Abstract
Ribotypes and toxin genotypes of clinical C. difficile isolates in Taiwan are rarely reported. A prospective surveillance study from January 2011 to January 2013 was conducted at the medical wards of a district hospital in southern Taiwan. Of the first toxigenic isolates from 120 patients, 68 (56.7%) of 120 isolates possessed both tcdA and tcdB. Of 52 (43.3%) with tcdB and truncated tcdA (tcdA-/tcdB+), all were ribotype 017 and none had binary toxin or tcdC deletion. Eighteen (15%) toxigenic isolates harbored binary toxins (cdtA and cdtB) and all had tcdC deletion, including Δ39 (C184T) deletion (14 isolates), Δ18 in-frame deletion (3 isolates), and Δ18 (Δ117A) deletion (1 isolate). Eleven of 14 isolates with Δ39 (C184T) deletion belonged to the ribotype 078 family, including ribotype 127 (6 isolates), ribotype 126 (4 isolates), and ribotype 078 (1 isolate). Among 8 patients with consecutive C. difficile isolates, these isolates from 6 (75%) patients were identical, irrespective of the presence or absence of diarrhea, suggestive of persistent fecal carriage or colonization. In conclusion in southern Taiwan, ribotype 017 isolates with a tcdA-/tcdB+ genotype were not uncommon and of C. difficile isolates with binary toxin, the ribotype 078 family was predominant.
Introduction
Clostridium difficile is the leading cause of nosocomial diarrhea with an increase in the incidence of sporadic outbreaks causing severe and fatal infections since the beginning of the century [1]. Most alarming is the outbreak of C. difficile infections (CDIs) in Quebec, Canada in 2003. During the outbreak involving 1,703 patients, CDI was the attributable cause of death in 117 (6.9%) cases and a contributing factor in additional 127 (7.5%) deaths [2]. The hypervirulent strain had been assigned as the North American pulse-field type 1 (NAP1), restriction endonuclease analysis (REA) group BI, and polymerase chain reaction (PCR) ribotype 027 (sometimes referred to as BI/NAP1/027). Three bacterial factors have been found in the epidemic C. difficile strain, including in vitro increased production of toxin A and B, fluoroquinolone resistance, and production of binary toxin [1]. Toxin A and B are transcribed from a pathogenicity locus comprised of five genes: tcdA (toxin A), tcdB (toxin B), and three regulatory genes. One of the latter, tcdC, is a negative regulator of toxin production. Binary toxin is transcribed from cdtA and cdtB [1].
Changing C. difficile epidemiology is noted worldwide. In the European Study Group of C. difficile (ESGCD) between 2002 and 2005, major toxigenic ribotypes were 001 (13%), 014 (9%), 002, 012, 017, 020, and 027 (each about 6%), with the CDI incidence ranging from 0.13 to 7.1 cases per 10,000 patient-days in different countries [3,4]. In Japan, a shift of the predominant ribotype, from PCR ribotype a in 2000 (15/33, 45%) to PCR ribotype f (type smz) in 2004 (18/28, 64%), was noted in a teaching hospital between 2000 and 2004 [5,6]. In Korea, tcdA-/tcdB+ C. difficile strains accounted for <7% in 2002, but increased to 13.2% in 2003 and 50.3% in 2004. In Taiwan ribotype 027, 078, or 001 isolates were not reported [7–9], until 2012 when the first case of CDI due to ribotype 027 was reported [10]. Nevertheless the information regarding toxin genotype and ribotype distribution in Taiwan remains scarce.
Genetic relationship of the C. difficile isolates causing colonization, infection, or recurrence in the same individual remains variable in several studies. Among 20 recurrent cases, Oka found 16 (80%) cases were identical between the strains at initial infection and at recurrence [11]. Nevertheless Barbut et al. reported that of the strains from 93 hospitalized patients with recurrent CDI between 1994 and 1997, 48.4% of clinical recurrences were caused by different strains compared with initial strains [12]. However, the question of whether the initial colonized C. difficile strain was the same as or different from the strain causing subsequent infection was not answered.
In our previously published data, we reported the clinical impact and risk factor of C. difficile colonization and infection in a prospective study from January 2011 to June 2012 [8,13,14]. We analyzed the clinical C. difficile isolates during the study period and found the first hypervirulent C. difficile ribotype 126 strain in Taiwan [15]. We further extended the prospective study to January 2013. In this study we aimed to investigate the toxin gene content and ribotype distribution of C. difficile isolates with tcdC deletion collected from previous studies [13–15].
