Abstract
Members of the RING E3 ubiquitin ligase family bind to both substrate and ubiquitin-charged E2 enzyme, promoting transfer of ubiquitin from the E2 to substrate. Either a single ubiquitin or one of several types of polyubiquitin chains can be conjugated to substrate proteins, with different types of ubiquitin modifications signaling distinct outcomes. E2 enzymes play a central role in governing the nature of the ubiquitin modification, although the essential features of the E2 that differentiate mono- versus polyubiquitinating E2 enzymes remain unclear. RNF4 is a compact RING E3 ligase that directs ubiquitination of polySUMO chains in concert with several different E2 enzymes. RNF4 monoubiquitinates polySUMO substrates in concert with RAD6B and polyubiquitinates substrates together with UBCH5B, a promiscuous E2 that can function with a broad range of E3 ligases. We find that the divergent ubiquitination activities of RAD6B and UBCH5B are governed by differences at the RING-binding surface of the E2. By mutating the RAD6B RING-binding surface to resemble that of UBCH5B, RAD6B can be transformed into a highly active UBCH5B-like E2 that polyubiquitinates SUMO chains in concert with RNF4. The switch in RAD6B activity correlates with increased affinity of the E2 for RNF4. These results point to an important role of the affinity between an E3 and its cognate E2 in governing the multiplicity of substrate ubiquitination.
Keywords: ubiquitin, E3 ubiquitin ligase, E2 ubiquitin-conjugating enzyme, RING finger protein 4 (RNF4), RAD6B, UBCH5B, UBE2B, UBE2D2
Graphical Abstract
Introduction
The covalent attachment of ubiquitin (Ub) to substrates plays a central role in regulating a broad range of biological processes in eukaryotes [1, 2]. Substrates can be modified with a single ubiquitin (monoubiquitination) or with varying types of polyubiquitin chains (polyubiquitination) that are distinguished by the particular lysine through which one ubiquitin is joined to the next [3, 4]. Ubiquitin can also be conjugated to protein N-termini to form an N-terminal ubiquitin fusion [5, 6] or linear polyubiquitin chain [7]. The functional consequences of ubiquitination are determined by the distinct nature and topology of the ubiquitin modification [3, 8–10]. Ubiquitin is conjugated to substrates via the concerted action of E1, E2 and E3 enzymes [11]. An E1 ubiquitin-activating enzyme is charged with ubiquitin and then transfers its ubiquitin to the active site cysteine of an E2 ubiquitin-conjugating enzyme to yield a charged E2~Ub thioester. The E3 ligase binds to both substrate and the E2~Ub thioester and catalyzes the transfer of ubiquitin to the target substrate, resulting in an isopeptide bond between the C-terminus of ubiquitin and the epsilon-amino group of the substrate lysine [3, 9] or, in select cases, a peptide bond with the N-terminal alpha-amino group of the substrate [5, 6, 12]. In the case of RING/U-box E3 ligases, which comprise the largest class of E3s [13], the E3 binds to both E2~Ub and substrate and catalyzes attack of the substrate lysine on the E2~Ub thioester to yield the ubiquitinated substrate [14]. RING domains bind to E2 enzymes in a conserved manner, with the RING domain contacting a surface that includes the E2 N-terminal helix 1 (H1) as well as loops 4 and 7 [15]. In addition, the RING domain also contacts the donor ubiquitin, stabilizing a conformation of the E2~Ub conjugate [16–18] that promotes a nucleophilic attack by the substrate lysine on the thioester linkage, yielding an isopeptide bond between the lysine and the ubiquitin C-terminus [19]. Most RING E3 ligases can ubiquitinate substrates in conjunction with multiple E2 enzymes. Depending upon the identity of the E2, these modifications can take on a variety of forms. In many cases, the E2 itself appears to govern the nature of the ubiquitin modification, both in terms of the multiplicity of the modifications and, in the case of polyubiquitin chains, linkage type [3, 9, 14]. While much of the core enzymology and structures of the ubiquitin conjugation machinery are widely conserved, it is clear that individual E2-E3 pairs have evolved an array of mechanisms to generate distinct ubiquitin modifications.
One of the most fundamental distinctions between ubiquitin signals is substrate monoubiquitination versus polyubiquitination. With the exception of the E2, UBE2W, which represents a special case because it only ubiquitinates the flexible N-termini of substrates [5, 6, 20], most examples of monoubiquitination studied to date involve a role for the E3 ligase in either suppressing intrinsic polyubiquitination activity of the E2 [21] or in directing the E2 to a preferred target lysine in the substrate [22, 23]. The E3 ligase, APC/C is capable of driving both the direct multi-monoubiquitination of substrate in conjunction with UBE2C, and the extension of polyubiquitin chains on substrate with UBE2S, by interacting with each E2 using two distinct binding architectures [24]. UBCH5C contains a non-covalent ubiquitin-binding site on its “backside”, distal from the active site, that mediates polyubiquitination as assayed in E3 autoubiquitination assays [25]. However, the Bmi1-RING1 E3 complex targets UBCH5C to monoubiquitinate histone H2A-Lys119 [26–28] by juxtaposing the UBCH5C~Ub thioester and the target lysine [22, 23]. The E2, RAD6B, can synthesize polyubiquitin chains on its own in vitro [21, 29], but monoubiquitinates PCNA together with the E3, RAD18 [30, 31]. RAD18 suppresses intrinsic RAD6B polyubiquitination activity through a special domain called the R6BD, which binds to the RAD6B backside and suppresses its polyubiquitination activity [21]. Yeast Rad6 also has intrinsic polyubiquitinating activity [32] that is mediated, in part through backside interactions [33]. Rad6 monoubiquitinates histone H2B-Lys123 in conjunction with the E3 ligase, Bre1 [32]. Although Bre1, like human RAD18, contains a special domain that contacts the Rad6 backside, the Bre1 RING domain alone is sufficient to direct monoubiquitination of nucleosomal H2B [34], potentially through substrate targeting as recently observed in the structure of Bmi1-RING1/UBCH5C bound to a nucleosome [23]. While backside interactions and direct targeting of the E2 to its substrate are clearly important in governing mono- versus polyubiquitination, there is evidence that the molecular interface between the E2 and the RING domain may also govern substrate ubiquitination [35]. Indeed, a screen for mutants of the U-box E3, UBE4B, that enhanced auto-polyubiquitination of UBE4B in conjunction with the E2, UBCH5C, yielded a subclass of activating point mutations in UBE4B that increased its affinity for the E2 [36]. The activating mutations in UBE4B, while increasing the rate at which polyubiquitin accumulated, did not impact the multiplicity of autoubiquitination. Interestingly, whether the E2 enzyme, UBCM2 (UBE2E3), mono- or polyubiquitinates a RING E3 partner in autoubiquitination assays correlates with the ability of GST-tagged UBCM2 to interact with the E3 in a pull-down assay: GST-UBCM2 pulls down AO7T, which is polyubiquitinated in conjunction with UBCM2, but not BD/BC, which is monoubiquitinated in conjunction with UBCM2 [37]. Since the E3 is also a substrate in these reactions, it is not possible to separate the potential contributions of substrate versus E3 affinity to the observed differences in ubiquitination.
While a number of studies have helped elucidate the principles of target amino group specificity, UBL selectivity, and ubiquitin linkage specificity [3, 4, 9, 10, 14, 19, 20, 23, 24, 33, 34, 37–41], the role of the canonical E2-RING interface in governing substrate ubiquitination is less well-understood. We therefore investigated how differences in E2-E3 RING interactions affect substrate ubiquitination. In order to separate the role of interactions between the RING domain and the E2 versus the interactions between the charged E2 and the substrate, we utilized a system in which we could monitor ubiquitination of a substrate other than the E3 ligase itself. RNF4 is a compact 190-residue E3 ligase that belongs to the SUMO-targeted ubiquitin ligase (STUbL) subfamily and directs ubiquitination of polySUMO chains [42, 43]. RNF4 contains a C-terminal RING domain that binds the E2 and N-terminal SUMO-interacting motifs (SIMs) that bind to the polySUMO substrate [44–46]. RNF4 monoubiquitinates polySUMO substrates in concert with RAD6B and robustly polyubiquitinates substrate together with UBCH5B, a promiscuous E2 that can function with a broad range of E3 ligases [46–49]. We find that the ubiquitinating activities of RAD6B and UBCH5B in concert with RNF4 are governed by interactions between the E2 and the RNF4 RING domain. By reengineering the RAD6B RING-binding surface to resemble that of UBCH5B, we transformed RAD6B into a UBCH5B-like E2 that polyubiquitinates polySUMO in the presence of RNF4. The switch from weak monoubiquitinating activity to robust polyubiquitinating activity correlates with increased affinity of the E2 for RNF4. Our results shed new light on the characteristics of E2-RING interactions that govern the activity and nature of substrate ubiquitination.
Results
Mono- versus polyubiquitination of polySUMO by RNF4 is E2-dependent
To ask how differences in E2-E3 interactions affect RNF4 substrate ubiquitination, we monitored RNF4 activity in the presence of several different E2 enzymes using linear tetraSUMO2 (4xSUMO2) as a model substrate [5, 46, 50]. Consistent with a previous study [5], UBCH5A (UBE2D1), UBCH5B (UBE2D2) and UBCH5C (UBE2D3) efficiently polyubiquitinated 4xSUMO2 (Fig. 1a, Supplementary Fig. 1, and Supplemental Fig. 2a). The presence of polyubiquitin chains is evident even at early times points, when the high molecular weight smear characteristic of polyubiquitination begins to appear (compare five min. time point for UBCH5B with 2.5 hr. time point for RAD6B in Fig. 1a). By contrast, we found that RAD6A (UBE2A) and RAD6B (UBE2B) monoubiquitinated the substrate, with no evidence of high-molecular weight species, and had lower overall activity (Fig. 1b, Supplementary Fig. 1, and Supplementary Fig. 2b). The monoubiquitinating activity of RAD6A and RAD6B is consistent with their activity with other E3 ligases such as RAD18 and hBRE1 [21, 31, 34, 51, 52].
