1 Introduction
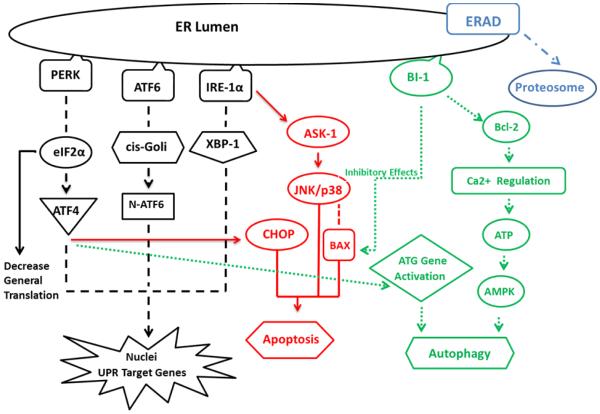
ER plays an essential role in the secretory pathways and is the destination where proteins are distributed into the endo or exocytotic pathways. Assembly, folding and disulfidation of proteins occur in the ER before exposure to the extracellular space. Concentration of proteins in the ER lumen is extremely high (Stevens and Argon, 1999) and there is about 13 million secretory proteins per minute in hepatocytes (Snapp, 2005). The liver cell consumes enormous amounts of energy to perform this transportation accurately. Energy imbalance occurs for many reasons such as starvation, changes in oxidation-reduction balance, calcium level, post translational adjustments or secretory protein synthesis (Rutkowski et al., 2004). This imbalance creates a lipotoxic environment by accumulation of lipids and misfolded proteins in the ER lumen (Lake et al., 2014) and leads to the activation of BiP/GRP78 (immunoglobulin heavy chain-binding protein/glucose-regulated protein of molecular weight 78 kDa) a chaperone member of heat-shock protein 70 (Hsp70) (Wu and Kaufman, 2006). This is part of a compensatory program called “ER stress signaling” and the subsequent “Unfolded Protein Response (UPR)”. There are three determined sub-pathways involve in the UPR response, PKR-like ER protein kinase (PERK), Inositol-requiring enzyme 1 (IRE1) and activating transcription factor 6 (ATF6), associate with BiP in their inactive states. In severe ER stress, BiP loses its bond (Ma et al., 2002) from UPR components simultaneously. Each sub-pathway initiates a special reaction in the metabolic processes, but the immediate response starts by activation of PERK (Shi et al., 1998, Harding et al., 1999) which phosphorylates eucaryotic translation initiation factor 2 (eIF2α). Phosphorylated eIF2α decreases the rate of mRNA translation to attenuate general protein synthesis and also activates transcription factor 4 (ATF4) (Harding et al., 2000). BiP detachment from IRE1α, or binding unfolded protein itself to IRE1α (Sano and Reed, 2013), activates the IRE1α site-specific endoribonuclease (RNase). This RNase activity splices X-box-binding protein 1 (XBP1) mRNA (Shen et al., 2001). Activated ATF6 translocates to the cis- Golgi compartment and splits up into fragments. N-terminal fragment of ATF6, along with ATF4 and spliced XBP1, migrate to the nucleus and act as active transcription factors to enhance the gene expression of proteins that augment ER protein folding capacity (Cinaroglu A et al., 2011, Tirasophon et al., 1998, Haze et al., 1999, Ye at al., 2000, Yoshida et al., 2000, Shen et al., 2001, Yoshida et al., 2001, Calfon at al., 2002, Lee et al., 2002).
ER stress, under a severe insult, such as chronic alcohol consumption, activates two protein degradation pathways to preserve cell function and viability (Bernales et al., 2006). 1) The ER-assisted degradation process (ERAD) accumulates increased stress proteins, e.g. HSPs, and other components of the increased metabolic processes in the cytosol for ubiquitination and degradation by the 26S proteasome. Mallory- Denk Body (MDB) contains many ubiquitated misfolded proteins, mostly located in ballooned hepatocytes might produce ERAD (Basaranoglu et al., 2011, Liu et al., 2014) 2) Autophagy which is mediated by lysosomal derived catalytic enzymes digestion.