Materials and Methods
Study design
We collected clinical C. difficile isolates from stool culture of the prospective study from January 2011 to June 2012 as described before [8,13,14]. Briefly a prospective investigation was conducted in the medical wards of the Tainan Hospital, Ministry of Health and Welfare, a district hospital in southern Taiwan. The prospective clinical study was further extended to January 2013. The study was approved by the institutional review board of the Tainan Hospital, Ministry of Health and Welfare, and written informed consents were obtained from enrolled patients. Patients with age of at least 20 years old, and admitting to medical wards with expected hospital stays of at least 5 days were included. Exclusion criteria were patients with previous metronidazole or oral vancomycin therapy within three months, colectomy, or CDI at admission [16–18]. We retrieved demographic information, laboratory data, medication history, and underlying disease from medical records. Stool samples from the patients included in the study from January 2011 to January 2013 were sent for C. difficile culture. Stool samples were plated on cycloserine–cefoxitin-fructose agar (CCFA) under anaerobic conditions. C. difficile colonization (CdC) is defined as an asymptomatic patient with the presence of C. difficile in stool and CDI as a patient with diarrhea and the detection of toxigenic C. difficile in stool. Recurrence was defined as the resurgence of clinical symptoms after cessation of antimicrobial therapy, at least 10 days after the first episode [12].
Bacterial strains
C. difficile CCUG4938T (ribotype 001 with wild type tcdC, toxinotype 0, purchased from the Culture Collections of the University of Goteborg, Sweden), a ribotype 078 strain (with a 39-bp deletion in tcdC, provided by Prof. EJ Kuijper at Leiden University Medical Center, the Netherlands), and ATCC BAA1805 (ribotype 027 with an 18-bp deletion of tcdC, purchased from American Type Culture Collection, USA), ribotype 106 and ribotype 001/072 strain (provided by Prof. Ellie JC Glodstein at UCLA, USA), were used as reference strains.
Genomic DNA
C. difficile strains were grown anaerobically in Brain Heart Infusion broth (Becton, Dickinson and Company) with 5 mg/ml yeast extract (MO BIO Laboratories, Inc.) and 0.1% L-cysteine (AMRESCO®) at 37°C for two days. After harvesting the bacteria, C. difficile genomic DNA was extracted with a genomic DNA mini kit (Geneaid, Ltd, Taiwan).
Detection of toxin genes
The extracted DNA was amplified for the 16s rDNA, tcdA, tcdB, cdtA, cdtB, and tcdC genes of C. difficile in a single multiplex PCR, as described in [19]. The strains containing a truncated tcdC profile were further examined through tcdC sequencing, as previously described in [20]. Sequencing was performed by Mission Biotech Co., Ltd. Amplification was performed with a BigDye terminator 3.1 kit (Applied Biosystems) according to the manufacturer’s instructions. Capillary sequence analysis was also performed by Mission Biotech, Taiwan with an ABI 3730xl DNA sequencer (Applied Biosystems).
Antimicrobial susceptibility
Overnight cultures of C. difficile strains were inoculated onto Brucella agar (Oxoid) plates containing vitamin K1 (0.5 mg/L), haemin (5 mg/L) and 5% defibrinated sheep red blood cells. Minimum inhibitory concentrations (MICs) of moxifloxacin (MX), metronidazole (MZ), and vancomycin (VA) were evaluated by Etest (AB Biodisk, Solna, Sweden). Quality control strains included Bacteroides fragilis ATCC25285, Bacteroides thetaiotaomicron ATCC 29741, and C. difficile ATCC700057. The breakpoint used for three tested antibiotics was 8 mg/L, 32 mg/L and 16 mg/L, respectively, in accordance with the guideline established by the Clinical and Laboratory Standards Institute (CLSI).
Sequence analysis of gyrA and gyrB was performed as previously described [21]. Briefly, the DNA region was amplified using the primer pairs, gyrA1–gyrA2 for gyrA and gyrB1–gyrB2 for gyrB. PCR products were purified and sequenced. Pairwise alignments of DNA sequences were carried out using the BLAST server of the National Center for Biotechnology Information.
PCR ribotyping
The isolates containing tcdB and determined as toxigenic strains were further examined by polymerase chain reaction (PCR) ribotyping. The ribotyping method and PCR primers used were as described previously [22]. After PCR amplification, the samples were concentrated using the Gel/PCR DNA Fragments Extraction Kit (Geneaid, Ltd, Taiwan) and separated by the QIAxcel capillary electrophoresis system (Qiagen, Hilden, Germany) using the “OM500” method and QX Alignment Marker 15 bp/3 kb (Qiagen, Hilden, Germany).
Repetitive sequence-based polymerase chain reaction (Rep-PCR)
The identity of the consecutive isolates collected beyond 28 days from the same patient was determined by Rep-PCR. Rep-PCR was performed as described by Versalovic et al. [23], and the PCR products were analyzed using the QIAxcel system.
Statistical analysis
Statistical analyses were performed by statistical software (SPSS, version 13.0). Continuous data were expressed as the means ± standard deviations. The χ2 test or Fisher’s test was used to compare categorical variables, and Student’s t-test was used to compare continuous variables. A two-tailed P value of less than 0.05 was considered to be statistically significant.
