set of primer pairs was performed at RainDance Technologies according to the manufacturer’s specification. In summary, that assay uses a microfluidic chip to perform multiplex amplification of bisulfite treated DNA using the set of primers designed to amplify the selected set of genomic regions. This step is followed by sequencing of PCR product. Read mapping and methylation level calling was performed using Bismark (Krueger and Andrews, 2011). Target regions were sequenced on average to 200X coverage. For all subsequent analyses, DNA methylation levels for all CpGs overlapping each of the target regions were averaged, giving an average methylation value for each region of interest. For eight of the breast cancer samples profiled using this assay, 450k arrays were also performed by the TCGA group. We observed over 0.9 correlation between methylation levels measured by both platforms over the 614 regions overlapping 450K array probes for all samples analyzed.
TCGA data processing
Methylation array data
The breast cancer TCGA DNA methylation data was generated using either the Infinium HumanMethylation450 BeadChip (450K array) or the Infinium HumanMethylation27 BeadChip (27K array). We used the TCGA Assembler (Zhu et al., 2014) to download level 3 data (fully processed) for all 27K and 450K profiles. Since most 27K probes are present in the 450K array, we merged the two datasets and included only overlapping probes in our analysis. We also removed any probe with a detection p-value less than 0.05 in at least one sample, those that overlapped known SNPs, and those that were previously reported as cross reactive (Chen et al., 2013). The final number of probes passing these criteria was 17,907. We also corrected for platform biases using an Empirical Bayes-based approach (ComBat) (Johnson et al., 2007), implemented in the SVA package in R (Leek, J.T. et al., 2015).
RNA-Seq data
TCGA Assembler (Zhu et al., 2014) was used to download normalized (RNA-seq v2 - RNA-seq by Expectation Maximization) gene transcript abundance measurements from the TCGA database. PAM50 classification (Parker et al., 2009) based on RNA-seq for 1030 breast cancer samples generated by the TCGA Analysis Working Group were obtained from the UCSC Cancer Genomics Browser (Goldman et al., 2013). Of the TCGA breast cancer samples that had RNA-sequencing data and associated PAM50 classification, 1005 also had DNA methylation data. For normal breast samples, 100 had both DNA methylation and RNA-sequencing data. Therefore, the final set of RNA-sequencing samples contained 1105 samples.
Code and dataset availability
The EDec software is available as an R package. It can be downloaded from https://github.com/BRL-BCM/EDec. Documentation and usage examples are also available on that same page. All datasets associated with this publication can be found at http://genboree.org/theCommons/projects/edec. Primary human breast tumor tissue and adjacent normal tissue were obtained with local Institutional Review Board (IRB# PRO11090404) from the University of Pittsburgh’s Health Science Tissue Bank.
Accession Numbers
The accession number for the targeted bisulfite sequencing data reported in this paper is GEO: GSE87297.
Supplementary Material
Highlights.
EDec infers cell types within tissues and molecular profiles of constituent cells
EDec deconvolutes molecular profiles of breast tumors within the TCGA collection
EDec-estimated immune infiltration predicts prognosis for basal-like breast tumors
Switch from adipose to fibrous stroma enhances oxidative metabolism of cancer cells
Acknowledgments
This work was supported in part by a grant from the Common Fund of the National Institutes of Health (Roadmap Epigenomics Program, grant number U01 DA025956) to AM and the Scientific Advisory Council award from Susan G. Komen for the Cure and the Hillman Foundation Fellow award to AVL. This study used UPCI Cancer Tissue and Research Pathology services supported by NIH award P30CA047904. We also acknowledge the support of the Health Sciences Tissue Bank (HSTB) at the University of Pittsburgh. We thank Christina Kline and the other HSTB team members. We would also like to thank the University of Pittsburgh Cancer Institute (UPCI), and the clinicians, staff, and patients at UPMC for making this study possible.
Footnotes
Supplemental information includes Extended Experimental Procedures, six figures and 2 tables.