Autophagy, initiated by ER stress mechanisms, varied in the current studies. one of the proposed mechanisms is related to Ca2+ release from ER which activates the CamKK/AMPK-dependent pathway. This complex inhibits mTOR to induce autophagy (Høyer-Hansen et al., 2007). PERK also regulates autophagy on ATG genes via ATF-4 transcriptional effects (Harding et al., 2000). There are other cell protective pathways during ER stress that act as apoptotic suppressors. Bax-inhibitor1 (BI-1), that located in the ER membrane, provides protection during ER stress by a) regulates ER Ca2+ homeostasis through Bcl-2 family members and b) inhibits IRE1α RNase activity (Xu et al., 2008).
When compensatory pathways fail after chronic and prolonged ER stress, cell dysfunction and cell death develop. Based on the longevity and severity of the stress, IRE1α activates downstream proteins resulting in cell death and apoptosis, such as Jun-N-terminal kinase, ASK1 (apoptosis signal-regulating kinase1) and p38 MAPK (Pincus et al., 2010, Ron and Hubbard, 2008). p38 MAPK activated by IRE1α phosphorylates and activates transcription factor CHOP (transcriptional factor C/EBP homologous protein). CHOP increases the expression of genes facilitating apoptosis (Puthalakath et al., 2007). CHOP is also one of the targets activated with ATF4 and is known as one of the most important regulators of ER stress induced apoptosis (Oyadomari and Mori, 2004, Cullinan and Diehl, 2007). Bax/Bcl-2 complex is another mediator inducing apoptosis via controlling ER Ca2+ release (Chae et al., 2004 , Ishikawa et al., 2011). Bax also will be activated as pro-apoptotic agents by p38MAPK and JNK (Kim et al., 2006).
Chronic alcohol consumption is a well-known stressor and induces ER stress signaling /UPR. Since the mechanisms and causes that induce ASH and NASH are different, we surveyed the expressions of proteins and mediators that take part in ER stress pathway (Fig. 1) in the liver of alcoholic hepatitis and NASH.
Fig. 1.
Cross reaction of cytoprotective pathways and cell death pathways under ER stress
2 Materials and Methods
Formalin-fixed paraffin embedded (FFPE) human liver biopsies from patients who were diagnosed with ASH and NASH from Harbor–UCLA hospital archived and a clinical trial funded by NIH/NIAAA grant, i.e., “Alcoholic hepatitis pathogenesis as determined from human liver tissue analysis”(exempted as determined by the iRIS system) were compared to normal controls. In this study we studied 3-5 ASH, 3-5 NASH and 3 normal liver controls. The slides were double stained for ubiquitin plus PERK, IRE1, ATF4, ATF6 and BAX. Texas Red (Millipore, Temecula, CA) was used to detect ubiquitin. The other proteins were detected as green fluorescence by using either donkey-anti mouse or anti rabbit Alex Fluor for the secondary antibody labeled (Jackson Labs, West Grove, PA). The nuclei were stained by DAPI. The slides were all stained together at the same time to provide accurate comparisons between the different groups. We obtained the intensity of the fluorescent staining in 3 different areas on each slide with the 40x objective and 800ms standard exposure time by using a Nikon 400 fluorescent microscope. The Nikon morphometric system was used to a quantitate measurement of the fluorescent intensity. The mean, standard error and statistical differences of data achieved from the Nokia were analyzed by Graphpad statistical software. Controls vs. ASH, controls vs NASH and ASH vs. NASH were compared by unpaired t-test with p < 0.05. (See table 1)
Table 1.