Results
Toxin gene content distribution of clinical C. difficile isolates
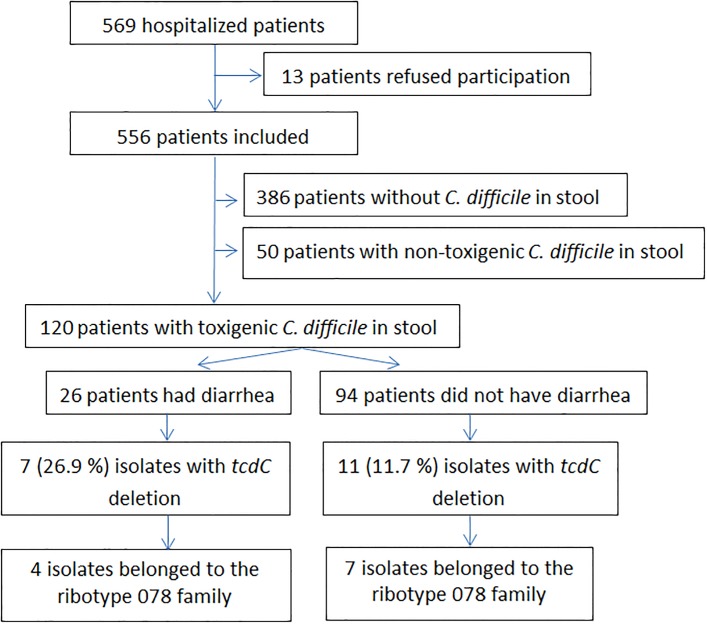
Of 569 hospitalized patients, 556 fulfilling the inclusion criteria were enrolled. C. difficile was found in 170 patients. Overall 120 (70.6%) patients had toxigenic C. difficile isolates harvested from stool and 50 (29.4%) harboured non-toxigenic C. difficile isolates (Fig 1). Of 120 patients, 26 (21.7%) developed diarrhea and were regarded as having CDI while 94 patients had toxigenic C. difficile colonization (tCdC). Of 120 toxigenic C. difficile isolates, 68 (56.7%) possessed both tcdA and tcdB and 52 (43.3%) had tcdB and truncated tcdA (Table 1). Among 68 tcdA+/tcdB+ isolates, 18 (26.4%) harbored binary toxin (cdtA and cdtB) and tcdC deletion, including Δ39 (C184T) deletion (14 isolates), Δ18 in-frame deletion (3 isolates), and Δ18 (Δ117A) deletion (1 isolate). Of 52 tcdA-/tcdB+ isolates, none had binary toxin or tcdC deletion.
Fig 1. Flowchart of the patients enrolled in this study.
Table 1. Toxin gene contents of 120 toxigenic clinical Clostridium difficile isolates.
| Toxin genes, isolate No. (%) | CDT | tcdC pattern | Isolate No. (%) |
|---|---|---|---|
| tcdA+/tcdB+, 68 (56.7) | CDT+ | Δ18 bp, in-frame | 3 (2.5) |
| CDT+ | Δ18 bp, Δ117A | 1 (0.8) | |
| CDT+ | Δ39 bp, C184T | 14 (11.7) | |
| CDT- | Wild type | 50 (41.7) | |
| tcdA-/tcdB+, 52 (43.3) | CDT- | Wild type | 52 (43.3) |
CDT = C. difficile binary toxin; bp = base-pair.
Genetic relationship of consecutive toxigenic C. difficile isolates from the same individual
Genetic relationship of consecutive toxigenic isolates from the same individual collected beyond 28 days was examined by Rep-PCR (Table 2). Of 8 patients, consecutive isolates were identical in 6 (75.0%) patients, irrespective of the presence or absence of diarrhea. Genetic similarity among consecutive fecal C. difficile isolates obtained from 2 patients was illustrated (Fig 2).
Table 2. The toxin gene content and ribotype of consecutive toxigenic Clostridium difficile isolates.
| Patient No. | Follow-up period | Clinical condition | Strain shift* | Toxin gene content | Ribotype (RT) |
|---|---|---|---|---|---|
| 1 | 2011/11/9-2012/2/15 | C→D→C | Yes | A+B+CDT-→ A-B+CDT- | unknown→RT 017 |
| 2 | 2012/4/17-2012/11/30 | C→D→D | Yes | A+B+CDT-→ A+B+CDT- → A+B+CDT- | RT 106→RT 001/072 →unknown |
| 3 | 2012/2/2-2012/8/20 | D→C | No | A+B+CDT+ | RT 126 |
| 4 | 2012/3/16-2012/4/16 | D→C→D | No | A+B+CDT- | RT 106 |
| 5 | 2012/4/20-2012/10/11 | C→D | No | A-B+CDT- | RT 017 |
| 6 | 2012/5/28-2012/10/1 | C→D | No | A+B+CDT- | RT 106 |
| 7 | 2012/6/20-2012/7/18 | C→D | No | A+B+CDT- | unknown |
| 8 | 2012/8/9-2012/9/7 | C→D→C→D | No | A+B+CDT- | unknown |
A = tcdA; B = tcdB; C = colonization (C. difficile colonization); CDT = cdtA/cdtB; D = disease (C. difficile infection).
* Indicates the detection of other rep-PCR profiles.
Fig 2. Molecular characteristics of consecutive fecal isolates of Clostridium difficile obtained from two patients, TNHP 173~TNHP 328 (ribotype 126) and TNHP 269~ TNHP 356 (ribotype 106), respectively.