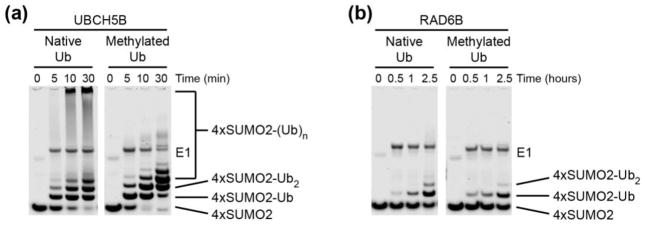
Figure 1. RNF4-catalyzed ubiquitination of a polySUMO substrate by UBCH5B and RAD6B enzymes.
(a) Time course comparing the in vitro ubiquitination of 4xSUMO2, using both native and methylated Ub, by RNF4 in the presence of UBCH5B. Reactions contained 200 nM E1, 5 μM E2, 1 μM RNF4, 15 μM 4xSUMO-2 and 100 μM of native or methylated Ub, and were incubated at 37°C. Samples were analyzed by SDS-PAGE, followed by Coomassie Brilliant Blue stain. (b) Time course comparing the in vitro ubiquitination of 4xSUMO2, using both native and methylated Ub, by RNF4 in the presence of RAD6B. Reactions contained 200 nM E1, 5 μM E2, 1 μM RNF4, 15 μM 4xSUMO-2 and 100 μM of native or methylated Ub, and were incubated at 37°C. Samples were analyzed by SDS-PAGE, followed by Coomassie Brilliant Blue stain.
To better compare the reaction products and distinguish between multi-monoubiquitination and polyubiquitination activity, we characterized the behaviors of RAD6B and UBCH5B in ubiquitination assays using methylated ubiquitin (UbMe). Methylated ubiquitin can be conjugated to free amines on a substrate but cannot form polyubiquitin chains because its surface amines are blocked. As shown in Fig. 1b, RAD6B exhibits a virtually identical pattern of substrate ubiquitination whether or not ubiquitin is methylated, as expected from its monoubiquitination activity. By contrast, the higher molecular weight smears characteristic of polyubiquitin chain synthesis by UBCH5B disappear when ubiquitin is methylated, replaced by multi-monoubiquitinated substrate (Fig. 1a and Supplementary Fig. 2b), as has been previously observed [46, 47]. The differing ubiquitinated product patterns generated in the presence of native versus methylated ubiquitin indicate that UBCH5B polyubiquitinates 4xSUMO2. Even after 2.5 hours of incubation, RAD6B yields only monoubiquitinated 4xSUMO2, while UBCH5B robustly polyubiquitinates the substrate within 30 minutes (Fig. 1a–b, Supplementary Fig. 2a–b).
Interactions at the E2-RING interface govern mono- versus polyubiquitination
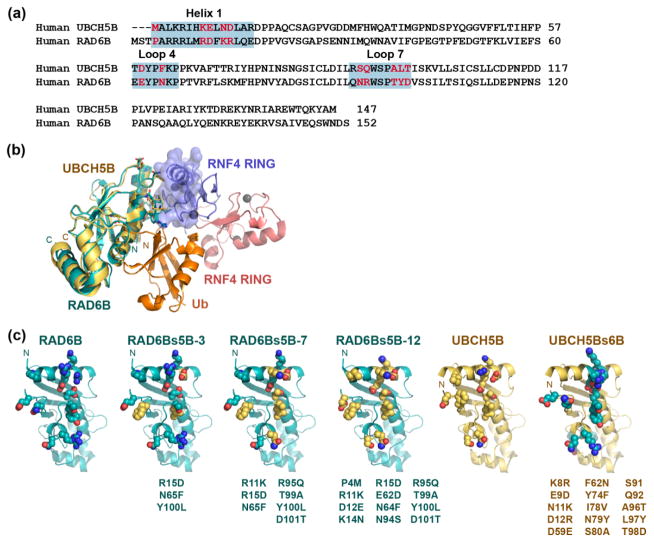
RAD6B and UBCH5B were previously shown to exhibit similar E1-E2 transthiolation kinetics [53–55], which suggests that differences in 4xSUMO2 ubiquitination behavior do not result from the E1-E2 transthiolation step, but rather from later RNF4-mediated substrate ubiquitination steps. While RAD6B and UBCH5B are very similar in fold and overall structure, a comparison of the canonical RING-interacting surfaces of RAD6B and UBCH5B shows that many of the residues that lie at the RING-E2 interface are not well conserved between the two E2s (Fig. 2a). Based on these observations, we considered the possibility that differences in RING-interacting residues might govern mono- versus polyubiquitination by RAD6B and UBCH5B, respectively. To test whether differences in the RING-contacting residues in the E2 could account for the different properties of RAD6B and UBCH5B, we generated a set of RAD6B mutants with amino acid substitutions designed to convert the canonical RING-binding surface of RAD6B into one resembling that of UBCH5B. Twelve side chains were identified at the canonical RING-binding surface of RAD6B that differ from those found in UBCH5B (Fig. 2a–b). Three RAD6B-surface conversion mutants of UBCH5B (RAD6Bs5B) were generated containing three (RAD6Bs5B-3), seven (RAD6Bs5B-7) or twelve (RAD6Bs5B-12) side chain substitutions, respectively (Fig. 2c). As shown in Fig. 3a, RAD6Bs5B conversion mutants yield products corresponding to polyubiquitinated 4xSUMO2. These patterns indicate that the conversion substitutions not only increase the overall activity of the mutated RAD6B, but that they also conferred the ability to polyubiquitinate the 4xSUMO2 substrate. The conversion mutant with the largest number of UBCH5B-like residues, RAD6Bs5B-12, robustly polyubiquitinated the 4xSUMO2 substrate in a manner very similar to that of UBCH5B, while the intermediate variants (RAD6Bs5B-3 and RAD6Bs5B-7) displayed a gradient of UBCH5B-like ubiquitination of the polySUMO substrate (Fig. 3a, Supplementary Fig. 3a, and Supplementary Fig. 4).
Figure 2. Mutating the RAD6B RING-binding surface to resemble UBCH5B.
(a) Sequence alignment of human UBCH5B and RAD6B. RING-binding surface regions are highlighted in blue. RAD6B residues targeted for mutagenesis shown in red. (b) Docking of the structure of RAD6B (cyan, PDB ID: 2YB6) and UBCH5B (yellow, PDB ID: 2ESK) on the crystal structure of UBCH5A~Ub in complex with an RNF4 RING dimer (PDB ID: 4AP4). The donor ubiquitin is shown in orange and the RNF4 RING dimer is shown in blue and red. (c) Residues at the RING-binding surfaces of RAD6B, the RAD6Bs5B variants, UBCH5Bs6B, and UBCH5B. RAD6B is shown in cyan, UBCH5B is shown in yellow, substituted residues in the RAD6Bs5B conversion mutants are shown as yellow spheres, and substituted residues in the UBCH5Bs6B conversion mutant are shown as cyan spheres.
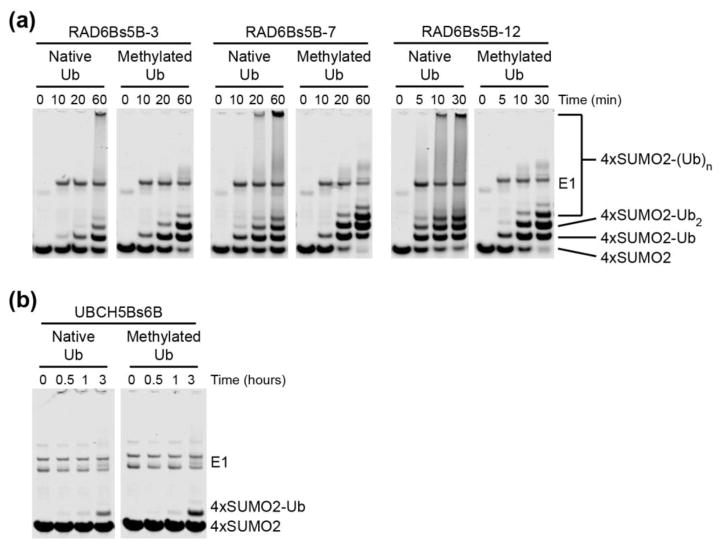
Figure 3. RNF4-catalyzed ubiquitination of polySUMO by RAD6B and UBCH5B conversion mutants.
(a) Time course comparing the in vitro ubiquitination of 4xSUMO2, using both native and methylated Ub, by RNF4 in the presence of the RAD6Bs5B variants. Reactions contained 200 nM E1, 5 μM E2, 1 μM RNF4, 15 μM 4xSUMO-2 and 100 μM of native or methylated Ub, and were incubated at 37°C. Samples were analyzed by SDS-PAGE, followed by Coomassie Brilliant Blue stain. (b) Time course comparing the in vitro ubiquitination of 4xSUMO2, using both native and methylated Ub, by RNF4 in the presence of UBCH5Bs6B. Reactions contained 200 nM E1, 5 μM E2, 1 μM RNF4, 15 μM 4xSUMO-2 and 100 μM of native or methylated Ub, and were incubated at 37°C. Samples were analyzed by SDS-PAGE, followed by Coomassie Brilliant Blue stain.
To verify that the RING-binding surfaces of RAD6B and UBCH5B can indeed account for their differing polySUMO ubiquitination behaviors, we performed the reciprocal experiment in which we the mutated canonical RING-binding surface of UBCH5B into one resembling that of RAD6B. Eleven side chains were identified at the canonical RING-binding surface of UBCH5B that differ from those found in RAD6B, and substituted to generate the UBCH5B-surface conversion mutant, UBCH5Bs6B (Fig. 2c). In conjunction with RNF4, UBCH5Bs6B monoubiquitinated 4xSUMO2 and had lower overall activity than wild type UBCH5B (Fig. 3b, Supplementary Fig. 3b, and Supplementary Fig. 4). This result is consistent with a model in which the interactions between the E2 and RING domain play an important role in governing the overall ubiquitination activity in an E2-dependent manner.