Author Contributions
Conceptualization, V.O., R.H., D.N.B, M.E.R., M.L.S., S.O., W.M.W., V.D.G., A.V.L., A.M.; Methodology, V.O., R.H., D.N.B, M.E.R., M.L.S., A.V.L., A.M.; Software, V.O.; Validation, V.O., R.H, D.N.B; Formal Analysis, V.O.; Investigation, V.O., R.H., D.N.B., A.V.L., A.M.; Resources, A.V.L., M.L.S., A.M.; Data Curation, V.O., D.N.B., R.H., R.Y.P.; Writing – Original Draft, V.O.; Writing – Review & Editing, V.O., R.H, D.N.B., M.E.R., M.L.S., A.V.L, A.M.; Visualization, V.O.; Supervision, A.V.L, A.M.; Project Administration, V.O., M.E.R. A.V.L, A.M.; Funding Acquisition, A.V.L, M.L.S., A.M.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams S, et al. Prognostic value of tumor-infiltrating lymphocytes in triple-negative breast cancers from two phase III randomized adjuvant breast cancer trials: ECOG 2197 and ECOG 1199. J Clin Oncol. 2014;32:2959–66. doi: 10.1200/JCO.2013.55.0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aran D, Sirota M, Butte AJ. Systematic pan-cancer analysis of tumour purity. Nat Commun. 2015;6:8971. doi: 10.1038/ncomms9971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochet L, et al. Adipocyte-derived fibroblasts promote tumor progression and contribute to the desmoplastic reaction in breast cancer. Cancer Res. 2013;73:5657–68. doi: 10.1158/0008-5472.CAN-13-0530. [DOI] [PubMed] [Google Scholar]
- Chen YA, Lemire M, Choufani S, Butcher DT, Grafodatskaya D, Zanke BW, Gallinger S, Hudson TJ, Weksberg R. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 2013;8:203–9. doi: 10.4161/epi.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coussens LM, Zitvogel L, Palucka AK. Neutralizing tumor-promoting chronic inflammation: a magic bullet? Science. 2013;339:286–91. doi: 10.1126/science.1232227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debey S, Schoenbeck U, Hellmich M, Gathof BS, Pillai R, Zander T, Schultze JL. Comparison of different isolation techniques prior gene expression profiling of blood derived cells: impact on physiological responses, on overall expression and the role of different cell types. Pharmacogenomics J. 2004;4:193–207. doi: 10.1038/sj.tpj.6500240. [DOI] [PubMed] [Google Scholar]
- Dirat B, et al. Cancer-associated adipocytes exhibit an activated phenotype and contribute to breast cancer invasion. Cancer Res. 2011;71:2455–65. doi: 10.1158/0008-5472.CAN-10-3323. [DOI] [PubMed] [Google Scholar]
- Finak G, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14:518–27. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- Gaujoux R, Seoighe C. Semi-supervised Nonnegative Matrix Factorization for gene expression deconvolution: A case study. Infection, Genetics and Evolution. 2011;12:913–21. doi: 10.1016/j.meegid.2011.08.014. [DOI] [PubMed] [Google Scholar]
- Gentles Newman, Liu Bratman. The prognostic landscape of genes and infiltrating immune cells across human cancers. 2015 doi: 10.1038/nm.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman M, et al. The UCSC Cancer Genomics Browser: update 2013. Nucleic Acids Res. 2013;41:D949–54. doi: 10.1093/nar/gks1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg R. Hallmarks of Cancer: The Next Generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hofmann AD, Beyer M, Krause-Buchholz U, Wobus M, Bornhäuser M, Rödel G. OXPHOS supercomplexes as a hallmark of the mitochondrial phenotype of adipogenic differentiated human MSCs. PLoS ONE. 2012;7:e35160. doi: 10.1371/journal.pone.0035160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman E, Accomando W, Koestler D, Christensen B, Marsit C, Nelson H, Wiencke J, Kelsey K. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman EA, Ince TA. Normal cell-type epigenetics and breast cancer classification: a case study of cell mixture-adjusted analysis of DNA methylation data from tumors. Cancer Inform. 2014;13:53–64. doi: 10.4137/CIN.S13980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman EA, Kile ML, Christiani DC, Ince TA, Kelsey KT, Marsit CJ. Reference-free deconvolution of DNA methylation data and mediation by cell composition effects. BMC Bioinformatics. 2016;17:259. doi: 10.1186/s12859-016-1140-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman EA, Molitor J, Marsit CJ. Reference-free cell mixture adjustments in analysis of DNA methylation data. Bioinformatics. 2014;30:1431–9. doi: 10.1093/bioinformatics/btu029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M, Yao J, Cai L, Bachman KE, Brûle F, van den Velculescu V, Polyak K. Distinct epigenetic changes in the stromal cells of breast cancers. Nat Genet. 2005;37:899–905. doi: 10.1038/ng1596. [DOI] [PubMed] [Google Scholar]
- Huang DWaW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009a;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang D, Wa W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009b;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–27. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Jung YY, Lee YK, Koo JS. Expression of cancer-associated fibroblast-related proteins in adipose stroma of breast cancer. Tumour Biol. 2015;36:8685–95. doi: 10.1007/s13277-015-3594-9. [DOI] [PubMed] [Google Scholar]