A = tcdA; B = tcdB; CDT = binary toxin; bp = base-pair; del. = deletion; Rep-PCR = repetitive sequence-based polymerase chain reaction.
Ribotype and antimicrobial susceptibility of C. difficile isolates
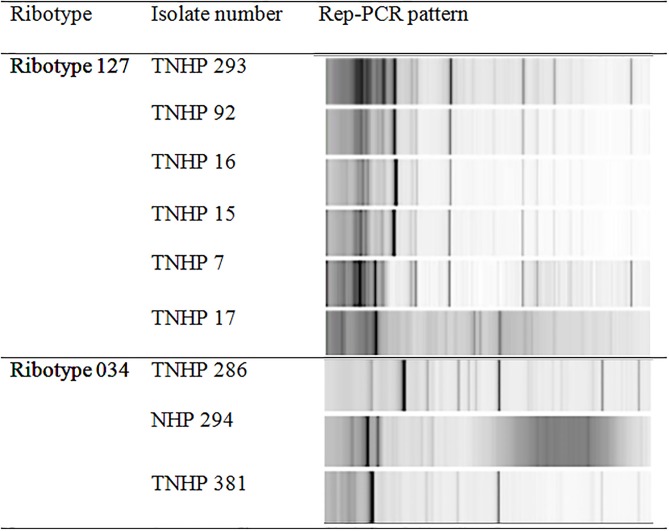
The ribotypes, antimicrobial susceptibility, and partial sequences of gyrA/gyrB of 18 toxigenic isolates with binary toxin and tcdC deletion were investigated (Table 3). Eleven (61.1%) isolates belonged to the ribotype 078 family, including ribotype 078 (1 isolate), ribotype 126 (4 isolates), and ribotype 127 (6 isolates) (S1 Fig). Of the 52 isolates with tcdA-/tcdB+ genotype, all were identified to be ribotype 017. Prof. M Wilcox at the Leeds Teaching Hospitals NHS Trust confirmed ribotyping of the above clinical isolates. For genetic relationship studied using Rep-PCR, 4 isolates of ribotype 126 were found to be identical as reported previously [15], and 6 isolates of ribotype 127 exhibited 3 distinct subtypes with a predominant Rep-PCR subtype (4 isolates). However, 3 isolates of ribotype 034 were genetically different (Fig 3). All 18 toxigenic isolates with binary toxin were susceptible to metronidazole and vancomycin in vitro, and of 10 (55.6%) moxifloxacin-resistant isolates (MIC >32 mg/L), 9 had gyrA mutation (Thr82Ile) and 1 had gyrB mutation (Asp426Asn).
Table 3. Ribotypes, gyrA and gyrB mutations, and antimicrobial susceptibility of 18 Clostridium difficile isolates with binary toxin.
| Isolate No. | Ribotype (RT) | gyrA mutation | gyrB mutation | MIC, mg/L | ||
|---|---|---|---|---|---|---|
| MX | MZ | VA | ||||
| 2 | RT 328 | - | - | 0.5 | 0.032 | 0.5 |
| 286 | RT 034 | - | - | 0.5 | 0.064 | 0.5 |
| 294 | - | Asp426Asn | >32 | 0.125 | 0.5 | |
| 381 | - | - | 0.5 | 0.032 | 0.25 | |
| 80 | RT 078 | - | - | 0.25 | 0.032 | 0.38 |
| 61 | RT 126 | Thr82Ile | - | >32 | 0.032 | 0.25 |
| 173 | Thr82Ile | - | >32 | 0.032 | 0.5 | |
| 203 | - | - | 0.5 | 0.032 | 0.75 | |
| 264 | - | - | 0.5 | 0.047 | 0.75 | |
| 7 | RT 127 | Thr82Ile | - | >32 | 0.032 | 0.38 |
| 15 | Thr82Ile | - | >32 | 0.032 | 0.38 | |
| 16 | Thr82Ile | - | >32 | 0.032 | <0.016 | |
| 17 | Thr82Ile | - | >32 | <0.016 | 0.38 | |
| 92 | - | - | 0.38 | 0.032 | 0.38 | |
| 293 | Thr82Ile | - | >32 | 0.023 | 0.38 | |
| 9 | Unknown | Thr82Ile | - | >32 | 0.032 | 0.38 |
| 13 | Thr82Ile | - | >32 | 0.032 | 0.38 | |
| 14 | - | - | 0.1 | 0.047 | <0.016 | |
MX: moxifloxacin; MZ: metronidazole; VA: vancomycin.
Fig 3. Repetitive sequence-based polymerase chain reaction (Rep-PCR) gel patterns of 6 toxigenic Clostridium difficile isolates of ribotype 127 (3 subtypes) and ribotype 034 isolates (3 subtypes).