To better compare the substrate ubiquitination activity of UBCH5Bs6B, and the RAD6Bs5B variants to that of RAD6B and UBCH5B, we again assayed ubiquitination using methylated ubiquitin, which makes it possible to distinguish between multi-monoubiquitinating and polyubiquitinating activity. We then asked whether the RAD6Bs5B conversion mutants, whose E3-binding surfaces were mutated to resemble that of UBCH5B, polyubiquitinated substrate as well. As shown in Fig. 3a, RAD6Bs5B-12, containing the most UBCH5B-like RING-binding surface, polyubiquitinated the substrate in the presence of unmodified ubiquitin but produced the characteristic ladder of multi-monoubiquitinated 4xSUMO2 substrate in the presence of reductively methylated Ub. The product patterns generated by RAD6Bs5B-12 with native Ub versus UbMe (Fig. 3a and Supplementary Fig. 3a) are similar to those produced by UBCH5B (Fig. 1a and Supplementary Fig. 2a). In contrast, UBCH5Bs6B, containing a RAD6B-like RING-binding surface, produced monoubiquitinated 4xSUMO2 in the presence of both native and methylated Ub (Fig. 3b and Supplementary Fig. 3b), similar to the monoubiquitination activity observed with RAD6B (Fig. 1b and Supplementary Fig. 2b). Taken together, RAD6Bs5B-3, RAD6Bs5B-7, and RAD6Bs5B-12 exhibited a gradient of monoubiquitination, multi-monoubiquitination, and UBCH5B-like polyubiquitination of the 4xSUMO2 substrate, producing polyubiquitinated substrate smears in the presence of native Ub, while yielding ladders of multi-monoubiquitinated 4xSUMO2 substrate in the presence of methylated Ub (Fig. 3a and Supplementary Fig. 3a). Although RAD6Bs5B-12 clearly exhibited polySUMO ubiquitination behavior similar to its counterpart, UBCH5B, comparison of substrate ubiquitination over a time course showed that substrate ubiquitination by RAD6Bs5B-12 (Fig. 3a and Supplementary Fig. 3a) was still slower than that of UBCH5B (Fig. 1a and Supplementary Fig. 2a). Similarly, UBCH5Bs6B and RAD6B resemble each other in substrate monoubiquitination behavior, but when compared in a time course UBCH5Bs6B (Fig. 3b and Supplementary Fig. 3b) monoubiquitinated polySUMO at slower rate than that of its counterpart, RAD6B (Fig. 1b and Supplementary Fig. 2b).
Since the E2 surface that contacts the E3 RING domain overlaps with the surface of the E2 that binds to the E1 ubiquitin activating enzyme [56], we performed assays to check whether mutations in either E2 impact E1-E2 transthiolation activity. In assays of E1-catalyzed E2~Ub thioester formation, we found that RAD6B, UBCH5B, UBCH5Bs6B, and the RAD6Bs5B conversion variants were indeed capable of accepting activated Ub from E1 (Supplementary Fig. 5).
E2-RING affinity governs substrate mono- versus polyubiquitination and sensitivity to RNF4
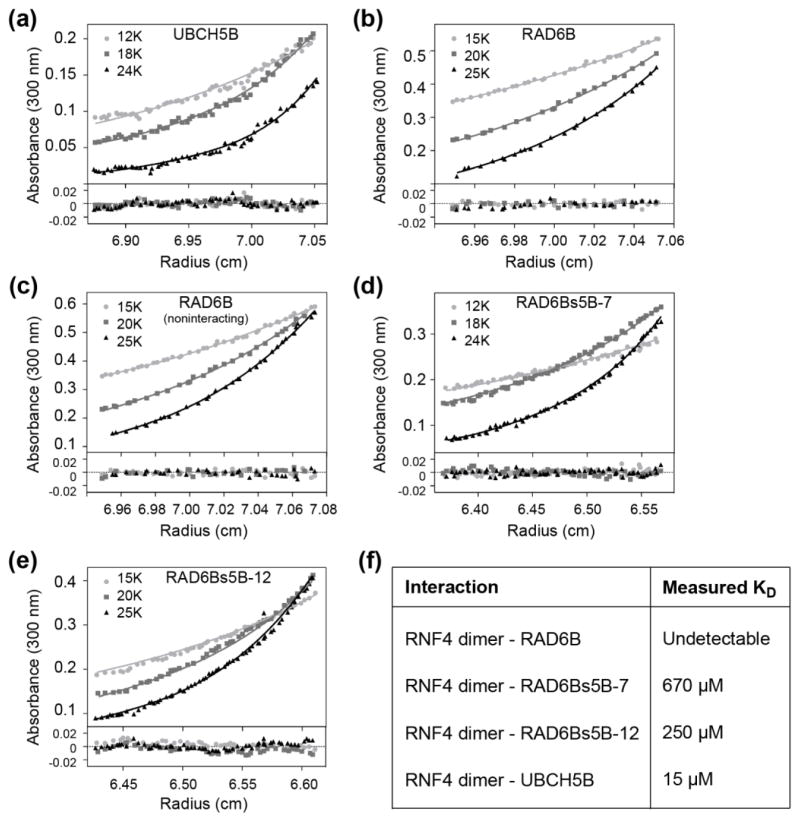
The ability to convert RAD6B into a UBCH5B-like E2 simply by mutating residues at the E2 RING-binding interface suggested that features of the E2-E3 interaction are likely to play an important role in determining ubiquitination behavior. One potential effect of the mutations at the RING-E2 interface could be to alter the affinity of the RNF4 RING for RAD6B. We therefore determined the effect of the mutations on E2 affinity for RNF4 using sedimentation equilibrium (SE) analytical ultracentrifugation. We first confirmed that RNF4 was a dimer, as has been previously reported [57], under all assay conditions (Supplementary Figs. 6–7, and Supplementary Table 1). Sedimentation equilibrium experiments were then used to determine the dissociation constants for binding of RNF4 to UBCH5B, RAD6B, RAD6Bs5B-7, and RAD6Bs5B-12. Wild-type UBCH5B binds to the RNF4 dimer with an equilibrium dissociation constant (Kd) of 15 μM, while RNF4 binding to RAD6B was undetectable (Figs. 4a–c, Supplementary Figs. 8–10, and Supplementary Table 1). RAD6Bs5B-7, and RAD6Bs5B-12 bound to RNF4 dimer with a Kd of 670 μM and 250 μM, respectively (Figs. 4d–e, Supplementary Figs. 11–12, and Supplementary Table 1). Overall, these affinities indicate a trend in which increasing UBCH5B-like character at the RING-binding surface correlates with increased affinity for RNF4 (Fig. 4f), and a corresponding increase in UBCH5B-like polyubiquitination activity (Fig. 3a).
Figure 4. Affinity of RNF4 for E2 enzymes.
Sedimentation equilibrium analytical ultracentrifugation (AUC) was used to monitor E2 association with the RNF4 homodimer. Absorbance (300 nm) profiles at three different rotor speeds are shown. Data were fit to heteroassociation model of the interaction between E2 and the RNF4 homodimer, except where noted as otherwise. (a) UBCH5B (50 μM) with RNF4 (75 μM), (b) RAD6B (50 μM) with RNF4 (75 μM), (c) RAD6B (50 μM) with RNF4 (75 μM) fit to a noninteracting species model for two species, RAD6B and the RNF4 homodimer, (d) RAD6Bs5B-7 (50 μM) with RNF4 (50 μM), (e) and RAD6Bs5B-12 (50 μM) with RNF4 (50 μM). (f) Table summarizing E2-RNF4 affinities measured by sedimentation equilibrium AUC.
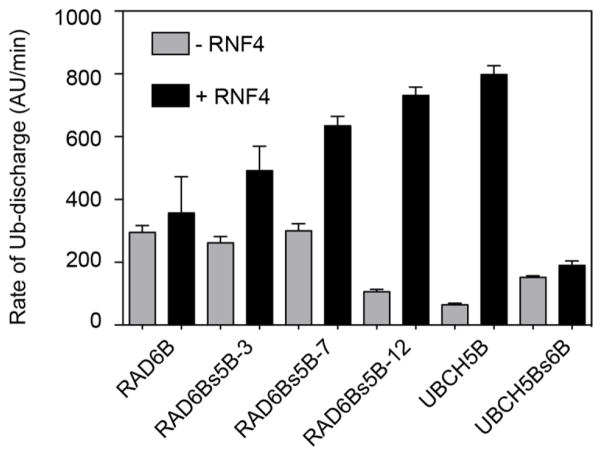
We next used the E2 Ub-discharge assay to assess the stability of the E2~Ub thioester complex, and the ability of RNF4 to stimulate ubiquitin discharge from the RAD6Bs5B and UBCH5Bs6B conversion mutants as compared to wild type RAD6B and UBCH5B. To assay E2 ubiquitin discharge, each E2 was incubated for a fixed time with E1, fluorescein-labeled ubiquitin (UbFL), and ATP to form E2~UbFL, and the E1-E2 charging reaction was then quenched by hydrolyzing the remaining ATP with the enzyme, apyrase. Ubiquitin discharge from E2~UbFL was initiated by adding an excess of lysine, and then monitored by non-reducing SDS-PAGE. The rate of ubiquitin discharge to lysine was monitored in the presence and absence of RNF4. In the absence of RNF4, the rates of ubiquitin discharge are similar for each of the E2s, while RNF4 stimulates ubiquitin discharge. The fold-stimulation of ubiquitin discharge from the E2 enzymes shows a correlation between increasing discharge rate and increased rate of substrate polyubiquitination (Fig. 5 and Supplementary Fig. 13). RNF4 has little effect on discharge of ubiquitin from RAD6B~UbFL thioester, whereas RNF4 substantially stimulates discharge from UBCH5B~UbFL. RAD6Bs5B conversion mutants (−3, −7, and 12) display a trend whereby increasing UBCH5B-like character at the RING-binding surface correlates with greater sensitivity to stimulation of ubiquitin discharge by RNF4. Similarly, addition of RNF4 has a negligible effect on discharge of ubiquitin from UBCH5s6B~Ub (Fig. 5 and Supplementary Fig. 13a), consistent with the RAD6B-like ubiquitinating behavior of this UBCH5B mutant.