Antibodies used in the study with animal source and company/Vendor
| Antibody | Company/Vendor |
|---|---|
| PERK | Abcam, Cambridge, MA 02139 |
| IRE-1 | Novus Biological, Littleton, CO 80120 |
| ATF4 | Abcam, Cambridge, MA 02139 |
| ATF6 | Life Span, Seattle, WA 98121 |
| BAX | Abcam, Cambridge, MA 02139 |
3 Results
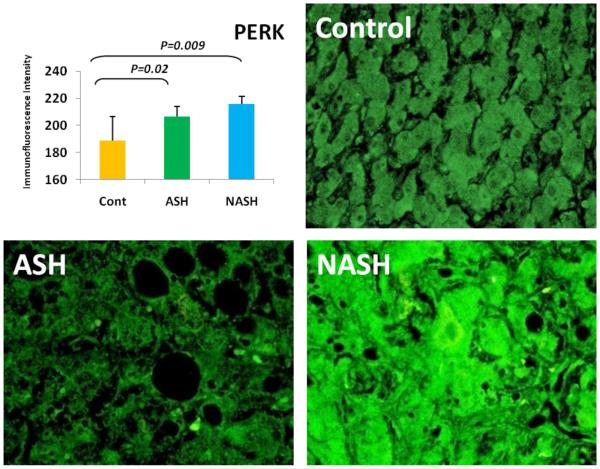
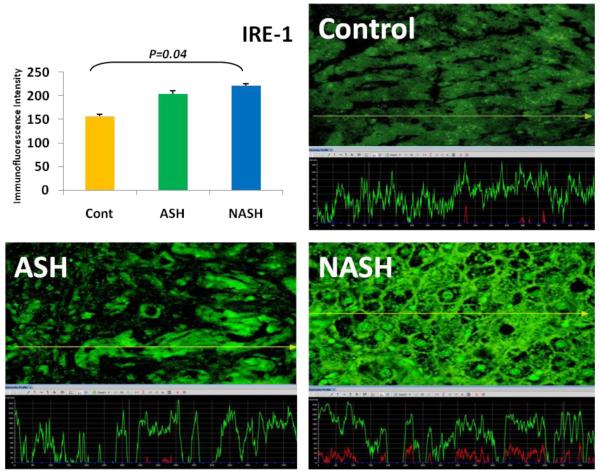
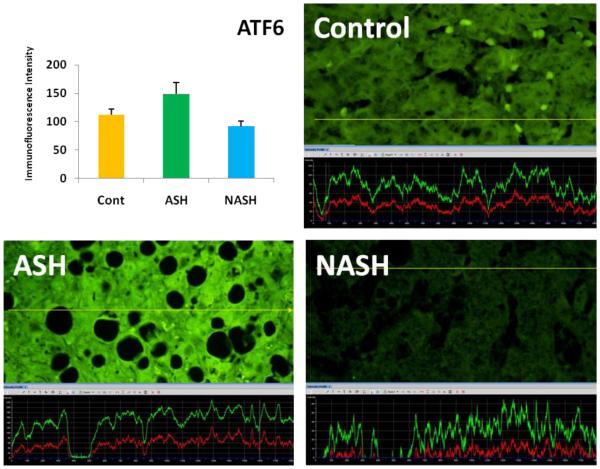
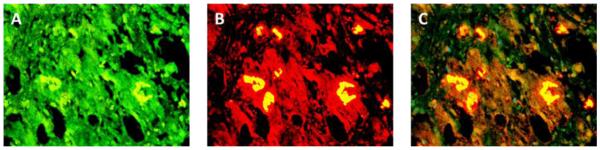
3.1 Alcoholic Steatohepatitis (ASH) causes severe liver injury due to excessive alcohol consumption. Chronic alcohol intake and exhausting cytoprotective pathways push the liver cells to lose cell function through different pathways, e.g. ERAD, which leads to MDB formation and apoptosis. The pathology of ASH, the presence of MDBs, excess fat and misfolded proteins, ballooning degeneration and degrees of fibrosis indicated a severe ER stress response. In this study the expression of PERK was significantly increased in ASH compared to the controls (Fig. 2). The expression of IRE-1α, ATF6 and ATF4 were tending to be increased compared to the controls (Fig. 4, 5, 6). Bax, tended to be increased specifically in ASH samples with MDB formation (Fig. 7). PERK was found to be colocalized with ubiquitin in in Mallory Bodies (Fig. 3).
Fig. 2.
PERK in ASH and NASH were significantly upregulated (p < 0.05) compared to the Controls. The expressions of PERK tend to be elevated in NASH in comparison to ASH. (×690)
Fig. 4.
IRE-1α expression in NASH was significantly (p < 0.05) elevated compared to controls and tend to be elevated in comparison to ASH. IRE-1α expressions in ASH tend to be increased compared to the controls. The yellow tracer line converted the fluorecence intensity in to the green line in the graph. (×690)
Fig. 5.