Clinical characteristics of patients with C. difficile ribotype 078 family
Of 26 isolates causing CDI, 7 (26.9%) had binary toxin and 4 (15.4%) belonged to the ribotype 078 family. In contrast, 11 (11.7%) of 94 tCdC isolates had binary toxin and 7 (7.4%) isolates belonged to the ribotype 078 family (S1 Table). Clinical characteristics of 26 patients with CDI and 94 patients with tCdC were compared (Table 4). Patients with CDI were less likely to be nursing home residents (57.7% vs. 77.7%; P = 0.049), and more likely to have nasogastric tube (77.3% vs. 46.8%, P = 0.016) or diabetes mellitus (61.5% vs. 39.4%, P = 0.049). However, there were no differences in underlying disease, laboratory findings, ribotype distribution, and moxifloxacin resistance in two groups (Tables 4 and 5).
Table 4. Clinical characteristics of 120 patients with toxigenic Clostridium difficile in stool, stratified by the presence (C. difficile infection, CDI) or absence (C. difficile colonization, CdC) of diarrhea.
| Characteristics | CDI, n = 26 | CdC, n = 94 | P values |
|---|---|---|---|
| Male | 17 (65.4) | 51 (54.8) | 0.377 |
| Age, years | 70.5 ± 12.7 | 73.6 ± 13.5 | 0.274 |
| Body weight, kg | 50.2 ± 12.1 | 50.6 ± 11.2 | 0.404 |
| Nursing home residents | 15 (57.7) | 73 (77.7) | 0.049 |
| Recent hospitalization within three months | 9 (34.6) | 28 (29.8) | 0.638 |
| Nasogastric tube use | 17 (77.3) | 44 (46.8) | 0.016 |
| Prior exposure to antibiotic* | 7 (30.4) | 23 (24.5) | 0.598 |
| Prior exposure to proton pump inhibitor* | 2 (8.7) | 5 (5.3) | 0.622 |
| Underlying medical diseases | |||
| Hypertension | 10 (38.5) | 45 (47.9) | 0.506 |
| Diabetes mellitus | 16 (61.5) | 37 (39.4) | 0.049 |
| Old stroke | 11 (42.3) | 28 (29.8) | 0.244 |
| Chronic kidney disease (Ccr <60 ml/min) | 3 (11.5) | 19 (20.2) | 0.400 |
| On hemodialysis | 1 (3.8) | 8 (8.5) | 0.682 |
| Malignancy | 4 (15.4) | 9 (9.6) | 0.475 |
| Laboratory data, mean ± standard deviation | |||
| White blood count, 103/mm3 | 11.4 ± 5.8 | 11.5 ± 5.9 | 0.977 |
| Neutrophils, % | 75.5 ± 12.6 | 76.1 ± 15.1 | 0.856 |
| Hemoglobin, g/dL | 10.4 ± 2.3 | 11.92 ± 2.1 | 0.117 |
| Platelet, 103/mm3 | 253.7 ± 105.6 | 219.2 ± 107.5 | 0.152 |
| Alanine aminotransferase, U/L | 22.5 ± 24.3 | 39.9 ± 70.5 | 0.058 |
| Creatinine, mg/dL | 1.5 ± 1.0 | 2.6 ± 2.8 | 0.214 |
Data are no. (%) of patients, unless otherwise indicated; Ccr = creatinine clearance.
*Medication within three months before admission.
Table 5. Ribotype (RT), binary toxin, and gyrA/B mutation of initial toxigenic Clostridium difficile isolates from 120 patients, stratified by the presence (C. difficile infection, CDI) or absence (C. difficile colonization, CdC) of diarrhea.
| Characteristics | CDI, n = 26 | CdC, n = 94 | P values |
|---|---|---|---|
| Binary toxin | 7 (26.9) | 11 (11.7) | 0.067 |
| RT 078 family | 4 (15.4) | 7 (7.4) | 0.250 |
| RT 078 | 0 | 1 | |
| RT 126 | 3 | 1 | |
| RT 127 | 1 | 5 | |
| RT 034 | 2 | 1 | |
| RT 328 | 0 | 1 | |
| Unknown ribotype | 1 | 2 | |
| gyr mutation | |||
| gyrA mutation | 5 (19.2) | 9 (9.6) | 0.181 |
| gyrB mutation | 6 (23.1) | 8 (8.5) | 0.077 |
Data are no. (%) of patients, unless otherwise indicated.
We assessed the seasonal distribution of toxigenic C. difficile isolates stratified by binary toxin and ribotype 078 family (S2 Fig). More C. difficile isolates with binary toxin, particularly those belonging to the ribotype 078 family, were harvested at the first and second quarters of 2011 and 2012.
Discussion
In the present study, the ribotype 078 family was dominant among toxigenic C. difficile isolates with binary toxin and tcdC deletion in Taiwan. Moreover the ribotype 078 family in Taiwan often displayed a high level of moxifloxacin resistance. Several retrospective studies in Taiwan investigated the prevalence of hypervirulent C. difficile strains, such as ribotype 027 or 078, but no hypervirulent isolates had been discovered before 2012 [7–9]. To our knowledge, this is the first study describing ribotype and toxin genotype distribution of clinical C. difficile isolates in Taiwan. Our study revealed that among the isolates with binary toxin, more than half belonged to the ribotype 078 family, including ribotypes 078, 126, and 127. The result was unique in that ribotype 078 has been reported from Korea and China [24–26], but it not the dominant ribotype. Clinical impact and origin of the dominant ribotype 078 family among binary toxin producers in Taiwan warrant further evaluations.