Figure 5. Reactivity of wild type and mutant E2~Ub thioester in the presence and absence of RNF4.
The rates of ubiquitin discharge from the E2~Ub to lysine in the presence (black columns) and absence (grey columns) of RNF4, with native RAD6B and UBCH5B, UBCH5Bs6B, as well as with the RAD6Bs5B conversion constructs. E2 was pre-charged with fluorescein-tagged ubiquitin, UbFL, and then added to the E2~UbFL discharge reaction. E2~UbFL discharge reactions contained 50 mM lysine, 10 μM RNF4, and 5 μM E2~UbFL. Ub-discharge was monitored by SDS-PAGE, followed by fluorescent imaging, and fluorescence signal densitometry.
Discussion
Many studies have shown that intrinsic properties of the E2 enzyme govern the nature of the ubiquitin modification, including both the multiplicity of the ubiquitin modification as well as the specific linkage type in the case of polyubiquitin chains [3, 4, 9, 14]. Whereas previous studies of both UBCH5 and RAD6 isoforms have pointed primarily to interactions with the so-called E2 backside in governing mono- versus polyubiquitination [21, 25], we find that the nature of the RING binding to the E2 plays a critical role in determining the overall activity and multiplicity of substrate ubiquitination. By mutating the RING-binding surface of RAD6B and UBCH5B (Figs. 2), a monoubiquitinating E2 and polyubiquitinating E2, respectively, to resemble that of the other, we were able to convert RAD6B into a robust polyubiquitinating enzyme and UBCH5B into a weaker, monoubiquitinating enzyme (Fig. 3). Moreover, the polyubiquitinating activity correlated with the affinity of the E2 for the RNF4 RING domain, with increasing polyubiquitinating activity observed with increasing E2-E3 RING affinity. Our results are also consistent with the finding that tighter binding of the RING domain correlates with higher overall activity, as had been previously observed for gain-of-function mutations in the U-box E3, UBE4B [36]. Since the present study monitored substrate ubiquitination rather than autoubiquitination, we were able to separate the contribution of tighter interactions with the E2 – and higher reactivity of the E2~Ub thioester (Fig. 5) – from tighter binding to the substrate.
While our findings are, to our knowledge, the first to show a direct relationship between the affinity of the E2 for the RING domain and substrate mono- versus polyubiquitination, our results are consistent with observations made in a number of previous studies. As mentioned above, a possible correlation between the strength of E3-E2 interactions and mono- versus polyubiquitination was suggested by the observation that the multiplicity of E3 autoubiquitination in conjunction with the E2, UBCM2 (UBE2E3), correlated with whether the E3 binding could be detected in pull-down assays with GST-tagged UBCM2 [37]. While the relative contribution of E3-E2 versus E3-substrate binding affinity could not be uncoupled because the E3 ligases in that study were both the substrate and the enzyme, our results point to a role for E3 affinity for the E2 enzyme as a determinant of mono- versus polyubiquitinating activity by UBCM2. In an earlier study, Haas and colleagues [58] described a similar phenomenon in an investigation of E3-catalized substrate ubiquitination by the E2s, CDC34 and RAD6. While the identity of the E3 was unclear because they used a crude reticulocyte fraction containing E3 activity, they found that RAD6 exhibited a significantly higher E2-E3 Km than did CDC34, an E2 known to polyubiquitinate substrates [59], suggesting that differences in E3 affinity might play a role in determining the differing substrate ubiquitination behavior of RAD6 and CDC34 [58]. A role for RING interactions in directing substrate monoubiquitination has also been suggested for yeast Rad6 [34], which monoubiquitinates nucleosomal histone H2B in conjunction with the RING E3 ligase, Bre1 [32]. The Bre1 RING also has very low affinity for Rad6 [32, 34]. A recent study found that a minimal domain comprising the Bre1 RING and a coiled-coil dimerization domain were sufficient to direct monoubiquitination of histone H2B [34]. Interestingly, directly fusing the coiled-coil-RING fragment of Bre1 to Rad6 increased the rate of histone ubiquitination without significantly altering the overall pattern; the primary product was still monoubiquitinated H2B, with only slight increase of a secondary H2B ubiquitination site [34].
How does an increase in E2 affinity for the RING domain favor higher activity and polyubiquitination by the E2? Binding of the RING domain to the E2 positions the donor ubiquitin in the E2~Ub conjugate in a closed complex [16–18] that increases the reactivity of the thioester [60]. A higher affinity of the RING for the E2 would promote the closed E2~Ub complex and thus make the thioester more reactive. Indeed, we observed a correlation between affinity of RNF4 for the E2 and reactivity of the E2~Ub thioester to lysine discharge (Fig. 5). Formation of polyubiquitin chains, in which ubiquitin itself is the substrate, may depend upon a more reactive E2~Ub conjugate as compared to monoubiquitination of other substrates. We cannot rule out the possibility that the mutant RAD6B enzymes, as well as UBCH5B, are allosterically activated by the RNF4 RING in a different manner than wild type RAD6B. Either a larger set of favorable interactions or a longer E2-E3 complex lifetime could contribute to different allosteric activation of the E2, although how this would translate into different multiplicity or activity is unclear.
A role for the E2-E3 interface in governing mono- versus polyubiquitination is not mutually exclusive with that of the E2 backside, which has previously been shown to mediate polyubiquitination through its ability to bind to ubiquitin [21, 25, 33]. Brzovic and colleagues first demonstrated the importance of backside Ub-binding by UBCH5-family E2s to processive auto-polyubiquitination of BRCA1 [25]. A recent study of UBCH5B-RNF38 suggested that backside ubiquitin binding promotes polyubiquitination by stabilizing the E3-E2~Ub complex in catalytically competent conformation [41]. Conversely, low affinity E3 binding, as observed here for the RNF4-RAD6B interaction and as has been reported for yeast Rad6 [32, 34], would be expected to have the converse effect and be less effective at stabilizing the E2~Ub conjugate.
It has long been clear that a simple model of ubiquitination, whereby the E2 simply accepts activated ubiquitin from the E1 and assists in the E3-directed ubiquitination of substrate, significantly underestimates the crucial role played by E2 enzymes writing the ubiquitin code. Numerous studies have demonstrated the important role of E2s in determining the topologically distinct ubiquitin modifications that form the basis of that code [3, 9, 14]. While E2 family members clearly enlist a variety of approaches to regulating ubiquitination, their general ability to harness multiple binding interactions to dictate ubiquitination specificity and topology has arisen as a common mechanistic theme within the family [3, 20, 21, 25, 35, 37]. Our finding that the affinity of the RNF4 for its cognate E2 plays an important role in determining the topology and multiplicity of substrate ubiquitination, demonstrates the importance of yet another E2 binding interaction in governing ubiquitination. Going forward, it will be important to ask whether the E2-RING interaction plays a role in governing substrate ubiquitination by other E2-E3 pairs. Additionally, further investigation into potential crosstalk between the E2-RING interface and other E2-binding interactions, such as backside ubiquitin binding, could provide a clearer understanding of how E2 enzymes govern the nature of ubiquitination.
Materials and Methods
Cloning, site-directed mutagenesis and expression
Human RNF4 was PCR amplified from a human cDNA library and subcloned into the pET32a vector, which directs expression of a fusion protein containing thioredoxin (TRX) followed by a hexahistidine tag and a Tobacco Etch Virus (TEV) protease cleavage site fused to the N-terminus of full-length RNF4. The TRX tag increased overall solubility during overexpression while the TEV site facilitated removal of the TRX-His tag after the first purification step. The genes encoding RAD6B and UBCH5B were PCR amplified from a human cDNA library and cloned into a modified pET32a vector, called pETSUMO2. In the pETSUMO2 vector the TRX tag, His tag, Thrombin cleavage site, S tag, and enterokinase site coding regions of the original pET32a vector were replaced with an N-terminal His-SUMO2 tag using the Infusion ligase-free cloning kit (Clonetech). Use of the N-terminal His tag allowed efficient purification of the fusion proteins, while the SUMO2 tag was used to enhance solubility and enable precise removal of the complete tag using the SENP2 protease [61]. RAD6B and UBCH5B mutants were generated by site-directed mutagenesis of the wild-type construct using the Quick Change mutagenesis kit (Agilent). The polymeric SUMO2 substrate construct was a fusion of four SUMO2 coding regions: full-length SUMO2 was used for the N-terminal SUMO2, while a truncated SUMO2 (corresponding to residues 11-92) was used for the second, third and fourth. The polymeric 4xSUMO2 substrate was expressed as a linear fusion protein, with an N-terminal His tag and C-terminal strep-tag to allow for easy purification.
All proteins were expressed in E. coli Rosetta2(DE3) cells using M9ZB media. Cultures (1 L) were inoculated using 1% (v/v) overnight saturated cultures in MDG media [62] and grown at 37°C to an O.D600 of ~1.5. Protein expression was induced by addition of 500 μM Isopropyl βD-1-thiogalactopyranoside (IPTG) and cells were grown overnight at 16°C. Cells were harvested by centrifugation at 4000 rpm and cell pellets were stored at −80°C until later purification.
Protein purification
Cells expressing RNF4 were lysed in buffer containing 40 mM Na phosphate pH 8.0, 500 mM NaCl, 25 mM imidazole, 25 μM ZnSO4, 10 mM β-mercaptoethanol (BME), and 0.1 mM phenylmethylsulfonyl fluoride (PMSF) using a Microfluidizer (Microfluidics). The lysate was centrifuged to remove cell debris and then subjected to immobilized metal affinity chromatography (IMAC) using a 5 mL HisTrap column (GE Life Sciences). The protein was eluted using a 20–400 mM linear imidazole gradient. Fractions containing purified protein were pooled and dialyzed overnight at 4°C against 20 mM Na phosphate pH 8.0, 300 mM NaCl, 5 mM BME. TEV protease was added to the protein pool to cleave off the TRX-His tag. Cleaved protein was then subjected to a second round of IMAC and the flow-through containing the cleaved protein was collected. Following the second round of IMAC, RNF4 was dialyzed into 50 mM MES pH 5.6, 100 mM NaCl, 10 mM BME, and purified by anion exchange chromatography on a 5 mL HiTrap SP HP column (GE Life Sciences) running in the same buffer. RNF4 was eluted from the column with a linear NaCl gradient of 100 mM to 1 M over 20 column volumes. Fractions containing RNF4 were pooled, concentrated, and then subjected to size exclusion chromatography using a Superdex 75 26/60 running in 25 mM HEPES pH 7.5, 150 mM NaCl, 1 mM dithiothreitol (DTT).