ATF6 expressions in ASH tend to be increased compared to Controls and NASH. Its expression in NASH was decreased in comparison to the controls. (×690)
Fig. 6.
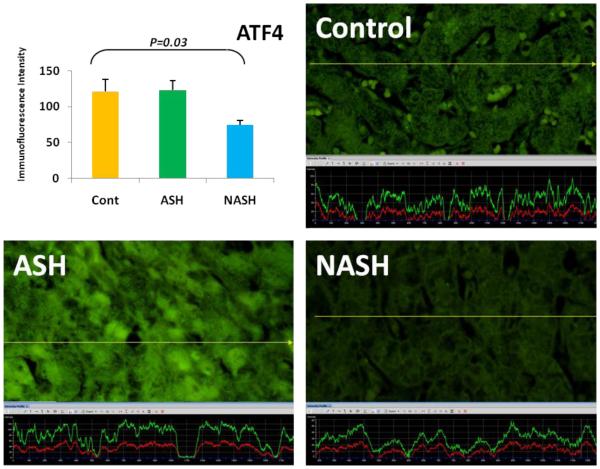
The expression of ATF4 in NASH was significantly (p < 0.05) downregulated compared to the controls. Its expression in ASH tended to be elevated in comparison to NASH. (×690)
Fig. 7.
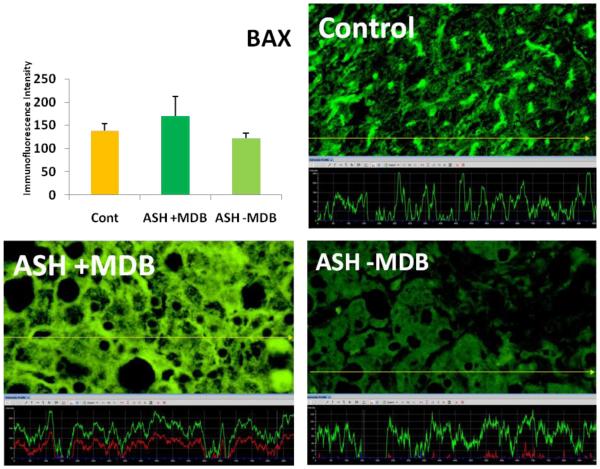
The expression of BAX in ASH with the presence of MDB tended to be elevated in comparison to the controls and ASH without MDB.
Fig. 3.
The double stain for PERK and UB is shown (A, B). Colocalization of PERK and UB is seen within the Mallory body (C). Also shown is an increase in the fluorescent intensity in the adjacent liver cells. (×520)
3.2 Non-Alcoholic Steatohepatitis (NASH) is one of the causes of fatty liver, occurring when fat is increased and stored in the liver due to obesity, metabolic syndrome or insulin resistance. Steatosis is usually combined with ballooning degeneration, MDB formation, inflammation and fibrosis. The progression of NASH to cirrhosis is slower than ASH due to presence of alcohol abuse. In NASH the expression of the first line ER stress regulators PERK and IRE-1 were significantly increased in comparison to controls (Fig. 2). ATF6 and ATF4 expressions both were decreased in NASH compared to controls (Fig. 4, 5).
3.3 ASH vs. NASH The expression of PERK and IRE-1 tended to be increased in NASH compared to ASH (Fig. 2, 4). The level of ATF4, ATF6 tended to be decreased in NASH (Fig. 5, 6).
4 Discussion
ER stress under a severe insult, such as chronic alcohol consumption in hepatocytes, activates a different protein degradation pathways to preserve cell function and viability. Significant upregulation of the first line proteins in ER stress response in NASH compared to controls and the increasing of the level of the downstream proteins of the ER response in ASH compared to controls, indicates that alcohol toxicity causes more severe cell responses in ASH in comparison to NASH. More gene expression in the downstream UPR pathway in ASH induces autophagocytosis and apoptosis (Fig. 1). In our previous study (Masouminia et al., 2016) we showed an increased level of autophagocytosis and apoptosis in ASH compared to controls and NASH.