In Hangzhou, China in 2013, the predominant C. difficile ribotypes in hospitalized cancer patients included 001, 017/1, and 017 [27]. At another hospital in Hunan, China between April 2009 and February 2010, the dominant ribotype was 017, followed by 046 and 012 [28]. Ribotypes 018, 017 and 001 were prevalent in Seoul, Korea from September 2008 to January 2010 [29]. In general, ribotype 017 isolates were commonly present in China and Korea, and here we found that this ribotype accounted for 43% of toxigenic isolates, suggestive of its widespread in Asia countries. An international surveillance of toxigenic C. difficile isolates is essential to disclose ribotype distribution and clinical significance of toxigenic C. difficile isolates in Asia.
Of the patients in our study, identical C. difficile isolates can be obtained from the same individuals with tCdC, CDI, or recurrent CDI. The result was similar to the study conducted by Oka et al., who noted 80% were identical between the strains at initial infection and at recurrence [11]. Moreover Kim et al. found relapse rates of ribotype 017 and 018 isolates were higher than those of other ribotypes (63.6% and 63.6% vs. 22.2%, respectively) [30]. The clinical impact of C. difficile ribotypes on CDI recurrence warrants more studies. However, in our study the same C. difficile isolate persisted from CdC to CDI or from CDI to CdC, suggestive of long-term carriage of identical C. difficile strains, irrespective of the initial presence or absence of diarrhea. Furthermore, the role of asymptomatic carriers in C. difficile transmission has been supported by a recent study in which active surveillance detection and isolation of C. difficile carriers was associated with a significant decrease in the incidence of healthcare-associated CDI [31]. Clinical significance and management of asymptomatic C. difficile carriers warrant more attention.
Patients with CDI were characterized as less likely to be nursing home residents, more likely to use nasogastric tube and to have diabetes mellitus, than those with CdC. Nasogastric tube use [32,33] and diabetes mellitus [34] had been regarded as risk factors for CDI before. It is notable that CDI in our study was inversely correlated with nursing home residing, though the prevalence of C. difficile colonization or infection among residents in long-term care facilities had been increasing in recent years due to advanced age, the recipients of multiple courses or longer duration of antibiotics, and previous CDI history [35,36].
There are some limitations in our study. Firstly, we only analyzed the ribotypes of toxigenic C. difficile isolates with binary toxin and tcdC deletion and tcdA-/tcdB+ isolates. Secondly, the C. difficile isolates studied were only obtained from a district hospital in southern Taiwan. Their representativeness is of course questionable. Thirdly, it is likely that the boundary of C. difficile colonization and infection is blurred. With longer followed-up periods, the isolates of tCdC and CDI will be exchangeable. For example, the isolates initially causing colonization can later be related to CDI, and vice versa. Finally, though a total of 13 patients were excluded from the study due to expected hospital stay for less than 5 days, recent metronidazole or vancomycin therapy, colectomy, or CDI at admission, the estimated number of toxigenic C. difficile isolates obtained from them was 2 or 3. Therefore, the exclusion of these patients will not change significantly our study results.
In conclusion, ribotype 017 isolates were not uncommon in toxigenic C. difficile isolates southern Taiwan and the ribotype 078 family predominated among clinical C. difficile isolates with binary toxin.
Supporting Information
(TIF)
(TIF)
(XLS)
Acknowledgments
We acknowledge Prof. M. Wilcox at the Leeds Teaching Hospitals NHS Trust for typing our clinical strains. This study was supported by the grants from the Ministry of Science and Technology (MOST 102-2628-B-006-015-MY3, 105-2321-B-006-012 and 105-2321-B-006-013) and Ministry of Health and Welfare, Taiwan (MOHW 104-CDC-C-114-11 and 105-TDU-B-211-133016).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by the grants from the Ministry of Science and Technology (MOST 102-2628-B-006-015-MY3, 105-2321-B-006-012 and 105-2321-B-006-013) and Ministry of Health and Welfare, Taiwan (MOHW 104-CDC-C-114-11 and 105-TDU-B-211-133016). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kelly CP, LaMont JT (2008) Clostridium difficile—more difficult than ever. N Engl J Med 359: 1932–1940. 10.1056/NEJMra0707500 [DOI] [PubMed] [Google Scholar]