E2 enzymes were expressed as fusion proteins containing an N-terminal His-SUMO2 tag. Cells were lysed in 40 mM Na phosphate pH 8.0, 400 mM NaCl, 20 mM imidazole, 10 mM BME and clarified by centrifugation. Protein samples were purified by IMAC using a 5 mL HisTrap column as described for RNF4, and eluted using a 20–400 mM linear imidazole gradient. Peak fractions were pooled and dialyzed overnight against 20 mM Na phosphate pH 8.0, 150 mM NaCl and 5 mM BME. The His-SUMO2 tag was removed by digesting the fusion proteins with an N-terminal His-tagged catalytic domain construct of SENP2 protease. The cleaved proteins were passed through a 5 mL HisTrap column, which retained the cleaved His-SUMO2 tag and the SENP2 protease and allowed the purified E2 to flow through. All purified E2 enzymes were dialyzed against 20 mM Tris-HCl pH 8.0, 100 mM NaCl, 5% glycerol, 2.5 mM BME at 4°C, concentrated and stored at −80°C.
The 4xSUMO2 protein was purified on a 5 mL HisTrap column as described for the RNF4 constructs and eluted using a 20–400 mM linear imidazole gradient. Fractions containing the 4xSUMO2 were then pooled and further purified using a 5mL StrepTrap column (GE Life Sciences). The protein was eluted with 40 mM Tris-HCl pH 8.0, 300 mM NaCl buffer containing 10 mM d-desthiobiotin. Purified 4xSUMO2 was finally dialyzed against 20 mM Tris-HCl pH 8.0 containing 300 mM NaCl, concentrated with to about 30 mg/ml and stored at −80°C.
Human ubiquitin was expressed and purified as previously described [63]. Wild type human ubiquitin was methylated using a previously established reductive lysine methylation protocol [64]. Fluorescein-labeled ubiquitin (UbFL) was purchased from Life Sensors (Life Sensors, Product # SI270F).
In vitro 4xSUMO2 ubiquitination assay
Reactions contained 200 nM E1, 5 μM E2, 1 μM RNF4, 15 μM 4xSUMO2 and 100 μM Ub were incubated at 37°C, unless otherwise noted, for the indicated time. Reactions were carried out in a buffer containing 50 mM HEPES pH 7.5, 100 mM NaCl, 0.5 mM DTT, 0.005% Tween-20, 10 mM MgCl2, 5 mM ATP. Samples were analyzed on SDS-PAGE gels stained with either Coomassie Brilliant Blue or SYPRO Ruby stain, as indicated.
E2 ubiquitin charging assays
E2~Ub charging reactions contained 500 nM E1, 10 μM E2, and 100 μM Ub, and were incubated at 37°C for the indicated time. Reactions were carried out in a buffer containing 50 mM HEPES pH 7.5, 100 mM NaCl, 0.5 mM DTT, 0.005% Tween-20, 10 mM MgCl2, 5 mM ATP. E1 was pre-charged with Ub, and then E2 was added to initiate the charging reaction. E2~Ub formation was monitored by SDS-PAGE followed by staining with Coomassie Brilliant Blue.
E2 ubiquitin discharge assays
Discharge of Ub from E2~Ub was monitored by the method of Pickart and Rose [65]. E2 was first pre-charged with UbFL, and then added to the E2~UbFL discharge reaction. E2~UbFL charging reactions contained 500 nM E1, 25 μM E2, and 150 μM UbFL, and were incubated at 37°C for 15 minutes. Charging reactions were carried out in a buffer containing 50 mM HEPES pH 7.5, 100 mM NaCl, 0.5 mM DTT, 0.005% Tween-20, 10 mM MgCl2, 5 mM ATP. Apyrase was then added to the charging reaction to quench the reaction. Charging reactions were incubated with apyrase at 37°C for 15 minutes. To initiate discharge, assay the charging reaction was diluted by five into a discharge reaction containing 50 mM lysine, 10 μM RNF4, and ~5 μM E2~UbFL. E2~UbFL discharge was monitored by SDS-PAGE, followed by fluorescence imaging and fluorescence signal densitometry.
Sedimentation equilibrium analytical ultracentrifugation
Sedimentation equilibrium experiments were performed at 10°C in an An60-Ti rotor at three different speeds. Protein samples were dialyzed overnight at 4°C into buffer containing 50 mM sodium phosphate pH 7.4, 100 mM NaCl, 200 nM tris(2-carboxyethyl)phosphine (TCEP). Samples were loaded into six-sector cells with a 12 mm path length using the dialysis buffer as the reference. Absorbance scans were collected at two hour intervals for 12 to 24 hours for at each speed. The approach to equilibrium was confirmed using SEDFIT [66]. Only datasets on samples that had reached equilibrium were used for further data analysis. Datasets were fit globally in SEDPHAT [66, 67] to determine equilibrium association constants.
For the analysis of RNF4 alone, RNF4 was loaded into two six-sector cells at concentrations of 10 μM, 20 μM, 40 μM, 50 μM, 100 μM, and 150 μM. RNF4-E2 mixes were loaded into six-sector cells at concentrations of 50 μM, 100 μM, or 150 μM RNF4 in addition to 50 μM E2. All experiments were carried out at 15,000, 20,000, and 25,000 rpm, except for those experiments analyzing RNF4 with UBCH5B and RAD6Bs5B-7, which were carried out 12,000, 18,000, and 24,000 rpm. Experiments analyzing RNF4 alone were monitored at an absorbance at 280 nm, while all other experiments were monitored at 300 nm. Extinction coefficients were experimentally determined and used for data analysis in SEDFIT/SEDPHAT.
RNF4 was determined to be a constitutive dimer, as it could be modeled using a noninteracting single species model as a dimer, or using a self-associating monomer-dimer equilibrium model with an effective self-association Kd of zero. RNF4-E2 data were fit using a hetero-association model to determine the best fit Kd for each interaction. No interaction between RNF4 and RAD6B was observed, as the data could be modeled using a noninteracting two species model, or using a hetero-association equilibrium with an effective hetero-association Kd of zero.
Supplementary Material
Supplementary figure 1. In vitro ubiquitination of 4xSUMO2 by RNF4 in the presence of a panel of E2 enzymes. Endpoint reactions contained 200 nM E1, 5 μM E2, 1 μM RNF4, 15 μM 4xSUMO-2 and 100 μM Ub, and were incubated at 37°C for 90 minutes. SDS-PAGE gels stained with Coomassie Brilliant Blue. (b) Time course comparing the in vitro ubiquitination of 4xSUMO2, using both native and methylated Ub, by RNF4 in the presence of UBCH5B. Reactions contained 200 nM E1, 5 μM E2, 1 μM RNF4, 15 μM 4xSUMO-2 and 100 μM of native or methylated Ub, and were incubated at 37°C. Samples were analyzed by SDS-PAGE, followed by Coomassie Brilliant Blue stain. (c) Time course comparing the in vitro ubiquitination of 4xSUMO2, using both native and methylated Ub, by RNF4 in the presence of UBCH5B. Reactions contained 200 nM E1, 5 μM E2, 1 μM RNF4, 15 μM 4xSUMO-2 and 100 μM of native or methylated Ub, and were incubated at 37°C. Samples were analyzed by SDS-PAGE and stained with Coomassie Brilliant Blue stain.
Supplementary figure 2. (a) Time course comparing the in vitro ubiquitination of 4xSUMO2, using both native and methylated Ub, by RNF4 in the presence of UBCH5B. Reactions contained 200 nM E1, 5 μM E2, 1 μM RNF4, 15 μM 4xSUMO-2 and 100 μM of native or methylated Ub, and were incubated at 37°C. Samples were analyzed by SDS-PAGE, followed by Coomassie Brilliant Blue stain. (b) Time course comparing the in vitro ubiquitination of 4xSUMO2, using both native and methylated Ub, by RNF4 in the presence of RAD6B. Reactions contained 200 nM E1, 5 μM E2, 1 μM RNF4, 15 μM 4xSUMO-2 and 100 μM of native or methylated Ub, and were incubated at 37°C. Samples were analyzed by SDS-PAGE and stained with Coomassie Brilliant Blue stain.
Supplementary figure 3. (a) Time course comparing the in vitro ubiquitination of 4xSUMO2, using both native and methylated Ub, by RNF4 in the presence of the RAD6Bs5B variants. Reactions contained 200 nM E1, 5 μM E2, 1 μM RNF4, 15 μM 4xSUMO-2 and 100 μM of native or methylated Ub, and were incubated at 37°C. Samples were analyzed by SDS-PAGE, followed by Coomassie Brilliant Blue stain. (b) Time course comparing the in vitro ubiquitination of 4xSUMO2, using both native and methylated Ub, by RNF4 in the presence of UBCH5Bs6B. Reactions contained 200 nM E1, 5 μM E2, 1 μM RNF4, 15 μM 4xSUMO-2 and 100 μM of native or methylated Ub, and were incubated at 37°C. Samples were analyzed by SDS-PAGE and stained with Coomassie Brilliant Blue stain.
Supplementary figure 4. Coomassie-stained SDS-PAGE gel showing products of in vitro ubiquitination of 4xSUMO2 by RNF4 in the presence of RAD6B, RAD6Bs5B conversion variants, UBCH5B, and UBCH5Bs6B. Reactions contained 200 nM E1, 5 μM E2, 1 μM RNF4, 15 μM 4xSUMO-2 and 100 μM Ub, and were incubated at 37°C for 60 minutes. SDS-PAGE gels were stained with Coomassie Brilliant Blue.