- Kalluri R, Zeisberg M. Fibroblasts in cancer. Nature Reviews Cancer. 2006 doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- Komori HK, et al. Application of microdroplet PCR for large-scale targeted bisulfite sequencing. Genome Res. 2011;21:1738–45. doi: 10.1101/gr.116863.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F, Andrews S. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Method Biochem Anal. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn A, Thu D, Waldvogel H, Faull R, Luthi-Carter R. Population-specific expression analysis (PSEA) reveals molecular changes in diseased brain. Nature Methods. 2011;8:945–7. doi: 10.1038/nmeth.1710. [DOI] [PubMed] [Google Scholar]
- Kundaje A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–30. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek JT, Johnson WE, Parker HS, Fertig EJ, Jaffe AE, Storey JD. sva: Surrogate Variable Analysis. R package version 3.19.0 2015 [Google Scholar]
- Li Y, Xie X. A mixture model for expression deconvolution from RNA-seq in heterogeneous tissues. BMC Bioinformatics. 2013;14(Suppl 5):S11. doi: 10.1186/1471-2105-14-S5-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Foulkes WD, Leung S, Gao D, Lau S, Kos Z, Nielsen TO. Prognostic significance of FOXP3+ tumor-infiltrating lymphocytes in breast cancer depends on estrogen receptor and human epidermal growth factor receptor-2 expression status and concurrent cytotoxic T-cell infiltration. Breast Cancer Res. 2014;16:432. doi: 10.1186/s13058-014-0432-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X-JJ, Dahiya S, Richardson E, Erlander M, Sgroi DC. Gene expression profiling of the tumor microenvironment during breast cancer progression. Breast Cancer Res. 2009;11:R7. doi: 10.1186/bcr2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Outschoorn UE, Lisanti MP, Sotgia F. Catabolic cancer-associated fibroblasts transfer energy and biomass to anabolic cancer cells, fueling tumor growth. Semin Cancer Biol. 2014;25:47–60. doi: 10.1016/j.semcancer.2014.01.005. [DOI] [PubMed] [Google Scholar]
- Martinez-Outschoorn UE, Sotgia F, Lisanti MP. Caveolae and signalling in cancer. Nat Rev Cancer. 2015;15:225–37. doi: 10.1038/nrc3915. [DOI] [PubMed] [Google Scholar]
- Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–7. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JS, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–7. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul DS, Guilhamon P, Karpathakis A, Butcher LM, Thirlwell C, Feber A, Beck S. Assessment of RainDrop BS-seq as a method for large-scale, targeted bisulfite sequencing. Epigenetics. 2014;9:678–84. doi: 10.4161/epi.28041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlides S, et al. The reverse Warburg effect: aerobic glycolysis in cancer associated fibroblasts and the tumor stroma. Cell Cycle. 2009;8:3984–4001. doi: 10.4161/cc.8.23.10238. [DOI] [PubMed] [Google Scholar]
- Rahmani E, et al. Sparse PCA corrects for cell type heterogeneity in epigenome-wide association studies. Nat Methods. 2016 doi: 10.1038/nmeth.3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen-Orr S, et al. Cell type–specific gene expression differences in complex tissues. Nature Methods. 2010 doi: 10.1038/nmeth.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tlsty TD, Coussens LM. Tumor stroma and regulation of cancer development. Annu Rev Pathol. 2006;1:119–50. doi: 10.1146/annurev.pathol.1.110304.100224. [DOI] [PubMed] [Google Scholar]
- Toss A, Cristofanilli M. Molecular characterization and targeted therapeutic approaches in breast cancer. Breast Cancer Research. 2015;17:60. doi: 10.1186/s13058-015-0560-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venet Pecasse, Maenhaut Bersini. Separation of samples into their constituents using gene expression data. Bioinformatics. 2001;17(Suppl 1):S279–87. doi: 10.1093/bioinformatics/17.suppl_1.s279. [DOI] [PubMed] [Google Scholar]
- Wallace DC. Mitochondria and cancer: Warburg addressed. Cold Spring Harb Symp Quant Biol. 2005;70:363–74. doi: 10.1101/sqb.2005.70.035. [DOI] [PubMed] [Google Scholar]
- Wilson-Fritch L, Burkart A, Bell G, Mendelson K, Leszyk J, Nicoloro S, Czech M, Corvera S. Mitochondrial biogenesis and remodeling during adipogenesis and in response to the insulin sensitizer rosiglitazone. Mol Cell Biol. 2003;23:1085–94. doi: 10.1128/MCB.23.3.1085-1094.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara K, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nature Communications. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, et al. MethylPurify: tumor purity deconvolution and differential methylation detection from single tumor DNA methylomes. Genome Biol. 2014;15:419. doi: 10.1186/s13059-014-0419-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, Wan YW, Pang K, Chow L, Liu Z. Digital sorting of complex tissues for cell type-specific gene expression profiles. BMC bioinformatics. 2013;14:89. doi: 10.1186/1471-2105-14-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Qiu P, Ji Y. TCGA-assembler: open-source software for retrieving and processing TCGA data. Nat Methods. 2014;11:599–600. doi: 10.1038/nmeth.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Lippert C, Heckerman D, Aryee M, Listgarten J. Epigenome-wide association studies without the need for cell-type composition. Nature Methods. 2014;11:309–11. doi: 10.1038/nmeth.2815. [DOI] [PubMed] [Google Scholar]
- Zu XL, Guppy M. Cancer metabolism: facts, fantasy, and fiction. Biochem Biophys Res Commun. 2004;313:459–65. doi: 10.1016/j.bbrc.2003.11.136. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.