- 2.Loo VG, Poirier L, Miller MA, Oughton M, Libman MD, Michaud S, et al. (2005) A predominantly clonal multi-institutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality. N Engl J Med 353: 2442–2449. 10.1056/NEJMoa051639 [DOI] [PubMed] [Google Scholar]
- 3.Barbut F, Delmee M, Brazier JS, Petit JC, Poxton IR, Rupnik M, et al. (2003) A European survey of diagnostic methods and testing protocols for Clostridium difficile. Clin Microbiol Infect 9: 989–996. [DOI] [PubMed] [Google Scholar]
- 4.Barbut F, Mastrantonio P, Delmee M, Brazier J, Kuijper E, Poxton I. (2007) Prospective study of Clostridium difficile infections in Europe with phenotypic and genotypic characterisation of the isolates. Clin Microbiol Infect 13: 1048–1057. 10.1111/j.1469-0691.2007.01824.x [DOI] [PubMed] [Google Scholar]
- 5.Freeman J, Bauer MP, Baines SD, Corver J, Fawley WN, Goorhuis B, et al. (2010) The changing epidemiology of Clostridium difficile infections. Clin Microbiol Rev 23: 529–549. 10.1128/CMR.00082-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sawabe E, Kato H, Osawa K, Chida T, Tojo N, Arakawa Y, et al. (2007) Molecular analysis of Clostridium difficile at a university teaching hospital in Japan: a shift in the predominant type over a five-year period. Eur J Clin Microbiol Infect Dis 26: 695–703. 10.1007/s10096-007-0355-8 [DOI] [PubMed] [Google Scholar]
- 7.Liao CH, Ko WC, Lu JJ, Hsueh PR (2012) Characterizations of clinical isolates of Clostridium difficile by toxin genotypes and by susceptibility to 12 antimicrobial agents, including fidaxomicin (OPT-80) and rifaximin: a multicenter study in Taiwan. Antimicrob Agents Chemother 56: 3943–3949. 10.1128/AAC.00191-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hung YP, Tsai PJ, Hung KH, Liu HC, Lee CI, Lin HJ, et al. (2012) Impact of toxigenic Clostridium difficile colonization and infection among hospitalized adults at a district hospital in southern Taiwan. PLoS One 7: e42415 10.1371/journal.pone.0042415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin YC, Huang YT, Tsai PJ, Lee TF, Lee NY, Liao CH, et al. (2011) Antimicrobial susceptibilities and molecular epidemiology of clinical isolates of Clostridium difficile in Taiwan. Antimicrob Agents Chemother 55: 1701–1705. 10.1128/AAC.01440-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hung YP, Cia CT, Tsai BY, Chen PC, Lin HJ, Liu HC, et al. (2015) The first case of severe Clostridium difficile ribotype 027 infection in Taiwan. J Infect 70: 98–101. 10.1016/j.jinf.2014.08.003 [DOI] [PubMed] [Google Scholar]
- 11.Oka K, Osaki T, Hanawa T, Kurata S, Okazaki M, Manzoku T, et al. (2012) Molecular and microbiological characterization of Clostridium difficile isolates from single, relapse, and reinfection cases. J Clin Microbiol 50: 915–921. 10.1128/JCM.05588-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barbut F, Richard A, Hamadi K, Chomette V, Burghoffer B, Petit JC. (2000) Epidemiology of recurrences or reinfections of Clostridium difficile-associated diarrhea. J Clin Microbiol 38: 2386–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hung YP, Lin HJ, Wu TC, Liu HC, Lee JC, Lee CI,et al. (2013) Risk factors of fecal toxigenic or non-toxigenic Clostridium difficile colonization: impact of Toll-like receptor polymorphisms and prior antibiotic exposure. PLoS One 8: e69577 10.1371/journal.pone.0069577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin HJ, Hung YP, Liu HC, Lee JC, Lee CI, Wu YH, et al. (2015) Risk factors for Clostridium difficile-associated diarrhea among hospitalized adults with fecal toxigenic C. difficile colonization. J Microbiol Immunol Infect 48: 183–189. 10.1016/j.jmii.2013.08.003 [DOI] [PubMed] [Google Scholar]
- 15.Hung YP, Lin HJ, Tsai BY, Liu HC, Liu HC, Lee JC, et al. (2014) Clostridium difficile ribotype 126 in southern Taiwan: a cluster of three symptomatic cases. Anaerobe 30: 188–192. 10.1016/j.anaerobe.2014.06.005 [DOI] [PubMed] [Google Scholar]
- 16.Ackermann G, Tang YJ, Jang SS, Silva J, Rodloff AC, Cohen SH. (2001) Isolation of Clostridium innocuum from cases of recurrent diarrhea in patients with prior Clostridium difficile associated diarrhea. Diagn Microbiol Infect Dis 40: 103–106. [DOI] [PubMed] [Google Scholar]
- 17.Satomura H, Odaka I, Sakai C, Kato H (2009) [Antibiotic-associated diarrhea due to Clostridium perfringens]. Kansenshogaku Zasshi 83: 549–552. [DOI] [PubMed] [Google Scholar]
- 18.Hunley TE, Spring MD, Peters TR, Weikert DR, Jabs K (2008) Clostridium septicum myonecrosis complicating diarrhea-associated hemolytic uremic syndrome. Pediatr Nephrol 23: 1171–1175. 10.1007/s00467-008-0774-5 [DOI] [PubMed] [Google Scholar]