Supplementary figure 5. E1 to E2 transthiolation assay showing E2~Ub formation for RAD6B, UBCH5B, the UBCH5Bs6B conversion mutant, and the RAD6Bs5B conversion variants. E1 was pre-charged with Ub, and then added to the E2~Ub charging reaction. E2~Ub charging reactions contained 500 nM E1, 10 μM E2, and 100 μM Ub. E2~Ub formation was monitored by SDS-PAGE followed by staining with Coomassie Brilliant Blue.
Supplementary figure 6. Sedimentation equilibrium AUC analysis of RNF4 fit to a monomer-dimer equilibrium model. Absorbance measurements were taken at 280 nm. Data were analyzed and fit in SEDFIT/SEDPHAT. Data were fit to a model based on a Kd of ~10−21 μM, which suggests that there is effectively no monomer present, and that RNF4 behaves as a constitutive dimer under our conditions.
Supplementary figure 7. Sedimentation equilibrium analytical ultracentrifugation analysis of RNF4 fit to a single species model as a constitutive dimer. Absorbance measurements were taken at 280 nm. Data were analyzed and fit in SEDFIT/SEDPHAT. The data fits well to a model in which RNF4 behaves as a single constitutive dimer species.
Supplementary figure 8. Sedimentation equilibrium analytical ultracentrifugation analysis of UBCH5B with RNF4 fit to an equilibrium heteroassociation model. Absorbance was recorded at 300 nm. Data were analyzed and fit in SEDFIT/SEDPHAT. Data were fit to a model based on a Kd of 15 μM for the interaction between UBCH5B and the RNF4 homodimer.
Supplementary figure 9. Sedimentation equilibrium analytical ultracentrifugation analysis of RAD6B with RNF4 fit to an equilibrium heteroassociation model. Absorbance was recorded at 300 nm. Data were analyzed and fit in SEDFIT/SEDPHAT. Data were fit to a model based on a Kd of ~1026 μM for the interaction between RAD6B and the RNF4 homodimer, which suggests that there is effectively no detectable interaction between the two under our conditions.
Supplementary figure 10. Sedimentation equilibrium analytical ultracentrifugation analysis of RAD6B with RNF4 fit to a two non-interacting species model. Absorbance was recorded at 300 nm. Data were analyzed and fit in SEDFIT/SEDPHAT. The data fits well to a model where RAD6B and the RNF4 homodimer behaves as two non-interacting species.
Supplementary figure 11. Sedimentation equilibrium analytical ultracentrifugation analysis of RAD6Bs5B-7 with RNF4 fit to an equilibrium heteroassociation model. Absorbance was recorded at 300 nm. Data were analyzed and fit in SEDFIT/SEDPHAT. Data were fit to a model based on a Kd of 670 μM for the interaction between RAD6Bs5B-7 and the RNF4 homodimer.
Supplementary figure 12. Sedimentation equilibrium analytical ultracentrifugation analysis of RAD6Bs5B-12 with RNF4 fit to an equilibrium heteroassociation model. Absorbance was recorded at 300 nm. Data were analyzed and fit in SEDFIT/SEDPHAT. Data were fit to a model based on a Kd of 250 μM for the interaction between RAD6Bs5B-12 and the RNF4 homodimer.
Supplementary figure 13. Representative data from Ub-discharge assays used to calculate the rates of Ub-discharge from E2~Ub to lysine in the presence and absence of RNF4, with RAD6B, UBCH5B, UBCH5Bs6B, as well as with the RAD6Bs5B conversion constructs in Fig. 5. (a) The top figure shows an example where fluorescein-tagged ubiquitin, UbFL, was used to assay E2~Ub discharge. E2 was pre-charged with UbFL and then added to the E2~UbFL discharge reaction. E2~UbFL discharge reactions contained 50 mM lysine, 10 μM RNF4, and 5 μM E2~UbFL. Ub-discharge was monitored by SDS-PAGE, followed by fluorescent imaging, and fluorescence signal densitometry. (b) Representative data showing a Ub-discharge assay carried out under the same conditions, but using native Ub rather than UbFL, analyzed by SDS-PAGE followed by Coomassie Brilliant Blue staining is shown in the bottom figure. E2~UbFL discharge assays, in which the fluorescein-tagged ubiquitin signal was followed, were used to calculate the rates of Ub-discharge from E2~Ub and generate the data shown in Fig. 5. Monitoring E2~Ub discharge using UbFL was particularly useful because the RAD6B~Ub, RAD6Bs5B-3~Ub, RAD6Bs5B-7~Ub, and RAD6Bs5B-12~Ub bands overlap with the RNF4 band at the same position when analyzed by SDS-PAGE, which complicated the assessment of E2~Ub discharge. Monitoring E2~UbFL discharge by following the UbFL signal rendered the RNF4 band effectively invisible, allowing us to assess E2~Ub discharge without the RNF4 band overlap.
Supplementary table 1. Constants and statistics from fits of sedimentation equilibrium analytical ultracentrifugation data, analyzed in SEDFIT/SEDPHAT.
Highlights.
Ubiquitination activity on polySUMO substrates by RNF4 is E2-dependent.
Reengineered RAD6B RING-binding surface transforms substrate ubiquitination activity.
Increased UBCH5B-like RING-binding surface drives polyubiquitination.
Increased UBCH5B-like RING-binding surface confers greater E2-RNF4 affinity.
E2-E3 affinity governs multiplicity of RNF4-mediated polyubiquitination of substrates.
Acknowledgments
We thank Mario Amzel, Joel Tolman, Michael Matunis, Jie Xiao, Albert Lau, Dan Leahy, Ananya Majumdar, Reuven Wiener, and Chris Berndsen for valuable feedback, advice, discussions, and assistance. Supported by a grant from the National Institute of General Medical Sciences (GM109102).
Abbreviations
- RNF4
RING finger protein 4
- E1
ubiquitin-activating enzyme
- E2
ubiquitin-conjugating enzyme
- E3
ubiquitin ligase
- UBC domain
ubiquitin-conjugating domain
- Ub
ubiquitin
- UBE2D2
ubiquitin-conjugating enzyme E2 D2
- UBE2B
ubiquitin-conjugating enzyme E2 B
- RAD6Bs5B
human RAD6B carrying UBCH5B-like substitutions to its RING-binding surface
- UbMe
lysine-methylated ubiquitin
- UbFL
fluorescein-labeled ubiquitin
- SIM
SUMO-interacting motif
- RING
relay interesting new gene
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–80. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 2.van der Veen AG, Ploegh HL. Ubiquitin-like proteins. Annu Rev Biochem. 2012;81:323–57. doi: 10.1146/annurev-biochem-093010-153308. [DOI] [PubMed] [Google Scholar]
- 3.Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–29. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 4.Swatek KN, Komander D. Ubiquitin modifications. Cell Res. 2016;26:399–422. doi: 10.1038/cr.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tatham MH, Plechanovova A, Jaffray EG, Salmen H, Hay RT. Ube2W conjugates ubiquitin to alpha-amino groups of protein N-termini. Biochem J. 2013;453:137–45. doi: 10.1042/BJ20130244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scaglione KM, Basrur V, Ashraf NS, Konen JR, Elenitoba-Johnson KS, Todi SV, et al. The ubiquitin-conjugating enzyme (E2) Ube2w ubiquitinates the N terminus of substrates. J Biol Chem. 2013;288:18784–8. doi: 10.1074/jbc.C113.477596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kirisako T, Kamei K, Murata S, Kato M, Fukumoto H, Kanie M, et al. A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J. 2006;25:4877–87. doi: 10.1038/sj.emboj.7601360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramanathan HN, Ye Y. Cellular strategies for making monoubiquitin signals. Crit Rev Biochem Mol Biol. 2012;47:17–28. doi: 10.3109/10409238.2011.620943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mattiroli F, Sixma TK. Lysine-targeting specificity in ubiquitin and ubiquitin-like modification pathways. Nat Struct Mol Biol. 2014;21:308–16. doi: 10.1038/nsmb.2792. [DOI] [PubMed] [Google Scholar]
- 10.Yau R, Rape M. The increasing complexity of the ubiquitin code. Nat Cell Biol. 2016;18:579–86. doi: 10.1038/ncb3358. [DOI] [PubMed] [Google Scholar]
- 11.Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta. 2004;1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 12.Tokunaga F, Sakata S, Saeki Y, Satomi Y, Kirisako T, Kamei K, et al. Involvement of linear polyubiquitylation of NEMO in NF-kappaB activation. Nat Cell Biol. 2009;11:123–32. doi: 10.1038/ncb1821. [DOI] [PubMed] [Google Scholar]
- 13.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 14.Berndsen CE, Wolberger C. New insights into ubiquitin E3 ligase mechanism. Nat Struct Mol Biol. 2014;21:301–7. doi: 10.1038/nsmb.2780. [DOI] [PubMed] [Google Scholar]
- 15.Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–9. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
- 16.Plechanovova A, Jaffray EG, Tatham MH, Naismith JH, Hay RT. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature. 2012;489:115–20. doi: 10.1038/nature11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pruneda JN, Littlefield PJ, Soss SE, Nordquist KA, Chazin WJ, Brzovic PS, et al. Structure of an E3:E2~Ub complex reveals an allosteric mechanism shared among RING/U-box ligases. Mol Cell. 2012;47:933–42. doi: 10.1016/j.molcel.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dou H, Buetow L, Sibbet GJ, Cameron K, Huang DT. BIRC7-E2 ubiquitin conjugate structure reveals the mechanism of ubiquitin transfer by a RING dimer. Nat Struct Mol Biol. 2012;19:876–83. doi: 10.1038/nsmb.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scott DC, Sviderskiy VO, Monda JK, Lydeard JR, Cho SE, Harper JW, et al. Structure of a RING E3 trapped in action reveals ligation mechanism for the ubiquitin-like protein NEDD8. Cell. 2014;157:1671–84. doi: 10.1016/j.cell.2014.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vittal V, Shi L, Wenzel DM, Scaglione KM, Duncan ED, Basrur V, et al. Intrinsic disorder drives N-terminal ubiquitination by Ube2w. Nat Chem Biol. 2015;11:83–9. doi: 10.1038/nchembio.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hibbert RG, Huang A, Boelens R, Sixma TK. E3 ligase Rad18 promotes monoubiquitination rather than ubiquitin chain formation by E2 enzyme Rad6. Proc Natl Acad Sci U S A. 2011;108:5590–5. doi: 10.1073/pnas.1017516108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bentley ML, Corn JE, Dong KC, Phung Q, Cheung TK, Cochran AG. Recognition of UbcH5c and the nucleosome by the Bmi1/Ring1b ubiquitin ligase complex. EMBO J. 2011;30:3285–97. doi: 10.1038/emboj.2011.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGinty RK, Henrici RC, Tan S. Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature. 2014;514:591–6. doi: 10.1038/nature13890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown NG, VanderLinden R, Watson ER, Weissmann F, Ordureau A, Wu KP, et al. Dual RING E3 Architectures Regulate Multiubiquitination and Ubiquitin Chain Elongation by APC/C. Cell. 2016;165:1440–53. doi: 10.1016/j.cell.2016.