MDB formation in hepatocytes is one of the signs of prolonged ER stress and gradual failure of the cytoprotective pathways. Increased expression of BAX as a proapoptotic regulator in the hepatocytes that formed MDBs, indicating an increase in cell death in exhausted hepatocytes.
Acknowledgements
This study was funded by NIH/NIAAA grant # UO-21898-04
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement:
The authors declare that there are no conflicts of interest.
References
- Basaranoglu M, Turhan N, Sonsuz A, Basaranoglu G. Mallory-Denk Bodies in chronic hepatitis. World J. Gastroenterol. 2011;17(17):2172–2177. doi: 10.3748/wjg.v17.i17.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernales S, McDonald KL, Walter P. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS. Biol. 2006;4(12):e423. doi: 10.1371/journal.pbio.0040423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- Chae HJ, Kim HR, Xu C, Bailly-Maitre B, Krajewska M, Krajewski S, Banares S, Cui J, Digicaylioglu M, Ke N, Kitada S, Monosov E, Thomas M, Kress CL, Babendure JR, Tsien RY, Lipton SA, Reed JC. BI-1 regulates an apoptosis pathway linked to endoplasmic reticulum stress. Mol. Cell. 2004;15(3):355–366. doi: 10.1016/j.molcel.2004.06.038. [DOI] [PubMed] [Google Scholar]
- Cinaroglu A, Gao C, Imrie D, Sadler KC. Activating transcription factor 6 plays protective and pathological roles in steatosis due to endoplasmic reticulum stress in zebrafish. Hepathology. 2011;54(2):495–508. doi: 10.1002/hep.24396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullinan SB, Diehl JA. PERK-dependent activation of Nrf2 contributes to redox homeostasis and cell survival following endoplasmic reticulum stress. J. Biol. Chem. 2007;279(19):20108–20117. doi: 10.1074/jbc.M314219200. [DOI] [PubMed] [Google Scholar]
- Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell. 2000;6(5):1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol. Cell. 2000;5(5):897–904. doi: 10.1016/s1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum- resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol. Cell. 1999;10:3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Høyer-Hansen M, Bastholm L, Szyniarowski P, Campanella M, Szabadkai G, Farkas T, Bianchi K, Fehrenbacher N, Elling F, Rizzuto R, Mathiasen IS, Jäättelä M. Control of macroautophagy by calcium, calmodulin-dependent kinase kinase-beta, and Bcl-2. Mol. Cell. 2007;25(2):193–205. doi: 10.1016/j.molcel.2006.12.009. [DOI] [PubMed] [Google Scholar]
- Ishikawa T, Watanabe N, Nagano M, Kawai-Yamada M, Lam E. Bax inhibitor-1: a highly conserved endoplasmic reticulum-resident cell death suppressor. Cell Death Differ. 2011;18(8):1271–1278. doi: 10.1038/cdd.2011.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BJ, Ryu SW, Song BJ. JNK- and p38 kinase-mediated phosphorylation of Bax leads to its activation and mitochondrial translocation and to apoptosis of human hepatoma HepG2 cells. J. Biol. Chem. 2006;281(30):21256–21265. doi: 10.1074/jbc.M510644200. [DOI] [PubMed] [Google Scholar]
- Lake AD, Novak P, Hardwick RN, Flores-Keown B, Zhao F, Klimecki WT, Cherrington NJ. The adaptive endoplasmic reticulum stress response to lipotoxicity in progressive human nonalcoholic fatty liver disease. Toxicol. Sci. 2014;137(1):26–35. doi: 10.1093/toxsci/kft230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Tirasophon W, Shen X, Michalak M, Prywes R, Okada T, Yoshida H, Mori K, Kaufman RJ. IRE1- mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 2002;16:452–466. doi: 10.1101/gad.964702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Gong M, French BA, Li J, Tillman B, French SW. Mallory-Denk Body (MDB) formation modulates Ufmylation expression epigenetically in alcoholic hepatitis (AH) and non-alcoholic steatohepatitis (NASH) Exp. Mol. Pathol. 2014;97(3):477–483. doi: 10.1016/j.yexmp.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma K, Vattem KM, Wek RC. Dimerization and release of molecular chaperone inhibition facilitate activation of eukaryotic initiation factor-2 kinase in response to endoplasmic reticulum stress. J. Biol. Chem. 2002;277(21):18728–18735. doi: 10.1074/jbc.M200903200. [DOI] [PubMed] [Google Scholar]