- 19.Persson S, Jensen JN, Olsen KE (2011) Multiplex PCR method for detection of Clostridium difficile tcdA, tcdB, cdtA, and cdtB and internal in-frame deletion of tcdC. J Clin Microbiol 49: 4299–4300. 10.1128/JCM.05161-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Curry SR, Marsh JW, Muto CA, O'Leary MM, Pasculle AW, Harrison LH. (2007) tcdC genotypes associated with severe TcdC truncation in an epidemic clone and other strains of Clostridium difficile. J Clin Microbiol 45: 215–221. 10.1128/JCM.01599-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dridi L, Tankovic J, Burghoffer B, Barbut F, Petit JC (2002) gyrA and gyrB mutations are implicated in cross-resistance to ciprofloxacin and moxifloxacin in Clostridium difficile. Antimicrob Agents Chemother 46: 3418–3421. 10.1128/AAC.46.11.3418-3421.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stubbs SL, Brazier JS, O'Neill GL, Duerden BI (1999) PCR targeted to the 16S-23S rRNA gene intergenic spacer region of Clostridium difficile and construction of a library consisting of 116 different PCR ribotypes. J Clin Microbiol 37: 461–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Versalovic J, Koeuth T, Lupski JR (1991) Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res 19: 6823–6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim H, Jeong SH, Roh KH, Hong SG, Kim JW, Shin MG, et al. (2010) Investigation of toxin gene diversity, molecular epidemiology, and antimicrobial resistance of Clostridium difficile isolated from 12 hospitals in South Korea. Korean J Lab Med 30: 491–497. 10.3343/kjlm.2010.30.5.491 [DOI] [PubMed] [Google Scholar]
- 25.Huang H, Fang H, Weintraub A, Nord CE (2009) Distinct ribotypes and rates of antimicrobial drug resistance in Clostridium difficile from Shanghai and Stockholm. Clin Microbiol Infect 15: 1170–1173. 10.1111/j.1469-0691.2009.02992.x [DOI] [PubMed] [Google Scholar]
- 26.Collins DA, Hawkey PM, Riley TV (2013) Epidemiology of Clostridium difficile infection in Asia. Antimicrob Resist Infect Control 2: 21 10.1186/2047-2994-2-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fang WJ, Jing DZ, Luo Y, Fu CY, Zhao P, Qian J, et al. (2014) Clostridium difficile carriage in hospitalized cancer patients: a prospective investigation in eastern China. BMC Infect Dis 14: 523 10.1186/1471-2334-14-523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hawkey PM, Marriott C, Liu WE, Jian ZJ, Gao Q, Ling TK, et al. (2013) Molecular epidemiology of Clostridium difficile infection in a major chinese hospital: an underrecognized problem in Asia? J Clin Microbiol 51: 3308–3313. 10.1128/JCM.00587-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim J, Kang JO, Kim H, Seo MR, Choi TY, Pai H, et al. (2013) Epidemiology of Clostridium difficile infections in a tertiary-care hospital in Korea. Clin Microbiol Infect 19: 521–527. 10.1111/j.1469-0691.2012.03910.x [DOI] [PubMed] [Google Scholar]
- 30.Kim J, Seo MR, Kang JO, Kim Y, Hong SP, Pai H. (2014) Clinical characteristics of relapses and re-infections in Clostridium difficile infection. Clin Microbiol Infect 20: 1198–1204. 10.1111/1469-0691.12704 [DOI] [PubMed] [Google Scholar]
- 31.Longtin Y, Paquet-Bolduc B, Gilca R, Garenc C, Fortin E, Longtin J, et al. Effect of Detecting and isolating Clostridium difficile carriers at hospital admission on the incidence of c difficile infections: a quasi-experimental controlled study. JAMA Intern Med. 2016;176(6):796–804. 10.1001/jamainternmed.2016.0177 [DOI] [PubMed] [Google Scholar]
- 32.O'Keefe SJ (2010) Tube feeding, the microbiota, and Clostridium difficile infection. World J Gastroenterol 16: 139–142. 10.3748/wjg.v16.i2.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bliss DZ, Johnson S, Savik K, Clabots CR, Willard K, O'Shea M, et al. (1998) Acquisition of Clostridium difficile and Clostridium difficile-associated diarrhea in hospitalized patients receiving tube feeding. Ann Intern Med 129: 1012–1019. [DOI] [PubMed] [Google Scholar]
- 34.Wong-McClure RA, Guevara-Rodriguez M, Abarca-Gomez L, Solano-Chinchilla A, Marchena-Picado M, O'Shea M, et al. (2012) Clostridium difficile outbreak in Costa Rica: control actions and associated factors. Rev Panam Salud Publica 32: 413–418. [PubMed] [Google Scholar]
- 35.Chopra T, Goldstein EJ (2015) Clostridium difficile Infection in long-term care facilities: a call to action for antimicrobial stewardship. Clin Infect Dis 60 Suppl 2: S72–76. [DOI] [PubMed] [Google Scholar]
- 36.Jarvis WR, Schlosser J, Jarvis AA, Chinn RY (2009) National point prevalence of Clostridium difficile in US health care facility inpatients, 2008. Am J Infect Control 37: 263–270. 10.1016/j.ajic.2009.01.001 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
(XLS)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.