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brzovic PS, Lissounov A, Christensen DE, Hoyt DW, Klevit RE. A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol Cell. 2006;21:873–80. doi: 10.1016/j.molcel.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 26.Buchwald G, van der Stoop P, Weichenrieder O, Perrakis A, van Lohuizen M, Sixma TK. Structure and E3-ligase activity of the Ring-Ring complex of polycomb proteins Bmi1 and Ring1b. EMBO J. 2006;25:2465–74. doi: 10.1038/sj.emboj.7601144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell. 2004;7:663–76. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 28.Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–8. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 29.Hwang CS, Shemorry A, Auerbach D, Varshavsky A. The N-end rule pathway is mediated by a complex of the RING-type Ubr1 and HECT-type Ufd4 ubiquitin ligases. Nat Cell Biol. 2010;12:1177–85. doi: 10.1038/ncb2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parker JL, Ulrich HD. Mechanistic analysis of PCNA poly-ubiquitylation by the ubiquitin protein ligases Rad18 and Rad5. EMBO J. 2009;28:3657–66. doi: 10.1038/emboj.2009.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–41. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 32.Kim J, Roeder RG. Direct Bre1-Paf1 complex interactions and RING finger-independent Bre1-Rad6 interactions mediate histone H2B ubiquitylation in yeast. J Biol Chem. 2009;284:20582–92. doi: 10.1074/jbc.M109.017442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar P, Magala P, Geiger-Schuller KR, Majumdar A, Tolman JR, Wolberger C. Role of a non-canonical surface of Rad6 in ubiquitin conjugating activity. Nucleic Acids Res. 2015;43:9039–50. doi: 10.1093/nar/gkv845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turco E, Gallego LD, Schneider M, Kohler A. Monoubiquitination of histone H2B is intrinsic to the Bre1 RING domain-Rad6 interaction and augmented by a second Rad6-binding site on Bre1. J Biol Chem. 2015;290:5298–310. doi: 10.1074/jbc.M114.626788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Christensen DE, Brzovic PS, Klevit RE. E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat Struct Mol Biol. 2007;14:941–8. doi: 10.1038/nsmb1295. [DOI] [PubMed] [Google Scholar]
- 36.Starita LM, Pruneda JN, Lo RS, Fowler DM, Kim HJ, Hiatt JB, et al. Activity-enhancing mutations in an E3 ubiquitin ligase identified by high-throughput mutagenesis. Proc Natl Acad Sci U S A. 2013;110:E1263–72. doi: 10.1073/pnas.1303309110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nguyen L, Plafker KS, Starnes A, Cook M, Klevit RE, Plafker SM. The ubiquitin-conjugating enzyme, UbcM2, is restricted to monoubiquitylation by a two-fold mechanism that involves backside residues of E2 and Lys48 of ubiquitin. Biochemistry. 2014;53:4004–14. doi: 10.1021/bi500072v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Banka PA, Behera AP, Sarkar S, Datta AB. RING E3-Catalyzed E2 Self-Ubiquitination Attenuates the Activity of Ube2E Ubiquitin-Conjugating Enzymes. J Mol Biol. 2015;427:2290–304. doi: 10.1016/j.jmb.2015.04.011. [DOI] [PubMed] [Google Scholar]
- 39.Kelly A, Wickliffe KE, Song L, Fedrigo I, Rape M. Ubiquitin chain elongation requires E3-dependent tracking of the emerging conjugate. Mol Cell. 2014;56:232–45. doi: 10.1016/j.molcel.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 40.Brown NG, Watson ER, Weissmann F, Jarvis MA, VanderLinden R, Grace CR, et al. Mechanism of polyubiquitination by human anaphase-promoting complex: RING repurposing for ubiquitin chain assembly. Mol Cell. 2014;56:246–60. doi: 10.1016/j.molcel.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buetow L, Gabrielsen M, Anthony NG, Dou H, Patel A, Aitkenhead H, et al. Activation of a primed RING E3-E2-ubiquitin complex by non-covalent ubiquitin. Mol Cell. 2015;58:297–310. doi: 10.1016/j.molcel.2015.02.017. [DOI] [PubMed] [Google Scholar]
- 42.Sriramachandran AM, Dohmen RJ. SUMO-targeted ubiquitin ligases. Biochim Biophys Acta. 2014;1843:75–85. doi: 10.1016/j.bbamcr.2013.08.022. [DOI] [PubMed] [Google Scholar]
- 43.Prudden J, Pebernard S, Raffa G, Slavin DA, Perry JJ, Tainer JA, et al. SUMO-targeted ubiquitin ligases in genome stability. EMBO J. 2007;26:4089–101. doi: 10.1038/sj.emboj.7601838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moilanen A-m, Poukka H, Karvonen U, Jänne OA, Palvimo JJ. Identification of a Novel RING Finger Protein as a Coregulator in Steroid Receptor-Mediated Gene Transcription. Molecular and cellular biology. 1998;18:5128–39. doi: 10.1128/mcb.18.9.5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hakli M, Karvonen U, Janne OA, Palvimo JJ. SUMO-1 promotes association of SNURF (RNF4) with PML nuclear bodies. Exp Cell Res. 2005;304:224–33. doi: 10.1016/j.yexcr.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 46.Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG, et al. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol. 2008;10:538–46. doi: 10.1038/ncb1716. [DOI] [PubMed] [Google Scholar]
- 47.Kirkpatrick DS, Hathaway NA, Hanna J, Elsasser S, Rush J, Finley D, et al. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat Cell Biol. 2006;8:700–10. doi: 10.1038/ncb1436. [DOI] [PubMed] [Google Scholar]
- 48.Simkus C, Makiya M, Jones JM. Karyopherin alpha 1 is a putative substrate of the RAG1 ubiquitin ligase. Mol Immunol. 2009;46:1319–25. doi: 10.1016/j.molimm.2008.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Soss SE, Yue Y, Dhe-Paganon S, Chazin WJ. E2 conjugating enzyme selectivity and requirements for function of the E3 ubiquitin ligase CHIP. J Biol Chem. 2011;286:21277–86. doi: 10.1074/jbc.M111.224006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Plechanovova A, Jaffray EG, McMahon SA, Johnson KA, Navratilova I, Naismith JH, et al. Mechanism of ubiquitylation by dimeric RING ligase RNF4. Nat Struct Mol Biol. 2011;18:1052–9. doi: 10.1038/nsmb.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hwang WW, Venkatasubrahmanyam S, Ianculescu AG, Tong A, Boone C, Madhani HD. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol Cell. 2003;11:261–6. doi: 10.1016/s1097-2765(02)00826-2. [DOI] [PubMed] [Google Scholar]
- 52.Wood A, Krogan NJ, Dover J, Schneider J, Heidt J, Boateng MA, et al. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol Cell. 2003;11:267–74. doi: 10.1016/s1097-2765(02)00802-x. [DOI] [PubMed] [Google Scholar]
- 53.Kumar B, Lecompte KG, Klein JM, Haas AL. Ser(120) of Ubc2/Rad6 regulates ubiquitin-dependent N-end rule targeting by E3{alpha}/Ubr1. J Biol Chem. 2010;285:41300–9. doi: 10.1074/jbc.M110.169136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tokgoz Z, Siepmann TJ, Streich F, Jr, Kumar B, Klein JM, Haas AL. E1-E2 interactions in ubiquitin and Nedd8 ligation pathways. J Biol Chem. 2012;287:311–21. doi: 10.1074/jbc.M111.294975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Siepmann TJ, Bohnsack RN, Tokgoz Z, Baboshina OV, Haas AL. Protein interactions within the N-end rule ubiquitin ligation pathway. J Biol Chem. 2003;278:9448–57. doi: 10.1074/jbc.M211240200. [DOI] [PubMed] [Google Scholar]
- 56.Wenzel DM, Stoll KE, Klevit RE. E2s: structurally economical and functionally replete. Biochem J. 2011;433:31–42. doi: 10.1042/BJ20100985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rojas-Fernandez A, Plechanovova A, Hattersley N, Jaffray E, Tatham MH, Hay RT. SUMO chain-induced dimerization activates RNF4. Mol Cell. 2014;53:880–92. doi: 10.1016/j.molcel.2014.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haas AL, Reback PB, Chau V. Ubiquitin conjugation by the yeast RAD6 and CDC34 gene products. Comparison to their putative rabbit homologs, E2(20K) AND E2(32K) J Biol Chem. 1991;266:5104–12. [PubMed] [Google Scholar]
- 59.Petroski MD, Deshaies RJ. Mechanism of lysine 48-linked ubiquitin-chain synthesis by the cullin-RING ubiquitin-ligase complex SCF-Cdc34. Cell. 2005;123:1107–20. doi: 10.1016/j.cell.2005.09.033. [DOI] [PubMed] [Google Scholar]
- 60.Berndsen CE, Wiener R, Yu IW, Ringel AE, Wolberger C. A conserved asparagine has a structural role in ubiquitin-conjugating enzymes. Nat Chem Biol. 2013;9:154–6. doi: 10.1038/nchembio.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reverter D, Lima CD. A basis for SUMO protease specificity provided by analysis of human Senp2 and a Senp2-SUMO complex. Structure. 2004;12:1519–31. doi: 10.1016/j.str.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 62.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–34. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 63.Pickart CM, Raasi S. Controlled synthesis of polyubiquitin chains. Methods Enzymol. 2005;399:21–36. doi: 10.1016/S0076-6879(05)99002-2. [DOI] [PubMed] [Google Scholar]
- 64.Kim Y, Quartey P, Li H, Volkart L, Hatzos C, Chang C, et al. Large-scale evaluation of protein reductive methylation for improving protein crystallization. Nat Methods. 2008;5:853–4. doi: 10.1038/nmeth1008-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pickart CM, Rose IA. Functional heterogeneity of ubiquitin carrier proteins. J Biol Chem. 1985;260:1573–81. [PubMed] [Google Scholar]
- 66.Vistica J, Dam J, Balbo A, Yikilmaz E, Mariuzza RA, Rouault TA, et al. Sedimentation equilibrium analysis of protein interactions with global implicit mass conservation constraints and systematic noise decomposition. Anal Biochem. 2004;326:234–56. doi: 10.1016/j.ab.2003.12.014. [DOI] [PubMed] [Google Scholar]
- 67.Ghirlando R. The analysis of macromolecular interactions by sedimentation equilibrium. Methods. 2011;54:145–56. doi: 10.1016/j.ymeth.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure 1. In vitro ubiquitination of 4xSUMO2 by RNF4 in the presence of a panel of E2 enzymes. Endpoint reactions contained 200 nM E1, 5 μM E2, 1 μM RNF4, 15 μM 4xSUMO-2 and 100 μM Ub, and were incubated at 37°C for 90 minutes. SDS-PAGE gels stained with Coomassie Brilliant Blue. (b) Time course comparing the in vitro ubiquitination of 4xSUMO2, using both native and methylated Ub, by RNF4 in the presence of UBCH5B. Reactions contained 200 nM E1, 5 μM E2, 1 μM RNF4, 15 μM 4xSUMO-2 and 100 μM of native or methylated Ub, and were incubated at 37°C. Samples were analyzed by SDS-PAGE, followed by Coomassie Brilliant Blue stain. (c) Time course comparing the in vitro ubiquitination of 4xSUMO2, using both native and methylated Ub, by RNF4 in the presence of UBCH5B. Reactions contained 200 nM E1, 5 μM E2, 1 μM RNF4, 15 μM 4xSUMO-2 and 100 μM of native or methylated Ub, and were incubated at 37°C. Samples were analyzed by SDS-PAGE and stained with Coomassie Brilliant Blue stain.