- Masouminia M, Samadzadeh S, Mendoza AS, French BA, Tillman B, French SW. Upregulation of Autophagy Components in Alcoholic Hepatitis and Nonalcoholic Steatohepatitis. Exp. Mol. Pthol. 2016 doi: 10.1016/j.yexmp.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyadomari S, Mori M. Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ. 2004;11(4):381–389. doi: 10.1038/sj.cdd.4401373. [DOI] [PubMed] [Google Scholar]
- Pincus D, Chevalier MW, Aragón T, Van Anken E, Vidal SE, El-Samad H, Walter P. BiP binding to the ER-stress sensor Ire1 tunes the homeostatic behavior of the unfolded protein response. PLoS. Biol. 2010 doi: 10.1371/journal.pbio.1000415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthalakath H, O'Reilly LA, Gunn P, Lee L, Kelly PN, Huntington ND, Hughes PD, Michalak EM, McKimm-Breschkin J, Motoyama N, Gotoh T, Akira S, Bouillet P, Strasser A. ER stress triggers apoptosis by activating BH3-only protein Bim. Cell. 2007;129(7):1337–1349. doi: 10.1016/j.cell.2007.04.027. [DOI] [PubMed] [Google Scholar]
- Ron D, Hubbard SR. How IRE1 reacts to ER stress. Cell. 2008;132(1):24–26. doi: 10.1016/j.cell.2007.12.017. [DOI] [PubMed] [Google Scholar]
- Rutkowski DT, Kaufman RJ. A trip to the ER: coping with stress. Trends Cell Biol. 2004;14(1):20–28. doi: 10.1016/j.tcb.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Sano R, Reed JC. ER stress-induced cell death mechanisms. Biochim. Biophys. Acta. 2013;1833(12):3460–3470. doi: 10.1016/j.bbamcr.2013.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Ellis RE, Lee K, Liu CY, Yang K, Solomon A, Yoshida H, Morimoto R, Kurnit DM, Mori K, Kaufman RJ. Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell. 2001;107:893–903. doi: 10.1016/s0092-8674(01)00612-2. [DOI] [PubMed] [Google Scholar]
- Shi Y, Vattem KM, Sood R, An J, Liang J, Stramm L, Wek RC. Identification and characterization of pancreatic eukaryotic initiation factor 2 alpha-subunit kinase, PEK, involved in translational control. Mo.l Cell Biol. 1998;18:7499–7509. doi: 10.1128/mcb.18.12.7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapp E. Endoplasmic reticulum biogenesis proliferation and differentiation. Biogenesis of Cellular Organelles Mullins C. 2005:63–95. [Google Scholar]
- Stevens FJ, Argon Y. Protein folding in the ER. Semin. Cell Dev. Biol. 1999;10:443–454. doi: 10.1006/scdb.1999.0315. [DOI] [PubMed] [Google Scholar]
- Tirasophon W, Welihinda AA, Kaufman RJ. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 1998;12:1812–1824. doi: 10.1101/gad.12.12.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Kaufman RJ. From acute ER stress to physiological roles of the Unfolded Protein Response. Cell Death Differ. 2006;13(3):374–384. doi: 10.1038/sj.cdd.4401840. [DOI] [PubMed] [Google Scholar]
- Xu C, Xu W, Palmer AE, Reed JC. BI-1 regulates endoplasmic reticulum Ca2+ homeostasis downstream of Bcl-2 family proteins. J. Biol. Chem. 2008;283(17):11477–11484. doi: 10.1074/jbc.M708385200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Rawson RB, Komuro R, Chen X, Dave UP, Prywes R, Brown MS, Goldstein JL. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell. 2000;6:1355–1364. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Okada T, Haze K, Yanagi H, Yura T, Negishi M, Mori K. ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol. Cell Biol. 2000;20:6755–6767. doi: 10.1128/mcb.20.18.6755-6767.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]