Supplementary figure 2. (a) Time course comparing the in vitro ubiquitination of 4xSUMO2, using both native and methylated Ub, by RNF4 in the presence of UBCH5B. Reactions contained 200 nM E1, 5 μM E2, 1 μM RNF4, 15 μM 4xSUMO-2 and 100 μM of native or methylated Ub, and were incubated at 37°C. Samples were analyzed by SDS-PAGE, followed by Coomassie Brilliant Blue stain. (b) Time course comparing the in vitro ubiquitination of 4xSUMO2, using both native and methylated Ub, by RNF4 in the presence of RAD6B. Reactions contained 200 nM E1, 5 μM E2, 1 μM RNF4, 15 μM 4xSUMO-2 and 100 μM of native or methylated Ub, and were incubated at 37°C. Samples were analyzed by SDS-PAGE and stained with Coomassie Brilliant Blue stain.
Supplementary figure 3. (a) Time course comparing the in vitro ubiquitination of 4xSUMO2, using both native and methylated Ub, by RNF4 in the presence of the RAD6Bs5B variants. Reactions contained 200 nM E1, 5 μM E2, 1 μM RNF4, 15 μM 4xSUMO-2 and 100 μM of native or methylated Ub, and were incubated at 37°C. Samples were analyzed by SDS-PAGE, followed by Coomassie Brilliant Blue stain. (b) Time course comparing the in vitro ubiquitination of 4xSUMO2, using both native and methylated Ub, by RNF4 in the presence of UBCH5Bs6B. Reactions contained 200 nM E1, 5 μM E2, 1 μM RNF4, 15 μM 4xSUMO-2 and 100 μM of native or methylated Ub, and were incubated at 37°C. Samples were analyzed by SDS-PAGE and stained with Coomassie Brilliant Blue stain.
Supplementary figure 4. Coomassie-stained SDS-PAGE gel showing products of in vitro ubiquitination of 4xSUMO2 by RNF4 in the presence of RAD6B, RAD6Bs5B conversion variants, UBCH5B, and UBCH5Bs6B. Reactions contained 200 nM E1, 5 μM E2, 1 μM RNF4, 15 μM 4xSUMO-2 and 100 μM Ub, and were incubated at 37°C for 60 minutes. SDS-PAGE gels were stained with Coomassie Brilliant Blue.
Supplementary figure 5. E1 to E2 transthiolation assay showing E2~Ub formation for RAD6B, UBCH5B, the UBCH5Bs6B conversion mutant, and the RAD6Bs5B conversion variants. E1 was pre-charged with Ub, and then added to the E2~Ub charging reaction. E2~Ub charging reactions contained 500 nM E1, 10 μM E2, and 100 μM Ub. E2~Ub formation was monitored by SDS-PAGE followed by staining with Coomassie Brilliant Blue.
Supplementary figure 6. Sedimentation equilibrium AUC analysis of RNF4 fit to a monomer-dimer equilibrium model. Absorbance measurements were taken at 280 nm. Data were analyzed and fit in SEDFIT/SEDPHAT. Data were fit to a model based on a Kd of ~10−21 μM, which suggests that there is effectively no monomer present, and that RNF4 behaves as a constitutive dimer under our conditions.
Supplementary figure 7. Sedimentation equilibrium analytical ultracentrifugation analysis of RNF4 fit to a single species model as a constitutive dimer. Absorbance measurements were taken at 280 nm. Data were analyzed and fit in SEDFIT/SEDPHAT. The data fits well to a model in which RNF4 behaves as a single constitutive dimer species.
Supplementary figure 8. Sedimentation equilibrium analytical ultracentrifugation analysis of UBCH5B with RNF4 fit to an equilibrium heteroassociation model. Absorbance was recorded at 300 nm. Data were analyzed and fit in SEDFIT/SEDPHAT. Data were fit to a model based on a Kd of 15 μM for the interaction between UBCH5B and the RNF4 homodimer.
Supplementary figure 9. Sedimentation equilibrium analytical ultracentrifugation analysis of RAD6B with RNF4 fit to an equilibrium heteroassociation model. Absorbance was recorded at 300 nm. Data were analyzed and fit in SEDFIT/SEDPHAT. Data were fit to a model based on a Kd of ~1026 μM for the interaction between RAD6B and the RNF4 homodimer, which suggests that there is effectively no detectable interaction between the two under our conditions.
Supplementary figure 10. Sedimentation equilibrium analytical ultracentrifugation analysis of RAD6B with RNF4 fit to a two non-interacting species model. Absorbance was recorded at 300 nm. Data were analyzed and fit in SEDFIT/SEDPHAT. The data fits well to a model where RAD6B and the RNF4 homodimer behaves as two non-interacting species.
Supplementary figure 11. Sedimentation equilibrium analytical ultracentrifugation analysis of RAD6Bs5B-7 with RNF4 fit to an equilibrium heteroassociation model. Absorbance was recorded at 300 nm. Data were analyzed and fit in SEDFIT/SEDPHAT. Data were fit to a model based on a Kd of 670 μM for the interaction between RAD6Bs5B-7 and the RNF4 homodimer.
Supplementary figure 12. Sedimentation equilibrium analytical ultracentrifugation analysis of RAD6Bs5B-12 with RNF4 fit to an equilibrium heteroassociation model. Absorbance was recorded at 300 nm. Data were analyzed and fit in SEDFIT/SEDPHAT. Data were fit to a model based on a Kd of 250 μM for the interaction between RAD6Bs5B-12 and the RNF4 homodimer.
Supplementary figure 13. Representative data from Ub-discharge assays used to calculate the rates of Ub-discharge from E2~Ub to lysine in the presence and absence of RNF4, with RAD6B, UBCH5B, UBCH5Bs6B, as well as with the RAD6Bs5B conversion constructs in Fig. 5. (a) The top figure shows an example where fluorescein-tagged ubiquitin, UbFL, was used to assay E2~Ub discharge. E2 was pre-charged with UbFL and then added to the E2~UbFL discharge reaction. E2~UbFL discharge reactions contained 50 mM lysine, 10 μM RNF4, and 5 μM E2~UbFL. Ub-discharge was monitored by SDS-PAGE, followed by fluorescent imaging, and fluorescence signal densitometry. (b) Representative data showing a Ub-discharge assay carried out under the same conditions, but using native Ub rather than UbFL, analyzed by SDS-PAGE followed by Coomassie Brilliant Blue staining is shown in the bottom figure. E2~UbFL discharge assays, in which the fluorescein-tagged ubiquitin signal was followed, were used to calculate the rates of Ub-discharge from E2~Ub and generate the data shown in Fig. 5. Monitoring E2~Ub discharge using UbFL was particularly useful because the RAD6B~Ub, RAD6Bs5B-3~Ub, RAD6Bs5B-7~Ub, and RAD6Bs5B-12~Ub bands overlap with the RNF4 band at the same position when analyzed by SDS-PAGE, which complicated the assessment of E2~Ub discharge. Monitoring E2~UbFL discharge by following the UbFL signal rendered the RNF4 band effectively invisible, allowing us to assess E2~Ub discharge without the RNF4 band overlap.
Supplementary table 1. Constants and statistics from fits of sedimentation equilibrium analytical ultracentrifugation data, analyzed in SEDFIT/SEDPHAT.